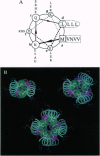
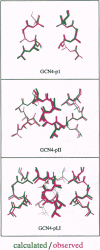
Abstract
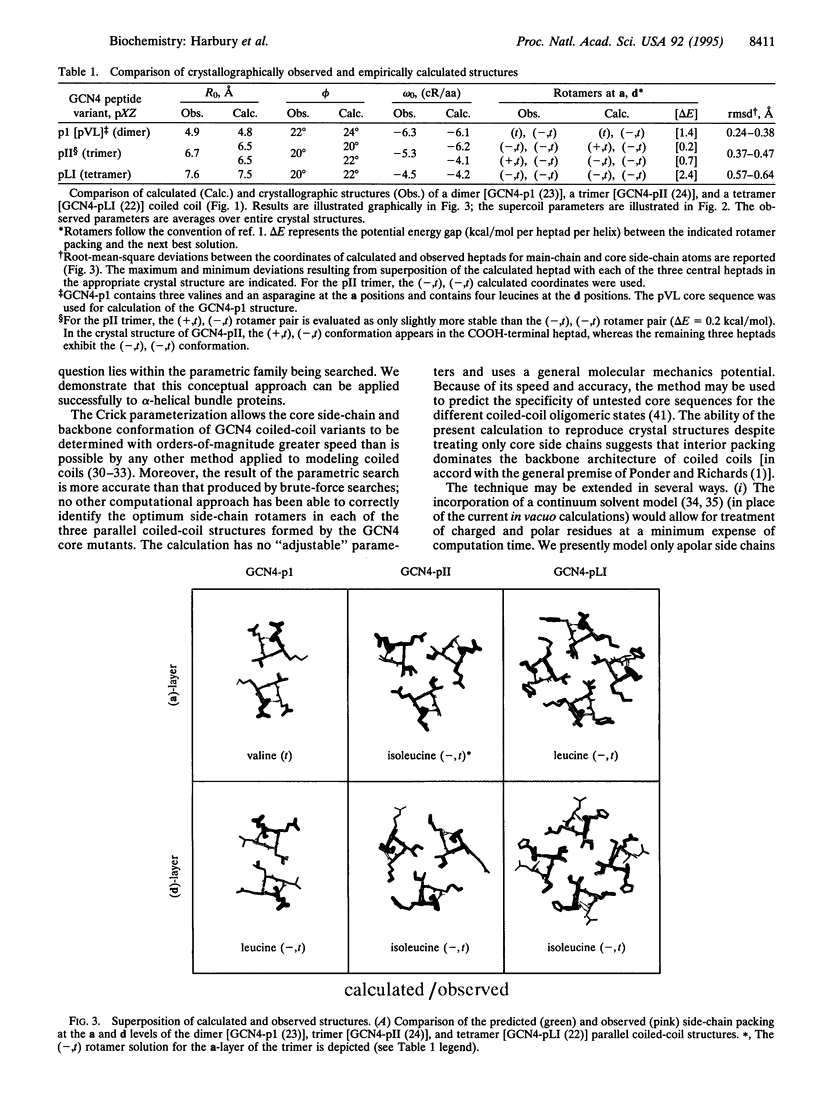
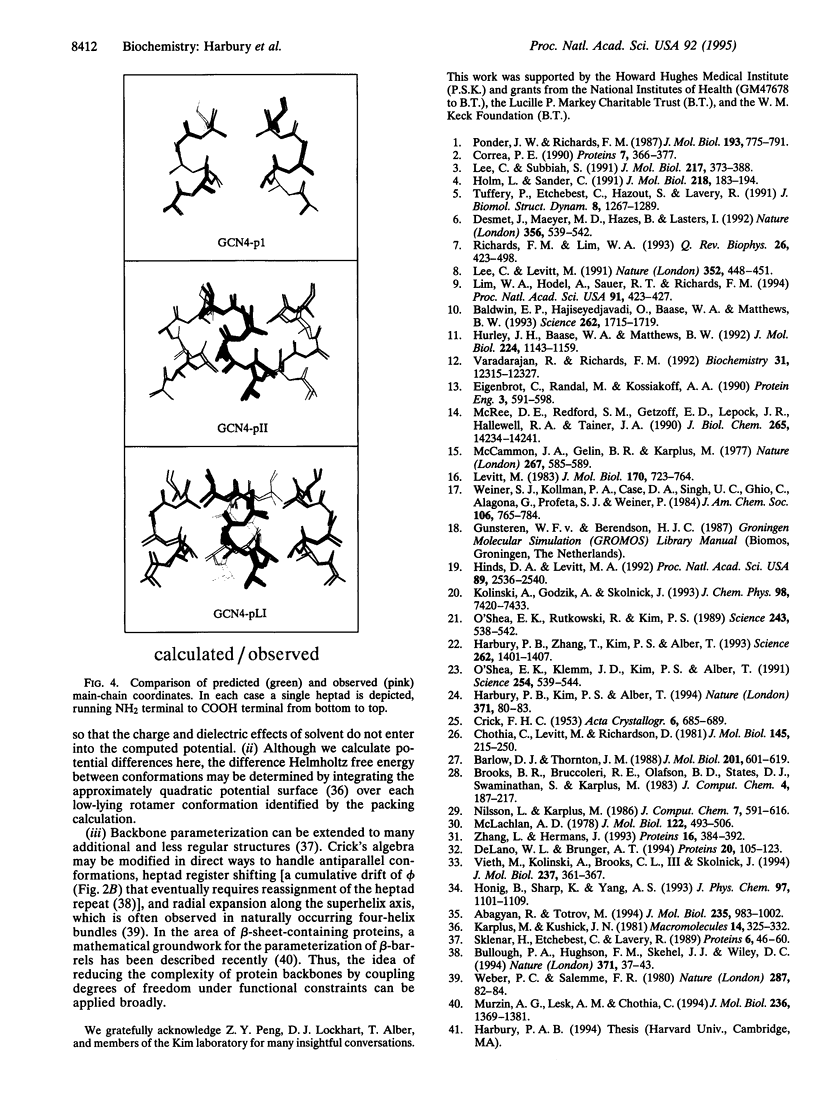
Progress in homology modeling and protein design has generated considerable interest in methods for predicting side-chain packing in the hydrophobic cores of proteins. Present techniques are not practically useful, however, because they are unable to model protein main-chain flexibility. Parameterization of backbone motions may represent a general and efficient method to incorporate backbone relaxation into such fixed main-chain models. To test this notion, we introduce a method for treating explicitly the backbone motions of alpha-helical bundles based on an algebraic parameterization proposed by Francis Crick in 1953 [Crick, F. H. C. (1953) Acta Crystallogr. 6, 685-689]. Given only the core amino acid sequence, a simple calculation can rapidly reproduce the crystallographic main-chain and core side-chain structures of three coiled coils (one dimer, one trimer, and one tetramer) to within 0.6-A root-mean-square deviations. The speed of the predictive method [approximately 3 min per rotamer choice on a Silicon Graphics (Mountain View, CA) 4D/35 computer] permits it to be used as a design tool.
Full text
PDF
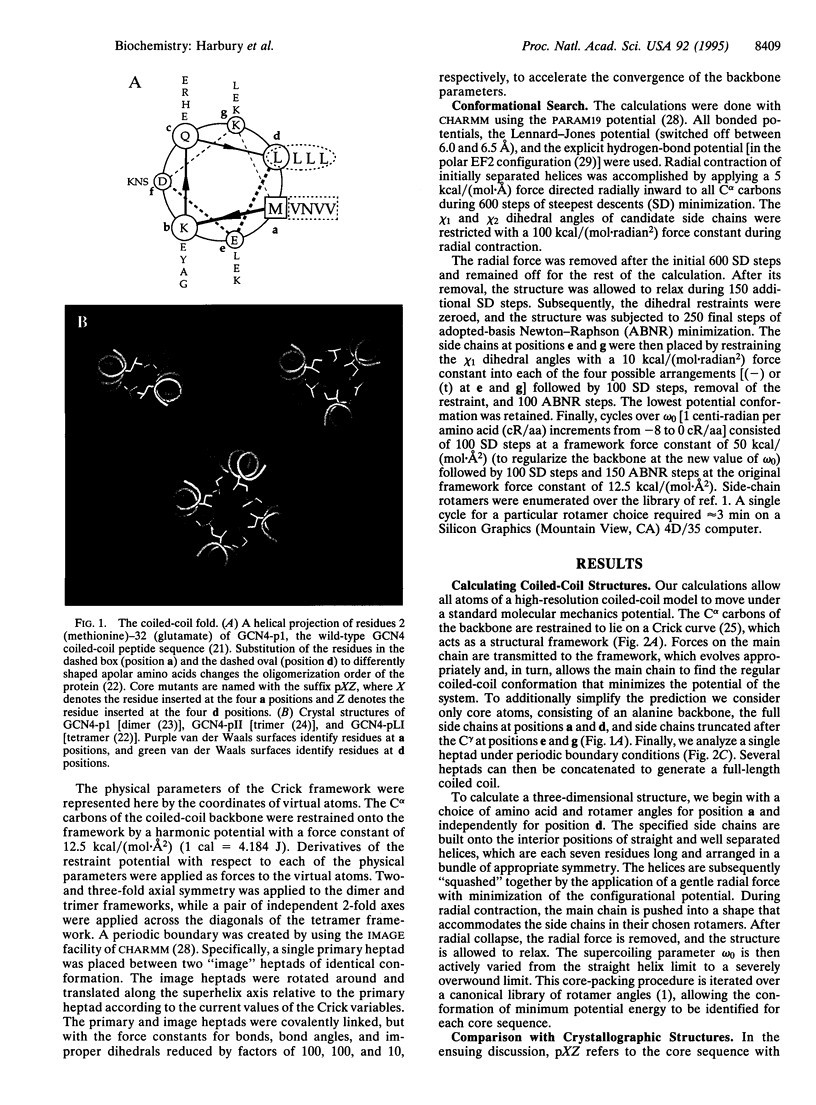
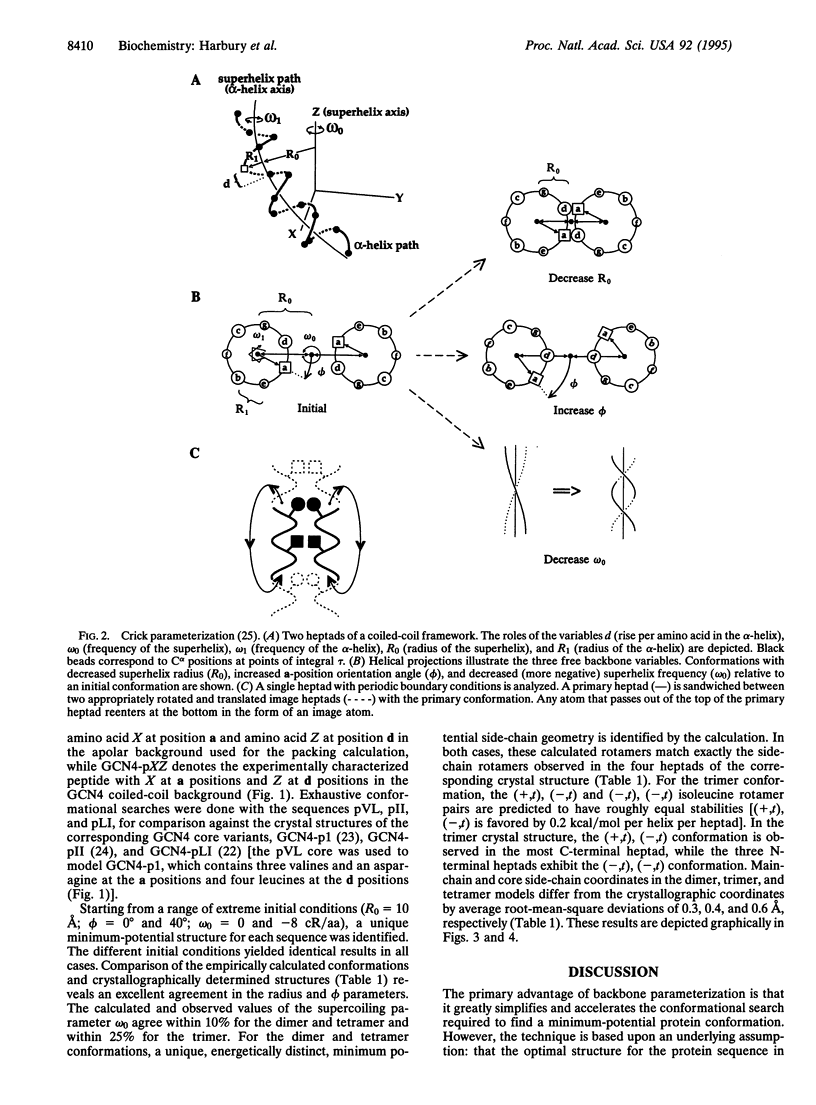
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abagyan R., Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994 Jan 21;235(3):983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- Baldwin E. P., Hajiseyedjavadi O., Baase W. A., Matthews B. W. The role of backbone flexibility in the accommodation of variants that repack the core of T4 lysozyme. Science. 1993 Dec 10;262(5140):1715–1718. doi: 10.1126/science.8259514. [DOI] [PubMed] [Google Scholar]
- Barlow D. J., Thornton J. M. Helix geometry in proteins. J Mol Biol. 1988 Jun 5;201(3):601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- Bullough P. A., Hughson F. M., Skehel J. J., Wiley D. C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994 Sep 1;371(6492):37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Chothia C., Levitt M., Richardson D. Helix to helix packing in proteins. J Mol Biol. 1981 Jan 5;145(1):215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- Correa P. E. The building of protein structures from alpha-carbon coordinates. Proteins. 1990;7(4):366–377. doi: 10.1002/prot.340070408. [DOI] [PubMed] [Google Scholar]
- DeLano W. L., Brünger A. T. Helix packing in proteins: prediction and energetic analysis of dimeric, trimeric, and tetrameric GCN4 coiled coil structures. Proteins. 1994 Oct;20(2):105–123. doi: 10.1002/prot.340200202. [DOI] [PubMed] [Google Scholar]
- Eigenbrot C., Randal M., Kossiakoff A. A. Structural effects induced by removal of a disulfide-bridge: the X-ray structure of the C30A/C51A mutant of basic pancreatic trypsin inhibitor at 1.6 A. Protein Eng. 1990 Jul;3(7):591–598. doi: 10.1093/protein/3.7.591. [DOI] [PubMed] [Google Scholar]
- Harbury P. B., Kim P. S., Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994 Sep 1;371(6492):80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- Harbury P. B., Zhang T., Kim P. S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993 Nov 26;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Hinds D. A., Levitt M. A lattice model for protein structure prediction at low resolution. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2536–2540. doi: 10.1073/pnas.89.7.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J Mol Biol. 1991 Mar 5;218(1):183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Baase W. A., Matthews B. W. Design and structural analysis of alternative hydrophobic core packing arrangements in bacteriophage T4 lysozyme. J Mol Biol. 1992 Apr 20;224(4):1143–1159. doi: 10.1016/0022-2836(92)90475-y. [DOI] [PubMed] [Google Scholar]
- Lee C., Levitt M. Accurate prediction of the stability and activity effects of site-directed mutagenesis on a protein core. Nature. 1991 Aug 1;352(6334):448–451. doi: 10.1038/352448a0. [DOI] [PubMed] [Google Scholar]
- Lee C., Subbiah S. Prediction of protein side-chain conformation by packing optimization. J Mol Biol. 1991 Jan 20;217(2):373–388. doi: 10.1016/0022-2836(91)90550-p. [DOI] [PubMed] [Google Scholar]
- Levitt M. Protein folding by restrained energy minimization and molecular dynamics. J Mol Biol. 1983 Nov 5;170(3):723–764. doi: 10.1016/s0022-2836(83)80129-6. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Hodel A., Sauer R. T., Richards F. M. The crystal structure of a mutant protein with altered but improved hydrophobic core packing. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):423–427. doi: 10.1073/pnas.91.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon J. A., Gelin B. R., Karplus M. Dynamics of folded proteins. Nature. 1977 Jun 16;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D. The double helix coiled coil structure of murein lipoprotein from Escherichia coli. J Mol Biol. 1978 Jun 5;121(4):493–506. doi: 10.1016/0022-2836(78)90396-0. [DOI] [PubMed] [Google Scholar]
- McRee D. E., Redford S. M., Getzoff E. D., Lepock J. R., Hallewell R. A., Tainer J. A. Changes in crystallographic structure and thermostability of a Cu,Zn superoxide dismutase mutant resulting from the removal of a buried cysteine. J Biol Chem. 1990 Aug 25;265(24):14234–14241. doi: 10.2210/pdb3sod/pdb. [DOI] [PubMed] [Google Scholar]
- Murzin A. G., Lesk A. M., Chothia C. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J Mol Biol. 1994 Mar 11;236(5):1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991 Oct 25;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Kim P. S. Evidence that the leucine zipper is a coiled coil. Science. 1989 Jan 27;243(4890):538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Richards F. M., Lim W. A. An analysis of packing in the protein folding problem. Q Rev Biophys. 1993 Nov;26(4):423–498. doi: 10.1017/s0033583500002845. [DOI] [PubMed] [Google Scholar]
- Sklenar H., Etchebest C., Lavery R. Describing protein structure: a general algorithm yielding complete helicoidal parameters and a unique overall axis. Proteins. 1989;6(1):46–60. doi: 10.1002/prot.340060105. [DOI] [PubMed] [Google Scholar]
- Tuffery P., Etchebest C., Hazout S., Lavery R. A new approach to the rapid determination of protein side chain conformations. J Biomol Struct Dyn. 1991 Jun;8(6):1267–1289. doi: 10.1080/07391102.1991.10507882. [DOI] [PubMed] [Google Scholar]
- Varadarajan R., Richards F. M. Crystallographic structures of ribonuclease S variants with nonpolar substitution at position 13: packing and cavities. Biochemistry. 1992 Dec 15;31(49):12315–12327. doi: 10.1021/bi00164a005. [DOI] [PubMed] [Google Scholar]
- Vieth M., Kolinski A., Brooks C. L., 3rd, Skolnick J. Prediction of the folding pathways and structure of the GCN4 leucine zipper. J Mol Biol. 1994 Apr 8;237(4):361–367. doi: 10.1006/jmbi.1994.1239. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Salemme F. R. Structural and functional diversity in 4-alpha-helical proteins. Nature. 1980 Sep 4;287(5777):82–84. doi: 10.1038/287082a0. [DOI] [PubMed] [Google Scholar]
- Zhang L., Hermans J. Molecular dynamics study of structure and stability of a model coiled coil. Proteins. 1993 Aug;16(4):384–392. doi: 10.1002/prot.340160407. [DOI] [PubMed] [Google Scholar]