Abstract
T cell development in the thymus produces multiple lineages of cells, including innate T cells such as γδ TCR+ cells, iNKT cells, MAIT cells, and H2-M3-specific cells. Although innate cells are generally a minor subset of thymocytes, in several strains of mice harboring mutations in T cell signaling proteins or transcriptional regulators, conventional CD8+ T cells develop as innate cells with characteristics of memory T cells. Thus, in Itk-deficient mice, mature CD4-CD8+ (CD8SP) thymocytes express high levels of the transcription factor, Eomesodermin, and are dependent on IL-4 being produced in the thymic environment by a poorly-characterized subset of CD4+ thymocytes expressing the transcriptional regulator, PLZF. Here we show that a sizeable proportion of mature CD4+CD8- (CD4SP) thymocytes in itk-/- mice also develop as innate Eomesodermin-expressing T cells. These cells are dependent on MHC class II and IL-4 signaling for their development, indicating that they are conventional CD4+ T cells that have been converted to an innate phenotype. Surprisingly, neither CD4SP nor CD8SP innate Eomes+ thymocytes in itk-/- or SLP-76(Y145F) mice are dependent on γδ T cells for their development. Instead, we find that the predominant population of Eomes+ innate itk-/- CD4SP thymocytes is largely absent in mice lacking CD1d-specific iNKT cells, with no effect on innate itk-/- CD8SP thymocytes. In contrast, both subsets of innate Eomes+ itk-/- T cells require the presence of a novel PLZF-expressing, SAP-dependent thymocyte population that is essential for the conversion of conventional CD4+ and CD8+ T cells into innate T cells with a memory phenotype.
Introduction
T cell development in the thymus produces a wide range of T cell subsets with varying functions in immune responses. In addition to conventional naïve CD4+ and CD8+ T cells, which require prolonged activation and differentiation to acquire protective effector functions, several subsets of T cells with innate effector functions are now known to develop in the thymus (1, 2). This latter group includes several distinct categories of γδT cells, CD1d-specific invariant natural killer (iNKT) cells, MR1-specific mucosal-associated invariant T (MAIT) cells, H2-M3-specific CD8+ T cells, and Foxp3+ regulatory T cells, among others (1, 2). While a thorough understanding of the signals giving rise to each of these T cell lineages has not yet been achieved, recent studies indicate a role for the strength of TCR signaling, extrinsic signals provided by cytokines, as well as components of intrinsic developmental programming in this process (3-8).
One important clue to dissecting the signals regulating T cell lineage development has come from studies of genetically-altered mice. In the absence of the Tec kinase, Itk, as well as in mice lacking the transcription factors Krüppel-like factor 2 (Klf2), Inhibitor of DNA-binding 3 (Id3), and CREB-binding protein (CBP), conventional CD8+ T cells developing in the thymus are converted into innate/memory-like T cells expressing high levels of the effector-promoting transcription factor, Eomesodermin (Eomes) (6, 8-10). While the involvement of Itk indicates a role for TCR signaling in this process, further evidence supporting this conclusion comes from studies demonstrating an identical phenotype in mice expressing a mutant form of the adapter protein SLP-76, SLP-76(Y145F), which lacks the ability to recruit Itk in response to TCR stimulation (7). Together, these data indicate that intact TCR signaling pathways are critical for the normal development of conventional CD8+ T cells.
Interestingly, impaired TCR signaling is not the only requirement for the development of innate/memory CD8+ thymocytes expressing high levels of Eomes. Studies by Hogquist and colleagues first demonstrated a requirement for exogenous IL-4 to induce Eomes expression in CD8+ αβ T cells (3, 6-8). Thus, in the absence of the IL-4Rα (CD124), Eomes is no longer expressed in itk-/-CD8+ T cells (8). Further evidence indicates that a subset of T cells expressing the transcription factor promyelocytic leukemia zinc finger (PLZF) are involved in this pathway and are the likely source of the excess IL-4 (6-8).
Data from our lab and others have shown that in the absence of Itk, there is an increase in PLZF+γδ T cells expressing the Vγ1.1 Vδ6.3 TCR, a subset known as γδ NKT cells (11, 12). In general, it is thought that these γδ NKT cells are responsible for the excess IL-4 in the absence of Itk, and thus for the development of Eomes+ innate/memory CD8+ T cells (13). While it has been demonstrated that γδ NKT cells are responsible for the hyper-IgE syndrome seen in itk-/- mice (11, 12), it is not currently known whether γδ NKT cells are required to induce Eomes expression in CD8+ T cells. Multiple cell types (i.e. αβ iNKT cells, MAIT cells, etc.) are capable of IL-4 production. Therefore, even though γδ NKT cells are the most likely candidate for the excess IL-4 acting on CD8+ thymocytes, it remains possible that other cell types are contributing to this process.
Additionally, the effect of thymicIL-4 on the development of conventional CD4+ αβ T cells has not been addressed. While numerous studies have documented that itk-/- mice have an increased frequency of activated CD4+ αβ T cells, it is not known whether these cells are expressing Eomes similarly to itk-/-CD8+ αβ T cells (14). In this study, we examined itk-/-CD4+ αβ thymocytes and found a substantial subset of conventional T cells that are upregulating Eomes in response to the environment. Using these innate/memory Eomes+ CD4+ and CD8+ thymocytes to track the environmental contribution to innate T cell development in itk-/- mice, we demonstrate that this phenotype is not induced by γδ T cells.
Materials and Methods
Mice
Wild-type (WT) C57Bl/6 mice were purchased from either Taconic Farms, Inc. (Hudson, NY), Jackson Laboratories (Bar Harbor, ME), or Charles River Laboratories International, Inc. (Wilmington, MA). itk-/- mice were previously described (15-17) and housed at the University of Massachusetts Medical School in accordance with the institutional animal care and use committee (IACUC) and in a specific-pathogen free environment. IL-4 reporter (4get) mice were a gift from Markus Mohrs (Trudeau Institute, Saranac Lake, NY) and were crossed to itk-/- at the University of Massachusetts Medical School. cd1d-/- mice were a gift from the laboratory of Raymond Welsh and were crossed to itk-/- at the University of Massachusetts Medical School. mr1-/- and il-15-/- mice were a gift from Joonsoo Kang and were also crossed to itk-/- mice at the University of Massachusetts Medical School. sh2d1a-/- were previously described (18) and were crossed to itk-/- mice at the University of Massachusetts Medical School. H2dlAb1-Ea mice were purchased from Jackson Laboratories (Bar Harbor, ME) and were crossed to itk-/- at the University of Massachusetts Medical School. tcrd-/- were a gift from the laboratory of Raymond Welsh and were crossed to itk-/- mice at the University of Massachusetts Medical School as previously described (11). SLP-76(Y146F) mice were previously described (19) and housed at the University of Pennsylvania in accordance with their IACUC. tcrd-/- mice were purchased from Jackson Laboratories and crossed with SLP-76(Y146F) mice to generate Y145F/tcrd-/- mice. il4ra-/- mice were a gift from Kristin Hogquist (University of Minnesota, Minneapolis, MN) and were bred to itk-/- mice at the National Institute of Health (NIH) in accordance with their IACUC to generate itk/il4ra-/- mice.
Cell Preparation
Thymi were harvested from mice and stored in RPMI (Gibco by Invitrogen, Grand Island, New York) supplemented with fetal bovine serum (FBS), L-glutamine, penicillin, streptomycin, β-mercaptoethanol, and Hepes (RPMI-10). Thymi were processed using forceps and frosted microscope slides. Thymocytes were lysed with red blood cell (RBC) lysis buffer prior to single cell suspension with RPMI-10.
Cell Stimulations
Cells were plated at 106-107 cells per well prior to stimulating with phorbol myristate acetate (PMA, 10 ng/mL) and ionomycin (1 μg/mL) for six hours at 37°C in the presence of brefeldin A and monensin.
Extracellular/Intracellular Staining
Cells were plated at 106-107 cells per well prior to washing with FACS buffer (1X PBS supplemented with 2% FBS). Fc receptors were blocked using supernatant from 2.4G2 hybridoma grown in the lab prior to staining with the CD1d tetramer loaded with PBS57 (a gift from the NIH). Cells were stained with various combinations of the cell surface antibodies against CD4 (RM4-5), CD8 (53-6.7 or 5H10), TCRβ (H57-597), TCRδ (GL3), Heat-shock antigen (HSA, CD24) (30-F1 or M1/69), CD44 (IM7), CD62L (MEL-14), CD124 (IL-4Rα) (mIL-4R-M1), CD122 (IL-2Rβ) (5H4 or TM-Beta 1), and CXCR3 (CXCR3-173). Cells were then permeabilized using the eBioscience (San Diego, CA) Foxp3/transcription factor staining kit according to the manufacturer's protocol prior to staining with antibodies against the transcriptions factors, Eomesodermin (Dan11mag) and promyelocytic leukemia zinc finger protein (PLZF: D-9, IgG1: A85-1), and the cytokines, IL-4 (BVD6-24G2) and IFNγ (XMG1.2). Antibodies were purchased from BD Biosciences (San Jose, CA), eBioscience (San Diego, CA) or Santa Cruz Biotechnology, Inc (Dallas, TX).
Statistical analysis
Statistics were analyzed using GraphPad Prism (La Jolla, CA) software using student's t test or one-way ANOVAs, as applicable.
Results
CD4+ Eomesodermin+ αβ T cells Develop in the Absence of Itk
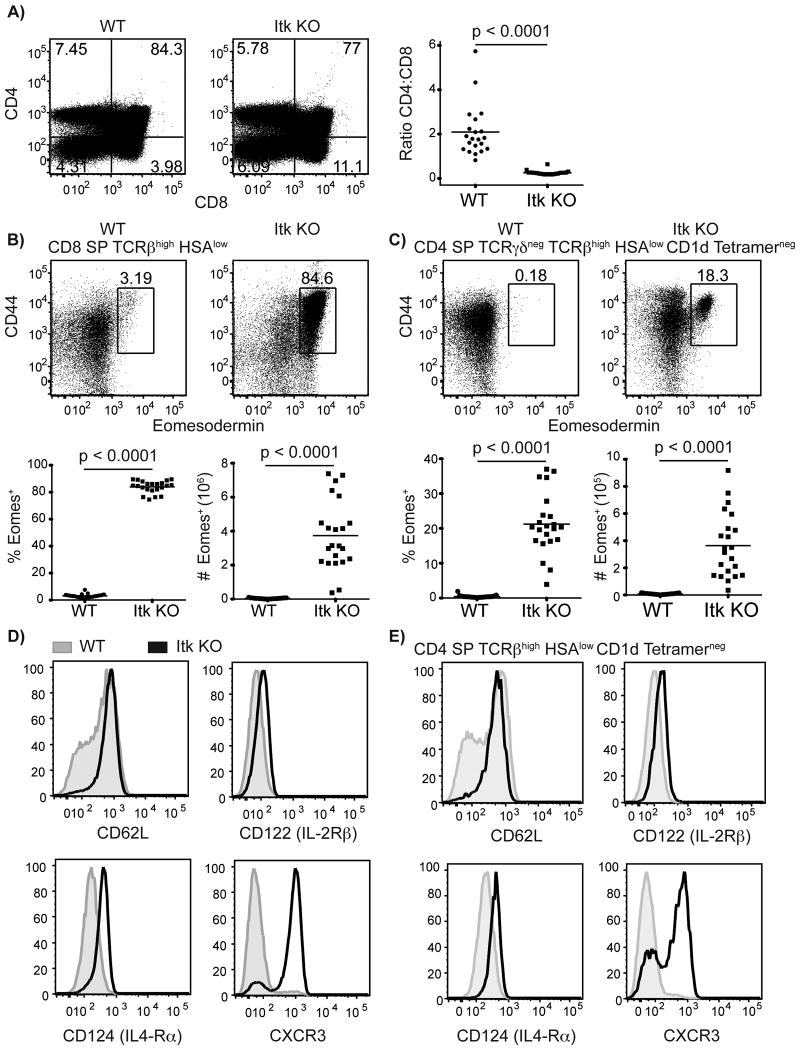
As previously described (15, 20), thymocytes from itk-/- mice have an increased frequency of CD4-CD8+ (CD8SP) cells compared to wild-type (WT) thymocytes, leading to a decreased ratio of CD4:CD8 SP thymocytes (Figure 1A). In addition, the predominant population of CD8SP thymocytes in itk-/- mice express high levels of Eomes, due to increased IL-4 signaling (8, 15) (Figure 1B). itk-/- CD8SP thymocytes also express increased levels of the IL-4Rα (CD124) and have a memory-like phenotype characterized by high expression of CD44, CD62L, IL-2Rβ (CD122), and CXCR3 (Figure 1B,D).
Figure 1. Mature Eomesodermin+ CD8 and CD4 T cells develop in the absence of Itk.
Thymocytes from WT and itk-/- mice were isolated and stained with CD1d-tetramer and antibodies to CD4, CD8, TCRβ, TCRδ, HSA (CD24), CD44, CD62L, CD122 (IL-2Rβ), CXCR3, CD124 (IL-4Rα), and Eomesodermin.
(A) CD4:CD8 ratio of total thymocytes in WT versus itk-/- mice.
(B-C) Eomesodermin versus CD44 staining of CD8SP (B) and CD4SP (C) thymocytes, gated as indicated. The numbers indicate the percentages of CD44high Eomes+ cells in each subset. The graphs below show compilations of all data indicating percentages and absolute numbers of Eomes+ cells in each subset. n = 20-22 mice per group from more than five independent experiments. Statistical analysis was done using a Mann-Whitney test.
(D-E) Histograms show staining of CD62L, CD122, CD124, and CXCR3 on mature CD8SP (D) or mature CD4SP (E) thymocytes. WT thymocytes are shown in gray filled histograms, itk-/- thymocytes are shown in black. (D) WT and itk-/- thymocytes are gated on TCRβ+ CD24low CD8 SP cells. (E) WT thymocytes gated on CD4 SP TCRβ+ CD1d-tetramerneg HSAlow thymocytes, itk-/- thymocytes gated on CD4 SP TCRβ+ CD1d-tetramerneg HSAlow Eomesodermin+ thymocytes. Results are representative of at least three independent experiments.
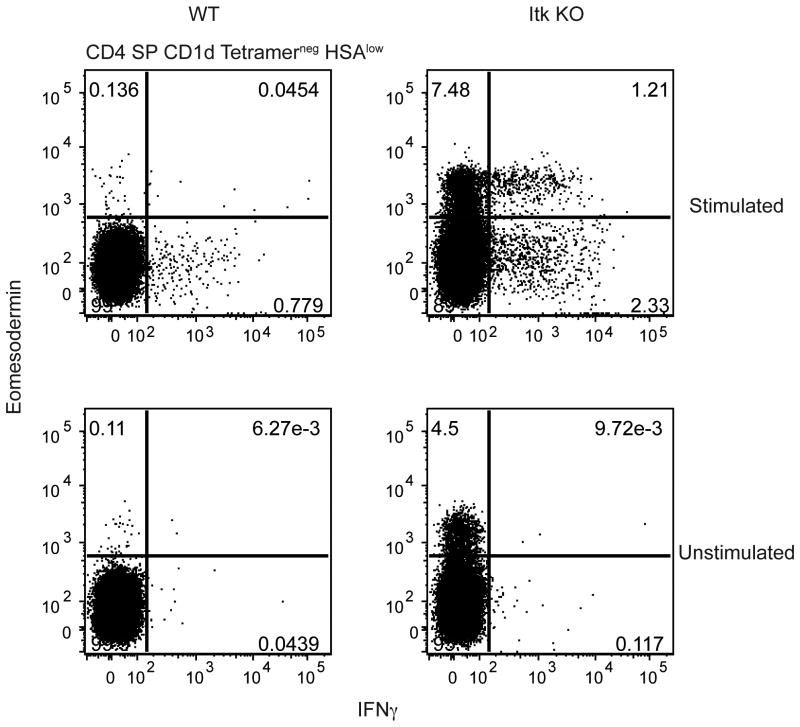
To determine whether itk-/- CD4+ αβ T cells also underwent altered development, we characterized CD4+CD8-(CD4SP) thymocytes from itk-/- mice. For this analysis, we excluded invariant αβ NKT (iNKT) cells, which are known to have a memory-like phenotype (21). When conventional, mature (TCRβhigh HSAlow) CD1d tetramerneg CD4SP thymocytes were analyzed, we found an increased population of itk-/- cells expressing Eomes compared to WT CD4SP thymocytes (Figure 1C). Like the CD8SP thymocytes, Eomes+ itk-/- CD4SP thymocytes also expressed increased levels of CD44, CD62L, IL-2Rβ (CD122), IL-4Rα (CD124), and CXCR3 (Figure 1C,E). In addition to the upregulated expression of Eomes, CD44, CD62L, IL-2Rβ, IL-4Rα, and CXCR3, itk-/- Eomes+ CD8SP thymocytes produce IFNγ upon ex vivo stimulation (15, 20). This effector function is shared with itk-/- CD4SP thymocytes (Figure 2). Together, these data indicate that innate-like CD4+αβ T cells expressing Eomes develop in the absence of Itk.
Figure 2. Eomesodermin+ CD4SP thymocytes produce IFNγ in response to stimulation.
Thymocytes from WT and itk-/- mice were stimulated with 10 ng/mL PMA and 1 μg/mL ionomycin for six hours, and then stained with CD1d-tetramer and antibodies against CD4, CD8, HSA, IFNγ, and Eomesodermin. Dot-plots are gated on CD4SP CD1d-tetramerneg HSAlow thymocytes, and show IFNγ versus Eomes staining. Results are representative of two independent experiments involving 2-4 mice per group.
IL-15 regulates the expansion of Eomesodermin+ T cells
Previously, we and others demonstrated the importance of IL-15 for the development and/or maintenance of itk-/- CD44high peripheral CD8+ T cells (15, 22). Additionally, we have observed a slight reduction in the overall thymic cellularity of itk-/-il15-/- mice (Supplemental Figure 1A). Further, as shown in Supplemental Figure 1B, itk-/- Eomes+ CD8SP thymocytes are substantially reduced in number in the absence of IL-15. However, the presence of a significant residual population of itk-/- Eomes+ CD8SP cells, even in the absence of IL-15, indicates that IL-15 is critical for the maintenance and/or expansion of these cells, rather than for their development per se. In contrast, the frequency and number of itk-/- Eomes+ CD4SP thymocytes increased dramatically in the absence of IL-15 (Supplemental Figure 1C). This increase is likely due to the reduced number of Eomes+ itk-/- CD8SP cells that would compete with the CD4+ T cells for survival cytokines, such as IL-7 or IL-4. These data indicate that CD4+ Eomes+ T cells are independent of IL-15 for their development, maintenance and expansion. Thus, while both of these subsets of innate T cells in itk-/- mice express Eomes, they are differentially regulated by the cytokine, IL-15.
Conventional CD4+ T cells promote the expansion and development of Eomesodermin expressing αβ T cells
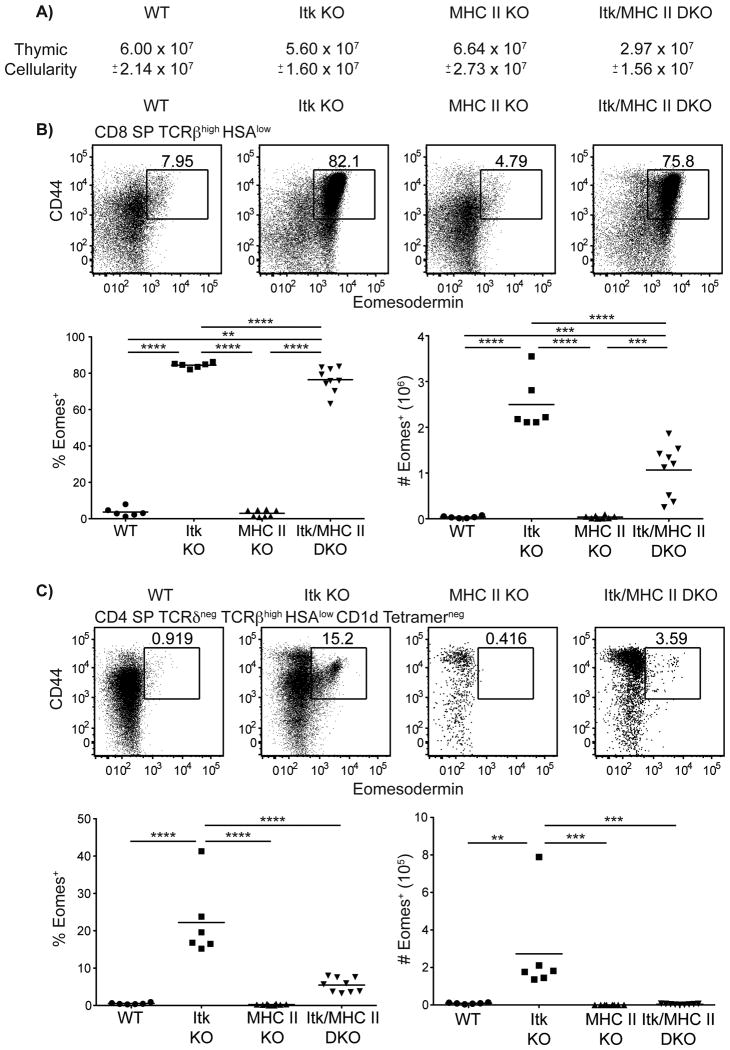
Eomes expressing itk-/- CD8+ T cells are conventional αβ T cells that are converted to this innate-like phenotype by IL-4 (6, 8). To determine if Eomes+ CD4+ T cells were also conventional T cells that are converted to an innate-like phenotype, we examined the development of these cells in itk-/- mice lacking all MHC class II expression (H2dlAb1-Ea). Unexpectedly, we first observed that, in the absence of MHC class II, there is an overall reduction in the thymic cellularity of Itk-deficient mice lacking MHC class II (Figure 3A). Further, the frequency and number of Eomes+ CD8+ αβ T cells were significantly decreased (Figure 3B), and lack of MHC class II expression led to a near disappearance of itk-/- Eomes+ CD4SP thymocytes (Figure 3C). These latter data support the conclusion that itk-/- Eomes+ CD4SP thymocytes are conventional CD4+ T cells that are converted to an innate-like phenotype. The reason for the reduced numbers of itk-/- innate-like CD8SP in mice lacking conventional αβ CD4+ T cells is unclear, but may reflect an unforeseen contribution of IL-4 from this conventional T cell subset. Alternatively, itk-/- CD4SP thymocytes may promote the expansion of thymocytes during the DN to DP transition since we saw a significant decrease in thymic cellularity.
Figure 3. CD4+ Eomes+ T cells are MHC class II-dependent.
Thymocytes from WT, itk-/-, H2dlAb1-Ea, and itk-/-/H2dlAb1-Ea mice were stained with CD1d-tetramer and antibodies to CD4, CD8, TCRδ, TCRβ, HSA, CD44, and Eomesodermin. Dot-plots show Eomes versus CD44 staining, and graphs show compilations of the percentages and absolute numbers of Eomes+ cells.
(A) Total thymic cellularity. Significant differences were seen between H2dlAb1-Ea and itk-/-/H2dlAb1-Ea mice (p < 0.005).
(B) Gated on CD8SP TCRβhigh HSAlow thymocytes.
(C) Gated on CD4SP TCRδneg TCRβhigh HSAlow CD1d-tetramerneg thymocytes. n = 6-8 mice per group. Results are representative of three independent experiments. Statistical analysis was performed using a one-way ANOVA. **p < 0.005 ***p < 0.0005 ****p < 0.00001
Eomesodermin+ αβ T cells develop independently of MAIT cells
The data shown above indicate that both conventional CD8+ and CD4+ αβ T cells aberrantly upregulate Eomes expression in itk-/- mice. For the CD8SP thymocytes, this altered development is dependent on excess IL-4 in the environment. One potential source of IL-4 is a subset of innate T cells called mucosal-associated invariant T (MAIT) cells. MAIT cells express an invariant TCR (mouse Vα19-Jα33, human Vα7.2-Jα33) that recognizes the non-classical MHC class Ib molecule, MR1; furthermore, these cells preferentially home to the mesenteric lymph node (mLN) or gut lamina propria (LP) (23). Vα19 transgenic cells can produce copious amounts of IL-4, consistent with the finding that human Vα7.2+ cells express the transcription factor, PLZF, a key regulator of cytokine production by iNKT cells (24, 25). As shown in Supplemental Figure 2, a deficiency in MR1, which has been shown to block the development of MAIT cells (23), had no effect on the numbers of itk-/- total thymocytes nor on the numbers of CD8SP or CD4SP thymocytes expressing high levels of Eomes.
iNKT cells promote the development of Eomesodermin+ αβ T cells
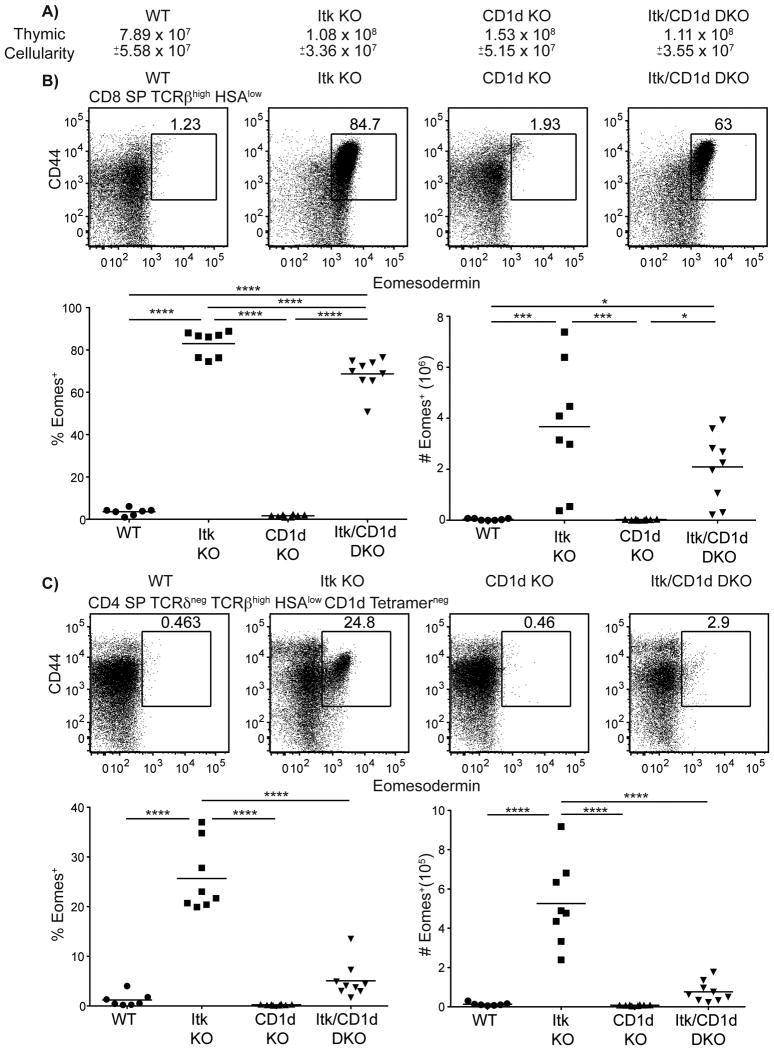
A second potential source of excess IL-4 in itk-/- mice is iNKT cells. Although mature iNKT cells predominantly produce IFNγ, with little IL-4 production, immature iNKT cells produce high levels of IL-4 (26). Additionally, previous studies showed that iNKT cell maturation in the absence of Itk is impaired, leading to increased proportions of the immature IL-4-producing subset in these mice (27, 28). To test for a role of iNKT cells, we crossed itk-/- mice to CD1d-deficient mice, and examined the thymocytes in these mice. In the absence of iNKT cells, while we observed no overall decrease in thymic cellularity between itk-/- and itk-/-cd1d-/- mice (Figure 4A), we did observe a slight decrease in the frequency and number of CD8SP Eomes+ cells (Figure 4B). These data indicate that while iNKT cells do not appear to contribute to the development of Eomes+ CD8+ thymocytes, iNKT cells may promote the expansion of itk-/- Eomes+ CD8+ thymocytes. In contrast, CD4SP Eomes+ cells were substantially reduced in Itk/CD1d double-deficient mice when compared to Itk-deficient mice (Figure 4C). Further, Itk/CD1d double-deficient CD4SP thymocytes resembled WT and CD1d-deficient mice CD4 SP thymocytes with no statistical significance between these three groups (Figure 4C). Detailed analysis revealed that the percentages and absolute numbers of total CD4SP thymocytes were identical between itk-/- and itk-/-cd1d-/- mice (Supplemental Figure 3A,B), indicating that iNKT cells contributed to the conversion of conventional CD4SP to Eomes+ CD4SP, and not to their development per se. These data show that iNKT cells are significantly promoting Eomesup regulation in conventional CD4+ T cells, but iNKT cells are not contributing significantly to the development of itk-/- Eomes+ CD8+ thymocytes.
Figure 4. αβ NKT cells promote the development of Eomesodermin+ T cells.
Thymocytes from WT, itk-/-, cd1d-/-, and itk-/-cd1d-/- mice were stained with CD1d-tetramer and antibodies against CD4, CD8, TCRδ, TCRβ, HSA, CD44, and Eomesodermin. Dot-plots show Eomes versus CD44 staining, and graphs show compilations of the percentages and absolute numbers of Eomes+ cells.
(A) Total thymic cellularity. Significant differences were seen between WT and CD1d KO mice (p < 0.05).
(B) Gated on CD8SP TCRβhigh HSAlow thymocytes.
(C) Gated on CD4SP TCRδneg TCRβhigh HSAlow CD1d-tetramerneg thymocytes. n = 7-9 mice per group. Results are from four independent experiments. Statistical analysis performed using a one-way ANOVA. *p < 0.05 ***p < 0.0005 ****p < 0.0001
If iNKT cells are contributing significant amounts of IL-4 to the thymic environment of itk-/- mice, then IL-4Rα (CD124) expression should also be decreased on itk-/- CD8SP and CD4SP thymocytes in the absence of iNKT cells, as IL-4 signaling is known to induce a positive feedback loop leading to upregulation of the IL-4R (29). Although there was a slight decrease in the geometric MFI of IL-4Rα on mature Itk/CD1d double-deficient CD8SP and CD4SP cells compared to those in itk-/- mice, IL-4Rα was still highly expressed on itk-/- mature SP thymocytes in the absence of iNKT cells (Supplemental Figure 3C,D). One possible explanation for these data is that the loss of IL-4 normally contributed by iNKT cells in itk-/- mice is not sufficient to impact CD124 expression, but is enough to prevent Eomes up-regulation. Alternatively, the absence of αβ iNKT cells may enhance the development of γδ NKT cells, which has been demonstrated in WT mice (30). Indeed, we saw that PLZF+ γδ T cells were increased in both percentage and number in the absence of αβ iNKT cells (Supplemental Figure 3E,F), which may account for the maintained high expression of IL-4Rα. Therefore, iNKT cells may regulate innate T cell development in itk-/- mice by a mechanism independent of IL-4 production.
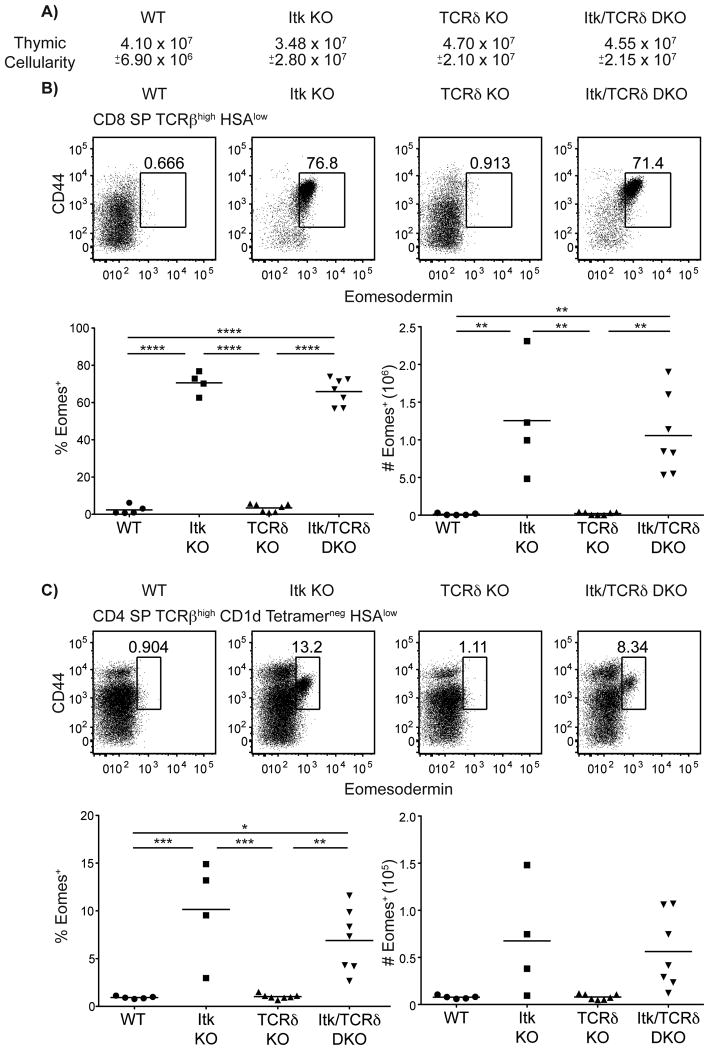
Eomesodermin+ T cells develop independently of T γδ cells
The data shown above indicate that iNKT cells are essential for the development of innate Eomes+ CD4+ αβ T cells in itk-/- mice. Nonetheless, the Itk/CD1d double-deficient mice still had a substantial population of Eomes+ CD8+ T cells. In addition to αβ iNKT cells, a closely-related subset of NKT cells has been described that expresses an invariant γδ TCR, utilizing Vγ1.1/Vδ6.3 (30, 31). Recent studies have reported a dramatic expansion in this population in itk-/- mice, as well as in other lines of mice that develop Eomes+ innate CD8+ T cells, including SLP-76(Y145F) mice (6, 7, 11, 12).γδ NKT cells also produce copious amounts of IL-4 in response to stimulation (11, 12), and have been proposed as a possible cellular source of the excess IL-4 driving innate T cell development in both itk-/- and SLP-76(Y145F) mice (13).
To address the role of γδ T cells, we crossed itk-/- mice to TCRδ-deficient (tcrd-/-) mice. To our surprise, thymocytes in the itk-/- mice lacking γδ T cells were indistinguishable from those in single itk-/- mice. Overall thymic cellularity and both the CD8SP and CD4SP subsets had comparable proportions and numbers of cells expressing Eomes when itk-/- were compared to itk-/- tcrd-/- thymocytes (Figure 5). While we did see significant increases in the frequency and cell number of αβ iNKT cells in tcrd-/- mice, there were no differences in αβ iNKT cells between itk-/- and itk-/- tcrd-/- mice (Supplementary Figure 4A-B). Thus, in contrast to recent studies in WT mice (30), it seems unlikely that itk-/- αβ iNKT cells are overcompensating in the absence of itk-/- γδ NKT cells. Similar findings were observed in SLP-76(Y145F) mice that lacked γδ T cells, such that the frequencies and numbers of Eomesodermin+ CD8SP and CD4SP thymocytes were similar in SLP-76(Y145F) mice and tcrd-/- SLP-76(Y145F) mice (Supplementary Figure 4C,D). Additionally, the absence of γδ NKT cells did not influence the frequency or number of αβ NKT cells in SLP-76(Y145F) mice (Supplementary Figure 4E). Interestingly, although γδ T cells constitute half of the PLZF+ thymocytes in SLP-76(Y145F) mice, the total number of PLZF+ thymocytes was not reduced in tcrd-/- SLP-76(Y145F) mice (Supplementary Figure 4F) leaving open the possibility that the contribution of γδ T cells to the development of innate-like thymocytes might be masked by a compensatory increase in other PLZF+ populations. Thus, these data demonstrate that, although γδ NKT cells contribute significantly to the hyper-IgE syndrome seen in Itk-deficient mice (11, 12), these cells are not required for the development of Eomes+ αβ T cells.
Figure 5. Eomesodermin+ T cells develop independently of γδ T cells.
Thymocytes from WT, itk-/-, tcrd-/-, and itk-/-tcrd-/- mice were stained with CD1d-tetramer and antibodies to CD4, CD8, TCRβ, HSA, CD44, and Eomesodermin. Dot-plots show Eomes versus CD44 staining, and graphs show compilations of the percentages and absolute numbers of Eomes+ cells.
(A) Total thymic cellularity. No significant differences were detected.
(B) Gated on CD8SP TCRβhigh HSAlow thymocytes.
(C) Gated on CD4SP TCRβhigh HSAlow CD1d-tetramerneg thymocytes.
n = 4-7 mice per group. Results are representative of two independent experiments. Statistical analysis was performed using a one-way ANOVA. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001
SAP-dependent PLZF+ CD1d tetramerneg αβ T cells develop in the absence of Itk
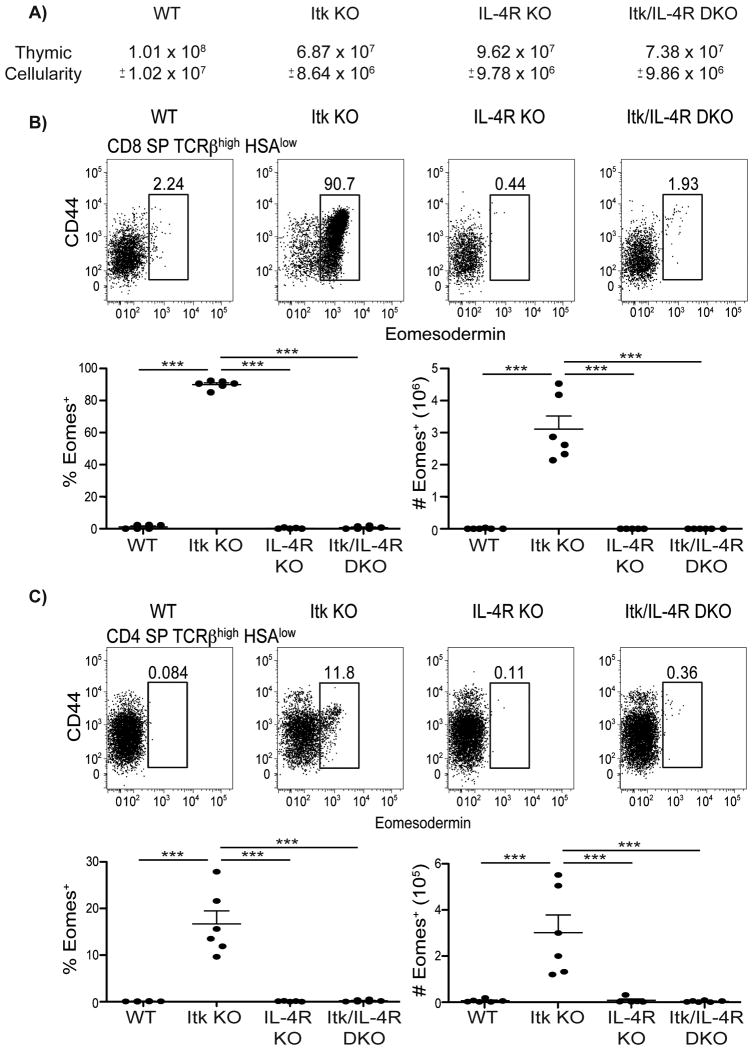
Previous studies have shown a dependence of itk-/- Eomes+ innate CD8SP thymocytes on IL-4 for their development (8). However, we were unsure if the same was true for itk-/- Eomes+ innate CD4SP thymocytes, since we observed differential requirements for IL-15 signaling (Supplementary Figure 1) and the presence of iNKT cells (Figure 4) between these two subsets. To further examine the dependence on IL-4 for the development of itk-/- Eomes+ innate CD4SP thymocytes, Itk-deficient mice were crossed to mice deficient in IL-4Rα XΔ124. While we saw reduced thymic cellularity in itk-/- and itk-/-il4ra-/- mice, these populations did not differ significantly from each other (Figure 6A). As previously seen for innate CD8SP thymocytes, the Eomes-expressing population was eliminated in the absence of IL-4 signaling (Figure 6B). Eomes+ innate CD4SP thymocytes also failed to develop in the absence of IL-4 signaling (Figure 6C). Thus, although Eomes+ innate CD8SP and CD4SP thymocytes do have some differential requirements for their development, both populations require IL-4.
Figure 6. CD4+ and CD8+ Eomes+ T cells are dependent on IL-4.
Thymocytes from WT, itk-/-, il4ra-/-, and itk-/-il4ra-/- mice were stained with antibodies to CD4, CD8, TCRβ, HSA, CD44, and Eomesodermin. Dot-plots show Eomes versus CD44 staining, and graphs show compilations of the percentages and absolute numbers of Eomes+ cells.
(A) Total thymic cellularity. Significant differences were detected between WT and itk-/- (p < 0.0001), WT and itk-/-il4ra-/- (p < 0.0005), itk-/- and il4ra-/-(p < 0.0005), and il4ra-/- and itk-/-il4ra-/- (p < 0.005).
(B) Gated on CD8SP TCRβhigh CD24(HSA)low thymocytes.
(C) Gated on CD4SP TCRβhigh CD24(HSA)low thymocytes.
n = 3-6 mice per group. Results are representative of three independent experiments. Statistical analysis was performed using one-way ANOVA. ***p < 0.0001
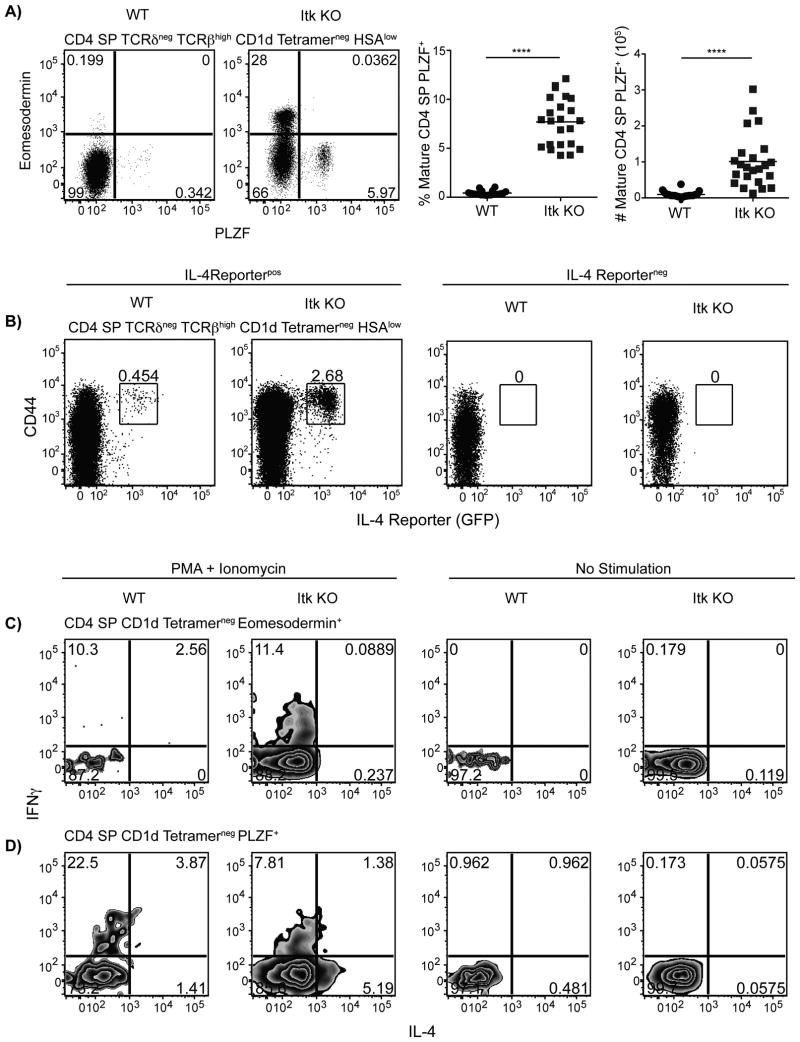
The data presented above ruled out a role for well-defined IL-4-producing CD4+ T cells populations, such as γδ NKT cells and MR-1-dependent MAIT cells, in the development of Eomes+ αβ T cells in itk-/- mice. Additionally, the data indicated that CD1d-dependent αβ iNKT cells were not essential. This led us to consider whether itk-/- mice might have an additional population of PLZF+ cells capable of producing IL-4. Therefore, we examined mature itk-/- CD4SP αβ thymocytes (TCRδneg, TCRβhigh, HSAlow, CD1d-tetramerneg) and found an increased population of PLZF+ cells compared to WT mature CD4SP thymocytes (Figure 7A). Further, when Itk KO mice were crossed to IL-4 reporter mice that have a bicistronic promoter adding an IRES-GFP component to the untranslated region of the IL-4 locus (32), we found an increased population of mature CD4SP thymocytes expressing IL-4 mRNA (Figure 7B). IL-4 mRNA appears to only be present in cells expressing PLZF because CD4SP CD1d-tetramerneg thymocytes expressing PLZF produce IL-4 and IFNγ in response to stimulation while CD4SP CD1d-tetramerneg thymocytes expressing Eomes only produce IFNγ (Figure 7C,D). Thus, in the absence of Itk, a population of mature CD4SP PLZF+ αβ T cells with capabilities of producing IL-4 develops in the thymus.
Figure 7. IL-4 producing CD1d-tetramerneg PLZF+ αβ T cells develop in the absence of Itk.
(A) Thymocytes from WT or itk-/- mice were stained with CD1d-tetramer and antibodies to CD4, CD8, TCRδ, TCRβ HSA, Eomesodermin, and PLZF.
Dot-plots (left) show PLZF versus Eomes staining on CD4SP TCRδneg TCRβhigh HSAlow CD1d-tetramerneg thymocytes.
Graphs (right) show frequencies and absolute numbers of CD4SP TCRδneg TCRβhigh CD1d-tetramerneg HSAlow PLZF+ thymocytes.
n = 20-23 mice per group from 8 independent experiments. Statistical analysis was performed using a Mann-Whitney student t test. ****p < 0.0001
(B) Itk KO mice were crossed to IL-4 reporter (4get) mice as previously described (31). Thymocytes from WT and Itk KO expressing the IL-4 reporter (positive) or without the transgene (negative) were harvested, processed, and stained with CD1d tetramer and with antibodies against CD4, CD8, TCRδ, TCRβ, HSA, and CD44. Flow cytometry plots are gated on CD4 SP TCRδneg TCRβhigh CD1d tetramerneg HSAlow thymocytes.
Results are representative of four independent experiments involving 2-3 mice per group per experiment.
(C-D) Thymocytes from WT or Itk KO mice were harvested, processed, and stimulated with PMA and ionomycin (right) or remained unstimulated (left) in the presence of brefeldin A and monensin for 5-6 hours at 37°C before staining with CD1d tetramer and with antibodies against CD4, CD8, Eomesodermin, PLZF, IFNγ, and IL-4.
(C) Gated on CD4SP CD1d-tetramerneg Eomesodermin+ thymocytes.
(D) Gated on CD4 SP CD1d-tetramerneg PLZF+ thymocytes.
Results are representative of two independent experiments involving 5-7 mice per group.
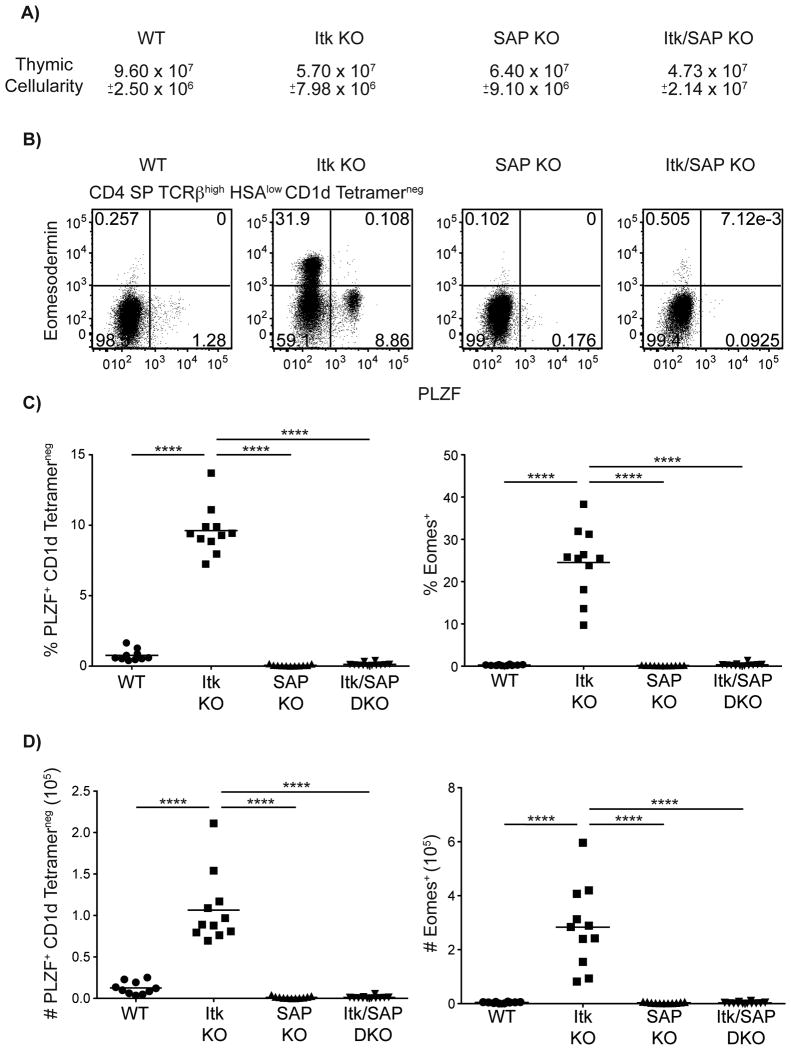
Eomes+ αβ T cells in itk-/- mice are dependent on the SLAM family receptor signaling molecule, SAP, for their development (6, 33), an observation that has been linked to the requirement for SAP in the development of PLZF+ cells in other mouse models (6). To determine if itk-/- CD4SP PLZF+ CD1d-tetramerneg αβ thymocytes were also dependent on SAP, we examined Itk/SAP double-deficient mice. As shown, SAP was essential for the development of PLZF+ CD4SP CD1d-tetramerneg αβ thymocytes in itk-/- mice (Figure 8). Furthermore, all itk-/- Eomes+ CD4SP thymocytes reverted to a conventional CD4SP phenotype in the absence of SAP. These data identify a novel population of SAP-dependent PLZF+ CD4+ T cells that is essential for the development of innate-like Eomes+ αβ T cells in itk-/- mice.
Figure 8. Development of all innate T cells in itk-/- is dependent on the SLAM-family receptor adapter protein, SAP.
Thymocytes from WT, itk-/-, sh2d1a-/-, and itk-/- sh2d1a-/- mice were stained with CD1d-tetramer and antibodies against CD4, CD8, TCRβ, HSA, Eomesodermin, and PLZF.
(A) Total thymic cellularity. Significant differences were seen between WT and itk-/- mice (p < 0.005), WT and sh2d1a-/- mice (p < 0.05), and WT and itk-/- sh2d1a-/- mice (p < 0.0005).
(B) Dot-plots show PLZF versus Eomes staining on CD4SP TCRβhigh HSAlow CD1d-tetramerneg thymocytes.
(C-D) Graphs show frequencies (C) and absolute numbers (D) of CD4 SP TCRβhigh HSAlow CD1d-tetramerneg cells expressing PLZF (left) or Eomes (right). n = 10-12 mice per group. Results are representative of four independent experiments. Statistical analysis was performed using a one-way ANOVA. ****p < 0.0001
Discussion
The data presented here show that innate T cell development is not restricted to the CD8+ T cell subset in itk-/- mice. We demonstrate that a sizeable proportion of itk-/- CD4SP thymocytes develop as innate-like cells expressing high levels of Eomesodermin. While there is a sizeable population of Eomes+ innate itk-/- CD4SP thymocytes, we found that only a small proportion (5.5-11.4%) of these cells produce IFNγ upon stimulation. However, this finding is similar to results obtained with mature innate itk-/- CD8SP thymocytes where only a portion (∼13%) produce IFNγ upon stimulation, in spite of the fact that the vast majority express Eomes (8, 15). We also saw a sizeable proportion of Eomes- CD4SP thymocytes that produced IFNγ. This is most likely due to an increase in PLZF+ CD4SP thymocytes, since PLZF allows αβ NKT cells to produce both IL-4 and IFNγ(25). Altogether, our data support the conclusion that the Eomes+ CD4SP cells in itk-/- mice are conventional MHC class II-dependent cells that have been induced to upregulate Eomes by the cell-extrinsic factor IL-4. When considering both the CD4+ and the CD8+ subsets of Eomes-expressing innate T cells in itk-/- mice, we have identified two independent cell populations that are critical for the dramatic conversion of conventional T cells into innate T cells in these mice.
First, we discovered that IL-4 signaling is crucial for the development of Eomes+ innate CD4SP thymocytes, similarly to what has been previously demonstrated for itk-/- CD8SP thymocytes (8). Furthermore, we found that a SAP-dependent population of PLZF+ T cells is essential for the development of Eomes+ CD8+ T cells in itk-/- mice, data that is consistent with several previous studies showing that both SAP and PLZF are necessary for this process (6, 8). Second, our data reveal a surprising role for iNKT cells in the development of itk-/- Eomes+ CD4+, but not CD8+ T cells. This latter finding indicates that the two subsets of Eomes+ innate T cells in itk-/- mice have distinct requirements and/or signaling pathways that regulate their development. One possibility is that CD4SP thymocytes need a higher concentration of IL-4 to induce Eomes upregulation relative to CD8SP thymocytes. If true, this might necessitate the combined cytokine production of several PLZF+ populations to achieve this threshold. While possible, this scenario seems unlikely, as we observed no effect of eliminating γδ T cells on the Eomes+ CD4SP population, in spite of the fact that the itk-/- γδ NKT cells produce enough IL-4 to induce a hyper-IgE phenotype in these mice (11, 12). An alternative possibility is that different cell populations reside in distinct compartments in the thymus, thus limiting access of cytokines to cells in the immediate environment. Thus, Eomes+ CD4SP thymocytes in itk-/- mice might co-localize with iNKT cells to a greater extent than CD8SP thymocytes. However, while we find that iNKT cells do not appear to regulate the development of itk-/- Eomes+ CD8SP thymocytes, iNKT cells may contribute to the expansion of this innate T cell population. Further, it appears that conventional CD4SP thymocytes may also have a role in the expansion of itk-/- Eomes+ CD8SP thymocytes. Our lab has previously described that Itk-deficient mice have a slight proliferative defect at the DN to DP transitions (34). Thus, SP thymocytes may aid in the proliferation of developing thymocytes via cytokine secretion.
One unexpected finding from our studies was the lack of importance of γδ NKT cells in the development of innate Eomes+ CD4+ and CD8+ T cells in Itk-/- mice. As this cell population is greatly expanded in numbers in itk-/- mice, and is a significant producer of IL-4, it was a reasonable hypothesis that these cells would contribute to, or possibly be completely responsible for, the IL-4 production that generates Eomes+ innate T cells. Yet, we observed no detectable change in either CD4SP or CD8SP thymocytes in itk-/- mice lacking γδ T cells, a result that was confirmed in SLP-76(Y145F) mice. Instead, our data suggest that the PLZF+ T cells responsible for converting itk-/- CD4+ and CD8+ thymocytes into innate Eomesodermin-expressing cells are primarily αβ TCR+ cells. Furthermore, for the development of itk-/- CD8+ innate T cells, our data rule out a role for the other known subsets of PLZF+ T cells, such as iNKT cells and MAIT cells. Although it is possible that these subsets could be sufficient to induce the development of memory/innate-like thymocytes in some contexts, our findings indicate that these subsets are not required. Instead, our data argue for a novel PLZF+ CD4+ αβ T cell subset capable of producing IL-4 that is expanded in itk-/- mice, similar to the situation for γδ NKT cells.
One interesting possibility is that the population of itk-/- PLZF+ CD4+ αβ T cells responsible for inducing innate T cell development is related to the ‘T-CD4’ cells found in the thymus of mice engineered to express MHC class II proteins on their thymocytes (35, 36). These T-CD4 cells develop as a result of SAP-dependent thymocyte-thymocyte interactions that lead to upregulation of PLZF, and to an innate capacity to secrete IL-4 (37-39). In addition, the CD8SP thymocytes in mice with T-CD4 cells develop as innate cells expressing high levels of Eomesodermin (39). Further, these mice also have a population of CD4SP thymocytes expressing Eomesodermin (39). While there does appear to be few mature CD4SP thymocytes expressing Eomesodermin in WT mice, we did find significant differences in a population of mature PLZF+ CD4+ αβ T cells between WT and SAP-deficient mice (p < 0.0002 using a Mann-Whitney t test). Therefore, development of PLZF+ thymocytes may tightly regulated in WT mice to control the development of innate-like lymphocytes expressing Eomesodermin. Further, unlike mouse thymocytes, human thymocytes express MHC class II molecules, and studies from Chang and colleagues have found a substantial number of Eomesodermin+ CD8+ T cells in human fetal thymus and spleen (39). Based on these findings, Min et al have suggested that these innate T cells may function in protective immunity during the perinatal period (39). Thus, it is possible that a normal developmental pathway for human T cells has been revealed by the genetic ablation of key T cell signaling proteins and transcription factors in mice.
Supplementary Material
Acknowledgments
The authors would like to thank Regina Whitehead and Sharlene Hubbard for technical assistance and the NIH tetramer core facility for use of the mCD1d tetramer.
Footnotes
Funding for this work was provided by the NIH grant AI084987 to LJB and by the NIH grant 5K08 AI101008 to SAC.
References
- 1.Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- 2.Veillette A, Dong Z, Latour S. Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity. 2007;27:698–710. doi: 10.1016/j.immuni.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Nayar R, Enos M, Prince A, Shin H, Hemmers S, Jiang JK, Klein U, Thomas CJ, Berg LJ. TCR signaling via Tec kinase ITK and interferon regulatory factor 4 (IRF4) regulates CD8+ T-cell differentiation. Proceedings of the National Academy of Sciences. 2012;109:E2794–802. doi: 10.1073/pnas.1205742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. Journal of Experimental Medicine. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiao Y, Zhu L, Sofi H, Lapinski PE, Horai R, Mueller K, Stritesky GL, He X, Teh HS, Wiest DL, Kappes DJ, King PD, Hogquist KA, Schwartzberg PL, Sant'Angelo DB, Chang CH. Development of promyelocytic leukemia zinc finger-expressing innate CD4 T cells requires stronger T-cell receptor signals than conventional CD4 T cells. Proceedings of the National Academy of Sciences. 2012;109:16264–16269. doi: 10.1073/pnas.1207528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon SM, Carty SA, Kim JS, Zou T, Smith-Garvin J, Alonzo ES, Haimm E, Sant'Angelo DB, Koretzky GA, Reiner SL, Jordan MS. Requirements for eomesodermin and promyelocytic leukemia zinc finger in the development of innate-like CD8+ T cells. The Journal of Immunology. 2011;186:4573–4578. doi: 10.4049/jimmunol.1100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuyama T, Kasper LH, Boussouar F, Jeevan T, van Deursen J, Brindle PK. Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Molecular and Cellular Biology. 2009;29:3894–3904. doi: 10.1128/MCB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felices M, Yin CC, Kosaka Y, Kang J, Berg LJ. Tec kinase Itk in gammadelta T cells is pivotal for controlling IgE production in vivo. Proceedings of the National Academy of Sciences. 2009;106:8308–8313. doi: 10.1073/pnas.0808459106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi Q, Xia M, Hu J, Hicks E, Iyer A, Xiong N, August A. Enhanced development of CD4+ gammadelta T cells in the absence of Itk results in elevated IgE production. Blood. 2009;114:564–571. doi: 10.1182/blood-2008-12-196345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alonzo ES, Sant'Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol. 2011;23:220–227. doi: 10.1016/j.coi.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J, August A. Naive and innate memory phenotype CD4+ T cells have different requirements for active Itk for their development. J Immunol. 2008;180:6544–6552. doi: 10.4049/jimmunol.180.10.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Schaeffer EM, Yap GS, Lewis CM, Czar MJ, McVicar DW, Cheever AW, Sher A, Schwartzberg PL. Mutation of Tec family kinases alters T helper cell differentiation. Nat Immunol. 2001;2:1183–1188. doi: 10.1038/ni734. [DOI] [PubMed] [Google Scholar]
- 17.Lucas JA, Atherly LO, Berg LJ. The absence of Itk inhibits positive selection without changing lineage commitment. J Immunol. 2002;168:6142–6151. doi: 10.4049/jimmunol.168.12.6142. [DOI] [PubMed] [Google Scholar]
- 18.Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, Schwartzberg PL. Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proc Natl Acad Sci U S A. 2001;98:7449–7454. doi: 10.1073/pnas.131193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan MS, Smith JE, Burns JC, Austin JET, Nichols KE, Aschenbrenner AC, Koretzky GA. Complementation in trans of altered thymocyte development in mice expressing mutant forms of the adaptor molecule SLP76. Immunity. 2008;28:359–369. doi: 10.1016/j.immuni.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broussard C, Fleischacker C, Fleischecker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Kronenberg M, Engel I. On the road: progress in finding the unique pathway of invariant NKT cell differentiation. Curr Opin Immunol. 2007;19:186–193. doi: 10.1016/j.coi.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Dubois S, Waldmann TA, Müller JR. ITK and IL-15 support two distinct subsets of CD8+ T cells. Proc Natl Acad Sci U S A. 2006;103:12075–12080. doi: 10.1073/pnas.0605212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 24.Kawachi I, Maldonado J, Strader C, Gilfillan S. MR1-restricted V alpha 19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J Immunol. 2006;176:1618–1627. doi: 10.4049/jimmunol.176.3.1618. [DOI] [PubMed] [Google Scholar]
- 25.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gadue P, Stein PL. NK T cell precursors exhibit differential cytokine regulation and require Itk for efficient maturation. J Immunol. 2002;169:2397–2406. doi: 10.4049/jimmunol.169.5.2397. [DOI] [PubMed] [Google Scholar]
- 27.Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J Immunol. 2008;180:3007–3018. doi: 10.4049/jimmunol.180.5.3007. [DOI] [PubMed] [Google Scholar]
- 28.Au-Yeung BB, Fowell DJ. A key role for Itk in both IFN gamma and IL-4 production by NKT cells. J Immunol. 2007;179:111–119. doi: 10.4049/jimmunol.179.1.111. [DOI] [PubMed] [Google Scholar]
- 29.Perona-Wright G, Mohrs K, Mayer KD, Mohrs M. Differential regulation of IL-4Ralpha expression by antigen versus cytokine stimulation characterizes Th2 progression in vivo. The Journal of Immunology. 2010;184:615–623. doi: 10.4049/jimmunol.0902408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira P, Boucontet L. Innate NKTγδ and NKTαβ cells exert similar functions and compete for a thymic niche. Eur J Immunol. 2012;42:1272–1281. doi: 10.1002/eji.201142109. [DOI] [PubMed] [Google Scholar]
- 31.Yin CC, Cho OH, Sylvia KE, Narayan K, Prince AL, Evans JW, Kang J, Berg LJ. The Tec kinase ITK regulates thymic expansion, emigration, and maturation of γδ NKT cells. The Journal of Immunology. 2013;190:2659–2669. doi: 10.4049/jimmunol.1202531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 2001;15:303–311. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- 33.Horai R, Mueller KL, Handon RA, Cannons JL, Anderson SM, Kirby MR, Schwartzberg PL. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas JA, Felices M, Evans JW, Berg LJ. Subtle defects in pre-TCR signaling in the absence of the Tec kinase Itk. J Immunol. 2007;179:7561–7567. doi: 10.4049/jimmunol.179.11.7561. [DOI] [PubMed] [Google Scholar]
- 35.Li W, Kim MG, Gourley TS, McCarthy BP, Sant'Angelo DB, Chang CH. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–386. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, Simpson E, Park SH. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–396. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, Jung KC, Park SH. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. Journal of Experimental Medicine. 2010;207:237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min HS, Lee YJ, Jeon YK, Kim EJ, Kang BH, Jung KC, Chang CH, Park SH. MHC class II-restricted interaction between thymocytes plays an essential role in the production of innate CD8+ T cells. The Journal of Immunology. 2011;186:5749–5757. doi: 10.4049/jimmunol.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.