Abstract
Acute hyperammonemia (HA) causes cerebral edema and brain damage in children with urea cycle disorders (UCDs) and in patients in acute liver failure. Chronic HA is associated with developmental delay and mental retardation in children with UCDs, and with neuropsychiatric symptoms in patients with chronic liver failure. Astrocytes are a major cellular target of hyperammonemic encephalopathy, and changes occurring in these cells are thought to be causally related to the brain edema of acute HA. To study the effect of HA on astrocytes in vivo, we crossed the Otcspf mouse, a mouse with the X-linked UCD ornithine transcarbamylase (OTC) deficiency, with the hGFAP-EGFP mouse, a mouse selectively expressing green fluorescent protein in astrocytes. We used FACS to purify astrocytes from the brains of hyperammonemic and healthy Otcspf/GFAP-EGFP mice. RNA isolated from these astrocytes was used in microarray expression analyses and qRT-PCR. When compared with healthy littermates, we observed a significant downregulation of the gap-junction channel connexin 43 (Cx43) the water channel aquaporin 4 (Aqp4) genes, and the astrocytic inward-rectifying potassium channel (Kir) genes Kir4.1 and Kir5.1 in hyperammonemic mice. Aqp4, Cx43, and Kir4.1/ Kir5.1 are co-localized to astrocytic end-feet at the brain vasculature, where they regulate potassium and water transport. Since, ions can permeate water and K+-channels, downregulation of these three channels may be a direct effect of elevated blood ammonia levels. Our results suggest that alterations in astrocyte-mediated water and potassium homeostasis in brain may be key to the development of the brain edema.
Keywords: green fluorescent protein, cell sorting, ammonia, brain edema, Connexin 43, Aquaporin 4, Kir4.1, Kir5.1
INTRODUCTION
Urea cycle disorders (UCDs) are a group of devastating inborn errors of metabolism, which are caused by a deficiency of one of the enzymes or transporters that constitute the urea cycle. The urea cycle enables the excretion of nitrogen as urea. When the urea cycle does not function normally, nitrogen accumulates in the form of ammonia and hyperammonemic encephalopathy (HAE) ensues. Differently from UCDs, in acute and chronic liver failure the urea cycle is not the only pathway with impaired function, but hyperammonemia (HA) is still a major factor contributing to hepatic encephalopathy in these pathological conditions (Desjardins et al., 2001; Felipo and Butterworth, 2002).
Depending on the degree of the impairment of the urea cycle and on management, patients either develop acute HAE causing cerebral edema and severe brain damage, or chronic HAE causing significant morbidity. Chronic HAE is associated with developmental delay, mental retardation, attention deficit/hyperactivity disorder, and impaired executive function in children with UCDs (Batshaw et al., 1980, 1986; Gyato et al., 2004; Maestri et al., 1999; Msall et al., 1984, 1988). Chronic hepatic encephalopathy causes neuropsychiatric disturbances in patients with chronic liver dysfunction. By the time treatment normalizes the ammonia level in acute HAE, irreversible brain damage may have already occurred. Successful brain protection therapies are therefore needed to improve the overall outcome.
The pathophysiology of the encephalopathies associated with UCDs has not yet been elucidated. Astrocytes are considered to be the main site of ammonia toxicity in the brain (Jayakumar et al., 2006; Norenberg, 1977; Rao et al., 2005), and are the only brain cells that can detoxify ammonia through glutamine synthesis (Desjardins et al., 2001).
Many signaling pathways and cellular mechanisms have been under consideration as the cause of HAE, including: (i) excitotoxicity caused either by elevated extracellular glutamate levels, ammonia itself, or quinolinate; (ii) consequences of elevated glutamine levels in the brain, especially with regard to glutamine as an osmolyte; (iii) the role of water channels, such as Aqp4, in astrocyte swelling; (iv) disturbed energy metabolism, particularly glucose metabolism; (v) ion fluxes, including those of potassium ions; and (vi) altered nitric oxide (NO) synthesis. However, a main cause for HAE has not been defined and several mechanisms are likely to contribute to this pathology.
One of the animal models used to study the effect of ammonia in the brain is the sparse fur (spf) mouse, a mouse with the X-linked UCD ornithine transcarbamylase (OTC) deficiency (OTCD; DeMars et al., 1976). The Otcspf mouse has been widely used as a viable model for the analysis of HAE in UCDs. To define molecular changes that specifically occur in astrocytes during HAE, we crossed the Otcspf mouse with the hGFAPEGFP mouse, in which enhanced green fluorescent protein (EGFP) is selectively expressed in astrocytes (Nolte et al., 2001; Wehner et al., 2003). This enabled us to acutely purify astrocytes from hyperammonemic mouse brains by Fluorescence Activated Cell Sorting (FACS). We hypothesized that microarray analysis of astrocytes from ammonia-exposed brains would identify groups of astrocyte-specific genes displaying significant changes in their expression profiles. If specific sets of HA-sensitive genes would emerge from microarray analysis, identification of the main pathways affected by HA in astrocytes could then become possible. Identifying such pathways is the prerequisite for finding potential targets for pharmacological strategies aimed at arresting or preventing acute and chronic HAE.
MATERIALS AND METHODS
Generation and Characterization of the Otcspf/GFAP-EGFP+ Mouse
For this study, two 2 mouse colonies were established from breeding pairs, including a colony of Otcspf mice, carrying a mutation causing OTC deficiency, (DeMars et al., 1976; Jackson Laboratories) and a colony of mice expressing the hGFAP-EGFP transgene in astrocytes, FVB/NTgN(GFAP-EGFP) mice (F. Kirchhoff, Max Planck Institute of Experimental Medicine, Goettingen, Germany). The FVB/NTgN(GFAP-EGFP) mice were previously characterized to express EGFP in about 50% of their cortical astrocytes, 80% of their striatal astrocytes, and 100% of their cerebellar astrocytes at 8–12 weeks of age (Wehner et al., 2003).
The Otcspf/GFAP-EGFP mouse was generated by breeding heterozygous FVB/N-TgN(GFAP-EGFP) females to hemizygous B6EiC3 α/A Otcspf/J males and heterozygous B6EiC3 α/A Otcspf/J females to heterozygous FVB/NTgN(GFAP-EGFP) males, yielding an F1 generation of B6EiC3α/AOtcspf/J//FVB/N-TgN (GFAPEGFP) mice. Given the X-linked nature of OTC deficiency, progeny consisted of hemizygous affected Otcspf/Y males and heterozygous Otcspf females, as well as unaffected Otcspf hemizygous normal male and homozygous normal female mice, with or without the hGFAP-EGFP transgene. Transgene expressing progenies were identifed by transillumination of their skulls with ultraviolet light between postnatal Day 1 (P1) and P3. The point mutation causing the Otcspf phenotype (Veres et al., 1987) was detected in a Taqman allelic discrimination assay according to the manufacturer's suggestions (Applied Biosystems Foster City, CA). The assay was performed on DNA isolated from tail tips. Genotyping was primarily necessary for breeding purposes, because affected male pups can be phenotypically distinguished from their heterozygous or normal littermates by their considerably smaller size and their spars fur. Unaffected mice were weaned at 21 days of age, affected males are smaller and weaker than unaffected male or female mice, and were allowed to nurse 2–3 days longer.
The distribution of green fluorescent astrocytes in the brain of B6EiC3 α/A Otcspf/J//FVB/N-TgN mice was analyzed by immunohistochemistry. Because not all astrocytes were GFAP-positive in immunohistochemical analyses, expression of S100β was also used for comparison.
Induction of HA
To assure that affected mice were hyperammonemic at the time of brain harvest, mice were put on an arginine-free amino acid diet (Harlan TEKLAD, Madison, WI) rendered isonitrogenous with glycine. Arginine is the precursor for ornithine synthesis. Ornithine is the substrate of the second enzyme of the urea cycle. When eating an arginine-free diet susceptible animals became hyperammonemic. An arginine-free diet had been used to cause HA in other mammals (Stewart et al., 1981), and it was previously used by D'Hooge et al. (2000) to render spf mice hyperammonemic. We observed a severe clinical effect of the arginine-free diet within 24 hr (mice sitting stiff and still in their cage) in weaned 25-day-old affected mice (carrying or not carrying the EGFP trans-gene) after weaning. Therefore, we placed 27- to 33-dayold male mice carrying the spf mutation and unaffected male littermates, not carrying the spf mutation, on argi-nine-free diet for 18 hr. As only affected males display the disease phenotype of this X-linked trait, only affected and unaffected males were analyzed. Blood was collected by retro-orbital bleed from the mice before sacrifice, to document the plasma ammonia and the plasma glutamine levels at the time of sacrifice. Normal ammonia levels in weaned mice are close to those in humans (21–68 μM, mean 44 ± 10 μM; Koizumi et al., 1990). Unaffected mice had ammonia levels of 35–212 μM (most below 150 μM), and affected mice had levels of 420–960 μM after 18 hr on the diet. Plasma, as well as cerebellum and brain stem tissue, were collected from each mouse used in any experiment to measure gluta-mine levels. Glutamine levels correlate with ammonia levels, are much less sensitive to sampling artifacts, and can serve as a secondary marker for HA.
Measurement of Plasma Ammonia and Amino Acid Levels, and Brain Tissue Amino Acid Concentrations
Plasma ammonia measurements were performed in the clinical laboratory of Children's National Medical Center on a Dade Behring Dimension RXL clinical chemistry system (Dade Behring, Newark, DE), using an adaptation of the glutamate dehydrogenase (GLDH) enzymatic method of van Anken and Shiphorst which substitutes NADPH for NADH eliminating interference form other NADH-consuming reactions. The reaction was carried out in a Dimension® Ammonia (AMON) Flex reagent cartridge (K863840; Dade Behring, Newark, DE). The disappearance of NADPH was measured spectrophotometrically. Amino acid concentrations in mouse plasma and in hindbrain tissue extract were measured by ion-exchange chromatography on a Beckman 6300 amino acid analyzer in the Biochemical Genetics Laboratory at the University of Colorado at Denver and Health Sciences Center (UCDHSC). Brain was homogenized and sonicated in water (100 mg/mL), and amino acids were measured in the supernatant after precipitation with sulfosalicyclic acid (70 mg/mL). Tissue concentrations are reported in nanomoles per 100 mg wet weight.
Immunohistochemistry
Mice were anaesthetized by intraperitoneal injection of Nembutal (0.01 mL/g) and perfused intracardially with a solution of NaCl (9 g/L), followed by a fixative solution containing PFA 4% dissolved in PBS. Brains were dissected out and incubated overnight in a cryoprotective solution (PBS, sucrose 30%). Brains were then frozen in 2-methylbutane at −80°C, and 50 or 20 μm tissue sections were prepared by means of a cryostat. Sections were then incubated at RT for at least 30 min in blocking solution (PBS, gelatin 0.25%, Triton X-100 0.3%). Primary antibodies used were the monoclonal anti-GFAP antibody - clone G-A-5 from Sigma (St. Louis, MO; G3893; 1:500 dilution), and rabbit anti-human S100β antibody from DAKO (Carpinteria, CA; A510; 1:50 dilution) raised against purified, recombinant human S100β. Primary antibodies were diluted using a carrier solution (PBS, gelatin 0.25%, Triton X-100 0.3%). Brain sections were incubated in primary antibodies at 37°C for 2 hr or at 4°C overnight. Rinses were performed in PBS at RT, with three changes of solution after 1/3/10 min. All secondary antibodies (Jackson Immunoresearch Laboratories; Bar Harbor, Main) were diluted at 1:250. Incubation in secondary antibodies was performed for 2 hr at 37°C, followed by three washes, as described earlier. Sections were then rinsed in distilled water, mounted in Vectashield (Vector Laboratories; Burlingame, CA), and imaged using a confocal microscope.
Fluorescence-Activated Cell Sorting (FACS) of Acutely Isolated Astrocytes
Preparation of total cell suspensions and FACS performed as previously described (Belachew et al., 2002). In brief, brains from affected P27–30 mice and normal littermates on arginine-free diet were dissected, cerebellum and brain stem were collected and stored at −80°C for amino acid analysis. The remainder of the brain tissue was dissociated by incubation in enzyme solution (15 units/mL papain, 100 units/mL DNaseI). Cells were purified over a Percoll (Amersham Biosciences, Piscataway, NJ) gradient, washed, and strained prior to sorting. Cells were analysed for light forward- and side-scatter using a FACS Vantage SE instrument (Becton Dickinson, San Jose, CA). For EGFP fluorescence, excitation wavelength of the argon ion laser was set at 488 nm and emission was through a 530 nm bandpass filter. A size threshold was used to gate out erythrocytes and cellular debris. The sorting speed was 2,000–4,000 cells/s. FACS analyses lasted ~45 min and yielded 200–400,000 cells per brain, regardless of whether this was the brain of an affected or an unaffected mouse. Typically 12–24% of the total cells gated were green-fluorescent. The amount of cells sorted was related to the percentage of green-fluorescent cells in the total cells gated and not to the phenotype or genotype of the mouse. After FACS, the amount of EGFP positive viable cells was determined in an aliquot of the sorted sample by FACS-(re)analysis measuring green fluorescence and 7-Amino-actinomycin D (7-AAD) intercalation in sorted cells. In addition, to document purity, a fraction of sorted cells was plated onto coverslips after sorting, fixed after 3 hr and assessed for presence of EGFP, GFAP and S100β, as well as the neuron-, oligodendrocyte-, and microglia-specific proteins NeuN, O1 and Iba1, respectively. Primary antibodies used were the monoclonal anti-GFAP antibody clone G-A-5 from Sigma (see above), the rabbit anti-human S100B antibody from DAKO (see above), the monoclonal anti-mouse NeuN antibody from Chemicon (Temecula, CA; MAB377; 1:100 dilution), the O1 antibody from ATCC (Manassas, VA; 1:10 diliution) and the polyclonal rabbit anti-Iba1 antibody from Wako Chemicals USA, (Richmond, VA; 1:2000 dilution). As secondary antibodies rhodamine conjugated: goat-anti-mouse IgG from Jackson Laboratories (Bar Harbor, Main; 115-025-146; 1:200 dilution), goat-anti-mouse IgM from ICN pharmaceuticals (Aliso Viejo, CA; 1:200 dilution), and goat-anti-rabbit IgG from Jackson Laboratories (Bar Harbor, Main; 111-295-144; 1:100 dilution) were used.
RNA Isolation and Amplification and cDNA Synthesis
Astrocyte RNA was isolated using the Absolutely RNA Microprep Kit from Stratagene (La Jolla, CA). One astrocyte RNA sample was generated from each brain. Cortex RNA was prepared using TRIzol reagent (GIBCO BRL, Gaithersburg, MD), according to the manufacturer's suggestions. RNA concentration was determined using a NanoDrop spectrophotometer from NanoDrop Technologies (Rockland, DE), which allows accurate measurement of nanogram amounts of RNA. FACS-purified astrocytes yielded 130–350 ng total RNA, and a 30 ng aliquot of the total RNA was carried through a two-round amplification (Baugh et al., 2001; Sohn et al., 2006). In brief, double-stranded cDNA was synthesized from 30 ng of total RNA with an oligdT primer containing the T7 RNA polymerase promoter (Geneset, LaJolla, CA) and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). After purification by ethanol precipitation, the double-stranded cDNA was resuspended in 8 μL of DEPC H2O. First-round cRNA was synthesized from double-stranded cDNA by in vitro transcription using MegaScript T7 transcription kit (Ambion, Austin, TX). A portion (200 ng) of purified cRNA was subjected to a second round of cDNA synthesis using a random (N6) primer (Roche, Indianapolis, IN) to synthesize the first-strand cDNA and a T7-(dT)24 primer for the second-strand cDNA. The resulting double-stranded cDNA was purified via a Phase Lock Gel (PLG, Eppendorf, Brinkman Instrument, Westbury, NY), followed by ethanol precipitation. When starting with 5 μg of total RNA from cortex, double-stranded cDNA was synthesized using the cDNA synthesis kit from Affymetrix (Santa Clara, CA), according to the manufacturer's suggestions.
Generation of Biotinylated cRNA and GeneChip Array Analysis
Biotinylated cRNA was produced by in vitro transcription of first-round (when starting with 5 μg total RNA from cortex) or second-round (when starting with 30 ng of total RNA from sorted astrocytes) double-stranded cDNA, by using GeneChip Expression 3−-Amplification Reagents for IVT labeling (Affymetrix, SantaClara, CA). Biotinylated cRNA was purified and then fragmented. Fragmented cRNA (20 μg) was hybridized to mouse Genome 430 2.0 oligonucleotide microarrays (45,000 probe sets; Affymetrix, Santa Clara, CA), and signal intensities calculated using two distinct probe set algorithms (MAS 5.0, dCHIP difference model). Each microarray underwent a stringent quality control evaluation as reported previously (Natale et al., 2003). All values obtained from the microarrays in this experiment fell within the quality control range (Natale et al., 2003; Sohn et al., 2006). Only cRNA samples which represented at least a 400-fold amplification of the starting material were employed in the microarray analysis. When this is observed, the complexity of the amplified sample should reproduce that of the original sample (Natale et al., 2003). One sample representing one animal was used per each chip. Two-round amplification of individual samples and hybridization of one sample per microarray was chosen over pooling of samples from several mice, because inter-individual differences between mice can annihilate significant changes when samples are pooled (Tumor Analysis Best Practices Working Group, 2004).
Data Filtering and Statistical Analysis
To enrich for transcripts significantly expressed above noise (background), we employed a data filter requiring at least one “present call” (MAS5.0) in the microarrays in study (DiGiovanni et al., 2003; Natale et al., 2004). The levels for signal sensitivity detection and signal-to-noise ratio were optimized for the project (Seo et al., 2004; Tumor Analysis Best Practices Working Group, 2004). Signal intensity for each probe set was normalized to the median intensity across all chips in the experiment. A statistical filter was then employed, where we required each differentially expressed probe set to show P < 0.05 for both the MAS5.0 and dCHIP difference model probe set algorithms. Previous studies have found ~30–50% concordance of significant differences with different probe set algorithms, and our approach of requiring concordance is considered relatively stringent. We did not correct for multiple testing, but relied on validation by other methods.
After selection of significantly differentially expressed probe sets, gene lists were loaded into DAVID (Database for Annotation, Visualization and Integrated Discovery) of the DAVID and EASE bioinformatic resources of the National Institute of Allergy and Infectious Diseases (NIAID) and into Ingenuity Pathway Analysis (Ingenuity Systems, www.ingenuity.com), in order to identify affected pathways or networks. The significance of the association between the data set and the canonical pathway was measured in two ways: (i) A ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway; (ii) Fisher's exact test was used to calculate a P-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone. A number of the canonical pathways shown in the Ingenuity Pathways Analysis were adapted from the KEGG pathways (Kyoto Encyclopedia of Genes and Genomes available for free on the world wide web).
Quantitative RT-PCR
We used qRT-PCR as an independent method to confirm changes in gene expression first detected by micro-array analysis. The RNA samples used for qRT-PCR analyses were different from those used for microarray profiling. P27-30 Otcspf/GFAP-EGFP mice were exposed to arginine-free diet (six affected animals and five unaffected littermates) or to a diet containing 1% of the argi-nine of a normal mouse diet in the drinking water (six affected animals and four unaffected littermates) for 18 hr. Cx43, Aqp4, Kir4.1, and Kir5.1 RNA levels in FACS purified EGFP+ astrocytes of these mice were measured by qRT-PCR. The expression of the Cx43, Aqp4, Kir4.1, and Kir5.1 genes was compared with the expression of a control gene, GAPDH, which showed no significant changes in expression during HA. RNA samples were subjected to qRT-PCR as previously described (Zhao et al., 2002) using a LI-COR DNA analyzer (LI-COR Biosciences, Lincoln, NE). Briefly, ss cDNA was synthesized from total RNA by reverse transcription and then amplified in the same test tube using the SuperScriptTM One-Step RT-PCR System with Platinum® Taq DNA Polymerase kit (Invitrogen, Carlsbad, CA) and fluorescein labeled forward primers (Licor Biosciences, Lincoln, NE). The program Primer3 (available at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) was used for primer design. To avoid amplification of contaminating genomic DNA, exon-spanning primers were used for amplification. This primer design assures transcript-specific amplification. The qRT-PCR reaction also contained unlabeled reverse primers. Primer pairs allowed amplification of 170–235 bp fragments from each gene in a quantitative multiplex PCR reaction with three primer pairs (one for each of the two experimental genes and one for the control gene) for a total of 22 (Cx43, Aqp4, GAPDH) or 23 cycles (Kir4.1, Kir5.1, and GAPDH). Multiplex PCR products were run on a 7% gel and quantified by using Gene ImageIR 3.56 (LI-COR). Expression of the experimental genes was normalized to that of the control gene. An ANOVA t test was performed to generate P values for differences in the expression of each experimental gene in Otcspf/GFAP-EGFP affected mice versus their unaffected littermates. The qRT-PCR experiments were performed on 10–11 independent samples per group in triplicate (Cx43, Aqp4) or pent-plicate (Kir4.1, Kir5.1).
RESULTS
Generation and Characterization of the Otcspf/GFAP-EGFP Mouse
To isolate astrocytes from the brains of hyperammonemic mice, we crossed the Otcspf mouse and the hGFAPEGFP mouse. The progeny from the cross was termed B6EiC3α/AOtcspf/J//FVB/N-TgN(GFAP-EGFP) mouse or Otcspf/GFAP-EGFP mouse in short.
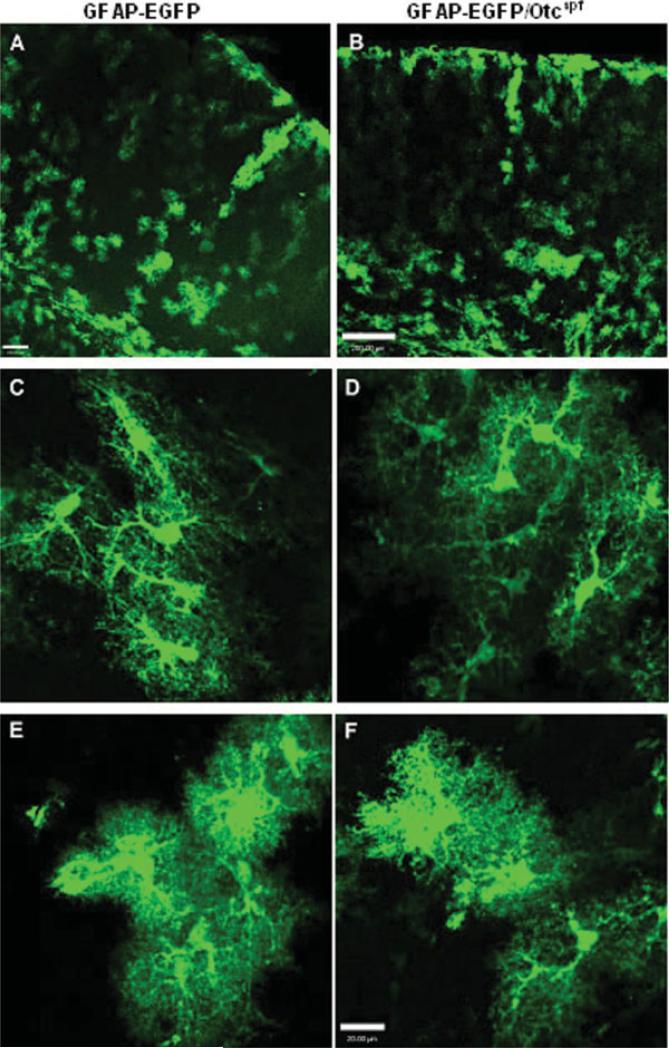
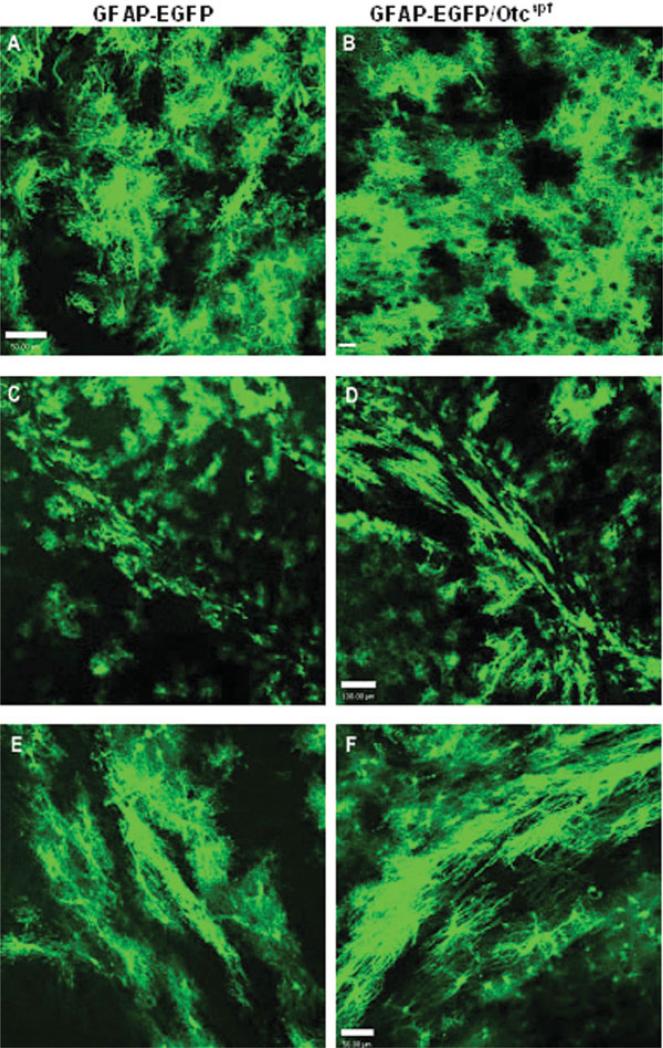
There was no phenotypic difference between progeny that carried the hGFAP-EGFP transgene and progeny that did not. The overall distribution of green fluores-cent astrocytes in P30 brains of Otcspf/GFAP-EGFP mice was similar to that of the original hGFAP-EGFP mice (Figs. 1,2). Analysis of cerebral cortex (see Fig. 1), diencephalon (Figs. 2A,B) and subcortical white matter (Figs. 2C–F) demonstrated the presence of GFAP-EGFP+ astrocytes with similar morphologies in Otcspf/ GFAP-EGFP and hGFAP-EGFP mice.
Fig. 1.
Comparative morphology and distribution of GFAP-EGFP+ cells in GFAP-EGFP and Otcspf/GFAP-EGFP mice. Morphology and distribution of EGFP+ cells in cerebral cortex of P30 FVB/N-TgN(GFAP-EGFP) transgenic mouse (A, C, and E) and Otcspf/GFAP-EGFP mouse (B, D, and F). (A and B) Low magnification images show similar percentages and similar distribution of GFAP-EGFP+ cells in the two mouse strains. Scale bars = 100 μm (A) and 200 μm (B). (C–F) Examples of cortical GFAP-EGFP+ astrocytes in the two mouse strains show cells with similar levels of EGFP expression and similar morphologies. Scale bars = 20 μm.
Fig. 2.
Comparative morphology and distribution of GFAP-EGFP+ cells in GFAP-EGFP and Otcspf/GFAP-EGFP mice. Morphology and distribution of EGFP+ cells in diencephalon and sub-cortical white matter of P30 FVB/NTgN(GFAP-EGFP) transgenic mouse (A, C, and E) and Otcspf/GFAP-EGFP mouse (B, D, and F). (A and B) GFAP-EGFP+ astrocytes in diencephalon. Scale bar = 50 μm in A, and 25 μM in B. (C–F) GFAP-EGFP+ astrocytes in subcortical white matter. Scale bar = 100 μm in C and D, and 50 μM in E and F.
The hGFAP-EGFP mice were previously characterized as expressing EGFP in about 50% of their cortical astrocytes, 80% of their striatal astrocytes, and 100% of their cerebellar astrocytes at 8–12 weeks of age (Wehner et al., 2003). Because not all astrocytes express EGFP in this mouse model and because not all astrocytes are GFAP-positive in situ, the presence of the astrocytic protein S100β in green fluorescent cells was used for comparison between mouse strains. In cerebral cortex, (90.9 ± 1.7)% of green fluorescent astrocytes were S100β+ in hGFAP-EGFP mice and (93.0 ± 2.6)% in Otcspf/GFAP-EGFP mice. In hippocampus, (91.5 ± 3.3)% were S100β+ in GFAP-EGFP mice versus (96.2 ± 1.8)% in Otcspf/hGFAP-EGFP mice. No significant differences in the percentages of these cells were observed between hGFAP-EGFP and Otcspf/GFAP-EGFP mice in either brain areas (three different brains for each group; total cells counted per brain region ranged between 165 and 216).
Affected Otcspf mice were consistently smaller and had considerably less hair than their Otcspf unaffected litter-mates. As only male mice show symptoms of this X-linked disease, only affected and unaffected males were analyzed. The life span of the affected Otcspf/GFAP-EGFP mice, with or without hGFAP-EGFP transgene, was consistent with the published life span for B6EiC3 α/A Otcspf/Y males (Robinson et al., 1995). Most affected mice not used in our experiments died spontaneously between one and two weeks after weaning (32–40 days old). Otcspf/Y mice may be hyperammonemic between weaning and 40–50 days of age (Gushiken et al., 1985; Inoue et al., 1987). Ammonia levels randomly measured in weaned mice were scattered over a wide range for healthy littermates (58–160 mmol/L; n = 8) and affected mice (251–404 mmol/L; n = 8). Reasons for blood ammonia levels to be artificially high in some normal mice are hemolysis and tissue damage caused by the instrument used for the blood draw.
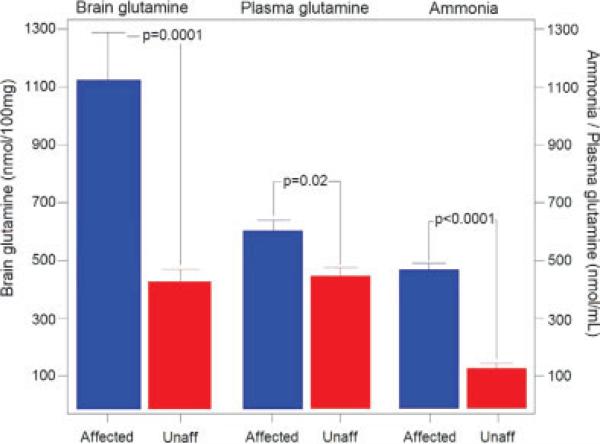
To assure that affected Otcspf/GFAP-EGFP mice were hyperammonemic at the time of brain harvest, affected mice and unaffected littermates were placed on an argi-nine-free diet for 18 hr. We tested different durations of arginine-free diet as well as 1% arginine diet. Mice were either very sick or dead after 24 hr on arginine-free diet, whereas the majority of the mice were viable after 18 hr on the diet. We chose 18 hr on diet as a time point where animals would be viable and results would not be biased by secondary effects of the terminal condition of the animal. Otcspf/GFAP-EGFP mice and unaffected littermates (27–30 day-old) both carrying the hGFAPEGFP transgene were used. After 18 hr on diet, the ammonia levels measured in unaffected mice ranged from 58 to 183 μmoles/L (mean 128.7 ± 16.4 μmoles/L), and those measured in affected mice from 420 to 960 lmoles/ L (mean 478.57 ± 20.2 μmoles/L; P < 0.0001; Fig. 3). Thus, affected mice were distinctly hyperammonemic at the time when astrocytes were isolated from their brains.
Fig. 3.
Glutamine levels in Otcspf/GFAP-EGFP affected mice and unaffected littermates. There was a significant difference between the brain glutamine levels, plasma glutamine levels, and plasma ammonia levels of affected and unaffected mice. Data are represented as averages ± SEM. A total of 28 P27-30 mice were analyzed from each strain for each parameter. The difference between strains was less significant for plasma glutamine levels than for brain glutamine levels or plasma ammonia levels.
Plasma as well as hindbrain was collected from each mouse to measure glutamine levels as an indirect measure for ammonia levels. There was a significant difference (P < 0.0001) between the brain glutamine levels of affected and unaffected mice (see Fig. 3). The difference was not as overt for the plasma glutamine levels, but it was also significant (P < 0.02). A similar observation was previously described in Otcspf mice by other authors (Batshaw et al., 1988).
Morphology of Green Fluorescent Astrocytes During HA
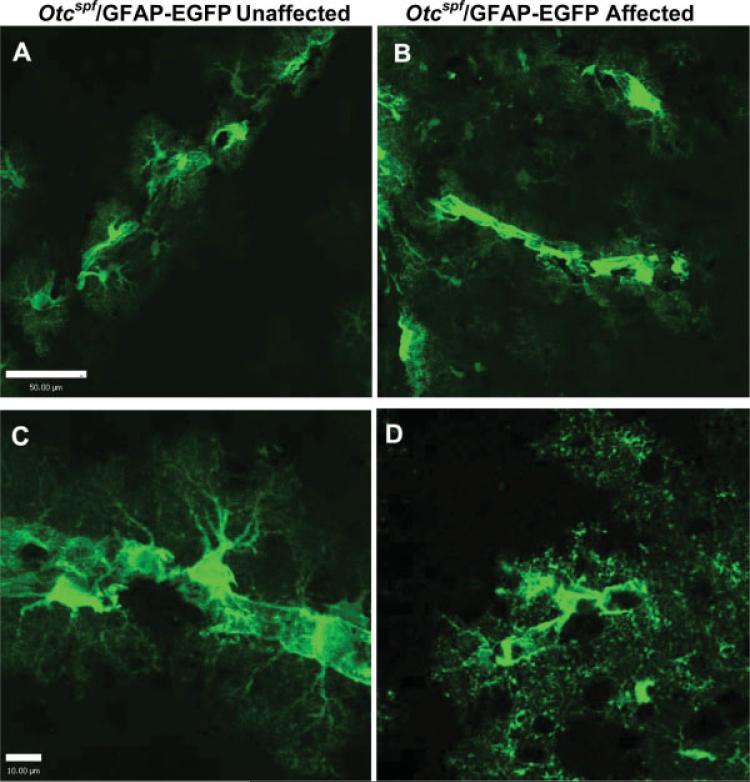
Based on previous findings that astrocyte changes were associated with HA (Jayakumar et al., 2006; Norenberg, 1977; Rao et al., 2005), brain slices of Otcspf/GFAP-EGFP mice were imaged in the hyperammonemic state to assess astrocyte morphology during HA. In hyperammonemic animals (Figs. 4B,D) green-fluorescent astrocytes at the brain vasculature displayed a different morphology than the ones of their unaffected littermates (Figs. 4A,C). Astrocytic process arborization was reduced and coarser in hyperammonemic mice, and they appeared to be clustered, rather than evenly distributed along blood vessels when compared with unaffected littermates (see Fig. 4).
Fig. 4.
Comparative morphology of EGFP+ astrocytes during hyperammonemia at brain vasculature. Morphology of green fluorescent astrocytes along brain vasculature in the cerebral cortex of (B, D) affected hyperammonemic Otcspf/GFAP-EGFP mice versus (A, C) unaffected littermates. There appears to be more distinct arborization and more connectivity between green fluorescent astrocytes along blood vessels in unaffected than in hyperammonemic animals. Scale bars = 50 μM in A and C, and 10 μM in B and D.
Gene Expression Profiling of FACS-Purified EGFP+ Astrocytes of Hyperammonemic mice
Expression profiling of FACS-purified astrocytes was performed on astrocytes isolated from P27-30 mice sacrificed after 18 hr of arginine-free diet. Re-analysis of an aliquot of the sorted cells by FACS immediately after sorting confirmed their purity and viability. Over 90% of the cells in the samples were green fluorescent cells, with a viability of 84–97% (Materials and Methods section). An aliquot of the FACS-purified cells from both unaffected and affected mice was seeded on glass coverslips and analyzed by immunohistochemical methods. There was no difference in the antigenic phenotype of cells purified from affected and unaffected animals. Immunocytochemical characterization demonstrated that 48–54% of the sorted cells expressed GFAP and 84% expressed S100β. Less than 1% of the cells expressed the oligodendrocyte marker O1, and 5% of the cells expressed the neuronal marker NeuN (average of two experiments, data not shown). Finally, no Iba1+ microglial cells were detected (data not shown).
Microarray analysis of RNA from purified astrocytes of hyperammonemic (n = 3) and unaffected mice (n = 3) identified changes in the expression of astrocyte specific genes (Bachoo et al., 2004) and in a number of canonical pathways. Several of these pathways were previously shown to be affected by HA (Felipo and Butterworth, 2002; Marcaggi and Coles, 2001; Rama Rao et al., 2005; and Norenberg, 2005; Ratnakumari et al., 1994; Tsacopoulos and Magistretti, 1996). They included glycolysis/gluconeogenesis pathways (Supplementary Fig. 1), oxidative phosphorylation (Supplementary Fig. 2), as well as the citric acid cycle and glutamate metabolism (data not shown).
We also found significant changes in the expression of a considerable number of genes representing pathways that had not been investigated before. These included calcium signaling (Supplementary Fig. 3), glutathione metabolism (Supplementary Fig. 4), as well as xenobiotic metabolism pathways (Supplementary Fig. 5).
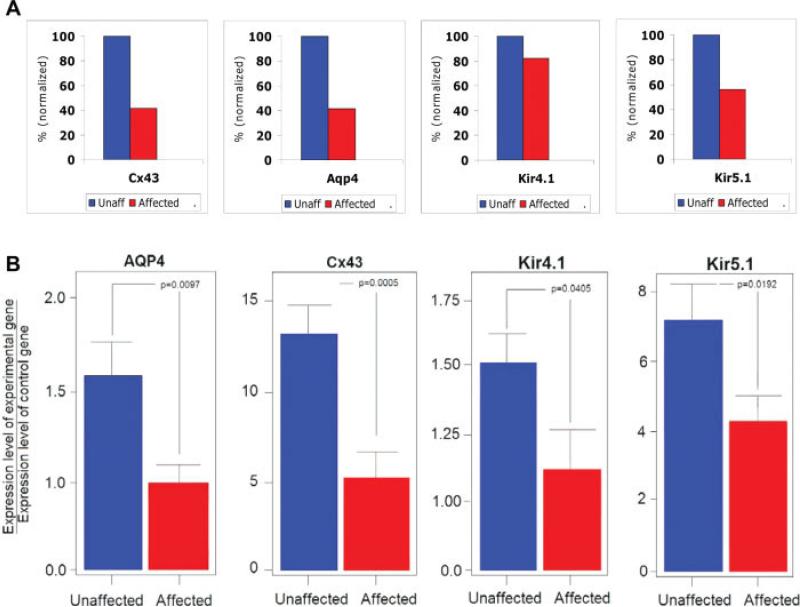
Besides these pathways, changes were detected in the expression levels of channels regulating water and potassium homeostasis in brain. Microarray analysis of FACS-purified astrocytes of hyperammonemic mice demonstrated that expression of the astrocytic gap junction channel gene connexin 43 (Cx43) was downregulated by 58% (P = 0.038) and expression of aquaporin 4 (Aqp4) by 57%, (P = 0.003), when compared with unaffected mice (Fig. 5A). The expression of potassium channel genes was also altered. Specifically, the expression of the Kir5.1 subunit of the astrocytic inward-rectifying potassium channel (Kir) was downregulated by 44% (P = 0.0125), while the expression of its counterpart Kir4.1 was downregulated by 18% (P = 0.044) (Fig. 5A). Changes in gene expression of the main astrocytic gap junction channel Cx43, the astrocytic inward rectifying potassium channel Kir4.1/Kir5.1, and the main astrocytic water channel Aqp4 became the focus of this investigation, because their function could be influenced directly by extracellular ammonium concentrations, and because these channels play a crucial role in potassium and water homeostasis in the brain and in inter-astrocyte signaling.
Fig. 5.
Changes in the expression of genes coding for astrocyte membrane channels during hyperammonemia. (A) Results of microarray analysis for Cx43, Kir4.1, Kir5.1, and Aqp4 genes. Data are averages from 3 to 5 mice for each group, and are expressed as percentage of unaffected mice. Blue: unaffected, red: affected. (B) qRT-PCR results for Aqp4, Cx43, Kir4.1, and Kir5.1. Histograms show expression levels of each gene normalized to an internal control gene (GAPDH). Bars represent s.e.m.
Gene Expression Profile in Cortex of Hyperammonemic Mice
Some short-term changes in gene expression induced by HA could be lost during the tissue dissociation and the FACS procedure, or the FACS purification itself might induce changes in astrocyte gene expression. To capture all changes in gene expression, RNA was isolated from the cortices of P27-30 affected (n = 3) and unaffected (n = 3) mice which had been on arginine-free diet for 18 hr. Therefore, these mice were of the same age and on the same diet as those used for astrocyte isolation by FACS. RNA isolated from the cortices of these mice was subjected to expression profiling. Using congruence between two algorithms and P-value ranking for data analysis (Tumor Analysis Best Practices Working Group, 2004), significant changes in the expression of genes involved in inter-astrocyte signaling and water homeostasis were detected; these included Cx43, Aqp4, the ryanodine receptor 2 (RYR2), and S100α. The Cx43 and Aqp4 genes showed a significant degree of downregulation (23%; P = 0.0025 and 47.5%; P = 0.012, respectively) in the cortex of affected mice, when compared with unaffected mice. RYR2 displayed a significant upregulation (17%, P = 0.0003) and S100α a significant downregulation (30%; P = 0.00036).
Quantitative RT-PCR Analysis
The differences in gene expression at the RNA level found by microarray analysis were confirmed by qRTPCR as a second independent method. RNA levels for Cx43, Kir4.1, Kir5.1, and Aqp4 were measured in 5 independent qRT-PCR experiments performed with new samples of FACS purified astrocytes from hyperammonemic (n = 6) and unaffected (n = 5) mice after 18 hr of arginine-free diet, and from hperammonemic (n = 6) and unaffected (n = 4) mice after 18 hr of a 1% arginine diet (Fig. 5B). Quantitative RT-PCR showed a 57% reduction in Cx43 expression (P = 0.0005), a 40% reduction in Aqp4 expression (P = 0.0097), a 23% reduction in Kir4.1 expression (P = 0.04), and a 41% reduction in Kir5.1 expression (P = 0.019) in affected mice, when compared with unaffected ones (Fig. 5B). We consistently observed a correlation between HA and changes in gene expression with both diets (arginine-free and 1% arginine). There was no significant difference in the results depending on the type of diet used, indicating that HA rather than the amount of arginine in the diet caused the changes in gene expression.
DISCUSSION
Animal models previously used to study the effect of ammonia in the brain (reviewed in Felipo and Butterworth, 2002) included: (i) portacaval shunted rats, as models for chronic liver failure; (ii) portacaval shunted rats with hepatic artery ligation, as models for acute liver failure (ALF); (iii) urease-treated or ammonium acetate-infused healthy rats; and (iv) sparse fur (spf) mice with OTC deficiency, as models for the HAE of UCDs. Making healthy animals persistently hyperammonemic requires very high concentrations of ammonia and remains difficult even then, because the rodent liver metabolizes ammonia very efficiently. Conversely, the genetic mouse models resemble human UCD patients in their biochemical and clinical phenotype (Felipo and Butterworth, 2002).
The animal model generated in this study (the Otcspf/GFAP-EGFP mouse) by crossing a mouse strain carrying the hGFAP-EGFP transgene and a mouse strain with a spontaneous mutation in its Otc gene displayed the same pathological phenotype as the original Otcspf mouse, which has a phenotype that resembles UCD patients. Use of an arginine-free diet ensured that affected Otcspf/GFAP-EGFP animals were hyperammonemic at the time of our molecular analysis and, although variability between individual affected animals was still present, Otcspf/GFAP-EGFP mice as a group had significantly higher plasma ammonia levels than unaffected littermates. Additionally, they also displayed significantly higher brain glutamine levels than unaffected littermate controls, with the glutamine level being an indirect measure of the ammonia level in brain. Feeding the arginine-free diet also to unaffected littermates controlled for the diet effect, but affected and unaffected animals could have still eaten different amounts of argi-nine-free diet. It was reassuring in this regard that statistical analysis of the qRT-PCR data from animals on arginine-free diet and animals on 1% arginine diet did not show a diet effect on the expression of the four plasma membrane channel genes under investigation (data not shown).
The distribution and cellular morphology of GFAPEGFP+ astrocytes was indistinguishable between Otcspf/ GFAP-EGFP and hGFAP-EGFP mice. The overall astrocyte number in cerebral cortex, diencephalon, and sub-cortical white matter was similar between the two mouse strains, and the cellular morphology of the GFAP-EGFP+ astrocytes appeared to be unaffected by the Otcspf mutation. Also, similar percentages of GFAPEGFP+ astrocytes that were stained with anti-S100β were found in the two mouse strains. These findings suggest that the Otcspf mutation itself did not cause detectable effects on astrocyte number and morphology.
A major goal of this study was to determine HA-induced molecular changes in astrocytes in vivo. Micro-array analyses of FACS-purified astrocytes showed that several molecular pathways are affected by HA, including genes encoding the plasma membrane channels Aqp4, Cx43 and Kir4.1/Kir 5.1.
A very important function of the astrocyte syncytium and its gap junction channels is the maintenance of potassium homeostasis in brain (Sontheimer et al., 1990). The downregulation of Cx43 observed in FACS-purified GFAP-EGFP+ astrocytes was analyzed further, because of: (i) the fundamental physiological role of Cx43 as the main gap junction channel in astrocytes (Dermietzel et al., 1991; Iacobas et al., 2004); (ii) the downregulation of Cx43 also found in cortical tissue of hyperammonemic mice (present study); (iii) the clustering and apparent loss of connectivity between EGFP+ astrocytes along the brain vasculature of hyperammonemic animals (present study); and (iv) the importance of Cx43 for potassium homeostasis in the brain (Kozoriz et al., 2006; Wallraff et al., 2006). In addition to Cx43 and gap junctions, potassium homeostasis is also regulated by astrocytic inward-rectifying potassium channels, which exist as Kir4.1 homodimers and Kir4.1/Kir5.1 heterodimers (Hibino et al., 2004; Kofuji et al., 2002; Kucheryavykh et al., 2007; Neusch et al., 2006). In the mouse neocortex, the heteromeric Kir4.1/Kir5.1 channel was detected perivascularly, and both the homomeric Kir4.1 and heteromeric Kir4.1/Kir5.1 channel were found in perisynaptic astrocytic processes (Hibino et al., 2004). Kir4.1 was also shown to be part of a plasma membrane dystroglycan complex that includes Aqp4 (Hibino et al., 2004; Kofuji and Newman, 2004), Agp4 the main astrocytic water channel at the brain vasculature and key to water homeostasis in brain (Badaut et al., 2002; Solenov et al., 2004). Aqp4, Cx43, and Kir4.1/Kir5.1 colocalize to astrocytic end-feet at the cerebral microvasculature (Badaut et al., 2002; Hibino et al., 2004; Rouach et al., 2002) and are involved in the regulation of potassium and water homeostasis in the brain. The similarity in the expression response of these three channels to HA in vivo shown in the present study may reflect a functional association between Cx43, Kir5.1 and Aqp4.
We found by microarray analysis that the expression level of a number of potassium channel genes was altered during HA. A disturbance of potassium homeostasis in the brain as a main effect of HA is consistent with the notion that has ionic properties similar to those of K+ ions. can cross the cell membrane via ion channels, exchangers, or membrane transporters, and can replace K+ or H+ on ion channels and transporters (Marcaggi and Coles, 2001). Recent studies demonstrated that glycoproteins of the Rh family are transporters in the kidney, where ammonium may be exchanged for H+ (Westhoff and Wylie, 2006). In some cell types, is also a substrate for NA+/K+-ATPases or K+-Cl− co-transporters (Marcaggi and Coles, 2001). In cultured neonatal mouse brain astrocytes, appears to permeate an inward rectifying K+ channel (Nagaraja and Brookes, 1998). Ammonia may thus compete with K+ ions at both membrane channels and transporters, thereby altering their function. It is conceivable that ammonium ions directly interfere with the function of the astrocytic gap junction channel Cx43 and the astrocytic potassium channels Kir4.1/Kir4.1 and Kir4.1/Kir5.1. As a result, ions could play an important role in the development of acute HAE by directly hampering potassium buffering.
A change in Aqp4 expression concomitant with a change in Cx43 expression has been previously described. K+ ions permeate Cx43 channels together with water, and water and K+ are separately expelled through Aqp4 and K+ channels from astrocytic endfeet into the brain vasculature (Nicchia et al., 2005). Through this mechanism, Aqp4, Kir4.1/Kir5.1 channels and Cx43 channels would act in concert to ensure proper transport of K+ from a site of neuronal activity to the vasculature. Downregulation of Kir4.1/Kir5.1, and Cx43 expression could consequently result in reduction of potassium ion siphoning during HA and cause elevated extracellular potassium levels and water accumulation. Consistent with this hypothesis, an elevated extracellular K+ concentration was measured in the parietal cortex of healthy rats made hyperammonemic by ammonium acetate infusion (Sugimoto et al., 1997).
Further evidence for a possible coupling of K+ and water transport in perivascular astrocytes stems from the observation that Aqp4 and Kir4.1 channels are interconnected via a dystrophin-glycoprotein complex (DGC) at the plasma membrane (PM) of perivascular retinal glia cells (Connors and Kofuji, 2006) and astrocytes (Guadagno and Moukhles, 2004; Hibino et al., 2004; Kofuji and Newman, 2004). The co-localization of Kir4.1, and Aqp4 to one PM complex in astrocytic end-feet, at capillaries, and at the pia further supports the notion that the regulation of extracellular potassium levels is coupled with water transport (Hibino et al., 2004; Nagelhus et al., 2004).
Finally, ammonium has also been found to permeate aquaporins (Aqp3, Aqp8, Aqp9) and it was speculated that ammonia-permeable aquaporins might constitute a link between metabolism and volume control (Holm et al., 2005). Downregulation of Cx43, Aqp4, and Kir4.1/Kir5.1 could be a direct effect of elevated blood levels. A decrease in the expression of these channels might reduce influx, and water and K+ efflux. The downregulation of these channels could thus reflect a protective response by astrocytes to elevated blood levels, however downregulation of channels responsible for K+ siphoning and water efflux would result in increased extracellular K+ and water levels in the brain. This could be a key mechanism of HA-associated brain injury, and ultimately result in altered neuronal activity and in the development of brain edema.
Our observation of decreased expression of Aqp4 after HA is at variance with previous reports in cultured cells. Rama Rao et al. (2003) demonstrated a time dependent upregulation of Aqp4 expression in astrocyte cultures exposed to ammonia. The discrepancies between our analysis and the cell culture studies are likely to be due to fundamental differences between in situ and in vitro systems, since the complex interplay between the brain vasculature and the perisynaptic space is maintained in intact tissue, but not in cultured astrocytes.
Astrocytic processes surround synapses and regulate the levels of signaling molecules in the extracellular space around synaptic contacts (Haydon, 2001). As a consequence, astrocytes are intimately involved in synaptic transmission, synaptic plasticity, and long term potentiation (Allen and Barres, 2005; Panatier et al., 2006; Robitaille, 1998). Astrocytes are also involved in a variety of brain insults and pathological states (Hansson et al., 2000; Kiening et al., 2002; Kimelberg, 2005; Nakase et al., 2003; Panickar and Norenberg, 2005), therefore these cells are very likely to play an important role in brain damage caused by acute HA and in brain injury caused by chronic HA.
Astrocytic gap junctions enable the transfer of K+ and other ions and small molecules, including Ca2+ and glutamate, between neighboring cells and throughout the syncytium. Through this mechanism, astrocytes can facilitate spatial buffering of K+ and spatial allocation of neurotransmitters and inter-astrocyte signalling over greater distances (Giaume et al., 1997; Hansson et al., 2000; Sontheimer et al., 1990; Wallraff et al., 2006). Therefore, the changes in Cx43, Kir channels and Aqp4 gene expression reported in this study are likely to have a significant impact on neuronal function and network activity during and after HA in the brain. Further studies are needed to demonstrate whether downregulation of these membrane channels is eventually deleterious to the brain. Pharmacological interventions at these channels could then potentially improve outcome.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Frank Kirchhoff for the gift of the GFAP-EGFP mouse. They are particularly thankful to Mark Batshaw and Mendel Tuchman for their support and for valuable discussion. They thank Tarik Haydar for help with confocal images and Linda Moses for help with microarray hybridization, Juan Cabrera for technical assistance, Andrea DeBiase for instructions in data analysis and software usage, and Teresa Hawley (George Washington University) for FACS.
Grant sponsor: NIH MRDDRC; Grant number: P30HD40677; Grant sponsor: NIH Rare Diseases Clinical Research Center; Grant number: 5U54RR019453; Grant sponsor: NIH; Grant number: R21NS050463; Grant sponsor: CURE (Citizens United for Research on Epilepsy); Grant sponsor: NIH NCMRR; Grant number: R24HD050846.
REFERENCES
- Allen NJ, Barres BA. Signaling between glia and neurons: Focus on synaptic plasticity. Curr Opin Neurobiol. 2005;15:542–548. doi: 10.1016/j.conb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Kim RS, Ligon KL, Maher EA, Brennan C, Billings N, Chan S, Li C, Rowitch DH, Wong WH, DePinho RA. Molecular diversity of astrocytes with implications for neurological disorders. Proc Natl Acad Sci USA. 2004;101:8384–8389. doi: 10.1073/pnas.0402140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badaut J, Lasbennes F, Magistretti PJ, Regli L. Aquaporins in brain: Distribution, physiology, and pathophysiology. J Cereb Blood Flow Metab. 2002;22:367–378. doi: 10.1097/00004647-200204000-00001. [DOI] [PubMed] [Google Scholar]
- Batshaw ML, Hyman SL, Coyle JT, Robinson MB, Qureshi IA, Mellits ED, Quaskey S. Effect of sodium benzoate and sodium phenyl-acetate on brain serotonin turnover in the ornithine transcarbamylase-deficient sparse-fur mouse. Pediatr Res. 1988;23:368–374. doi: 10.1203/00006450-198804000-00006. [DOI] [PubMed] [Google Scholar]
- Batshaw ML, Msall M, Trojak J. The risk of serious illness in carriers of ornithine transcarbamylase deficiency. J Pediatr. 1986;108:236–241. doi: 10.1016/s0022-3476(86)80989-1. [DOI] [PubMed] [Google Scholar]
- Batshaw ML, Roan Y, Jung AL, Rosenberg LA, Brusilow SW. Cerebral dysfunction in asymptomatic carriers of ornithine transcarbamylase deficiency. N Engl J Med. 1980;302:482–485. doi: 10.1056/NEJM198002283020902. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Brown EL, Hunter CP. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 2001;29:E29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belachew S, Aguirre AA, Wang H, Vautier F, Yuan X, Anderson S, Kirby M, Gallo V. Cyclin-dependent kinase-2 controls oligodendrocyte progenitor cell cycle progression and is downregulated in adult oligodendrocyte progenitors. J Neurosci. 2002;22:8553–8562. doi: 10.1523/JNEUROSCI.22-19-08553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors NC, Kofuji P. Potassium channel Kir4.1 macromolecular complex in retinal glial cells. Glia. 2006;53:124–131. doi: 10.1002/glia.20271. [DOI] [PubMed] [Google Scholar]
- DeMars R, LeVan SL, Trend BL, Russell LB. Abnormal ornithine carbamoyltransferase in mice having the sparse-fur mutation. Proc Natl Acad Sci USA. 1976;73:1693–1697. doi: 10.1073/pnas.73.5.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermietzel R, Hertberg EL, Kessler JA, Spray DC. Gap junctions between cultured astrocytes: Immunocytochemical, molecular, and electrophysiological analysis. J Neurosci. 1991;11:1421–1432. doi: 10.1523/JNEUROSCI.11-05-01421.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins P, Belanger M, Butterworth RF. Alterations in expression of genes coding for key astrocytic proteins in acute liver failure. J Neurosci Res. 2001;66:967–971. doi: 10.1002/jnr.10045. [DOI] [PubMed] [Google Scholar]
- D'Hooge R, Marescau B, Qureshi IA, De Deyn PP. Impaired cognitive performance in ornithine transcarbamylase-deficient mice on arginine-free diet. Brain Res. 2000;876:1–9. doi: 10.1016/s0006-8993(00)02589-0. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, Knoblach SM, Brandoli C, Aden SA, Hoffman EP, Faden AI. Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Ann Neurol. 2003;53:454–468. doi: 10.1002/ana.10472. [DOI] [PubMed] [Google Scholar]
- Felipo V, Butterworth RF. Neurobiology of ammonia. Prog Neurobiol. 2002;67:259–279. doi: 10.1016/s0301-0082(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Giaume C, Tabernero A, Medina JM. Metabolic trafficking through astrocytic gap junctions. Glia. 1997;21:114–123. [PubMed] [Google Scholar]
- Guadagno E, Moukhles H. Laminin-induced aggregation of the inwardly rectifying potassium channel, Kir41, and the water-permeable channel, AQP4, via a dystroglycan-containing complex in astrocytes. Glia. 2004;47:138–149. doi: 10.1002/glia.20039. [DOI] [PubMed] [Google Scholar]
- Gushiken T, Yoshimura N, Saheki T. Transient hyperammonemia during aging in ornithine transcarbamylase-deficient, sparse-fur mice. Biochem Int. 1985;11:637–643. [PubMed] [Google Scholar]
- Gyato K, Wray J, Huang ZJ, Yudkoff M, Batshaw ML. Metabolic and neuropsychological phenotype in women heterozygous for ornithine transcarbamylase deficiency. Ann Neurol. 2004;55:80–6. doi: 10.1002/ana.10794. [DOI] [PubMed] [Google Scholar]
- Hansson E, Muyderman H, Leonova J, Allansson L, Sinclair J, Blom-strand F, Thorlin T, Nilsson M, Ronnback L. Astroglia and glutamate in physiology and pathology: Aspects on glutamate transport, glutamate-induced cell swelling and gap-junction communication. Neurochem Int. 2000;37:317–329. doi: 10.1016/s0197-0186(00)00033-4. [DOI] [PubMed] [Google Scholar]
- Haydon PG. GLIA: Listening and talking to the synapse. Nat Rev Neurosci. 2001;2:185–193. doi: 10.1038/35058528. [DOI] [PubMed] [Google Scholar]
- Hibino H, Fujita A, Iwai K, Yamada M, Kurachi Y. Differential assembly of inwardly rectifying K1 channel subunits, Kir41 and Kir51, in brain astrocytes. J Biol Chem. 2004;279:44065–44073. doi: 10.1074/jbc.M405985200. [DOI] [PubMed] [Google Scholar]
- Holm LM, Jahn TP, Moller AL, Schjoerring JK, Ferri D, Klaerke DA, Zeuthen T. NH3 and NH14 permeability in aquaporin-expressing Xenopus oocytes. Pflugers Arch. 2005;450:415–428. doi: 10.1007/s00424-005-1399-1. [DOI] [PubMed] [Google Scholar]
- Iacobas DA, Scemes E, Spray DC. Gene expression alterations in connexin null mice extend beyond the gap junction. Neurochem Int. 2004;45:243–250. doi: 10.1016/j.neuint.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Inoue I, Gushiken T, Kobayashi K, Saheki T. Accumulation of large neutral amino acids in the brain of sparse-fur mice at hyperammonemic state. Biochem Med Metab Biol. 1987;38:378–386. doi: 10.1016/0885-4505(87)90102-2. [DOI] [PubMed] [Google Scholar]
- Jayakumar AR, Panickar KS, Murthy ChR, Norenberg MD. Oxidative stress and mitogen-activated protein kinase phosphorylation mediate ammonia-induced cell swelling and glutamate uptake inhibition in cultured astrocytes. J Neurosci. 2006;26:4774–84. doi: 10.1523/JNEUROSCI.0120-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiening KL, van Landeghem FK, Schreiber S, Thomale UW, von Deimling A, Unterberg AW, Stover JF. Decreased hemispheric Aqua-porin-4 is linked to evolving brain edema following controlled cortical impact injury in rats. Neurosci Lett. 2002;324:105–108. doi: 10.1016/s0304-3940(02)00180-5. [DOI] [PubMed] [Google Scholar]
- Kimelberg HK. Astrocytic swelling in cerebral ischemia as a possible cause of injury and target for therapy. Glia. 2005;50:389–397. doi: 10.1002/glia.20174. [DOI] [PubMed] [Google Scholar]
- Kofuji P, Biedermann B, Siddharthan V, Raap M, Iandiev I, Milenkovic I, Thomzig A, Veh RW, Bringmann A, Reichenbach A. Kir potassium channel subunit expression in retinal glial cells: Implications for spatial potassium buffering. Glia. 2002;39:292–303. doi: 10.1002/glia.10112. [DOI] [PubMed] [Google Scholar]
- Kofuji P, Newman EA. Potassium buffering in the central nervous system. Neuroscience. 2004;129:1045–1056. doi: 10.1016/j.neuroscience.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi T, Hayakawa J, Nikaido H. Blood ammonia concentration in mice: Normal reference values and changes during growth. Lab Anim Sci. 1990;40:308–311. [PubMed] [Google Scholar]
- Kozoriz MG, Bates DC, Bond SR, Lai CP, Moniz DM. Passing potassium with and without gap junctions. J Neurosci. 2006;26:8023–8024. doi: 10.1523/JNEUROSCI.2500-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucheryavykh YV, Kucheryavykh LY, Nichols CG, Maldonado HM, Baksi K, Reichenbach A, Skatchkov SN, Eaton MJ. Downregulation of Kir4.1 inward rectifying potassium channel subunits by RNAi impairs potassium transfer and glutamate uptake by cultured cortical astrocytes. Glia. 2007;55:274–281. doi: 10.1002/glia.20455. [DOI] [PubMed] [Google Scholar]
- Maestri NE, Clissold D, Brusilow SW. Neonatal onset ornithine transcarbamylase deficiency: A retrospective analysis. J Pediatr. 1999;134:268–272. doi: 10.1016/s0022-3476(99)70448-8. [DOI] [PubMed] [Google Scholar]
- Marcaggi P, Coles JA. Ammonium in nervous tissue: Transport across cell membranes, fluxes from neurons to glial cells, and role in signalling. Prog Neurobiol. 2001;64:157–183. doi: 10.1016/s0301-0082(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Msall M, Batshaw ML, Suss R, Brusilow SW, Mellits ED. Neurologic outcome in children with inborn errors of urea synthesis. N Engl J Med. 1984;310:1500–1505. doi: 10.1056/NEJM198406073102304. [DOI] [PubMed] [Google Scholar]
- Msall M, Monahan PS, Chapanis N, Batshaw ML. Cognitive development in children with inborn errors of urea synthesis. Acta Paediatr Jpn. 1988;30:435–441. doi: 10.1111/j.1442-200x.1988.tb02534.x. [DOI] [PubMed] [Google Scholar]
- Nagaraja TN, Brookes N. Intracellular acidification induced by passive and active transport of ammonium ions in astrocytes. Am J Physiol. 1998;274:C883–C891. doi: 10.1152/ajpcell.1998.274.4.C883. [DOI] [PubMed] [Google Scholar]
- Nagelhus EA, Mathiisen TM, Ottersen OP. Aquaporin-4 in the central nervous system: Cellular and subcellular distribution and coexpression with KIR4.1. Neuroscience. 2004;129:905–913. doi: 10.1016/j.neuroscience.2004.08.053. [DOI] [PubMed] [Google Scholar]
- Nakase T, Fushiki S, Naus CC. Astrocytic gap junctions composed of connexin 43 reduce apoptotic neuronal damage in cerebral ischemia. Stroke. 2003;34:1987–1993. doi: 10.1161/01.STR.0000079814.72027.34. [DOI] [PubMed] [Google Scholar]
- Natale JE, Ahmed F, Cernak I, Stoica B, Faden AI. Gene expression profile changes are commonly modulated across models and species after traumatic brain injury. J Neurotrauma. 2003;20:907–927. doi: 10.1089/089771503770195777. [DOI] [PubMed] [Google Scholar]
- Natale JE, Knight JB, Cheng Y, Rome JE, Gallo V. Metallothionein I, II mitigate age-dependent secondary brain injury. J Neurosci Res. 2004;78:303–314. doi: 10.1002/jnr.20265. [DOI] [PubMed] [Google Scholar]
- Neusch C, Papadopoulos N, Müller M, Maletzki I, Winter SM, Hirrlinger J, Handschuh M, Bahr M, Richter DW, Kirchoff F, Hülsmann S. Lack of the Kir4.1 channel subunit abolishes K1 buffering properties of astrocytes in the ventral respiratory group: Impact on extracellular K1 regulation. J Neurophysiol. 2006;95:1843–1852. doi: 10.1152/jn.00996.2005. [DOI] [PubMed] [Google Scholar]
- Nicchia GP, Srinivas M, Li W, Brosnan CF, Frigeri A, Spray DC. New possible roles for aquaporin-4 in astrocytes: cell cytoskeleton and functional relationship with connexin43. FASEB J. 2005;19:1674–1676. doi: 10.1096/fj.04-3281fje. [DOI] [PubMed] [Google Scholar]
- Nolte C, Matyash M, Pivneva T, Schipke CG, Ohlemeyer C, Hanisch UK, Kirchhoff F, Kettenmann H. GFAP promoter-controlled EGFP-expressing transgenic mice: A tool to visualize astrocytes and astrogliosis in living brain tissue. Glia. 2001;33:72–86. [PubMed] [Google Scholar]
- Norenberg MD. A light and electron microscopic study of experimental portal-systemic (ammonia) encephalopathy. Progression and reversal of the disorder. Lab Invest. 1977;36:618–627. [PubMed] [Google Scholar]
- Panatier A, Theodosis DT, Mothet JP, Touquet B, Pollegioni L, Poulain DA, Oliet SH. Glia-derived D-serine controls NMDA receptor activity and synaptic memory. Cell. 2006;125:775–784. doi: 10.1016/j.cell.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Panickar KS, Norenberg MD. Astrocytes in cerebral ischemic injury: Morphological and general considerations. Glia. 2005;50:287–298. doi: 10.1002/glia.20181. [DOI] [PubMed] [Google Scholar]
- Rama Rao KV, Chen M, Simard JM, Norenberg MD. Increased aquaporin-4 expression in ammonia-treated cultured astrocytes. Neuroreport. 2003;14:2379–2382. doi: 10.1097/00001756-200312190-00018. [DOI] [PubMed] [Google Scholar]
- Rama Rao KV, Jayakumar AR, Norenberg MD. Role of oxidative stress in the ammonia-induced mitochondrial permeability transition in cultured astrocytes. Neurochem Int. 2005;47:31–38. doi: 10.1016/j.neuint.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Ratnakumari L, Qureshi IA, Butterworth RF. Regional amino acid neurotransmitter changes in brains of spf/Y mice with congenital ornithine transcarbamylase deficiency. Metab Brain Dis. 1994;9:43–51. doi: 10.1007/BF01996073. [DOI] [PubMed] [Google Scholar]
- Robinson MB, Hopkins K, Batshaw ML, McLaughlin BA, Heyes MP, Oster-Granite ML. Evidence of excitotoxicity in the brain of the ornithine carbamoyltransferase deficient sparse fur mouse. Dev Brain Res. 1995;90:35–44. doi: 10.1016/0165-3806(96)83484-5. [DOI] [PubMed] [Google Scholar]
- Rouach N, Avignone E, Meme W, Koulakoff A, Venance L, Blomstrand F, Giaume C. Gap junctions and connexin expression in the normal and pathological central nervous system. Biol Cell. 2002;94:457–475. doi: 10.1016/s0248-4900(02)00016-3. [DOI] [PubMed] [Google Scholar]
- Robitaille R. Modulation of synaptic efficacy and synaptic depression by glial cells at the frog neuromuscular junction. Neuron. 1998;21:847–855. doi: 10.1016/s0896-6273(00)80600-5. [DOI] [PubMed] [Google Scholar]
- Seo J, Bakay M, Chen YW, Hilmer S, Shneiderman B, Hoffman EP. Interactively optimizing signal-to-noise ratios in expression profiling: Project-specific algorithm selection and detection p-value weighting in Affymetrix microarrays. Bioinformatics. 2004;20:2534–2544. doi: 10.1093/bioinformatics/bth280. [DOI] [PubMed] [Google Scholar]
- Sohn J, Natale J, Chew LJ, Belachew S, Cheng Y, Aguirre A, Lytle J, Nait-Oumesmar B, Kerninon C, Kanai-Azuma M, Kanai Y, Gallo V. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci. 2006;26:9722–9735. doi: 10.1523/JNEUROSCI.1716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solenov E, Watanabe H, Manley GT, Verkman AS. Sevenfold-reduced osmotic water permeability in primary astrocyte cultures from AQP-4-deficient mice, measured by a fluorescence quenching method. Am J Physiol Cell Physiol. 2004;286:C426–C432. doi: 10.1152/ajpcell.00298.2003. [DOI] [PubMed] [Google Scholar]
- Sontheimer H, Minturn JE, Black JA, Waxman SG, Ransom BR. Specificity of cell-cell coupling in rat optic nerve astrocytes in vitro. Proc Natl Acad Sci USA. 1990;87:9833–9837. doi: 10.1073/pnas.87.24.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PM, Batshaw M, Valle D, Walser M. Effects of arginine-free meals on ureagenesis in cats. Am J Physiol. 1981;241:E310–E315. doi: 10.1152/ajpendo.1981.241.4.E310. [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Koehler RC, Wilson DA, Brusilow SW, Traystman RJ. Methionine sulfoximine, a glutamine synthetase inhibitor, attenuates increased extracellular potassium activity during acute hyperammonemia. J Cereb Blood Flow Metab. 1997;17:44–49. doi: 10.1097/00004647-199701000-00006. [DOI] [PubMed] [Google Scholar]
- Tsacopoulos M, Magistretti PJ. Metabolic coupling between glia and neurons. J Neurosci. 1996;16:877–885. doi: 10.1523/JNEUROSCI.16-03-00877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumor Analysis Best Practices Working Group Expression profiling—Best practices for data generation and interpretation in clinical trials. Nat Rev Genet. 2004;5:229–237. doi: 10.1038/nrg1297. [DOI] [PubMed] [Google Scholar]
- Veres G, Gibbs RA, Scherer SE, Caskey CT. The molecular basis of the sparse fur mouse mutation. Science. 1987;237:415–417. doi: 10.1126/science.3603027. [DOI] [PubMed] [Google Scholar]
- Wallraff A, Kohling R, Heinemann U, Theis M, Willecke K, Steinhauser C. The impact of astrocytic gap junctional coupling on potassium buffering in the hippocampus. J Neurosci. 2006;26:5438–47. doi: 10.1523/JNEUROSCI.0037-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner T, Bontert M, Eyupoglu I, Prass K, Prinz M, Klett FF, Heinze M, Bechmann I, Nitsch R, Kirchhoff F, Kettenmann H, Dirnagl U, Priller J. Bone marrow-derived cells expressing green fluores-cent protein under the control of the glial fibrillary acidic protein promoter do not differentiate into astrocytes in vitro and in vivo. J Neurosci. 2003;23:5004–5011. doi: 10.1523/JNEUROSCI.23-12-05004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff CM, Wylie DE. Transport characteristics of mammalian Rh and Rh glycoproteins expressed in heterologous systems. Transfus Clin Biol. 2006;13:132–138. doi: 10.1016/j.tracli.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Zhao P, Iezzi S, Carver E, Dressman D, Gridley T, Sartorelli V, Hoffman EP. Slug is a novel downstream target of MyoD. Temporal profiling in muscle regeneration. J Biol Chem. 2002;277:30091–30101. doi: 10.1074/jbc.M202668200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.