Abstract
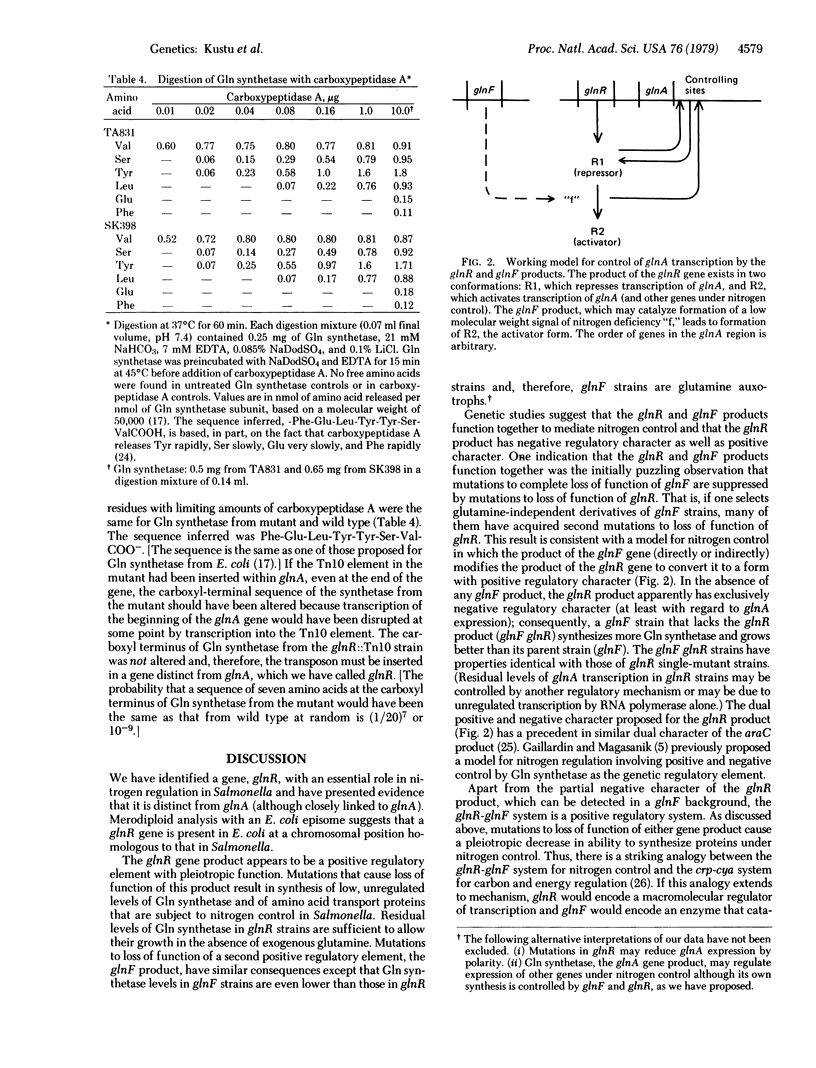
The product of the glnR gene is required for nitrogen regulation of the synthesis of glutamine synthesis (Gln synthetase) [L-glutamate:ammonia ligase (ADP-forming), EC 6.3.1.2] and two periplasmic transport proteins that are subject to nitrogen control in Salmonella. Strains with mutations to loss of function of the glnR product [e.g., a strain with a Tn10 insertion or one with an ICR-induced (frameshift) mutation in glnR] have about 3% as much Gln synthetase as a fully derepressed wild-type strain and are unable to increase synthesis of this enzyme or periplasmic transport proteins in response to nitrogen limitation. The structural gene for Gln synthetase, glnA, and those for the periplasmic transport proteins are unlinked on the chromosome; thus, glnR appears to encode a diffusible positive regulatory element. Consistent with this, the mutant glnR allele is recessive to the wild-type allele with regard to expression of glnA (synthesis of Gln synthetase). Although glnR is closely linked to glnA, strains with mutations to complete loss of function of the glnR product can be distinguished from glnA strains by their ability to produce detectable Gln synthetase and to grow in the absence of glutamine. To demonstrate unequivocally that glnR is distinct from glnA, we have purified and characterized Gln synthetase from a strain with a Tn10 insertion in glnR. Because the properties of Gln synthetase from the insertion mutant, most importantly the carboxyl-terminal sequence of amino acids, are the same as those of synthetase from wild type, the Tn10 insertion cannot be in glnA (if it were, the carboxyl terminus of Gln synthetase would have to be altered); therefore we conclude that the Tn10 insertion is in a regulatory gene, glnR, which is distinct from glnA. A model for the function of the glnR product together with the previously defined glnF product in mediating nitrogen control is discussed.
Keywords: regulation of protein synthesis, positive control, glutamine synthetase, amino acid transport components
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bancroft S., Rhee S. G., Neumann C., Kustu S. Mutations that alter the covalent modification of glutamine synthetase in Salmonella typhimurium. J Bacteriol. 1978 Jun;134(3):1046–1055. doi: 10.1128/jb.134.3.1046-1055.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann L., Baumann P. Studies of relationship among terrestrial Pseudomonas, Alcaligenes, and enterobacteria by an immunological comparison of glutamine synthetase. Arch Microbiol. 1978 Oct 4;119(1):25–30. doi: 10.1007/BF00407923. [DOI] [PubMed] [Google Scholar]
- Bender R. A., Magasanik B. Autogenous regulation of the synthesis of glutamine synthetase in Klebsiella aerogenes. J Bacteriol. 1977 Oct;132(1):106–112. doi: 10.1128/jb.132.1.106-112.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender R. A., Magasanik B. Regulatory mutations in the Klebsiella aerogenes structural gene for glutamine synthetase. J Bacteriol. 1977 Oct;132(1):100–105. doi: 10.1128/jb.132.1.100-105.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., Wilcox G. Regulation: positive control. Annu Rev Genet. 1974;8:219–242. doi: 10.1146/annurev.ge.08.120174.001251. [DOI] [PubMed] [Google Scholar]
- Gaillardin C. M., Magasanik B. Involvement of the product of the glnF gene in the autogenous regulation of glutamine synthetase formation in Klebsiella aerogenes. J Bacteriol. 1978 Mar;133(3):1329–1338. doi: 10.1128/jb.133.3.1329-1338.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E., Bancroft S., Rhee S. G., Kustu S. The product of a newly identified gene, gInF, is required for synthesis of glutamine synthetase in Salmonella. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1662–1666. doi: 10.1073/pnas.74.4.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kustu S. G., McFarland N. C., Hui S. P., Esmon B., Ames G. F. Nitrogen control of Salmonella typhimurium: co-regulation of synthesis of glutamine synthetase and amino acid transport systems. J Bacteriol. 1979 Apr;138(1):218–234. doi: 10.1128/jb.138.1.218-234.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu S. G., McKereghan K. Mutations affecting glutamine synthetase activity in Salmonella typhimurium. J Bacteriol. 1975 Jun;122(3):1006–1016. doi: 10.1128/jb.122.3.1006-1016.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lei M., Aebi U., Heidner E. G., Eisenberg D. Limited proteolysis of glutamine synthetase is inhibited by glutamate and by feedback inhibitors. J Biol Chem. 1979 Apr 25;254(8):3129–3134. [PubMed] [Google Scholar]
- Magasanik B. Classical and postclassical modes of regulation of the synthesis of degradative bacterial enzymes. Prog Nucleic Acid Res Mol Biol. 1976;17:99–115. doi: 10.1016/s0079-6603(08)60067-7. [DOI] [PubMed] [Google Scholar]
- Mendez E., Lai C. Y. Regeneration of amino acids from thiazolinones formed in the Edman degradation. Anal Biochem. 1975 Sep;68(1):47–53. doi: 10.1016/0003-2697(75)90677-6. [DOI] [PubMed] [Google Scholar]
- Niall H. D. Automated Edman degradation: the protein sequenator. Methods Enzymol. 1973;27:942–1010. doi: 10.1016/s0076-6879(73)27039-8. [DOI] [PubMed] [Google Scholar]
- Pahel G., Zelenetz A. D., Tyler B. M. gltB gene and regulation of nitrogen metabolism by glutamine synthetase in Escherichia coli. J Bacteriol. 1978 Jan;133(1):139–148. doi: 10.1128/jb.133.1.139-148.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastan I., Adhya S. Cyclic adenosine 5'-monophosphate in Escherichia coli. Bacteriol Rev. 1976 Sep;40(3):527–551. doi: 10.1128/br.40.3.527-551.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman E. R., Ginsburg A., Ciardi J. E., Yeh J., Hennig S. B., Shapiro B. M. Multiple molecular forms of glutamine synthetase produced by enzyme catalyzed adenylation and deadenylylation reactions. Adv Enzyme Regul. 1970;8:99–118. doi: 10.1016/0065-2571(70)90011-7. [DOI] [PubMed] [Google Scholar]
- Streicher S. L., Bender R. A., Magasanik B. Genetic control of glutamine synthetase in Klebiella aerogenes. J Bacteriol. 1975 Jan;121(1):320–331. doi: 10.1128/jb.121.1.320-331.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B. Regulation of the assimilation of nitrogen compounds. Annu Rev Biochem. 1978;47:1127–1162. doi: 10.1146/annurev.bi.47.070178.005403. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- White T. J., Ibrahimi I. M., Wilson A. C. Evolutionary substitutions and the antigenic structure of globular proteins. Nature. 1978 Jul 6;274(5666):92–94. doi: 10.1038/274092a0. [DOI] [PubMed] [Google Scholar]
- Woolfolk C. A., Shapiro B., Stadtman E. R. Regulation of glutamine synthetase. I. Purification and properties of glutamine synthetase from Escherichia coli. Arch Biochem Biophys. 1966 Sep 26;116(1):177–192. doi: 10.1016/0003-9861(66)90026-9. [DOI] [PubMed] [Google Scholar]