This study reveals that the chromatin-remodeling factor PICKLE (PKL) functions as a signaling convergent point that integrates BR and GA signals that regulate skotomorphogenesis. PKL interacts with key transcription factors and modulates the chromatin state of their targets. It provides a direct molecular link between trimethylation of histone H3 Lys-27 and the BR and GA signaling pathways.
Abstract
Plant cell elongation is controlled by endogenous hormones, including brassinosteroid (BR) and gibberellin (GA), and by environmental factors, such as light/darkness. The molecular mechanisms underlying the convergence of these signals that govern cell growth remain largely unknown. We previously showed that the chromatin-remodeling factor PICKLE/ENHANCED PHOTOMORPHOGENIC1 (PKL/EPP1) represses photomorphogenesis in Arabidopsis thaliana. Here, we demonstrated that PKL physically interacted with PHYTOCHROME-INTERACTING FACTOR3 (PIF3) and BRASSINAZOLE-RESISTANT1 (BZR1), key components of the light and BR signaling pathways, respectively. Also, this interaction promoted the association of PKL with cell elongation–related genes. We found that PKL, PIF3, and BZR1 coregulate skotomorphogenesis by repressing the trimethylation of histone H3 Lys-27 (H3K27me3) on target promoters. Moreover, DELLA proteins interacted with PKL and attenuated its binding ability. Strikingly, brassinolide and GA3 inhibited H3K27me3 modification of histones associated with cell elongation–related loci in a BZR1- and DELLA-mediated manner, respectively. Our findings reveal that the PKL chromatin-remodeling factor acts as a critical node that integrates light/darkness, BR, and GA signals to epigenetically regulate plant growth and development. This work also provides a molecular framework by which hormone signals regulate histone modification in concert with light/dark environmental cues.
INTRODUCTION
Plant growth and development, including cell elongation and seedling morphogenesis, are controlled by endogenous hormones, such as brassinosteroid (BR) and gibberellin (GA), and environmental signals, such as light/darkness. The individual signaling pathways that govern BR, GA, and light responses have been studied extensively (Schwechheimer and Willige, 2009; Arsovski et al., 2012; Wang et al., 2012). In the dark, plants undergo skotomorphogenesis (seedling etiolation), a process that involves the promotion of hypocotyl cell elongation, inhibition of cotyledon opening and expansion, and etioplast development (Von Arnim and Deng, 1996). After red light absorption by phytochrome converts inactive Pr to active Pfr, Pfr interacts with basic helix-loop-helix transcription factors named PIFs (for phytochrome-interacting factors) in the nucleus, resulting in the phosphorylation and degradation of PIFs (Chen and Chory, 2011; Leivar and Quail, 2011). PIF proteins accumulate in the nucleus in darkness to promote hypocotyl elongation (Leivar et al., 2008; Shin et al., 2009). BR exerts an opposite effect, as does light, on cell elongation. Upon binding to BR, the BRASSINOSTEROID-INSENSITIVE1 (BRI1) receptor kinase is activated and initiates a downstream signaling cascade, which leads to the dephosphorylation and activation of the BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS SUPPRESSOR1 (BES1) transcription factors. BZR1 and BES1 then move into the nucleus, where they bring about changes in target gene expression that result in cell growth (Kim and Wang, 2010; Wang et al., 2012). In the GA signaling pathway, binding of the GIBBERELLIN INSENSITIVE DWARF1 receptor to GA molecules results in the degradation of the master growth repressors, DELLA proteins, via a ubiquitin–proteasome pathway (Schwechheimer and Willige, 2009; Sun, 2011). Thus, GA promotes cell growth by destabilizing DELLA repressors. PIF, BZR1, and DELLA proteins are key components in their corresponding signaling pathways and modulate massive transcriptional reprogramming.
Accumulating evidence suggests that light, BR, and GA signaling pathways interact to coordinately regulate overlapping cellular responses. Light promotes the accumulation of DELLA proteins by reducing GA levels (Achard et al., 2007). Under light conditions, the phytochrome B photoreceptor destabilizes PIF3 and PIF4 and DELLAs attenuate PIF3/4 transcriptional activity by binding to their DNA recognition domains. GA triggers the ubiquitination and degradation of DELLA proteins, thus releasing PIF3/4 from the negative effect of DELLAs (de Lucas et al., 2008; Feng et al., 2008). Therefore, light and GA antagonistically regulate seedling growth. Light and BR exert a similar antagonistic effect on seedling development. PIF4 and BZR1 interact directly to regulate a core transcription network that governs cell elongation, facilitating hypocotyl growth in response to BR and darkness (Oh et al., 2012). BR and GA also function interdependently through a direct interaction between BZR1 and DELLA proteins. This interaction suppresses the DNA binding activity of BZR1, similar to the effect of DELLAs on PIF4 (Bai et al., 2012b; Gallego-Bartolomé et al., 2012; Li et al., 2012). Therefore, the interaction among PIF4, BZR1, and DELLA proteins defines a core transcriptional cascade that mediates the coordinated regulation of growth by light, BR, and GA signaling (Bai et al., 2012a, 2012b).
Chromatin remodeling is a crucial regulator of gene expression in eukaryotes (Jarillo et al., 2009; Ho and Crabtree, 2010). We previously identified PICKLE (PKL)/ENHANCED PHOTOMORPHOGENIC1 (EPP1) as a negative regulator in the light signaling pathway (Jing et al., 2013; Jing and Lin, 2013). PKL/EPP1 encodes an ATP-dependent CHROMODOMAIN HELICASE-DNA BINDING3 (CHD3) type chromatin-remodeling factor of the SWITCH/SUCROSE NONFERMENTING (SWI/SNF) family. This factor is thought to modify interactions between DNA and histone octamers, allowing the transcriptional complex to come into contact with the DNAs (Ogas et al., 1999; Kwon and Wagner, 2007). PKL/EPP1 interacts with ELONGATED HYPOCOTYL5 (HY5), a master transcription factor of light signaling, and negatively regulates its activity by repressing the trimethylation of histone H3 Lys-27 (H3K27me3) at loci involved in cell elongation (Jing et al., 2013). PKL/EPP1 has also been implicated in responses to various plant hormones, such as auxin, abscisic acid, GA, and cytokinin (Fukaki et al., 2006; Perruc et al., 2007; Zhang et al., 2008; Furuta et al., 2011). However, it is unclear precisely how chromatin remodeling regulates hormone signaling.
In this study, we demonstrate that PKL/EPP1 interacts directly with PIF3 and BZR1 to promote hypocotyl growth in Arabidopsis thaliana by repressing the H3K27me3 modification of cell elongation–related genes. DELLA proteins physically interact with PKL/EPP1 and negatively regulate its activity. We found that BR and GA signaling regulate the H3K27me3 modification status. Thus, this study reveals a crucial role for PKL/EPP1 in the crosstalk of BR and GA signaling in regulating the skotomorphogenic response and demonstrates a direct link between BR and GA signaling and histone modification.
RESULTS
PKL/EPP1 Interacts with PIF3
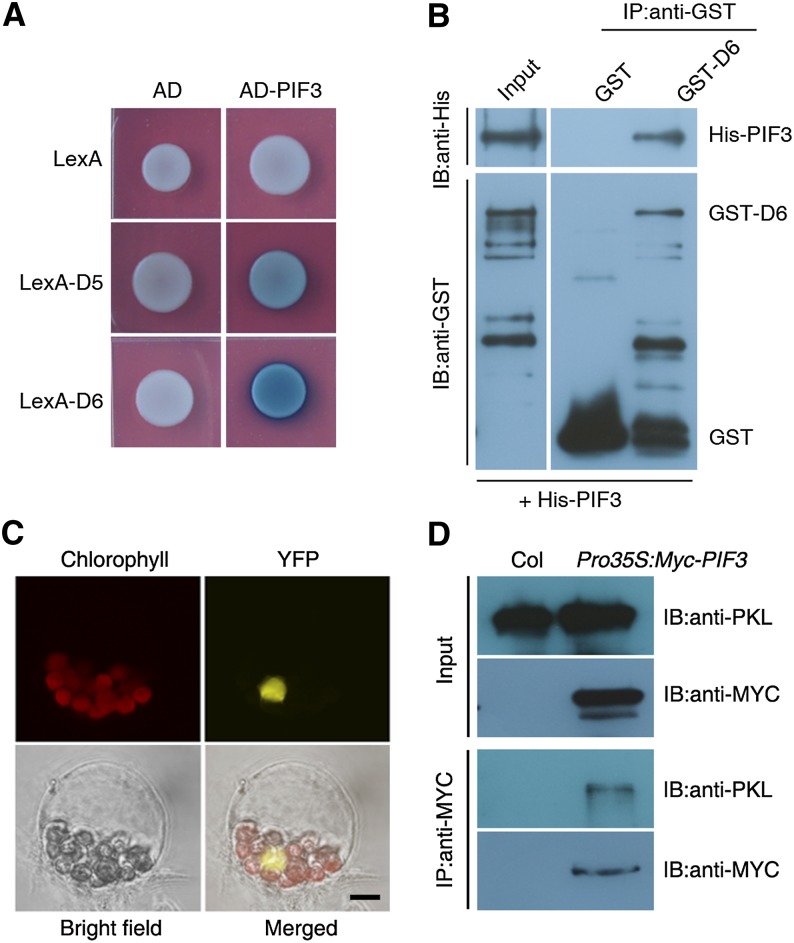
We previously showed that epp1 mutants (allelic to pkl) display a weak constitutive photomorphogenic phenotype in darkness and that PKL/EPP1 (hereafter referred to as PKL) interacts with HY5 to regulate light responses (Jing et al., 2013). However, HY5 is largely degraded in darkness (Osterlund et al., 2000; Chen et al., 2013). We reasoned that other protein(s) in addition to PKL might regulate responses in the dark. Because members of the PIF family, including PIF1, -3, -4, and -5, play essential roles in promoting skotomorphogenesis (Leivar et al., 2008; Shin et al., 2009), we tested whether PKL interacts with the PIF proteins. Using various truncated fragments of PKL developed previously (Supplemental Figure 1A; Jing et al., 2013), we found that the D5 (containing the chromo and ATPase domains) and D6 (containing the ATPase domain) fragments, but not other fragments (D1, D3, and D4) of PKL, strongly interacted with PIF3 and PIF1 and weakly interacted with PIF4 and PIF5 in yeast two-hybrid assays (Figure 1A; Supplemental Figure 1B). This indicates that the ATPase domain of PKL is responsible for the interaction with PIF proteins. In this study, we focused on the relationship between PKL and PIF3.
Figure 1.
PKL Physically Interacts with PIF3.
(A) Yeast two-hybrid assay using PIF3 and PKL constructs. AD, the B42 activation domain alone; AD-PIF3, PIF3 fused with the B42 activation domain; LexA, the LexA DNA binding domain alone; LexA-D5 and LexA-D6, the LexA DNA binding domain fused to either the D5 (amino acids 80 to 765) or D6 (amino acids 250 to 765) fragment of PKL, respectively (as shown in Supplemental Figure 1A).
(B) In vitro pull-down assay between GST-D6 and His-PIF3 recombinant proteins. IB, immunoblot; IP, immunoprecipitation.
(C) BiFC analysis of the interaction between YFPN-PKL and PIF3-YFPC in the nucleus of Arabidopsis protoplasts. Chlorophyll autofluorescence is shown in red. Chlorophyll, chlorophyll autofluorescence; YFP, YFP fluorescence; merged, chlorophyll and YFP fluorescence. Bar = 5 μm.
(D) In vivo Co-IP assay between PKL and PIF3. Extracts from Pro35S:Myc-PIF3 or Col wild-type seedlings grown in darkness for 5 d (input; top panels) were immunoprecipitated (IP:anti-MYC; bottom panels) and then immunoblotted (IB) using anti-PKL or anti-MYC antibody as indicated.
To confirm the direct interaction between PKL and PIF3, we performed a pull-down assay using recombinant 6× His–tagged PIF3 (His-PIF3) and a glutathione S-transferase–tagged D6 fragment of PKL (GST-D6). GST-D6, but not GST alone, precipitated His-PIF3 in vitro (Figure 1B). Next, we monitored the in vivo interaction using a bimolecular fluorescence complementation (BiFC) assay. Coexpression of the N terminus of yellow fluorescent protein fused to PKL (YFPN-PKL) and PIF3 linked to the C terminus of YFP (PIF3-YFPC) reconstituted functional YFP fluorescence in the nucleus (Figure 1C). Furthermore, we conducted a coimmunoprecipitation (Co-IP) assay using wild-type and transgenic plants expressing Pro35S:Myc-PIF3 (Chen et al., 2013). Myc-tagged PIF3 precipitated PKL in planta (Figure 1D). Together, these results demonstrate that PKL indeed physically interacts with PIF3.
PKL and PIF3 Coregulate Skotomorphogenesis
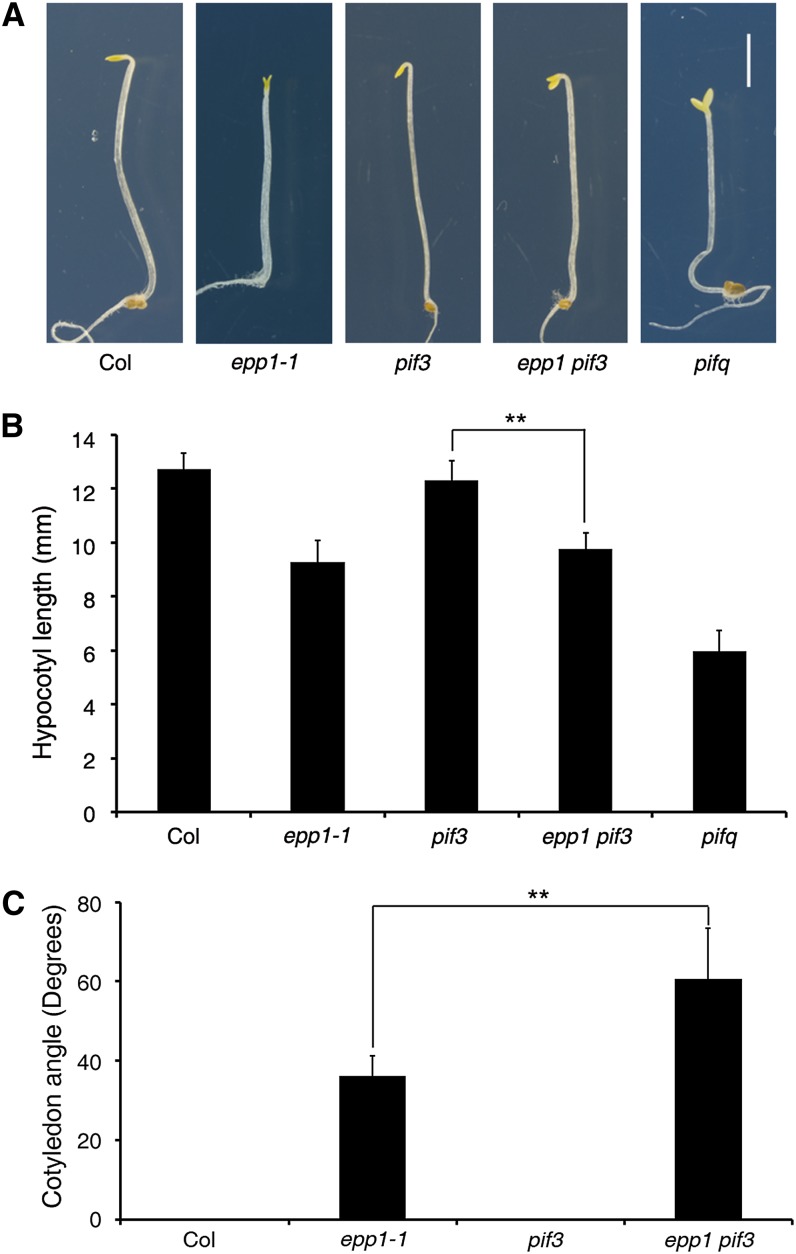
To investigate the genetic relationship between PKL and PIF3, we first generated the epp1 pif3 double mutant crossing epp1-1 and pif3-1 and then examined the hypocotyl growth phenotype of the double mutant in darkness. Whereas the epp1 seedlings exhibited reduced hypocotyl growth and pif3 was similar to the wild type in the dark, the epp1 pif3 plants had significantly shorter hypocotyls than pif3 (Figures 2A and 2B). As a control, quadruple mutant pifq (loss of PIF1, PIF3, PIF4, and PIF5) seedlings displayed a constitutive photomorphogenic response. Moreover, compared with the cotyledons of wild-type and pif3 seedlings grown in darkness, those of epp1 pif3 were wide open, whereas those of epp1 were slightly open, as reported previously (Figures 2A and 2C) (Jing et al., 2013). These data indicate that PKL and PIF3 additively regulate hypocotyl elongation and synergistically modulate cotyledon opening. In addition, when 6-d-old etiolated seedlings were exposed to light for 1 d, the epp1 mutant turned green faster than did the wild type, while the photobleached phenotype of pif3 (Chen et al., 2013) was inhibited by EPP1 mutation in the epp1 pif3 double mutant (Supplemental Figure 2A). We also constructed transgenic plants that expressed Pro35S:Myc-PIF3 in the epp1 mutant background. The hypocotyl lengths of the doubly homozygous plants were significantly longer than epp1 but shorter than pif3 (Supplemental Figures 2B and 2C), supporting the overlapping function of PIF3 and PKL in regulating hypocotyl growth.
Figure 2.
Skotomorphogenic Phenotypes of pkl and pif3 Mutants.
(A) Seedling phenotypes of etiolated plants. The panels show seedlings of 5-d-old Col wild type and epp1-1, pif3, epp1 pif3, and pifq mutants grown in the dark. Bar = 2 mm.
(B) Hypocotyl lengths of the seedlings in (A).
(C) Cotyledon angles of seedlings grown in darkness for 5 d.
Data in (B) and (C) represent means ± sd of at least 20 seedlings. Double asterisks indicate significant differences at P < 0.01 using Student’s t test.
[See online article for color version of this figure.]
PKL Binding to Target Genes Primarily Requires PIF3
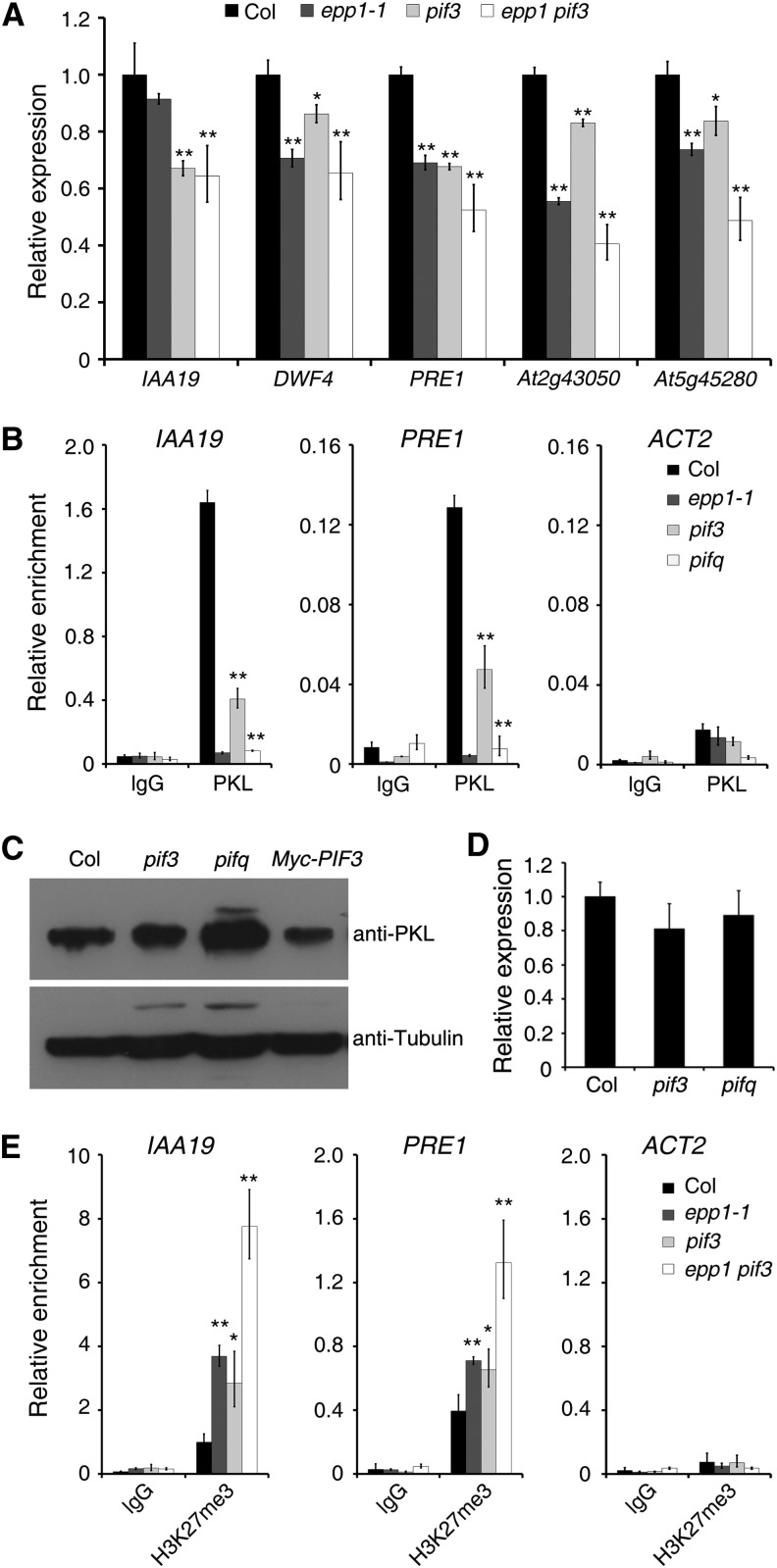
Next, we investigated how PKL and PIF3 regulate hypocotyl growth by examining the expression of cell elongation–related genes, including INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19), PACLOBUTRAZOL RESISTANCE1 (PRE1), DWARF4 (DWF4), and two uncharacterized genes (At2g43050 and At5g45280) that are putatively involved in cell wall modification (Oh et al., 2012; Jing et al., 2013). The transcript levels of these genes were lower in epp1, pif3, and the double mutant than in the wild-type plants, and PRE1, At2g43050, and At5g45280 mRNA levels were lower in epp1 pif3 than in either of the single mutants (Figure 3A).
Figure 3.
The Binding of PKL to Targets Is Largely Dependent on PIF3.
(A) Relative expression of cell elongation–related genes in Col wild-type, epp1, pif3, and epp1 pif3 mutant plants. The amounts of mRNA were quantified by RT-qPCR, and the relative expression levels were normalized to that of a UBQ control.
(B) ChIP-qPCR assay. Data show the relative enrichment of IAA19, PRE1, and ACT2 (negative control) promoter fragments upon precipitation with anti-PKL antibody or the IgG control.
(C) Immunoblot assay showing PKL protein levels in the pif3 and pifq mutants and PIF3 overexpression plants. Immunoblotting using an anti-tubulin antibody served as the loading control.
(D) Relative PKL expression in the pif3 and pifq mutants. The amounts of mRNA were quantified by RT-qPCR, and the relative expression levels were normalized to that of a UBQ control.
(E) ChIP-qPCR assay showing the relative enrichment of IAA19, PRE1, and ACT2 (negative control) promoter fragments upon precipitation with H3K27me3 antibody.
For all experiments, the wild-type and various mutant seedlings were grown in darkness for 5 d. In (A), (B), (D), and (E), data represent means ± sd of biological triplicates. Asterisks indicate significant differences from the wild type at P < 0.05 (*) or P < 0.01 (**) using Student’s t test.
A putative G-box (CACGTG) motif and the core of the BZR1 binding site (CGTG) were identified in the promoter regions of IAA19, PRE1, DWF4, and At5g45280 (Supplemental Figure 3A). Chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) analysis was conducted using Pro35S:Myc-PIF3 plants. For this, DNA samples were first immunoprecipitated with an anti-PKL antibody followed by a second round of chromatin immunoprecipitation with an anti-Myc antibody. As shown in Supplemental Figure 3B, the promoter regions of the cell elongation–related genes were enriched after immunoprecipitation, indicating that PKL and PIF3 form a complex on the promoter of these target genes. In the following studies, we further investigated the regulation of IAA19 and PRE1.
We next performed a ChIP-qPCR assay in the pif3 and pifq mutant backgrounds and found that, compared with the wild type, the binding ability of PKL to IAA19 and PRE1 promoters was remarkably reduced in pif3 and was compromised in pifq, as it was in epp1 (Figure 3B). To test whether the reduced binding ability was due to a lower level of PKL in these mutants, we performed an immunoblot assay with an anti-PKL antibody. Surprisingly, PKL levels were greatly increased in the pifq mutant compared with the Columbia (Col) wild type but were reduced in the transgenic plants overexpressing PIF3 (Figure 3C). However, the transcript level of PKL was not affected by mutations in pif3 and pifq (Figure 3D). These data indicate that PKL is likely modulated by PIFs at the posttranslational level and that the association of PKL with the target genes is largely dependent on the presence of PIF proteins.
Loss of PKL was shown previously to cause an increase in H3K27me3 levels at the regulatory region of IAA19 (Jing et al., 2013). A similar increase was observed for the PRE1 locus (Figure 3E). To examine whether PIF3 affects the enrichment of H3K27me3 at target loci, we performed a ChIP-qPCR analysis using an anti-H3K27me3 antibody. H3K27me3 levels on IAA19 and PRE1 were increased in pif3, as they were in epp1 (Figure 3E). Intriguingly, disruption of both PKL and PIF3 in the epp1 pif3 double mutant increased H3K27me3 levels (Figure 3E), suggesting that PKL and PIF3 act additively to repress the recruitment of H3K27me3 to loci corresponding to cell elongation genes.
PKL Interacts with BZR1 and Regulates the BR-Mediated Response
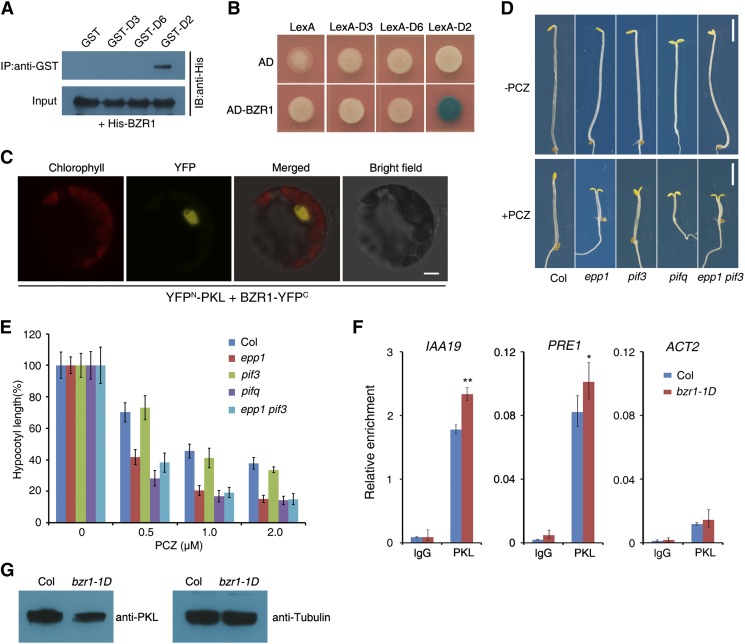
Previous studies documented that PIF3 and PIF4 interact with DELLA proteins during seedling development and that PIF4 is part of a transcription cascade with BZR1 and DELLAs that regulates hypocotyl growth (de Lucas et al., 2008; Feng et al., 2008; Bai et al., 2012b; Oh et al., 2012). Our in vitro and in vivo analyses revealed that PIF3 interacts directly with BZR1 via their C-terminal regions (Supplemental Figure 4). This result led us to hypothesize that PKL also interacts with BZR1 and/or DELLAs. A pull-down assay showed that the GST-tagged D2 fragment of PKL (amino acids 761 to 1384; GST-D2), but not GST alone or the D3 or D6 fragment, pulled down His-tagged BZR1 (His-BZR1) recombinant proteins in vitro (Figure 4A; Supplemental Figure 1A). Consistently, the D2 fragment fused with the LexA DNA binding domain (LexA-D2) interacted with the activation domain–tagged BZR1 (AD-BZR1) in yeast cells (Figure 4B). Furthermore, overexpression of the YFPN-PKL and BZR1-YFPC constructs in protoplasts reconstituted a functional YFP in the nucleus (Figure 4C). These data verify that PKL indeed interacts directly with BZR1.
Figure 4.
PKL Interacts with BZR1.
(A) In vitro pull-down assay between recombinant His-BZR1 and various PKL fragments tagged with GST. His-BZR1 proteins were incubated with immobilized GST or GST-PKL, and immunoprecipitated fractions were probed with an anti-His antibody. IB, immunoblot; IP, immunoprecipitation.
(B) Yeast two-hybrid assay between BZR1 fused with the B42 activation domain (AD) and the D2, D3, or D6 fragment of PKL (Supplemental Figure 1A) fused with the LexA DNA binding domain.
(C) BiFC assay showing that YFPN-PKL and BZR1-YFPC interact to form a functional YFP in the nucleus of Arabidopsis protoplasts. Chlorophyll, chlorophyll autofluorescence; YFP, YFP fluorescence; merged, chlorophyll and YFP fluorescence. Bar = 5 μm.
(D) Effect of BR biosynthesis inhibition on seedling growth. The panels show phenotypes of Col wild-type, epp1, pif3, and epp1 pif3 seedlings grown in medium with or without 1 μM of the BR biosynthesis inhibitor PCZ in darkness for 5 d. Bars = 2 mm.
(E) Relative hypocotyl lengths of seedlings grown in various concentrations of PCZ for 5 d. Data represent means ± sd of at least 20 seedlings.
(F) ChIP-qPCR assay showing the relative enrichment of IAA19, PRE1, and ACT2 (negative control) genomic fragments upon precipitation with an anti-PKL antibody. Col and bzr1-1D seedlings were grown in darkness for 5 d. Data represent means ± sd of three biological replicates. Asterisks indicate significant differences from the wild type at P < 0.05 (*) or P < 0.01 (**) using Student’s t test.
(G) Immunoblot assay for PKL in the bzr1-1D mutant. Immunoblotting against tubulin antibody served as the loading control.
We then asked whether BR-mediated hypocotyl growth requires PKL and PIF3. To address this question, epp1, pif3, epp1 pif3, and pifq mutants and wild-type seedlings were grown in darkness for 5 d in medium supplemented with various concentrations of propiconazole (PCZ), a BR biosynthesis inhibitor (Hartwig et al., 2012). Similar to the pifq mutant, epp1 and epp1 pif3 were more sensitive to PCZ, which inhibits hypocotyl growth (Figures 4D and 4E), suggesting that loss of PKL compromises the BR response. However, pif3 had a similar response to PCZ as the wild type (Figures 4D and 4E). The bzr1-1D gain-of-function mutant is constitutively dephosphorylated and exhibits a BR-sensitive phenotype (Wang et al., 2002). In the presence of PCZ, bzr1-1D seedlings displayed very long hypocotyls compared with wild-type seedlings. However, the hypocotyl length of epp1 bzr1-1D double mutant seedlings was significantly reduced compared with that of bzr1-1D (Supplemental Figure 5). This suggests that the function of BZR1 partially requires PKL.
A ChIP-qPCR assay showed greater enrichment of PKL at IAA19 and PRE1 promoters in the bzr1-1D mutant than in wild-type plants grown in darkness (Figure 4F). However, PKL was drastically reduced in the bzr1-1D mutant (Figure 4G). These results indicate that BZR1 enhances the association ability of PKL with the promoters of target genes. Because PKL interacts with both PIF3 and BZR1, we tested the PIF3- and/or BZR1-dependent binding ability of PKL using a strong BR-deficient mutant, bri1-5, and its Wassilewskija (Ws) wild type (Supplemental Figure 6). After being pulled down with an anti-PKL antibody, the amounts of DNA of IAA19 and PRE1 (common targets of PIF3 and BZR1) were only slightly reduced in bri1-5, whereas the levels of LONG HYPOCOTYL IN FAR-RED1 (HFR1) and PIF6 (PIF3-specific targets) (Zhang et al., 2013) DNAs were not changed between bri1 and Ws. Intriguingly, the relative enrichment of two BZR1-specific targets, TOUCH4 (TCH4) and At4g02330 (Oh et al., 2012), was almost abolished by the BRI1 mutation (Supplemental Figure 6). Together with the PKL–BZR1 interaction results, these data suggest that PKL is associated with BR-dependent targets that require its interaction with BZR1 and that PIF proteins play a dominant role in recruiting PKL to PIF and BZR1 common targets.
DELLAs Interact with PKL and Attenuate Its Activity
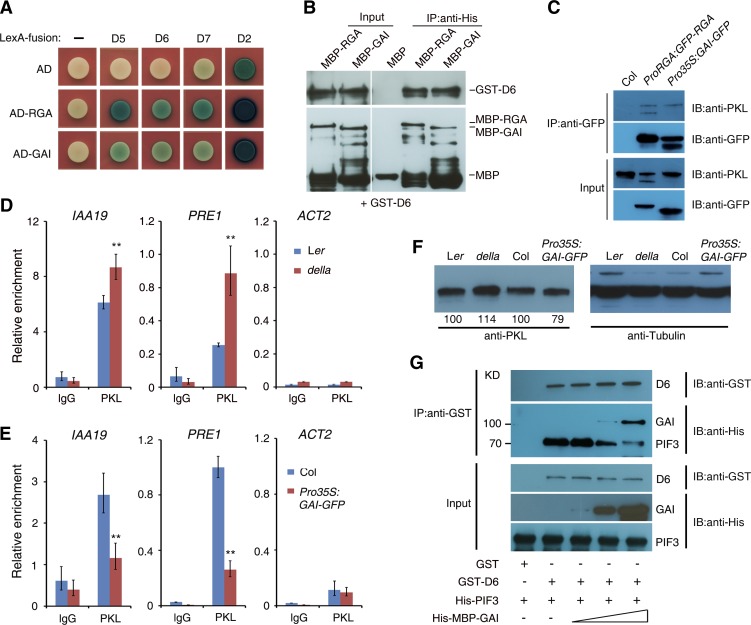
We next tested the interaction between PKL and DELLA proteins, including REPRESSOR OF ga1-3 (RGA), GIBBERELLIC ACID INSENSITIVE (GAI), RGA-LIKE1 (RGL1), RGL2, and RGL3. The yeast two-hybrid assays showed that both the N-terminal (D5 and D6) and C-terminal (D2 and D7) portions of PKL strongly interacted with all five DELLA proteins (Figure 5A; Supplemental Figures 1A and 7). The interaction between PKL and RGA and GAI was confirmed by pull-down and Co-IP assays. As shown in Figure 5B, recombinant RGA fused to maltose binding protein (MBP-RGA) or MBP-GAI pulled down GST-tagged PKL (GST-D6). We next conducted an in vivo Co-IP analysis using ProRGA:GFP-RGA and Pro35S:GAI-GFP transgenic plants (Supplemental Figure 8). The anti-GFP (for green fluorescent protein) antibody immunoprecipitated PKL in both transgenic lines (Figure 5C). We thus conclude that PKL interacts directly with DELLA proteins.
Figure 5.
RGA and GAI Interact with PKL and Inhibit Its Binding Activity.
(A) Yeast two-hybrid analysis of either RGA or GAI fused to the B42 activation domain (AD) and different fragments of PKL (Supplemental Figure 1A) fused to LexA-BD.
(B) In vitro pull-down assay. Recombinant proteins of GST-D6 and MBP-RGA or MBP-GAI (input; left panels) were immunoprecipitated with an anti-His antibody and then immunoblotted using either anti-GST (top right panel) or anti-MBP (bottom right panel). IB, immunoblot; IP, immunoprecipitation.
(C) Co-IP assay between PKL and RGA or GAI. Col wild-type, ProRGA:RGA-GFP, and Pro35S:GAI-GFP seedlings were grown in darkness for 5 d. Total protein extracts (input; bottom panels) were immunoprecipitated with an anti-GFP antibody (top panels) and then immunoblotted using either anti-GFP or anti-PKL antibody.
(D) and (E) ChIP-qPCR assay showing the relative enrichment of IAA19, PRE1, and ACT2 (negative control) promoter fragments upon precipitation with anti-PKL antibody in the della mutant (D) or the Pro35S:GAI-GFP transgenic line (E). Seedlings were grown in darkness for 5 d. Data represent means ± sd of biological triplicates. Asterisks indicate significant differences from the wild type at P < 0.01 using Student’s t test. Ler, Landsberg erecta.
(F) Immunoblot assay showing PKL protein levels in the della mutant and GAI overexpression plants and their corresponding wild types. Immunoblotting against tubulin antibody served as the loading control. Values denote relative amounts of PKL normalized to the tubulin control, and values in the wild type are set as 100.
(G) Pull-down assay showing that GAI blocks the PKL–PIF3 interaction. Recombinant protein GST-D6 was preincubated with His-MBP-GAI for 1 h. His-PIF3 was then added and incubated for an additional 1 h. It should be noted that the GST band was not shown due to its low molecular mass.
RGA and GAI can abrogate the DNA binding activity of BZR1 (Bai et al., 2012b; Li et al., 2012). Since PKL interacts with DELLAs, we examined the role of DELLAs in PKL function. When pulled down by an anti-PKL antibody, the enrichment of IAA19 and PRE1 promoters was increased in DNA samples prepared from a della mutant in which all five DELLA genes were disrupted. By contrast, it was reduced in DNA samples extracted from Pro35S:GAI-GFP transgenic plants compared with the wild-type control (Figures 5D and 5E). The amount of PKL protein was increased slightly in the della mutant but was decreased in GAI overexpression plants (Figure 5F).
To further dissect the molecular relevance of the PKL–DELLA interaction, we tested the competitive binding ability among PKL, PIF3, and GAI using an in vitro pull-down assay. GST-D6 was preincubated with increasing amounts of His-MBP-GAI and then His-PIF3 was added to the reaction. After precipitation with Glutathione Sepharose 4B beads, the amount of His-PIF3 was decreased gradually along with the increasing level of His-MBP-GAI, suggesting that GAI blocks the PKL–PIF3 interaction in vitro (Figure 5G). Therefore, these results together indicate that DELLA proteins attenuate PKL function, at least in part, through their interaction.
PKL Is Regulated by BR and GA Signaling
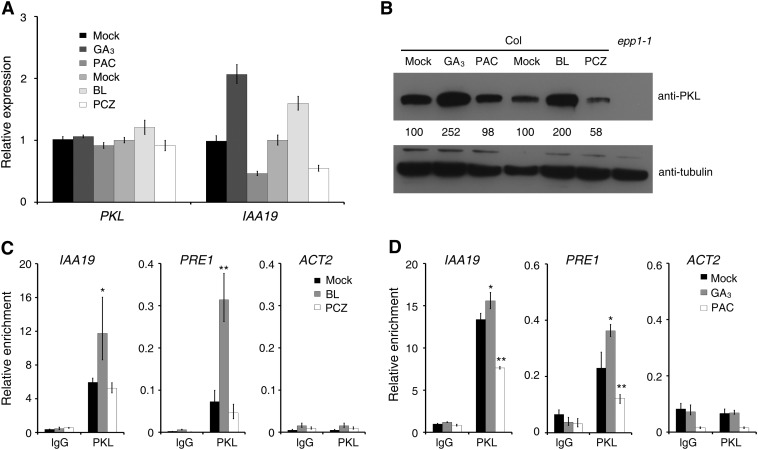
We then tested how PKL is regulated by BR and GA hormones. Col wild-type seedlings were treated with brassinolide (BL; the most active form of BR), PCZ, gibberellic acid (GA3; an active form of GA), and paclobutrazol (PAC; a GA biosynthesis inhibitor) in the dark. RT–quantitative PCR (RT-qPCR) analysis revealed that IAA19 was induced by BL (0.2 μM) and GA3 (10 μM) and inhibited by PCZ (1 μM) and PAC (0.1 μM). By contrast, the expression of PKL was not influenced by these chemical treatments (Figure 6A). Intriguingly, PKL protein levels were markedly increased in samples treated with GA3 or BL but were reduced in seedlings treated with PCZ but not PAC (Figure 6B). This suggests that PKL is largely regulated at the protein level by BR and GA hormones.
Figure 6.
PKL Protein Accumulation and Binding Ability Are Regulated by BR and GA.
(A) Relative expression levels of PKL and IAA19, as determined by quantitative RT-PCR. Data represent means ± sd of biological triplicates.
(B) Immunoblot analysis showing PKL protein levels in Col seedlings treated with BR and GA and their inhibitors. Immunoblotting against tubulin antibody served as a loading control. Values denote relative amounts of PKL normalized to the tubulin control, and values in the mock controls are set as 100.
(C) and (D) ChIP-qPCR assay showing the relative enrichment of IAA19, PRE1, and ACT2 genomic fragments by PKL when the Col seedlings were treated with BR (C) or GA (D). Data represent means ± sd of biological triplicates. For the BR treatment, Col wild-type seeds were germinated on MS medium for 1 d and were then transferred to MS plates without (mock) or with BL (0.2 μM) or PCZ (1 μM) and grown for an additional 4 d. For the GA treatment, 3-d-old Col seedlings were transferred to medium without (mock) or with GA3 (10 μM) or PAC (0.1 μM) and grown for an additional 2 d. All treatments were performed in darkness. Asterisks indicate significant differences from the mock treatment at P < 0.05 (*) or P < 0.01 (**) using Student’s t test.
We next examined how BR and GA affect the ability of PKL to bind to promoters of its downstream genes. A ChIP-qPCR assay revealed that the promoter fragments of IAA19 and PRE1 were greatly enriched in samples prepared from seedlings grown in medium with BL, compared with those grown in Murashige and Skoog (MS) medium that lacked BL (mock; Figure 6C). However, the enrichment of neither of these gene promoters was significantly affected by PCZ treatment (Figure 6C). Furthermore, we found that GA3 treatment significantly enhanced the binding of PKL to IAA19 and PRE1, whereas PAC inhibited the binding (Figure 6D). Notably, the binding efficiency of PKL was not altered by these chemical treatments (Supplemental Figure 9). These data together indicate that BR and GA promote the binding of PKL to cell elongation–related genes by regulating the level of PKL protein.
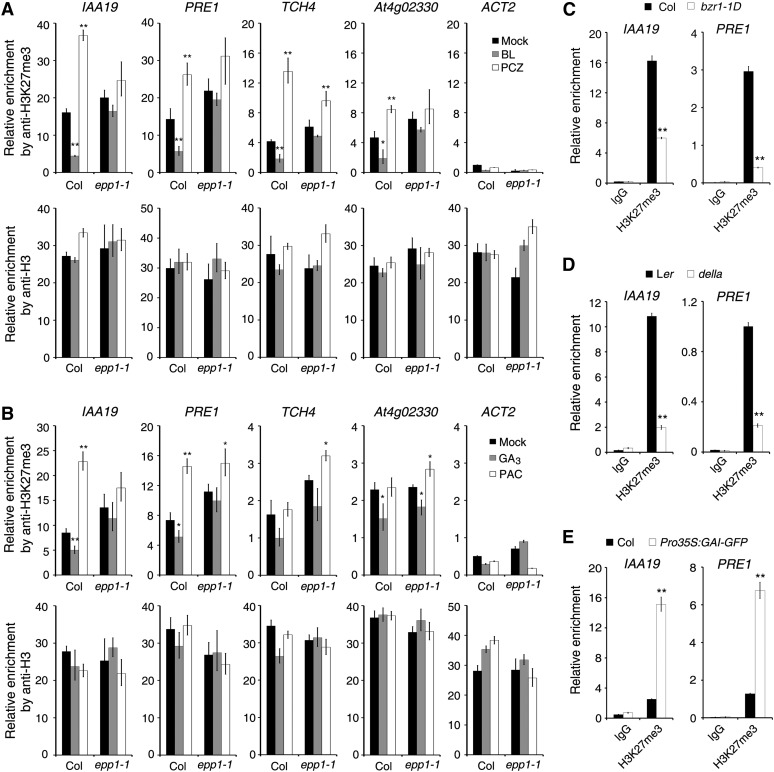
BR and GA Inhibit H3K27me3 Modification of Cell Elongation–Related Genes
Considering that BR and GA affect the protein level and binding ability of PKL, and that PKL regulates H3K27me3 histone modification, we predicted that BR and GA regulate H3K27me3 levels on cell elongation–related genes such as IAA19, PRE1, TCH4, and At4g02330. To this end, we performed ChIP-qPCR assays using H3K27me3 antibody and Col and epp1-1 mutant seedlings grown in medium containing various hormones or their inhibitors. Application of BL to the Col wild type significantly reduced the recruitment of IAA19, PRE1, TCH4, and At4g02330 promoters pulled down by the H3K27me3 antibody as compared with the untreated control (mock), whereas inhibition by PCZ increased the enrichment of these targets (Figure 7A). Most remarkably, in the epp1-1 mutant, the difference of DNAs immunoprecipitated by H3K27me3 between samples of BL or PCZ treatment and the mock control was mostly diminished or even abolished (Figure 7A). Similarly, we found that GA3 significantly reduced the recruitment of H3K27me3 to the promoters of IAA19, PRE1, and At4g02330, whereas PAC promoted the recruitment of H3K27me3 to IAA19 and PRE1 in the wild-type seedlings (Figure 7B). However, in the epp1 mutant, less difference was observed between GA3 treatment and the mock control, although PAC treatment caused slightly increased enrichment of PRE1, TCH4, and At4g02330 (Figure 7B). Taken together, these observations indicate that BR- and GA-regulated recruitment of H3K27me3 to the cell elongation–related targets largely requires PKL.
Figure 7.
BR and GA Signaling Regulate H3K27me3 Modification.
(A) and (B) ChIP-qPCR assays showing the relative enrichment of genomic fragments pulled down by the H3K27me3 and H3 (control) antibodies when seedlings were treated with BL and the inhibitor PCZ (A) or with GA3 and the GA inhibitor PAC (B). Data represent means ± sd of biological triplicates. Seedlings were treated as described in the legend to Figure 6.
(C) ChIP-qPCR assay. Col and bzr1-1D seedlings were grown in medium containing 1 μM PCZ in darkness for 5 d.
(D) ChIP-qPCR assay. Landsberg erecta (Ler) wild-type and della mutant seedlings were grown in medium containing 0.1 μM PAC in darkness for 5 d.
(E) ChIP-qPCR assay. Col wild-type and Pro35S:GAI-GFP transgenic seedlings were grown in darkness for 5d.
For (C) to (E), the protein-DNA complexes were pulled down by the H3K27me3 antibodies or IgG control, and the DNA fragments were quantified by qPCR. For all panels, data represent means ± sd of biological triplicates. Asterisks indicate significant differences from the wild type or mock treatment at P < 0.05 (*) or P < 0.01 (**) using Student’s t test.
To further investigate the regulation of H3K27me3 by BR and GA signaling, we performed ChIP-qPCR assays using mutants of key BR or GA signaling components. H3K27me3 levels at the IAA19 and PRE1 promoters were significantly lower in bzr1-1D than in the Col wild type (Figure 7C), indicating that BZR1 inhibits the recruitment of H3K27me3 to cell elongation–related targets. Similarly, the relative amount of IAA19 and PRE1 promoter DNA pulled down by H3K27me3 antibody was markedly decreased in the della mutant but drastically increased in the GAI overexpression plants, relative to their corresponding wild types (Figures 7D and 7E), suggesting that DELLAs promote H3K27me3 association with downstream loci. Together, our results show that BR and GA inhibit H3K27me3 modification on cell elongation–related genes and that this modification is largely mediated by BZR1 and DELLAs, respectively.
DISCUSSION
PKL Mediates Crosstalk Involving Darkness, BR, and GA Signaling
Several lines of evidence support our conclusion that PKL integrates darkness, BR, and GA signaling pathways to regulate hypocotyl cell elongation in plants. First, the protein level and possible binding activity of PKL are promoted by BL and GA3 (Figure 6). We previously showed that light represses PKL both at the mRNA and protein levels (Jing et al., 2013). Second, PKL physically interacts with PIF3 (a light signaling factor) and BZR1 (a BR signaling factor) through its N-terminal ATPase domain and the C-terminal portion, respectively (Figures 1 and 4). PIF3 also interacts with BZR1 (Supplemental Figure 4). Furthermore, both the N- and C-terminal regions of PKL bind to DELLA proteins of the GA pathway (Figure 5). The amount of target DNAs pulled down by PKL was greatly reduced in pif3 but was increased in the bzr1-1D and della mutants (Figures 3 to 5), suggesting that PIF3, BZR1, and DELLA proteins affect the recruitment of PKL to the promoters of cell elongation–related genes. We would have hypothesized that PKL was degraded without an association with PIF3 or BZR1. However, our immunoblot analyses showed that PKL protein accumulation is enhanced in the pifq mutant and reduced in the bzr1-1D mutant (Figure 6). This negative regulation of PKL might provide a way to prevent the overactivation of downstream genes by PIF3 and BZR1. The mechanisms by which these factors inhibit PKL deserve further investigation. Nevertheless, these data indicate that the recruitment of PKL to target genes largely requires its interacting with PIF3 and BZR1.
It has been shown previously that PIF4 and BZR1 interdependently and independently regulate downstream gene expression and that DELLAs interfere with the DNA binding activity of PIF4 and BZR1 (Bai et al., 2012b; Li et al., 2012; Oh et al., 2012). Our study further substantiates that PIF3/4, BZR1, and DELLAs interact to regulate a core transcription module that strongly regulates cell elongation (Wang et al., 2012). PKL also interacts with PIF1, PIF4, and PIF5, whereas BZR1 interacts with PIF1 (Supplemental Figure 1; Oh et al., 2012). Thus, all PIF proteins, including PIF1, -3, -4, and -5, appear to act together with PKL and BZR1. The ability of PKL to interact with either PIFs or BZR1 likely allows PKL to modulate the transcription of common and distinct targets and regulate multiple physiological responses.
PIF3, BZR1, and DELLA transcription factors/regulators respond to dark/light, BR, and GA, respectively, and the responses are integrated by the coordinated regulation of transcription of the downstream genes (Bai et al., 2012b; Oh et al., 2012; Wang et al., 2012). However, as a chromatin-remodeling factor, PKL is simultaneously regulated by various stimuli and acts in concert with multiple transcription factors that might underlie its regulatory efficiency and flexibility in facilitating histone modification and altering gene expression in plants grown in nature (Figure 8A). Therefore, we propose that PIF3 (and other PIFs), BZR1, DELLA, and PKL proteins act cooperatively to regulate skotomorphogenic growth in the dark. PIF3 and BZR1 interdependently or independently bind to their common or specific targets involved in cell elongation and, thereby, attract PKL through physical interaction. This interaction inhibits the recruitment of the repressive histone mark H3K27me3 on the corresponding chromatin, leading to the activation of cell elongation and etiolation (Figure 8B). GA molecules trigger the degradation of DELLAs, which attenuate the DNA binding ability of PIF3 and BZR1 and the interaction of PKL with these transcription factors. In agreement with this, GAI could block the PKL–PIF3 interaction (Figure 5F). This finding thus elucidates an important regulatory layer that involves the epigenetic control of plant growth and development through the integration of multiple signals.
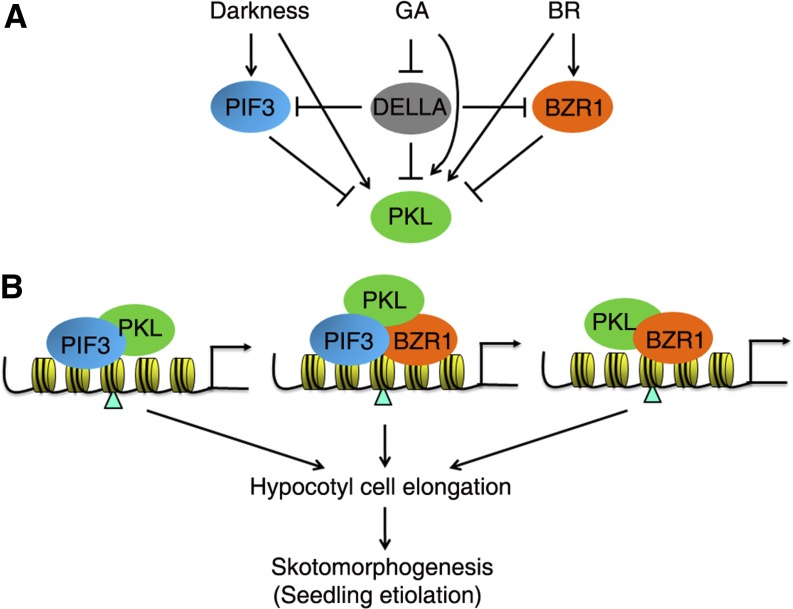
Figure 8.
A Proposed Model of PKL, PIF3, BZR1, and DELLAs in Regulating Skotomorphogenesis.
(A) Regulation of PKL by light, GA, and BR. The level of PKL is increased by darkness, GA, and BR. Meanwhile, PIF3, BZR1, and DELLA repress the level of PKL by unknown mechanisms. PIF3 is stabilized in the dark, and BZR1 is activated upon BR stimulation. GA promotes the degradation of DELLAs, thus releasing their inhibitory role on the binding of PIF3 and BZR1 and the recruitment of PKL to the target DNAs.
(B) PIF3 (and other PIFs) and BZR1 transcription factors interdependently and independently bind to the promoter regions of cell elongation–related genes and recruit PKL through direct interaction. The PKL chromatin-remodeling factor thereafter represses the recruitment of H3K27me3 marks (indicated by triangles) on target loci. Both the activation activity of PIF3 and BZR1 and the reduced H3K27me3 level lead to the activation of cell elongation–related genes and the promotion of skotomorphogenesis (seedling etiolation). Arrows, positive regulation; bars, negative effect.
We recently demonstrated that PKL interacts with the photomorphogenesis-promoting transcription factors HY5 and HYH in modulating hypocotyl growth in the light (Jing et al., 2013). In the dark, PIF proteins are stabilized and predominantly promote hypocotyl elongation, and loss of PIF1, -3, -4, and -5 leads to a constitutive photomorphogenic response (Leivar et al., 2008; Shin et al., 2009). However, in the light, PIFs are degraded but HY5 and HYH accumulate, repressing hypocotyl growth. Thus, PKL functions differentially by interacting with positive or negative factors: PKL acts together with PIFs and/or BZR1 to promote skotomorphogenesis in the dark, whereas it interacts with HY5 to dampen HY5/HYH activity and to prevent inadvertent entry into photomorphogenesis. It will be of great interest to research whether and how PKL integrates BR and GA signals to regulate responses in the light.
PKL is also involved in auxin, cytokinin, and abiotic stress responses (Fukaki et al., 2006; Perruc et al., 2007; Furuta et al., 2011). DELLA levels are affected by auxin, abscisic acid, ethylene, and abiotic stresses (Achard et al., 2003, 2008; Fu and Harberd, 2003), whereas PIF3 and PIF4 are regulated by ethylene, auxin, temperature, and the circadian clock (Nozue et al., 2007; Koini et al., 2009; Franklin et al., 2011; Zhong et al., 2012; Sun et al., 2013). We anticipate that PKL-based transcriptional machinery might integrate a variety of endogenous factors and environmental stimuli to control key physiological processes.
Regulatory Mechanism of the PKL Chromatin-Remodeling Factor
Chromatin-remodeling factors, which alter histone–DNA contacts and thereby render genomic regions accessible to the transcriptional machinery or transcription factors, serve as spatial and temporal regulators of gene expression (Clapier and Cairns, 2009). After being recruited to specific loci, the chromatin-remodeling factors mediate the activity of histone modification enzymes involved in (de)acetylation or (de)methylation. Our previous and current studies support the notion that chromatin-remodeling proteins likely recognize the target and regulatory specificity by interacting with particular transcription factors (such as HY5, PIF3, and BZR1) in a spatial and temporal manner. Mutations in histone acetyltransferases or histone deacetylases also lead to defects in light-responsive gene expression and seedling growth (Benhamed et al., 2006). Similarly, the Arabidopsis SWI2/SNF2 ATP-dependent chromatin-remodeling enzymes, SPLAYED and BRAHMA, regulate floral organ identity by interacting with LEAFY and SEPALLATA3 transcription factors (Wu et al., 2012). A similar situation occurs in animal systems. For instance, the Drosophila melanogaster CHD3 chromatin remodeler Mi-2 interacts with the transcription factor dDREF to modulate cell development (Hirose et al., 2002). In mice (Mus musculus), BRG1 mediates Schwann cell differentiation and myelination by interacting with the Sox10 transcription factor (Weider et al., 2012).
Hence, transcription factors/regulators, chromatin-remodeling factors, and histone modification enzymes appear to act together to modulate the dynamic transcription of downstream genes. PKL regulates other plant developmental processes such as embryonic development (Ogas et al., 1999), seed germination (Fukaki et al., 2006; Perruc et al., 2007), and root meristem activity (Aichinger et al., 2011). It is possible that a similar regulatory mechanism, in which PKL interacts with a corresponding transcription factor(s), is involved in the regulation of these responses.
BR and GA Signaling Regulate Histone Modification
The BR- and GA-mediated regulation of histone modification has been largely uncharacterized, although a few studies suggested the existence of this phenomenon. For example, EARLY FLOWERING6 (ELF6) and its homolog RELATIVE OF ELF6 (REF6) interact with BES1 (Yu et al., 2008). These proteins are closely related to the mammalian jmj family, members of which have been shown to demethylate different methyl-lysines, including H3K9me3 (Klose et al., 2006). Loss of either ELF6 or REF6 significantly increased H3K9me3 levels at the promoter of TCH4, which is a direct target of BES1 (Yu et al., 2008). More recently, a chromatin immunoprecipitation sequencing analysis revealed increased H3K27me3 levels at six BR-responsive loci, including TCH4. TCH4 expression was downregulated in ref6 mutant plants but was upregulated in REF6 overexpression plants, in which the H3K27me3 level at this locus was decreased (Lu et al., 2011). Moreover, H3K36 methylation was shown to regulate BR-related gene expression and plant development in rice (Oryza sativa; Sui et al., 2012). In addition, a proteomic study identified several BR-responsive nucleus-enriched proteins, including nucleosome assembly proteins, HISTONE DEACETYLASE 2B, and VERNALIZATION INDEPENDENCE3, suggesting that BR-induced gene expression is regulated by chromatin remodeling (Shigeta et al., 2011a, 2011b). Furthermore, a genome-wide analysis in Arabidopsis of histone modifications during photomorphogenesis demonstrated that H3K27me3 is a major regulator of the GA metabolic pathway, because dioxygenase genes are highly targeted by H3K27me3 and the level of H3K27me3 modification is correlated with expression changes (Charron et al., 2009). However, the functions and underlying mechanisms of these modifications are poorly understood.
BR and GA are central regulators of cell elongation and plant growth (Depuydt and Hardtke, 2011). Accordingly, several cell elongation–related genes are directly regulated by BZR1, DELLAs, PIFs, and PKL (de Lucas et al., 2008; Feng et al., 2008; Bai et al., 2012b; this study). Here, we found that exogenous application of BL or GA3 reduced the recruitment of H3K27me3 to cell elongation–related targets, whereas their inhibitors, PCZ or PAC, had an opposite effect, and that this regulation is largely dependent on PKL. Most intriguingly, the H3K27me3 status is regulated by BZR1 and DELLA proteins (Figure 7). This is in agreement with the effects of H3K27me3 status, which is regulated by BR and GA hormones/inhibitors and signaling components, on hypocotyl elongation. Thus, our study reveals that BR and GA signaling regulate H3K27me3 modification, providing a direct molecular link between histone modification and these signaling pathways.
Cellular hormone levels fluctuate in tissues in response to developmental stages and environmental conditions. In addition, the dynamic nature of histone methylation and acetylation states provides increased regulatory flexibility. A recent study suggested that a dynamic and multilevel relationship exists between BR and GA pathways during photomorphogenesis (Lilley et al., 2013). Light-regulated changes in histone modification may be an important component of light-controlled gene transcription (Guo et al., 2008). Therefore, through the PIF-BZR1-PKL-DELLA module, BR, GA, and exogenous signals dynamically regulate histone modifications to ensure that appropriate cellular responses are achieved. This complex mechanism likely underlies the developmental plasticity of plant lineages.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana epp1-1, pif3-1, pifq, and bzr1-1D mutants, and Pro35S:Myc-PIF3 transgenic plants, are of the Col ecotype (Wang et al., 2002; Leivar et al., 2008; Chen et al., 2013; Jing et al., 2013). The bri1-5 mutant is of the Ws ecotype (Noguchi et al., 1999). The della mutant and ProRGA:GFP-RGA are of the Landsberg erecta ecotype (Silverstone et al., 2001; Feng et al., 2008). Double mutants or transgenic plants were generated by standard genetic crossing and were verified by phenotype inspection, antibiotic selection, PCR genotyping, and/or sequencing. After sterilization, seeds were sown on MS medium containing 1% Suc and 0.8% agar and incubated at 4°C in darkness for 3 d. Seedlings were grown in the dark at 22°C for different times as indicated in the text.
Hypocotyl Length and Cotyledon Angle Measurements
Seedlings were grown on MS medium in the absence or presence of hormones or inhibitors for 5 d as stated in the text. At least 20 seedlings for each genotype were photographed, and their hypocotyl lengths and cotyledon angles were measured by using ImageJ software (http://rsb.info.nih.gov/ij).
Plasmid Construction
To obtain the open reading frames of PIF4, PIF5, BZR1, RGA, GAI, RGL1, RGL2, and RGL3, RT-PCR was performed using the primers listed in Supplemental Table 1. The resulting fragments were cloned into the pEASY-Blunt vector (TransGen), resulting in pEASY-PIF4/PIF5/BZR1/RGA/GAI/RGL1/RGL2/RGL3, respectively. The D7 fragment of PKL was amplified from the pEASY-PKL plasmid and ligated into pEASY-Blunt to generate pEASY-D7. Restriction sites were included in the primers to facilitate cloning. All clones were validated by sequencing. The pEASY-PKL, pEASY-D2, pEASY-D3, pLexA-PKL/D1/D2/D3/D4/D5/D6, and pGEX-D6 plasmids were constructed previously (Jing et al., 2013). The pAD-PIF1, pAD-PIF3, pAD-PIF3N, pAD-PIF3C, pSPYNE-PIF3, and pHis-PIF3 plasmids were described by Chen et al. (2013). To generate constructs for the yeast two-hybrid assay, pEASY-PIF4, pEASY-BZR1, and pEASY-GAI were digested with EcoRI and SalI, and the corresponding fragments were cloned into pB42AD (Clontech) cut with EcoRI and XhoI, to produce pAD-PIF4, pAD-BZR1, and pAD-GAI, respectively. pEASY-PIF5 was digested with EcoRI and XhoI, and PIF5 was inserted into the EcoRI-XhoI sites of pB42AD, to generate pAD-PIF5. pEASY-RGA, pEASY-RGL1, pEASY-RGL2, and pEASY-RGL3 were cut with MfeI and SalI, and the corresponding genes were ligated into the EcoRI-XhoI sites of pB42AD, resulting in pAD-RGA, pAD-RGL1, pAD-RGL2, and pAD-RGL3, respectively. pEASY-BZR1, pEASY-BZR1N, and pEASY-BZR1C were digested with EcoRI and SalI, and the corresponding fragments were cloned into the EcoRI-XhoI sites of the pLexA vector (Clontech), to give rise to pLexA-BZR1, pLexA-BZR1N, and pLexA-BZR1C, respectively.
To construct vectors for the generation of recombinant proteins, pEASY-D2 and pEASY-D3 were digested with MfeI and XhoI, and the fragments were cloned into the pGEX-5X-1 vector (GE Healthcare) cut with EcoRI and XhoI, to yield pGEX-D2 and pGEX-D3, respectively. pEASY-RGA and pEASY-GAI were digested with BamHI and NotI, and the fragments were ligated into the BamHI-NotI sites of pETMALc-H (Pryor and Leiting, 1997), resulting in pMBP-RGA and pMBP-GAI, respectively. The pEASY-BZR1 plasmid was cut with EcoRI and SalI, and BZR1 was cloned into pET-28a (Novagen) digested with EcoRI and XhoI, to give rise to pHis-BZR1.
To prepare vectors for the BiFC assay, BZR1 was released from pEASY-BZR1 by digestion with EcoRI and SalI, and cloned into the EcoRI-XhoI sites of pUC-SPYCE (Walter et al., 2004), to generate pSPYCE-BZR1.
For binary vector construction, the GFP gene was amplified from pCAMBIA1302 (http://www.cambia.org/daisy/cambia/585) and cloned into pEASY-Blunt to generate pEASY-GFP. The fragment of GFP was released from pEASY-GFP, and then inserted into the XbaI-StuI–digested pVIP96 vector (Chen et al., 2010), to produce pVIP-N-GFP, in which the EcoRI and XhoI sites were introduced at the N terminus of GFP. To prepare the Pro35S:GAI-GFP binary vector, pEASY-GAI was cut with EcoRI and SalI, and then ligated into the EcoRI-XhoI sites of pVIP-N-GFP, to generate the destination construct. The binary constructs were electroporated into Agrobacterium tumefaciens GV3101 and then introduced into destination plants via the floral dip method (Clough and Bent 1998). Transgenic plants were selected on MS plates in the presence of 50 mg/L kanamycin.
Gene Expression Analysis
Plant total RNA was isolated using an RNA Extraction Kit (Tiangen), and the first-strand cDNA was synthesized from 2 μg of RNA by reverse transcriptase (Invitrogen). cDNA was diluted 1:100 into quantitative PCR in a volume of 15 μL with SYBR Premix ExTaq Mix (Takara) and a LightCycler 480 thermal cycler (Roche), following the manufacturer’s instructions. Three biological replicates were performed for each sample, and the expression level was normalized to that of a UBQ control. Primers are listed in Supplemental Table 1.
Yeast Two-Hybrid Assay
The LexA DNA binding domain (LexA-BD) fusion plasmids were cotransformed with a LexAop:LacZ (Clontech) reporter into the yeast strain EGY48, and B42 activation domain (AD) fusion constructs were transformed into strain Y864. Different combinations of LexA and AD fusions as described in the text were generated by mating. The yeast colonies were grown on SD/-Trp-Ura-His dropout plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for color development.
ChIP-qPCR Assay
Chromatin immunoprecipitation assays were performed as described (Bowler et al., 2004). Briefly, seedlings were cross-linked by submerging in 37 mL of a 1% formaldehyde solution with vacuum infiltration for 10 min, followed by neutralization with Gly to a final concentration of 0.125 M. Chromatin complexes were isolated and sonicated to reduce the average DNA fragment size to ∼500 bp. The chromatin suspensions were incubated with anti-PKL (homemade; 1:250), anti-MYC (Abcam, ab32; 1:500), and anti-H3K27me3 (Millipore, 07-449; 1:500) antibodies or anti-H3 (Millipore, 07-690; 1:500) polyclonal antibodies or IgG serum overnight at 4°C with gentle agitation, followed by incubation with 40 μL of protein A agarose beads (Roche) for at least 1 h. After several rounds of washing, the pellets were eluted and the supernatants were incubated with proteinase K to digest proteins. The precipitated DNA fragments were recovered and quantified by quantitative PCR with SYBR Premix ExTaq Mix and the primers shown in Supplemental Table 1. Relative enrichment of a corresponding gene was normalized to the respective input DNA samples (100× to 500× dilution). All experiments were performed at least three times with similar results.
Recombinant Protein Production, Pull-Down, and Co-IP
GST, His, and MBP fusion recombinant proteins were induced by isopropyl β-d-1-thiogalactopyranoside and expressed in Escherichia coli BL21 (DE3). The proteins were then purified using Glutathione Sepharose 4B beads (GE Healthcare; for GST fusions) or Ni-NTA Agarose (Qiagen; for His and MBP fusions containing the His tag), following the manufacturers’ instructions. The procedures used for pull-down, Co-IP, and immunoblot assays were described previously (Jing et al., 2013). The input and precipitated proteins were size-fractionated on an 8 to 10% SDS-PAGE gel and immunoblotted with anti-His (Abcam, ab14923; 1:1000), anti-GST (Abcam, ab19256; 1:1000), anti-GFP (Abcam, ab1218; 1:1000), anti-PKL, or anti-tubulin (homemade; 1:5000) antibodies.
BiFC Assay
Plasmids of the corresponding N- and C-terminal fusions of YFP were cotransformed into Arabidopsis Col wild-type protoplasts as described previously (Walter et al., 2004). The protoplasts were incubated in darkness for 12 to 16 h, and the fluorescence was determined using a confocal microscope (Leica). The YFP fluorescence was excited by a 514-nm laser and captured at 523 to 600 nm, and the chlorophyll autofluorescence was captured at 650 to 750 nm.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library under the following accession numbers: PKL/EPP1 (At2g25170), PIF1 (At2g20180), PIF3 (At1g09530), PIF4 (At2g43010), PIF5 (At3g59060), BZR1 (At1g75080), RGA (At2g01570), GAI (At1g14920), RGL1 (At1g66350), RGL2 (At3g03450), RGL3 (At5g17490), IAA19 (At3g15540), PRE1 (At5g39860), DWF4 (At3g50660), TCH4 (At5g57560), HFR1(At1g02340), PIF6 (At3g62090), UBQ1 (At3g52590), and ACT2 (At3g18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Yeast Two-Hybrid Assay Showing the Interaction between PKL and PIFs.
Supplemental Figure 2. Phenotypes of epp1 pif3 and epp1/Myc-PIF3 Plants.
Supplemental Figure 3. PIF3 and PKL Bind to the Promoters of Cell Elongation–Related Genes.
Supplemental Figure 4. PIF3 Interacts with BZR1.
Supplemental Figure 5. Phenotype of epp1/bzr1-1D Plants.
Supplemental Figure 6. ChIP-qPCR Assay in the Wild Type and the bri1-5 Mutant.
Supplemental Figure 7. Interaction between PKL and DELLA Proteins in Yeast.
Supplemental Figure 8. Characterization of Pro35S:GAI-GFP Transgenic Plants.
Supplemental Figure 9. Relative Binding Activity of PKL to the Downstream Genes.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Xing Wang Deng (Yale University) for providing della seeds, Zhiyong Wang (Stanford University) for bzr1-1D and bri1-5 seeds, and Tai-Ping Sun (Duke University) for the ProRGA:GFP-RGA line. This work was supported by the National Science Foundation of China (Grants 31325002 and 31370023) and the Chinese Academy of Sciences.
AUTHOR CONTRIBUTIONS
R.L. and Y.J. conceived the research. D.Z. and R.L. designed the experiments. D.Z., Y.J., and Z.J. performed the experiments. D.Z., Y.J., and R.L. analyzed the data. R.L. wrote the article.
Glossary
- BR
brassinosteroid
- GA
gibberellin
- BiFC
bimolecular fluorescence complementation
- Co-IP
coimmunoprecipitation
- ChIP-qPCR
chromatin immunoprecipitation followed by quantitative PCR
- Col
Columbia
- PCZ
propiconazole
- Ws
Wassilewskija
- BL
brassinolide
- PAC
paclobutrazol
- GA3
gibberellic acid
- RT-qPCR
RT–quantitative PCR
- MS
Murashige and Skoog
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Achard P., Liao L., Jiang C., Desnos T., Bartlett J., Fu X., Harberd N.P. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143: 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Renou J.P., Berthomé R., Harberd N.P., Genschik P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660. [DOI] [PubMed] [Google Scholar]
- Achard P., Vriezen W.H., Van Der Straeten D., Harberd N.P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E., Villar C.B.R., Di Mambro R., Sabatini S., Köhler C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23: 1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski A.A., Galstyan A., Guseman J.M., Nemhauser J.L. (2012). Photomorphogenesis. The Arabidopsis Book 10: e0147, /10.1199/tab.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Fan M., Oh E., Wang Z.Y. (2012a). A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012b). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M., Bertrand C., Servet C., Zhou D.X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J. (2004). Chromatin techniques for plant cells. Plant J. 39: 776–789. [DOI] [PubMed] [Google Scholar]
- Charron J.B., He H., Elling A.A., Deng X.W. (2009). Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Xu G., Tang W., Jing Y., Ji Q., Fei Z., Lin R. (2013). Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang Z., Teng K., Lai J., Zhang Y., Huang Y., Li Y., Liang L., Wang Y., Chu C., Guo H., Xie Q. (2010). Up-regulation of LSB1/GDU3 affects geminivirus infection by activating the salicylic acid pathway. Plant J. 62: 12–23. [DOI] [PubMed] [Google Scholar]
- Chen M., Chory J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21: 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C.R., Cairns B.R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Depuydt S., Hardtke C.S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21: R365–R373. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S., Yu P., Breen G., Cohen J.D., Wigge P.A., Gray W.M. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 108: 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Harberd N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743. [DOI] [PubMed] [Google Scholar]
- Fukaki H., Taniguchi N., Tasaka M. (2006). PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 48: 380–389. [DOI] [PubMed] [Google Scholar]
- Furuta K., Kubo M., Sano K., Demura T., Fukuda H., Liu Y.G., Shibata D., Kakimoto T. (2011). The CKH2/PKL chromatin remodeling factor negatively regulates cytokinin responses in Arabidopsis calli. Plant Cell Physiol. 52: 618–628. [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Zhou J., Elling A.A., Charron J.B., Deng X.W. (2008). Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 147: 2070–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T., Corvalan C., Best N.B., Budka J.S., Zhu J.Y., Choe S., Schulz B. (2012). Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS ONE 7: e36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose F., Ohshima N., Kwon E.J., Yoshida H., Yamaguchi M. (2002). Drosophila Mi-2 negatively regulates dDREF by inhibiting its DNA-binding activity. Mol. Cell. Biol. 22: 5182–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Crabtree G.R. (2010). Chromatin remodelling during development. Nature 463: 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo J.A., Piñeiro M., Cubas P., Martínez-Zapater J.M. (2009). Chromatin remodeling in plant development. Int. J. Dev. Biol. 53: 1581–1596. [DOI] [PubMed] [Google Scholar]
- Jing Y., Lin R. (2013). PICKLE is a repressor in seedling de-etiolation pathway. Plant Signal. Behav. 8: e25026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Zhang D., Wang X., Tang W., Wang W., Huai J., Xu G., Chen D., Li Y., Lin R. (2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006). The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442: 312–316. [DOI] [PubMed] [Google Scholar]
- Koini M.A., Alvey L., Allen T., Tilley C.A., Harberd N.P., Whitelam G.C., Franklin K.A. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19: 408–413. [DOI] [PubMed] [Google Scholar]
- Kwon C.S., Wagner D. (2007). Unwinding chromatin for development and growth: A few genes at a time. Trends Genet. 23: 403–412. [DOI] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.F., Wang C., Jiang L., Li S., Sun S.S., He J.X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5: ra72. [DOI] [PubMed] [Google Scholar]
- Lilley J.L.S., Gan Y., Graham I.A., Nemhauser J.L. (2013). The effects of DELLAs on growth change with developmental stage and brassinosteroid levels. Plant J. 76: 165–173. [DOI] [PubMed] [Google Scholar]
- Lu F., Cui X., Zhang S., Jenuwein T., Cao X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43: 715–719. [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Yoshida S., Yuan H., Feldmann K.A., Tax F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121: 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Perruc E., Kinoshita N., Lopez-Molina L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52: 927–936. [DOI] [PubMed] [Google Scholar]
- Pryor K.D., Leiting B. (1997). High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expr. Purif. 10: 309–319. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Willige B.C. (2009). Shedding light on gibberellic acid signalling. Curr. Opin. Plant Biol. 12: 57–62. [DOI] [PubMed] [Google Scholar]
- Shigeta T., Yasuda D., Mori T., Yoshimitsu Y., Nakamura Y., Yoshida S., Asami T., Okamoto S., Matsuo T. (2011a). Characterization of brassinosteroid-regulated proteins in a nuclear-enriched fraction of Arabidopsis suspension-cultured cells. Plant Physiol. Biochem. 49: 985–995. [DOI] [PubMed] [Google Scholar]
- Shigeta T., Yoshimitsu Y., Nakamura Y., Okamoto S., Matsuo T. (2011b). Does brassinosteroid function require chromatin remodeling? Plant Signal. Behav. 6: 1824–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui P., Jin J., Ye S., Mu C., Gao J., Feng H., Shen W.H., Yu Y., Dong A. (2012). H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J. 70: 340–347. [DOI] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Zhai Q., Li C. (2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25: 2102–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21: R338–R345. [DOI] [PubMed] [Google Scholar]
- Von Arnim A., Deng X.W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 215–243. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Weider M., Küspert M., Bischof M., Vogl M.R., Hornig J., Loy K., Kosian T., Müller J., Hillgärtner S., Tamm E.R., Metzger D., Wegner M. (2012). Chromatin-remodeling factor Brg1 is required for Schwann cell differentiation and myelination. Dev. Cell 23: 193–201. [DOI] [PubMed] [Google Scholar]
- Wu M.F., Sang Y., Bezhani S., Yamaguchi N., Han S.K., Li Z., Su Y., Slewinski T.L., Wagner D. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. USA 109: 3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Li L., Guo M., Chory J., Yin Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 7618–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Rider S.D., Jr, Henderson J.T., Fountain M., Chuang K., Kandachar V., Simons A., Edenberg H.J., Romero-Severson J., Muir W.M., Ogas J. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283: 22637–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wang L., Xi Y., Li J., Quail P.H., Deng X.W., Guo H. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22: 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.