Abstract
Cryptosporidium is one of the most important parasites in poultry, and this pathogen can infect more than 30 avian species. The present study investigated the infection rate of Cryptosporidium among broiler chicken flocks. A total of 385 fecal samples from broiler chickens in 7 regions of Zhejiang Province collected from November 2010 to January 2012 were examined by microscopy. Thirty-eight (10%) samples were positive for Cryptosporidium infection, and 3 genotypes (Cryptosporidium baileyi, Cryptosporidium meleagridis, and avian genotype II) were identified by PCR and sequencing. A phylogenetic tree of the isolates was analyzed. These results suggest that cryptosporidiosis is widespread in poultry in Zhejiang Province, and is a potential threat to public health as well as the economy. This is the first report about the infection rate and molecular characterization of Cryptosporidium in broiler chickens in Zhejiang.
Keywords: Cryptosporidium, Infection rate, China, Chicken, Epidemiology
Abstract
Cryptosporidium est l’un des parasites les plus importants de la volaille, et cet agent pathogène peut infecter plus de 30 espèces aviaires. La présente étude a examiné le taux d’infection de Cryptosporidium chez les poulets de chair. Un total de 385 échantillons fécaux de poulets de chair dans 7 régions de la province du Zhejiang, recueillis de novembre 2010 à janvier 2012, ont été examinés par microscopie. Trente-huit (10 %) étaient positifs pour l’infection à Cryptosporidium, et 3 génotypes (Cryptosporidium baileyi, Cryptosporidium meleagridis, génotype aviaire II) ont été identifiés par PCR et séquençage. Un arbre phylogénétique des isolats a été analysé. Ces résultats suggèrent que la cryptosporidiose est répandue chez les volailles dans la province du Zhejiang, et est une menace potentielle pour la santé publique ainsi que l’économie. Ceci est le premier rapport sur le taux d’infection et la caractérisation moléculaire de Cryptosporidium dans les poulets de chair dans le Zhejiang.
Introduction
Cryptosporidiosis is caused by Cryptosporidium, which has a broad range of hosts and worldwide distribution. The Cryptosporidium species can infect human beings as well as other animals; it is mainly located in the epithelial cells of the gastrointestinal tract, and is likely in the respiratory tract [7], particularly in the case of infection by C. meleagridis, a bird species. The main clinical manifestation is persistent diarrhea, and respiratory disturbances can occasionally be observed [5, 12].
Cryptosporidium is one of the most common and important parasites in poultry, and this pathogen can infect more than 30 avian species. Traditionally, three different species of Cryptosporidium (C. baileyi, C. meleagridis, and C. galli) were considered to be the major species in birds [4, 6, 17, 20, 23, 30]. Recently, more than 11 novel genotypes have been described which are genetically different from the above three species by means of molecular analysis, including avian genotypes (I–V), goose genotypes (I–IV), the Eurasian woodcock genotype and the black duck genotype [1, 9, 14–17, 20, 31]. C. meleagridis, C. parvum, and C. hominis carried by poultry may infect human beings through the oocysts in contaminated water or vegetables eaten uncooked [1, 13, 20, 31, 32]. Considering that the possibility of exposure to Cryptosporidium of human beings still exists, the prevention of Cryptosporidiosis is important for public health [10].
Up to now, there is little information regarding the distribution of Cryptosporidium spp. in Zhejiang Province, and also limited data about its molecular characterization in poultry, particularly in chickens. The aim of the present study was to estimate the infection rate of Cryptosporidium in poultry in Zhejiang Province, and to determine the genotypes in the area.
Materials and methods
Sample collection
Three hundred and eighty-five fecal samples were randomly collected from broiler chickens that were around 90 days old in seven regions (Hangzhou, Huzhou, Jiaxing, Jinhua, Ningbo, Quzhou, and Shaoxing) in Zhejiang Province during the period from November 2010 to January 2012. Chickens were reared in steel cages; each contained 5 ~ 7 birds whose feces were collected on the tray under the cages gathered as a mixture. Therefore, the feces in one cage were considered as one sample, collected by the use of disposable plastic gloves, marked with the region name, serial number, and collection date. The sampling process was conducted to collect the fresh droppings to the best of our ability. The weight of each sample was approximately equal to about 50 g. Samples were kept in ice boxes until they were transported to the laboratory, then stored in the refrigerator at 4 °C and processed as soon as possible.
Microscopy detection
Samples were handled by Sheather’s sugar flotation method as previously described by Huber et al. [8], and Cryptosporidium oocysts were examined by optical microscopy observation under 400× magnification based on the shape of oocysts and the shape index measured. Subsequently, the positive samples containing oocysts were stored in 2.5% potassium dichromate and kept at 4 °C until DNA extraction.
DNA extraction
Oocysts in positive samples were purified by discontinuous sucrose density gradient centrifugation [3]. For genomic DNA extraction, 100 μL of suspension liquid containing oocysts was frozen-thawed for 5 min in liquid nitrogen and then kept in a 65 °C water bath kettle for 5 min. The process was repeated three times, then the treated samples were centrifuged at 12 000 × g for 5 min. The genomic DNA was extracted using a Genomic DNA Extraction Kit (TaKaRa Biotechnology (Dalian) Co. Ltd., Dalian, China) in accordance with the manufacturer’s instructions, and kept at −20 °C until detected by the PCR method.
Nested-PCR amplification and sequencing
A nested PCR was done in order to amplify a fragment of approximately 830 bp [30]. Then, the secondary purified PCR product was sequenced (BGI sequencing) to confirm the species/genotype identification.
Cryptosporidium genotyping and phylogenetic analysis
The acquired sequences were submitted to a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to initially define the species/genotypes and to confirm the high similarity and homology with other known sequences of Cryptosporidium spp. in GenBank. All sequences were multiple-aligned and analyzed by Bioedit and MEGA 4.0 software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html and http://www.megasoftware.net/). A neighbor-joining cladogram was built using MEGA 4.0. To assess the reliability of this tree, bootstrap analysis was done with 1000 replicates using the Kimura 2-parameter logarithm.
The partial 18S rRNA nucleotide sequences obtained in this study have been deposited in the GenBank database under accession numbers JX548291–JX548300.
Statistical analysis
The infection rate of Cryptosporidium was analyzed using the chi-square test. Differences were considered significant when P < 0.05.
Results
Infection rate of Cryptosporidium in Zhejiang Province
Of the 385 fecal samples from broiler chickens examined in this study, 38 samples were positive, and the overall infection rate of Cryptosporidium was 9.9%. The infection rate of Cryptosporidium varied among the seven regions. The highest infection rate was 16.9% in Huzhou, followed by 16.7% in Hangzhou, 12.1% in Ningbo, 7.7% in Shaoxing, 5.3% in Jiaxing, 4.0% in Quzhou, and 2.3% in Jinhua (Table 1). Significant statistical differences were not found in relation to the geographical provenance of chickens and infection rates in each region.
Table 1.
Infection rate of Cryptosporidium in different source supplied for the native chicken.
| Region | No. of cages examined | No. of cages Cryptosporidium positive | Infection rate (%) | Cryptosporidium species/genotypes (No. of isolates) |
|---|---|---|---|---|
| Hangzhou | 66 | 11 | 16.7 | C. baileyi (10), C. meleagridis (1) |
| Huzhou | 59 | 10 | 16.9 | C. baileyi (7), C. meleagridis (1), Avian genotype II (2) |
| Shaoxing | 52 | 4 | 7.70 | C. baileyi (4) |
| Ningbo | 58 | 7 | 12.1 | C. baileyi (6), Avian genotype II (1) |
| Jiaxing | 57 | 3 | 5.26 | C. baileyi (3) |
| Jinhua | 43 | 1 | 2.33 | C. baileyi (1) |
| Quzhou | 50 | 2 | 4.00 | C. baileyi (2) |
| Total | 385 | 38 | 9.87 | C. baileyi (33), C. meleagridis (2), Avian genotype II (3) |
Distribution of Cryptosporidium species
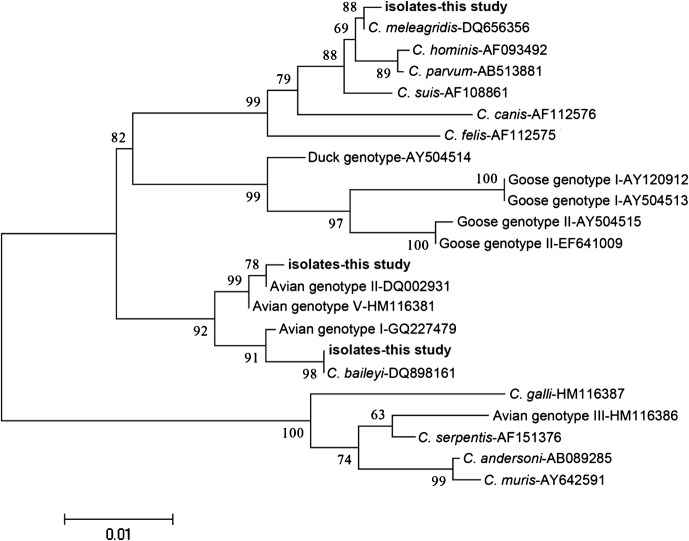
Two species and one genotype, including C. baileyi, C. meleagridis, and the avian genotype II, were identified through morphological observation and sequencing. C. baileyi was widely distributed in the seven areas. 33 of 38 positive samples were for C. baileyi infection (86.8%), only two samples for C. meleagridis infection (5.3%), and three samples for avian genotype II infection (7.9%). Moreover, the average size of oocysts for C. baileyi was 5.96 μm × 4.73 μm by measuring 35 oocysts, for C. meleagridis it was 4.96 μm × 4.15 μm, and for avian genotype II it was 6.02 μm × 5.47 μm. The neighbor-joining among these species/genotypes and other previously described species and genotypes of Cryptosporidium is shown in Figure 1.
Figure 1.
Phylogenetic tree of Cryptosporidium spp. built by the neighbor-joining method using Mega 4.0, based on 18S rRNA. To assess the reliability of this tree, bootstrap analysis was done with 1000 replicates.
Sequence and phylogenetic analysis
The 18S rRNA gene of C. baileyi isolates showed high sequence homology with each other by direct sequence alignment, and exhibited 99% sequence similarity with a GenBank sequence of quail origin (DQ898161). Two C. meleagridis isolates showed 99.5% sequence similarity with a GenBank sequence from Iran (DQ656356). Three avian genotype II isolates showed 99.2% sequence similarity with a GenBank sequence (DQ002931).
Discussion
Cryptosporidium is widespread around the world except Antarctica, and is most common in developing countries and less developed countries [22]. Thus far, there are few data available about the infection rate of Cryptosporidium in poultry, especially in chickens in China. It was reported that the infection rate of Cryptosporidium was 3.4% in Henan and 23.8% in Shandong, China [26, 27]. Moreover, the rate was 3.75% in the Garmsar region, Iran, and 4.5% in Tunisia [21, 24]. It was clear that the overall rate in this study was much higher than in other countries. We speculate that this difference may be attributed to the different approaches in the sample collection. In our study, the fecal sample was collected from a pool of animals; thus, it is possible to find a higher rate of infection when compared with individual samples. Moreover, the various environmental factors and host species may also account for the difference [2]. The differences between different studies may be due to various factors such as seasonality, the age of chickens, the breeding environment, etc. Given the similar climate of the regions in Zhejiang Province, the interregional variation in infection rates observed could be due to the nutrient levels, breeding conditions and the density of animals [11–25].
In China, the infection rate of Cryptosporidium derived from poultry has been reported so far in almost 10 provinces. However, most researchers were focused on the prevalence in the region through traditional morphological observation, rather than identifying the species or genotypes of the Cryptosporidium by modern molecular biology approaches. One exception is Henan Province. In more recent studies, the prevalence of Cryptosporidium spp. has been reported in detail in quails, chickens, Pekin ducks, pet birds, and ostriches in Henan, and most species have been identified as C. baileyi and C. meleagridis [18, 27–29]. The epidemiological investigation of Cryptosporidium in poultry in Zhejiang Province was carried out in our research.
Among the three Cryptosporidium species/genotypes identified in this study, C. baileyi was the primary species of the broiler chickens, for the reason that it was not only widely distributed in the seven areas, but also had the highest percentage of positive samples (33/38). Thus, the phenomenon of C. baileyi infection in broiler chickens in this study confirmed the following views. C. baileyi, which is the most common species in poultry, has more than 20 kinds of hosts including chickens [19]. In addition, studies in China showed that most cases in chickens were diagnosed as C. baileyi infections, which can slow down the growth rate in broilers and decrease egg production of layer chickens. Because the poultry industry plays a critical role in the agricultural economy in China, it is essential to pay sufficient attention to this pathogen. Only two isolates were identified as C. meleagridis in our investigation. Meanwhile, C. meleagridis had extensive hosts too, and it could cause deadly diseases in immunocompromised persons [19]. People could be infected with C. meleagridis by potential zoonotic transmission when the chickens carry the pathogen. Therefore, it has a great influence on public health. Three isolates were identified as avian genotype II in the present study. Avian genotype II was first described in turkey and was reported in parrot in the same year [14, 16]. Afterward, avian genotype II was found in ostriches [17]. Recently, chickens have also appeared to be a new host for avian genotype II. Perhaps we could make a bold conjecture that avian genotype II also has a range of hosts like C. baileyi and C. meleagridis. In addition, according to the phylogenetic analyses of 18S rRNA, together with the analysis based on actin and HSP70 in a previous study by Abe and Makino [1], it is further assumed that avian genotype II has a close relationship with avian genotype V at a genetic level. The cladogram in the present study proved the above points, which demonstrated the reliability of the tree.
In conclusion, this is the first report about the infection rate and molecular characterization of Cryptosporidium in broiler chickens in Zhejiang. The results suggest that Cryptosporidium is widespread in poultry in Zhejiang Province, and may pose a potential threat to public health as well as the economy.
Acknowledgments
This work was supported by a grant from the Science and Technology Department of Zhejiang Province (Grant number 2012C12009-2). The authors thank the team of the Parasite Laboratory, College of Animal Sciences, Zhejiang University.
Cite this article as: Wang L, Xue X, Li J, Zhou Q, Yu Y & Du A: Cryptosporidiosis in broiler chickens in Zhejiang Province, China: molecular characterization of oocysts detected in fecal samples. Parasite, 2014, 21, 36.
References
- 1.Abe N, Makino I. 2010. Multilocus genotypic analysis of Cryptosporidium isolates from cockatiels, Japan. Parasitology Research, 106, 1491–1497 [DOI] [PubMed] [Google Scholar]
- 2.Amer S, Zidan S, Adamu H, Ye J, Roellig D, Xiao L, Feng Y. 2013. Prevalence and characterization of Cryptosporidium spp. in dairy cattle in Nile River delta provinces, Egypt. Experimental Parasitology, 135, 518–523 [DOI] [PubMed] [Google Scholar]
- 3.Arrowood MJ, Donaldson K. 1996. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. Journal of Eukaryotic Microbiology, 43, S89–S89 [DOI] [PubMed] [Google Scholar]
- 4.Current WL, Upton SJ, Haynes TB. 1986. The life cycle of Cryptosporidium baileyi n. sp. (Apicomplexa, Cryptosporidiidae) infecting chickens. Journal of Protozoology, 33, 289–296 [DOI] [PubMed] [Google Scholar]
- 5.Das G, Changkija B, Sarkar S, Das P. 2011. Genotyping of Cryptosporidium parvum isolates in bovine population in Kolkata and characterization of new bovine genotypes. Research in Veterinary Science, 91, 246–250 [DOI] [PubMed] [Google Scholar]
- 6.Fayer R. 2010. Taxonomy and species delimitation in Cryptosporidium. Experimental Parasitology, 124, 90–97 [DOI] [PubMed] [Google Scholar]
- 7.Gomes RS, Huber F, da Silva S, do Bomfim TC. 2012. Cryptosporidium spp. parasitize exotic birds that are commercialized in markets, commercial aviaries, and pet shops. Parasitology Research, 110, 1363–1370 [DOI] [PubMed] [Google Scholar]
- 8.Huber F, Bomfim TC, Gomes RS. 2005. Comparison between natural infection by Cryptosporidium sp., Giardia sp. in dogs in two living situations in the West Zone of the municipality of Rio de Janeiro. Veterinary Parasitology, 130, 69–72 [DOI] [PubMed] [Google Scholar]
- 9.Jellison KL, Distel DL, Hemond HF, Schauer DB. 2004. Phylogenetic analysis of the hypervariable region of the 18S rRNA gene of Cryptosporidium oocysts in feces of Canada geese (Branta canadensis): evidence for five novel genotypes. Applied and Environmental Microbiology, 70, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karanis P, Kourenti C, Smith H. 2007. Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. Journal of Water and Health, 5, 1–38 [DOI] [PubMed] [Google Scholar]
- 11.Lai M, Zhou RQ, Huang HC, Hu SJ. 2011. Prevalence and risk factors associated with intestinal parasites in pigs in Chongqing, China. Research in Veterinary Science, 91, e121–e124 [DOI] [PubMed] [Google Scholar]
- 12.Langsley G, van Noort V, Carret C, Meissner M, de Villiers EP, Bishop R, Pain A. 2008. Comparative genomics of the Rab protein family in Apicomplexan parasites. Microbes and Infection, 10, 462–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majewska AC, Graczyk TK, Slodkowicz-Kowalska A, Tamang L, Jedrzejewski S, Zduniak P, Solarczyk P, Nowosad A, Nowosad P. 2009. The role of free-ranging, captive, and domestic birds of Western Poland in environmental contamination with Cryptosporidium parvum oocysts and Giardia lamblia cysts. Parasitology Research, 104, 1093–1099 [DOI] [PubMed] [Google Scholar]
- 14.Meireles MV, Soares RM, dos Santos MM, Gennari SM. 2006. Biological studies and molecular characterization of a Cryptosporidium isolate from ostriches (Struthio camelus). Journal of Parasitology, 92, 623–626 [DOI] [PubMed] [Google Scholar]
- 15.Morgan UM, Monis PT, Xiao L, Limor J, Sulaiman I, Raidal S, O’Donoghue P, Gasser R, Murray A, Fayer R, Blagburn BL, Lal AA, Thompson RC. 2001. Molecular and phylogenetic characterisation of Cryptosporidium from birds. International Journal for Parasitology, 31, 289–296 [DOI] [PubMed] [Google Scholar]
- 16.Ng J, Pavlasek I, Ryan U. 2006. Identification of novel Cryptosporidium genotypes from avian hosts. Applied and Environmental Microbiology, 72, 7548–7553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen ST, Fukuda Y, Tada C, Huynh VV, Nguyen DT, Nakai Y. 2013. Prevalence and molecular characterization of Cryptosporidium in ostriches (Struthio camelus) on a farm in central Vietnam. Experimental Parasitology, 133, 8–11 [DOI] [PubMed] [Google Scholar]
- 18.Qi M, Wang R, Ning C, Li X, Zhang L, Jian F, Sun Y, Xiao L. 2011. Cryptosporidium spp. in pet birds: genetic diversity and potential public health significance. Experimental Parasitology, 128, 336–340 [DOI] [PubMed] [Google Scholar]
- 19.Ryan U. 2010. Cryptosporidium in birds, fish and amphibians. Experimental Parasitology, 124, 113–120 [DOI] [PubMed] [Google Scholar]
- 20.Ryan UM, Xiao L, Read C, Sulaiman IM, Monis P, Lal AA, Fayer R, Pavlasek I. 2003. A redescription of Cryptosporidium galli Pavlasek, 1999 (Apicomplexa: Cryptosporidiidae) from birds. Journal of Parasitology, 89, 809–813 [DOI] [PubMed] [Google Scholar]
- 21.Shemshadi B, Rangbar Bahadori S, Mozafari A. 2010. Study on cryptosporidiosis incidence in broilers in Garmsar region, Iran. Comparative Clinical Pathology, 20, 143–149 [Google Scholar]
- 22.Shirley D, Moonah S, Kotloff K. 2012. Burden of disease from cryptosporidiosis. Current Opinion in Infectious Diseases, 25, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slavin D. 1955. Cryptosporidium meleagridis (sp. nov.). Journal of Comparative Pathology, 65, 262–266 [DOI] [PubMed] [Google Scholar]
- 24.Soltane R, Guyot K, Dei-Cas E, Ayadi A. 2007. Prevalence of Cryptosporidium spp. (Eucoccidiorida: Cryptosporiidae) in seven species of farm animals in Tunisia. Parasite, 14, 335–338 [DOI] [PubMed] [Google Scholar]
- 25.Taylor M, Webster K. 1998. Recent advances in the diagnosis in livestock of Cryptosporidium, Toxoplasma, Giardia and other protozoa of veterinary importance. Research in Veterinary Science, 65, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ti-sen X. 2009. Epidemiological investigation of Cryptosporidium from poultry in Shandong province. Chinese Journal of Veterinary Medicine, 4, 3–5 [Google Scholar]
- 27.Wang R, Jian F, Sun Y, Hu Q, Zhu J, Wang F, Ning C, Zhang L, Xiao L. 2010. Large-scale survey of Cryptosporidium spp. in chickens and Pekin ducks (Anas platyrhynchos) in Henan, China: prevalence and molecular characterization. Avian Pathology, 39, 447–451 [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Qi M, Zhu J, Sun D, Ning C, Zhao J, Zhang L, Xiao L. 2011. Prevalence of Cryptosporidium baileyi in ostriches (Struthio camelus) in Zhengzhou, China. Veterinary Parasitology, 175, 151–154 [DOI] [PubMed] [Google Scholar]
- 29.Wang R, Wang F, Zhao J, Qi M, Ning C, Zhang L, Xiao L. 2012. Cryptosporidium spp. in quails (Coturnix coturnix japonica) in Henan, China: Molecular characterization and public health significance. Veterinary Parasitology, 187, 534–537 [DOI] [PubMed] [Google Scholar]
- 30.Xiao L, Fayer R, Ryan U, Upton SJ. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clinical Microbiology Reviews, 17, 72–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou L, Kassa H, Tischler ML, Xiao L. 2004. Host-adapted Cryptosporidium spp. in Canada geese (Branta canadensis). Applied and Environmental Microbiology, 70, 4211–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou P, Chen N, Zhang R-L, Lin R-Q, Zhu X-Q. 2008. Food-borne parasitic zoonoses in China: perspective for control. Trends in Parasitology, 24, 190–196 [DOI] [PubMed] [Google Scholar]