Significance
Desorption electrospray ionization mass spectrometric imaging (DESI-MSI) has been shown to be particularly powerful for identifying lipids and metabolites directly from tissue sections and providing a chemical map of their distribution. We applied DESI-MSI to investigate changes in lipid profiles that occur in animal and human lymphomas associated with the overexpression of the v-myc avian myelocytomatosis viral oncogene homolog (MYC). Using statistical analysis, we found 86 lipids that were either increased or decreased in MYC-induced transgenic mouse models of lymphoma. Most of the increased lipids were glycerophosphoglycerols and cardiolipins with a higher content of monounsaturated fatty acids when compared with control tissue. The lipid profiles of MYC associated human lymphomas with overexpression of MYC resemble closely those observed in MYC-induced transgenic mouse models.
Keywords: biostatistics, transgenic models, lipidomics, metabolomics, cancer
Abstract
Overexpression of the v-myc avian myelocytomatosis viral oncogene homolog (MYC) oncogene is one of the most commonly implicated causes of human tumorigenesis. MYC is known to regulate many aspects of cellular biology including glucose and glutamine metabolism. Little is known about the relationship between MYC and the appearance and disappearance of specific lipid species. We use desorption electrospray ionization mass spectrometry imaging (DESI-MSI), statistical analysis, and conditional transgenic animal models and cell samples to investigate changes in lipid profiles in MYC-induced lymphoma. We have detected a lipid signature distinct from that observed in normal tissue and in rat sarcoma-induced lymphoma cells. We found 104 distinct molecular ions that have an altered abundance in MYC lymphoma compared with normal control tissue by statistical analysis with a false discovery rate of less than 5%. Of these, 86 molecular ions were specifically identified as complex phospholipids. To evaluate whether the lipid signature could also be observed in human tissue, we examined 15 human lymphoma samples with varying expression levels of MYC oncoprotein. Distinct lipid profiles in lymphomas with high and low MYC expression were observed, including many of the lipid species identified as significant for MYC-induced animal lymphoma tissue. Our results suggest a relationship between the appearance of specific lipid species and the overexpression of MYC in lymphomas.
The v-myc avian myelocytomatosis viral oncogene homolog (MYC) is commonly overexpressed in human neoplasia (1, 2) and has been strongly associated with the clinical aggressiveness of human cancers (3, 4). MYC is particularly associated with the pathogenesis of hematopoietic tumors such as lymphomas and of epithelial tumors (5, 6). In Burkitt’s lymphoma, for example, the c-Myc gene is translocated to one of the Ig loci in virtually all tumors (5, 7, 8). The MYC oncogene contributes to tumorigenesis by functioning as a global regulator of transcription involving many cellular programs, including cellular growth, metabolism, and lipid synthesis (9, 10). MYC induces a global shift in metabolism associated with anaerobic glycolysis, a phenomenon known as the Warburg effect (11). Several studies have shown a relationship between MYC regulation and metabolism in lymphomas, especially as related to the processes of glycolysis and glutaminolysis (12, 13). Some reports suggest that MYC regulates fatty acid synthesis, specifically palmitate (14); however, little is known about the relationship between MYC expression and the up-regulation or down-regulation of various lipid species in lymphomas.
We have generated transgenic mice to conditionally regulate expression of MYC oncogenes alone or in combination with oncogenes such as rat sarcoma (RAS), BCR-ABL, and BCL2 to produce many transgenic models of cancers, including T-cell acute lymphocytic leukemia, acute myeloid leukemia, osteosarcoma, lung adenocarcinoma, and hepatocellular carcinoma (HCC) (15–18). Recently, we described the use of desorption electrospray ionization mass spectrometry imaging (DESI-MSI) and transgenic mouse models to examine the lipid profiles of MYC-induced HCC (19). Here, we use our conditional transgenic mouse models, high mass resolution and high mass accuracy DESI-MSI, statistical analysis, and molecular biology approaches to investigate and identify the alterations in lipid profiles in MYC-induced lymphomas and in human lymphomas.
DESI-MSI is an ambient ionization technique in mass spectrometry (20, 21) that has recently emerged for imaging biological samples without the need of extensive sample preparation. DESI-MSI has been particularly powerful for investigating the distribution of diagnostic lipids and metabolites in a variety of human cancers directly from tissue sections (22–24). Samples are bombarded with microdroplets that dissolve hundreds of lipids and metabolites. The splash forms secondary microdroplets that enter a mass spectrometer, providing a detailed chemical map of the distribution of molecules within the sample surface. Using DESI-MSI, we observed a lipid signature characteristic of MYC-induced lymphoma. Because MSI provides such a wealth of chemical information, we used the statistical method called significance analysis of microarrays (SAM) (25, 26) to select the molecules with increased or decreased abundance in MYC-induced lymphomas that were statistically significant. Although SAM has been traditionally used to analyze gene expression microarrays, similarly, in DESI-MSI, hundreds of data points are obtained from a single imaging experiment. In our study, SAM selected 104 molecular ions with increased or decreased abundance in MYC-induced lymphoma compared with control tissue with a false discovery rate (FDR) of less than 5%. The majority of the selected molecular ions were identified by tandem and high-resolution mass spectrometry as complex glycerophospholipids of biological relevance including many different subclasses. We also analyzed 15 human lymphoma samples and, similarly to the animal model results, observed distinct lipid profiles in lymphomas with high expression of MYC, in comparison with lymphomas with low expression of MYC. Our results suggest that there may be a relationship between the alterations in specific lipid metabolism and pathways and the expression of MYC in lymphomas. We specifically identified 86 molecules that have potential value as diagnostic and prognostic markers.
Results
Specific Lipid Signatures from MYC-Induced Lymphomas.
We have previously described the generation of a conditional transgenic lymphoma model that uses the Tet-off system to regulate oncoprotein expression (15). Briefly, the transgene is expressed in the absence of doxycycline, whereas the transgene is turned off in the presence of doxycycline. We used negative ion mode DESI-MSI to investigate the distribution and abundance of molecules in thymus tissue of transgenic Emu-tTA/TetO-MYC mice and normal control thymus tissue. In particular, we focused our negative ion mode analysis in the m/z 700–1,100 range, in which a broad variety of complex glycerophospholipids of different classes is commonly detected compared with positive ion mode in which glycerophosphocholines and sphingomyielines are most abundantly detected (27). Three samples of MYC-induced lymphomas (lymphoma A, B, and C) and three samples of normal thymus (TY, T01, and T02) were imaged using DESI-MSI coupled to a high mass resolution–mass accuracy mass spectrometer (LTQ-Orbitrap XL) using the same experimental conditions. A histologically compatible solvent system, composed of dimethylformamide:acetonitrile (1:1) was used in the DESI-MSI experiments to allow histological evaluation to be performed on the same tissue section. This method allowed H&E staining to be performed on the same tissue section or cells previously analyzed by DESI-MSI, providing unambiguous correlation between lipid signatures and tissue disease state (28). Details on the materials and methods can be found in SI Materials and Methods.
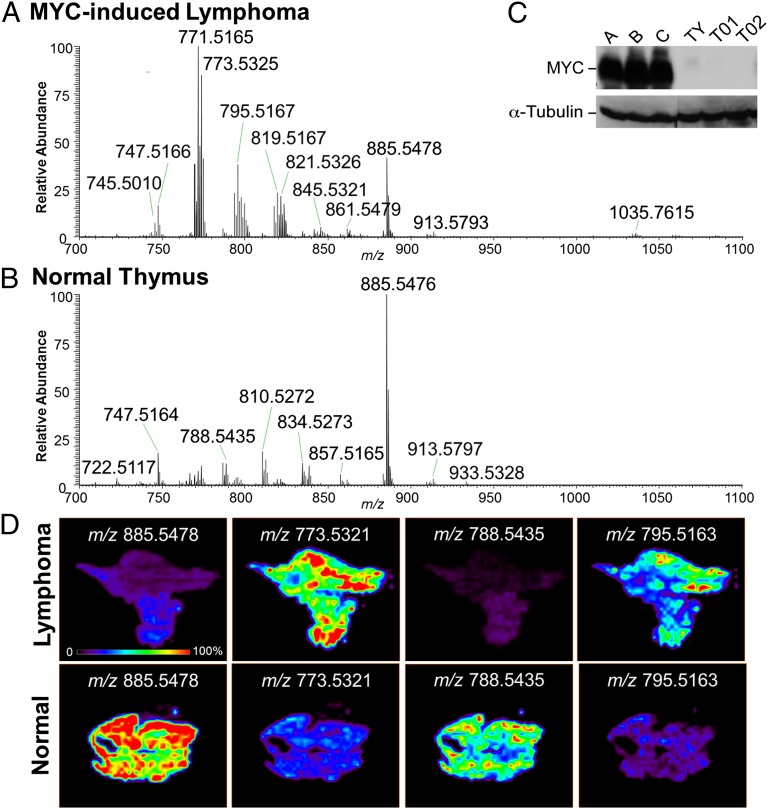
Fig. 1 shows representative mass spectra and selected 2D ion images obtained from a tissue sample of MYC-induced lymphoma (lymphoma A), and a tissue sample of control normal thymus (thymus T02). Striking differences in the spectra of MYC-induced lymphoma and normal thymus tissues are clearly observed by inspection. For example, high relative abundances of m/z 771.5161, m/z 773.5321, and m/z 795.5163 were observed in the lymphoma A sample (Fig. 1A), whereas high relative abundances of m/z 810.5275 and m/z 834.5275 are observed in the control thymus 1 tissue (Fig. 1B). Highly reproducible results were obtained for the other mice tissue samples analyzed by DESI-MSI (Fig. S1). The expression of MYC in the thymus tissue samples was evaluated by Western blot analysis, as shown in Fig. 1C, as well as the expression of α-tubulin, which was used as the loading control. The results confirm the high overexpression of MYC in the lymphoma A, B, and C tissue samples in comparison with the control thymus tissue samples TY, T01, and T02. Two-dimensional DESI ion images for lymphoma A and control thymus T02 are shown in Fig. 1D, displaying the intensity of selected m/z values throughout the tissue sections. Histopathologic evaluation of the hematoxylin and eosin (H&E) stained tissue sections confirmed that sample lymphoma A was completely comprised of tumor cells, whereas the control sample was fully composed of normal thymus cells. Some heterogeneity in the distribution of ions throughout the tissue section was observed in the 2D ion images, which is associated with regions having slight differences in lymphoma cell concentration as confirmed by histopathologic evaluation.
Fig. 1.
DESI-MSI of MYC-induced lymphomas shows specific lipid signature. Representative negative ion mode DESI mass spectra of (A) MYC-induced lymphoma (sample lymphoma A) and (B) normal control thymus (samples T02) tissues in the m/z 700–1,100 range are shown. Results for Western blot analysis of MYC and α-tubulin are shown in C for MYC-induced lymphomas samples A, B, and C, and normal thymus controls samples TY, T01, T02. DESI-MS ion images of selected molecular ions at m/z 885.5478, m/z 773.5321, m/z 788.5435, and m/z 795.5163 are shown in D for lymphoma and control samples.
Identification of Statistically Significant Lipids with Altered Abundance in MYC-Induced Lymphomas Using SAM.
The general differences in the relative abundances of specific ions between MYC-induced lymphoma and normal thymus can be evaluated by inspection of 2D DESI ion images. The complexity and wealth of information obtained in a single mass spectra by high mass resolution DESI-MSI for each pixel in the ion images calls for the use of refined biostatistical tools. We applied SAM to the set of DESI-MSI data obtained from all of the tissue samples analyzed to identify if the changes in abundance of molecular ions in MYC-induced lymphoma were statistically significant. SAM is a statistical technique conventionally used to analyze gene expression microarrays that include thousands of data points in a single experiment (25). SAM determines whether the change in the abundance of a molecule is statistically significant between different phenotypes or experimental conditions.
In DESI-MSI data, hundreds of data points are obtained from a single imaging experiment. Hence, we adapted SAM to identify statistically significant ions (m/z) by computing a score, d, that measures the average change in the normalized peak abundance for that m/z between MYC-induced lymphoma and normal thymus. Repeated permutations were used to determine whether the change is significantly related to the phenotype and estimate the percentage of molecular ions identified by chance, which is called the FDR. From all of the ions detected in the m/z 700–1,200 range for all of the samples analyzed, 179 different m/zs were selected as being altered in abundance in MYC-induced lymphomas in comparison with control samples, with an FDR of less than 5%. To identify these species, we used tandem mass spectrometry analysis and high mass accuracy measurements. The fragmentation patterns obtained were compared with that reported in the literature as characteristic for the different lipid classes (29), and were used in combination with high mass accuracy measurements for lipid identification. The high mass resolution–accuracy measurements allow us to separate and identify lipids of different classes with the same nominal mass without the need of chromatographic separation. For example, both m/z 766.5015 and m/z 766.5379 were selected by SAM as down-regulated in MYC-induced lymphoma tissue. These m/z values agree with the chemical formula of the deprotonated lipids PS(P-18:0/18:4) and PE(18:0/20:4), with a mass error of 1.70 and −1.76 ppm, respectively. Tandem mass spectrometry analysis of m/z 766.5 showed a mixture of fragments that characterize both the PS and PE lipids and their respective fatty acid chains.
A search through the LipidMaps database further confirmed that these m/zs can be assigned to these specific lipids. Note, however, that many of the 179 m/z values selected by SAM as statistically significant with an FDR of <5% corresponded to the 13C isotopes peaks of the same lipid species. Therefore, after careful evaluation of the mass spectra data, a total of 104 different m/zs were identified as the monoisotopic m/z of different lipid species, within which 86 different m/zs were identified as glycerophospholipids, 40 with increased abundance in MYC-induced lymphomas, and 46 with decreased abundance, as shown in Table S1. Lipids were identified with an absolute mass error of less than 3 ppm. Note that isomerism of the double bonds in the fatty acid (FA) chains of the glycerophospholipids complicates precise structural assignment, which is why FA chains are only tentatively assigned. Exact m/z values, mass errors, chemical formulas, and statistical scores for each lipid identified are given in Tables S2 and S3.
As seen in Table S1, the majority of the glycerophospholipid species found to be down-regulated are glycerophosphoserines (PS, 21 lipids) and glycerophosphoethanolamines (PE, 14 lipids), within a few glycerophosphoinositols (PI, 8 lipids), a glycerophosphate (PA), a glycerophosphoglycerol (PG, 1 lipid), and a cardiolipin (CL, 1 lipid). PS is a minor membrane phospholipid that makes up to 2–10% of the total phospholipids in mammalian cells. One of the biological roles of PS is as a cofactor that activates several signaling proteins, such as protein kinase C (30). PE is the second most abundant mammalian membrane phospholipid, constituting 20–30% of the total cell phospholipid, and PE lipids are known to play a role in membrane fusion and fission events. In contrast, the majority of the glycerophospholipids found to be up-regulated (Table S1) are PG (23 lipids), acyl-glycerophospholipids (acyl-PG, 4 lipids), and CL (4 lipids). A few PE (three lipids), PI (three lipids), PS (two lipids), and a PA (one lipid) were also observed. Acyl-PG are unusual lipids commonly found in bacterial cells, which have been reported to accumulate in mitochrondrial fractions of mammalian apoptotic cells (31), and to be present at significant levels in Golgi membranes (32). PG lipids are present at low levels (1–2%) in most animal tissue and can serve as the precursor of CL found in mitochrondrial membranes. CL is a complex phospholipid found almost exclusively in the inner mitochondrial membrane and is intimately involved in maintaining mitochondrial functionality and membrane integrity. We have recently reported the detection of a doubly deprotonated form of CL(72:8) at m/z 723.4768 in increased abundance in normal gastric tissue in comparison with gastric adenocarcinoma (24). Interestingly, this same CL was selected by SAM in its doubly deprotonated form in decreased abundance in lymphoma, whereas four other CL species with one to three fewer double bonds (72:7, 72:6, 72:5, and 74:7) were selected as in higher abundance in lymphoma tissue compared with normal thymus control. These findings are consistent with earlier studies showing the presence of immature CL with lower instauration and shorter FA chains in tumor cells (33).
Besides the alterations in the abundance of certain lipid classes in MYC-induced lymphomas, we also observed changes in the saturation of the FA chain constituents of the glycerophospholipids identified in higher abundance in lymphoma tissue in comparison with control tissue. When evaluating the saturated stearic acid FA(18:0) and the monounsaturated oleic acid FA(18:1) chain constituents of the glycerophospholipids, a total of 32 were identified as chains in glycerophospholipids found in decreased abundance in lymphoma, 27 FA(18:0) and 5 FA(18:1). Strikingly, out of the 35 identified in increased abundance in lymphoma tissue, only 7 were saturated FA(18:0) and 28 instead were FA(18:1). The monounsaturated fatty acid oleic acid is produced from the saturated FA stearic acid by the enzyme stearoyl-CoA desaturase-1 (SCD1), which catalyzes the synthesis of monounsaturated FAs from saturated FAs. SCD1 has been reported to be a target of the MYC oncogene as documented in the MYC target gene database and in other studies (34). Increased SCD1 gene expression has been detected in various malignant human tissues such as colonic and esophageal carcinomas, hepatocellular adenoma, as well as in human lymphoma cell lines (35). Similarly to oleic acid, we also observed a higher number of the monounsaturated palmitoleic acid FA(16:1) as FA chains in the glycerophospholipids found in increased abundance in lymphoma tissue than the saturated palmitic acid FA(16:0) compared with control tissue (Fig. S2).
Different Lipid Profiles Are Observed in RAS- Versus MYC-Induced Lymphoma Cell Lines.
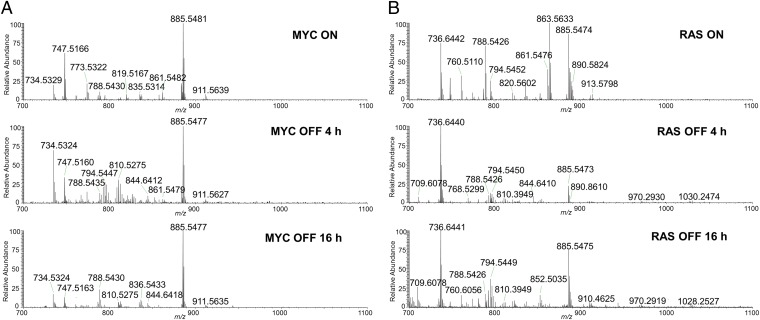
To evaluate if the lipid signature of lymphoma identified through DESI-MSI and SAM is oncogene specific, we examined if the alterations in the abundances of the glycerophospholipids are causally related to MYC or RAS oncogene activation through in vitro analysis. Tumor-derived cell lines isolated from MYC- or RAS-induced lymphoma models were subjected to doxycycline to shut down oncogene expression. Tumor cells were harvested when MYC or RAS was activated (MYC ON or RAS ON), and at two different time points following oncogene suppression (4- and 16-h MYC OFF). Cell samples were analyzed under the same experimental conditions used for lymphoma and thymus tissue imaging. Fig. 2 shows the negative ion mode DESI-MSI mass spectra obtained from MYC ON and RAS ON cells samples. Clearly distinct mass spectra profiles are observed in the mass range displaying glycerophospholipids for each lymphoma cell line. Consistently with what was observed in lymphoma tissue, many of the glycerophospholipids identified by SAM and DESI-MSI as being increased in MYC-induced lymphomas, such as the glycerophosphoglycerols PG(18:1/18:1) at m/z 773.5321 and PG(18:1/22:6) at m/z 819.5163, and the glycerophosphoinositols PI(18:0/18:2) at m/z 861.5475 and PI(18:0/20:3) at m/z 887.5679 are also observed in the mass spectra obtained from the MYC ON lymphoma cells. Interestingly, the relative intensities of these specific PG and PI species are observed to decrease with MYC inactivation, as found in the mass spectra obtained of MYC OFF 4 h, and even more drastically in MYC OFF 16 h (Fig. 2A). Concomitantly, the relative intensities of the glycerophosphoserines PS(18:0/22:5) at m/z 836.5440 and PS(18:0/18:1) at m/z 788.5435, which were identified in lower abundance in MYC-induced lymphoma compared with normal control tissue, increase with MYC inactivation. In the case of the RAS ON lymphoma cell line, the species observed at high relative intensities at m/z 861.5471, m/z 863.5631, and m/z 835.5314, identified as glycerophosphoinositols PI(18:1/18:1), PI(18:0/18:1), and PI(16:0/18:1), respectively, are observed to decrease in relative intensity in the mass spectra obtained of RAS OFF 4 h, and RAS OFF 16 h (Fig. 2B). These results support our findings using MYC-induced transgenic mouse models. We infer the abundance of glycerophospholipids in lymphomas is casually related to the expression of oncogenes rather than solely related to disease state.
Fig. 2.
Different lipid profiles are observed in MYC-induced versus RAS-induced lymphoma cell lines. Representative negative ion mode DESI mass spectra of (A) MYC-induced lymphoma cells: (Top) MYC ON; (Middle) MYC OFF 4 h; (Bottom) MYC OFF 16 h and (B) RAS-induced lymphoma cells: (Top) RAS ON; (Middle) RAS OFF 4 h; (Bottom) RAS OFF 16 h.
Lipid Profiles of Human Lymphomas with High and Low MYC Expression.
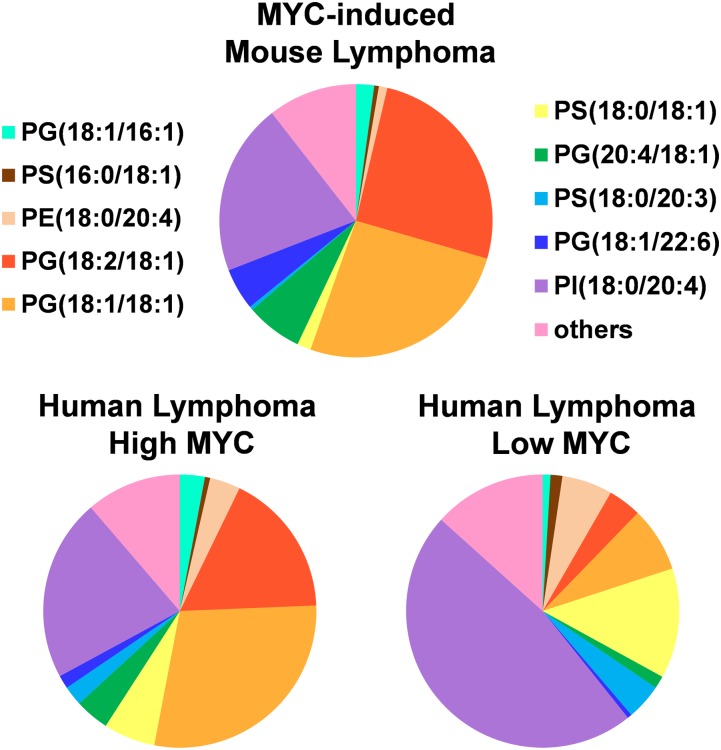
We evaluated whether the lipid signature identified in our mouse models of MYC-induced lymphomas could be observed in human lymphoma tissue specimens with different levels of MYC protein expression. A set of 15 human lymphoma samples of different subtypes including 3 diffuse large B-cell lymphomas (DLBCL), 7 follicular lymphomas, and 5 Burkitt’s lymphomas was investigated by DESI-MSI with the same experimental parameters used for murine samples. Many of the human lymphoma samples showed spatial heterogeneity, including regions with necrotic tissue, adjacent normal tissue, and blood vessels, as identified by histopathologic evaluation of the H&E stained tissue sections. The DESI mass spectra evaluated were extracted from selected regions with clear accumulation of tumor cells. To quantify MYC levels in human lymphoma, we used nanofluidic proteomic immunoassay (NIA), a highly sensitive method that we have previously developed for determining oncoprotein expression in clinical specimens (36). NIA has been used to detect and quantify MYC in human lymphoma specimens with results comparable to Western blot analysis (36). We have shown that the cutoff value of 0.2 relative luminescence units (RLU) is statistically significant for characterizing samples with high and low MYC expression levels. In the set of human lymphoma samples used for this study, nine samples of human lymphomas, including all DLBCL and four Burkitt’s lymphomas, had high expression levels of MYC (>0.2 RLU), whereas the remaining six samples, two follicular lymphomas and one Burkitt’s lymphoma, had low expression levels of MYC (Fig. S3). Fig. S4 shows the mass spectra for HL02, human DLBCL lymphoma with high expression of MYC (0.90 RLU), sample HL18, human Burkitt’s lymphoma with high expression of MYC (0.95 RLU), sample HL07, human follicular lymphoma with low expression of MYC (0.13 RLU) and sample HL08, and human follicular lymphoma with low expression of MYC (0.17 RLU). When comparing the mass spectral data obtained for the human samples with the murine data, strong similarities are observed between the human lymphoma samples with high MYC expression and MYC-induced lymphomas. Many of the lipid species identified in higher abundance in MYC-induced lymphoma by DESI-MSI and SAM are also observed in higher relative abundances in the human lymphoma samples with high MYC expression. To demonstrate the overall trends in lipid expression, we selected a total of 18 representative lipid species found by SAM (FDR = 0%, and statistical score |d| > 2) as either increased or decreased in MYC-induced lymphoma, and calculated their normalized abundance in three samples of MYC-induced lymphomas analyzed, nine samples of human lymphomas with high expression of MYC, and six samples of human lymphomas with low expression of MYC. Fig. 3A presents pie charts showing the average normalized abundance of the lipids for each group. Overall, there is a similar lipid expression pattern that is observed in the MYC-induced mouse lymphoma and the human lymphoma samples with high MYC expression rather than with the human lymphoma samples with low MYC expression.
Fig. 3.
Lipid profiles of human lymphomas with high MYC expression are similar to that observed in MYC-induced mouse lymphomas. Pie charts display the average normalized abundance of selected lipid species for three samples of MYC-induced lymphoma, nine samples of human lymphomas (HL01, HL02, HL04, HL10, HL12, HL14, HL15, HL17, and HL18) with high MYC expression (>0.2 RLU) and six samples of human lymphomas (HL05, HL06, HL07, HL08, HL09, and HL13) with low MYC expression (<0.2 RLU). The lipids selected were found to be increased [PG(18:1/16:1), PG(18:2/18:1), PG(18:1/18:1), PG(20:4/18:1), and PG(18:1/22:6)] or decreased [PS(16:0/18:1), PE(18:0/20:4), PS(18:0/18:1), PS(18:0/20:3), and PI(18:0/20:4)] in MYC-induced lymphoma by SAM with FDR = 0% and statistical scores |d| > 2. The “others” category refer to the sum of the average normalized abundances of the less abundant lipids: PE(P-16:0/20:4), CL(20:3/18:1/18:1/16:1), PE(18:0/18:2), PE(P-16:0/22:6), PG(18:2/20:4), PS(P-18:0/20:4), PS(18:0/22:6), and PI(18:0/18:2).
Discussion
We have combined DESI-MSI with SAM to identify MYC-associated lipid species in both transgenic mice and human lymphomas. Unlike traditional approaches used for lipid analysis, such as high-performance liquid chromatography mass spectrometry (HPLC-MS) and gas chromatography mass spectrometry (GC-MS), DESI-MSI allows for the direct and rapid analysis of lymphoma tissue and cells without the need for extensive sample preparation. A multitude of lipids are detected, and an unambiguous correlation between their 2D distribution and disease state has been established. We also used an advanced statistical technique, SAM, in a previously unemployed way to identify statistically significant molecular signatures within the complex MSI data for MYC-induced lymphoma. In combination with high mass resolution, high mass accuracy, and tandem mass spectrometry approaches, we were able to identify a total of 86 different glycerophospholipids, including species in classes PS, PI, PG, PE, PA, acyl-PG, and CL, with varied important biological roles. The majority of the lipid species selected by SAM as being significantly increased in abundance in lymphoma were identified as PG and CL, whereas lipids of the classes PE, PS, and PI were identified to be decreased in abundance.
The MYC oncogene regulates FA import for beta oxidation, which may play an important role in sustaining the survival of lymphoma cells (37, 38). The specific lipid species of various classes identified in our study with statistically significant altered abundance in MYC-induced lymphomas could have diagnostic and prognostic value, and could provide insight into the role of MYC in lymphoma tumorigenesis. PGs are of particular interest owing to their increased abundance in MYC-induced lymphomas and their major role as the precursors of CL, complex lipids that are found almost exclusively in mitochondrial membranes (39). This increase in PGs would be consistent with MYC’s induction of mitochondrial biogenesis (40). Abnormalities in CL can impair mitochondrial function and bioenergetics and cause irreversible respiratory injuries in tumors, as related to the Warburg cancer theory (33). CL content and distribution of molecular CL species has been reported to differ between tumor and nontumor cells (41) and in highly and slowly proliferating cells from leukemia patients (42). We have found that specific CL species with a smaller number of double bonds in MYC-induced lymphomas are significantly increased compared with normal control tissue. In vitro and in vivo studies both indicate that MYC has many connections to mitochondrial biogenesis and function (43, 44), although a correlation between MYC expression and CL composition in lymphomas appears, to our knowledge, not to have been reported previously. Besides identifying changes in the abundance of lipids of certain classes, our approach also enabled the identification of marked changes in FA chain composition in MYC-induced lymphoma compared with normal tissue, with a larger number of monounsaturated FA in lymphoma tissue compared with their saturated FA counterpart.
We also investigated lipid profiles of MYC-induced and RAS-induced lymphoma cells, and found them to be markedly different. The relative abundances of lipids in each cell lines also varied after 4 and 16 h of oncogene inactivation. RAS-induced lymphoma contained higher relative abundance of PI as opposed to PG. This finding could relate to the canonical RAS-PI3K-Akt pathway that requires phosphoinositides for signaling. PIs, which are phosphoinositide metabolites, are catalyzed by phospholipase A2 and a lysophospholipase, which are known to be RAS-dependent (45). Lysophospholipase catalyzes the production of PI from phosphoinositides, a process which is greatly augmented by RAS transformation in both fibroblasts and other cells, indicating the importance of glycerophospholipid metabolism in RAS (46–48). Our results could reflect that generally specific oncogenes will be associated with unique lipid signatures. Note that some of the changes we observe in the lipid profiles could reflect differences between naive thymic and activated tumor-derived T cells. However, because the lipid signature we identify appears to distinguish MYC from RAS-induced lymphomas, we presume that many of these changes are causally specific to MYC, but this remains to be examined (please see SI Discussion for additional discussion).
Our transgenic mouse models of MYC-induced cancers recapitulate many properties of human cancer, thereby serving as a tractable way to dissect how MYC regulates metabolism in cancer cells (49). We examined 15 samples of different types of human lymphomas using DESI-MSI and found that many of the lipid species identified by SAM to be overexpressed in MYC-induced lymphoma were also observed in higher relative abundances in human lymphoma samples having a high expression of MYC. These results suggest that there is a correlation between lipid abundance and MYC oncogene expression in human lymphomas.
Our approach provides a means for in situ analysis of specific metabolic signatures that may be useful to detect, diagnose, and prognosticate MYC-associated human tumors. The lipid species found in our study might provide new biological insights into how MYC regulates cellular metabolism in cancer. Future studies will identify the biological mechanisms by which MYC changes lipid profiles and evaluate whether these lipid signatures can serve as biomarkers for cancer detection and therapy.
Supplementary Material
Acknowledgments
We are grateful to Stephanie Casey and Yulin Li for animal samples provided and to Jialing Zhang for the assistance with MS experiments. We thank Mari Prieto Conaway and Iman Mohtashemi from Thermo Fisher Scientific for the custom-built software tools provided. R.J.T. is supported by National Science Foundation Grant DMS-9971405 and National Institutes of Health (NIH) Grant N01-HV-28183. A.C.F. is supported by NIH Grant K23CA140722. We also acknowledge a subcontract with the University of Utah (Agreement 10029173-S2), for which the Air Force Office of Scientific Research (Grant FA9550-12-1-0481) is the prime sponsor, which made possible our use of an Orbitrap mass spectrometer. This work was supported by NIH Grant 1R01CA184384-01 and by the 2012 Stanford Hospital and Clinics Cancer Innovation Fund Award. L.S.E. is grateful to the Center of Molecular Analysis and Design for her postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409778111/-/DCSupplemental.
References
- 1.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15(21):6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcu KB, Bossone SA, Patel AJ. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 3.Wolfer A, et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci USA. 2010;107(8):3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, Chen Y, Olopade OI. MYC and breast cancer. Gene Cancer. 2010;1(6):629–640. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalla-Favera R, et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79(24):7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawate S, Fukusato T, Ohwada S, Watanuki A, Morishita Y. Amplification of c-myc in hepatocellular carcinoma: Correlation with clinicopathologic features, proliferative activity and p53 overexpression. Oncology. 1999;57(2):157–163. doi: 10.1159/000012024. [DOI] [PubMed] [Google Scholar]
- 7.Dang CV, O’donnell KA, Juopperi T. The great MYC escape in tumorigenesis. Cancer Cell. 2005;8(3):177–178. doi: 10.1016/j.ccr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20(40):5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 9.Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28(27):2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harbor Perspect Med. 2013;3(8):a014217. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 12.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19(1):1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrish F, et al. Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J Biol Chem. 2010;285(47):36267–36274. doi: 10.1074/jbc.M110.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4(2):199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 16.Jain M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297(5578):102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 17.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 18.Tran PT, et al. Combined inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PLoS ONE. 2008;3(5):e2125. doi: 10.1371/journal.pone.0002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry RH, et al. Characterization of MYC-induced tumorigenesis by in situ lipid profiling. Anal Chem. 2013;85(9):4259–4262. doi: 10.1021/ac400479j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alberici RM, et al. Ambient mass spectrometry: Bringing MS into the “real world.”. Anal Bioanal Chem. 2010;398(1):265–294. doi: 10.1007/s00216-010-3808-3. [DOI] [PubMed] [Google Scholar]
- 21.Harris GA, Galhena AS, Fernández FM. Ambient sampling/ionization mass spectrometry: Applications and current trends. Anal Chem. 2011;83(12):4508–4538. doi: 10.1021/ac200918u. [DOI] [PubMed] [Google Scholar]
- 22.Dill AL, et al. Multivariate statistical differentiation of renal cell carcinomas based on lipidomic analysis by ambient ionization imaging mass spectrometry. Anal Bioanal Chem. 2010;398(7-8):2969–2978. doi: 10.1007/s00216-010-4259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberlin LS, et al. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012;72(3):645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberlin LS, et al. Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc Natl Acad Sci USA. 2014;111(7):2436–2441. doi: 10.1073/pnas.1400274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eberlin LS, et al. Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Mol Cell Biol Lipids. 2011;1811(11):946–960. doi: 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberlin LS, et al. Nondestructive, histologically compatible tissue imaging by desorption electrospray ionization mass spectrometry. ChemBioChem. 2011;12(14):2129–2132. doi: 10.1002/cbic.201100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan S, Reinhold VN. Detailed structural characterization of lipid A: Electrospray ionization coupled with tandem mass spectrometry. Anal Biochem. 1994;218(1):63–73. doi: 10.1006/abio.1994.1141. [DOI] [PubMed] [Google Scholar]
- 30.Berry KAZ, et al. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem Rev. 2011;111(10):6491–6512. doi: 10.1021/cr200280p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorice M, et al. Cardiolipin and its metabolites move from mitochondria to other cellular membranes during death receptor-mediated apoptosis. Cell Death Differ. 2004;11(10):1133–1145. doi: 10.1038/sj.cdd.4401457. [DOI] [PubMed] [Google Scholar]
- 32.Cluett EB, Kuismanen E, Machamer CE. Heterogeneous distribution of the unusual phospholipid semilysobisphosphatidic acid through the Golgi complex. Mol Biol Cell. 1997;8(11):2233–2240. doi: 10.1091/mbc.8.11.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiebish MA, Han X, Cheng H, Chuang JH, Seyfried TN. Cardiolipin and electron transport chain abnormalities in mouse brain tumor mitochondria: Lipidomic evidence supporting the Warburg theory of cancer. J Lipid Res. 2008;49(12):2545–2556. doi: 10.1194/jlr.M800319-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: Identification of direct genomic targets. Genome Biol. 2003;4(10):R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robetorye RS, et al. Microarray analysis of B-cell lymphoma cell lines with the t(14;18) J Mol Diagn. 2002;4(3):123–136. doi: 10.1016/S1525-1578(10)60693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan AC, et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med. 2009;15(5):566–571. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pacilli A, et al. Carnitine-acyltransferase system inhibition, cancer cell death, and prevention of myc-induced lymphomagenesis. J Natl Cancer Inst. 2013;105(7):489–498. doi: 10.1093/jnci/djt030. [DOI] [PubMed] [Google Scholar]
- 38.Ambrosio MR, et al. The alteration of lipid metabolism in Burkitt lymphoma identifies a novel marker: Adipophilin. PLoS ONE. 2012;7(8):e44315. doi: 10.1371/journal.pone.0044315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li F, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25(14):6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartz JW, Morton RE, Waite MM, Morris HP. Correlation of fatty acyl composition of mitochondrial and microsomal phospholipid with growth rate of rat hepatomas. Lab Invest. 1982;46(1):73–78. [PubMed] [Google Scholar]
- 42.Schild L, et al. Composition of molecular cardiolipin species correlates with proliferation of lymphocytes. Exp Biol Med (Maywood) 2012;237(4):372–379. doi: 10.1258/ebm.2011.011311. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11(5):407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS ONE. 2008;3(3):e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berrie CP, Iurisci C, Corda D. Membrane transport and in vitro metabolism of the Ras cascade messenger, glycerophosphoinositol 4-phosphate. FEBS J. 1999;266(2):413–419. doi: 10.1046/j.1432-1327.1999.00870.x. [DOI] [PubMed] [Google Scholar]
- 46.Alonso T, Morgan RO, Marvizon JC, Zarbl H, Santos E. Malignant transformation by ras and other oncogenes produces common alterations in inositol phospholipid signaling pathways. Proc Natl Acad Sci USA. 1988;85(12):4271–4275. doi: 10.1073/pnas.85.12.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alonso T, Santos E. Increased intracellular glycerophosphoinositol is a biochemical marker for transformation by membrane-associated and cytoplasmic oncogenes. Biochem Biophys Res Commun. 1990;171(1):14–19. doi: 10.1016/0006-291x(90)91349-w. [DOI] [PubMed] [Google Scholar]
- 48.Valitutti S, Cucchi P, Colletta G, Di Filippo C, Corda D. Transformation by the k-ras oncogene correlates with increases in phospholipase A2 activity, glycerophosphoinositol production and phosphoinositide synthesis in thyroid cells. Cell Signal. 1991;3(4):321–332. doi: 10.1016/0898-6568(91)90061-x. [DOI] [PubMed] [Google Scholar]
- 49.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16(4):313–317. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.