Using the FATTY ACID ELONGATION1 (FAE1) gene as a case study, this work reveals that transposable element insertion and epigenetic modification generate multiple heritable alleles varying in expression level and modulating trait phenotype from qualitative to quantitative variation.
Abstract
Naturally occurring heritable variation provides a fundamental resource to reveal the genetic and molecular bases of traits in forward genetic studies. Here, we report the molecular basis of the differences in the four alleles E1, E2, E3, and e of the FATTY ACID ELONGATION1 (FAE1) gene controlling high, medium, low, and zero erucic content in yellow mustard (Sinapis alba). E1 represents a fully functional allele with a coding DNA sequence (CDS) of 1521 bp and a promoter adjacent to the CDS. The null allele e resulted from an insertional disruption in the CDS by Sal-PIF, a 3100-bp PIF/Harbinger-like DNA transposon, whereas E2 and E3 originated from the insertion of Sal-T1, a 4863-bp Copia-like retrotransposon, in the 5′ untranslated region. E3 was identical to E2 but showed cytosine methylation in the promoter region and was thus an epiallele having a further reduction in expression. The coding regions of E2 and E3 also contained five single-nucleotide polymorphisms (SNPs) not present in E1, but expression studies in Saccharomyces cerevisiae indicated that these SNPs did not affect enzyme functionality. These results demonstrate a comprehensive molecular framework for the interplay of transposon insertion, SNP/indel mutation, and epigenetic modification influencing the broad range of natural genetic variation in plants.
INTRODUCTION
Genetic and molecular mechanisms of many traits in plants have been revealed based on naturally occurring heritable variation. Allelic differences in nucleotide sequences resulting from single-nucleotide polymorphisms (SNPs), indels (insertions or deletions), and transposable element (TE) insertions are primarily responsible for contrasting trait phenotypes. TEs represent a large proportion of the nuclear genomes in many plant species (Vicient et al., 1999; Meyers et al., 2001). They are classified into class I (RNA) and II (DNA) based on their different modes of mobility (Charlesworth et al., 1994). Class I elements are retroelements that use reverse transcriptase to transpose via an RNA intermediate, whereas class II elements can transpose directly from DNA to DNA. TEs and the movement of TEs provide an important source of new genetic variation in both plants and animals (Kidwell and Lisch, 1997). Insertions of TEs in exons, introns, and 5′ flanking sequences of genes have resulted in new alleles having no or reduced function. Allelic variation of the knotted1 locus resulted from the insertion of Mu1 or Mu8 within a 310-bp region of the kn1 third intron in maize (Zea mays) (Greene et al., 1994). In the blood orange (Citrus sinensis), the expression of Ruby (regulator of anthocyanin biosynthesis) was found to be modified by the expression of a Copia-like retrotransposon inserted in its upstream region (Butelli et al., 2012).
Epigenetic modifications including DNA cytosine methylation, histone variants, and small RNAs provide another molecular mechanism for regulating gene expression and generate a potential source of natural heritable variation (for reviews, see Kalisz and Purugganan, 2004; Richards, 2006, 2011; Becker and Weigel, 2012; Weigel and Colot, 2012). The clark kent (clk) epiallele described in Arabidopsis thaliana contains hypermethylated cytosines in the coding region of the SUPERMAN gene (Jacobsen and Meyerowitz, 1997). The clk epiallele showed a phenotype similar to loss-of-function mutants of SUPERMAN, having increased numbers of stamens and carpels. Genes controlling ecologically important traits such as floral symmetry in toadflax (Linaria vulgaris) and pathogen resistance in Arabidopsis show differences in expression due to differential methylation (Cubas et al., 1999; Stokes et al., 2002). All natural epialleles reported to date have involved a gain or loss of DNA methylation (for a review, see Weigel and Colot, 2012).
Erucic acid (cis-13 docosenoic acid [C22:1]) is one of the major fatty acids produced in the seeds of many plant species, including Brassica species and yellow mustard (Sinapis alba, SS, 2n=24). High erucic oil and erucic acid derivatives have many potential industrial applications, including as lubricants, surface coatings, and pharmaceuticals (Sonntag, 1991, 1995; Leonard, 1994). However, erucic acid is undesirable in edible oil due to its antinutritional effects (Sauer and Kramer, 1983). The biosynthesis of erucic acid occurs in the cytoplasm via an elongation complex containing a seed-specific condensing enzyme, β-ketoacyl-CoA synthase (KCS) (Stumpf and Pollard, 1983). KCS is encoded by the FATTY ACID ELONGATION1 (FAE1) gene, which has been cloned from Arabidopsis (James et al., 1995), jojoba (Simmondsia chinensis) (Lassner et al., 1996), turnip rape (Brassica rapa), cabbage (Brassica olereacea) (Das et al., 2002), oilseed rape (Brassica napus) (Han et al., 2001), and many other plant species. Additionally, FAE1 genes from a number of high and low erucic acid cultivars of B. napus have been cloned and sequenced. In the FAE1 gene from the A genome of B. napus, a single base change of nucleotide 845 from cytosine to thymine resulted in a loss-of-function allele (Han et al., 2001; Roscoe et al., 2001; Katavic et al., 2002; Wu et al., 2007, 2008). A second mutation consisting of a 4-bp deletion between T1366 and G1369 in the FAE1 gene, which led to a frameshift mutation, was predominantly found in the C genome (Wu et al., 2007, 2008). Thus, the known erucic acid variants in B. napus have resulted from SNP/indel mutations of FAE1.
Erucic acid variants have been identified by inbreeding of open-pollinated plants in yellow mustard, which is an obligate outcrossing crop due to its self-incompatible reproductive system (Raney et al., 1999; Cheng et al., 2012). The high (52.9%), medium (24.4%), low (1.4%), and zero (0.1%) erucic contents of the yellow mustard lines Y517, Y496, Y1130, and Y514 were conditioned by the four FAE1 alleles E1, E2, E3, and e, respectively (Javidfar and Cheng, 2013). Here, we describe the TE insertion and epigenetic variation in FAE1 that lead to the observed changes in erucic acid content in yellow mustard.
RESULTS
The Four FAE1 Alleles E1, E2, E3, and e Show Variation in Coding DNA Sequence
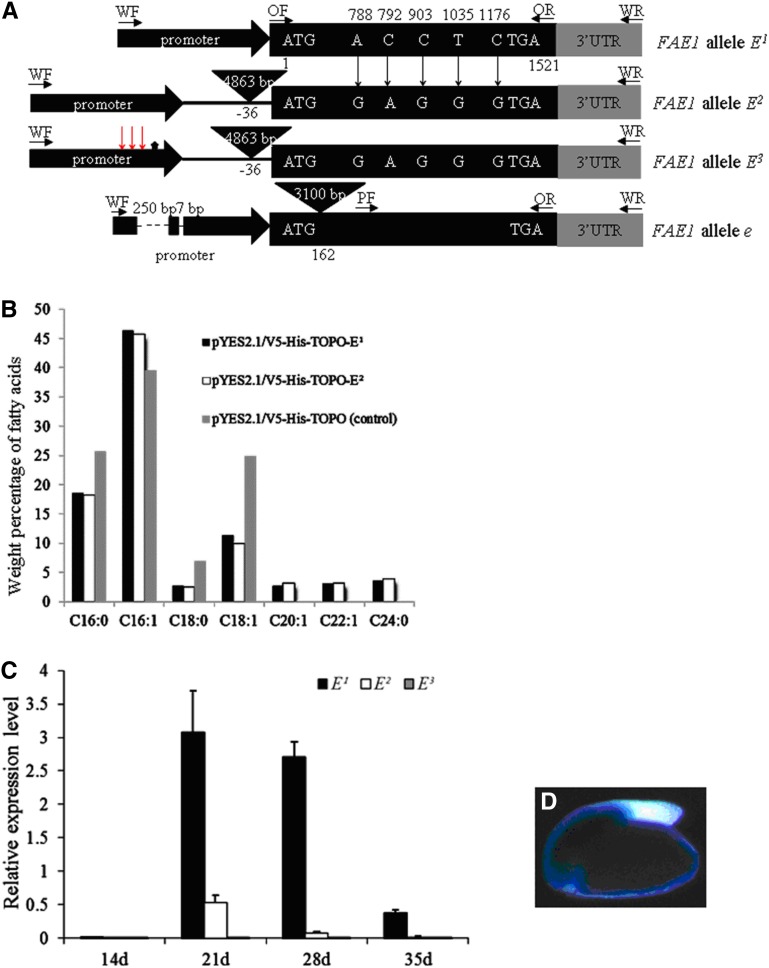
The coding regions of E1, E2, and E3 were successfully cloned from lines Y517 (E1E1), Y496 (E2E2), and Y1130 (E3E3), respectively, using primer pair No 1 (Supplemental Table 1; Figure 1A). E1 had a coding DNA sequence (CDS) of 1521 bp encoding a polypeptide of 507 amino acids, which contained six conserved cysteine residues, four conserved histidine residues, and a conserved serine residue compared with the polypeptides encoded by the FAE1 genes in Arabidopsis, B. napus, Brassica juncea, B. rapa, and B. oleracea (Supplemental Figure 1). The sequences of FAE1 alleles E2 and E3 were identical in the coding region. Sequence alignment with E1 allowed the identification of five point mutations at positions 788, 792, 903, 1035, and 1176 in the CDS of E2 and E3 (Figure 1A). Only the mutation at position 788 (A of E1 to G of E2 and E3) led to an amino acid change; the arginine residue at position 263 in the protein sequence of E1 was substituted by lysine in the protein encoded by the E2 and E3 alleles (Supplemental Figure 1).
Figure 1.
The Structure and Function of the Four FAE1 Alleles E1, E2, E3, and e in Yellow Mustard.
(A) Schematic representation of the structure of the four FAE1 alleles E1, E2, E3, and e. Black rectangle, the entire coding region; gray rectangle, 300-bp 3′ UTR; solid black arrow, promoter; vertical long arrow, point mutation; inverted triangle, insertion; asterisk, putative transcription start site at position −4935; red vertical long arrow, cytosine methylation at positions −5146, −5095, and −4943; dashed lines, deletion; OF/OR, primer set No 1 for cloning the entire coding region; PF/OR, Primer set No 2 for cloning the partial coding region of e; WF/WR, primer set No 14 for cloning the entire FAE1 gene.
(B) Gas chromatography analysis of fatty acid composition of yeast cells containing the construct pYES2.1/V5-His-TOPO-E1 with the FAE1 allele E1 (black), the construct pYES2.1/V5-His-TOPO-E2 with the FAE1 allele E2 (white), and the empty vector pYES2.1/V5-His-TOPO (gray color, control).
(C) Real-time qPCR analysis of expression levels of the FAE1 alleles E1, E2, and E3 in 14- to 35-d-old embryos. Error bars indicated sd of the mean. Actin2 (FG576123) was used as an internal control.
(D) Histochemical localization of GUS activity in the 9-d-old embryos of transgenic Arabidopsis plants containing the construct pBI101-ProE1-GUS with the promoter of the FAE1 allele E1.
However, primer pair No 1 failed to amplify the CDS of the FAE1 allele e of line Y514 (ee). A 1328-bp fragment from nucleotide 194 to 1521 was successfully obtained for the allele e using primer pair No 2 (Supplemental Table 1; Figure 1A). Sequence alignment with E1 indicated that eight point mutations occurred at positions 439, 532, 601, 922, 1040, 1097, 1295, and 1470 in the partial CDS of e, but none of these led to amino acid changes. An additional 1447-bp DNA fragment was cloned from allele e by PCR walking using primer pair No 3 (Supplemental Table 1 and Supplemental Figure 2C). This fragment included another 32 bp from the CDS (nucleotide 162 to 193) and an unknown DNA fragment of 1415 bp. Thus, although 1360 bp of the CDS from nucleotide 162 to 1521 for the FAE1 allele e was cloned, the remaining 161 bp CDS from nucleotide 1 to 161 was not obtainable using these primer sets. BLAST analysis of the unknown 1415-bp fragment against the sequence database of Arabidopsis (TAIR) revealed that it had high similarity to a transposable element (At3G15310, E value = 9e−85). Alignment with the E1 allele indicated that the transposable element was inserted into the CDS of e at position 162 (Figure 1A). Therefore, it was inferred that the insertion of the 1415 bp transposable element in the coding region of FAE1 led to the occurrence of the loss-of-function allele e. However, the use of these primer sets did not allow us to determine whether the entire sequence of the transposable element had been obtained.
Heterologous Expression of FAE1 in Yeast Indicates That Enzymes Encoded by Alleles E1 and E2 Are Functional and Have Similar Expression Level
Transgenic yeast (Saccharomyces cerevisiae) cultures containing the empty construct pYES2.1/V5-His-TOPO produced the typical fatty acids, i.e., C16:0, C16:1, C8:0, and C18:1, found in untransformed yeast cells (Figure 1B). Transgenic yeast cells harboring the yeast expression vector pYES2.1/V5-His-TOPO-E1 carrying E1 coding sequence or pYES2.1/V5-His-TOPO-E2 with E2 coding sequence produced the novel long-chain fatty acids C20:1, C22:1, and 24:0, and erucic acid content was not significantly different between cultures expressing E1 versus E2 (Figure 1B). These data indicated that the cloned alleles E1 and E2 encoded functional enzymes capable of elongation of C18:1 to C20:1 and C22:1 and had similar activity levels. Therefore, the reduction of erucic acid content in the yellow mustard lines Y496 and Y1130 did not appear to be caused by the nucleotide mutations in the CDS of E2 and E3.
The Three FAE1 Alleles E1, E2, and E3 Exhibit Significant Differences in Transcript Levels
Transcripts of the FAE1 were only detected in 14- to 35-d-old embryos and were absent in leaf, stem, flower bud, and open flower tissue of Y517, Y496, and Y1130 (Supplemental Figure 3 A ), suggesting that E1, E2, and E3 were specifically expressed in the embryo. Transcripts were not detected in immature embryo tissue of Y514 since Y514 carried the loss-of-function FAE1 allele e (Supplemental Figure 3 A ).
Peak transcript levels of E1, E2, and E3 were observed in 21-d-old embryos (Figure 1C). E1 had the highest expression level (Figure 1C), and the expression levels of E2 and E3 were 21.6% (sd ± 2.3%) and 1.1% (sd ± 0.1%) of that of E1, respectively. E3 had the lowest expression level, at 4.8% (sd ± 0.1%) of E2. The transcript levels of E1, E2, and E3 were positively correlated with erucic acid contents in the seeds of Y517, Y496, and Y1130. Thus, although heterologous expression in yeast indicated that the expressed proteins encoded by E1, E2, and E3 had the same activity level in the elongation of C18:1 to C20:1 and C22:1, significant differences at the transcript level were detected among these alleles by quantitative PCR (qPCR).
Histochemical GUS Assays Indicate That the 877-bp 5′ Flanking Fragment of E1 Functions as a Seed-Specific Promoter
DNA fragments comprising the 300 bp downstream from the translation stop codon were cloned for E1, E2, and E3 using primer pair No 4 (Supplemental Table 1 and Supplemental Figure 2) and had the same nucleotide sequence, indicating that they were not related to the reduction of transcript levels seen for E2 and E3. To determine if the 5′ upstream (promoter) region was involved in the differential transcription of the FAE1 alleles, the 5′ flanking sequences of E1, E2, and E3 were cloned by PCR walking using primer pair No 5 (Supplemental Table 1 and Supplemental Figure 2). The 1095-bp 5′ flanking fragment cloned for E1 contained a 703-bp putative promoter, which showed 87% identity to that of the FAE1 promoter region from the high erucic acid B. napus line Askari. The 1221-bp 5′ flanking fragments cloned for E2 and E3 shared the same nucleotide sequence, but did not exhibit any similarity with the putative promoter sequence of E1.
To test if the isolated 5′ flanking fragments included functional promoters, the 877-bp region upstream from the translational start codon of E1 and the 1003-bp upstream region of E2 and E3 were cloned using primer pairs No 16 and 17, respectively (Supplemental Table 1), then placed in front of a β-glucuronidase (GUS) reporter gene in a plant expression vector. The resulting constructs pBI101-ProE1-GUS and pBI101-ProE2-GUS were transformed into Arabidopsis. In transgenic plants containing the construct pBI101-proE1-GUS, histochemical assays led to the detection of GUS activity in the embryo (Figure 1D), but not in the leaf, stem, bud, or flower tissues (Supplemental Figures 3 B to 3E ), indicating that the 877-bp 5′ flanking sequence of E1 functioned as a seed-specific promoter. In contrast, no GUS activity was detected in the embryo or any vegetative tissues of the transgenic plants containing the construct pBI101-proE2-GUS, suggesting that the 1003-bp 5′ flanking sequence of E2 and E3 did not function as a promoter.
Insertion of a Copia-Like Retrotransposon in the 5′ Untranslated Region Separates the FAE1 Promoter from the CDS in Alleles E2 and E3
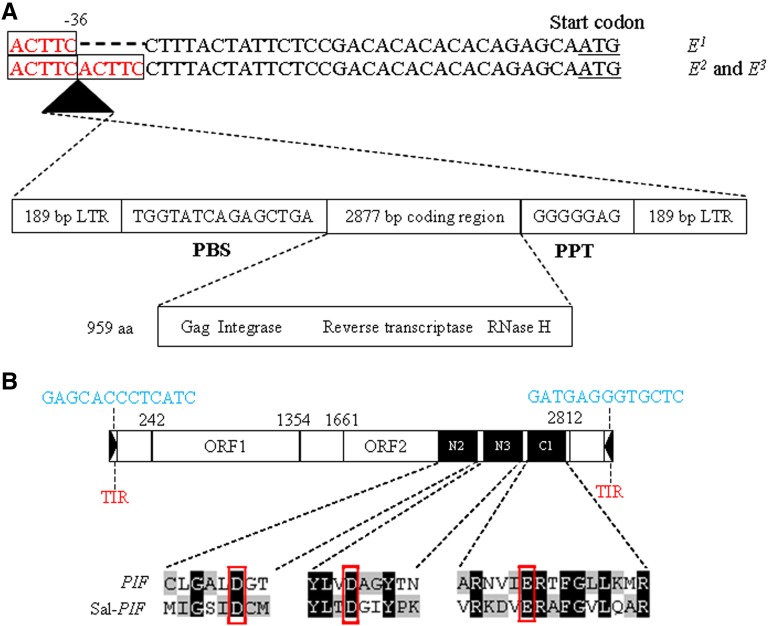
To determine if E2 and E3 had promoters that were located further away from the coding region, additional upstream regions were cloned by PCR walking. Four rounds of PCR walking using primer pairs No 6, 7, 8, and 9 (Supplemental Table 1 and Supplemental Figure 2B) allowed the isolation of four additional 5′ upstream fragments of 673, 741, 791, and 903 bp, respectively, from both E2 and E3. Sequence alignments indicated that the 3337-bp 5′ flanking fragments of E2 and E3 obtained using primer pairs No 5-9 shared the same nucleotide sequence but did not exhibit any similarity with the promoter sequence of E1. However, the promoters of E2 and E3 were successfully cloned using primer pair No 10, which was designed based on the promoter sequence of E1 (Supplemental Table 1 and Supplemental Figure 2). The E2 and E3 promoters had the same nucleotide sequence and length (703 bp) as the promoter of E1. The gap between the E2/E3 promoter region and the 3337-bp 5′ flanking fragment was filled with a 1717-bp sequence obtained using primer pair No 11 (Supplemental Table 1 and Supplemental Figure 2B). Sequence alignments of the promoter and the 1717- and 3337-bp 5′ flanking fragments revealed an insertion of 4863 bp between the coding region and promoter in both alleles. Sequence alignment with E1 indicated that the 4863-bp insertion was located in the 5′ untranslated region (UTR) of E2 and E3 at position −36 (Figure 1A). An additional 600 bp upstream of the promoter region was obtained for E1 and E2/E3 using primer pair No 12 (Supplemental Table 1 and Supplemental Figure 2B), and the sequence of this region was the same in these alleles.
The 4863-bp insertion contained a primer binding site (PBS) and a polypurine tract (PPT), with 189-bp long terminal repeats (LTRs) on each side (Figure 2A; Supplemental Figure 4). It was flanked by target site duplication sequence (Figure 2A). These characteristics implied that the inserted 4863-bp DNA fragment was a retrotransposon. The protein encoded by this retrotransposon had several conserved domains including regions with sequence similarity to retrotransposon gag, reverse transcriptase, and the Ty1/Copia family of RNase H (Supplemental Figure 5) and exhibited 58% similarity to a putative retroelement pol polyprotein in Arabidopsis. Therefore, the 4863-bp insertion in the 5′ UTR of E2 and E3 appears to be a Copia-like retrotransposon and was designated as Sal-T1. The Sal-T1 is oriented in the antisense transcriptional direction (Figure 2A). The two LTRs of Sal-T1 had the same nucleotide sequence, suggesting that the insertion of Sal-T1 in the FAE1 could be a relatively recent event. The increased physical distance between the promoter and the CDS due to the insertion of Sal-T1 in the 5′ UTR might lead to the observed reduction in mRNA transcription of E2 and E3 alleles, thereby resulting in the decrease of erucic acid content seen in lines Y496 and Y1130.
Figure 2.
Structures of the Copia-Like Retrotransposon Sal-T1 and the DNA Transposon Sal-PIF in Yellow Mustard.
(A) Structure of the Copia-like retrotransposon Sal-T1 inserted in the 5′ UTR of FAE1 alleles E2 and E3. Sal-T1 comprised a PBS, a PPT, LTRs on each side, and a 2877-bp coding region. The target site duplication sequence is in red font. Sal-T1 was inserted in reverse orientation at position −36 as indicated by a black triangle.
(B) Structure of the DNA transposon Sal-PIF inserted in the coding region of FAE1 allele e. Black triangle, inverted repeats (TIRs); nucleotide sequence of the TIR is in blue font; ORF1, predicted coding region from position 242 to 1354; ORF2, predicted coding region from 1661 to 2812; putative N2, N3, and C1 catalytic domains are indicated by a black box; amino acid sequences of the catalytic domains of maize PIF and Sal-PIF are shown.
A 3100-bp PIF/Harbinger-Like DNA Transposon Is Inserted into the CDS of FAE1 Allele e
The promoter of the FAE1 allele e was cloned using primer pair No 10 (Supplemental Table 1 and Supplemental Figure 2). It had a size of 446 bp due to a 7-bp deletion at position −350 and a 250-bp deletion at position −410 compared with the promoter sequences of E1, E2, and E3 (Figure 1A). The gap between the promoter and the 1415-bp transposable element inserted in the CDS of e was bridged by a 2082-bp DNA fragment obtained using primer pair No 13 (Supplemental Table 1 and Supplemental Figure 2C). The 2082-bp fragment contained the remaining 161 bp (nucleotide 1 to 161) of the coding region and an additional 1685 bp of transposable element sequence. In summary, the mutant allele e had a 1521-bp CDS with a 3100-bp transposable element inserted at position 162 and a promoter adjacent to the start codon of the CDS (Figure 1A).
The 3100-bp insertion had a 13-bp terminal inverted repeat on each end (Figure 2B; Supplemental Figure 6) and was flanked by a target site duplication sequence TAA. These characteristics suggested that the 3100-bp insertion was a DNA transposon. The insertion contained two open reading frames (ORF1 and ORF2) (Figure 2B; Supplemental Figure 6). The protein encoded by ORF1 was 103 amino acids in length (Supplemental Figure 7A) and did not have any similarity with amino acid sequences in the National Center for Biotechnology Information protein database. The protein encoded by ORF2 was 383 amino acids in length (Supplemental Figure 7B). It had a putative Asp-Asp-Glu (DDE) motif (referred to as N2, N3, and C1) similar to that found in the PIF element of maize (Zhang et al., 2001) (Figure 2B). ORF2 encoded a domain with high similarity (E = 3.87e−129) to the transposases of the PIF/Harbinger superfamily. This suggested that the inserted DNA fragment in the coding region of the FAE1 allele e was a DNA element putatively encoding a transposase belonging to the PIF/Harbinger superfamily, and it was therefore designated as Sal-PIF.
Thus, the entire nucleotide sequence for each of the four FAE1 alleles E1, E2, E3, and e with sizes of 2524, 7424, 7424, and 5369 bp, respectively, was obtained by PCR walking. To validate these results, the four FAE1 alleles were each cloned in their entirety using primer pair No 14 (Supplemental Table 1; Figure 1A). As expected, the resulting DNA fragments of the four FAE1 alleles had the same sizes (Supplemental Figure 8) and nucleotide sequences as those obtained from PCR walking.
The FAE1 Allele E3 Is an Epiallele
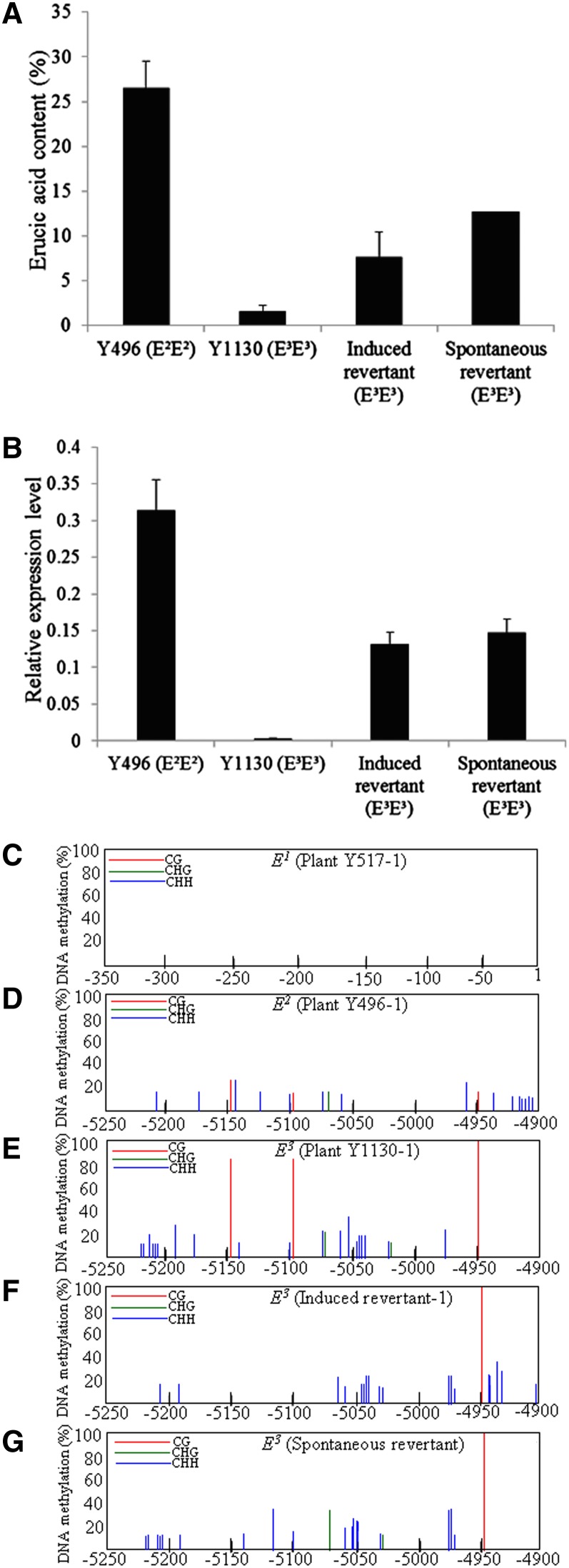
The FAE1 alleles E2 and E3 had the same coding and promoter sequences and contained a Copia-like retrotransposon Sal-T1 in the 5′ UTR, but exhibited significant differences in transcription level (Figure 3B), and seeds of plants carrying these alleles showed significant differences in erucic acid content (Figure 3A). To determine if DNA methylation was involved in regulating the expression of E3, we analyzed the methylation status of the coding and promoter regions of the three alleles E1, E2, and E3 in 21-d-old embryos from different plants for each of the three lines Y517, Y496, and Y1130 using a bisulfite sequencing procedure. None of the three alleles were methylated in the coding regions (nucleotides 1 to 1515) (Supplemental Figure 10). As expected, the allele E1 of line Y517 did not show methylation in its promoter region (nucleotides −335 to 1) (Figure 3C; Supplemental Figure 11) nor was DNA methylation detected in the promoter region (nucleotides −5234 to −4900) of E2 (Figure 3D; Supplemental Figure 12). However, CG methylation was detected at positions −5146, −5095, and −4943 in the promoter region (nucleotides −5234 to −4900) of the allele E3 (Figure 3E; Supplemental Figure 13). These data suggested that the FAE1 allele E3 is an epiallele.
Figure 3.
Seed Erucic Acid Content, FAE1 Expression Level, and DNA Methylation Status of FAE1 Alleles of Lines Y496 (E2E2), Y1130 (E3E3), 5-azaC–Induced Revertants (E3E3), and the Spontaneous Revertant (E3E3) of Y1130 in Yellow Mustard.
(A) Erucic acid contents of Y496, Y1130, 5-azaC–induced revertants, and spontaneous revertant of Y1130.
(B) FAE1 expression in the 21-d-old embryos of Y496, Y1130, 5-azaC–induced revertants, and spontaneous revertants of Y1130.
(C) DNA methylation was not detected in the promoter region (nucleotides −335 to 1) of E1 in Y517.
(D) DNA methylation was not detected in the promoter region (nucleotides −5234 to −4900) of E2 in Y496.
(E) DNA methylation was detected at positions −5146, −5095, and −4943 in the promoter region (nucleotides −5234 to −4900) of E3 in Y1130.
(F) and (G) DNA methylation was detected at position −4943 in the promoter region (nucleotides −5234 to −4900) of E3 in the 5-azaC induced and spontaneous revertants of Y1130.
To confirm this, 45 seeds of line Y1130 were treated with 1 mM 5-azacytidine (5-azaC), a demethylation agent, for 3 d. Twenty-five plants were obtained from the 45 5-azaC–treated seeds. Of the 25 plants, 13 (52%) plants were revertants induced by 5-azaC treatment and produced seeds with significantly higher erucic content (average, 7.6%; range, 4.3 to 15.4%) than the control plants of Y1130 (average, 1.6%; range, 0.7 to 3.4%) (t = 13.8, P = 0) (Figure 3A). The expression level of E3 in these revertants was also higher than that of E3 in the control plants of Y1130 (Figure 3B). DNA methylation analysis of the revertants indicated that the FAE1 allele E3 was demethylated at positions −5146 and −5095 but remained methylated at position −4943 (Figure 3F; Supplemental Figure 14). These results confirmed that E3 is an epiallele and the methylation in the promoter region of the FAE1 allele E3 led to the reduction in transcription, thereby decreasing the erucic acid content.
Furthermore, 100 untreated progeny seeds of Y1130 were analyzed for erucic content using the half-seed technique (Downey and Harvey, 1963). Ninety-nine seeds had low erucic content (range: 0.7 to 3.4%), whereas one seed (Y1130-16) contained 12.7% erucic acid and was a spontaneous revertant. The plant germinated from the seed Y1130-16 produced seeds with an average erucic content of 12.3% (range: 11.6 to 12.7%). Analysis by qPCR indicated that the expression level of E3 in this revertant was higher than that of E3 in Y1130 (control) (Figure 3B). DNA methylation analysis of this plant showed that the FAE1 allele E3 was also demethylated at positions −5146 and −5095, but remained methylated at position −4943 (Figure 3G; Supplemental Figure 15), as did those revertants induced by 5-azaC. These results indicated the metastability of the epiallele E3. Thus, we concluded that the FAE1 allele E3 is an epiallele and designated it as Eepi3 to differentiate it from the nonepialleles E1, E2, and e.
The Allele-Specific Markers for e and E2 Cosegregate with the Zero and Medium Erucic Acid Contents in F2 Populations of Y514 (ee) × Y517 (E1E1) and Y496 (E2E2) × Y517 (E1E1), Respectively
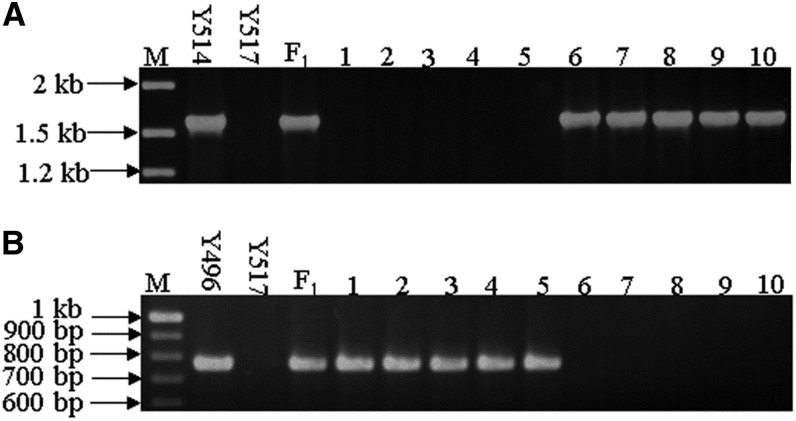
Primer pair No 31 (Supplemental Table 1) generated one dominant marker of 1616 bp specific for the e locus carrying the transposon Sal-PIF. Cosegregation of the dominant marker with the zero erucic acid trait was studied in 96 F2 plants from the cross Y514 (ee) × Y517 (E1E1). The dominant marker was present in all F2 plants with erucic acid content (range: 0.0 to 0.2%) resembling the zero erucic acid parent Y514 (0.0 to 0.1%), but absent in the F2 plants with high erucic content (range: 39.5 to 58.0%) resembling levels found in Y517 (range: 50.3 to 55.5%) (Figure 4A). The cosegregation of this marker with the zero erucic trait suggested that the loss-of-function FAE1 allele e was due to the insertion of the DNA transposon Sal-PIF in its coding region.
Figure 4.
Cosegregation of the FAE1 Allele-Specific Markers with Erucic Acid Contents.
Cosegregation of marker for e with the zero erucic acid trait in the F2 population of Y514 (ee) × Y517 (E1E1) (A) and of the marker for E2 with the medium erucic content trait in the F2 population of Y496 (E2E2) × Y517 (E1E1) (B).
(A) M, DNA ladder; Y514 (ee), zero erucic line; Y517 (E1E1), high erucic line; F1, Y514 × Y517; 1-5, F2 plants with high erucic content (39.5 to 58.0%); 6 to 10, F2 plants with zero erucic content (0.0 to 0.2%).
(B) M, DNA ladder; Y496 (E2E2), medium erucic line; Y517 (E1E1), high erucic line; F1, Y496 × Y517; 1-5, F2 plants with medium erucic content (20.0 to 29.8%); 6-10, F2 plants with high erucic content (44.4 to 49.2%).
Primer pair No 32 (Supplemental Table 1) produced a dominant marker of 768 bp specific for the region carrying the retrotransposon Sal-T1 in the 5′ UTR of E2. Cosegregation of the dominant marker with the medium erucic content was studied in 70 F2 plants of the cross of Y496 (E2E2) × Y517 (E1E1). The dominant marker was present in all the F2 plants with erucic acid content (range: 20.0 to 29.8%) similar to that of the medium erucic acid parent Y496 (21.7 to 27.1%), but absent in the F2 plants with high erucic content (range: 44.4 to 49.2%) similar to that of Y517 (50.3 to 55.5%) (Figure 4B), implying that the reduction of erucic content in Y496 was due to the Sal-T1 insertion in the 5′ UTR of E2.
DISCUSSION
In this study, we investigated the molecular and epigenetic mechanisms leading to the occurrence of four different alleles of the FAE1 in yellow mustard. The FAE1 allele E1 conditioning high erucic content had a coding sequence of 1521 bp, which was in good agreement with lengths of the FAE1s of B. napus and Arabidopsis (James et al., 1995; Barret et al., 1998; Fourmann et al., 1998). The predicted polypeptide of 507 amino acids encoded by the FAE1 allele E1 also showed high similarity with the proteins encoded by the FAE1s in B. juncea (97% identity), B. napus (96% identity), B. rapa (96% identity), B. oleracea (96% identity), and Arabidopsis (86% identity) (Supplemental Figure 1). Four conserved histidine residues in the polypeptide (Supplemental Figure 1) were demonstrated to be essential for the function of the FAE1 in Arabidopsis (Ghanevati and Jaworski, 2001), while a conserved serine residue at position 282 (Supplemental Figure 1) was shown to be essential for the FAE1 functionality in B. napus (Han et al., 2001; Katavic et al., 2002). The promoter of E1 was 703 bp and proved to be seed specific and functional. It had high similarity to the promoter of the wild-type FAE1 in Brassica species (Han et al., 2001). Yellow mustard is phylogenetically related to Brassica and Arabidopsis species (Warwick and Black, 1991), and the high level of similarity between the FAE1 genes of these species likely reflects their common evolutionary origin.
This report reveals that transposable element insertion and epigenetic modification are involved in modulating the expression of FAE1 in the erucic acid–containing species of Brassicaceae. In B. napus, SNP/indel mutations in the coding region of the FAE1 resulted in loss-of-function alleles (Han et al., 2001; Roscoe et al., 2001; Katavic et al., 2002; Wu et al., 2007, 2008). In contrast, the loss-of-function FAE1 allele e in yellow mustard was caused by an insertional disruption in its coding region by Sal-PIF, a PIF/Harbinger-like DNA transposon. The FAE1 allele E2, controlling the medium erucic acid trait, originated from the insertion of a Copia-like retrotransposon Sal-T1 in its 5′ UTR. The low erucic allele E3 was identical to E2 in nucleotide sequence but was shown to be an epiallele due to DNA methylation in its promoter region. The different molecular mechanisms underpinning the allelic variation of FAE1 in yellow mustard and Brassica species suggested that the mutations of FAE1 occurred after the divergence of the species.
Insertions of DNA transposons or retrotransposons within or near genes have been reported to negatively affect the expression of genes by decreasing or abolishing transcription (Kumar and Bennetzen, 1999; Hollister and Gaut, 2009; Ahmed et al., 2011; Eichten et al., 2012, 2013). For example, the null wx-ml allele originated from the insertion of the 409-bp DNA transposon Dissociation 1 in the 9th exon of the waxy (wx) gene in maize (Wessler, 1991). The gypsy-like retrotransposon Gret1 inserted upstream of the mybA1 coding sequence blocked gene expression, causing the loss of pigmentation in white cultivars of grape (Vitis vinifera) (Kobayashi et al., 2004; Fournier-Level et al., 2010). Here, our data further confirmed that mutations induced by TE insertions can have negative effects on gene expression. The insertion of Sal-PIF in the coding region of the FAE1 caused the loss-of-function FAE1 allele e. The insertion of the Copia-like retrotransposon Sal-T1 in the 5′ UTR of E2 and E3 resulted in reduced transcription levels. There are three possible explanations for this: First, the promoters of E2 and E3 are farther from the CDS due to the insertion of Sal-T1, which might result in a reduction in transcription efficiency compared with E1, where the promoter is adjacent to the coding region. Second, the inserted Sal-T1 might disturb the interaction of the promoter and other transcriptional factors, thereby affecting FAE1 expression. Third, the transcription direction of Sal-T1 was opposite to that of E2 and E3. Therefore, transcripts of E2 and E3 might form sense-antisense pairs by pairing with the transcript of Sal-T1 on the opposite strand, causing a modulation of gene expression.
Jacobsen and Meyerowitz (1997) reported that the clk epialleles were extensively methylated, covering the start of transcription and most of the transcribed region in Arabidopsis. In tomato (Solanum lycopersicum), the colorless nonripening (Cnr) epimutant exhibited high levels of cytosine methylations in the SPL-CNR promoter region (Manning et al., 2006). However, our study revealed that the expression levels of the FAE1 epiallele E3 were associated with the presence/absence of only three methylated cytosines in the promoter region. This could be due to that the methylated cytosines are critical for the FAE1 promoter activity. Smale and Kadonaga (2003) reported that transcription initiation of genes generally relies on a simple core promoter sequence that can extend ∼35 bp upstream and/or downstream of the transcription initiation site in eukaryotes. The four nucleotides “ACGT” in the promoter can act as the transcription factor recognition site (Jakoby et al., 2002). In the promoter of FAE1 allele E3, the four nucleotides “ACGT” were positioned from −5145 to −5148 and the cytosine at position −5146 was methylated. In addition, methylation was observed at position -4943C, which is very close to the predicted transcription start site (nucleotide −4935) in the promoter of Eepi3 (Figure 1A) (http://www.fruitfly.org/seq_tools/promoter.html). Methylation at these critical positions in the promoter of Eepi3 could have a negative effect on binding of the transcription preinitiation complex and therefore result in reduction of transcription efficiency and lowered erucic acid content. The revertants of Y1130 had increased erucic acid content due to demethylation at positions −5146 and −5095, confirming that the methylation level in the promoter of Eepi3 is negatively correlated with the erucic content in the seed. Interestingly, the methylated cytosine at position −4943 was close to the retrotransposable element Sal-T1 (nucleotide −4899 to −37), and this position was not demethylated either by 5-azaC treatment or spontaneously. It has been reported that DNA methylation on and near transposons is highly stable, whereas it is less stable within genes at a distance from transposons (Weigel and Colot, 2012). The genetic and molecular mechanisms underlying the methylation status of E3 remains to be studied.
Our previous genetic studies indicated that E3 was heritable and acted in an additive manner with E2, but fit to a partial dominance model with E1 in yellow mustard (Javidfar and Cheng, 2013). The present molecular analysis revealed that E3 is an epiallele differing from E2 due to DNA methylation in its promoter region. Heritable epialleles have been reported in many plant species such as Arabidopsis (Jacobsen and Meyerowitz, 1997), tomato (Messeguer et al., 1991), and toadflax (Cubas et al., 1999). Recently, genome-wide DNA methylation analyses have further confirmed the occurrence of epigenome-wide inheritance of methylation variants in soybean (Glycine max; Schmitz et al., 2013) and maize (Regulski et al., 2013). However, it is well documented that epialleles are not stable and occasionally revert phenotypically. The clk-3 mutant spontaneously reverted to wild-type or nearly wild-type phenotype at a rate of 2.9% in Arabidopsis (Jacobsen and Meyerowitz, 1997). The Cnr mutant is very stable with only 0.1% reversion rate in tomato (Manning et al., 2006). The Epi-df causes a dwarf stature and various floral defects in rice (Oryza sativa) (Zhang et al., 2012). The reversion of Epi-df occurred in the df progeny at frequencies of 1.97, 0.31, and 0.20%, respectively, in different growing seasons (Zhang et al., 2012). In our study, the epiallele Eepi3 partially reverted with a frequency of 1.0%. The partial revertant Y1130-16 contains only one methylated cytosine and produced seeds with an average of 12.3% erucic acid, suggesting that this partial revertant is a new heritable epimutant. This indicates that DNA methylation can generate multiple epialleles with various expression levels, thereby leading to continuous quantitative variation of a trait. Epigenetic modifications may have played a significant role in quantitative modulation of trait variation.
The epigenetic modification observed here and the mutations induced by the TEs Sal-PIF and Sal-T1 revealed in this study had deleterious effects on the function of FAE1 in yellow mustard. However, such mutants have been maintained in the populations during the speciation and evolution of yellow mustard. Likely, erucic acid content is a neutral trait for plant viability and is not subject to elimination by natural selection. Furthermore, yellow mustard is an obligate outcrossing species in which high levels of heterozygosity could help maintain not only neutral but even slightly deleterious mutations in the species.
To conclude, we have shown that TE insertion and epigenetic modification cause the multiallelic variation in FAE1 in yellow mustard. Our results have demonstrated a comprehensive molecular framework for the interplay of TE insertion, SNP/indel mutation, and epigenetic variation accounting for the broad range of naturally occurring genetic variation in plants.
METHODS
Plant Materials
Four breeding lines Y517, Y496, Y1130, and Y514 with high (average: 52.9%), medium (average: 24.4%), low (average: 1.4%), and zero (average: 0.1%) erucic acid contents, respectively, were used in this study. The erucic genotypes of Y517, Y496, Y1130, and Y514 are E1E1, E2E2, E3E3, and ee (Javidfar and Cheng, 2013). The F2 populations of Y514 (ee) × Y517 (E1E1) and Y496 (E2E2) × Y517 (E1E1) developed by Javidfar and Cheng (2013) were used to study the cosegregation of the allele specific markers for the FAE1 alleles e and E2 with the zero and medium erucic acid contents, respectively, in this study.
DNA Extraction
Genomic DNA was extracted from young expanding leaves of the four parental lines Y517, Y496, Y1130, and Y514, F1 and F2 plants using a modified sodium dodecyl sulfate method (Somers et al., 1998).
Cloning the Coding Regions of the FAE1 Alleles E1, E2, E3, and e
Primers were designed using the software Primer3 (http://redb.croplab.org/modules/redbtools/primer3.php). Primer pair No 1 (Supplemental Table 1 and Supplemental Figure 2) was designed to clone the coding regions of E1, E2, E3, and e based on the FAE1 sequences in Brassica and the putative FAE1 sequence of yellow mustard. Primer pair No 2 (Supplemental Table 1 and Supplemental Figure 2C) was designed based on the sequence of the 3′ coding region of allele E1 to isolate the partial coding region of allele e. Primer pair No 3 (Supplemental Table 1 and Supplemental Figure 2C) was designed based on the sequence of the partial coding region of e and used to clone the remaining coding sequence of e. Genomic DNA of each of the four parental lines was used as template for PCR amplification with fideliTaq DNA polymerase (Affymetrix) in a thermocycler with 30 cycles of the following program: 94°C for 30 s, 56°C for 30 s, and 68°C for 2 min.
Cloning the 3′ and 5′ Flanking Sequences of the Coding Regions of the FAE1 Alleles E1, E2, E3, and e
The primer pairs used to clone the 3′ and 5′ flanking sequences of the coding regions of the FAE1 alleles E1, E2, E3, and e are presented in Supplemental Table 1 and Supplemental Figure 2. Primer pair No 4 was designed based on the sequence of the 3′ coding region and was used to clone the 3′ downstream sequence of each of the four alleles. Primer pair No 5, designed based on the 5′ coding region, was used to clone the 5′ upstream sequences of E1, E2, and E3. Primer pairs No 6 to 9 (Supplemental Figure 2B) were designed based on the 5′ upstream sequence obtained from each previous round of PCR walking to clone the further upstream sequences of E2 and E3. Primer pair No 10, designed based on the promoter sequence of allele E1, was used to clone the promoter region of E2, E3, and e. Primer pair No 11 was designed to clone the DNA fragment between the promoter and 5′ flanking sequence of E2 and E3 (Supplemental Figure 2 B ). Primer pair No 12 was designed based on the promoter sequences of E2 and E3 and was used to clone upstream sequences of these promoters. Primer pair No 13 was used to clone the DNA fragment between the promoter and 5′ flanking sequence of e (Supplemental Figure 2C). PCR walking was performed to clone the 5′ and 3′ flanking sequences of the coding region of E1, E2, E3, and e according to the protocol of Siebert et al. (1995). The standard protocol from the Clontech kit (Protocol PT 3042, version PR 03300) by Gwyneth Ingram and Karine Coenen was followed to facilitate the PCR walking. To validate the entire sequence including the coding region, 5′ and 3′ flanking sequences of each of the four alleles E1, E2, E3, and e obtained by PCR walking, primer pair No 14 (WF/WR) (Supplemental Table 1; Figure 1A) was designed to amplify each of the four alleles in their entirety. Genomic DNA of each of the four lines Y517, Y496, Y1130, and Y514 were used as templates with Phusion high-fidelity polymerase (NEB) with 30 cycles of the following program: 98°C for 10 s, 57°C for 30 s, and 72°C for 4 min.
Transformation of Yeast and Fatty Acid Analysis
The coding sequences of E1and E2 were amplified using primer pair No 15 (Supplemental Table 1) and cloned into the pYES2.1/V5-His-TOPO expression vector (Invitrogen), then sequenced to confirm the correct orientation of genes. To determine whether the cloned FAE1 alleles E1 and E2 from lines Y517 and Y496 could function as condensing enzymes and were capable of elongation of C18:1 to C20:1 and C22:1, the two constructs pYES2.1/V5-His-TOPO-E1 and pYES2.1/V5-His-TOPO-E2 were transformed into Saccharomyces cerevisiae strain Inv Sc1 (Invitrogen) by the lithium acetate method (Ausubel et al., 1995). Yeast cells transformed with the empty vector pYES2.1/V5-His-TOPO plasmid were used as a control. Transgenic cells were screened in complete minimal dropout uracil medium containing 2% raffinose as a carbon source at 30°C. The positive clones were grown at 30°C overnight in minimal media supplemented with 2% raffinose and lacking uracil. The transgenic cell culture was centrifuged, followed by washing, and then used to inoculate 20 mL induction media (minimal media lacking uracil and supplemented with 2% galactose and 1% raffinose) to an OD600 of 0.5. Cultures were grown overnight. For fatty acid analysis, the resulting cell cultures were pelleted by centrifugation, washed once with the induction media and once with water, and then freeze dried. The dried pellet was transferred to a glass insert. After adding 100 μL hexane and 100 μL of 4% sodium methoxide in methanol, the samples were grinded with a glass rod and then incubated at room temperature for 15 min. Then, 15 μL of 0.2 M sodium phosphate buffer was added to the solution followed by blowing with air stream for 2 min. About 200 μL of heptane was added to the insert and the resulting fatty acid methyl esters were analyzed for fatty acid composition on a Hewlett Packard 6890 gas chromatograph.
Transformation of Arabidopsis and Histochemical GUS Assays
The 5′ flanking fragments from the translation start codon for E1 and E2 were cloned using primer pairs No 16 and 17 (Supplemental Table 1) and were inserted into the plant expression vector pBI101 upstream of the GUS gene. The resulting constructs pBI101-ProE1-GUS with the 5′ flanking fragment of E1 and pBI101-ProE2-GUS with the 5′ flanking fragment of E2 were transformed into electrocompetent Agrobacterium tumefaciens cells GV3101 by electroporation according to the manufacturer’s instructions. Transformation of wild-type Arabidopsis thaliana plants was performed using the Agrobacterium-mediated transformation method described by Clough and Bent (1998). Transgenic plants were screened and analyzed according to Jako et al. (2001). Embryos of 9-d-old, leaf, stem, bud, and flower tissues of the transgenic plants were stained overnight in solution containing 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 50 mM Na phosphate buffer, pH 7.0, 1 mM K3Fe(CN)6, 1 mM K4Fe(CN)6, and 20 mM EDTA. The staining of the embryo and vegetative tissues was observed and photographed using a Zeiss stereomicroscope with a color CCD camera.
RT-PCR and Quantitative Real-Time PCR
Total RNA was extracted from leaf, stem, flower bud, flower, and embryos of Y517, Y496, Y1130, and Y514 using an RNeasy plant mini kit (Qiagen). Total RNA was extracted from only 21-d-old embryos of the 5-azaC–induced and spontaneous revertants of Y1130. Extracted RNA was treated with DNase I (Ambion), and cDNA was synthesized using the ReverTra Ace-a-First-Strand cDNA synthesis kit (Fermentas) according to the manufacturer’s instruction. Primer pair No 18 (Supplemental Table 1) was designed based on the coding sequences (position 1 to 928) of the FAE1 alleles E1, E2, and E3. The expression of the FAE1 alleles was then analyzed by RT-PCR using primer pair No 18 with 35 amplification cycles.
To detect differences in transcription levels among FAE1 alleles E1, E2, and E3, real-time qPCR analysis was conducted on embryos harvested at 14, 21, 28, and 35 d after pollination from plants of Y517, Y496, and Y1130. The 21-d-old embryos of 5-azaC–induced revertants and spontaneous revertant of Y1130 were also used for qPCR analysis to determine the expression level of FAE1 allele E3. Primer pair No 19 (Supplemental Table 1) specific for FAE1 was used to amplify ∼100-bp fragments (nucleotides 865 to 964) from the cDNA of 14- to 35-d-old embryos in Y517, Y496, and Y1130, the cDNA of 21-d-old embryos of the 5-azaC–induced revertants and the spontaneous revertant of Y1130. The qPCR analysis was performed with SsoFast EvaGreen supermix (Bio-Rad) according to the manufacturer’s instructions using a Bio-Rad CFX96 system. Primer pair No 20 (Supplemental Table 1) specific for Actin2 (FG576123) was used as an internal control for normalization. Three separate first-strand cDNA reactions were analyzed in duplicate for each sample, and expression levels were calculated as described by Livak and Schmittgen (2001).
Sequence Analysis of the Inserted DNA Fragments in the 5′ UTR of FAE1 Alleles E2 and E3 and in the Coding Region of FAE1 Allele e
The open reading frames and deduced amino acid sequences of the inserted fragments in the 5′ UTR of FAE1 alleles E2 and E3 and in the coding region of FAE1 allele e were predicted using the GenScan Web server at MIT (http://genes.mit.edu/GENSCAN.html). The conserved domains of the deduced amino acid sequences were identified using RPS-BLAST (reverse position-specific BLAST) (http://www.ncbi.nlm.nih.gov/Structure/cdd/docs/cdd_search.html). The PBS of the inserted fragment in the 5′ UTR of E2 and E3 was identified by aligning the sequence of the 5′ LTR to the 3′ region of tRNAs from a tRNA database (http://lowelab.ucsc.edu/GtRNAdb/). The PPT was determined by examining the purine-rich sequence in the upstream region of the 3′ LTR. The amino acid sequence alignment of maize PIF and the inserted fragment in the coding region of FAE1 allele e was performed by the ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Bisulfite Sequencing
Accurate analysis of DNA methylation by bisulfite sequencing depends on the complete conversion of all cytosines into uracil. Wang et al. (2011) developed a PCR-based assay for the mitochondrial gene ATP1 (ATPase SUBUNIT 1) and demonstrated its efficacy as a bisulfite conversion control in Brassica rapa and other plant taxa. The two primer pairs for the ATP1 provide a reliable control across a representative range of dicot and monocot angiosperm species. Therefore, the ATP1 of yellow mustard was used as a negative control to check if the bisulfite chemical reaction was complete. The ATP1 (227 bp) was amplified using primer pair No 21 (Supplemental Table 1; Wang et al., 2011). In the wild-type Arabidopsis Columbia-0, AtSN1 (A. thaliana SINE like element 1), a putative transposable retroelement located on chromosome 3, contains 18 methylated cytosines in the internal region between the long terminal repeats (Kuhlmann and Mette, 2012). AtSN1, with 18 known methylated cytosines, was used as a positive control in our bisulfite sequencing experiment. The AtSN1 (nucleotides 15,805,617 to 15,805,773, NCBI nucleotide sequence NC003074) was amplified using primer pair No 33 (Supplemental Table 1). Bisulfite sequence alignment of the AtSN1 (positive control) is shown in Supplemental Figure 9.
Bisulfite sequencing was performed on genomic DNA from three plants of both Y517 (E1E1) and Y496 (E2E2), four plants of Y1130 (E3E3), 5-azaC–induced revertants, and spontaneous revertant of Y1130. Genomic DNA from the embryos of each plant was bisulfite treated using the EpiTect Bisulfite kit (Qiagen). The candidate methylated coding and promoter regions of the FAE1 alleles were amplified using bisulfite primers No 22-27 and No 28-30, respectively, designed using MethPrimer software, and the resulting fragments were cloned into the pGEM-T vector (Promega). Primer pairs No 22-27 (Supplemental Table 1) were used to amplify the coding region (nucleotides 1 to 1515) of E1, E2, and E3. Primer pair No 28 (Supplemental Table 1) was designed to amplify the promoter sequence and partial coding region (nucleotides −143 to 56) of E1. Primer pair No 29 (Supplemental Table 1) was designed to amplify the promoter sequence (nucleotides −5042 to −4900) and retrotransposable element Sal-T1 (nucleotide −4899 to −4843) of E2 and E3. Primer pair No 30 (Supplemental Table 1) was designed based on the promoter sequence of E1 and was used to detect the methylation status of the promoter region (nucleotides −335 to −79) of E1 and the promoter regions (nucleotides −5234 to −4978) of E2 and E3. At least 10 independent clones of each fragment generated by each primer pair were sequenced to determine the number and position of methylated residues for each plant. Sequence analysis was performed according to the Web-based tool Kismeth (Gruntman et al., 2008).
5-Azacytidine Treatment
Forty-five seeds of the low erucic acid line Y1130 were treated with 1 mM 5-azaC (Sigma-Aldrich), a demethylation agent. A stock solution of 15 mM 5-azaC was prepared in 50 mM MES buffer (PH 6.3). The seeds were placed evenly on a filter paper in a Petri dish to which 4.5 mL 1 mM 5-azaC solution was added. The Petri dish was then incubated in the dark at room temperature for 3 d. The 5-azaC–treated seeds were transferred to 50-mL tubes, rinsed with water for eight times, and then transferred to soil for germination and growth in the greenhouse. The resulting 5-azaC–treated plants were bagged and bud-pollinated to produce self-pollinated seeds, since yellow mustard is an obligatory out-crossing species with a self-incompatible reproduction system. The self-pollinated 21-d-old embryos of each plant were sampled for DNA methylation and real-time qPCR analysis. The self-pollinated seeds of each plant were used for fatty acid analysis.
Cosegregation of the FAE1 Allele-Specific Markers with Erucic Acid Levels
Primer pair No 31 (Supplemental Table 1) was designed based on the sequences of the promoter and the transposon Sal-PIF inserted in the coding region of FAE1 allele e. Primer pair No 32 (Supplemental Table 1) was designed based on the promoter sequence and the sequence of the retrotransposon Sal-T1 in the 5′ UTR of FAE1 alleles E2 and E3. The cosegregation of the marker specific for the allele e and zero erucic content was studied in 96 F2 plants derived from the cross Y514 (ee) × Y517 (E1E1), while the cosegregation of the marker specific for the allele E2 with medium erucic content was studied in 70 F2 plants of the cross Y496 (E2E2) × Y517 (E1E1). Each PCR (20 µL) contained 1× PCR standard buffer (NEB), 200 µM each deoxynucleotide triphosphate, 0.1 µM of each forward and reverse primer, 1 unit of Taq polymerase (NEB), and 50 ng of genomic DNA. PCR was performed with an initial denaturation at 94°C for 3 min followed by 30 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, followed by a final extension cycle of 72°C for 7 min.
Fatty Acid Analysis
The seed erucic acid contents of lines Y517, Y496, Y1130, the 5-aza-C–induced revertants, and spontaneous revertant of Y1130 were determined using the half-seed technique (Downey and Harvey, 1963) and the gas chromatographic method of Thies (1971).
Accession Numbers
The GenBank accession numbers for the nucleotide sequences of the FAE1 allele E1, E2(E3), and e in yellow mustard are KJ817378, KJ817379, and KJ817380, respectively. The nucleotide sequence of Sal-PIF, a 3100-bp PIF/Harbinger-like DNA transposon, is KJ817381. The nucleotide sequence of Sal-T1, a 4863-bp Copia-like retrotransposon, is KJ817382. The GenBank accession number of the putative FAE1 sequence of yellow mustard is AY888040. The GenBank accession number of the FAE1 promoter region from the high erucic acid B. napus line Askari is AF275254. The GenBank accession number of the putative retroelement pol polyprotein is AAC67205. The GenBank accession numbers of the FAE1 genes in B. juncea, B. napus, B. rapa, B. oleracea, and Arabidopsis are AJ558198, AF490459, AF49041, AF490460, and Q38860, respectively. The GenBank accession number of the amino acid sequence maize PIF is AF412282.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Amino Acid Sequences Encoded by the FAE1 Alleles E1, E2, and E3 of Yellow Mustard, CAD90159 of B. juncea, AF490461 of B. rapa, AF490460 of B. oleracea, ACB55612 and AF274750 of B. napus, and NP_195178 of Arabidopsis.
Supplemental Figure 2. Schematic Diagram Showing the Position of Each Primer Used for Cloning of the FAE1 Alleles E1, E2, E3, and e.
Supplemental Figure 3. Expression Pattern of the FAE1 Alleles E1, E2, and E3.
Supplemental Figure 4. DNA Sequence of the Retrotransposable Element Sal-T1 Inserted in the 5′ UTR of the FAE1 Alleles E2 and E3.
Supplemental Figure 5. The 959 Amino Acids of the Deduced Protein of the Retrotransposable Element Sal-T1 Predicted Using GenScan at GeniusNet (http://genome.dkfz-heidelberg.de/cgi-in/GENSCAN/genscan.cgi).
Supplemental Figure 6. The Sequence of the DNA Transposon Sal-PIF Inserted in the Coding Region of the FAE1 Allele e.
Supplemental Figure 7. Deduced Amino Acid Sequences of the DNA Transposon Sal-PIF Predicted Using GenScan at GeniusNet (http://genome.dkfz-heidelberg.de/cgi-in/GENSCAN/genscan.cgi).
Supplemental Figure 8. PCR Amplification of the Entire FAE1 Alleles E1, E2, E3, and e Using the Primer Pair WF/WR No 14 (Supplemental Table 1).
Supplemental Figure 9. DNA Methylation in the Internal Region between the Long Terminal Repeats of AtSN1 (Positive Control).
Supplemental Figure 10. DNA Methylation in the Coding Regions of the FAE1 Alleles E1, E2, and E3.
Supplemental Figure 11. Bisulfite Sequence Alignment of ATP1 (Control Gene) and the FAE1 Allele E1 in Line Y517.
Supplemental Figure 12. Bisulfite Sequence Alignment of ATP1 (Control Gene) and the FAE1 Allele E2 in Line Y496.
Supplemental Figure 13. Bisulfite Sequence Alignment of ATP1 (Control Gene) and the FAE1 Allele E3 in Line Y1130.
Supplemental Figure 14. Bisulfite Sequence Alignment of ATP1 (Control Gene) and the FAE1 Allele E3 in the 5-azaC Induced Revertants of Y1130.
Supplemental Figure 15. Bisulfite Sequence Alignment of ATP1 (Control Gene) and the FAE1 Allele E3 in the Spontaneous Revertant Y1130-16.
Supplemental Table 1. Primer Pairs Used in This Study.
Supplementary Material
Acknowledgments
Breeding and research programs for the condiment yellow mustard are supported by the Growing Forward II-Agri-Innovation Program, Mustard 21 Canada and the Agriculture Development Fund of Saskatchewan, Canada. We thank Patricia Vrinten (National Research Council, Canada) and the two anonymous reviewers for their constructive comments on the article and Markus Kuhlmann (Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany) for his advice regarding bisulfite protocol and positive control. The vector pBI101 was kindly provided by Kevin Rozwadowski at Agriculture and Agri-Food Canada-Saskatoon Research Centre.
AUTHOR CONTRIBUTIONS
B.C. conceived the project. F.Z. and B.C. designed the research. F.Z. performed the research and analyzed data. F.Z. and B.C. wrote the article.
Glossary
- SNP
single-nucleotide polymorphism
- TE
transposable element
- CDS
coding DNA sequence
- qPCR
quantitative PCR
- UTR
untranslated region
- PBS
primer binding site
- PPT
polypurine tract
- LTR
long terminal repeat
- 5-azaC
5-azacytidine
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ahmed I., Sarazin A., Bowler C., Colot V., Quesneville H. (2011). Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 39: 6919–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K., Albright, L.M., Coen, D.M., and Varki, A. (1995). Current Protocols in Molecular Biology. (New York: John Wiley). [Google Scholar]
- Barret P., Delourme R., Renard M., Domergue F., Lessire R., Delseny M., Roscoe T.J. (1998). A rapeseed FAE1 gene is linked to the E1 locus associated with variation in the content of erucic acid. Theor. Appl. Genet. 96: 177–186. [Google Scholar]
- Becker C., Weigel D. (2012). Epigenetic variation: origin and transgenerational inheritance. Curr. Opin. Plant Biol. 15: 562–567. [DOI] [PubMed] [Google Scholar]
- Butelli E., Licciardello C., Zhang Y., Liu J., Mackay S., Bailey P., Reforgiato-Recupero G., Martin C. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24: 1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P., Stephan W. (1994). The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- Cheng B.F., Williams D.J., Zhang Y. (2012). Genetic variation in morphology, seed quality and self-(in)compatibility among the inbred lines developed from a population variety in outcrossing yellow mustard (Sinapis alba). Plants 1: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cubas P., Vincent C., Coen E. (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161. [DOI] [PubMed] [Google Scholar]
- Das S., Roscoe T.J., Delseny M., Srivastava P.S., Lakshmikumaran M. (2002). Cloning and molecular characterization of the Fatty Acid Elongase 1 (FAE1) gene from high and low erucic acid lines of Brassica campestris and Brassica oleracea. Plant Sci. 162: 245–250. [Google Scholar]
- Downey R.K., Harvey B.L. (1963). Methods of breeding for oil quality in rape. Can. J. Plant Sci. 43: 271–275. [Google Scholar]
- Eichten S.R., Ellis N.A., Makarevitch I., Yeh C.T., Gent J.I., Guo L., McGinnis K.M., Zhang X., Schnable P.S., Vaughn M.W., Dawe R.K., Springer N.M. (2012). Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 8: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., et al. (2013). Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmann M., Barret P., Renard M., Pelletier G., Delourme R., Brunel D. (1998). The two genes homologous to Arabidopsis FAE1 co-segregate with the two loci governing erucic acid content in Brassica napus. Theor. Appl. Genet. 96: 852–858. [Google Scholar]
- Fournier-Level A., Lacombe T., Le Cunff L., Boursiquot J.M., This P. (2010). Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.). Heredity (Edinb) 104: 351–362. [DOI] [PubMed] [Google Scholar]
- Ghanevati M., Jaworski J.G. (2001). Active-site residues of a plant membrane-bound fatty acid elongase β-ketoacyl-CoA synthase, FAE1 KCS. Biochim. Biophys. Acta 1530: 77–85. [DOI] [PubMed] [Google Scholar]
- Greene B., Walko R., Hake S. (1994). Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138: 1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E., Qi Y., Slotkin R.K., Roeder T., Martienssen R.A., Sachidanandam R. (2008). Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lühs W., Sonntag K., Zähringer U., Borchardt D.S., Wolter F.P., Heinz E., Frentzen M. (2001). Functional characterization of β-ketoacyl-CoA synthase genes from Brassica napus L. Plant Mol. Biol. 46: 229–239. [DOI] [PubMed] [Google Scholar]
- Hollister J.D., Gaut B.S. (2009). Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19: 1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S.E., Meyerowitz E.M. (1997). Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277: 1100–1103. [DOI] [PubMed] [Google Scholar]
- Jako C., Kumar A., Wei Y., Zou J., Barton D.L., Giblin E.M., Covello P.S., Taylor D.C. (2001). Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 126: 861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F., bZIP Research Group (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7: 106–111. [DOI] [PubMed] [Google Scholar]
- James D.W., Jr, Lim E., Keller J., Plooy I., Ralston E., Dooner H.K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javidfar F., Cheng B.F. (2013). Single locus, multiallelic inheritance of erucic acid content and linkage mapping of FAE1 gene in yellow mustard (Sinapis alba L.). Crop Sci. 53: 825–832. [Google Scholar]
- Kalisz S., Purugganan M.D. (2004). Epialleles via DNA methylation: consequences for plant evolution. Trends Ecol. Evol. (Amst.) 19: 309–314. [DOI] [PubMed] [Google Scholar]
- Katavic V., Mietkiewska E., Barton D.L., Giblin E.M., Reed D.W., Taylor D.C. (2002). Restoring enzyme activity in nonfunctional low erucic acid Brassica napus fatty acid elongase 1 by a single amino acid substitution. Eur. J. Biochem. 269: 5625–5631. [DOI] [PubMed] [Google Scholar]
- Kidwell M.G., Lisch D. (1997). Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA 94: 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Goto-Yamamoto N., Hirochika H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Kuhlmann M., Mette M.F. (2012). Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol. Biol. 79: 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Bennetzen J.L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Lassner M.W., Lardizabal K., Metz J.G. (1996). A jojoba β-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell 8: 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Leonard C. (1994). Sources and commercial applications of high-erucic vegetable oils. Lipid Technol. 4: 79–83. [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952. [DOI] [PubMed] [Google Scholar]
- Messeguer R., Ganal M.W., Steffens J.C., Tanksley S.D. (1991). Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol. Biol. 16: 753–770. [DOI] [PubMed] [Google Scholar]
- Meyers B.C., Tingey S.V., Morgante M. (2001). Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11: 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney, J.P., Rakow, G.F.W., and Olson, T.V. (1999). Selection for high oleic acid in “zero” erucic acid Sinapis alba (available online at www.regional.org.au/au/gcirc/4/78.htm). In Proceedings of the 10th International Rapeseed Congress, Canberra, Australia, N. Wratten and P.A. Salisbury, eds (Gosford, Australia: Regional Australia Institute). [Google Scholar]
- Regulski M., et al. (2013). The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E.J. (2006). Inherited epigenetic variation—revisiting soft inheritance. Nat. Rev. Genet. 7: 395–401. [DOI] [PubMed] [Google Scholar]
- Richards E.J. (2011). Natural epigenetic variation in plant species: a view from the field. Curr. Opin. Plant Biol. 14: 204–209. [DOI] [PubMed] [Google Scholar]
- Roscoe T.J., Lessire R., Puyaubert J., Renard M., Delseny M. (2001). Mutations in the fatty acid elongation 1 gene are associated with a loss of β-ketoacyl-CoA synthase activity in low erucic acid rapeseed. FEBS Lett. 492: 107–111. [DOI] [PubMed] [Google Scholar]
- Sauer, F.D., and Kramer, J.K.G. (1983). The problem associated with the feeding of high erucic acid rapeseed oils and some fish oils to experimental animals. In High and Low Erucic Acid Rapeseed Oils, Production, Usage, Chemistry and Toxicological Evaluation. J.K.G. Kramer, F.D. Sauer, and W.J. Pigden, eds (New York: Academic Press), pp. 253–292. [Google Scholar]
- Schmitz R.J., He Y., Valdés-López O., Khan S.M., Joshi T., Urich M.A., Nery J.R., Diers B., Xu D., Stacey G., Ecker J.R. (2013). Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 23: 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert P.D., Chenchik A., Kellogg D.E., Lukyanov K.A., Lukyanov S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23: 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S.T., Kadonaga J.T. (2003). The RNA polymerase II core promoter. Annu. Rev. Biochem. 72: 449–479. [DOI] [PubMed] [Google Scholar]
- Somers D.J., Friesen K.R.D., Rakow G. (1998). Identification of molecular markers associated with linoleic acid desaturation in Brassica napus. Theor. Appl. Genet. 96: 897–903. [Google Scholar]
- Sonntag N.O.V. (1991). Erucic, behenic: feedstocks of the 21st century. Inform 2: 449–463. [Google Scholar]
- Sonntag, N.O.V. (1995). Industrial utilization of long-chain fatty acids and their derivatives. In Brassica oilseeds. D.S. Kimber and D.I. Mcgregor, eds (Oxon, UK: CAB International), pp. 339–352. [Google Scholar]
- Stokes T.L., Kunkel B.N., Richards E.J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf, P.K., and Pollard, M.R. (1983). Pathways of fatty acid biosynthesis in higher plants with particular reference to developing rapeseed. In High and Low Erucic Acid Rapeseed Oils, J.K. Kramer, F. Sauer, and W.J. Pigden, eds (New York: Academic Press), pp. 131–141. [Google Scholar]
- Thies W. (1971). Schnelle und einfache Analysen der Fettsäurezusammensetzung in einzelnen Raps-Kotyledonen. 1. Gaschromatographische und papierchromatographische Methoden. Z. Pflanzenzüchtg. 65: 181–202. [Google Scholar]
- Vicient C.M., Suoniemi A., Anamthawat-Jónsson K., Tanskanen J., Beharav A., Nevo E., Schulman A.H. (1999). Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11: 1769–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang C., Long Y., Hopkins C., Kurup S., Liu K., King G.J., Meng J. (2011). Universal endogenous gene controls for bisulphite conversion in analysis of plant DNA methylation. Plant Methods 7: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick S.I., Black L.D. (1991). Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae) -chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82: 81–92. [DOI] [PubMed] [Google Scholar]
- Weigel D., Colot V. (2012). Epialleles in plant evolution. Genome Biol. 13: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler S.R. (1991). The maize transposable Ds1 element is alternatively spliced from exon sequences. Mol. Cell. Biol. 11: 6192–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Wu Y., Xiao L., Li X., Lu C. (2008). Zero erucic acid trait of rapeseed (Brassica napus L.) results from a deletion of four base pairs in the fatty acid elongase 1 gene. Theor. Appl. Genet. 116: 491–499. [DOI] [PubMed] [Google Scholar]
- Wu Y., Xiao L., Wu G., Lu C. (2007). Cloning of fatty acid elongase1 gene and molecular identification of A and C genome in Brassica species. Sci. China C Life Sci. 50: 343–349. [DOI] [PubMed] [Google Scholar]
- Zhang L., et al. (2012). Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24: 4407–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Feschotte C., Zhang Q., Jiang N., Eggleston W.B., Wessler S.R. (2001). P instability factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proc. Natl. Acad. Sci. USA 98: 12572–12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.