Abstract
Many delivery methods have been developed to improve the therapeutic efficacy and facilitate the clinical translation of nucleic acid-based therapeutics. A facile surface-mediated nucleic acid delivery by lipoplexes is prepared in a microwell array, which combines the advantages of lipoplexes as an efficient carrier system, surface-mediated delivery, and the control of surface topography. Uniform disc-like lipoplexes containing nucleic acids are formed in the microwell array with a diameter of ~ 818 nm and thickness of ~ 195 nm. The microwell array-mediated delivery of lipoplexes containing FAM-oligodeoxynucleotides is ~ 18.6 and ~ 10.6 times more efficient than the conventional transfection method in an adherent cell line (A549 non-small cell lung cancer cells) and a suspension cell line (KG-1a acute myelogenous leukemia cells), respectively. MicroRNA-29b is then used as a model nucleic acid to investigate the therapeutic efficacy of lipoplexes delivered by the microwell array. Compared to conventional transfection methods, the effective therapeutic dosage of microRNA-29b is reduced from the microgram level to the nanogram level by lipoplexes prepared in the microwell array. The microwell array is also a very flexible platform. Both nucleic acid therapeutics and imaging reagents are incorporated in lipoplexes and successfully delivered to A549 cells, demonstrating its potential applications in theranostic medicine.
1. Introduction
Nucleic acid based therapeutics, such as plasmid DNA, anti-sense oligodeoxynucleotides (ODN), small interfering RNA (siRNA), and microRNA, hold great potential for treating a variety of diseases, including many types of cancer and hereditary diseases. Currently, more than 1450 nucleic acid based therapeutic products are under clinical trial worldwide. [1] To improve the therapeutic efficacy of nucleic acid based therapeutics, many researchers have focused on the design of carrier systems, such as lipoplexes and polyplexes, the development of novel delivery routes, such as surface mediated delivery, and the control of substrate topography. Here, we report a novel and facile delivery method that efficiently delivers nucleic acids by lipoplexes prepared in a microwell array, which combines the advantages of the lipoplexes carrier system, the surface mediated delivery and the control of surface topography to achieve more efficient nucleic acid delivery that is not attainable by conventional transfection methods or other techniques.
Lipoplexes formed by the electrostatic interaction between cationic lipids and nucleic acids represent an important non-viral carrier system that has shown great potential in delivering nucleic acids in vitro and in vivo. [2] Lipoplexes are fairly non-toxic and have good biocompatibility. They can incorporate targeting ligands on the surface to provide targeted delivery. Multiple components can also be encapsulated in the lipoplexes to serve as theranostic nanomedicine.
Surface mediated delivery provides spatial and/or temporal control over the delivery of nucleic acid-based therapeutics in vitro and in vivo. [3,4] Currently, surface mediated transfection mainly relies on multilayer films created by an iterative layer-by-layer (LbL) assembly technique to deliver drugs/nucleic acids to adjacent cells or to release cargo into the surrounding media. Transfection through immobilized vectors on surfaces can prevent aggregation and improve delivery efficiency by presenting surface-bound nucleic acids directly to cells. [3–5] Surface medicated delivery plays important roles in biomedical applications that involve artificial transplants, cardiovascular stents and scaffolds, where therapeutic reagents including nucleic acids, antibiotic drugs and anti-inflammatory drugs are released from the surface directly at the site of action to achieve better therapeutic efficacy. [4–6] Recently surface mediated delivery has also been applied in guided differentiation of stem cells and transdermal delivery of vaccines and adjuvants. [6]
Micro/nanotopography has been found to induce changes in a wide range of cellular responses, including cell adhesion, spreading, migration, proliferation, gene regulation and cellular differentiation. [7] However, little investigation has been done to evaluate the effects of substrate topographies on cellular transfectability and nucleic acid delivery efficiency. [8,9] Teo et al. correlated the increased endocytosis caused by micro/nanotopography with increased transfection efficiency. [8] Specifically, they reported that human mesenchymal stem cells grown on 200 nm Poly(methyl methacrylate) (PMMA) pillars had a 2.5 fold increase in GFP expression relative to a smooth PMMA control following traditional Lipofectamine based transfection, which they attributed to differential protein adsorption and cytoskeletal arrangements that affected the plasma membrane tension and in turn, endocytosis rates. [8] Adler et al. investigated the effects of microscale topographies on transfection efficiency. [9] They determined that the spacing between microwells was a more important factor than the well diameter. On dense micropit patterns with 1 μm spacing between the features, as opposed to 2, 4, or 6 μm, GFP expression was increased 25% in primary human dermal fibroblasts after Lipofectamine transfection. These studies investigated topographical effects on endocytosis and transfection using the conventional transfection method. Our study is the first to investigate transfection of lipoplexes containing nucleic acids placed inside the microwell array.
We prepared lipoplexes inside the microwell array by a discontinuous dewetting technique. (Figure 1) This technique represents a straightforward way to quickly and uniformly fill very small wells with liquid by exploiting the differences in the interfacial free energies of the substrate and the liquid. [10] If the substrate bearing the small wells has a lower free energy than the liquid that is draining the surface, each well will be filled with an equal volume of liquid but the surface (or ridges) will dewet. As the receding liquid meets the top edge of the well, the droplet inside the well is pinned due to the abrupt change in contact angle. Then, as the liquid further drains the surface, the thin film at the bottom of the well is ruptured leaving the well filled with the liquid droplet. Taking advantage of this phenomenon, we filled the microwells with lipoplexes in a well-defined way, thereby systematically controlling the dosage to the cells.
Figure 1.
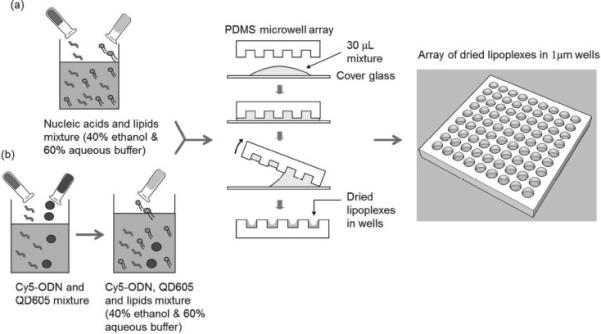
Distribution of (a) lipoplexes containing nucleic acids and (b) multifunctional lipoplexes containing Cy5-ODN/QD605 in the microwell array by discontinuous dewetting.
We first demonstrated microwell array mediated delivery of lipoplexes containing fluorescein amidite labeled ODN (FAM-ODN) in both an adherent cell line (A549 non-small cell lung cancer cells) and a suspension cell line (KG-1a acute myelogenous leukemia cells). MicroRNA-29b was then used as a model nucleic acid to investigate the therapeutic efficacy of lipoplexes delivered by the microwell array. Finally, both nucleic acid therapeutics and imaging reagents were incorporated in lipoplexes formed in the microwell array to evaluate the potential of this method for theranostic medicine.
2. Results
2.1. Lipoplexes Prepared in the Microwell Array
As shown in Figure 1, nucleic acids (FAM-ODN or miR-29b) and imaging reagents (Quantum dots 605, or QD605) in HEPES buffer were first mixed with lipids in ethanol solution. The dewetting process successfully and uniformly formed lipoplexes in the microwell array as shown in Figure 2 a. Our method is similar to the ‘ethanol dilution’ method introduced by Semple et al. [11] After the quick evaporation of ethanol and buffer, lipoplexes were formed inside the microwell array. The lipoplexes were transferred to the mica by gentle push and observed using atomic force microscopy (AFM). As shown in Figure 2 b, the AFM image showed that the resulting lipoplexes in the microwell array had ‘disc-like’ shape with the diameter of 817.6 ± 97.9 nm and the thickness of 194.5 ± 37.1 nm. When exposed to cell culture media (without cells), the lipoplexes were found to be quite stable at the first couple of hours, and then dissolved out of the microwell array and reached background levels after ~ 48 h. (Supporting Information Figure S1).
Figure 2.
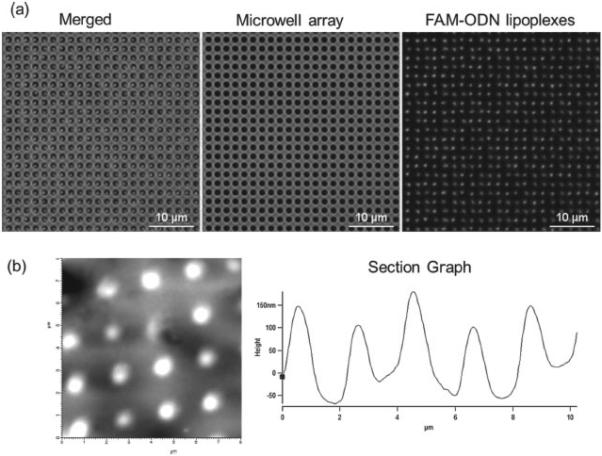
(a) Fluorescent microscopy images of lipoplexes containing FAM-ODN in the microwell array (b) AFM images of lipoplexes containing FAM-ODN.
2.2. Cellular Uptake of Lipoplexes Containing FAM-ODN by A549 and KG-1a Cells
Lipoplexes containing FAM-ODN were distributed in the microwell array at the FAM-ODN concentrations of 0, 4.4, 8.8, 16, 22.2, and 44.4 ng per device. A549 cells (adherent cells) and KG-1a cells (suspension cells) were seeded on top of the microwell array and incubated for 4 h. (Supporting Information Figure S2) The cell seeding density for the transfection experiments was optimized so that the majority of the polydimethlysiloxane (PDMS) stamp surface would be covered with a mono-layer of non-overlapping cells. For example, a 15 μm diameter cell would cover about 44 wells ( Figure 3 a). A high seeding density was desired to minimize material waste by ensuring that most of the microwells would be covered by cells and therefore most of the lipoplexes within the microwells were utilized for cellular uptake (Figure 3 b). The sticky PDMS reservoirs used in the experiments hold about 500 μL of liquid and for the 12 mm of exposed stamp surface, the optimized cell density was found to be ~ 500 000 cells/mL.
Figure 3.
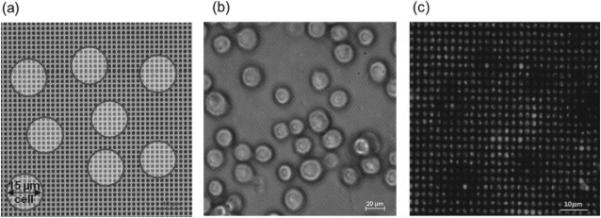
(a) Schematic diagram and (b) microscope image of cells located on top of microwell array during transfection (c) lipoplexes containing FAM-ODN in the microwell array 4 h after transfection with A549 cells removed. Cell outlines are evident on the microwell array stamp following cell removal.
Conventional transfection, i.e. bulk mixing of lipoplexes with cells, was also carried out as the control. Here, A549 cells and KG-1a cells were plated on empty microwell arrays, and the lipoplexes were then added to the cells to achieve the same concentration of nucleic acids as in the microwell array mediated transfection.
The cellular uptake of lipoplexes containing FAM-ODN delivered by the microwell array was analyzed 4 h post transfection using fluorescence microscopy and flow cytometry, and compared with conventional transfection method. Fluorescence microscopy provided qualitative analysis while flow cytometry provided quantitative analysis of the cellular uptake of lipoplexes. After transfection, the cells were removed from the microwell array by gentle pipetting and scraping. The outlines where the cells once were can clearly be seen on the device (Figure 3 c), indicating the successful delivery of lipoplexes by the microwell array. Figure 4 shows a typical set of A549 cellular uptake results by fluorescence microscopy (Figure 4 a) and flow cytometry (Figure 4 b) at FAM-ODN concentration of 4.4 ng per device.
Figure 4.
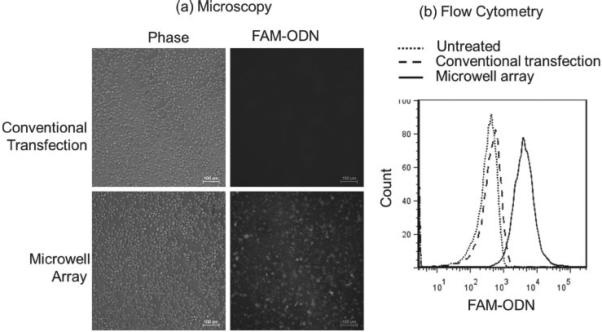
(a) Fluorescence microscopy images and (b) a representative set of flow cytometry data showed that microwell array mediated delivery of lipoplexes containing FAM-ODN to A549 cells was more efficient than conventional transfection at FAM-ODN concentration of 4.4 ng per device.
Fluorescence microscopy images showed that A549 cells transfected with the conventional method had little uptake of lipoplexes, whereas the cells on the microwell array showed a strong fluorescence signal. Flow cytometry also demonstrated higher cellular uptake of lipoplexes delivered by the microwell array compared to the conventional method.
The cellular uptake of lipoplexes at different FAM-ODN concentrations was quantified by flow cytometry using the mean fluorescence intensity (MFI) as shown in Figure 5 a (A549) and Figure 5 b (KG-1a). The microwell devices had much higher MFI than the equivalent conventional transfection variables. For example, The A549 cells from the microwell devices containing 22.2 ng FAM-ODN had ~ 18.6 times more fluorescence than the ones from the conventional transfection method, demonstrating that microwell array more efficiently delivered lipoplexes than its conventional counterpart.
Figure 5.
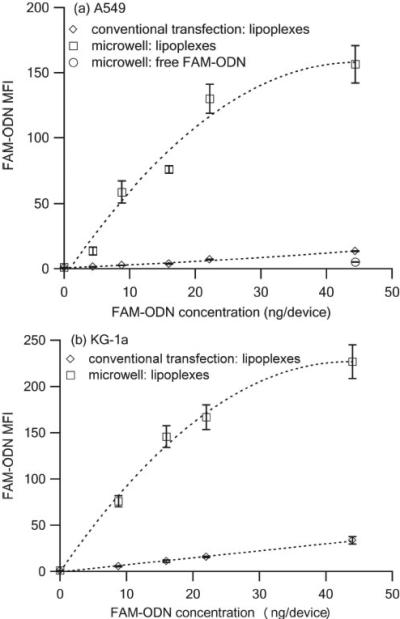
Mean fluorescence intensity (MFI) of (a) A549 cells and (b) KG-1a cells 4 h after transfection with lipoplexes containing FAM-ODN delivered via microwell array and conventional method. (n = 3)
In addition, for microwell array, at lower FAM-ODN concentrations ( < 22.2 ng/device), the MFI of A549 and KG-1a cells maintained a good linear relationship with the FAM-ODN concentrations, however, at higher FAM-ODN concentration, i.e. 44.4 ng per device, the FAM-ODN fluorescent signals of both A549 cells and KG-1a cells reached saturation, which may be due to the fluorescence saturation effect. For conventional transfection, no fluorescence saturation was ever met since the delivery efficiency was much lower, and therefore a good linear relationship between MFI and FAM-ODN concentration was observed for A549 cells and KG-1a cells.
Free FAM-ODN was also distributed in the microwell array with no lipids. Compared to its lipoplexes counterpart, the cellular uptake of free FAM-ODN in the microwell array was ~ 30.8 folds lower, which might be due to the poor interaction between FAM-ODN and negatively charged cell membrane and the strong association between FAM-ODN molecules and PDMS.
2.3. Mature miR-29b Expression and Target ID1 mRNA Down-Regulation in A549 Cells
In the United States, lung cancer is currently the leading cause of cancer death and the overall 5-year survival rate is only 15%. [12] Several studies to date have identified differential expression of microRNAs in lung tumors, microRNAs that carry prognostic information, and micro-RNAs that alter cancer cell phenotype by targeting biological pathways critical to tumorigenesis. [13,14] The microRNA-29 (miR-29) family is often down-regulated in lung cancer and appears to play an emerging role in tumor development and progression. [15] Re-expression of miR-29 homologues in lung cancer represents one potential therapeutic strategy. Compared with siRNA, microRNAs are able to target multiple genes, i.e. ‘one for all’. [16] DNMT3B, MCL1 and ID1 are all validated targets of miR-29b. [15,17,18] However, the ideal mode for miRNA delivery has yet to be determined. The therapeutic efficacy of lipoplexes containing miR-29b delivered by the microwell array and conventional method was evaluated by the down-regulation of ID1 expression in our study.
A549 cells were transfected by lipoplexes containing miR-29b delivered through the microwell array and the conventional method at miR-29b concentrations of 22.2, 27.75, 33.3, and 44.4 ng per device. Lipoplexes containing microRNA mimic negative control (miR-NC) were also delivered at 44.4 ng per device as the second negative control in addition to untreated cells. At 48 h post transfection, quantitative real time PCR (qRT-PCR) was used to measure the expression of mature miR-29b in order to quantitatively compare the delivery efficiency between the microwell array and the conventional method (Figure 6a). At the miR-29b concentration of 44.4 ng per device, mature miR-29b expression of the microwell array delivery was ~ 2463.88 fold higher than the untreated control, whereas conventional transfection only led to ~ 2.21 fold increase in mature miR-29b expression compared to the untreated control. As expected, A549 cells transfected with lipoplexes containing miR-NC did not show any mature miR-29b expression. We also observed a good linear relationship between the mature miR-29b expression and the concentrations of miR-29b deposited in the micro-well array (Figure 6 b), suggesting that the microwell array mediated delivery has the capacity to control the delivered dosage. The target ID1 mRNA expression was successfully down-regulated by lipoplexes containing miR-29b delivered by the microwell array. As shown in Figure 6 c, at a miR-29b concentration of 22.2 ng per device, ID1 mRNA expression was down-regulated ~ 38% compared to untreated control for microwell array mediated transfection. The down-regulation was further enhanced to ~ 52% when the miR-29b concentration increased to 44.4 ng per device. However, no significant down-regulation for ID1 mRNA expression was observed when lipoplexes containing miR-29b were delivered by the conventional transfection method.
Figure 6.
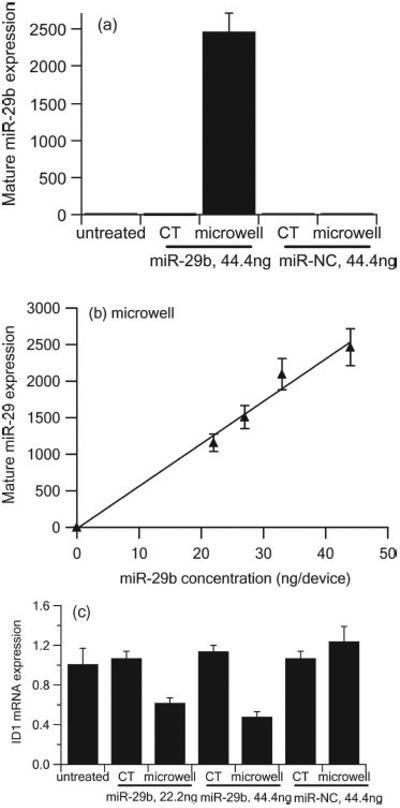
Expression of mature miR-29b (a,b) and target ID1 mRNA (c) in A549 cells 48 h after administration of lipoplexes containing miR-29b or miR-NC by microwell array and conventional transfection (CT) as measured by qRT-PCR and normalized to untreated control. (n = 3)
2.4. Microwell Array Mediated Delivery of Lipoplexes Containing QD605/Cy5-ODN in A549 Cells
Multifunctional theranostic particulates combining therapeutics with imaging/diagnostic reagents represent a new therapeutic approach to diagnose, prevent and treat life-threatening diseases. Here, we demonstrated that multifunctional lipoplexes were formed in the microwell array and delivered to cancer cells more efficiently than the conventional method. Cy5-ODN and QD605 were chosen as the model therapeutic reagent and the model imaging reagent. Cy5 and QD605 also formed the Föster resonance energy transfer (FRET) pair. Upon excitation at 405 nm, the excited donor (QD605) transfers energy to the acceptor (Cy5) when the donor is close to the acceptor, and therefore the observation of QD-FRET mediated Cy5 fluorescence indicates the compact and intact lipoplexes.
Lipoplexes containing QD605/Cy5-ODN were distributed in the microwell array and delivered to A549 cells. At 4 h post transfection, QD-FRET mediated Cy5 fluorescence was observed in A549 cells, indicating successful distribution of multifunctional lipoplexes in the microwell array (Figure 7a). In addition, the MFI of QD-FRET mediated Cy5 from A549 cells transfected by the lipoplexes in the microwell array was ~ 12.3 folds higher than the cells treated by conventional transfection, demonstrating that microwell array delivery was a more efficient transfection method (Figure 7 b).
Figure 7.
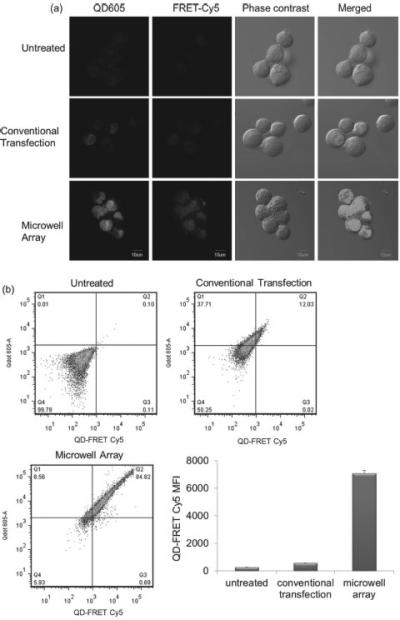
(a) Confocal microscopy images and (b) flow cytometry demonstrated that microwell array mediated delivery of multifunctional lipoplexes containing QD605/Cy5-ODN was ~ 12.8 fold more efficiently than conventional transfection. Transfection was done with A549 cells at Cy5-ODN concentration of 22.2 ng per device and QD605 concentration of 0.11 pmol per device. (n = 3)
3. Discussion and Conclusion
In this work, we distributed lipoplexes containing nucleic acid uniformly in the PDMS microwell array using the discontinuous dewetting technique without any surface modifications to the PDMS or chemical/biological modifications to the lipoplexes. The resulting lipoplexes had disc-like shape with the diameter of ~ 817.6 nm and the thickness of ~ 194.5 nm. Compared to other surfaced mediated delivery techniques, i.e. the LbL assembly technique and the microarray based technique, our method is easier and more cost-effective. The LbL assembly technique has been used to make surface-mediated transfection substrates [3,4,19–23] (also called ‘solid phase delivery’ [24] or ‘reverse transfection’ [25]). Surface properties can also be changed by adding another layer and surface patterning techniques can even be incorporated. [26] However, in practical terms, the repetitive dipping and washing is tedious and time consuming. [22] A single polyelectrolyte bilayer requires up to 6 dipping steps and could take 15 min. Successful transfections require up to 16 bilayers. This together with LbL polymeric pretreatments can result in up to 150 dips/washes for over 6 h. [23] With the LbL technique, both sides of the substrate are coated with the polyelectrolyte bilayers by virtue of the dipping process yet only one side is utilized for transfection, which reduces the yield in half right from the beginning. Many surface-mediated transfection methods used unconventional transfection protocols in which the substrate is placed on top of pre-cultured cells leaving one to wonder if this imposes unnecessary diffusion constraints for the nutrients in the media to the cells. [22,23,27] Furthermore, the liquid volume required to complete the dipping step is rather large in order to accommodate the dimensions of the glass slide which can become an economical hindrance for expensive, custom-designed biomolecules such as miRNA or siRNA. Moreover, without more complicated surface patterning techniques like microcontact printing, the LbL dip-coating technique can only incorporate the components on the substrate randomly with little control over how uniformly the DNA is distributed on the surface. With our microwell array, the well filling process requires just one step and the amount of the lipoplex components can be controlled based on the initial solution concentration and the volume constraints of each microwell.
Array-based nucleic acid delivery on substrates has been investigated previously. [25,28] Generally, a robotic arrayer is required to print the nucleic acids on a glass slide, which is then dip coated with lipids. Alternately, the nucleic acids/lipid complexes are pre-formed and then arrayed on the glass slide. The spot sizes of the array features are on the order of hundreds of microns as opposed to the 1 μm diameter wells used in this study. Due to the small size of the wells, our microwell array allows for more precise control over dosage to an individual cell based on the amount of lipoplexes in each well, the diameter of the cell and the corresponding number of wells the cell would cover. Lateral spread of the siRNA/lipid spots is an issue in the published literature, [28] which is a problem that may be obviated with the use of lipoplexes in our studies as they are fully contained within the microwells.
Arrays of empty liposomes have been achieved using biotin-avidin biological bonding. [29–31] One method described as ‘diffusion-limited, hierarchical self-assembly’ used microcontact printing to pattern biotinylated bovine serum albumin (BSA) into an array of 100 × 400 nm square dots with 800 nm spacing, followed by surface passivation of the non-printed areas using BSA (without biotin), conjugation of the biotin with streptavidin and finally, incubation of extremely dilute, pre-formed biotinylated liposomes on top of the biotin-streptavidin array, forming an array of liposomes on the surface. [29] In short, each ‘dot’ on the glass slide consisted of an initial biotin layer, a streptavidin layer, and finally a tethered biotinylated liposome. Another technique used an array of 1.2 μm diameter wells with 3 μm spacing and 0.55 μm depth etched into a silicon oxide substrate. [30] The inside of the silicon oxide wells was selectively surface-modified with neutravidin whereas the ridges were passivated with a microcontact printed self assembled monolayer of PEG before the surface was incubated for 3 h with pre-formed ~ 1 μm diameter biotinylated liposomes, resulting in an array of liposomes inside the microwells. Both of these methods to form liposome arrays require many surface modification steps and microcontact printing. In our studies, PDMS can be casted repeatedly on the silicon wafer to create micro-well arrays. Our dewetting method to fill the microwells does not require any time consuming and complicated hierarchical surface treatments and it can be accomplished in a single step without the biotin-avidin bonding chemistry and extended incubation times with the liposome solutions (over 3 h).
We have demonstrated that the microwell array can be used to transfect both adherent A549 cells and suspension KG-1a cells in a linear, dosage dependent manner. Cellular uptake of lipoplexes containing nucleic acids using the micro-well array mediated delivery was found to be superior to the conventional transfection method, which led to significant improvement of therapeutic efficacy. For lipoplexes containing FAM-ODN, the microwell array mediated delivery showed ~ 18.6 times more cellular uptake for A549 cells and ~ 10.6 times for KG-1a cells at the FAM-ODN concentration of 22.2 ng per device. When lipoplexes containing miR-29b were delivered by the microwell array to A549 cells at the miR-29b concentration of 44.4 ng per device, the mature miR-29b expression was ~ 1116 fold higher than the cells transfected by the conventional method. And the target ID1 mRNA expression was successfully down-regulated by ~ 52% in A549 cells treated by microwell array mediated transfection. On the other hand, no significant down-regulation was observed by the conventional transfection due the limited delivery of miR-29b. Since effective conventional transfections often require microgram concentrations of miRNA, [15] our microwell array mediated delivery of miRNA can provide significant savings on materials.
We believe that the efficient delivery of nucleic acids by the microwell array may be attributed to the combination of lipoplexes as the carrier system, the surface mediated delivery and the surface topography. When free FAM-ODN was distributed in the microwell array at concentration of 44.4 ng per device, A549 cellular uptake of free FAM-ODN from the microwell array was even less than conventional transfection, suggesting that free FAM-ODN molecules strongly associated with PDMS, making uptake more difficult. However, when FAM-ODN was distributed in the microwell array in the form of lipoplexes, the A549 cellular uptake increased ~ 30.8 fold, demonstrating that lipoplexes are an efficient carrier system for nucleic acid delivery. Surface mediated delivery also helped increase the cellular uptake of lipoplexes. In the micro-well array setup, both adherent cells and suspension cells were brought toward the surface by gravity and directly contacted with lipoplexes. In conventional transfection, lipoplexes were suspended in the solution and the chance for cells to contact with lipoplexes was much lower, and therefore therapeutic efficacy was only observed at much higher dosage. Compared with lipoplexes containing FAM-ODN delivered by the conventional transfection, microwell array mediated delivery increased A549 cellular uptake of lipoplexes by ~ 18.6 fold at FAM-ODN concentration of 22.2 ng per device, indicating that surface mediated delivery is a highly efficient transfection method. In addition, the substrate topography and chemistry may affect the transfection process through physical and biochemical cell-substrate interactions. [9] Cells usually interact with the substrates through integrins, which initiate the assembly of focal adhesion complexes. [32] Many studies have shown a strong correlation between the integrin function and the clathrin and caveolae medicated endocytosis, two of the major endocytic processes. [33–35] The substrate topography and chemistry may alter the density and conformation of integrin on the cells, and thus affect the clathrin and caveolae mediated endocytosis. [36,37] In our study, the microwell array created patterned substrate topography, while the lipoplexes in the microwells presented different surface chemistry to cells, which might have improved the endocytic uptake of lipoplexes and thus enhance the transfection efficiency.
Finally, microwell array is also a very flexible platform that can be used to deliver multifunctional lipoplexes for applications in theranostic medicine. Lipoplexes containing QD605 and Cy5-ODN were successfully distributed in the microwell array and delivered to A549 cells. QD-FRET mediated Cy5 fluorescence was ~ 12.3 fold higher in the cells treated by microwell array than conventional transfection.
In summary, microwell array mediated delivery of lipoplexes containing nucleic acids is a novel, simple and cost-effective method. It showed much more efficient delivery than the conventional transfection, and thus significantly enhanced the therapeutic efficacy at much lower effective dose of therapeutic reagents. In the future, the microwell array may be combined with various transfection reagents to improve in vitro transfection efficiency and achieve spatial and temporal controlled delivery, which may have great potential in applications such as controlled stem cell differentiation. In vivo, the microwell array delivery system may be incorporated with transplants, stents and scaffolds to deliver therapeutic reagents for tissue engineering, or be used in transdermal delivery of vaccine and therapeutics. In the future, automated discontinuous dewetting process (Supporting Information Figure S3) can be developed to deposit lipoplexes or other therapeutic drugs on almost all kinds of substrates as long as there are differences in the interfacial free energies of the substrate and the liquid. When the cells are in contact with the surface, the lipoplexes or other therapeutic drugs will be efficiently delivered to the cells to achieve better therapeutic efficacy.
4. Experimental Section
Fabrication of 1 μm PDMS Microwell Array
The 1 μm microwell array was prepared using standard soft lithography techniques by casting polydimethlysiloxane prepolymer (PDMS) and curing agent (Sylgard 184, Dow Corning, Midland, MI) at a 10:1 weight ratio onto patterned silicon wafers. Briefly, cleaned silicon wafers were spin coated with positive tone AZ5214E photoresist to a thickness of ~ 1 μm followed by a soft baking step at 90 °C for 1 min. The wafers were then loaded into a GCA I-line (365 nm) stepper and the photo resist was exposed through a photomask with 5 μ m features and reduced ~ 5 times by the stepper. The pattern was repeated with no spacing in between each square, resulting in ~ 52 squares that covered most of the wafer's surface. The optimum exposure dose was determined to be ~ 90 mJ/cm 2 based on a previously conducted exposure test. A subsequent image reversal bake (115 °C for 1 min), followed by a flood exposure was implemented. The processed photoresist was finally developed in MF319 developer for 1 min resulting in pillars with 1 μm diameter, 1 μm height, and 2 μm center-to-center distance (1 feature per 4 μm 2 ) and after casting the wafer, the PDMS wells had the same dimensions (with 1 μm depth instead of height). The wafers were treated with hexamethyldisilazane (HMDS, vapor phase) at 100 °C for 30 min prior to each casting to facilitate demolding. After the PDMS had cured at room temperature for 2 days, the patterns were demolded and sonicated in isopropanol and then washed with water and ethanol. Round microwell stamps with a 17 mm diameter for experiments were cut out from the cast PDMS. Round doughnut shaped reservoirs composed of ‘sticky PDMS’ (PDMS cast at a prepolymer to curing agent ratio of 30:1) with a ~ 25 mm diameter and a 12 mm diameter hole in the middle were also cut out and these were used to hold the volume containing the cells and media. The microwell stamp and reservoir setup were shown in Supporting Information Figure S2. The stamps and reservoirs were sterilized by washing with water, spraying with 70% ethanol and leaving under UV light overnight in the cell culture hood.
Prepare Lipoplexes Containing Nucleic Acids (FAM-ODN or miR-29b) in Microwell Array Using the Dewetting Method
FAM-ODN (FAM-G3139, Sequence 5’-FAM-TCT CCC AGC GTG CGC CAT-3′), a 18mer single stranded phosphorothioate oligodeoxyribonucleotide, was purchased from Alpha DNA Inc. (Montreal, Canada). MicroRNA-29b (miR-29b) mimic and microRNA mimic negative control #1 (miR-NC) were purchased from Dharmacon, Inc. (Chicago, IL). 1, 2-Dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP) was purchased from Avanti Polar Lipids Inc. (890890P, Alabaster, AL). Cholesterol was purchased from Sigma-Aldrich Inc. (C3045, St. Louis, MO). Methoxy-polyethylene glycol (MW ≈ 2000 Da)-distearoyl phosphatidylethanolamine (PEG-DSPE) were obtained from Lipoid (Newark, NJ). As shown in Figure 1 a, lipids in ethanol solution (DOTAP/Cholesterol/DSPE-PEG2k = 49/49/2 molar ratio) and nucleic acids (FAM-ODN or miR-29b) in HEPES buffer (20 mM, pH = 7.4) were mixed at varying concentrations to achieve lipid/nucleic acid weight ratio of 10, nucleic acids concentrations of 0.2, 0.4, 0.72, 1, 1.25, 1.5 and 2 μg/ μL, and 40% ethanol and 60% aqueous in the final mixture. After brief sonication, the mixture (30 μL) was applied to a cleaned and sterilized glass slide. A sterilized PDMS microwell stamp was immediately placed on top of the mixture. Care was taken to ensure that the liquid “cushion” below the stamp completely covered the entire stamp and that no air bubbles were trapped in the wells by applying firm but gentle pressure. When it was visually apparent that all wells were filled with the mixture, the stamp was peeled away from the droplet on the glass slide. If a small droplet of solution remained on the edge of the stamp after the dewetting process, it was gently blotted onto a Kimwipe to remove it. Due to the high surface to volume ratio of the microwells, the liquid evaporated almost immediately and lipoplexes containing nucleic acids were formed in the microwell array at nucleic acids concentrations of 4.4, 8.8, 16, 22.2, 27.75, 33.3 and 44.4 ng per device respectively (total volume of microwells was 22.2 nL).
Prepare Lipoplexes Containing Cy5-ODN and Quantum Dots 605 (QD605) in Microwell Array Using the Dewetting Method
As shown in Figure 1 b, Cy5-ODN (Cy5-G3139, 10 mg/mL, 10 μL, Sequence 5′-Cy5-TCT CCC AGC GTG CGC CAT-3′, Alpha DNA Inc. Montreal,Quebec, Canada) was mixed with QD605 (10 μM, 50 μL, Amine-Functionalized eFluor 605NC, 93-6366-33, eBioscience Inc. San Diego, CA) and incubated at room temperature for 30min, protected from light. The Cy5-ODN/QD605 mixture was then added into lipids in ethanol solution (25 mg/mL, 40 μL, DOTAP/Cholesterol/DSPE-PEG2k = 49/49/2 molar ratio). In the final mixture, the lipid/nucleic acid weight ratio was 10, Cy5-ODN concentration was 1 μg/ μL, and QD605 concentration was 5 μM. The mixture was sonicated briefly, and then deposited into the 1 μm micro-well array using the discontinuous dewetting procedure described above to form lipoplexes containing Cy5-ODN/QD605 in the micro-wells at Cy5-ODN concentration at 22.2 ng per device and QD605 concentration at 0.11 pmol per device.
Lipoplex Distribution in the Microwell Array Analyzed by Fluorescence Microscopy
Lipoplexes containing FAM-ODN were used to investigate the distribution and uniformity of lipoplexes in the microwell array by fluorescence microscopy. For imaging, stamps were inverted with the microwell array side down onto a glass cover slip with a few microliters of HEPES buffer to maintain the ‘wet’ conditions for the lipoplexes. The stamps were imaged on a Nikon Instruments Eclipse TiE inverted microscope (Tokyo, Japan) and captured with a Photometrics Evolve 512 EMCCD camera (Tucson, AZ). The fluorescent light source was a Nikon Intensilight with a 130 Watt short arc mercury bulb using a GFP filter cube with 450-490 nm excitation filter, 495 nm dichroic mirror, and 500 -550 nm emission filter.
Atomic Force Microscopy (AFM) Analysis of Lipoplexes
Lipoplexes containing FAM-ODN were transferred to the mica by gentle push and observed using AFM. AFM was performed in air environment using a MFP-3D-Bio-AFM (Asylum Research, Santa Barbara, CA). The image was acquired using a normal intermittent contact mode (AC mode) with a scan rate of 0.5 Hz, a resonant frequency of 315 kHz and a set point of 0.75 V (25% reduced from free amplitude of the AFM cantilever). Without foreside coated and with backside Al coated, a Si pyramidal cantilever with a nominal spring constant of 40 N/m (NSC-15, Micromash) was mounted in a standard cantilever holder. The Asylum software was used to analyze the surface profile and characterize the size and shape.
Cell Culture
A549 cells (non-small cell lung carcinoma cell line) and KG-1a cells (acute myelogenous leukemia cell line) obtained from the American type Culture Collection (ATCC) (Manassas, VA), were routinely cultured in a 75 cm2 T flask containing 15 mL of RPMI 1640 media supplemented with 10% fetal bovine serum (FBS, Gibco 160000). A549 cells were seeded into 75 cm2 T flasks at a concentration of 5 × 105 viable cells/flask. KG-1a cells were seeded into 75 cm2 T flasks at a concentration of 2 × 105 viable cells/mL. Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 and subcultured every 2 days.
Microwell Array Mediated Delivery of Lipoplexes
After lipoplexes were prepared in the microwell array, the sticky PDMS reservoir was then placed on top of the stamp. A549 cells or KG-1a cells (2.5 × 105 cells) in RPMI 1640 medium containing no serum (500 μL) were added into the reservoir on top of the microwell stamp surface (Supporting Information Figure S2). Conventional transfection, i.e. bulk mixing lipoplexes with cells, was also carried out as the control. In conventional transfection, empty microwell array was used and cells were added in the same way, i.e. the cell mixture (500 μL) was added to the reservoir on top of the empty microwell array, and then the lipoplexes was added to the cell mixture (500 μL) to achieve same concentration of nucleic acids as microwell array mediated transfection. At 4 h post transfection cells were removed from the microwell array by gentle pipetting and scraping, and then transferred to 6 well culture plates and cultured in RPMI 1640 medium containing serum (2 mL).
Cellular Uptake of Lipoplexes Analyzed by Flow Cytometry
Cellular uptake of lipoplexes containing FAM-ODN or lipoplexes containing Cy5-ODN/QD605 was analyzed by flow cytometry (BD LSR II, San Jose, CA). At 4 h post transfection, the cells were removed from the PDMS stamp by gentle pipetting and scraping. Cells were washed with PBS twice and fixed in 4% paraformaldehyde. The fluorescence signals of FAM-ODN were observed in the FITC channel. The fluorescence signals of QD605 and FRET-mediated Cy5 were observed in the QD605 and PE-Cy5 channels respectively. 10 000 events were collected for each sample, and the average results of 3 replicates were reported.
Cellular Uptake of Lipoplexes Analyzed by Fluorescence Microscopy
Cellular uptake of lipoplexes containing FAM-ODN was analyzed with fluorescence microscopy. Cells were placed in the reservoir and after 4 h, the cells were imaged through the PDMS stamp on the Nikon Instruments Eclipse TiE inverted microscope (Tokyo, Japan) for fluorescence activity. The fluorescent signal was observed using a GFP filter cube with 450–490 nm excitation filter, 495 nm dichroic mirror, and 500–550 nm emission filter.
Cellular uptake of lipoplexes containing QD605/Cy5-ODN was analyzed with laser scanning confocal microscopy (Olympus FV1000). At 4 h post transfection, the cells were removed from the PDMS stamp by gentle pipetting and scraping. Cells were washed with PBS twice, fixed in 4% paraformaldehyde, and mounted on glass slides for confocal microscopy analysis. The excitation wavelength was set at 405 nm and the fluorescence signals of QD605 and Cy5-ODN were observed in the QD605 (dichroic mirror 560-620nm) and Cy5 (dichroic mirror 655-755nm) channels respectively.
Expression of Mature miR-29b and ID1 mRNA in A549 Cells Measured by Quantitative Real Time PCR (qRT-PCR)
For miRNA-29b studies, at 48 h post transfection, the A549 cells were washed twice with cold PBS and then TRIzol reagent (1 mL, Invitrogen, 15596-018) was added. Chloroform was then added to extract the total RNA before further purification by isopropanol precipition and 75% ethanol wash. For mature miR-29b expression measurements, the TaqMan microRNA reverse transcription kit (Applied Biosystems, 4366596) was used to reverse transcribe the total RNA into cDNA. TaqMan microRNA assay (Applied Biosystems, Assay ID 000413) was then used for qRT-PCR where mature miR-29b expression was quantified by the ΔΔCT method and normalized to the endogenous control gene, RNU48 (Applied Biosystems, Assay ID 001006) relative to the untreated control cells.
For ID1 mRNA expression measurements, the Taqman high capacity cDNA reverse transcription kit (Applied Biosystems, 4368814) was used for cDNA synthesis from total RNA. The Taqman gene expression assay of ID1 (Applied Biosystems, Assay ID Hs03676575_s1) was then used for qRT-PCR where the expression of ID1 mRNA was quantified by the ΔΔCT method and normalized to the endogenous control gene, GAPDH, which was determined by the GAPDH Taqman gene expression assay (Applied Biosystems, Assay ID Hs02758991_m1) relative to the untreated control cells.
Supplementary Material
Acknowledgements
This work was supported by the National Science Foundation under grant No. EEC-0425625. The affiliations and author information were corrected on July 8, 2013
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Yun Wu, Nanoscale Science and Engineering Center for Affordable Nanoengineering The Ohio State University 174 W 18th Avenue, Room 1012, Columbus, Ohio 43210, USA.
Megan Cavanaugh Terp, Nanoscale Science and Engineering Center for Affordable Nanoengineering The Ohio State University 174 W 18th Avenue, Room 1012, Columbus, Ohio 43210, USA; William G. Lowrie Department of Chemical and Bimolecular Engineering The Ohio State University 140 W 19th Avenue, Room 125A Columbus, Ohio 43210, USA.
Kwang Joo Kwak, Nanoscale Science and Engineering Center for Affordable Nanoengineering The Ohio State University 174 W 18th Avenue, Room 1012, Columbus, Ohio 43210, USA.
Daniel Gallego-Perez, Nanoscale Science and Engineering Center for Affordable Nanoengineering The Ohio State University 174 W 18th Avenue, Room 1012, Columbus, Ohio 43210, USA.
Serge P. Nana-Sinkam, Division of Pulmonary, Allergy, Critical Care and Sleep Medicine DHLRI, 473 W 12th Avenue Room 201, Columbus, Ohio 43210, USA
L. James Lee, Nanoscale Science and Engineering Center for Affordable Nanoengineering The Ohio State University 174 W 18th Avenue, Room 1012, Columbus, Ohio 43210, USA; William G. Lowrie Department of Chemical and Bimolecular Engineering The Ohio State University 140 W 19th Avenue, Room 125A Columbus, Ohio 43210, USA.
References
- 1.Barton C. Products & Prospects to 2018. Vol. 3. Espicom; New York, NY, USA: 2009. Drug Delivery Technologies: Nucleic Acid Delivery Players. [Google Scholar]
- 2.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. Nat. Rev. Cancer. 2011;11:59. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jewell CM, Lynn DM. Curr. Opin. Colloid In. 2008;3:395. doi: 10.1016/j.cocis.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jewell CM, Lynn DM. Adv. Drug Deliver. Rev. 2008;60:979. doi: 10.1016/j.addr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bengali Z, Rea JC, Gibly RF, Shea LD. Biotechnol. Bioeng. 2009;102:1679. doi: 10.1002/bit.22212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zelikin AN. ACS Nano. 2010;4:2494. doi: 10.1021/nn100634r. [DOI] [PubMed] [Google Scholar]
- 7.Anselme K, Bigerelle M. Int. Mater. Rev. 2011;56:243. [Google Scholar]
- 8.Teo BKK, Goh SH, Kustandi TS, Loh WW, Low HY, Yim EKF. Biomater. 2011;32:9866. doi: 10.1016/j.biomaterials.2011.08.088. [DOI] [PubMed] [Google Scholar]
- 9.Adler AF, Speidel AT, Christoforou N, Kolind K, Foss M, Leong KW. Biomater. 2011;32:3611. doi: 10.1016/j.biomaterials.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackman RJ, Duffy DC, Ostuni E, Willmore ND, Whitesides GM. Anal. Chem. 1998;70:2280. doi: 10.1021/ac971295a. [DOI] [PubMed] [Google Scholar]
- 11.Semple SC, Klimuk SK, Harasym TO, Dos Santos N, Ansell SM, Wong KF, Maurer N, Stark H, Cullis PR, Hope MJ, Scherrer P. Biochim. Biophys. Acta. 2001;1510:152. doi: 10.1016/s0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- 12.Siegel R, Ward E, Brawley O, Jemal A. CA-Cancer J. Clin. 2011;61:212. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, Piper-Hunter MG, Crawford M, Nuovo GJ, Marsh CB, Otterson GA, Nana-Sinkam SP. J. Thorac. Oncol. 2009;4:1028. doi: 10.1097/JTO.0b013e3181a99c77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nana-Sinkam SP, Karsies T, Riscili B, Ezzie M, Piper M. Expert Rev. Resp. Med. 2009;3:373. doi: 10.1586/ers.09.30. [DOI] [PubMed] [Google Scholar]
- 15.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan K, Marcucci G, Calin GA, Huebner K, Croce CM. Proc. Natl. Acad. Sci. USA. 2007;104:15805. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garzon R, Marcucci G, Croce CM. Nat. Rev. Drug Discov. 2010;9:775. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mott JL, Kobayashi S, Bronk SF, Oncogene GJ. 2007;26:6133. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothschild SI, Tschan MP, Federzoni EA, Jaggi R, Fey MF, Gugger M, Gautschi O. Oncogene. 2012;31:4221. doi: 10.1038/onc.2011.578. [DOI] [PubMed] [Google Scholar]
- 19.Bengali Z, Shea LD. MRS Bull. 2005;30:659. doi: 10.1557/mrs2005.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bengali Z, Pannier AK, Segura T, Anderson BC, Jang JH, Mustoe TA, Shea LD. Biotechnol. Bioeng. 2005;90:290. doi: 10.1002/bit.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bengali Z, Rea JC, Shea LD. J. Gene Med. 2007;9:668. doi: 10.1002/jgm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jewell CM, Zhang J, Fredin NJ, Lynn DM. J. Control. Release. 2005;106:214. doi: 10.1016/j.jconrel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Jewell CM, Zhang J, Fredin NJ, Wolff MR, Hacker TA, Lynn DM. Biomacromolecules. 2006;7:2483. doi: 10.1021/bm0604808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bielinska AU, Yen A, Wu HL, Zahos KM, Sun R, Weiner ND, Baker Jr JR, Roessler BJ. Biomater. 2000;21:877. doi: 10.1016/s0142-9612(99)00229-x. [DOI] [PubMed] [Google Scholar]
- 25.Ziauddin J, Sabatini DM. Nature. 2001;411:107. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 26.Tang Z, Wang Y, Podsiadlo P, Kotov NA. Adv. Mater. 2006;18:3203. [Google Scholar]
- 27.Mehrotra S, Lee I, Chan C. Acta Biomater. 2009;5:1474. doi: 10.1016/j.actbio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mousses S, Caplen NJ, Cornelison R, Weaver D, Basik M, Hautaniemi S, Elkahloun AG, Lotufo RA, Choudary A, Dougherty ER, Suh E, Kallioniemi O. Genome Res. 2003;13:2341. doi: 10.1101/gr.1478703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamou D, Duschl C, Delamarche E, Vogel H. Angew. Chem. 2003;115:5738. doi: 10.1002/anie.200351866. [DOI] [PubMed] [Google Scholar]
- 30.Kalyankar ND, Sharma MK, Vaidya SV, Calhoun D, Maldarelli C, Couzis A, Gilchrist L. Langmuir. 2006;22:5403. doi: 10.1021/la0602719. [DOI] [PubMed] [Google Scholar]
- 31.Michel B, Bernard A, Bietsch A, Delamarche E, Geissler M, Juncker D, Kind H, Renault JP, Rothuizen H, Schmid H, Schmidt-Winkel P, Stutz R, Wolf H. Chimia. 2002;56:527. [Google Scholar]
- 32.Biggs MJP, Richards RG, Dalby MJ. Nanomed. Nanotechnol. 2010;6:619. doi: 10.1016/j.nano.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao WT, Kunz J. FEBS Lett. 2009;583:1337. doi: 10.1016/j.febslet.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Echarri A, Del Pozo MA. Cell Cycle. 2006;5:2179. doi: 10.4161/cc.5.19.3264. [DOI] [PubMed] [Google Scholar]
- 35.Del Pozo MA, Alderson NB, Kiosses WB, Chiang HH, Anderson RGW, Schwartz MA. Science. 2004;303:839. doi: 10.1126/science.1092571. [DOI] [PubMed] [Google Scholar]
- 36.Lord MS, Foss M, Besenbacher F. Nano Today. 2010;5:66. [Google Scholar]
- 37.Carpenter J, Khang D, Webster TJ. Nanotechnology. 2008;19:505103. doi: 10.1088/0957-4484/19/50/505103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


