Abstract
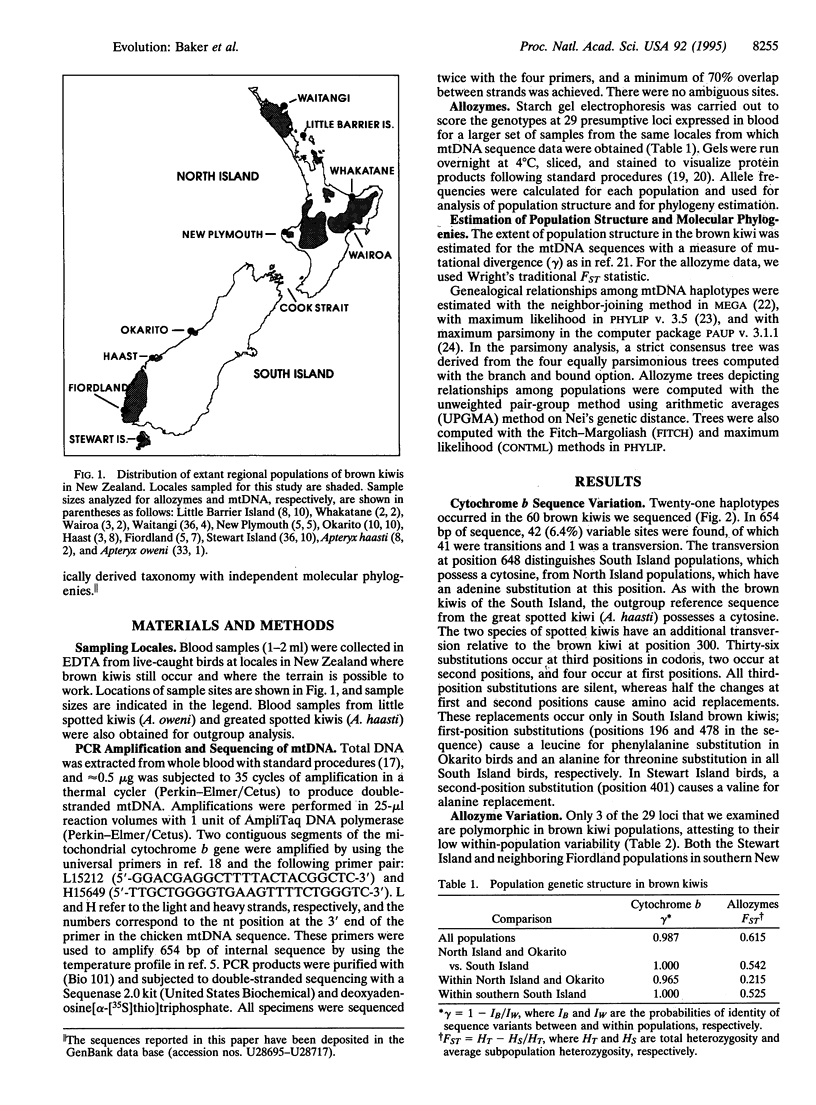
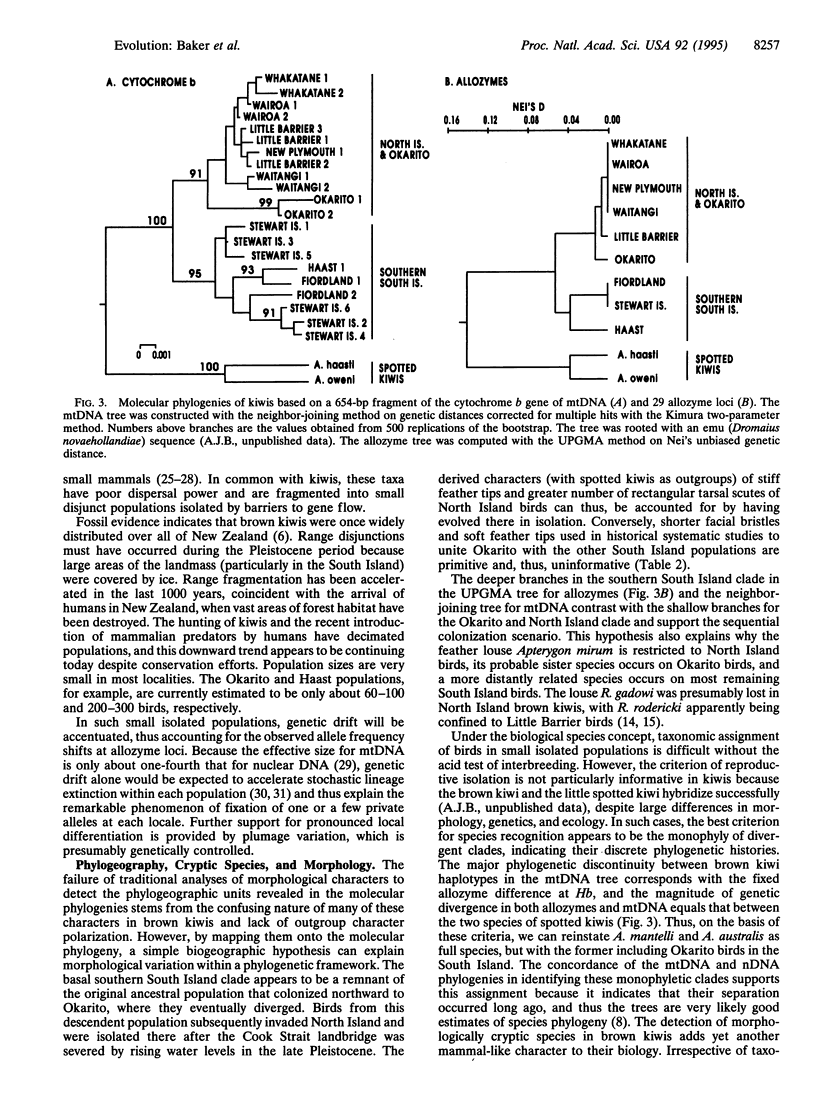
Using allozymes and mtDNA sequences from the cytochrome b gene, we report that the brown kiwi has the highest levels of genetic structuring observed in birds. Moreover, the mtDNA sequences are, with two minor exceptions, diagnostic genetic markers for each population investigated, even though they are among the more slowly evolving coding regions in this genome. A major unexpected finding was the concordant split in molecular phylogenies between brown kiwis in the southern South Island and elsewhere in New Zealand. This basic phylogeographic boundary halfway down the South Island coincides with a fixed allele difference in the Hb nuclear locus and strongly suggests that two morphologically cryptic species are currently merged under one polytypic species. This is another striking example of how molecular genetic assays can detect phylogenetic discontinuities that are not reflected in traditional morphologically based taxonomies. However, reanalysis of the morphological characters by using phylogenetic methods revealed that the reason for this discordance is that most are primitive and thus are phylogenetically uninformative. Shared-derived morphological characters support the same relationships evident in the molecular phylogenies and, in concert with the molecular data, suggest that as brown kiwis colonized northward from the southern South Island, they retained many primitive characters that confounded earlier systematists. Strong subdivided population structure and cryptic species in brown kiwis seem to have evolved relatively recently as a consequence of Pleistocene range disjunctions, low dispersal power, and genetic drift in small populations.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Neigel J. E., Arnold J. Demographic influences on mitochondrial DNA lineage survivorship in animal populations. J Mol Evol. 1984;20(2):99–105. doi: 10.1007/BF02257369. [DOI] [PubMed] [Google Scholar]
- Ball R. M., Freeman S., James F. C., Bermingham E., Avise J. C. Phylogeographic population structure of Red-winged Blackbirds assessed by mitochondrial DNA. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1558–1562. doi: 10.1073/pnas.85.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C. W., Jr, Maruyama T., Fuerst P. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics. 1983 Mar;103(3):513–527. doi: 10.1093/genetics/103.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher T. D., Thomas W. K., Meyer A., Edwards S. V., Päbo S., Villablanca F. X., Wilson A. C. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A., Wake D. B., Yanev K. P. Measuring gene flow among populations having high levels of genetic fragmentation. Genetics. 1984 Feb;106(2):293–308. doi: 10.1093/genetics/106.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latter B. D. The island model of population differentiation: a general solution. Genetics. 1973 Jan;73(1):147–157. doi: 10.1093/genetics/73.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenink P. W., Baker A. J., Tilanus M. G. Hypervariable-control-region sequences reveal global population structuring in a long-distance migrant shorebird, the Dunlin (Calidris alpina). Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):94–98. doi: 10.1073/pnas.90.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]


