Abstract
Background
About half of malignant hyperthermia (MH) cases are associated with skeletal muscle ryanodine receptor 1 (RYR1) and calcium channel, voltage-dependent, L type, α1S subunit (CACNA1S) gene mutations, leaving many with an unknown cause. We chose to apply a sequencing approach to uncover causal variants in unknown cases. Sequencing the exome, the protein-coding region of the genome, has power at low sample sizes and identified the cause of over a dozen Mendelian disorders.
Methods
We considered four families with multiple MH cases but in whom no mutations in RYR1 and CACNA1S had been identified by Sanger sequencing of complementary DNA. Exome sequencing of two affecteds per family, chosen for maximum genetic distance, were compared. Variants were ranked by allele frequency, protein change, and measures of conservation among mammals to assess likelihood of causation. Finally, putative pathogenic mutations were genotyped in other family members to verify cosegregation with MH.
Results
Exome sequencing revealed 1 rare RYR1 nonsynonymous variant in each of 3 families (Asp1056His, Val2627Met, Val4234Leu), and 1 CACNA1S variant (Thr1009Lys) in a 4th family. These were not seen in variant databases or in our control population sample of 5379 exomes. Follow-up sequencing in other family members verified cosegregation of alleles with MH.
Conclusions
Using both exome sequencing and allele frequency data from large sequencing efforts may aid genetic diagnosis of MH. In our sample, it was more sensitive for variant detection in known genes than Sanger sequencing of complementary DNA, and allows for the possibility of novel gene discovery.
Introduction
Malignant hyperthermia (MH, Online Mendelian Inheritance in Man #145600) is a life-threatening disorder of muscle metabolism which occurs after exposure to certain anesthetics, occurring in approximately 1 in 50,000 anesthetics.1,2 Its heritability was appreciated with the first reported case.3 Research on a fortuitously discovered animal form, followed by human linkage studies, led to the discovery that mutations in the skeletal muscle ryanodine receptor 1 gene (RYR1, chromosome 19q13) was involved in MH.4,5 Variants in the calcium channel, voltage-dependent, L type, α1S subunit gene (CACNA1S, chromosome 1q32) were later found to be linked.6 To date, variants in RYR1 and CACNA1S have been associated with only 50-70% of cases of MH.7–9 Variants in other genes presumably account for the remaining 30-50%, with linkage evidence pointing to regions in chromosomes 3q13 (logarithm of odds = 3.22 in 1 family10), 5p (logarithm of odds = 1.22 in 1 family11), 7q21 (logarithm of odds = 2.91 in 1 family12) and 17q11 (logarithm of odds = 3.26 in 5 families13).
Several methods are commonly used to detect pathogenic RYR1 variants.8 These often involve error-prone viral reverse transcriptase to create complementary DNA (cDNA) from messenger RNA (mRNA), which could increase the risk of false positive or negative results.14 Another drawback to using cDNA is the inability to detect either structural or expression-related changes, such as insertion-deletions, splice variants or promoter variants. Due to the large size of the gene (154 kb), Sanger sequencing remains costly and cumbersome for large-scale sequencing efforts. An alternative method is next-generation sequencing, which involves multiple determinations of the base at each position. Next-generation sequencing is being applied in a number of clinical contexts now, particularly in cancer, and has negligible false-positive rates when the quality of the sequence is high.15 One group demonstrated the feasibility of applying this technology to MH, which has the added benefit of novel gene mutation discovery.16
Once a missense RYR1 genomic variant is found, additional tests for determining pathologic significance are required. European Malignant Hyperthermia Group consensus guidelines * require that the segregating variant be found in at least two families and that either in vitro or ex vivo functional study results are consistent with the pathophysiology of MH. Of the >200 RYR1 coding variants identified in MH individuals so far, only 30 have passed this threshold.7,17 Since these steps add significant time and expense to identifying pathogenic variants, many have turned to evaluating genetic variants using statistical models for determining how damaging a mutation is to the protein. Here we used next-generation sequencing to find the potential pathogenic MH variants in individuals who were found mutation-negative in RYR1 and CACNA1S by cDNA sequencing. We also mined data from 5,379 control exome sequencing and current genome databases to evaluate the usefulness of predictive methods for inferring causality of RYR1 variants. As we show, the high genetic diversity of RYR1 and CACNA1S require additional data, such as segregation in large families and extremely low minor allele frequencies, to suggest pathogenicity.
Materials and Methods
MH DNA Samples
We considered 4 families with multiple individuals diagnosed with MH, but in whom no mutations in RYR1 and CACNA1S had been identified. To rule out RYR1 or CACNA1S variation, cDNA generated from probands’ muscle mRNA by reverse transcriptase polymerase chain reaction had undergone Sanger sequencing as previously described.18 Each subject in this study had undergone diagnostic testing for MH susceptibility at the MH Investigation Unit, Leeds, United Kingdom, using in vitro contracture testing (IVCT) according to the European Malignant Hyperthermia Group.19 IVCT and RYR1 and CACNA1S sequencing were previously performed for clinical purposes, and the results have not been previously reported. All subjects had given written consent for blood and skeletal muscle sample collection and storage for subsequent genetic analysis under a research protocol approved by the Leeds East Local Research Ethics Committee (Leeds, United Kingdom). DNA samples from both MH susceptible and MH negative individuals in each family were de-identified and sequenced at the University of Washington, after being granted approval by the University of Washington institutional review board (Seattle, WA). In three families, two individuals were chosen for exome sequencing, the proband and one distant affected relative, while in the remaining family only the proband was exome sequenced. All sequencing was blinded to MH diagnosis.
Control Exomes
The control group consisted of 5,379 individuals sequenced at the University of Washington and the Broad Institute as part of the National Heart, Lung and Blood Institute Grand Opportunity (GO) Exome Sequencing Project (ESP).20 The goal of the ESP was to discover variants in novel genes possibly causal for cardiovascular, pulmonary, and hematologic diseases. These subjects were selected for inclusion in their original studies based on these categories of disease, and were not screened for MH. The source study groups include the Seattle GO, Broad GO, Heart GO, the University of Washington Lung GO, and Women’s Health Initiative Sequencing Project GO. Data from this cohort were analyzed in aggregate at two genes, RYR1 and CACNA1S.
Library construction, capture, and sequencing
Exome shotgun libraries were prepared from whole blood extracted genomic DNA as previously described.21 Briefly, one microgram of genomic DNA was subjected to a series of shotgun library construction steps, including fragmentation through acoustic sonication (Covaris), end-polishing (NEBNext End Repair kit), A-tailing (NEBNext dA Tailing kit), and ligation of 8bp barcoded sequencing adaptors (Enzymatics Ultrapure T4 Ligase). Barcoded shotgun libraries were hybridized to capture probes targeting the coding exons (Roche Nimblegen SeqCap EZ Human Exome Library v2.0) with custom blockers complimentary to the full length of the flanking adaptor and barcodes. Enriched libraries were amplified via polymerase chain reaction before sequencing (BioRad iProof). Library quality was determined by examining molecular weight distribution and sample concentration (Agilent Bioanalyzer). Pooled, barcoded libraries were sequenced via paired-end 50 base pair reads with an 8 base pair barcode read on Illumina HiSeq sequencers.
De-multiplexed binary sequence files (.bam) were aligned to a human reference (hg19) using the Burrows-Wheeler Aligner.22, † Read data from a flow-cell lane were treated independently for alignment and quality control purposes in instances where the merging of data from multiple lanes was required. All aligned read data were subjected to: (1) removal of duplicate reads (Picard); (2) insertion/deletion variant (indel) realignment using the Genome Analysis ToolKit IndelRealigner; and (3) base qualities recalibration using Genome Analysis ToolKit TableRecalibration. Variant detection and genotyping were performed using the UnifiedGenotyper tool from Genome Analysis ToolKit v1.529.23, ‡ Variant data for each sample were formatted (variant call format .vcf) as “raw” calls that contained individual genotype data for one or multiple samples, and flagged using the filtration walker (Genome Analysis ToolKit) to mark sites that were of lower quality and potential false positives (e.g., quality scores (≤50), allelic imbalance (≥0.75), long homopolymer runs (>3), and/or low quality by depth (<5)). Variant data were annotated using the SeattleSeq Annotation Server.
Exome data analysis for single nucleotide variants (SNVs) and small indels
The exome definition was based on consensus coding sequence (CCDS 2009) of the human reference genome (hg19/GRCh37). All rare missense variants were viewed using the Integrative Genomics Viewer 1.5.37.24 In our effort to search for pathogenic MH variants in novel genes using exome sequencing, we adapted a filtering strategy that has been successful for other Mendelian disorders.25,26 This method works particularly well for SNVs and indels. We performed the following steps for each family. Our starting set included all genotypes with sufficient coverage (>7x) which differed from the reference homozygous genotype. Of these genotypes, we took those shared among all cases in that family. Next we removed all synonymous and intronic, nonsplice variants, which are least likely to cause this phenotype, based on the known coding mutations. To filter out variation that was too common to be highly penetrant for MH, we removed single nucleotide polymorphisms reported in the Single Nucleotide Polymorphism Database (dbSNP) 13127 or 1,000 Genomes28 phase 1 to have a minor allele frequency above 1%. We then looked at all remaining variants in RYR1 and CACNA1S. If these genes were devoid of variants, we would then look for genes which harbored a variant in more than one family. The Integrative Genome Viewer (Broad Institute, Cambridge, MA) was used to graphically display the sequence variants for inspection.24, §
Confirmation and segregation analysis of potentially causative variants
The presence of potentially causative variants was confirmed using conventional Sanger sequencing of genomic DNA, which was also used for segregation analysis of the variant within the family.
Conservation score analysis
For each base position in the exons of RYR1 and CACNA1S, we compared several conservation and protein change scores, described briefly here. The genomic evolutionary rate profiling (GERP++) score is a measure of evolutionary constraint at each base derived by aligning 29 mammalian genomes and comparing the observed to expected rejected substitution rate,29,30 where negative scores indicate a lack of conservation and high positive scores indicate strong conservation among multiple species. PhastCons is a conservation score that uses a phylogenetic hidden Markov model to generate a probability that a base-pair is located in a region that conforms to a conserved model.31 The Grantham score is a measure of similarity between two distinct amino acids based on chemical properties, taking polarity and stericity into consideration.32 PolyPhen2 is a method for predicting the potential damage that a sequence change will have on a protein. It takes into account both the likelihood of the sequence change based on homologous sequences, as well as its effect on the structure of the protein.33
The PhastCons and Grantham scores cited here were generated by the SeattleSeq Annotation Server 134.** GERP++ scores were pre-calculated by the Sidow group for chromosomes 1 and 19.†† PolyPhen2 scores were generated through the batch-query interface on the PolyPhen2 server version 2.2.2. ‡‡ Graphs and histograms displaying the conservation scores were generated using R (R Foundation for Statistical Computing, Vienna, Austria).34
Results
In a few filtering steps, we reduced the number of potential candidate variants from 6-7 thousand to 1-2 dozen (table 1). At this point, a quick scan of the known genes revealed that each of the four families had a rare missense variant in either RYR1 or CACNA1S (table 2). These exact variants are neither among the 30 known pathogenic variants, nor seen in the literature or in variant or exome databases, but alternate base substitutions at two of the same positions in RYR1 have been reported as potential pathological variants in the literature.35
Table 1.
Identifying Likely Pathologic Variants in Malignant Hyperthermia Families
| Filter | Family A | Family B | Family C | Subject G |
|---|---|---|---|---|
| Not homozygous reference | 15,259 | 16,477 | 15,344 | 22,973 |
| Missense/Nonsense/Splice Site/Indel | 6,499 | 6,998 | 6,538 | 9,220 |
| Not in databases | 139 | 208 | 132 | 347 |
| Not in control exomes | 7 | 23 | 2 | 103 |
| RYR1 or CACNA1S variants | 1 | 1 | 1 | 1 |
The numbers in each column represent counts of individual exome variants among affected members in each family. The first row indicates the number of variants in each family that were high quality (>7x coverage), shared by affected members, and were different from the reference genome. In the lower rows, each cell lists the number of variants that remain after applying the indicated filter.
calcium channel, voltage-dependent, L type, α1S subunit gene = CACNA1S; insertion or deletion = indel; ryanodine receptor 1 gene =RYR1.
Table 2.
Characteristics of Rare Exome Variants
| Family code: Gene |
Codon | AA change |
Base | Seen Before? |
Poly- Phen2 |
Grantham Score |
Phast- Cons |
GERP++ |
|---|---|---|---|---|---|---|---|---|
| A: RYR1 | 4234 | VAL/LEU | G > T | G > C | 0.994 | 32 | 1.0 | 3.07 |
| B: RYR1 | 1056 | ASP/HIS | G > C | G > A | 1.0 | 81 | 0.999 | 2.94 |
| G: RYR1 | 2627 | VAL/MET | G > A | No | 1.0 | 21 | 1.0 | 4.08 |
| C: CACNA1S | 1009 | THR/LYS | C > A | No | 0.998 | 78 | 0.987 | 5.12 |
Each row lists the characteristics of a novel rare variant suspected of being pathogenic. The letter to the left of the gene name indicates which family the variant was found in. All four variants have not been seen in any databases or in 5,379 UW ESP individuals. However, a different base change has been seen at the same position as the first two variants (Families A and B). The table includes scores that assess predicted protein change (PolyPhen2 and Grantham) and evolutionary conservation (PhastCons and GERP++), and highlights in bold either maximal scores (e.g., 1.0 for PhastCons and PolyPhen2) or extreme values.
Adenine = A; amino acid = AA; aspartic acid = ASP; calcium channel, voltage-dependent, L type, α1S subunit gene = CACNA1S; cytosine = C; exome sequencing project = ESP; genomic evolutionary rate profiling = GERP; guanine = G; histidine = HIS; leucine = LEU; lysine = LYS; methionine = MET; ryanodine receptor 1 gene = RYR1; threonine = THR; thymine = T; University of Washington = UW; valine = VAL.
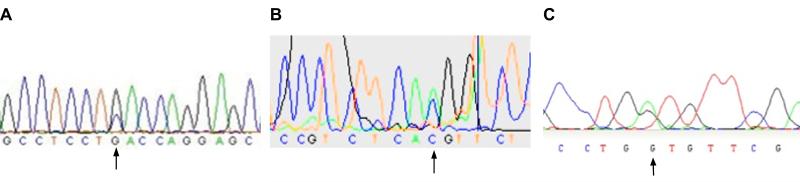
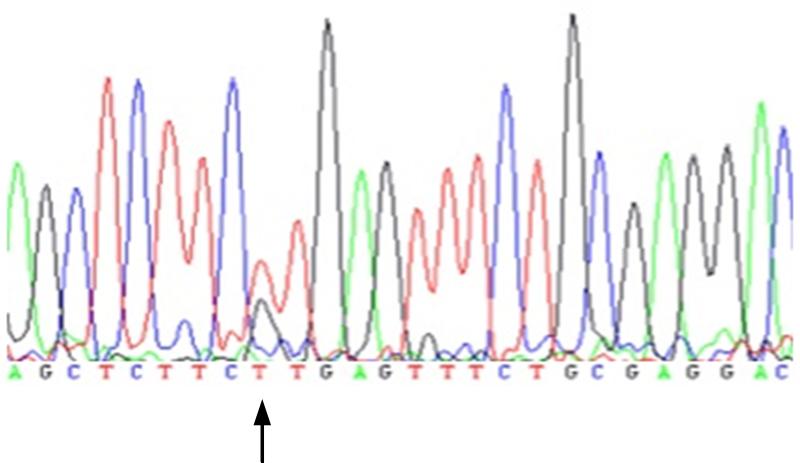
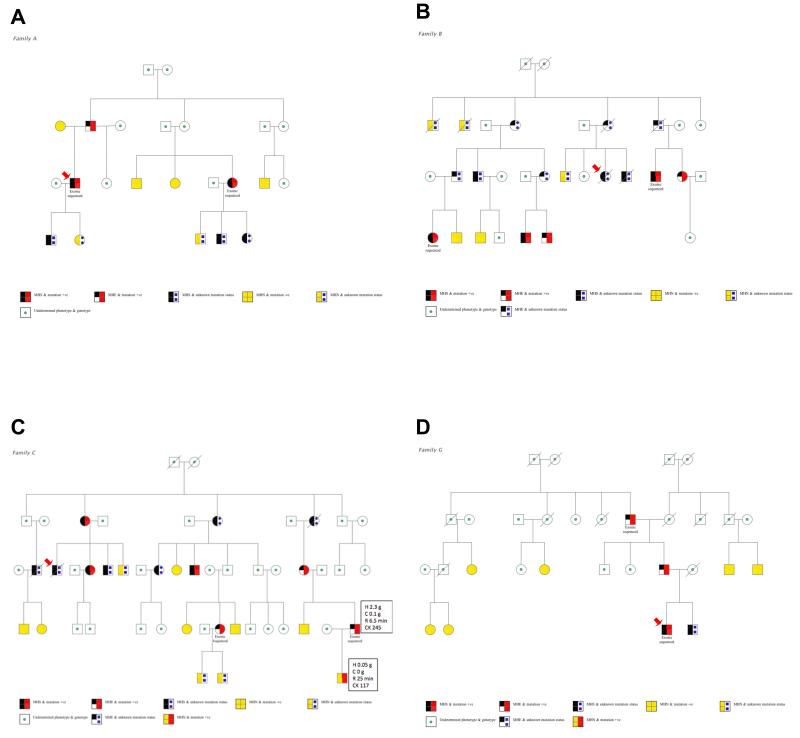
Follow-up analyses found that the variant in family A had not been detected by the Sanger sequencing because the mRNA for the RYR1 fragment containing exon 91 had been of insufficient quality to amplify during the polymerase chain reaction. Sanger sequencing had been complete for the samples from the other families but the original software base call (Gap4, Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom)36, §§ at each of the relevant cDNA positions was homozygous reference (fig. 1). The Integrative genomics viewer plots from the next-generation sequencing, however, show highly consistent reads at each location, with a near-perfect split of 49-51% of all calls representing the non-reference allele (fig. 2). This gives strong support for a heterozygous genotype at these loci, which is consistent with autosomal dominant inheritance. To ensure that these were not false positive sequencing results, we confirmed the presence of each variant using direct sequencing of genomic DNA and the electropherogram for the index case of Family A is shown in figure 3. We then genotyped the familial variant in all previously IVCT-tested family members where genomic DNA was available and segregation of the variants with IVCT status is shown in figure 4 for families A, B and C. In family G only two MH positive individuals have been screened for the variant. For each RYR1 variant there was complete segregation but for the variant in CACNA1S there were six IVCT positive individuals who carried the variant, seven IVCT negative individuals who did not carry the variant and one IVCT negative individual who did carry the variant. The IVCT results for the discordant individual and his father who carried the variant are shown in figure 4. To calculate the likelihood that a pedigree could have appeared the way it does by chance, we calculated each family’s logarithm of odds (LOD) score, using the software package Statistical Analysis for Genetic Epidemiology.*** A LOD score is the base 10 logarithm of the odds that a pedigree’s inheritance pattern occurred by chance recombination. For example, a LOD score of 3 indicates a 1 in 103 (or 1,000) odds of seeing the observed cosegregation of the trait and marker by chance. In order to perform these calculations, certain assumptions were made regarding the disease model: the mode of inheritance is autosomal dominant, the minor allele frequency for each disease allele is 1 × 10−6, penetrance is 0.99, and the sporadic rate of disease in noncarriers is 1 × 10−4. Although we had to make an assumption of disease allele frequency, the LOD scores were robust to variations in this frequency. In addition, positive MH status was indicated by either MH equivocal or MH susceptible IVCT results, in accordance with the European MH Group IVCT protocol. The LOD score was 1.32 for Family A, which corresponds to an odds of approximately 1 in 20. The LOD scores for Family B and Family C were much higher, at 2.52 and 2.12, which correspond to odds of less than 1 in 100. These high LOD scores are strong evidence that the variants we found are the causal variants or cosegregate with the causal variants in these families. The LOD score for Family G was much lower at 0.61, which is only a 1 in 4 odds. This is due to the fact that the three genotyped MH equivocal and susceptible individuals appeared within only one branch of the family, rendering the MH negative individuals in the other branches uninformative.
Fig. 1.
Original Sanger sequencingtraces of relevant ryanodine receptor 1 mutations.
Polymerase chain reactionelectropherograms showing sequencing traces from direct sequencing studies of muscle-derived complementary DNA in probands from three families. The base calls are those generated automatically by the software, and the arrow indicates the position of the erroneous homozygousreference base call. A) Trace for Family B proband; B) Trace for Family C proband; C) Trace for Family G proband.Adenine = A; cytosine = C; guanine = G; thymine = T.
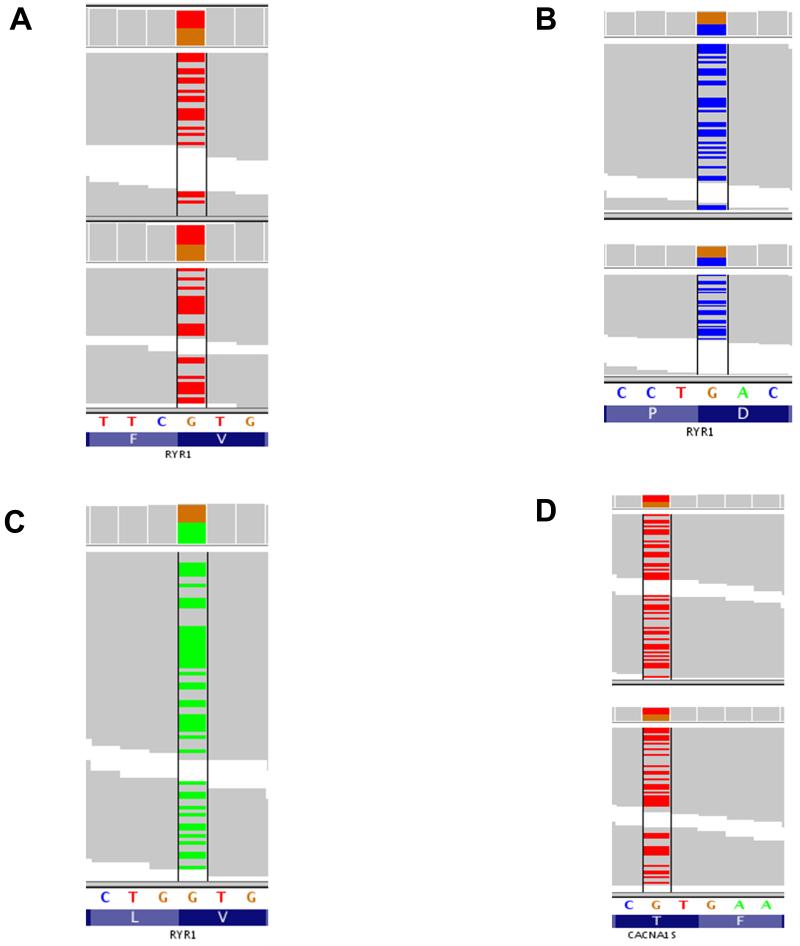
Fig. 2.
Rare variants found with exome sequencing.
In these Integrative Genomics Viewer figures, each column represents one position in the genome. The reference allele (which is color-coded) and amino acid is at the bottom, while colored bars above indicate the called allele for each read. Gray bar = reference allele; colored bar = alternate allele. The bar at the top of each column also shows the called alleles, with the height of each color being proportional to the number of reads at that position, which in each case were all >19x. A) Rare variant in two malignant hyperthermia susceptible subjects from family A at chromosome 19, position 39055674. B) Variant in twomalignant hyperthermia susceptible subjects from Family B at chromosome 19, position 38957026; C) Variant in onemalignant hyperthermia susceptible subject from Family G at chromosome 19, position 38993563; D) Variant in twomalignant hyperthermia susceptible subjects from Family C at chromosome 1, position201031099.
Adenine = A;aspartic acid = D;calcium channel, voltage-dependent, L type, α1S subunit gene = CACNA1S;cytosine = C; guanine = G;leucine = L;phenylalanine = F; proline = P;ryanodine receptor 1 gene = RYR1;threonine = T, blue background; thymine = T, white background; valine = V.
Fig. 3.
Follow-up confirmatory conventional sequencing trace.
Family A variant chromosome 19, position 39,055,674, complementary DNA position 12,700.Adenine = A; cytosine = C; guanine = G; thymine = T.
Fig. 4.
Pedigrees of all fourfamilies showing co-segregation of phenotype and genotype
These pedigrees are coded to indicate in vitro contracture test and mutation status. In all families except for C, all known genotypes were concordant with the phenotype. Circles represent females and squares males; diagonal line through symbol = deceased; diagonal arrows indicate clinical reaction. A) Family A; B) Family B; C) Family C. Here, one individual was discordant between genotype (variant +) and phenotype (MHN). For this individual and his MHE father the additional boxes provide details of in vitro contracture test and creatine kinase results: H = tension at 2% halothane; C = tension at 2 millimolar caffeine; R = onset time for ryanodine contracture test; CK = creatine kinase concentration in international units per liter (IU/L) (normal < 205); D) Family G.
malignant hyperthermia = MH; MH equivocal = MHE; MH negative = MHN; MH susceptible = MHS;negative = −ve; positive = +ve.
To provide more information regarding causal RYR1 variants, we analyzed all nonreference genotypes seen in 5,379 individuals of mixed ancestry from the ESP cohort. The ESP did not screen for MH status and this cohort can thus be viewed as a population sample. We found a total of 1,014 different RYR1 SNVs, 1,000 of which were in regions of high sequence quality (>7x fold coverage). Overall, 948 of these SNVs (94.8%) occurred at a frequency below 5% and 902 (90.2%) were below 1%. Among the low-frequency SNVs (<1%), 217 of 902 (24.1%) were synonymous, 312 (34.6%) were missense, 3 (0.3%) were nonsense, 2 (0.2%) were in putative splice sites, and 2 (0.2%) were in untranslated regions. Among all 317 deleterious SNVs (defined here as either missense, nonsense, or in a putative splice site) in RYR1, 127 of 317 (40.1%) were not previously reported in the dbSNP or 1,000 Genomes databases and of these 112 (35.3%) were seen in only 1 individual. Finally, if we look at this entire cohort, over a third (1,933/5,379 = 35.8%) had one or more potentially deleterious variants in RYR1. In summary, over a third of all people in our population sample had at least one protein-coding change in RYR1, and over a third of all protein-coding changes seen in RYR1 were rare and occurred in only one individual. This high number of rare protein-changing RYR1 variants in the population may be due to the large size of RYR1 (154 kb) and might suggest a relatively high de novo mutation rate. Given the low estimated incidence of MH, it follows that the majority of rare, missense RYR1 variants in the population are not causal for MH.
In order to help identify causal RYR1 variants, each variant’s conservation and protein prediction scores were compared. In order to facilitate the visual comparison, each score was transformed:
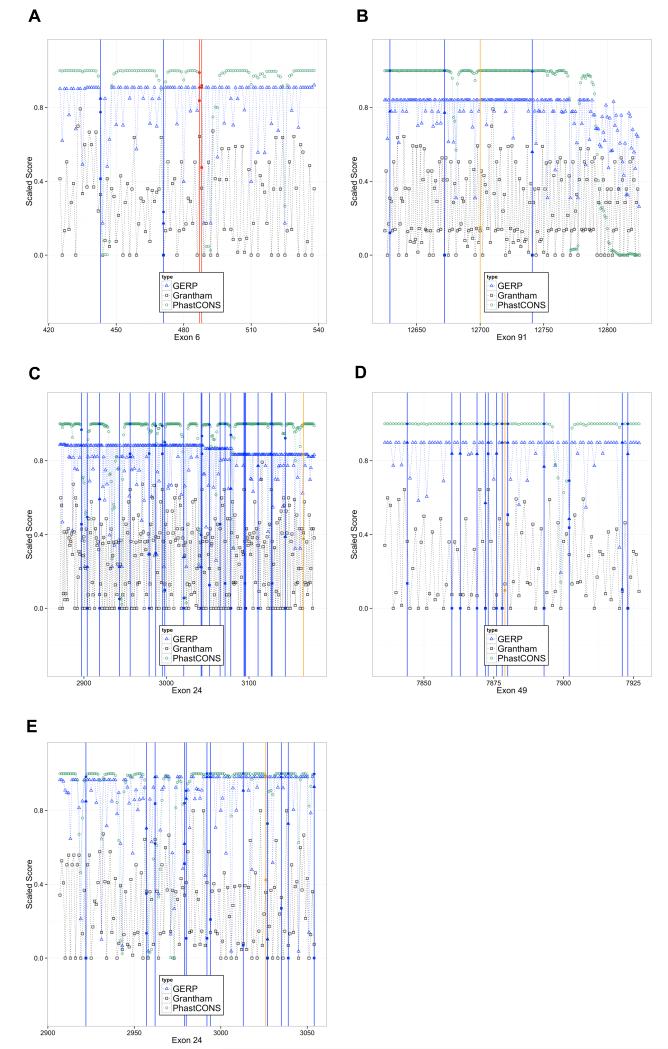
The result is a score, s, that is bounded by 0 and 1, with low conservation or benign protein prediction resulting in a score near 0 and high conservation or damaging protein prediction resulting in a score near 1. The results for RYR1 exons 6, 24, 49, and 91 and CACNA1S exon 24 are shown (fig. 5). RYR1 exon 6 demonstrates an idealized situation, where a known pathogenic variant in red has conservation and protein prediction scores that clearly distinguish it from the ESP exome variants in blue. However, in general, there was no clear cutoff value that could distinguish between known pathogenic and presumed benign, common variants. The pathogenic variants tend to have a distribution of conservation scores skewed towards the higher end, but the common variants have scores that span the range (fig. 6).
Fig. 5.
Gene conservation scores graphed for RYR1 and CACNA1S exons
GERP++, Grantham, and PhastCons scores are displayed for four RYR1 exons and oneCACNA1S exon. All scores were scaled to fit within a 0-1 scale on the vertical axis for ease of viewing. Blue vertical lines mark positions were variants were seen in 5,379 UW ESP exomes, red vertical lines mark an EMHG causal variant, and orange lines mark a rare variant found in one of four families in this study. A) RYR1 exon 6, showing two EMHG pathogenic variants and two UW ESP variants; B) RYR1 exon 91 which has the Family A variant; C) RYR1 exon 24 which has the Family B variant; D) RYR1 exon 49 which has the Family G variant; E) CACNA1S exon 24, which has the Family C variant.
Calcium channel, voltage-dependent, L type, α1S subunit gene= CACNA1S;European Malignant Hyperthermia Group = EMHG; Exome Sequencing Project = ESP; genomic evolutionary rate profiling = GERP; ryanodine receptor 1 gene= RYR1; University of Washington = UW.
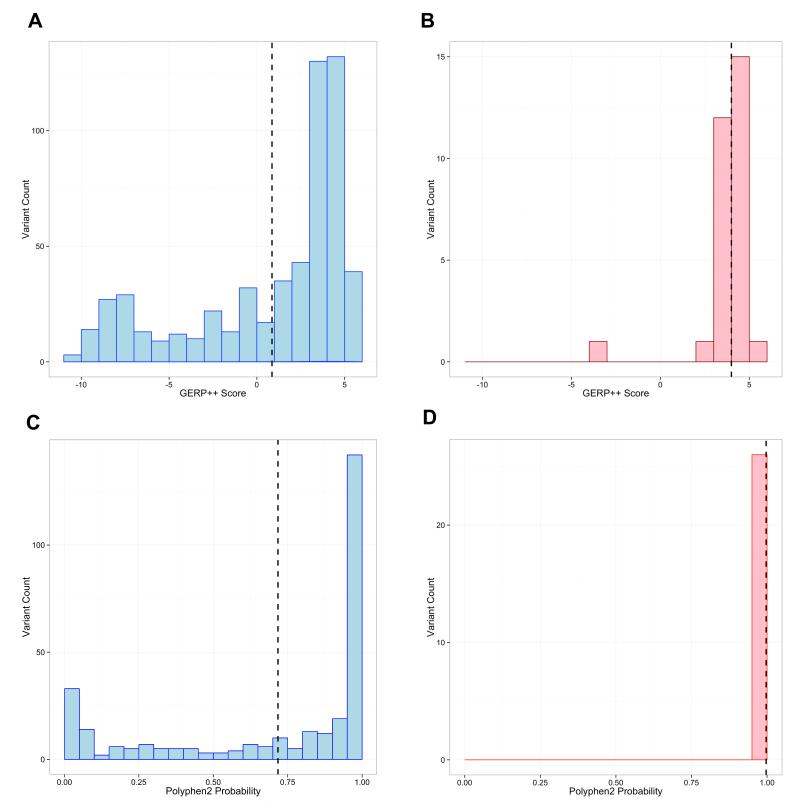
Fig. 6.
Histograms of GERP++ (conservation) and Polyphen2(prediction) scores of RYR1 variants in the UW ESP and EMHG data sets.
Each of these panels graphs the indicated conservation score for variants in RYR1 from either a subset of the 5,379 UW ESP exomes or those labeled pathogenic by the EMHG.A) GERP++ scores for thenonintronic UW ESP exome variants with MAF < 1%. Vertical dashed line indicates the mean score (1.12) of these 535 variants; B) GERP++ scores for the 30 EMHG pathogenic variants. Vertical dashed line indicates the mean score,3.99; C) PolyPhen2 scores for missense UW ESP variants with MAF < 1%.The vertical dashed line is the mean score (0.726) of these 317 variants; D) PolyPhen2 scores for the 30 EMHG pathogenic variants.The vertical dashed line is the mean score, 0.997.European Malignant Hyperthermia Group = EMHG; Exome Sequencing Project = ESP; genomic evolutionary rate profiling = GERP; minor allele frequency = MAF; ryanodine receptor 1 gene = RYR1; University of Washington = UW.
The distribution of prediction scores for known pathogenic variants versus population variants occurring at a frequency of ≤1% across the 5379 ESP exomes for GERP++ and PolyPhen2 2.2.2 scores is shown in figure 6. The GERP++ scores ranged from −10.70 to 5.84 with the average being 0.17 for all variants, and 1.12 for nonintronic variants. While the ESP variants tend to have a more uniform distribution centered near 0, the 30 European Malignant Hyperthermia Group pathogenic variants cluster at the higher end. These latter pathogenic variants had a much higher average of 3.99 GERP++ as well. The scores ranged from −3.09 to 5.08. Unfortunately, each of these plots demonstrates the difficulty in choosing a cut-off for any one of these scores as evidence of pathogenicity. In order to assess the use of all four scores evaluated in this study (GERP++, PhastCons, Grantham, PolyPhen2), we modeled the relationship between the conservation scores of the ESP and pathogenic variants using logistic regression. Since the lowest GERP++ score was the only negative score and a clear outlier, we performed our analysis with and without that variant and the results were identical. Case/control status was the dependent variable, with GERP++, PhastCons, Grantham, and Polyphen2 scores being the predictor variables. In the case of Polyphen2, when a mutation fell in multiple transcripts and the score differed, the maximum score was chosen. The regression equation was unable to predict with high sensitivity the likelihood of pathogenicity for either one of the confirmed mutations, or one of the newly discovered variants (data not shown).
We also examined the coding sequences of CACNA1S in the ESP exomes. In all, there were 478 SNVs, of which 474 were at high-quality sites. Again, a high proportion of these were uncommon and rare, with 434 of 474 (91.6%) ESP variants below 1%. Among those below 1% in frequency, 83 of 434 (19.1%) were synonymous, 152 (35.0%) were missense, 4 (0.9%) were nonsense, and 1 (0.2%) was predicted to be in a splice site, thus resulting in 157 rare, deleterious variants. Sixty-four of 157 (38.8%) were not seen in either the dbSNP or 1,000 Genomes, of which 54 (32.7%) were seen in only one individual. Finally, the most-striking finding in the ESP cohort was that 5,124 out of 5,379 individuals (95.6%) had one or more deleterious variants in CACNA1S. This is much higher than the proportion seen in RYR1 (35.8%), which may be unexpected since CACNA1S is half the size of RYR1 (73kb vs. 154kb).
Moving beyond variant counts and gene size comparisons, we turned next to calculating the average genetic diversity (pi) per base pair for RYR1 and CACNA1S across both populations included in the ESP dataset.37 This would help understand how diverse these two genes are compared to the rest of the genome, and whether we would expect to see this much variation in these genes. When compared to all other genes, we found that both CACNA1S and RYR1 are more diverse than the average gene in either population. Specifically, CACNA1S is more diverse than 76.8% of genes in African-Americans, and more diverse than 82.8% of genes in European-Americans; RYR1 was slightly less diverse but exhibited a similar pattern (more diverse than 78.8% of genes in African-Americans, 74.0% of genes in European-Americans). These data suggest that both of these genes can be expected to have a large amount of natural variation, which likely does not contribute to MH considering the rarity of the disease.
Discussion
In our search for novel mutations contributing to MH in four families from the United Kingdom, we detected three rare and likely pathogenic variants in RYR1 and 1 in CACNA1S. Each of these variants were missed by conventional, automated cDNA Sanger sequencing methods. Sanger sequencing these two genes in their entirely (using either cDNA or genomic DNA) is difficult because they are large (154kb and 73kb) and many times the length of the average human gene (~10-15kb). Also, the finding of a missense mutation on its own cannot be assumed to be causal. In our sample of 5,379 control exomes, we found 902 nonreference variants in RYR1 and 434 in CACNA1S that were below 1% in frequency, nearly all of which are presumed to be noncausal for MH. While the variants found in our families were novel and had not been seen in over 5,000 exomes or in variant databases, the same could be said for 112 RYR1 and 54 CACNA1S variants seen in the ESP dataset. However, the frequency data allowed us to quickly narrow our search to just one variant in each family. The subsequent finding of perfect or near-perfect segregation in each family confirmed our suspicions and provided strong evidence for their role in MH in these families.
The variants in RYR1 and CACNA1S could have been missed by Sanger sequencing the cDNA for a variety of reasons. First, Sanger sequencing for disorders that may be transmitted as dominant traits such as MH require accurate calling two bases at one position to call a mutation and even the best software available can sometimes miss a variant, which was the case in two of our families. In each of these cases, the traces at these newly variant positions were ambiguous enough to trigger a homozygous-reference genotype call by the software (Gap4). Upon closer examination of these traces, the minor allele trace could be seen, but was missed in the automated analysis. In Sanger sequencing, each individual base position is only covered by one or two sequencing reactions, and the 154kb gene and 15k cDNA length is large enough that the expense would limits the coverage. Therefore, calls could easily be missed from one or two poor or noisy sequencing reactions. Meanwhile, next-generation exome sequencing usually results in dozens if not hundreds of copies of DNA at any particular base position. This redundancy provides more consistent and confident calls, which are also less prone to errors of human interpretation. In addition, both real and artifactual differences may occur between the genomic DNA and cDNA sequences. Errors may be introduced by the poor fidelity of viral reverse transcriptase, which is an error-prone step introduced in generating cDNA from muscle tissue mRNA since splice variants or stop codons that lead to nonsense mediated decay could influence the amplification of both alleles from the mRNA. There is also the possibility that there are true differences between the genomic DNA and its mRNA product, but this is controversial.38,39 Schiemann and colleagues have applied next-generation sequencing to successfully uncover potentially pathogenic RYR1 variants in an MH family, but chose to target 32 candidate genes for reasons of feasibility, with cost presumably being the primary one. With costs of whole-exome and whole-genome sequencing decreasing in half every several years, this technology will be within reach for many research and clinical laboratories very soon. It will likely become influential in MH research and diagnosis, but the quality of the informatics and statistical analysis will help determine the success of its application.
In addition to successfully finding rare variants in RYR1 and CACNA1S in MH families, next-generation sequencing also has the valuable benefit of discovering variants in new genes not currently known to be involved in MH pathogenesis. Without a more complete knowledge of the involved genes and the characteristics of their variants, an effective and sensitive genetic screening test for MH is impossible. Eventually, developing an inexpensive and routine genetic screening test could save thousands of patients and families from invasive studies and unanticipated MH events. Toward this end, we conducted a genome-wide search and an in-depth analysis of protein prediction and sequence conservation scores in RYR1. However, the variability of these scores among the 30 confirmed pathogenic mutations was too great, and our attempts to find a deterministic algorithm to distinguish pathogenic from nonpathogenic variants were unsuccessful. Since RYR1 is one of the largest genes in the genome, and since both RYR1 and CACNA1S have a per-base genetic diversity that is in the top quartile of all genes, many nonpathogenic protein-coding variants can be expected in the population. To date, only 30 pathogenic mutations have been verified, but as more are added to this list, our prediction algorithms might improve over time. In addition, we have not yet modeled the strength of muscle contracture for the known pathogenic variants, which could enhance sensitivity. Further study into creating useful algorithms to predict a given variant’s pathogenicity is warranted.
Another issue that deserves attention is that of multiple protein isoforms. Many genes like RYR1 result in more than one transcript, and the protein prediction scores differ by transcript. Many of the 30 known pathogenic variants have >90% PolyPhen2 prediction of pathogenicity for one protein isoform, but not for the other. More basic research needs to be done on the role each transcript plays and the impact a variant has on each of them.
Finally, there are several unique advantages and disadvantages to using exome and next-generation sequencing. The main advantages are the breadth and depth of the data. For a relatively low price, all known protein-coding regions of the genome are included, and each region is sequenced in often 50-100 fold redundancy. Thus, if a patient with MH does not have an RYR1 or CACNA1S mutation, a search for a variant in another gene could be undertaken immediately. Since we and others40 have seen that the average individual has roughly 10,000 protein-coding nonreference variants, additional methods must be used to significantly reduce the target region for study, which could include standard linkage analysis or more complex inherited by descent sharing analysis.41 However, a limitation of this method is that more complex structural variation, such as copy-number changes (deletions, duplications), large indels (insertion or deletion of multiple bases), or epigenetic changes can be missed. If these were present they would require other approaches, such as array comparative genomic hybridization for copy-number changes or bisulfite sequencing for epigenetic changes.
Conclusion
In this study of four MH families, exome sequencing discovered novel coding variants in either RYR1 or CACNA1S in each of four families. Supporting evidence for their pathogenicity include the variants’ absence in population variant databases, absence in a sample of 5,379 exome sequences, and segregation with MH in our pedigrees. They were clearly present on exome sequencing, but were missed by automated Sanger cDNA sequencing. As whole exome or whole genome next-generation sequencing becomes increasingly available, its use on genomic DNA for MH research and diagnostics should be considered. Our data demonstrate the superior sensitivity of next-generation sequencing compared to cDNA Sanger sequencing, and the effectiveness of leveraging allele frequencies from exome sequence data to screen potentially pathogenic variants.
MS #201301054 – Final Boxed Summary Statement.
What we already know about this topic:
Known variants in the genes for the ryanodine receptor and a calcium channel are associated with about half of the cases of malignant hyperthermia
Problems with conventional DNA sequencing can miss certain mutations that can be detected using newer methods
What this article tells us that is new:
Exome sequencing of DNAs from four pedigrees associated with malignant hyperthermia identified novel genetic variant in each
The increased sensitivity of next-generation sequencing combined with allele frequency data is a powerful approach to identify rare variants associated with malignant hyperthermia
Acknowledgements
We would like to thank the Family Studies Project Team for their comparative data and the National Heart, Lung, and Blood Institute (Bethesda, Maryland) Grand Opportunity Exome Sequencing Project and its ongoing studies that produced and provided exome variant calls for comparison: the Lung Grand Opportunity Project (HL-102923), the WHI Sequencing Project (HL-102924), the Heart Grand Opportunity Project (HL-103010), the Broad Institute (HL-102925). Some of the results of this paper were obtained by using the software package Statistical Analysis for Genetic Epidemiology, which was supported by a U.S. Public Health Service Resource Grant (RR03655) from the National Center for Research Resources (Bethsda, Maryland).
Disclosure of Funding sources: Funding provided by the National Institutes of Health (Bethesda, Maryland): National Institute of General Medical Sciences (T32 GM086270: J.H.K.; P01 GM099568: B.L.B), National Center for Advancing Translational Sciences (KL2 TR000421: J.H.K.), National Institute of Arthritis, Musculoskeletal and Skin Diseases (2P01 AR05235: P.M.H), National Heart, Lung and Blood Institute Northwest Genomics Center Sequencing Project (1RC2 HL102926: D.A.N., M.J.R.), and National Human Genome Research Institute (R01 HG005701: B.L.B) Next Generation Mendelian Genetics Project (1RC2 HG005608: D.A.N.). Funding was also provided by an award from the Washington Life Sciences Discovery Fund (Seattle, Washington) to the Northwest Genomics Center (0905001: D.A.N) and the Northwest Institute for Genetic Medicine (2065508: G.P.J., J.H.K.).
Footnotes
http://www.emhg.org/documents, last accessed March 5, 2013.
http://bio-bwa.sourceforge.net/, last accessed March 5, 2013.
http://www.broadinstitute.org/gatk/, last accessed March 5, 2013.
http://www.broadinstitute.org/igv/, last accessed on March 5, 2013.
http://snp.gs.washington.edu/SeattleSeqAnnotation134/, last accessed on March 5, 2013.
http://mendel.stanford.edu/SidowLab/downloads/GERP/hg19.GERP_scores.tar.gz, last accessed on March 5, 2013.
http://genetics.bwh.harvard.edu/pph2/bgi.shtml, last accessed on March 5, 2013.
http://staden.sourceforge.net/, last accessed March 5, 2013.
http://darwin.cwru.edu/sage/, Statistical Analysis for Genetic Epidemiology 6.3 [2012], last accessed May 31, 2013.
Work Attributed to: Department of Anesthesiology & Pain Medicine, University of Washington, Seattle, Washington
The authors declare no competing interests.
Meetings Presented: Some of the data in this paper were presented at the European Malignant Hyperthermia Group Meeting, Nijmegen, Netherlands, on June 6, 2011, and the Annual Meeting of the American Society of Anesthesiologists, Chicago, Illinois, on October 19, 2011.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jerry H. Kim, Department of Anesthesiology & Pain Medicine, University of Washington.
Gail P. Jarvik, Department of Medicine, Division of Medical Genetics, University of Washington.
Brian L. Browning, Department of Medicine, Division of Medical Genetics, University of Washington.
Ramakrishnan Rajagopalan, Department of Medicine, Division of Medical Genetics, University of Washington.
Adam S. Gordon, Department of Genome Sciences, University of Washington.
Mark J. Rieder, Department of Genome Sciences, University of Washington.
Peggy D. Robertson, Department of Genome Sciences, University of Washington.
Deborah A. Nickerson, Department of Genome Sciences, University of Washington.
Nickla A. Fisher, Leeds Institute of Molecular Medicine, Leeds, United Kingdom.
Philip M. Hopkins, Leeds Institute of Molecular Medicine, Leeds, United Kingdom.
References
- 1.Ording H. Incidence of malignant hyperthermia in Denmark. Anesth Analg. 1985;64:700–4. [PubMed] [Google Scholar]
- 2.Brady JE, Sun LS, Rosenberg H, Li G. Prevalence of malignant hyperthermia due to anesthesia in New York State, 2001-2005. Anesth Analg. 2009;109:1162–6. doi: 10.1213/ane.0b013e3181ac1548. [DOI] [PubMed] [Google Scholar]
- 3.Denborough MA, Lovell RR. Anaesthetic deaths in a family. Lancet. 1960;276:45. doi: 10.1093/bja/34.6.395. [DOI] [PubMed] [Google Scholar]
- 4.Davies W, Harbitz I, Fries R, Stranzinger G, Hauge JG. Porcine malignant hyperthermia carrier detection and chromosomal assignment using a linked probe. Anim Genet. 1988;19:203–12. doi: 10.1111/j.1365-2052.1988.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 5.MacLennan DH, Duff C, Zorzato F, Fujii J, Phillips M, Korneluk RG, Frodis W, Britt BA, Worton RG. Ryanodine receptor gene is a candidate for predisposition to malignant hyperthermia. Nature. 1990;343:559–61. doi: 10.1038/343559a0. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter D, Ringrose C, Leo V, Morris A, Robinson RL, Halsall PJ, Hopkins PM, Shaw M-A. The role of CACNA1S in predisposition to malignant hyperthermia. BMC Med Genet. 2009;10:104. doi: 10.1186/1471-2350-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson R, Carpenter D, Shaw MA, Halsall J, Hopkins P. Mutations in RYR1 in malignant hyperthermia and central core disease. Hum Mutat. 2006;27:977–89. doi: 10.1002/humu.20356. [DOI] [PubMed] [Google Scholar]
- 8.Sambuughin N, Holley H, Muldoon S, Brandom BW, de Bantel AM, Tobin JR, Nelson TE, Goldfarb LG. Screening of the entire ryanodine receptor type 1 coding region for sequence variants associated with malignant hyperthermia susceptibility in the north american population. Anesthesiology. 2005;102:515–21. doi: 10.1097/00000542-200503000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Monnier N, Procaccio V, Stieglitz P, Lunardi J. Malignant-hyperthermia susceptibility is associated with a mutation of the α1-subunit of the human dihydropyridine-sensitive L-type voltage-dependent calcium-channel receptor in skeletal muscle. Am J Hum Genet. 1997;60:1316–25. doi: 10.1086/515454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudbrak R, Procaccio V, Klausnitzer M, Curran JL, Monsieurs K, van Broeckhoven C, Ellis R, Heyetens L, Hartung EJ, Kozak-Ribbens G. Mapping of a further malignant hyperthermia susceptibility locus to chromosome 3q13.1. Am J Hum Genet. 1995;56:684–91. [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson RL, Monnier N, Wolz W, Jung M, Reis A, Nuernberg G, Curran JL, Monsieurs K, Stieglitz P, Heytens L, Fricker R, van Broeckhoven C, Deufel T, Hopkins PM, Lunardi J, Mueller CR. A genome wide search for susceptibility loci in three European malignant hyperthermia pedigrees. Hum Mol Genet. 1997;6:953–61. doi: 10.1093/hmg/6.6.953. [DOI] [PubMed] [Google Scholar]
- 12.Iles DE, Lehmann-Horn F, Scherer SW, Tsui LC, Weghuis DO, Suijkerbuijk RF, Heytens L, Mikala G, Schwartz A, Ellis FR, Stewart AD, Deufel T, Wieringa B, Olde Weghuis D. Localization of the gene encoding the alpha 2/delta-subunits of the L-type voltage-dependent calcium channel to chromosome 7q and analysis of the segregation of flanking markers in malignant hyperthermia susceptible families. Hum Mol Genet. 1994;3:969–75. doi: 10.1093/hmg/3.6.969. [DOI] [PubMed] [Google Scholar]
- 13.Levitt RC, Olckers A, Meyers S, Fletcher JE, Rosenberg H, Isaacs H, Meyers DA. Evidence for the localization of a malignant hyperthermia susceptibility locus (MHS2) to human chromosome 17q. Genomics. 1992;14:562–6. doi: 10.1016/s0888-7543(05)80152-1. [DOI] [PubMed] [Google Scholar]
- 14.Katz RA, Skalka AM. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–43. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- 15.Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, Frazer K a. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiemann AH, Dürholt EM, Pollock N, Stowell KM. Sequence capture and massively parallel sequencing to detect mutations associated with malignant hyperthermia. Brit J Anaesth. 2012;110:122–7. doi: 10.1093/bja/aes341. [DOI] [PubMed] [Google Scholar]
- 17.Litman RS, Rosenberg H. Malignant hyperthermia: Update on susceptibility testing. JAMA. 2005;293:2918–24. doi: 10.1001/jama.293.23.2918. [DOI] [PubMed] [Google Scholar]
- 18.Carpenter D, Robinson RL, Quinnell RJ, Ringrose C, Hogg M, Casson F, Booms P, Iles DE, Halsall PJ, Steele DS, Shaw M-A, Hopkins PM. Genetic variation in RYR1 and malignant hyperthermia phenotypes. Brit J Anaesth. 2009;103:538–48. doi: 10.1093/bja/aep204. [DOI] [PubMed] [Google Scholar]
- 19.EMHG A protocol for the investigation of malignant hyperpyrexia (MH) susceptibility. The European Malignant Hyperpyrexia Group. Brit J Anaesth. 1984;56:1267–9. doi: 10.1093/bja/56.11.1267. [DOI] [PubMed] [Google Scholar]
- 20.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, Rieder MJ, Nickerson DA, Bernier R, Fisher SE, Shendure J, Eichler EE. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP, Interests CF. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2009;42:30–5. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, Mcmillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A, Program NCS. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–13. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 33.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Core Team . In: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, editor. Vienna; Austria: 2012. [Google Scholar]
- 35.Tammaro A, Martino A Di, Bracco A, Cozzolino S, Savoia G, Andria B, Cannavo A, Spagnuolo M, Piluso G, Aurino S, Nigro V. Novel missense mutations and unexpected multiple changes of RYR1 gene in 75 malignant hyperthermia families. Clin Genet. 2011;79:438–47. doi: 10.1111/j.1399-0004.2010.01493.x. [DOI] [PubMed] [Google Scholar]
- 36.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–30. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 37.Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Akey JM. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2012;493:216–20. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–8. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pickrell JK, Gilad Y, Pritchard JK. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng SB, Nickerson DA, Bamshad MJ, Shendure J. Massively parallel sequencing and rare disease. Hum Mol Genet. 2010;19:R119–24. doi: 10.1093/hmg/ddq390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browning BL, Browning SR. A fast, powerful method for detecting identity by descent. Am J Hum Genet. 2011;88:173–82. doi: 10.1016/j.ajhg.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]