Abstract
Polysialic acid (polySia) is a unique linear homopolymer of α2,8-linked sialic acid that has been studied extensively as a posttranslational modification of neural cell adhesion molecule in the central nervous system. Only two proteins are known to be polysialylated in cells of the immune system: CD56 on human natural killer cells and murine bone marrow (BM) leukocytes, and neuropilin-2 (NRP-2) on dendritic cells (DCs). We tested the hypothesis that polySia expression is regulated during maturation and migration of leukocytes and plays a role in functional activity. Using wild-type and NCAM−/− mice, we show that BM neutrophils express only polysialylated CD56, whereas a subset of BM monocytes expresses polysialylated CD56 and/or another polysialylated protein(s). We demonstrate that polysialylated CD56 expression is progressively down-regulated in wild-type monocytes and monocyte-derived cells during migration from BM through peripheral blood to pulmonary and peritoneal sites of inflammation. Freshly isolated monocyte-derived peritoneal macrophages are devoid of polySia yet re-express polySia on NRP-2 and an additional protein(s) after maintenance in culture. Removal of polySia from these cells enhances phagocytosis of Klebsiella pneumoniae, suggesting that down-regulation of polySia on macrophages facilitates bacterial clearance. Using wild-type and NRP-2−/− mice, we demonstrate that NRP-2 and an additional protein(s) are polysialylated by ST8 SiaIV in BM-derived DCs. We conclude that polySia expression in monocyte-derived cells is dynamically regulated by ST8 SiaIV activity and by expression of carrier proteins during recruitment to sites of inflammation and influences cellular interactions with microbes, contributing to innate and adaptive immune responses.
Keywords: macrophages, NCAM−/− and NRP-2−/− mice, neuropilin-2, phagocytosis, polysialic acid
Introduction
Changes in glycosylation of cell surface proteins modulate cellular activity. Given its terminal location on glycans and its hydrophilic and electronegative features, sialic acid (N-acetylneuraminic acid) plays an important role in regulating cellular interactions with ligands, microbes and neighboring cells and in controlling cellular activation, differentiation, transformation and migration (Schauer 2009; Miyagi and Yamaguchi 2012; Varki and Gagneux 2012). Cell surface glycoconjugates on diverse mammalian cells commonly are terminated with monomers of sialic acid that are α2-3 and α2-6 linked to penultimate residues of galactose. In contrast, polysialic acid (polySia), linear homopolymers of α2-8-linked sialic acid with a degree of polymerization greater than seven residues, thus far has been identified as a posttranslational modification on only a small number of mammalian proteins, mostly associated with cells of the central nervous system.
Originally described as a capsular carbohydrate of group B Neisseria meningitides and neuroinvasive K1-encapsulated Escherichia coli, polySia has been identified on only seven mammalian proteins: neural cell adhesion molecule (NCAM/CD56), synaptic cell adhesion molecule (SynCAM 1), neuropilin 2 (NRP-2), CD36, α-subunit of the voltage-gated sodium channel and the two sialyltransferases that polymerize polySia (ST8 SiaII and ST8 SiaIV) (Finne 1982; Zuber et al. 1992; Muhlenhoff et al. 1996; Close and Colley 1998; Yabe et al. 2003; Curreli et al. 2007; Galuska et al. 2010). Studied extensively in the developing central nervous system, polySia modification of NCAM plays a key role in cell migration, axonal guidance, synapse formation and functional plasticity by sterically preventing inappropriate homo- and heterophilic cellular interactions (Bruses and Rutishauser 2001; Bonfanti 2006). Heavily polysialylated in specific neurons during early ontogeny, NCAM loses its polySia content in the mature nervous system, except in a few nuclei that retain plasticity (Finne 1982; Rieger et al. 1985; Bruses and Rutishauser 2001; Bonfanti 2006). The polySia component of NCAM has also been reported to bind soluble neurotrophic growth factor (Kanato et al. 2008) and Siglec 11 (sialic acid-binding Ig superfamily lectin) (Wang and Neumann 2010), suggesting an additional role for polySia as a recognition site for ligand and receptor binding. Another protein (SynCAM 1) found in the central nervous system has recently been shown to be polysialylated (Galuska et al. 2010). As demonstrated for NCAM, the polySia moiety influences SynCAM 1-mediated homo- and heterophilic interactions (Galuska et al. 2010).
In light of numerous analogies between cells of the neuronal and immune systems, expression of the large electronegative polySia glycan on leukocytes is expected to modulate immune cell interactions with soluble and cell-associated ligands and thus, potentially regulate cell activation, differentiation and migration. In fact, we have described the presence of polySia on human dendritic cells (DCs) (Curreli et al. 2007) as a posttranslational modification of NRP-2, a receptor for semaphorin and vascular endothelial growth factor families in neurons and endothelial cells, respectively. We demonstrated that removal of polySia from the surface of DCs promotes DC-induced activation and proliferation of T lymphocytes (Curreli et al. 2007). Two recent reports showed that the polySia moiety on NRP-2 expressed on the surface of DCs binds CCL21, a ligand for the CCR7 chemokine receptor, and influences DC migration (Rey-Gallardo et al. 2010, 2011). Recently, murine DCs were also shown to express polysialylated NRP-2 (Rollenhagen et al. 2013). That polySia expression might similarly influence the function of other myeloid cells is suggested by our finding that human monocytes also express polySia on a protein other than NRP-2 that remains to be identified (Curreli et al. 2007) and by a report showing that polySia is expressed by myeloid cells in murine bone marrow (BM), with CD56 reported to be the polySia carrier on these cells (Drake et al. 2008). The importance of polySia in regulation of immune cell function is further suggested by impaired cellular immunity against tumor progression and by altered contact hypersensitivity observed in polysialyltransferase ST8 SiaIV-deficient mice (Drake et al. 2008).
In this report, we demonstrate that myeloid cell activation and differentiation that occur during cell recruitment and migration during an inflammatory response are associated with stage-specific polysialylation of select proteins. We show that murine BM neutrophils express polysialylated CD56 alone, whereas a subset of BM monocytes expresses polysialylated CD56 and/or another polysialylated protein(s). Furthermore, there is progressive loss of polysialylated CD56, partly mediated by protein cleavage, from the surface of monocytes and neutrophils as they migrate from BM through peripheral blood (PB) and to pulmonary and peritoneal sites of inflammation. We demonstrate that peritoneal macrophages can be induced to express polySia on NRP-2 and on an additional yet to be identified protein(s), but that polysialylation reduces the capacity of macrophages to phagocytize Klebsiella pneumoniae. Similarly, we show that a protein(s) in addition to NPR-2 is polysialylated in murine DCs. These data demonstrate that the dynamic regulation of polySia content on monocyte-derived cells is controlled by levels of expression of both ST8 SiaIV and carrier proteins and suggest that one potential role of the change in expression of polySia during monocyte maturation and migration is to help regulate the phagocytic activity of monocyte-derived cells during different stages of an immune response.
Results
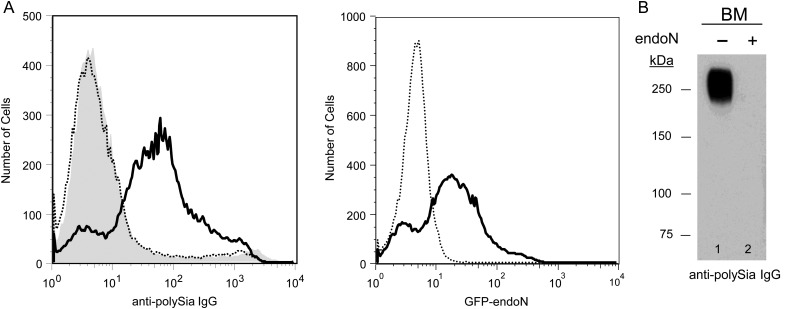
Polysia is expressed predominantly on monocytes/macrophages and neutrophils in murine bone marrow
We previously demonstrated that human monocyte-derived DCs express polysialylated NRP-2 (Curreli et al. 2007) but that a different protein(s) is modified by this unique glycan in monocytes. Subsequently, murine BM hematopoietic precursor and myeloid cells were also shown to express polySia, with CD56 the reported carrier protein (Drake et al. 2008). To characterize further the regulation of polySia expression on specific myeloid cell types during differentiation (Supplementary data, Figure S1), we analyzed murine BM cells for polySia content using two different reagents: anti-polySia mAb 735 (Frosch et al. 1985), used previously to identify polysialylated NRP-2 (Curreli et al. 2007), and a green fluorescent protein (GFP)-fused, inactive endoglycosidase N (GFP-endoN), that binds specifically to but does not cleave α2-8-linked polySia (Jokilammi et al. 2004). Both mAb 735 and GFP-endoN detect polySia on the surface of >60% of cells aspirated from murine BM (Figure 1A). On analysis by flow cytometry, the polySia-expressing cells comprise a heterogeneous but distinct group with higher side and forward scatter compared with cells without polySia (data not shown). In lysates prepared from BM cells, polySia is associated predominantly with an ∼270 kDa protein(s), with a molecular weight that is different from that of 130 kDa NRP-2 (Figure 1B). mRNA-encoding polysialyltransferases ST8SiaII and ST8SiaIV, the two enzymes responsible for synthesis of polySia, were detected in BM cells by RT-PCR analysis, albeit at relatively low copy number compared with DCs (data not shown). Thus, the presence of polySia on different proteins in murine BM cells and in human monocyte-derived DCs suggests that polySia is more widely expressed on cells of the immune system than previously appreciated and that the presence of polySia on specific proteins is regulated during myeloid cell differentiation, analogous to the developmentally controlled expression of polySia in the central nervous system (Finne 1982; Rieger et al. 1985; Bruses and Rutishauser 2001; Bonfanti 2006).
Fig. 1.
PolySia is present on the surface of a subset of murine bone marrow cells. Freshly isolated murine BM cells were stained with mAb 735 (left panel) or with GFP-endoN (right panel) and analyzed by flow cytometry (A); left panel: thin, dotted line—isotype IgG; bold line—mAb 735; right panel: dotted line—endoN-treated BM cells stained with GFP-endoN; bold line—mock-endoN-treated BM cells were stained with GFP-endoN. Cells that were depolysialylated by treatment with active endoN were incubated with mAb 735 (shaded region in left histogram) or exposed to GFP-endoN (right histogram) for use as controls. Five micrograms of protein from lysates of BM cells that were mock-treated (lane 1) or endoN-treated (lane 2) were separated by 8% SDS–PAGE and analyzed by immunoblot using mAb 735 (B).
To identify BM cells that express polySia, extracted BM cells were double-stained with polySia-specific GFP-endoN and a panel of hematopoietic lineage markers and analyzed by flow cytometry. PolySia was detected predominantly on the surface of neutrophils and monocytes/macrophages that express the myeloid lineage markers CD11b (Mac-1) and/or Ly6 G/C (Gr-1) (Figure 2). Two distinct populations of Ly6 G/C+ cells were found to be polysialylated. The Ly6 G/C+ cells of intermediate fluorescence intensity (Ly6 G/Cintermediate) correspond to BM monocytes/macrophages (Lagasse and Weissman 1996). This population of cells dimly expresses cell surface F4/80 as determined by flow cytometry, confirming the monocyte/macrophage commitment (data not shown). Monocytes/macrophages stain more brightly for polySia than BM neutrophils, the more intensely Ly6 G/C+ (Ly6G/Chigh)-expressing cells (Lagasse and Weissman 1996). PolySia was not detected on the surface of lymphocytes (recognized by cell surface CD3e and CD45R/B220) and erythroid cells (express TER-119) in the BM (Figure 2). These data were obtained with cells from C57/BL mice, and similar results were seen with cells from outbred mice (data not shown).
Fig. 2.
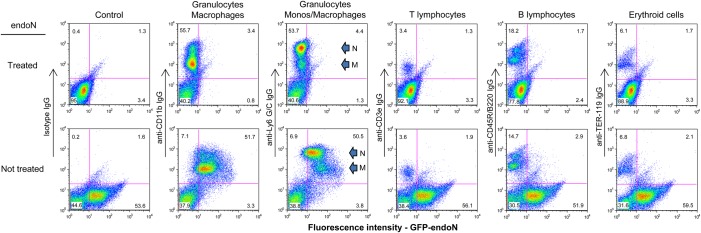
PolySia is expressed predominantly on myeloid cells in murine bone marrow. Freshly isolated murine BM cells were either exposed to endoN (top row) or mock-treated (bottom row), double-stained with GFP-endoN and an isotype control or various antibodies to hematopoietic lineage markers (CD11b, Ly6 G/C, CD3e, CD45R/B220 and TER-119), and analyzed by flow cytometry. In panels showing staining with Ly6 G/C, neutrophils (N) and monocytes/macrophages (M) are denoted by arrows.
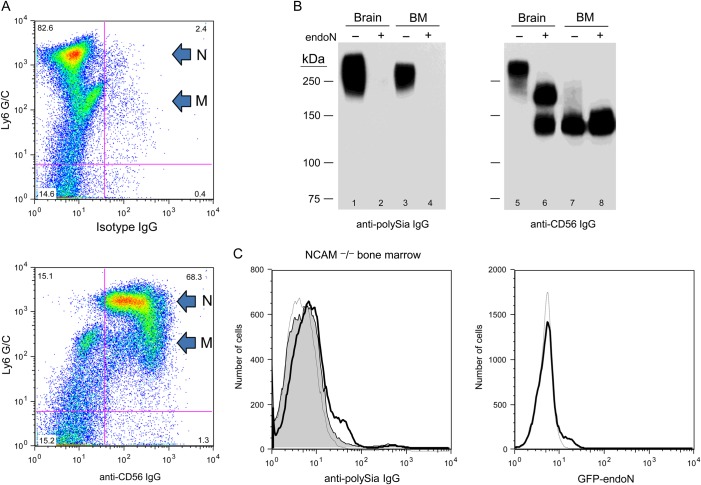
A small subset of CD56 expressed in myeloid cells in murine bone marrow is polysialylated
Although it was previously reported that CD56 is polysialylated in murine BM leukocytes (Drake et al. 2008), the polysialylated protein described in that study had a much smaller molecular weight (∼140 kDa) than the predominant ∼270 kDa polysialylated protein detected in our analysis (Figure 1), and the specific types of myeloid cells modified by polySia were not specified. We first identified the BM leukocytes that express CD56 and then evaluated whether CD56 in these cells is the ∼270 kDa polysialylated protein seen in Figure 1 using two methods; namely, by assessing the mobility shift of CD56 on immunoblot after treating cells with endoN and by analyzing the expression of polySia on the surface of BM leukocytes of NCAM−/− mice. By co-staining BM cells from wild-type mice with anti-Ly6 G/C and anti-CD56 IgGs, we found that most neutrophils (Ly6G/Chigh) and ∼50% of BM monocytes/macrophages (Ly6 G/Cintermediate) express CD56 (Figure 3A). Although the molecular weight of the major polysialylated protein from these BM cells is ∼270 kDa (Figures 1B and 3B, lane 3), the predominant form of CD56 in the same cell lysate migrated as a band at 140 kDa (Figure 3B, lane 7). A small amount of total CD56 was detected at a higher molecular weight, extending to the range of 250 kDa (Figure 3B, lane 7). After removal of polySia from the surface of these cells with endoN, though, the higher molecular weight forms of CD56 were not detected (Figure 3B, lanes 4 and 8), suggesting that only a small portion of CD56 is polysialylated in BM cells.
Fig. 3.
CD56 is the predominant polysialylated protein in bone marrow cells. Freshly isolated murine BM cells were co-stained with Ly6 G/C and isotype control (top panel) or anti-CD56 IgGs (bottom panel); neutrophils are indicated by an arrow labeled N and monocyte/macrophages are denoted by an M-labeled arrow (A). Protein from lysates of endoN-treated (lanes 2, 4, 6 and 8) and mock-treated (lanes 1, 3, 5 and 7) murine brain (lanes 1, 2, 5 and 6) and BM (lanes 3, 4, 7 and 8) cells were analyzed on immunblot using mAb 735 (lanes 1–4; 5 μg of protein from each sample) and anti-CD56 IgGs (lanes 5–8; 5 μg of protein was applied to lanes 5 and 6, 15 μg of protein was applied to lanes 7 and 8) (B). Bone marrow cells from NCAM−/− mice were stained with mAb 735 or with GFP-endoN and analyzed by flow cytometry (C); left panel, thin, dotted line—isotype IgG; bold line—BM cells stained with mAb 735; shaded region—endoN-treated BM cells stained with mAb 735; right panel, thin, dotted line—endoN-treated BM cells stained with GFP-endoN; bold line—mock-treated BM cells stained with GFP-endoN.
To confirm that polySia does not prevent binding of the anti-CD56 IgG used in our studies, we used the same antibody to analyze protein from lysates of untreated and endoN-treated brain cells on immunoblot. PolySia is expressed on NCAM/CD56 in these cells and migrates with a ∼270 kDa band (Figure 3B, lane 1). When protein from the same lysate of untreated brain cells is probed on immunoblot with anti-CD56 IgG, a broad ∼270 kDa protein is detected and converted into 180 and 140 kDa isoforms of NCAM/CD56 after endoN treatment (Figure 3B, lanes 5 and 6). These results demonstrate that the anti-CD56 IgGs bind to polysialylated CD56 and confirm that a very small amount of total CD56 on myeloid cells is decorated with polySia, in contrast to what occurs in neurons.
That CD56 is the predominant carrier of polySia on BM cells was corroborated by analysis of polySia expression in BM cells purified from NCAM−/− mice. The pattern of staining for polySia on BM cells from NCAM−/− mice was markedly different from that of wild-type mice. Whereas >60% of BM cells from wild-type mice express polySia as detected with GPF-endoN and mAb 735 (Figure 1), only ∼6% of cells from the BM of NCAM−/− mice express polySia (Figure 3C). Thus, although most myeloid cells in murine BM express polySia, and CD56 appears to be the predominant polysialylated protein in these cells, there is another carrier of polySia detected in BM cells from NCAM−/− mice.
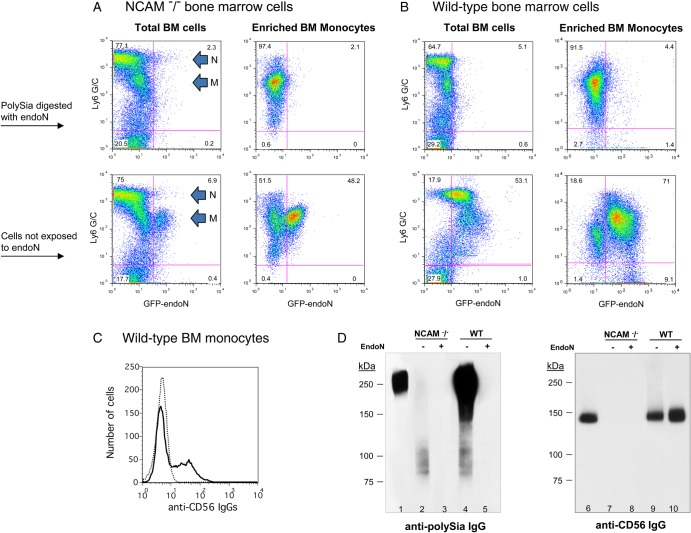
Bone marrow monocytes express polysialic acid on CD56 and another protein(s)
To determine whether CD56 and an additional protein are polysialylated in both BM monocytes and neutrophils, polySia expression on cells from NCAM−/− and wild-type mice was compared by flow cytometry and immunoblot. Neutrophils (Ly6G/Chigh) from the BM of NCAM−/− mice do not express polySia, as seen by identical staining patterns between untreated and endoN-treated neutrophils (Figure 4A, left top and bottom panels). This differs from the pattern of polySia expression in neutrophils from wild-type mice (Figure 4B, left top and bottom panels) and suggests that CD56 is the sole protein that is polysialylated in neutrophils. In contrast, the staining pattern of untreated- and endoN-treated monocytes from NCAM−/−and wild-type mice differed from that of neutrophils. Two major populations of monocytes (Ly6 G/Clow and Ly6 G/Cintermediate) were detected in wild-type mice, only one of which (Ly6 G/Cintermediate) is polysialylated (Figure 4B, left top and bottom panels). A subpopulation of monocytes (Ly6 G/Cintermediate) from NCAM−/− mice expresses polySia that was removed after exposure of cells to endoN (Figure 4A, left top and bottom panels). Thus the 6% of total BM cells from NCAM−/− mice that express polySia (Figure 3C) is comprised of a subset of monocytes.
Fig. 4.
Murine bone marrow monocytes display polySia on CD56 and on an additional protein(s). Unfractionated BM cells and enriched BM monocytes isolated from NCAM−/− (A) and wild-type (B) mice were either treated with endoN to remove cell surface polySia (top row) or mock-treated (lower row), were costained with anti-Ly6 G/C IgGs and GFP-endoN and were analyzed by flow cytometry (A and B). The Ly6 G/C staining pattern of neutrophils (N) and monocyte/macrophages (M) is indicated in the left panels of (A) and apply to (A) and (B). Purified monocytes were stained with anti-CD56 IgG (bold line) and with isotype control IgG (dotted line) and analyzed by flow cytometry (C). Five micrograms of protein from a lysate of unfractionated BM cells from wild-type mice (lane 1) or 20 μg of protein from lysates of enriched BM monocytes that had been exposed to endoN (lanes 3 and 5) or were mock-treated (lanes 2 and 4) from NCAM−/− (lanes 2 and 3) and wild-type (lanes 4 and 5) mice were evaluated on immunoblot using mAb 735 (left panel, D). Ten micrograms of protein from a lysate of unfractionated BM cells from wild-type mice (lane 6) or from lysates of enriched BM monocytes that had been exposed to endoN (lanes 8 and 10) or were mock-treated (lanes 7 and 9) from NCAM−/− (lanes 7 and 8) and wild-type (lanes 9 and 10) mice were evaluated on immunoblot using anti-CD56 IgGs (right panel, D).
To confirm these results, monocytes were enriched from BM cells of NCAM−/− and wild-type mice and were similarly analyzed on flow cytometry. Whereas >70% of monocytes from wild-type mice express polySia (Figure 4B, right top and bottom panels), ∼50% of purified monocytes (Ly6 G/Cintermediate) from NCAM−/− mice also express polySia (Figure 4A, right top and bottom panels). As only 40% of purified BM monocytes express cell surface CD56 (Figure 4C), a subpopulation of monocytes expresses only the additional polysialylated protein(s) and another subset of monocytes likely expresses multiple polysialylated proteins.
To characterize the additional polysialylated protein(s) in monocytes, proteins from lysates of NCAM−/− and wild-type monocytes were analyzed on immunoblot. By analyzing a larger amount of cell lysate than used in the immunoblots of Figures 1 and 2, a diffuse faint band with MW in the range of 80–105 kDa was detected using mAb735 (Figure 4D, lanes 2 and 4) that was not present in monocytes that were exposed to endoN (Figure 4D, lanes 3 and 5). In the monocyte lysate from wild-type mice, this 80–105 kDa protein(s) stained weakly compared with the predominant staining of CD56 (Figure 4D, lane 4). That this smaller molecular weight protein(s) was not a breakdown product of CD56 is shown by its presence in NCAM−/− mice (Figure 4D, lane 2). As seen in unfractionated BM cells (Figure 3B, lane 7), the predominant form of CD56 in monocytes has MW of 140 kDa (Figure 4D, lane 6) and only a small portion of CD56 is polysialylated in wild-type BM monocytes (Figure 4D, lanes 9 and 10). Thus, only a subpopulation of murine BM monocytes expresses polySia, and it is present on these cells as a modification of CD56 and/or an additional protein(s).
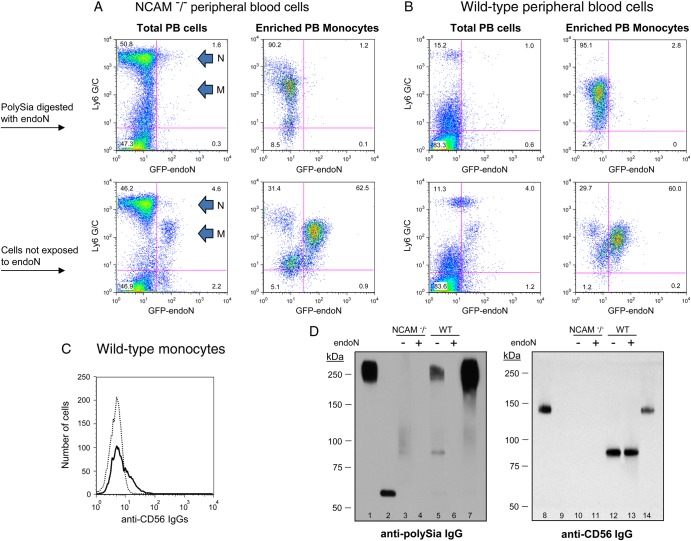
Egress of monocytes and neutrophils from bone marrow to peripheral blood is associated with reduced expression of polysialylated CD56
Recruitment of leukocytes to sites of infection is an essential step in host defense against microbial pathogens. To determine whether the content of polySia on the surface of monocytes and neutrophils changes during migration from BM to PB, a period of cell maturation that requires activation and increased deformability (Lichtman 1970), PB cells from NCAM−/− and wild-type mice were evaluated for polySia expression by flow cytometry as in Figure 4. A smaller percentage of PB neutrophils (Ly6 G/Chigh cells) compared with BM neutrophils from wild-type mice express polySia (Figures 4B vs. 5B, left lower panels). In contrast, the percentage of PB monocytes (Ly6 G/Cintermediate cells) from wild-type mice that express polySia is similar to that of BM monocytes (69 vs. 71%) (Figures 4B and 5B, left lower panels). Further analysis of enriched PB monocytes from NCAM−/− and wild-type mice, though, revealed a similar percentage (63 vs. 60%) of polysialylated cells (Figure 5A and B, right lower panels), suggesting that polysialylated CD56 contributes to less of the total polySia content of PB monocytes compared with BM monocytes. The fewer number of PB monocytes expressing CD56 (16%) compared with BM monocytes (40%) (Figures 5C vs. 4C) supports this finding.
Fig. 5.
Murine peripheral blood monocytes and neutrophils express less polysialylated CD56 than their bone marrow precursors. Unfractionated murine PB cells and enriched PB monocytes isolated from NCAM−/− (A) and wild-type (B) mice were either treated with endoN to remove cell surface polySia (top row) or mock-treated (lower row), were costained with anti-Ly6 G/C IgGs and GFP-endoN and were analyzed by flow cytometry (A and B). The Ly6 G/C staining pattern of neutrophils (N) and monocyte/macrophages (M) is indicated in the left panels of (A) and applies to (A) and (B). Purified monocytes were stained with anti-CD56 IgG (bold line) and with isotype control IgG (dotted line) and analyzed by flow cytometry (C). Five micrograms of protein from lysates of unfractionated BM (lane 1) or PB (lane 2) cells from wild-type mice or 10 μg of protein from lysates of enriched PB monocytes that had been exposed to endoN (lanes 4 and 6) or were mock-treated (lanes 3 and 5) from NCAM−/− (lanes 3 and 4) and wild-type (lanes 5 and 6) mice and from BM monocytes from wild-type mice (lane 7) were evaluated on immunoblot using mAb 735 (left panel, D). Ten micrograms of protein from lysates of these same samples were analyzed on a separate immunoblot that was probed with anti-CD56 IgGs (right panel, D): unfractionated BM (lane 8) and PB (lane 9) cells; NCAM−/− PB monocytes without (lane 10) or with (lane 11) endoN digestion; PB monocytes from wild-type mice before (lane 12) or after (lane 13) endoN digestion; BM monocytes from wild-type mice (lane 14).
The decreased expression of polysialylated CD56 in PB monocytes was confirmed by immunoblot analysis of protein from lysates of BM and PB cells. Whereas polysialylated CD56 (∼270 kDa) is a prominent band from lysates of purified BM monocytes probed with mAb 735 (Figure 5D, lane 7), a markedly less intense band with this molecular weight is detected in lysates from PB monocytes (Figure 5D, lane 5). This reduction in amount of CD56 in PB monocytes is associated with the loss of the 140 kDa form of CD56 and the appearance of a new 85 kDa band detected with the anti-CD56 IgGs (Figure 5D, lanes 12 and 13). Thus, not only is the expression of polysialylated CD56 reduced in PB monocytes compared with BM monocytes, but the molecular weight of the protein is reduced, suggesting protein cleavage during migration of monocytes from the BM to the peripheral circulation. Enriched PB monocytes from wild-type and NCAM−/− mice also reveal a faint polysialylated protein(s) in the range of 80–105 kDa (Figure 5D, lanes 3 and 5), as seen in BM monocytes (Figure 4D, lanes 2 and 4), but the amount of protein is not markedly different in BM and PB monocytes.
Cells in the PB contain less polySia in general compared with BM cells (Figure 5D, lanes 1 and 2). This likely reflects the relatively few myeloid cells in the PB compared with BM. An ∼60 kDa polysialylated protein is detected in PB leukocytes from wild-type mice, though (Figure 5D, lane 2), the identity of which remains to be determined.
Further loss of polySia during tissue migration of pulmonary and peritoneal monocytes/macrophages and neutrophils
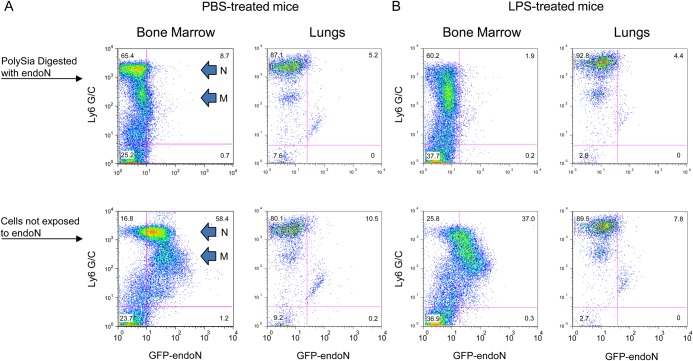
To determine whether migration of myeloid cells from the PB to tissue sites of inflammation is associated with further changes of cell surface polySia, we evaluated the polySia content of alveolar macrophages and neutrophils obtained after intratracheal injection of LPS and of peritoneal macrophages induced by injection of thioglycollate. Most of the cells obtained by bronchial alveolar lavage of control and LPS-treated wild-type mice stained intensely with Ly6 G/C (Figure 6A and B, right top and bottom panels) and had the general morphologic features of neutrophils when observed by light microscopy. An additional population of cells with intermediate Ly6 G/C staining intensity is also seen by flow cytometry, representing macrophages (Figure 6A and B, right top and bottom panels). Neither population of cells from bronchial alveolar lavage of control mice expresses polySia as seen by similar staining patterns with GFP-endoN in cells not exposed (Figure 6A, bottom right panel) and exposed (Figure 6A, top right panel) to active endoN. Intratracheal injection of LPS leads to a marked increase in the number of pulmonary neutrophils 24 h after inoculation (data not shown). Despite the induced mobilization of BM precursors under this condition, alveolar neutrophils and macrophages in LPS-treated mice also do not express polySia on the cell surface (Figure 6B, right top and bottom panels). The absence of polySia in these cell populations contrasts with the polysialylation of the respective BM precursor cells from the same mice (Figure 6A and B, left panels).
Fig. 6.
Pulmonary neutrophils and macrophages at sites of inflammation in mice express no cell surface polySia. Mice were injected intratracheally with PBS as control (A) or with 5 μg of LPS (B), sacrificed after 24 h and cells were collected from the bronchial alveolar washing and BM and analyzed by flow cytometry after staining with Ly6 G/C and GFP-endoN. Cells were exposed to endoN (top row) or mock-treated (lower row) prior to staining. The Ly6 G/C staining pattern of neutrophils (N) and monocyte/macrophages (M) is indicated in the left panels of (A) and applies to (A) and (B).
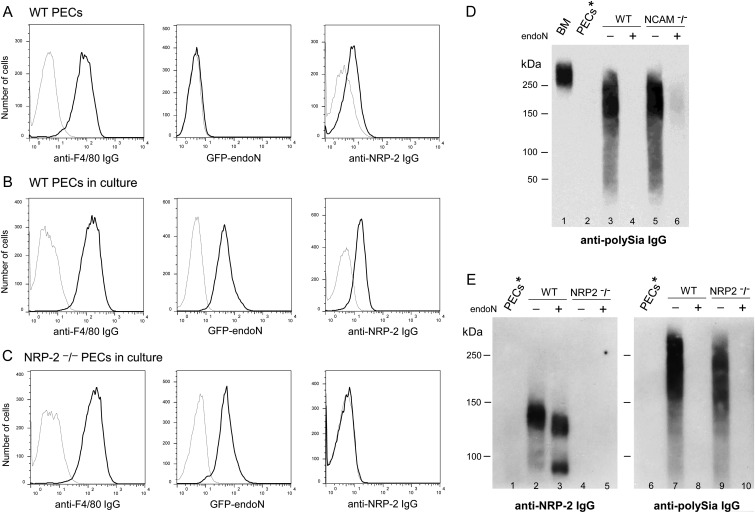
Loss of polySia on differentiating monocytes/macrophages in vivo was also demonstrated using a murine model of peritonitis. PB monocytes can be induced to migrate to the peritoneum and to differentiate into macrophages (peritoneal exudate cells or PECs) in response to intraperitoneal injection of thioglycollate (Gallily et al. 1964). After harvesting peritoneal cells and enriching for macrophages by adherence to tissue culture plates, >98% of adherent cells express the macrophage marker F4/80 (Figure 7A, left panel). These cells also express other phenotypic surface markers for murine macrophages such as CD11b, CD14, Ly6 G/C and CD86 (Supplementary data, Figure S2). A small percentage of these freshly isolated PECs express CD11c (Supplementary data, Figure S2). As seen with alveolar neutrophils and macrophages, PECs do not express polySia on the cell surface as determined by flow cytometry after staining with GFP-endoN (Figure 7A, middle panel) nor is polySia detected in cell lysates probed on immunoblot with mAb 735 (Figure 7D, lane 2 and E, lane 6). Thus, migration of monocytes/macrophages and neutrophils from the BM into the circulation and then to pulmonary and peritoneal sites of inflammation is associated with the progressive loss of cell surface polySia.
Fig. 7.
Peritoneal macrophages lose expression of polySia on the cell surface but can re-express polySia as a modification of NRP-2 and another protein(s). Peritoneal macrophages were isolated 72 h after injection of thioglycollate into the peritoneal cavity of wild-type, NCAM−/− and NRP-2−/− mice. Freshly-isolated PECs from wild-type mice were evaluated by flow cytometry after staining with anti-F4/80 and -NRP-2 IgGs and with GFP-endoN: thin lines—isotype IgGs or cells that were treated with endoN prior to staining with GFP-endoN, bold line—staining of cells with specific IgGs or GFP-endoN (A). After isolation from the peritoneal cavity, PECs were grown in culture in RPMI/10% FCS for 24 h, were harvested and were stained with F4/80 and NRP-2 IgGs and with GFP-endoN (B: cultured PECs from wild-type mice; C: cultured PECs from NRP-2−/− mice). Immunoblot of proteins from lysates of unfractionated BM cells (5 μg, lane 1), freshly-isolated peritoneal macrophages (25 μg, lane 2), macrophages grown in culture for an additional 24 h from wild-type (5 μg, lanes 3 and 4) or NCAM−/− (5 μg, lanes 5 and 6) mice without (lane 3 and 5) or with (lane 4 and 6) digestion with endoN probed for polySia expression with mAb735 (D). Immunoblots of proteins from lysates of freshly-isolated peritoneal macrophages (30 μg, lanes 1 and 6), macrophages grown in culture for an additional 24 h from wild-type (30 μg, lanes 2 and 3; 5 μg, lanes 7 and 8) or NRP-2−/− (30 μg, lanes 4 and 5; 5 μg, lanes 9 and 10) mice without (lanes 2, 4, 7 and 9) or with (lanes 3, 5, 8 and 10) digestion with endoN probed with antibody to NRP-2 (lanes 1–5) or to polySia (lanes 6–10) (E).
To determine whether peritoneal macrophages retain the capacity to repolysialylate the cell surface, PECs were maintained in culture in RPMI/10% FCS for 24 h to return macrophages to a more quiescent state of activation and then were re-evaluated for expression of polySia. Maintaining peritoneal macrophages in culture under these conditions leads to continued high cell surface expression of F4/80 (Figure 7B, left panel) and CD11b (Supplementary data, Figure S2). Cultured PECs express less CD14 and Ly6G/G, an increased amount of the DC marker CD11c and an unchanged level CD86 (Supplementary data, Figure S2). Cell surface expression of polySia is markedly up-regulated in PECs under these conditions (Figure 7B, middle panel). This up-regulation of polySia expression was confirmed by immunoblot analysis of protein from a lysate of cultured PECs that was probed with mAb 735 and reveals a diffuse pattern of staining with a center of intensity in the 150–250 kDa range (Figure 7D, lane 3) that is not present in endoN-treated cells (Figure 7D, lane 4). That CD56 is not a carrier of polySia in these cells is shown by an equivalent staining pattern in a lysate from cultured PECs purified from NCAM−/− mice (Figure 7D, lanes 5 and 6).
To determine whether NRP-2, a protein capable of being polysialylated in DCs is expressed in these cells and is polysialylated, PECs from WT mice were evaluated for polysialylated NRP-2 by flow cytometry and immunoblot. Neuropilin 2 is expressed at a low level in freshly isolated PECs (Figure 7A, right panel and E, lane 1) but is markedly up-regulated in cultured PECs (Figure 7B, right panel and E, lane 2). Treatment of cultured PECs with active endoN prior to analysis on immunoblot removes cell surface polySia (Figure 7E, lane 7 vs. 8) and leads to a faster migrating form of NRP-2 (Figure 7E, lane 3), demonstrating that NRP-2 is polysialylated in these cells.
To determine if NRP-2 is the sole carrier of polySia in cultured PECs, cells from NRP-2−/− mice were similarly analyzed. PECs from NRP-2−/− mice do not express detectable NRP-2 (Figure 7C, right panel and 7E, lane 4), yet they express a significant amount of polySia that migrates with a protein(s) in the same molecular weight range as seen in PECs from wild-type mice (Figure 7E, lanes 7 and 9). Thus, maturation of monocytes and macrophages and migration to tissue sites of inflammation lead to complete loss of polySia, yet these cells can be induced to express NRP-2 and have the capacity to polysialylate it and an additional protein(s) that remains to be identified.
Murine bone marrow-derived dendritic cells express polySia on neuropilin-2 and another protein(s)
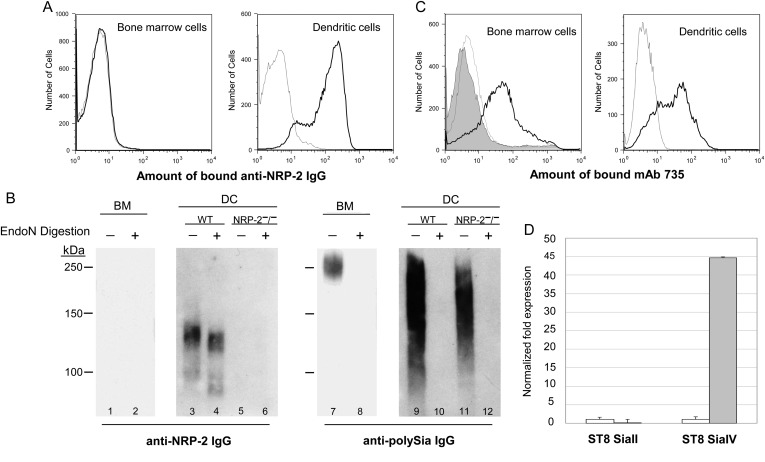
Human and murine DCs express polysialylated NRP-2 (Curreli et al. 2007; Rollenhagen et al. 2013). To determine whether polysialylated NRP-2 is synthesized de novo during differentiation of murine BM monocytes to DCs, as occurs in maturing human monocyte-derived DCs (Curreli et al. 2007), the expression of NRP-2 and polySia in these cells was determined by flow cytometry and immunoblot. NRP-2 is not expressed by any cells in murine BM (Figure 8A, left panel and B, lane 1). In contrast, most murine BM-derived DCs express surface NRP-2 (Figure 8A, right panel) that migrates as a diffuse band centered at ∼130 kDa on immunoblot (Figure 8B, lane 3). DCs also express polySia on the cell surface as seen by a heterogeneous pattern of staining with polySia-specific mAb 735 on flow cytometry (Figure 8C, right panel) and a diffuse 125–250 kDa band from a lysate of DCs on immunoblot (Figure 8B, lane 9). That NRP-2 is polysialylated is shown by a shift in mobility of the ∼130 kDa NRP-2 when cells are depolysialylated by treatment with endoN prior to analysis (Figure 8B, lanes 3 vs. 4 and 9 vs. 10). The expression of polySia by DCs is supported by the marked up-regulation of ST8 SiaIV in DCs compared with total BM cells (Figure 8D). Thus, as we demonstrated previously in human DCs, murine BM-derived DCs express polysialylated NRP-2 de novo on the cell surface during differentiation from BM precursor cells. BM cells also express polySia (Figures 1A and B and 8C, left panel and B, lanes 7 and 8), but as a modification of CD56 and another protein(s) that remains to be identified.
Fig. 8.
Murine bone marrow-derived DCs express polysialylated NRP-2. Freshly isolated murine BM cells and BM-derived DCs were stained with anti-NRP-2 IgG and analyzed by flow cytometry (A): thin, dotted line—isotype IgG; bold line—anti-NRP-2 IgG. Lysates of BM cells from wild-type mice (lanes 1, 2, 7 and 8) or DCs from wild-type (lanes 3, 4, 9 and 10) or NRP-2−/− (lanes 5, 6, 11 and 12) mice that were mock treated (lanes 1, 3, 5, 7, 9, 11) or endoN treated (lanes 2, 4, 6, 8, 10 and 12) were separated by 8% SDS–PAGE and analyzed by immunoblot using anti-NRP-2 IgG (left panel) or mAb 735 (right panel) (B). Bone marrow cells and BM-derived DCs were stained with anti-polySia mAb 735 and analyzed by flow cytometry (C): thin, dotted line—isotype IgG, shaded region—endoN-treated BM cells stained with mAb 735; bold line—mock-endoN-treated cells stained with mAb 735. The relative amount of RNA encoding ST8 SiaII and ST8 SiaIV in BM cells (open bars) and DCs (shaded bars) was determined by semi-quantitative PCR (D). The amount of RNA encoding each enzyme in BM cells was normalized to one and the fold change was in relation to the change in expression of 18S rRNA.
As a protein in addition to NRP-2 is modified by polySia in cultured PECs, we used DCs from NRP-2−/− mice to determine whether NRP-2 is the sole carrier of polySia in DCs. As expected, no NRP-2 was detected in DCs from NRP-2−/− mice (Figure 8B, lane 5). In spite of the absence of NRP-2, a diffuse endoN-sensitive band, spanning the same molecular weight range as in DCs from WT mice, was detected on immunoblot from a lysate of these cells (Figure 8B, lanes 11 and 12). Thus, NRP-2 and an additional protein(s) are polysialylated in both cultured PECs and DCs.
Loss of polySia from surface of peritoneal macrophages enhances phagocytosis
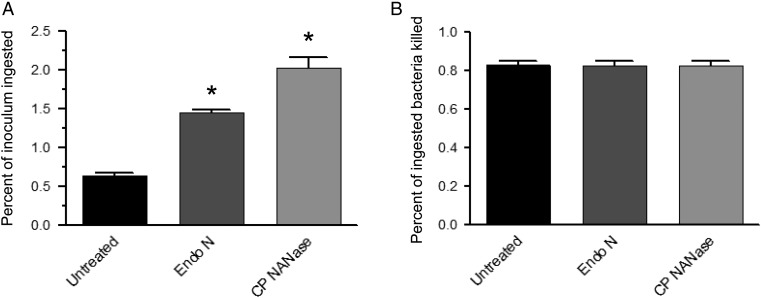
As removal of monomeric sialic acid from the surface of murine macrophages and DCs was shown to enhance phagocytosis of bacteria (Seyrantepe et al. 2010; Cabral et al. 2013), we determined whether changes in the polySia content of macrophages also plays a role in uptake and killing of bacterial pathogens. Polysialylated murine peritoneal macrophages were treated in vitro with endoN and with bacterial sialidase (Clostridium perfringens neuraminidase, CP NANase; an exoglycosidase that cleaves terminal α2-3- and α2-6-linked sialic acid from penultimate galactose residues) and were evaluated for their capacity to phagocytize and kill K. pneumoniae. As expected, removal of monomeric forms of sialic acid with CP NANase from the macrophage surface enhanced phagocytosis compared with untreated cells (Figure 9A). Similarly, removal of only the polySia form of sialic acid from the surface of macrophages enhanced phagocytosis >2-fold (Figure 9A). Despite increasing bacterial uptake, neither treatment with endoN nor with CP NANase affected the efficiency of killing ingested bacteria (Figure 9B). These data suggest that the loss of polySia from monocytes as they mature into macrophages in vivo during recruitment to inflammatory sites plays an important role in optimizing the ability of the macrophages to clear microorganisms.
Fig. 9.
Removal of polySia from the surface of peritoneal macrophages enhances phagocytosis of K. pneumoniae. Mouse peritoneal macrophages (0.5 × 105/well) were prepared as indicated above and were left untreated or were treated with endoN or 100 mU/mL bacterial neuraminidase (CP NANase) for 1 h at 37°C before being exposed to 1 × 108 cfu/well of K. pneumoniae. Macrophages were collected and lysed 1 h (A, phagocytosis assay) or 3.5 h (B, bacteriocidal efficiency) after exposure to the bacteria. The number of intracellular bacteria at each time point was quantitated as described in Materials and Methods. Phagocytosis values (A) are presented as percentage of cfu per well divided by input number of cfu. Bacterial killing values (B) were determined by subtracting the percentage of cfu remaining after the additional 2.5 h incubation divided by the initial cfu phagocytosed (value after 1 h) from 1. Statistical significance compared with untreated cells (*P < 0.0001) is shown by asterisks and was determined by a two-tailed t-test using GraphPad Prism 5 software. Data represent mean ± SEM from three wells for each condition and are representative of five different experiments.
Discussion
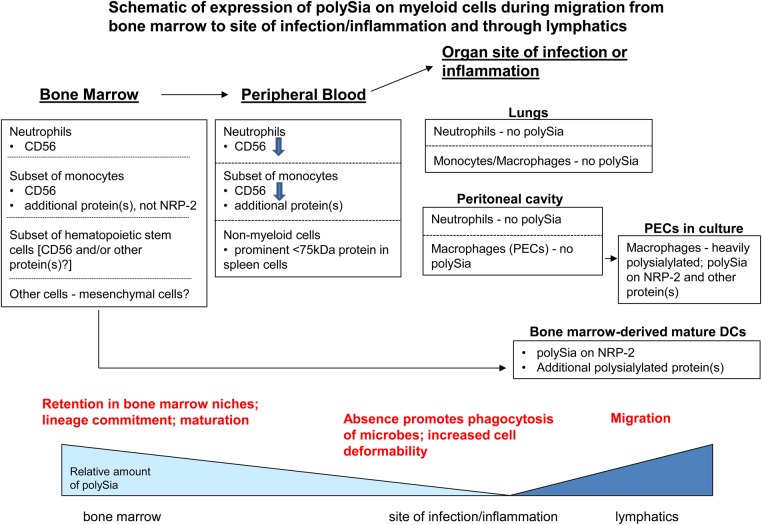
PolySia is a unique glycan modification of a limited number of mammalian proteins that carries great potential for regulating cellular activity. We are the first to show in this report that polysialylation of murine monocytes and monocyte-derived macrophages and DCs is dynamically regulated during an inflammatory response (Figure 10). Murine BM-derived DCs express polysialylated NRP-2 on the cell surface (Rollenhagen et al. 2013), as do human DCs (Curreli et al. 2007). Using DCs from NRP-2−/− mice, we show here that a protein(s) in addition to NRP-2 is polysialylated in these cells. Murine BM monocytes that differentiate into DCs also express surface polySia but, in contrast to DCs, do not express NRP-2 and display polySia on CD56 and another protein(s) that remains to be identified. PolySia is shed from the surface of monocytes by cleavage of CD56 as they are released from BM into the PB during early stages of monocyte differentiation. Further migration to a pulmonary site of inflammation is associated with complete loss of polySia from monocyte-derived cells, as well as from neutrophils. Thioglycollate-induced peritoneal macrophages, that also are derived from BM monocytes, also are devoid of polySia. When placed in culture though, these cells re-express polySia on NRP-2 and on an additional cell surface protein(s) other than CD56 that remains to be identified. The findings in this report of changes in polysialylation of murine myeloid cells at different stages of maturation both in vivo and in vitro (Figure 10) parallel our results with human monocytes and monocyte-derived DCs (Curreli et al. 2007) and macrophages (unpublished results), where different stages of monocyte differentiation are associated with the dynamic modulation of polySia on the cell surface.
Fig. 10.
Schematic of changes in polySia content and modified proteins of myeloid cells during differentiation of myeloid precursor cells from recruitment from BM to migration through lymphatics. Bold print above bottom scale—potential functions of polySia during various stages of myeloid cell maturation while in different anatomic compartments.
These data demonstrate that expression of polySia in monocytes is down-regulated during an early stage of immune responses (e.g. during recruitment from BM to site of inflammation/infection) and then re-expressed on monocyte-derived cells that reflect a later stage of the immune response (e.g. after encounter with microbe and during migration through lymphatics) (Figure 10). It is possible, though, that polysialylated macrophages and DCs are derived from precursors that do not express polySia. We have identified two populations of BM monocytes by the presence (high Ly6G/C cells) or absence (low Ly6G/C cells) of polySia. It will be of interest to determine whether the specific macrophage and DC progenitor cell (Fogg et al. 2006) in the BM matures into cells in one or both of these populations as we have found that only a subset of hematopoietic precursor cells in the BM is polysialylated (unpublished results). Expression of polySia on monocytes might also be a useful way to discriminate cells on functional grounds. Murine monocytes have been described as “classical” inflammatory cells (high Ly6G/C) that are rapidly recruited to sites of inflammation and “non-classical” resident monocytes (low Ly6G/C) that persist for extended periods in blood and normal tissue (Strauss-Ayali et al. 2007). Only the former subset is polysialylated, and polySia on these cells may contribute to their distinct functional characteristics.
PolySia was previously shown to be expressed on murine BM cells differentiating along a myeloid lineage (Drake et al. 2008). In this report, we characterize the pattern of polySia expression more specifically on BM monocytes, macrophages and neutrophils. Most BM neutrophils express polySia and CD56 is the sole carrier of polySia in those cells. In contrast, only a subset of BM monocytes is polysialylated, with CD56 the carrier on only some of those cells and an additional polySia carrier that remains to be identified on others. Given the nearly exclusive expression of polySia on myeloid cells in murine BM, it will be of interest to determine whether this glycan plays a role in lineage commitment. In addition, as murine NK cells do not express polySia (Drake et al. 2008), whereas human NK cells do, it will be important to establish whether this pattern of exclusive myeloid cell polySia expression extends to human BM cells.
PolySia has been a useful handle for identifying two proteins (NCAM/CD56 and NRP-2) in monocytes and monocyte-derived cells, respectively, that were previously recognized as “neuronal proteins” that are involved in cell migration and cell–cell interactions. We have also found that another protein (i.e. SynCAM 1), that is polysialylated in a subset of NG+ progenitor cells in the brain and plays a role in homophilic–heterophilic cell interactions, is also expressed in monocytes and derived cells, but its polysialylation status and function in these cells remain unclear (unpublished results). Do these polysialylatable proteins play similar roles in the migration and effector functions of monocytes, macrophages and DCs as they do in neurons? Does the polySia modification regulate the function of these particular proteins or are these proteins selected by the cell as carriers for polySia to achieve a more general cellular function? We have already shown that polySia on human DCs affects their ability to activate lymphocytes in a mixed lymphocyte reaction (Curreli et al. 2007). It has also been shown that polySia on NRP-2 enhances attraction of the CCR7 chemokine ligand CCL21, thus influencing DCs migration to lymph nodes (Rey-Gallardo et al. 2010, 2011). In both cases, though a direct role of polySia on NRP-2 activity (i.e., binding of semaphorins and signaling through plexin co-receptors) has yet to be shown. That polySia influences cell function independently of an effect on NRP-2 is supported by a report that the migration of T lymphocytes that do not express NRP-2 (unpublished results) to the thymus is influenced by polySia (Drake et al. 2009). As this report did not demonstrate directly that murine lymphocytes express polySia and we have shown that murine BM lymphocytes do not express polySia, it is possible that the effect of polySia on lymphocyte migration is related to polysialylation of other cells with which lymphocytes interact (e.g. endothelial cells).
It is possible that polySia assumes a different role on the surface of myeloid cells at various stages of maturation. For instance, the presence of this bulky, electronegative glycan on early monocytes in BM may prevent cell–cell interactions that are needed for cell maturation and may be involved in retention of progenitor cells in specific BM niches. Alternatively, polySia could facilitate cell detachment from BM stroma to initiate migration from BM and/or may serve to attract growth factors or cytokines (e.g. macrophage colony-stimulating factor, M-CSF) that are needed for cell maturation, as has been demonstrated for nerve growth factor (Kanato et al. 2008). It will be of interest to determine whether the loss of polySia from BM monocytes and neutrophils promotes migration through BM endothelium by changing cell deformability and plasticity. This concept is supported by the loss of total sialic acid from the surface of neutrophils as they migrate from the BM (Lichtman 1970). It is also possible that polySia modification of different proteins on the same cell may have different roles.
We demonstrate in this report that the phagocytic activity of macrophages is enhanced by removal of cell surface polySia. This finding adds to the growing list of potential functions of this unique glycan in diverse cells. The loss of polySia during maturation of monocytes into peritoneal and pulmonary macrophages is physiologically relevant as enhanced phagocytosis can lead to improved microbial clearance at sites of infection (Figure 10). Although the mechanism for the enhanced uptake of live K. pneumoniae is unclear, possible explanations include unmasking of specific cell surface pathogen receptors or nonspecific removal of repulsive negative charge on the cell surface that promote microbe–cell interactions and/or activation of intracellular signaling pathways. The effect on bacterial phagocytosis that is mediated by cell surface polySia appears to be related to polysialylated moieties that are independent of glycans modified by monomeric sialic acid that also control phagocytosis (Seyrantepe et al. 2010; Cabral et al. 2013) (CP NANase used under the conditions of our study does not efficiently cleave α2-8-linked polySia; unpublished results), and it will be of interest to understand the mechanism(s) governing each. Just as enhanced macrophage phagocytosis is important at sites of infection, macrophages must also have the capacity to down-regulate phagocytosis to proceed through the next stages of an inflammatory response (Figure 10), and addition of polySia may be one such mechanism.
The macrophages that were used for the phagocytosis studies were derived from freshly isolated PECs that were devoid of polySia at time of harvest but that expressed large amounts of surface polySia after maintenance in culture. It is common practice by investigators to maintain freshly isolated, thioglycollate-induced PECs in short-term culture prior to further work with these cells. This brief culture period is intended to return these activated macrophages to a more quiescent state (unpublished communication from Dr. Stefanie Vogel). It is of interest that these cells express more CD11c (DC phenotypic marker) and less CD14 and Ly6 G/C (monocytes/macrophage phenotypic marker) while up-regulating the expression of NRP-2 and polySia. The pattern of expression of these proteins resembles that of mature DCs (Curreli et al. 2007; Rollenhagen et al. 2013), exposing the phenotypic and functional plasticity of macrophages. Further characterization of these cells will determine whether ST8 SiaIV is also up-regulated in these cells. The in vivo relevance of these cultured macrophages with this phenotype remains to be determined by analysis of macrophages in vivo as they mature while migrating from a site of inflammation or infection through the draining lymphatic system.
The limited number of mammalian proteins known to be modified by polySia suggests that the expression and activity of polysialyltransferases ST8 SiaII and ST8 SiaIV are tightly controlled. Indeed, specific amino acid sequences in the first fibronectin type III repeat and in the Ig5 domain of NCAM are necessary for binding of ST8 SiaIV and for subsequent addition of polySia to N-linked glycans in the Ig5 domain (Close et al. 2003; Thompson et al. 2013). To date, similar sequences have not been identified in the other polysialylated proteins. In addition, polySia is known to be O-linked to NRP-2 (Curreli et al. 2007; Rollenhagen et al. 2013), rather than N-linked as it is to NCAM. Although a specific cluster of mucin-type O-glycans has been identified as a site for in vitro polysialylation of human NRP-2, factors controlling recognition and polysialylation of glycans at this site in NRP-2 remain to be determined. Polysialylation of different proteins could also be influenced by posttranslational modifications of ST8 SiaII and ST8 SiaIV. As both proteins are capable of being polysialylated (Muhlenhoff et al. 1996; Close and Colley 1998), it will be of interest to determine whether activity and protein recognition are influenced by the extent of polySia on these enzymes.
Although addition of polySia is known to occur by action of ST8 SiaII and ST8 SiaIV, little is known related to how polySia is down-regulated in eukaryotic cells. Clearly, the bacteriophage endoN cleaves polySia effectively (Finne and Makela 1985; Rutishauser et al. 1985), but an equivalent eukaryotic enzyme has not yet been identified. In forced expression systems, the mammalian exosialidase Neu4 is able to degrade polySia from NCAM in neurons (Takahashi et al. 2012), but the physiological relevance of this finding is unclear. We have found that the protein scaffold of CD56 is cleaved into an 85 kDa cell-associated protein as monocytes migrate from BM to PB and this is associated with loss of polySia. Thus, proteolysis of CD56 resulting in release of the polysialylated domain may be one mechanism for regulated loss of polySia from this protein. There is precedent for this concept. Regulated metalloprotease-induced ectodomain shedding of NCAM, resulting in a 110–115 kDa fragment, has been described (Diestel et al. 2005; Hinkle et al. 2006). More recently, it was shown that activated murine pulmonary epithelial cells release a metalloprotease truncated, polysialylated 110 kDa portion of NCAM, with a proposed role in neutralizing the cytotoxic effect of “neutrophil extracellular traps” during infection (Ulm et al. 2013). The larger size of the cell-associated portion of NCAM in our studies raises the possibility that the 180 kDa isoform of NCAM, in addition to NCAM-140, is polysialylated in murine myeloid cells.
The presence of polySia on cells of the immune system has implications for the development of vaccines. The polySia component of the Neisseria meningitidis group B capsule and of the surface of malignant cells is the target of efforts to develop therapeutic vaccines. In the case of the meningococcal group B vaccine, there has been concern that potential cross-reactivity of generated antibodies with polySia on NCAM in the CNS would be deleterious to the host (Finne et al. 1983). The presence of polySia on cells in the immune system would increase concern over more systemic binding of Abs generated by these experimental vaccines, requiring a careful analysis of the antigenicity of polySia on these cells.
Our work shows that the expression of the unique glycan polySia is more widespread in mammalian cells than was previously appreciated and supports the potentially significant role for polySia and its carrier proteins in the immune system (Curreli et al. 2007; Drake et al. 2008, 2009; Bax et al. 2009; Rey-Gallardo et al. 2010, 2011). As the details of the role of polySia modification of NCAM/CD56 and NRP-2 in immune cell function are unraveled, we expect to identify additional proteins whose expression was previously unrecognized in monocytes and monocyte-derived macrophages and DCs that likely control critical steps in the immune response. The murine system appears to be a good model to study the changing expression of polySia on myeloid cells during maturation as both human and murine DCs express polysialylated NRP-2 de novo and monocytes from both express polySia on a protein other than NRP-2. The availability of NCAM, NRP-2, ST8 SiaII and ST8 SiaIV knock-out mice will provide excellent models to assess the impact of polysialylated proteins during an immune response to viral and bacterial pathogens.
Materials and methods
Preparation of murine cells
BM cells were harvested from the femurs of 8- to 12-week-old C57/BL wild type, NCAM−/− (Cremer et al. 1994) and NRP-2−/− (Giger et al. 2000) mice and were used as a source of monocytes, purified by negative selection using an EasySep® Mouse Monocyte Enrichment Kit (Stem Cell Technologies, Vancouver, BC, Canada), or to generate mature DCs as described previously (Stamatos et al. 2010). Wild-type and NCAM−/− mice were purchased from Jackson Laboratories, Bar Harbor, ME; NRP-2−/− mice were a kind gift from Dr. David Ginty (Johns Hopkins School of Medicine, Baltimore, MD). PB cells were collected by cardiac puncture of anesthetized mice and monocytes were isolated as indicated for BM monocytes. Bronchoalveolar lavage fluid was collected by instilling 2.0 mL of normal saline via a transtrancheal incision in anesthetized mice that had received an intratracheal injection of 5 μg of lipopolysaccharide (from E. coli J5 (RC), List Biological Laboratories, Campbell, CA) or normal saline 24 h earlier. Mouse peritoneal macrophages were harvested from the peritoneal cavity of wild-type, NCAM−/− and NRP-2−/− mice 3 days after the intraperitoneal injection of 3% thioglycollate and were placed in culture for 24 h in Corning Costar 6-well tissue culture plates in RPMI, 10% FCS (GIBCO, Invitrogen, Carlsbad, CA) in a CO2 incubator. To purify macrophages from freshly isolated cells for phenotypic analysis, 2.5 × 106 total peritoneal cells were seeded in wells of 6-well tissue culture plates for 30 min, nonadherent cells were removed by two washes with medium, and adherent macrophages were collected by gentle pipetting. Breeding of NRP-2−/− mice and all experiments with mice were performed under protocols approved by the University of Maryland Medical School Institutional Animal Care and Use Committee.
Immunofluorescent staining of cell surface proteins and analysis by flow cytometry
Proteins on the surface of cells were detected by incubating cells at 1 × 106 cells/mL in PBS containing 2% heat-inactivated human AB serum (Gemini Bioproducts, Calabasas, CA) and 2.5% rat IgG (Stem Cell Technologies) at 4°C for 15 min to minimize nonspecific binding of reagents. Prior to staining peritoneal macrophages, cells were incubated in PBS containing 2% heat-inactivated normal donkey serum (Jackson ImmunoResearch, West Grove, PA) and 2% bovine serum albumin (Calbiochem, San Diego, CA). Cells were stained with 10 μg/mL mouse anti-polySia mAb 735 (Frosch et al. 1985; Curreli et al. 2007) followed by incubation with biotinylated rabbit anti-mouse IgG and phycoerythrin (PE)-conjugated streptavidin (both from Dako, Carpinteria, CA). Alternatively, surface polySia was detected by staining cells with 8 μg/mL of a catalytically inactive endoN that binds specifically to but does not degrade polySia acid and is fused to GFP-endoN (Jokilammi et al. 2004). As control, cells were first treated with an active endoN (kindly provided by Karen Colley, University of Illinois, Chicago, IL) that specifically cleaves α2-8-linked polySia (Finne and Makela 1985; Rutishauser et al. 1985). Cells were also stained with rabbit anti-mouse NRP-2 mAb (Cell Signaling Technology, Danvers, MA) at 5 μg/mL or with rabbit anti-NCAM/CD56 IgG (EMD Millipore Corporation, Billerica, MA) at 2 μg/mL followed by 4 μg/mL fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit F(ab′)2 fragment (Alexa Fluor 488, Molecular Probes, Eugene, OR), with allophycocyanin (APC)-labeled anti-mouse F4/80 (clone BM8, eBiosciences, San Diego, CA), APC-labeled anti-mouse CD11b (clone M1/70, eBiosciences), FITC-labeled anti-mouse CD14 (clone rmC5-3, BD Pharmingen, San Diego, CA), APC-labeled anti-mouse CD11c (clone HL3, BD Pharmingen), PE-labeled anti-mouse CD86 (clone GL1, BD Pharmingen) and with biotin-conjugated hamster anti-mouse CD3e (clone 145-2C11), rat anti-mouse CD11b (clone M1/70), rat anti-mouse CD45R/B220 (clone RA3-6B2), rat anti-mouse Ly-6G/C (clone RB6-8C5) and rat anti-mouse TER119 (clone TER-119) (all mAbs from BD PharMingen) followed by PE-conjugated streptavidin (Dako). Following incubation at 4°C for 30 min with antibodies or GFP-endoN, cells were washed with 2 mL PBS containing 2% human AB serum and fixed with 1% paraformaldehyde. Cells were acquired using a Becton Dickinson FACSCaliber (Mountain View, CA), and data were analyzed using FlowJo data analysis software (Tree Star, Ashland, OR).
Isolation of RNA and real-time RT-PCR
Total RNA was isolated from BM cells and monocyte-derived DCs using an RNeasy mini kit (Qiagen, Valencia, CA) following the protocol suggested by the manufacturer. The RNA preparation was treated with DNase I (Invitrogen) at 37°C for 30 min to remove contaminating DNA. DNase was then removed by binding to Blue Sorb DNase affinity slurry (Clonogene, St. Petersburg, Russia). Semi-quantitative real-time RT-PCR was performed using a QuantiTect SYBR green RT-PCR Kit (Qiagen) with an ABI Sequence Detection System (ABI PRISM 5700) to detect gene expression of murine ST8 SiaII (Genbank accession NM_009181) and ST8 SiaIV (GenBank Accession BC060112) using purified RNAs. Gene expression of 18S rRNA (GenBank accession NR_003278) was also measured for use as an internal control. Ten nanograms (10 ng) of total RNA were used to quantitate expression of each gene using the following primers (Operon Biotechnologies, Huntsville, AL): ST8 SiaII (forward, nt 688–706, 5′-GGCTGTGGCCAGGAGATTG-3′, and reverse, nt 759–737, 5′-GGCATACTCCTGAACTGGAGCC-3′) yielding a 71-bp product; ST8 SiaIV (forward, nt 400–421, 5′-GCACCAAGAGACGTCAACTCATC-3′, and reverse, nt 467-444, 5′-CAGAGCTGTTGACAAGTGATCTGC-3′) yielding a 67-bp product; 18S rRNA (forward, nt 1202–1222, 5′-TTGACGGAAGGGCACCACCAG-3′, and reverse, nt 1331–1310, 5′-GCACCACCACCCACGGAATCG-3′), yielding a 129-bp product. The fold change in expression of ST8 SiaII and of ST8 SiaIV in DCs was normalized to the fold increase of each relative to the amount of 18S RNA. All reactions were run in triplicate and the accuracy of each reaction was monitored by analysis of melting curves and product size on gel electrophoresis.
Western blot analysis of cellular proteins
Cells were collected and solubilized as described previously (Curreli et al. 2007) and protein concentration was determined using a Bicinchoninic Acid Kit (Sigma-Aldrich Co., St. Louis, MO). Five to 30 μg (5–30 μg) of protein from each cell lysate were resolved by electrophoresis on a 8% SDS–polyacrylamide gel using Tris–glycine–SDS running buffer (gel and running buffer from Invitrogen), electrotransferred by a semi-wet method to a Sequi-Blot PVDF membrane (Bio-Rad, Hercules, CA) and probed with 0.5 μg/mL mouse mAB 735 against polySia, 0.5 μg/mL rabbit anti-mouse NRP-2 mAb (Cell Signaling Technology) and 1 μg/mL rabbit anti-NCAM IgG (EMD Millipore Corporation). The respective blots were incubated with a 1:5000 dilution of horseradish peroxidase-conjugated anti-mouse or -rabbit IgGs (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), developed using an ECL chemiluminescence substrate kit (Amersham Biosciences, Piscataway, NJ) and exposed to Kodak X-ray film.
Phagocytosis of bacteria by peritoneal macrophages
Thioglycollate-induced mouse peritoneal macrophages were prepared as indicated above and placed in culture in 0.5 mL RPMI/10% FCS at 5 × 105 cells per well in 24-well Corning Costar tissue culture plates. After 24 h in culture, macrophages were exposed to endoN or 100 mU/mL bacterial neuraminidase (CP NANase, crystalline, type X, from Clostridium perfringens, Sigma-Aldrich) or left untreated (6 wells per condition) for 1 h at 37°C before being exposed to 1 × 108 cfu (50 μL) of K. pneumoniae (strain B5055, O1:K2; WHO Collaborating Centre for Reference and Research on Escherichia and Klebsiella, Statens Serum Institut, Copenhagen S, Denmark) in a final volume of 0.550 mL RPMI/10% FCS (MOI of 200:1). Prior to use, K. pneumoniae was grown to the exponential phase in 10 mL LB, collected by centrifugation (2700 × g, 10 min, 25°C), washed once with PBS and resuspended at 2 × 109 cfu/mL in RPMI/10% FCS. To promote bacterial contact with the macrophage monolayer, plates were centrifuged at 500 × g for 10 min. After incubation for 30 min at 37°C, medium was removed, cells were washed twice with PBS and were incubated an additional 30 min with 0.500 mL RPMI 1640/10% FCS containing gentamycin (50 μg/mL) (AMRESCO, Solon, OH) to eliminate extracellular bacteria. Cells were then washed two times with PBS and lysed with 1 mL 1% Triton X-100 in PBS for 10 min at 37°C or placed back in culture for an additional 2.5 h in medium containing 10 μg/mL gentamicin. Following this incubation, cells were washed with PBS and lysed as indicated above. Ten microliters of 10-fold serial dilutions were plated on LB plates and incubated at 37°C for 15 h to quantify the number of intracellular bacteria at each time point. Phagocytosis data are presented as percentage of cfu per well divided by input number of cfu and bacterial killing data as 1 minus the percentage of cfu remaining after the additional 2.5 h incubation divided by the initial cfu phagocytosed. Exposure of macrophages to the amount of K. pneumoniae used in these experiments did not induce a cytotoxic effect as determined by trypan blue dye exclusion.
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported in part by institutional funds provided by the Institute of Human Virology and a grant from the Baltimore Research and Education Foundation to N.M.S., by National Institutes of Health grant HL086933-01A1 to A.S.C. and L.Z. and by grants from the Academy of Finland to J.F. and A.J.
Conflict of interest statement
None declared
Abbreviations
BM, bone marrow; CP NANase, Clostridium perfringens neuraminidase; DC, dendritic cell; endoN, endoglycosidase N; GFP-endoN, green fluorescent protein fused to inactive endoN; NCAM/CD56, neural cell adhesion molecule; NRP-2, neuropilin-2; PB, peripheral blood; polySia, polysialic acid; ST8 SiaII and ST8 SiaIV, polysialyltransferases II and IV; SynCAM 1, synaptic cell adhesion molecule.
Supplementary Material
Acknowledgements
N.M.S. is grateful to Stefanie Vogel and Kari Ann Shirey for assistance in characterizing the phenotype of peritoneal macrophages and to Joseph Bryant, Harry Davis and Janine Davenport for assistance in breeding NRP-2−/− mice.
References
- Bax M, van Vliet SJ, Litjens M, Garcia-Vallejo JJ, van Kooyk Y. Interaction of polysialic acid with CCL21 regulates the migratory capacity of human dendritic cells. PLoS ONE. 2009;4:e6987. doi: 10.1371/journal.pone.0006987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfanti L. PSA-NCAM in mammalian structural plasticity and neurogenesis. Prog Neurobiol. 2006;80:129–164. doi: 10.1016/j.pneurobio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Bruses JL, Rutishauser U. Roles, regulation, and mechanism of polysialic acid function during neural development. Biochimie. 2001;83:635–643. doi: 10.1016/s0300-9084(01)01293-7. [DOI] [PubMed] [Google Scholar]
- Cabral MG, Silva Z, Ligeiro D, Seixas E, Crespo H, Carrascal MA, Silva M, Piteira AR, Paixao P, Lau JT, et al. The phagocytic capacity and immunological potency of human dendritic cells is improved by alpha2,6-sialic acid deficiency. Immunology. 2013;138:235–245. doi: 10.1111/imm.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close BE, Colley KJ. In vivo autopolysialylation and localization of the polysialyltransferases PST and STX. J Biol Chem. 1998;273:34586–34593. doi: 10.1074/jbc.273.51.34586. [DOI] [PubMed] [Google Scholar]
- Close BE, Mendiratta SS, Geiger KM, Broom LJ, Ho LL, Colley KJ. The minimal structural domains required for neural cell adhesion molecule polysialylation by PST/ST8Sia IV and STX/ST8Sia II. J Biol Chem. 2003;278:30796–30805. doi: 10.1074/jbc.M305390200. [DOI] [PubMed] [Google Scholar]
- Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, et al. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- Curreli S, Arany Z, Gerardy-Schahn R, Mann D, Stamatos NM. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J Biol Chem. 2007;282:30346–30356. doi: 10.1074/jbc.M702965200. [DOI] [PubMed] [Google Scholar]
- Diestel S, Hinkle CL, Schmitz B, Maness PF. NCAM140 stimulates integrin-dependent cell migration by ectodomain shedding. J Neurochem. 2005;95:1777–1784. doi: 10.1111/j.1471-4159.2005.03475.x. [DOI] [PubMed] [Google Scholar]
- Drake PM, Nathan JK, Stock CM, Chang PV, Muench MO, Nakata D, Reader JR, Gip P, Golden KP, Weinhold B, et al. Polysialic acid, a glycan with highly restricted expression, is found on human and murine leukocytes and modulates immune responses. J Immunol. 2008;181:6850–6858. doi: 10.4049/jimmunol.181.10.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake PM, Stock CM, Nathan JK, Gip P, Golden KP, Weinhold B, Gerardy-Schahn R, Bertozzi CR. Polysialic acid governs T-cell development by regulating progenitor access to the thymus. Proc Natl Acad Sci USA. 2009;106:11995–12000. doi: 10.1073/pnas.0905188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finne J. Occurrence of unique polysialosyl carbohydrate units in glycoproteins of developing brain. J Biol Chem. 1982;257:11966–11970. [PubMed] [Google Scholar]
- Finne J, Leinonen M, Makela PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;2:355–357. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- Finne J, Makela PH. Cleavage of the polysialosyl units of brain glycoproteins by a bacteriophage endosialidase. Involvement of a long oligosaccharide segment in molecular interactions of polysialic acid. J Biol Chem. 1985;260:1265–1270. [PubMed] [Google Scholar]
- Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- Frosch M, Gorgen I, Boulnois GJ, Timmis KN, Bitter-Suermann D. NZB mouse system for production of monoclonal antibodies to weak bacterial antigens: Isolation of an IgG antibody to the polysaccharide capsules of Escherichia coli K1 and group B meningococci. Proc Natl Acad Sci USA. 1985;82:1194–1198. doi: 10.1073/pnas.82.4.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallily R, Warwick A, Bang FB. Effect of cortisone of genetic resistance to mouse hepatitis virus in vivo and in vitro. Proc Natl Acad Sci USA. 1964;51:1158–1164. doi: 10.1073/pnas.51.6.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galuska SP, Rollenhagen M, Kaup M, Eggers K, Oltmann-Norden I, Schiff M, Hartmann M, Weinhold B, Hildebrandt H, Geyer R, et al. Synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in postnatal mouse brain. Proc Natl Acad Sci USA. 2010;107:10250–10255. doi: 10.1073/pnas.0912103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giger RJ, Cloutier JF, Sahay A, Prinjha RK, Levengood DV, Moore SE, Pickering S, Simmons D, Rastan S, Walsh FS, et al. Neuropilin-2 is required in vivo for selective axon guidance responses to secreted semaphorins. Neuron. 2000;25:29–41. doi: 10.1016/s0896-6273(00)80869-7. [DOI] [PubMed] [Google Scholar]
- Hinkle CL, Diestel S, Lieberman J, Maness PF. Metalloprotease-induced ectodomain shedding of neural cell adhesion molecule (NCAM) J Neurobiol. 2006;66:1378–1395. doi: 10.1002/neu.20257. [DOI] [PubMed] [Google Scholar]
- Jokilammi A, Ollikka P, Korja M, Jakobsson E, Loimaranta V, Haataja S, Hirvonen H, Finne J. Construction of antibody mimics from a noncatalytic enzyme-detection of polysialic acid. J Immunol Methods. 2004;295:149–160. doi: 10.1016/j.jim.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Kanato Y, Kitajima K, Sato C. Direct binding of polysialic acid to a brain-derived neurotrophic factor depends on the degree of polymerization. Glycobiology. 2008;18:1044–1053. doi: 10.1093/glycob/cwn084. [DOI] [PubMed] [Google Scholar]
- Lagasse E, Weissman IL. Flow cytometric identification of murine neutrophils and monocytes. J Immunol Methods. 1996;197:139–150. doi: 10.1016/0022-1759(96)00138-x. [DOI] [PubMed] [Google Scholar]
- Lichtman MA. Cellular deformability during maturation of the myeloblast. Possible role in marrow egress. N Engl J Med. 1970;283:943–948. doi: 10.1056/NEJM197010292831801. [DOI] [PubMed] [Google Scholar]
- Miyagi T, Yamaguchi K. Mammalian sialidases: Physiological and pathological roles in cellular functions. Glycobiology. 2012;22:880–896. doi: 10.1093/glycob/cws057. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff M, Eckhardt M, Bethe A, Frosch M, Gerardy-Schahn R. Autocatalytic polysialylation of polysialyltransferase-1. EMBO J. 1996;15:6943–6950. [PMC free article] [PubMed] [Google Scholar]
- Rey-Gallardo A, Delgado-Martin C, Gerardy-Schahn R, Rodriguez-Fernandez JL, Vega MA. Polysialic acid is required for neuropilin-2a/b-mediated control of CCL21-driven chemotaxis of mature dendritic cells and for their migration in vivo. Glycobiology. 2011;21:655–662. doi: 10.1093/glycob/cwq216. [DOI] [PubMed] [Google Scholar]
- Rey-Gallardo A, Escribano C, Delgado-Martin C, Rodriguez-Fernandez JL, Gerardy-Schahn R, Rutishauser U, Corbi AL, Vega MA. Polysialylated neuropilin-2 enhances human dendritic cell migration through the basic C-terminal region of CCL21. Glycobiology. 2010;20:1139–1146. doi: 10.1093/glycob/cwq078. [DOI] [PubMed] [Google Scholar]
- Rieger F, Grumet M, Edelman GM. N-CAM at the vertebrate neuromuscular junction. J Cell Biol. 1985;101:285–293. doi: 10.1083/jcb.101.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollenhagen M, Buettner FF, Reismann M, Jirmo AC, Grove M, Behrens GM, Gerardy-Schahn R, Hanisch FG, Muhlenhoff M. Polysialic acid on neuropilin-2 is exclusively synthesized by the polysialyltransferase ST8SiaIV and attached to mucin-type o-glycans located between the b2 and c domain. J Biol Chem. 2013;288:22880–22892. doi: 10.1074/jbc.M113.463927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser U, Watanabe M, Silver J, Troy FA, Vimr ER. Specific alteration of NCAM-mediated cell adhesion by an endoneuraminidase. J Cell Biol. 1985;101:1842–1849. doi: 10.1083/jcb.101.5.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer R. Sialic acids as regulators of molecular and cellular interactions. Curr Opin Struct Biol. 2009;19:507–514. doi: 10.1016/j.sbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyrantepe V, Iannello A, Liang F, Kanshin E, Jayanth P, Samarani S, Szewczuk MR, Ahmad A, Pshezhetsky AV. Regulation of phagocytosis in macrophages by neuraminidase 1. J Biol Chem. 2010;285:206–215. doi: 10.1074/jbc.M109.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatos NM, Carubelli I, van de Vlekkert D, Bonten EJ, Papini N, Feng C, Venerando B, d'Azzo A, Cross AS, Wang LX, et al. LPS-induced cytokine production in human dendritic cells is regulated by sialidase activity. J Leukoc Biol. 2010;88:1227–1239. doi: 10.1189/jlb.1209776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss-Ayali D, Conrad SM, Mosser DM. Monocyte subpopulations and their differentiation patterns during infection. J Leukocyte Biol. 2007;82:244–252. doi: 10.1189/jlb.0307191. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Mitoma J, Hosono M, Shiozaki K, Sato C, Yamaguchi K, Kitajima K, Higashi H, Nitta K, Shima H, et al. Sialidase NEU4 hydrolyzes polysialic acids of neural cell adhesion molecules and negatively regulates neurite formation by hippocampal neurons. J Biol Chem. 2012;287:14816–14826. doi: 10.1074/jbc.M111.324186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MG, Foley DA, Colley KJ. The polysialyltransferases interact with sequences in two domains of the neural cell adhesion molecule to allow its polysialylation. J Biol Chem. 2013;288:7282–7293. doi: 10.1074/jbc.M112.438374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm C, Saffarzadeh M, Mahavadi P, Muller S, Prem G, Saboor F, Simon P, Middendorff R, Geyer H, Henneke I, et al. Soluble polysialylated NCAM: A novel player of the innate immune system in the lung. Cell Mol Life Sci. 2013;70:3695–3708. doi: 10.1007/s00018-013-1342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Gagneux P. Multifarious roles of sialic acids in immunity. Ann N Y Acad Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Neumann H. Alleviation of neurotoxicity by microglial human Siglec-11. J Neurosci. 2010;30:3482–3488. doi: 10.1523/JNEUROSCI.3940-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe U, Sato C, Matsuda T, Kitajima K. Polysialic acid in human milk. CD36 is a new member of mammalian polysialic acid-containing glycoprotein. J Biol Chem. 2003;278:13875–13880. doi: 10.1074/jbc.M300458200. [DOI] [PubMed] [Google Scholar]
- Zuber C, Lackie PM, Catterall WA, Roth J. Polysialic acid is associated with sodium channels and the neural cell adhesion molecule N-CAM in adult rat brain. J Biol Chem. 1992;267:9965–9971. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.