FKBP is a nuclear peptidyl-proline isomerase. This study shows that FKBP interacts with nucleolin in an RNA-dependent way and interacts with immature but not mature large ribosomal subunits. It seems likely that FKBP participates in some way in ribosome biogenesis.
Keywords: FK506 binding protein, ribosome biogenesis, nucleolin, nucleus
Abstract
Peptidyl-proline isomerases of the FK506-binding protein (FKBP) family belong to a class of enzymes that catalyze the cis–trans isomerization of prolyl-peptide bonds in proteins. A handful of FKBPs are found in the nucleus, implying that the isomerization of proline in nuclear proteins is enzymatically controlled. FKBP25 is a nuclear protein that has been shown to associate with chromatin modifiers and transcription factors. In this study, we performed the first proteomic characterization of FKBP25 and found that it interacts with numerous ribosomal proteins, ribosomal processing factors, and a small selection of chromatin modifiers. In agreement with previous reports, we found that nucleolin is a major FKBP25-interacting protein and demonstrated that this interaction is dependent on rRNA. FKBP25 interacts with the immature large ribosomal subunit in nuclear extract but does not associate with mature ribosomes, implicating this FKBP's action in ribosome biogenesis. Despite engaging nascent 60S ribosomes, FKBP25 does not affect steady-state levels of rRNAs or its pre-rRNA intermediates. We conclude that FKBP25 is likely recruited to preribosomes to chaperone one of the protein components of the ribosome large subunit.
INTRODUCTION
Peptidyl-proline can exist in either a cis or trans conformation, in which the Ω-angle of the peptide bond differs by 180 degrees between the two states. Although proline isomerization occurs spontaneously, the inter-conversion is slow, and peptidyl-prolyl isomerases accelerate this process by orders of magnitude. Altering the shape or dynamics of proteins via cis–trans exchange is a means to regulate their function. As such, isomerases participate in the coordination of the cell cycle (Lu et al. 1996; Yeh and Means 2007) and transcription (Nelson et al. 2006; Wang et al. 2010; Dilworth et al. 2012), which is presumably accomplished through the recruitment to, and isomerization of, substrate proteins. However, there are examples in which this is not the case, and prolyl isomerases function independent of catalytic activity (Arévalo-Rodríguez et al. 2004; Riggs et al. 2007). Of the 18 annotated FKBP prolyl isomerases in humans, few have been extensively characterized, limiting the resolution of FKBP interactions, substrates, and biological functions.
The human prolyl isomerase, FKBP25, is a nuclear enzyme comprised of an amino terminal basic tilted helix bundle (Helander et al. 2014) and a carboxyl terminal canonical FKBP isomerase domain. Previous reports implicate FKBP25 in chromatin biology and transcription: It regulates the auto-ubiquitination and degradation of MDM2 (Ochocka et al. 2009) and inhibits the DNA binding of the transcription factor YY1 (Yang et al. 2001). Additionally, FKBP25 associates with two proteins that localize in the nucleolus: casein kinase II (CK2) and nucleolin (Jin and Burakoff 1993).
The nucleolus is the major site of ribosome biogenesis within the eukaryotic cell, containing multiple tandem arrays of ribosomal DNA (rDNA) repeats, as well as a variety of factors involved in transcription and preribosomal RNA (rRNA) processing. Nucleolin is integral to ribosomal biogenesis and has extensive roles in rDNA transcriptional regulation (Roger et al. 2003; Rickards et al. 2007), pre-rRNA processing (Ginisty et al. 1998), and preribosome assembly (Bouvet et al. 1998). Although it is established that FKBP25 and nucleolin associate, this interaction has not been further characterized, and the biological consequence of this interaction is not known.
In this study, we perform the first unbiased proteomic screen to define FKBP25's interactome. We find that the majority of interacting proteins are nuclear and nucleolar, which supports the predictions that this FKBP is dedicated to nuclear substrates. Most FKBP25 interactors are either ribosomal proteins or preribosomal processing factors, including nucleolin, which binds to FKBP25 when engaged with rRNA. Since FKBP25 only cofractionates with the 60S subunit in nuclear extract, our data support a model in which this FKBP participates in biogenesis of the large ribosomal subunit. To our knowledge, this is the first report to show an involvement of a human nuclear FKBP in the process of ribosome biogenesis.
RESULTS AND DISCUSSION
FKBP25 localizes to the nucleus and nucleolus and associates with ribosomal proteins
To gain insight into the cellular functions of FKBP25, we performed a proteomic characterization of FKBP25-interacting proteins. To this end, we stably expressed 3×-flag-tagged FKBP25 in HEK293 cells and purified FKBP25 and its associated proteins via Flag affinity chromatography (Fig. 1A). From this material, we performed two separate mass spectrometry methods: an in-gel trypsin digestion on 18 prominent bands followed by MALDI-TOF-TOF, as well as an in-solution trypsin digestion followed by LC/MS analysis. These methods identified 18 and 104 proteins respectively, which represents 113 FKBP25-interacting factors (Supplemental Tables S2, S3). We next performed a functional annotation and enrichment analysis using Database for Annotation, Visualization and Integrated Discovery version 6.7 (DAVID) (Supplemental Table S1; Huang da et al. 2009a,b), which is summarized in Figure 1B. From this analysis, we observe an abundance of ribosomal-related ontologies derived from 59 ribosomal and RNA-processing proteins present in our FKBP25 interactome. Nucleolin was the only protein identified in this screen that has previously been shown to associate with FKBP25 (Jin and Burakoff 1993). The remaining protein interactions identified are novel. These include 35 structural ribosomal proteins mostly of the large subunit (26 RPLs and 9 RPSs) and multiple proteins that are involved in rDNA transcriptional regulation or pre-rRNA processing. The abundance of ribosomal factors suggests an involvement of FKBP25 in ribosome biogenesis.
FIGURE 1.
FKBP25 localizes to the nucleus and nucleolus and associates with proteins in these compartments. (A) Coomassie-stained SDS-PAGE of FKBP25-FLAG immunoprecipitations performed from HEK293 cells. (B) Functional annotation and enrichment analysis of FKBP25-interacting proteins identified in two mass spectrometry experiments. (C) Western blots of FKBP25-FLAG immunoprecipitations from HEK293 cells. (D) Cellular fractionation of HEK293 cells displaying the localization of FKBP25 by Western blot. Tubulin is a cytoplasmic marker, H4 is a nuclear/nucleolar marker, and UBF is a nucleolar marker.
In addition to ribosomal factors, we find a collection of chromatin-associated proteins in FKBP25 enriched material from the in-gel analysis: poly ADP-ribose polymerase 1 (PARP1), replication protein A1 (RPA1), and KRAB-associated protein 1 (KAP1). The remaining chromatin-associated interactions identified by in-solution digestion will be described in a separate publication. This link is notable in light of previous findings that FKBP25 regulates p53 levels via MDM2 (Ochocka et al. 2009), and KAP1, PARP1, and RPA1 all have links to the regulation of p53 in DNA damage (Valenzuela et al. 2002; Bochkareva et al. 2005; Wang et al. 2005). In support of a potential role for FKBP25 in chromatin biology, histone deacetylaces (HDAC) 1 and 2 have been identified as FKBP25-associated proteins (Yang et al. 2001), and Mybbp1a and KAP1 are found in complexes containing HDAC1 and 2 (Schultz et al. 2001; Tan et al. 2012). Although nucleolin was the only reported FKBP25-associated protein identified in our screen, the lack of others being identified may be explained by a cell type dependency. Although we used HEK293 cells for this study, Ochocka et al. (2009) used a combination of MCF-7, U2OS, H1299 and HTC116 cells, whereas Yang et al. (2001) used Jurkat and HeLa cells. Thus, in addition to a possible role in ribosome biology, FKBP25 may also regulate chromatin dynamics and the response to DNA damage.
To validate the mass spectrometry results, we performed FLAG IPs followed by Western blots on the most (nucleolin) and least (KAP1) abundant proteins from the in-gel digestion analysis based on the number of peptides detected. This biological replicate confirms nucleolin and KAP1 copurify with FKBP25 in HEK293 cells (Fig. 1C). As we have not confirmed all interactions by Western blot, there is the possibility that some represent nonspecific interactions. However, the overlapping functional nature of the proteins serves as a validation of the approach taken. Additionally, we purified FLAG-FKBP25 complexes from whole cell, cytoplasmic, and nuclear extracts (Supplemental Fig. S1A). In support of FKBP25's interactions occurring mainly in the nucleus, we observe very few interacting proteins with cytoplasmic FKBP25, whereas the nuclear FKBP25 interactions are similar to that occurring from whole cell extract. This is further supported by FKBP25 only interacting with nucleolin in the nucleus and not the cytoplasmic fraction (Supplemental Fig. S1B).
Given the abundance of nucleolar factors identified as FKBP25-associated proteins, we wanted to confirm that FKBP25 does indeed localize to the nucleolus. Therefore, we performed a cellular fractionation, which revealed FKBP25 localizes to the cytoplasmic, nuclear, and nucleolar compartments (Fig. 1D). Densitometric analysis of FKBP25 ratios in the cytoplasm, nucleus, and nucleolus reveal FKBP25 is mainly localized to the cytoplasm (62%), whereas 23% is nuclear and 15% is nucleolar (Supplemental Fig. S2). In support of this, FKBP25 has an exposed nuclear localization signal (NLS), accumulates in the nucleus upon treatment with the nuclear export inhibitor Leptomycin B (LMB) (Ochocka et al. 2009), and its reported functions are carried out in the nucleus. Additionally, we performed chromatin immunoprecipitation (ChIP) using anti-sera raised against the N terminus of FKBP25. Our data confirm the presence of FKBP25 at rDNA and exhibits a similar occupancy as observed for nucleolin (Supplemental Fig. S3). The presence of FKBP25 in the nucleolus is intriguing because there are no reports of FKBP25 having nucleolar functions; and although FKBP25 has been shown to associate with nucleolin, it has only been speculated to be involved in ribosome biogenesis.
The interaction between FKBP25 and nucleolin is dependent on 28S rRNA
With the abundance of ribosome biogenesis factors associating with FKBP25, we next set out to determine the nature of the interaction between FKBP25 and the ribosomal protein complex. We consistently observe a strong association between FKBP25 and nucleolin as seen by Western blot as well as the number of peptides identified by mass spectrometry, indicating the potential of a direct interaction.
Nucleolin contains a highly acidic N terminus, four RNA recognition motifs (RRMs), and a C-terminal Glycine-Arginine rich region (RGG). In our assays, we used a nucleolin construct containing RRM1-4 and the RGG motif (referred to as nucleolin hereafter); we and others (Yang et al. 1994; Haluska et al. 1998) were unable to express full-length nucleolin protein in bacteria. This is likely due to the highly acidic N terminus and size of full-length nucleolin.
Pulldown assays with GST-FKBP25 and 6His-nucleolin demonstrate that these two proteins interact directly, albeit weakly (Fig. 2A). This interaction requires full-length FKBP25 because GST-fusions to either the amino terminal basic helical bundle (amino acids 1–107; NTD) or to the carboxyl terminal peptidyl prolyl isomerase FKBP domain (107–224; PPI) do not interact with 6His-nucleolin. To identify the minimal FKBP25-interaction domain on nucleolin, we performed reciprocal GST pulldowns using a GST-deletion series of nucleolin domains (Becherel et al. 2006) against full-length FKBP25 (Supplemental Fig. S4B). This experiment identifies the RNA recognition motifs 1 and 2 (amino acids 290–480) of nucleolin as the minimal FKBP25 binding surface.
FIGURE 2.
FKBP25 interacts with nucleolin RRMs 1 and 2 in the presence of rRNA. (A) GST pulldown assays of GST, GST-FKBP25 N-terminal domain (NTD), GST-FKBP25 PPI and GST-FKBP25 FL against 6-His nucleolin RRM1-4 + RGG. Pulldowns were performed with and without rRNA. (B) GST pulldown assays using GST-NCL RRM1-2 (aa 290–480) (see Supplemental Fig. S4) against full-length 6His-FKBP25 in the presence and absence of RNase and supplemented with total RNA from HEK293 cells. (C) FLAG Immunoprecipitations from HEK293 cellular extract with and without pretreatment of RNase A. (D) Trizol extracted RNA from FLAG immunoprecipitations from HEK293 nuclear material electrophoresed on a denaturing formaldehyde-agarose gel. (E) Schematic of rDNA transcript. The position of primer sets used for qPCR is indicated below the schematic. (F) qPCR analysis of reverse transcribed RNA extracted from FLAG immunoprecipitations. GAPDH is used as an mRNA control.
A weak interaction between FKBP25 and nucleolin in vitro (Fig. 2A) is not in agreement with the relatively strong enrichment of nucleolin in FKBP25 immunoprecipitates (Fig. 1). We speculated that either a nucleic acid or protein was necessary to bridge the interaction. Given the number of RNA-binding or modifying proteins associated with FKBP25, we hypothesized that rRNA may promote the interaction between these proteins.
We first tested this hypothesis by supplementing in vitro binding reactions with RNA. GST-pulldown assays were repeated, but they were supplemented with 18S and 28S rRNA purified by sucrose gradient ultracentrifugation and TRIzol extraction. We observe an increase in the amount of nucleolin recovered with GST-FKBP25 (Fig. 2A, right). Even in the presence of rRNA, only full-length FKBP25, and not its composite domains in isolation, interact with nucleolin. To address whether rRNA, and not any cellular RNA, mediates the FKBP25/nucleolin interaction, we performed the same assays in the presence of an equal molar amount of tRNA. Since tRNA cannot promote the nucleolin-FKBP25 interaction (Supplemental Fig. S4c), we conclude that the nucleolin-FKBP25-RNA complex is RNA sequence or structure dependent. We also performed reciprocal pulldowns with GST-NCL290-480 (encompassing RRMs1-2), FKBP25, and rRNA (Fig. 2B). rRNA also promotes this interaction, confirming that rRNA, the RRMs 1 and 2 of nucleolin, and FKBP25 interact in a complex. It is noteworthy that we do observe weak interactions in these GST-pulldown assays without supplemented rRNA. The fact that addition of RNase ablates this weak association (Fig. 2B) implies that recombinant nucleolin may copurify with low amounts of bacterial RNAs, and this facilitates FKBP25 interaction.
To confirm that RNA contributes to the enrichment of nucleolin in FKBP25 immunoprecipitations, we treated nuclear extracts with RNase A prior to immunoprecipitations for FKBP25 (Fig. 2C). In support of our hypothesis, we observe a complete loss of the FKBP25/nucleolin interaction in RNase treated conditions (Fig. 2C). Nucleolin protein levels in the input control were unaffected by RNase treatment, ruling out protein instability or degradation. Thus, RNA is also required for the FKBP25/nucleolin interaction in vivo.
In order to map the RNA species that facilitates the FKBP25-nucleolin interaction, RNA immunoprecipitations were performed. 3×-FLAG FKBP25 was immunprecipitated, and copurifying RNA was separated on an agarose gel, revealing a band the same size as the 28S rRNA (Fig. 2D). The presence of 28S rRNA was not unexpected because we identified 26 ribosomal proteins of the large subunit by mass spectrometry; however, the abundance of other processing factors indicates this may be an immature 28S species. Additionally, we observed a faint slower migrating species, which we suspect represents a pre-28S rRNA transcript that has not been fully processed. To address the possibility that FKBP25 is interacting nonspecifically with 28S rRNA, we also performed immunoprecipitations with another nuclear prolyl isomerase, PPWD1, with no known function in ribosome biogenesis. PPWD1 does not interact with nucleolin or RNA (Supplemental Fig. S1B,C); thus, the interaction between FKBP25 and the preribosome is not an artifact of the system.
To confirm the RNA species we observe in Figure 2D is in fact 28S rRNA and to directly test if preprocessed 28S rRNA species is also bound FKBP25, the abundance of these rRNA transcripts was evaluated by qPCR. We failed to detect any enrichment with the 5′ETS and 18S regions and only a small enrichment of ITS1. The lack of enrichment of 18S rRNA is in agreement with our proteomic data that show FKBP25 interacts predominantly with proteins of the 60S preribosomal subunit. In contrast, we detected enrichment of both ITS2 and 28S, although the 28S was significantly more enriched than ITS2 (Fig. 2F). This relative distribution is expected as the amount of processed 28S in the nucleus significantly outnumbers the amount of the ITS2/28S preprocessed transcript. Thus, FKBP25 can be found on pre-28S rRNA. Together with the data described above and the fact that FKBP25 can be found on rDNA chromatin, we speculate that FKBP25 is recruited to the 47S transcript quite early, and possibly cotranscriptionally. Since the processing machinery is present and potentially functional during FKBP25 enrichment, it is possible that processing of rRNA can still occur, resulting in the greater enrichment of the processed 28S transcript.
These data show that FKBP25 is present at an early step in ribosome biogenesis. They also independently support our mass spectrometry-based prediction that FKBP25 performs a role in ribosome biogenesis of the large 60S subunit.
FKBP25 transiently associates with the pre-60S ribosomal subunit
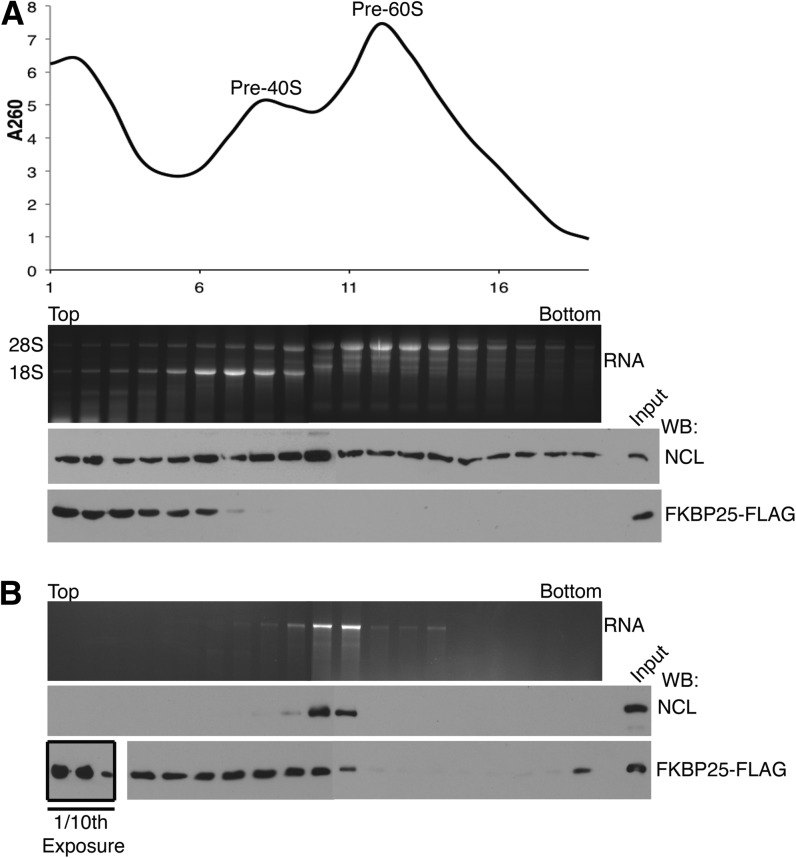
Ribosome biogenesis and rRNA processing factors transiently interact with structural ribosomal proteins and rRNA in the nucleus during 40S and 60S subunit assembly. In contrast, structural ribosomal proteins remain integral to functional ribosomes in the cytoplasm. To resolve whether FKBP25 associates with the mature 60S ribosomal subunit or a pre-60S ribosomal species, we used sucrose density gradient ultracentrifugation to separate the large macromolecular complexes from the smaller protein complexes. First, we subjected a cytoplasmic extract to sucrose gradient ultracentrifugation, followed by Western blotting of fractions for FKBP25 and nucleolin. We were unable to identify an association between FKBP25 and mature ribosomes in cytoplasmic extract (Supplemental Fig. S5A). Similarly, no interaction was observed for nucleolin and mature ribosomes, which supports previous reports that nucleolin associates with preribosomes (Herrera and Olson 1986). To separate pre-40S and pre-60S ribosomal subunits, the same experiment was performed on nuclear extract to enrich for preribosomal species before nuclear export. Nucleolin was broadly distributed and found in every sucrose gradient fraction collected (Fig. 3A). This is in agreement with a previous report which shows nucleolin can be UV cross-linked to both the 18S and 28S transcripts (Herrera and Olson 1986). Although nucleolin evidently associates with the preribosomes, we were unable to identify FKBP25 in the fractions encompassing the pre-40S or pre-60S particles (Fig. 3A).
FIGURE 3.
FKBP25 interacts with the pre-60S ribosome in the presence of nucleolin. (A) Sucrose density gradient ultracentrifugation of HEK293 nuclear extract. RNA and protein were extracted from each fraction and were resolved by denaturing-formaldehyde gel electrophoresis or SDS-PAGE and Western blot. (B) Sucrose density gradient ultracentrifugation of FLAG-FKBP25 immunoprecipitate from HEK293 cells. RNA and protein were extracted from each fraction and were resolved by denaturing formaldehyde gel electrophoresis or SDS-PAGE and Western blot.
We speculated that the interaction between the pre-60S particles might be transient or disrupted by centrifugation, which may explain the absence of FKBP25 entering the gradient. To address this possibility, FKBP25-enriched material was separated by sucrose gradient ultracentrifugation. FKBP25 was mainly found at the top of the gradient (fractions 1–3); however, a longer exposure reveals fractions that contain FKBP25, nucleolin, and the 28S rRNA (fractions 9–11) (Fig. 3B). This confirms that under the conditions of profiling in vitro, a fraction of FKBP25 associates with the pre-60S ribosomal subunit, whereas the remainder sediments with lower molecular weight material. We also performed immunoprecipitations of FKBP25 from cytoplasmic extract (Supplemental Fig. 5B). Although, we do observe a trace amount of 28S rRNA associating with cytoplasmic FKBP25, we speculate that FKBP25 may transit with the 60S subunit to the cytoplasm and is displaced before the final maturation. Regardless, the lack of 18S rRNA indicates that mature ribosomes do not interact with FKBP25. These data support a model in which FKBP25 transiently associates with the pre-60S subunit in the nucleus but not with mature translating ribosomes in the cytoplasm.
FKBP25 does not affect steady-state levels of ribosomal RNA
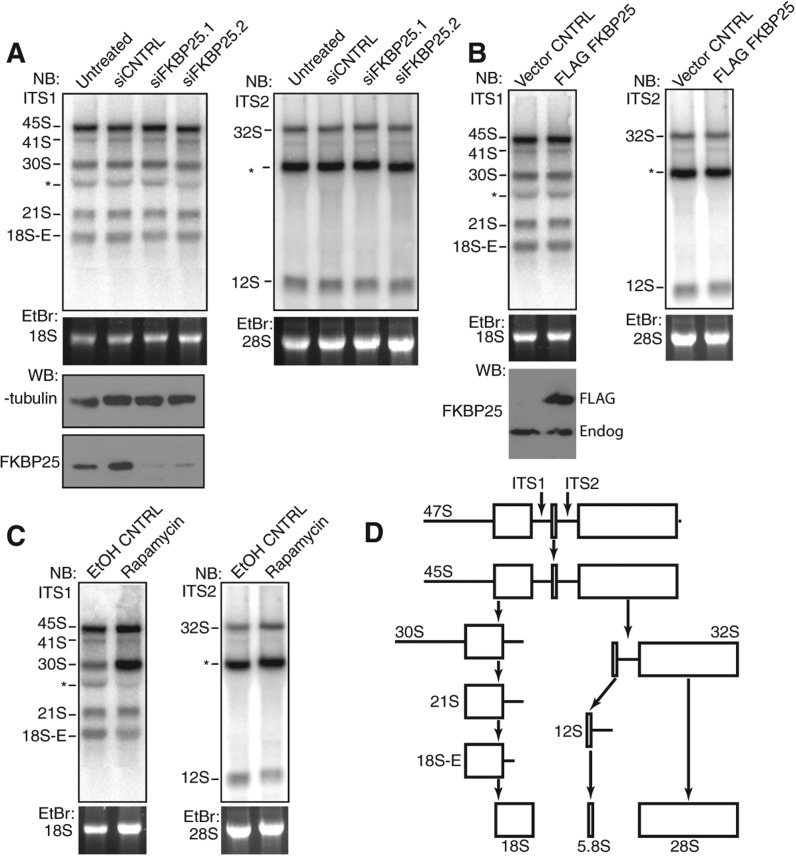
The presence of FKBP25 associating with a pre-60S ribosomal species led us to hypothesize that FKBP25 may affect the processing of rRNA. To explore this, we performed Northern blots on total RNA isolated from cells depleted of FKBP25 (Fig. 4A). We detected no major change of any processed transcript level upon knockdown of FKBP25. Thus, the presence of FKBP25 is not essential for the processing of rRNA. Similarly, we observed no major differences in transcript levels of any processed stage upon overexpression of FKBP25 in comparison to the control (Fig. 4B). Thus, the overexpression of FKBP25 does not affect processing of rRNA.
FIGURE 4.
Overexpression or knockdown of FKBP25 does not affect steady-state levels of pre-rRNA intermediates. (A) Northern blot of RNA extracted from untransfected HEK293 cells or HEK293 cells transfected with control siRNA or FKBP25-targeting siRNA. Blots were probed for ITS1 (left) or ITS2 (right). Extracts were Western blotted for FKBP25 to verify knockdown. (B) Northern blot of RNA extracted from HEK293 from control and cells overexpressing FLAG-FKBP25. Blots were probed for ITS1 (left) or ITS2 (right). (C) Northern blot of HEK293 cells treated with vehicle or rapamycin probed for ITS1 (left) or ITS2 (right). (D) Schematic of rRNA processing steps. (*) Nonspecific probe binding.
As a control to ensure that we could detect changes in rRNA processing, we treated cells with rapamycin, which is well documented to affect rRNA processing through mTOR (Iadevaia et al. 2012). Consistent with previous reports (Iadevaia et al. 2012), we observe a rapamycin-induced accumulation of the 30S transcript control (Fig. 4C).
We conclude that FKBP25 does not have an effect on steady-state levels of pre-rRNA. Neither the depletion or overexpression of FKBP25 had any effect on processing; thus, FKBP25's role within the nucleolus remains elusive.
CONCLUSIONS
Although we did not identify a function for FKBP25 in rRNA processing, the presence of a prolyl isomerase in ribosome biogenesis is intriguing. Fpr4, the S. cerevisiae ortholog of FKBP25, also associates with a number of ribosomal proteins and regulators (Ho et al. 2002; Saveanu et al. 2003; Krogan et al. 2006); however, the role of Fpr4 in ribosome biogenesis has yet to be explored. Moreover, S. pombe FKBP39, a related prolyl isomerase to FKBP25 and Fpr4, acts as a histone chaperone for ribosomal DNA silencing (Kuzuhara and Horikoshi 2004). The involvement of FKBP25 orthologs in ribosome biogenesis, combined with the data presented in this report, strongly suggests a function of FKBP25 in the maturation of ribosomes.
Interestingly, bacteria contain a ribosome-associated chaperone, trigger factor, which is a member of the FKBP family. Although FKBP25 and trigger factor do share a canonical FKBP fold, they have low sequence identity (13.7%) outside the isomerase domain. The N-terminal domain of trigger factor, which engages the translating ribosome (Ferbitz et al. 2004), is not structurally similar to the N-terminal domain of FKBP25 (PDB code 2KFV). Domain I of trigger factor is composed of a four-stranded anti-parallel β-sheet and two α-helices, whereas the N-terminal domain of FKBP25 is composed of five α-helices.
In this study, we have provided evidence that FKBP25 interacts with the pre-60S ribosome. Although we confirmed the presence of FKBP25, nucleolin, and the pre-60S ribosomal subunit in the same fractions by sucrose gradient, only a small proportion of total cellular FKBP25 is stably associated with this ribosome complex under these experimental conditions. This may be a consequence of a weak/transient interaction that is affected by centrifugation. Alternatively, this may reflect the in vivo conditions, and only a small portion of nuclear FKBP25 may be engaged with preribosomes in asynchronous cells.
The catalytic action of prolyl-isomerases is important for the proper folding and function of proteins. Although we cannot fully rule out that FKBP25 may regulate the kinetics of rRNA processing, given the “foldase” action of FKBPs, we speculate that FKBP25 likely functions to chaperone proteins, or the numerous protein–rRNA interactions within the nascent pre-60S ribosomal subunit. Identifying the proline targets within this large nucleo–protein complex is now an important question to address in order to understand the biological function of FKBP25 in ribosome biogenesis.
MATERIALS AND METHODS
Cell lines and transfections
HEK293 Flp-In T-Rex cells (Life Technologies) were maintained in DMEM containing 10% FBS and Pen/Strep at 37°C in 5% CO2. For tetracycline inducible cells, FKBP25 and PPWD1 were cloned into pcDNA5/FRT/TO (Life Technologies) containing a tetracycline-regulated, hybrid CMV/TetO2 promoter, and a triple FLAG epitope tag. Stable cell lines were constructed as per manufacturer's recommendations. Cells were transfected overnight using Lipofectamine 2000 (Life Technologies) in six-well dishes as per manufacturer's recommendations with either 60 nM siRNA or pcDNA5/FRT/TO 3×-FLAG FKBP25 vector. Cells were split in the morning 1:4 and incubated for 48 h. In the case of cells transfected with pcDNA5/FRT/TO 3×-FLAG-FKBP25, 0.1 μg/mL tetracycline was added to the media to induce expression of the transgene.
Cellular fractionation
Cellular fractionation to isolate cytoplasmic, nuclear, and nucleolar components was performed as described previously (Andersen et al. 2005) (http://www.lamondlab.com/pdf/CellFractionation.pdf). Briefly, cells were lysed in a hypotonic buffer with dounce homogenization yielding the cytoplasmic fraction. The nuclear fraction was isolated using a series of centrifugations over sucrose cushions and sonication. Finally, nucleoli were isolated by centrifugation of the nuclear fraction. Extracts were normalized for total protein and resolved by SDS-PAGE and Western blotted as described below.
Immunoprecipitation and Western blot
For whole cell extraction, HEK293 cells (∼3 × 107 cells) were lysed in 0.75 mL immunoprecipitation (IP) buffer (50 mM Tris pH8, 150 mM NaCl, 0.5% IGEPAL, 0.5% Triton X100, 2 mM EDTA, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 μg/mL pepstatin), vortexed for 3 sec, and incubated on ice for 10 min. Insoluble material was pelleted at 10,000 rpm for 10 min. Cytoplasmic and nuclear extractions are described below for sucrose density ultracentrifugation. Extract were normalized for total protein, and buffers were compensated to wash buffer conditions prior to immunoprecipitation. Soluble extracts were added to prewashed EZ-view Red ANTI-FLAG M2 Affinity gel beads (Sigma F2426). For RNase A-treated extracts, 100 μg RNase A (Qiagen) was added to extracts and incubated for 5 min at 37°C prior to addition of FLAG beads. IPs were incubated for 1.5 h at 4°C with nutating. After binding, beads were washed three times in IP buffer (0.75 mL IP buffer was added to beads and nutated for 5 min, then centrifuged at 1000 rpm for 1 min). FLAG-complexes were eluted with 1.25 μg/μL 3×-FLAG peptide (Sigma F4799) by nutating for 15 min at 4°C. For Western blotting, eluted proteins were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Membranes were blocked in 10% skim milk for 30 min and probed in primary antibody overnight at 4°C. Antibodies and dilutions used were the following: 1/50,000 anti-FLAG M2 antibody (Sigma F1804); 1/5,000 anti-Nucleolin (Abcam ab22758); 1/1000 anti-TIF1β (KAP1) (Santa Cruz sc-33186); 1/1000 anti-His probe (Santa Cruz sc-803) anti-UBF (Santa Cruz sc-13125X); anti-H3 (Abcam ab1791), anti-αTubulin (Rockland 600-401-880), and anti-FKBP25 (raised against amino acids 3–18). Blots were washed three times for 10 min in TBST (TBS + 0.1% Tween-20) and incubated in secondary antibody at room temperature for 1–2 h. Horseradish peroxidase conjugated anti-mouse (GE NXA931) or anti-rabbit (GE NA934) secondary antibody was used at 1:5000. Proteins were detected by chemiluminescence (Millipore WBLUF0500) and exposed to film.
Protein expression and GST pulldown assays
Escherichia coli BL21 RIL strains containing pGEX5 × 1 encoding: GST; GST-FKBP25 FL (1-224); GST-FKBP25 NTD (1-107); GST-FKBP25 PPI (108-224); GST-NCL 1-55, GST-NCL 1-140, GST-NCL 130-300; GST-NCL 290-480; GST-NCL 470-650; GST-NCL 640-707 (NCL series a gift from O. Becherel), pET 6-His FKBP25 (1-224), and pET15b encoding 6-His nucleolin RRM-RGG (308-710) were diluted 1:20 from overnight cultures into LB broth. Cells were grown to an OD600 of approximately 0.8. Protein expression was induced with 1 mM IPTG for 4 h at 37°C. Cells were pelleted and resuspended in binding buffer (50 mM Tris pH 8, 150 mM NaCl, 0.5% IGEPAL, 5 mM EDTA, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 μg/mL pepstatin). Cells were sonicated with five 30-sec bursts on high using a Bioruptor (Diagenode). Extracts were subjected to centrifugation at 10,000 rpm for 10 min, and proteins were normalized for loading by SDS-PAGE. For each pulldown reaction, 500 μL binding buffer, 100 μg BSA, RNase inhibitors, and GST/6-His protein extracts were added. Equal molar amounts of tRNA or rRNA containing 18S and 28S transcripts purified from sucrose gradient ultracentrifugation or 100 ng RNase A were added to the corresponding samples. Samples were nutated for 1.5 h at 4°C, then added to prewashed Glutathione-Agarose beads (Qiagen) and allowed to bind for another hour. Protein-bound beads were washed three times and resuspended in SDS loading buffer. Proteins were separated by SDS-PAGE and immunoblotted as described above.
Sucrose density gradient ultracentrifugation
Sucrose density gradient ultracentrifugation was performed as previously described (Yoshikawa et al. 2011). Cell pellets were resuspended in 1 mL buffer A (16.7 mM Tris pH 8, 50 mM NaCl, 1.67 mM MgCl2, 0.1% Triton X-100, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 μg/mL pepstatin), vortexed 10 sec, and incubated on ice for 5 min. Extracts were centrifuged at 1000g for 5 min. The pellet was washed once more in buffer A as above. To extract the nuclear material, the pellet was sonicated 3 × 20 sec in 0.5 mL sonication buffer containing 25 mM Tris pH 8, 100 mM KCl, 2 mM EDTA, 1 mM DTT, 0.05% IGEPAL, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 1 μg/mL pepstatin, and 10 units RNase inhibitor. Nuclear material was overlaid on 10%–30% sucrose gradients containing 25 mM Tris pH 8, 100 mM KCl, 2 mM EDTA, and 1 mM DTT. Gradients were centrifuged at 36,000 rpm for 3 h at 4°C. Immunoprecipitated FLAG-FKBP25 complexes were prepared as above and overlaid on a 10%–30% sucrose gradient with buffer conditions the same as IP buffer minus detergents. Following centrifugation, sucrose gradients were fractioned into 0.5 mL volumes. Absorbance at 260 nm was measured by Nano-Drop spectrophotometry for each fraction and graphed in Excel. Half of each fraction was used to analyze protein and RNA. Proteins from each fraction were precipitated by the addition of one-fourth volume of 100% TCA and incubated for 10 min at 4°C. Precipitated proteins were pelleted at 15,000g for 5 min, and pellets were washed two times with cold acetone. Proteins were subjected to SDS-PAGE and transferred to nitrocellulose membranes as described above. RNA was extracted from each fraction with TRIzol (Life Technologies) as per standard protocols. RNA was visualized by a 1% denaturing agarose-formaldehyde gel electrophoresis and stained with ethidium bromide.
RNA immunoprecipitation and qPCR
FLAG immunoprecipitations were performed as described above. RNA was extracted from eluates with TRIzol as per manufacturer's recommendations. Purified RNA was reverse transcribed using a cDNA kit (Applied Biosystems) with random hexamers as per manufacturer's recommendations. qPCR reactions (20 μL per reaction) contained 10 μL 2X Maxima SYBR Green qPCR Master Mix (Thermo Scientific K0253), 1.25 μM ROX (Thermo Scientific R1371), 0.5 μM oligos, 7.5 μL dsH2O, and 2 μL diluted template cDNA. qPCR reactions were run on a Stratagene MX3000p real-time qPCR system (Agilent Technologies). The thermocycle program included an initial activation step at 95°C (10 min) and 40 cycles of 95°C denaturation (15 sec) and 60°C annealing (30 sec). Specificity of target amplification was analyzed by no DNA template controls and subjecting completed runs to melting curve analysis. Data were analyzed by normalizing IP samples to 10% input samples and then calculating the fold change over the FLAG (-) control sample. Oligonucleotides used for qPCR are described in Table 1.
TABLE 1.
Oligonucleotide sequences used for qPCR and Northern blotting

Northern blot
Total RNA was extracted with TRIzol. Northern blot analysis (adapted from PerkinElmer protocols) was performed by separation on a 1% agarose-formaldehyde gel, run at 200V for 1.5 h, and transferred overnight by capillary transfer to a Nylon membrane (Whatman, Inc., 10416230). The blot was rinsed in 2× SSPE and UV crosslinked (auto crosslink setting, 254 nm, Stratgene, Stratalinker). Membranes were prehybridized for 4–6 h at 42°C in prehybridization solution (5× SSPE, 50% deionized formamide, 5× Denhardt's Solution, 1% SDS, 10% dextran sulfate, and 100 μg/mL sheared salmon sperm DNA). Oligonucleotides were end-labeled with γ 32P (PerkinElmer) and T4 polynucleotide kinase (NEB M0201) for 1 h at 37°C and heat inactivated. Membranes were then incubated overnight with labeled probe. The blots were washed twice with 2× SSPE, 0.1% SDS for 10 min at room temperature and then with 0.2× SSPE, 0.1% SDS until background signal was minimal. Blots were then exposed to a phosphor screen (Molecular Dynamics) and imaged using a Phosphorimager (STORM, GE Healthcare).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Drs. Juan Ausio and Perry Howard for helpful discussions. This work was supported by a grant to C.J.N. from the Canadian Cancer Society Research Institute (CCSRI:2010-700447). G.G. is supported by an NSERC CGS-D scholarship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.042648.113.
REFERENCES
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M 2005. Nucleolar proteome dynamics. Nature 433: 77–83 [DOI] [PubMed] [Google Scholar]
- Arévalo-Rodríguez M, Pan X, Boeke JD, Heitman J 2004. FKBP12 controls aspartate pathway flux in Saccharomyces cerevisiae to prevent toxic intermediate accumulation. Eukaryot Cell 3: 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherel OJ, Gueven N, Birrell GW, Schreiber V, Suraweera A, Jakob B, Taucher-Scholz G, Lavin MF 2006. Nucleolar localization of aprataxin is dependent on interaction with nucleolin and on active ribosomal DNA transcription. Hum Mol Genet 15: 2239–2249 [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, et al. 2005. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci 102: 15412–15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F 1998. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem 273: 19025–19029 [DOI] [PubMed] [Google Scholar]
- Dilworth D, Gudavicius G, Leung A, Nelson CJ 2012. The roles of peptidyl-proline isomerases in gene regulation. Biochem Cell Biol 90: 55–69 [DOI] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N 2004. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431: 590–596 [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J 17: 1476–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolova KV, Nicoloso M, Mazan S, Hadjiolov AA, Bachellerie JP 1993. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur J Biochem 212: 211–215 [DOI] [PubMed] [Google Scholar]
- Haluska P Jr, Saleem A, Edwards TK, Rubin EH 1998. Interaction between the N-terminus of human topoisomerase I and SV40 large T antigen. Nucleic Acids Res 26: 1841–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander S, Montecchio M, Lemak A, Fares C, Almlof J, Li Y, Yee A, Arrowsmith CH, Dhe-Paganon S, Sunnerhagen M 2014. Basic Tilted Helix Bundle—a new protein fold in human FKBP25/FKBP3 and HectD1. Biochem Biophys Res Commun 447: 26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera AH, Olson MO 1986. Association of protein C23 with rapidly labeled nucleolar RNA. Biochemistry 25: 6258–6264 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Iadevaia V, Zhang Z, Jan E, Proud CG 2012. mTOR signaling regulates the processing of pre-rRNA in human cells. Nucleic Acids Res 40: 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YJ, Burakoff SJ 1993. The 25-kDa FK506-binding protein is localized in the nucleus and associates with casein kinase II and nucleolin. Proc Natl Acad Sci 90: 7769–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Kuzuhara T, Horikoshi M 2004. A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat Struct Mol Biol 11: 275–283 [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T 1996. A human peptidyl–prolyl isomerase essential for regulation of mitosis. Nature 380: 544–547 [DOI] [PubMed] [Google Scholar]
- Nelson CJ, Santos-Rosa H, Kouzarides T 2006. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126: 905–916 [DOI] [PubMed] [Google Scholar]
- Ochocka AM, Kampanis P, Nicol S, Allende-Vega N, Cox M, Marcar L, Milne D, Fuller-Pace F, Meek D 2009. FKBP25, a novel regulator of the p53 pathway, induces the degradation of MDM2 and activation of p53. FEBS Lett 583: 621–626 [DOI] [PubMed] [Google Scholar]
- Rickards B, Flint SJ, Cole MD, LeRoy G 2007. Nucleolin is required for RNA polymerase I transcription in vivo. Mol Cell Biol 27: 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs DL, Cox MB, Tardif HL, Hessling M, Buchner J, Smith DF 2007. Noncatalytic role of the FKBP52 peptidyl-prolyl isomerase domain in the regulation of steroid hormone signaling. Mol Cell Biol 27: 8658–8669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger B, Moisand A, Amalric F, Bouvet P 2003. Nucleolin provides a link between RNA polymerase I transcription and pre-ribosome assembly. Chromosoma 111: 399–407 [DOI] [PubMed] [Google Scholar]
- Saveanu C, Namane A, Gleizes PE, Lebreton A, Rousselle JC, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M 2003. Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol 23: 4449–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ 3rd 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: The PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev 15: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Yang CC, Hsieh CL, Chou YH, Zhong CZ, Yung BY, Liu H 2012. Epigeneitc silencing of ribosomal RNA genes by Mybbp1a. J Biomed Sci 19: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela MT, Guerrero R, Núñez MI, Ruiz De Almodóvar JM, Sarker M, de Murcia G, Oliver FJ 2002. PARP-1 modifies the effectiveness of p53-mediated DNA damage response. Oncogene 21: 1108–1116 [DOI] [PubMed] [Google Scholar]
- Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ III, Chen J 2005. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J 24: 3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ 2010. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell 141: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TH, Tsai WH, Lee YM, Lei HY, Lai MY, Chen DS, Yeh NH, Lee SC 1994. Purification and characterization of nucleolin and its identification as a transcription repressor. Mol Cell Biol 14: 6068–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Yao YL, Seto E 2001. The FK506-binding protein 25 functionally associates with histone deacetylases and with transcription factor YY1. EMBO J 20: 4814–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ES, Means AR 2007. PIN1, the cell cycle and cancer. Nat Rev Cancer 7: 381–388 [DOI] [PubMed] [Google Scholar]
- Yoshikawa H, Komatsu W, Hayano T, Miura Y, Homma K, Izumikawa K, Ishikawa H, Miyazawa N, Tachikawa H, Yamauchi Y, et al. 2011. Splicing factor 2-associated protein p32 participates in ribosome biogenesis by regulating the binding of Nop52 and fibrillarin to preribosome particles. Mol Cell Proteomics 10: M110.006148 [DOI] [PMC free article] [PubMed] [Google Scholar]