Abstract
The Cus system of Escherichia coli aids in protection of cells from high concentrations of Ag(I) and Cu(I). The histidine kinase CusS of the CusRS two-component system functions as a Ag(I)/Cu(I)-responsive sensor kinase and is essential for induction of the genes encoding the CusCFBA efflux pump. In this study, we have examined the molecular features of the sensor domain of CusS in order to understand how a metal-responsive histidine kinase senses specific metal ions. We find that the predicted periplasmic sensor domain of CusS directly interacts with Ag(I) ions and undergoes a conformational change upon metal binding. Metal binding also enhances the tendency of the domain to dimerize. These findings suggest a model for activation of the histidine kinase through metal binding events in the periplasmic sensor domain.
Keywords: CusS, Cus system, Histidine kinase, Silver, Copper, Two-component system
1. Introduction
Bacteria thrive in a variety of environments which necessitates their ability to respond to changing environments in order to acquire nutrients or limit effects of potential toxins. Some transition metals are essential for cellular systems as these elements can play important structural and catalytic roles in proteins and enzymes. However, some metal ions, such as silver, play no cellular roles and their presence can be harmful [1, 2]. Even for necessary metal ions like copper, an overabundance can lead to toxicity through a variety of mechanisms [3], including disruption of iron-sulfur clusters [4]. In bacteria, appropriate levels of metal ions are maintained through a variety of homeostatic systems [5]. To properly respond to a given metal ion, systems are needed which will specifically detect a metal ion and initiate gene expression of the correct homeostatic system [6]. Bacterial two-component systems are commonly used to detect and respond to environmental challenges [7], though two-component systems that specifically respond to monovalent metal ions such as silver and copper have not been previously characterized.
Among the cellular systems that function to prevent metal toxicity in Escherichia coli, the Cus system was first identified as a silver resistance system [8] It was later determined that the Cus system is also involved in anaerobic copper homeostasis [9, 10] which is not unexpected due to the similar chemical properties of silver and copper. The Cus system consists of the CusR/CusS two-component system (TCS) and the CusCFBA efflux complex responsible for transporting copper/silver ions into the extracellular space [8, 9]. The genes encoding these proteins are present on two divergently transcribed operons on the genome of E. coli K12. While the structural and functional properties of the CusCFBA efflux pump have been extensively studied, the metal responsive CusR/CusS system remains largely uncharacterized. Previous work has shown that the CusR/CusS TCS is essential for growth in medium containing silver and copper, as deletion of the cusR and cusS genes leads to a silver and copper sensitive phenotype in the bacterium [10]. It has also been shown that cusR and cusS are required for the silver/copper inducible expression of the promoter region of cusC [10, 11].
The cusS gene encodes a histidine kinase which is predicted to sense elevated levels of silver/copper in the periplasmic space of E. coli. Copper shock causes a two-fold increase in the cusS transcript [12], and the absence of cusS is shown to increase copper accumulation in the cells [11]. From sequence analysis, CusS is predicted to be a prototypical periplasmic sensing histidine kinase (HK). In prototypical histidine kinases, a periplasmic sensor domain is flanked by transmembrane α-helices which link it to conserved cytoplasmic catalytic domains [7]. These proteins function as membrane receptors using a periplasmic (membrane external) domain that regulates activity of the histidine kinase (membrane internal) domain. Most of the variability in histidine kinases is in the periplasmic sensor domains, as they are specific to particular stimulants [13]. The CusS histidine kinase has overall sequence identity to putative metal-sensing HKs such as SilS (56%), CopS (42%), PcoS (38%) and CinS (35%) [10].
Although CusS has an important role in silver resistance and regulation of copper homeostasis [11], little is known about the mechanism of metal sensing and signal transmission. Based on studies on HKs CitA, DcuS and PhoQ, which sense citrate, dicarboxylates and divalent cations, respectively, ligand binding in the periplasmic domain of the proteins results in conformational changes that activate the cytoplasmic kinase domain which in turn leads to altered activity [14–19]. However, the structural and functional features of metal-responsive sensory domains are largely uncharacterized.
This study characterizes the putative periplasmic sensor domain of the histidine kinase CusS from the Gram-negative bacterium Escherichia coli. We have utilized an array of biochemical and biophysical techniques to determine the effect of Ag(I) and other metal ions on the periplasmic domain of CusS.
2. Materials and Methods
2.1 Strain and Plasmid Construction
The residues constituting the two transmembrane helices of CusS were predicted using the HMMTOP and TMHMM transmembrane topology prediction software [20, 21], and were used to determine the boundaries of the CusS periplasmic sensor domain (CusSs) that lies between these helices. The pTXB3CusSs plasmid was constructed for expression of the putative periplasmic domain of CusS for in vitro studies. For this plasmid, the portion of the E. coli W3110 cusS gene encoding amino acids 39-187 was amplified using oligonucleotides containing sites for NcoI and XhoI restriction endonucleases. The amplified PCR product was digested using NcoI/XhoI, then ligated into the NcoI/XhoI sites in vector pTXB3 (NEB). The correct product was verified by DNA sequencing. The final construct, pTXBCusSs, contains the in-frame insertion of the cusS gene encoding the putative periplasmic domain of the protein (residues 39-187) upstream of the Mxe intein and chitin binding domain (CBD) affinity tag [22]. After cleavage of the intein, this construct has a C-terminal EGSS sequence as a cloning artifact.
2.2 Protein Expression, Purification and Sample Preparation
E. coli BL21 (DE3) cells containing the pTXB3CusSs plasmid with the gene encoding CusS39-187 (CusSs) were grown in LB medium containing 100 μg/L ampicillin until an OD600 of 0.8–1.0 was achieved. The cells were induced for CusSs expression with 0.5 mM IPTG and grown at 30 °C for 5–7 hours. Cells were harvested by centrifugation and resuspended in 100 mM Tris pH 8.0 and 150 mM NaCl (column buffer). Lysis was achieved using a French press pressure cell and the cell debris was removed by centrifugation. The clarified supernatant was loaded onto a chitin bead column (NEB) and thiol-induced cleavage was initiated by washing the column with column buffer containing 50 mM DTT. The column was incubated at 16 °C for 24 hours to allow efficient on-column cleavage. CusSs was eluted by washing the column with column buffer. Aliquots of the eluted fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). The fractions containing CusSs were pooled, concentrated to 2 mL, and loaded on a HiPrep Sephacryl S-100 26/60 column previously equilibrated with 100 mM Tris pH 8.0 and 250 mM NaCl. CusSs containing fractions from the column were pooled and dialyzed against 50 mM MES buffer pH 7.0. The purification strategy yielded more than 95% pure protein as confirmed by SDS-PAGE and MALDI-TOF mass spectrometry. Protein concentrations were determined using Bradford (BioRad) and BCA assays (Thermo).
CusSs samples for Inductively Coupled Plasma Mass Spectrometry (ICP-MS) were prepared by dialyzing purified CusSs against Chelex-treated 25 mM MES buffer pH 7.0. To obtain Ag(I)-CusSs, dialysis was performed at 4 °C for 12 hours in 25 mM MES buffer pH 6.0 buffer containing a 5-fold molar excess of AgNO3. As controls for ICP-MS, CusSs samples were dialyzed against buffer containing 5-fold molar excess of ZnCl2 and NiCl2. After equilibration, excess and non-specifically bound metal was removed by dialysis twice against 100 mL of 25 mM MES buffer pH 6.0 (sample buffer) at 4 °C for 3 hours using mini-dialysis units (Thermo Scientific). After dialysis the protein concentration was determined to be 30 μM using Bradford Assay (BioRad).
To obtain protein for NMR analysis a similar protocol was followed except that BL21 (DE3) cells containing the pTXB3CusSs plasmid were grown in M9 minimal medium containing 1 gm/L 15N ammonium chloride as the nitrogen source. Cells were induced for CusSs expression with IPTG and incubated at 37 °C with aeration for 7–9 hours. After purification following the protocol described above, the protein samples were concentrated to 100 μM in 25 mM MES buffer pH 6.0 using an Amicon Ultra 5000 Da concentrator.
CusSs samples for analytical ultracentrifugation experiments were prepared by dialysis against 5-fold molar excesses of AgNO3, ZnCl2 or NiCl2 similarly to that described above for the ICP-MS samples. The concentration of the samples after dialysis was determined to be 1.5 mg/mL using Bradford assay (BioRad).
2.3 Inductively Coupled Plasma Mass Spectrometry
Protein samples (30 μM) were diluted with 1% nitric acid and injected into an Elan DRC II ICP-MS instrument (Perkin Elmer) calibrated using a multi-element stock solution (AccuStandard). All glassware and plastic ware used for these experiments were washed with 10% nitric acid to remove contaminating metal. The protein and metal ion concentrations were determined as an average of three independent experiments.
2.4 Nuclear Magnetic Resonance Spectroscopy
All NMR experiments were performed on a Varian INOVA 600 MHz spectrometer equipped with a cryogenically-cooled, triple-resonance, Z-axis gradient probe. Standard pulse sequences from the Varian BioPak were used to obtain 1H – 15N gradient enhanced heteronuclear single quantum coherence (HSQC) spectra. The HSQC data were processed and analyzed using the NMRPipe/NMRDraw [23] and NMRView programs [24].
2.5 Circular Dichroism Spectroscopy
Circular dichroism of apo and Ag (I)-CusSs were performed in an Olis DSM 20 double beam spectrometer. These experiments were performed at 25 °C in 25 mM MES buffer pH 7.0, using protein concentrations of 25 μM and a 0.1 cm path length cuvette. Spectra were acquired using 1 nm steps from 200 to 260 nm. Five independent sets of data were obtained for each. Separate spectra were obtained for sample buffer alone and the average of these data points was used for baseline correction in the protein spectra.
2.6 Equilibrium Dialysis
Equilibrium dialysis units were obtained from The Nest Group (Southborough, MA). The apparatus consisted of two 500 μL chambers separated by a 5000 Da cellulose membrane (Harvard Apparatus, Holliston, MA). Control experiments were performed to determine the time required to achieve equilibrium (Supplementary table 1). The protein and buffers used in the experiment were Chelex treated to prior to setting up the dialysis experiment. Protein concentrations for each sample were measured at the beginning and end of the experiment by Bradford assay. One chamber contained 500 μL of 6.5 μM CusSs in 25 mM MES buffer pH 6.0 and the other contained 500 μL AgNO3 (at varying concentrations) in 25 mM MES buffer pH 6.0. Twelve different dialysis experiments were performed by varying the concentration of AgNO3 (0 – 200 μM). The dialysis units were allowed to equilibrate at 4 °C on a plate rotator for 72 hours. Samples from both chambers of the dialysis units were analyzed for Ag(I) content using ICP-MS, using the procedure described above. To determine the amount of Ag(I) bound to the protein, the Ag(I) from the buffer-only chamber was subtracted from the protein-containing chamber. The binding data was fit with GraphPad Prism using a single-site model accounting for ligand depletion to determine dissociation constant (Kd) and the total number of binding sites, Bmax, as described by Swillens [25]. The specific binding was calculated for 4 binding sites using a 6.5 μM protein concentration (i.e. 26 μM binding sites). In this case, the term β, which accounts for the use of radioligands, was set to 1, since the ligand (Ag(I)) used in these experiments was non-radioactive.
2.7 Analytical Ultracentrifugation
Sedimentation velocity experiments were performed using a Beckman XL-I ultracentrifuge. Apo CusSs,, Ag(I)-CusSs, Zn(II)+CusSs and Ni(II)+CusSs (1.5 mg/mL each) were centrifuged at 47,000 rpm overnight at 20 °C and the data were analyzed using the Sedfit program using a continuous c(s) distribution model. Sedimentation coefficients were determined using the Svedberg program and using extrapolation to infinite dilution, the coefficients were converted to standard conditions (S 20,W) [26, 27].
2.8 Chemical Crosslinking and Peptide Sequence Analysis
All experiments were performed at 4 °C. Apo and Ag(I)-CusSs were prepared as described above in 50 mM HEPES buffer pH 7.0. Crosslinking reactions were carried out in a total reaction volume of 100 μL using the amine reactive crosslinker BS3 (Thermo). A 100 mM stock of the crosslinker BS3 was prepared in DMF. A 50-fold molar excess of crosslinker (1 mM) was added to 20 μM CusSs and Ag(I)-CusSs. The samples were incubated on ice for 1 hour and the reaction was quenched using 1 M ammonium bicarbonate. Proteins were visualized after separation by SDS-PAGE and silver staining, and quantified using ImageJ [28]. To analyze the peptide sequence of oligomers in the CusSs and Ag(I)-CusSs samples, the proteins were separated on a SDS-PAGE gel, stained using Coomassie Brilliant Blue (Sigma) and bands of interest were excised. Proteins were extracted from the bands using in-gel trypsin digest and extraction using trifluoroacetic acid and acetonitrile. The samples were purified using a C18 mini-column prior to mass spectrometric analysis. The digested peptide samples were run on a LTQ-Orbitrap instrument (Thermo Scientific) and ionized by electrospray ionization in a positive ion mode. The peptide sequence was analyzed using the database search algorithm SEQUEST [29].
3. Results
3.1 Ag(I) ions bind to the periplasmic domain of CusS
CusS is predicted to be a canonical histidine kinase [7] with a periplasmic sensor domain connected by transmembrane helices to the catalytic cytoplasmic domains (Supplementary Figure 1). The periplasmic sensor domain of CusS is predicted to consist of residues 39-187. To analyze the properties of the putative sensor domain, we expressed and purified this domain of CusS (CusSs) for analysis.
The predicted periplasmic domain of CusS contains numerous histidine and methionine residues (9 and 7, respectively) that could serve as potential ligands for Cu(I) and Ag(I), and therefore we expected that CusSs may bind metal ions directly. However, addition of concentrated metal ion solutions to the CusSs sample caused protein precipitation; therefore a dialysis technique was employed to obtain metal-bound protein. Purified CusSs was dialyzed against buffer containing AgNO3, followed by extensive dialysis against metal ion-free buffer to remove excess metal ions. Analysis of these CusSs samples by inductively coupled plasma mass spectrometry (ICP-MS) revealed a significant amount of silver in the samples. Quantitation of the amounts of metal ions in the protein-containing samples shows approximately 4 metal ions per CusSs domain, which is primarily silver along with trace amounts of copper (Table 1). ICP-MS of the buffer without protein does not indicate any significant amount of copper, silver, or other metal ions (data not shown). To determine the specificity of CusSs, Zn(II) and Ni(II) were added by dialysis as described for Ag(I). No significant amounts of these metal ions were associated with CusSs (Table 1).
Table 1.
Metal content of CusSs analyzed by ICP-MS. Metal ion content in CusSs (30 μM) was determined after initial purification (“as purified”) and after dialysis against AgNO3, NiCl2, or ZnCl2 followed by dialysis to remove excess unbound metal. Values indicated are an average of three independent measurements along with their standard deviations.
| Metal Ion | Number of metal ions per molecule of CusSs |
|---|---|
| As purified | |
| 107Ag | 0.00 |
| 63Cu | 0.14 ± 0.023 |
| 60Ni | 0.00 |
| 64Zn | 0.00 |
| After addition of each metal by dialysis | |
| 107Ag | 3.82 ± 0.32 |
| 63Cu | 0.23 ± 0.069 |
| 60Ni | 0.009 ± 0.005 |
| 64Zn | 0.00 |
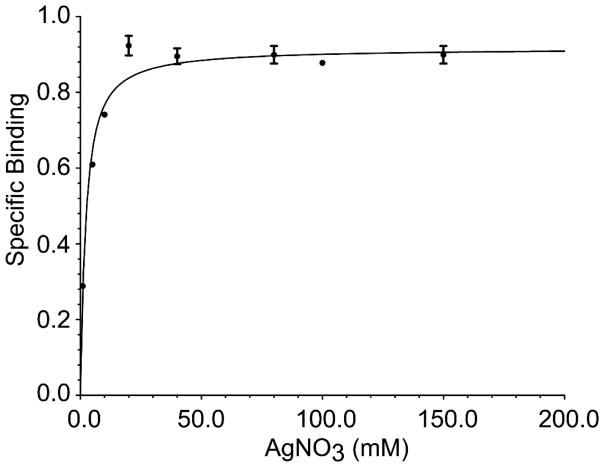
We employed equilibrium dialysis against varying AgNO3 concentrations followed by ICP-MS analysis to determine binding constants (Figure 1), since other experimental approaches were limited due to the observation that CusSs precipitates after direct addition of concentrated metal ion solutions. The apparent affinity of CusSs for Ag(I) was measured to be 8.23 μM ± 0.00057 μM, with a Bmax of 3.62 μM Ag(I)/μM CusSs. Overall, these data suggests that the total of four metal ions (Ag(I) + Cu(I)) bound per molecule of CusSs.
Figure 1. Ag(I) binding by the periplasmic domain of CusS measured by equilibrium dialysis and ICP-MS.
The smooth line represents the best fit model using a dissociation constant of 8.23 μM and a Bmax of 3.62 μM Ag(I)/μM CusSs. Data points are an average of three independent experiments.
3.2 Binding to Ag(I) ions causes changes in spectral properties of CusSs
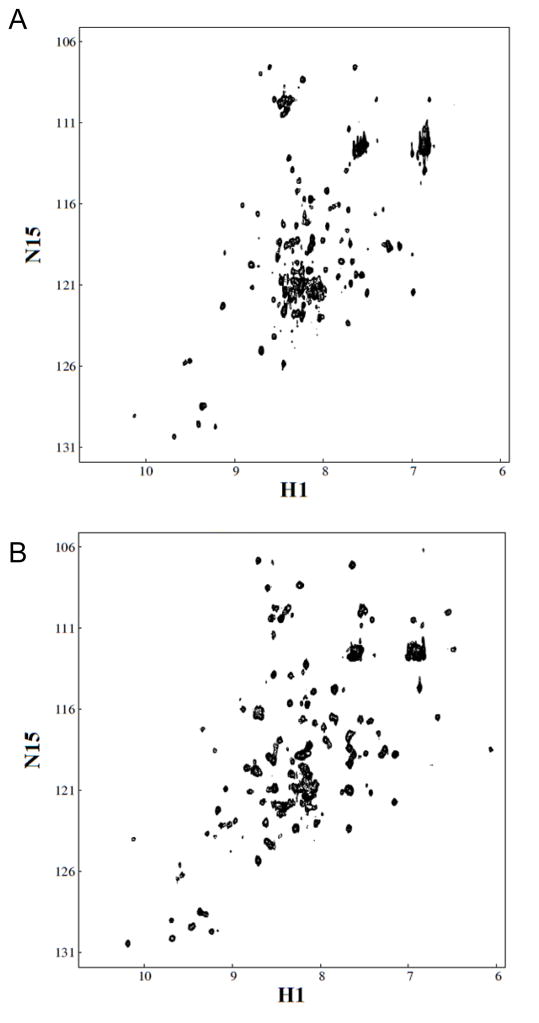
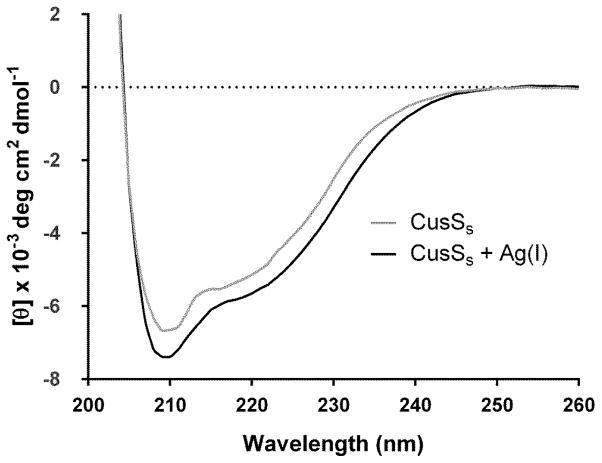
To investigate the effects of Ag(I) on the protein, we collected 1H – 15N correlation spectra of CusSs both in the absence and presence of Ag(I). The peak positions in the HSQC experiments reflect the chemical environment of 1H-15N nuclei for each amino acid residue. The peaks in the NMR spectra of Apo-CusSs are mostly well-dispersed. Addition of Ag(I) to CusSs causes numerous changes in the positions of peaks in the HSQC spectra (Figure 2), and overall shows somewhat increased dispersion. Far-UV circular dichroism (CD) spectra for Apo-CusSs and Ag(I)-CusSs show negative ellipticity with minima near 208 nm and 222 nm (Figure 3).
Figure 2. 1H-15N HSQC NMR spectra of CusSs.
A. Apo-CusSs, B. Ag(I)-CusSs.
Figure 3. Far-UV CD spectra of CusSs (black), and Ag(I)-CusSs (gray).
Circular dichroism experiments were performed in an Olis DSM 20 double beam spectrometer at 25 °C in 25 mM MES buffer pH 7.0, using protein concentrations of 25 μM and a 0.1 cm path length cuvette. Spectra were acquired using 1 nm steps from 200 to 260 nm. Data points from five independent experiments are plotted and the smooth line represents the average of these data points.
3.3 The putative sensor domain of CusS shows increased oligomerization in the presence of Ag(I) ions
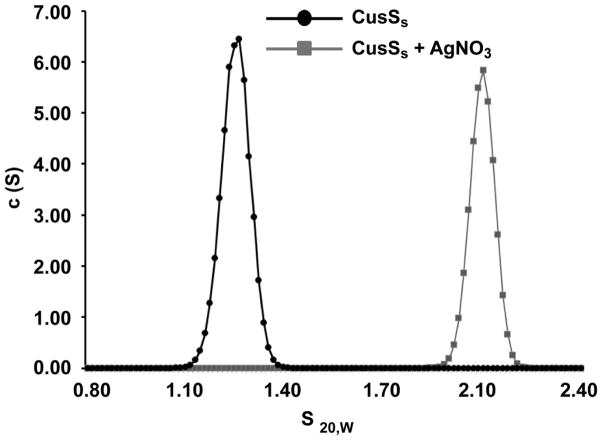
Prototypical histidine kinases function as dimers, with dimerization mediated through cytoplasmic domains. However, some periplasmic sensor domains are also dimeric [19], though it is not clear what role this plays in sensor domain function. To investigate the oligomeric state of CusSs, sedimentation velocity analytical ultracentrifugation (SV-AUC) experiments were performed on apo and Ag(I)-bound CusSs. In these experiments, apo-CusSs sedimented as a single elongated species with a molecular weight consistent with the monomeric form of the protein (Figure 4). For Ag(I)-bound CusSs, there was a distinct shift in the sedimentation coefficient and the frictional ratio (f/f0) represented a molecule more elongated than the monomeric CusSs. These data for Ag(I)-CusSs suggest dimer formation in the presence of metal (Table 2). No significant changes in the SV patterns were observed upon addition of other metals such as zinc or nickel (Supplementary Figure 2, Table 2).
Figure 4. Sedimentation velocity analytical ultracentrifugation analysis of Apo (black) and Ag(I)-bound (gray) samples of CusSs.
The c(s) distribution plots show one sedimentation species for each sample.
Table 2.
Sedimentation velocity parameters for CusSs in the presence of different metal ions. The data were analyzed using the Sedfit program using a continuous c(S) distribution model. Sedimentation coefficients were determined using the Svedberg program and using extrapolation to infinite dilution, the coefficients were converted to standard conditions (s20,w).
| Protein | s20,w | f/f0 |
|---|---|---|
| CusSs | 1.28 | 1.34 |
| CusSs + Ag(I) | 2.1 | 1.63 |
| CusSs + Ni(II) | 1.32 | 1.30 |
| CusSs + Zn(II) | 1.29 | 1.33 |
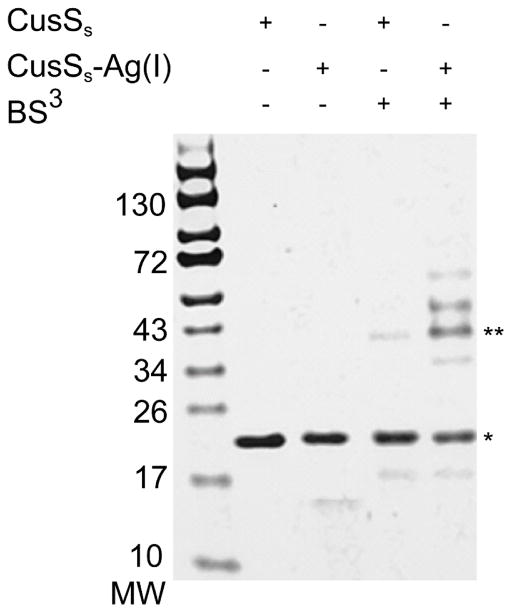
The oligomeric state of CusSs was also probed with in vitro chemical crosslinking using the homobifunctional amine-reactive crosslinker BS3 followed by gel electrophoresis (Figure 5). The cross-linking experiments show that apo-CusSs predominantly migrates at the same position in the denaturing gel in both the absence and presence of crosslinker, though a small amount of a higher molecular weight species is captured in the presence of the crosslinker. However, in the presence of Ag(I), significant higher molecular weight species are captured by the crosslinker. The major higher molecular weight band is of appropriate size for a dimer of CusSs. Using ImageJ analysis, this band was quantified to be 32% of the original sample used for the experiment. The protein in this band, as well as that in the minor bands of higher molecular weight that are seen upon addition of Ag(I), was confirmed to be CusSs using MS/MS analysis and peptide sequence identification using SEQUEST (data not shown).
Figure 5. Cross-linking reactions of Apo and Ag(I)- CusSs.
Samples of Apo and Ag(I)-CusSs (20 μM) in the presence and absence of the BS3 cross-linker were analyzed using SDS-PAGE. The expected monomer of CusSs is indicated by (*) and the expected dimer is indicated by (**)
4. Discussion
Metal ions can exert toxic effects through a variety of mechanisms. The primary mechanisms by which silver ions present toxicity is by binding to sulfhydryl and nitrogen containing groups in biomolecules and by competing for copper sites in proteins [30]. Low amounts of silver ions induce leakage of protons leading to cell death [1]. There is no known biological function for silver ions, and therefore the presence of silver ions must be detected to elicit the appropriate response to reduce its toxicity.
Genetic and molecular analyses have shown that the E. coli cusCFBA genes are upregulated by the CusR/CusS TCS in response to silver and copper [10, 11]. At present, it is not well understood how these ions enter bacterial cells (reviewed in [31]), and no uptake systems in E. coli have been described. The mechanism of sensing silver ions and signal transmission by CusS is unclear. In order to investigate its role in signal recognition, we purified the periplasmic region of CusS and characterized its Ag(I) binding properties.
Many sensor domains of histidine kinases directly bind to the ligands they sense (reviewed in [13]). Since the periplasmic domain of CusS has several His and Met residues that could act as potential ligands to coordinate Ag(I), we tested for Ag(I) binding by this domain. By equilibrium dialysis and ICP-MS, we determined that approximately four metal ions are directly bound to one molecule of the periplasmic domain of CusS with an apparent affinity of 8.23 μM, though the affinities of the individual metal binding sites have not been measured. This apparent affinity is similar to the affinities of several other sensor kinases for their ligands. For example, the sensor kinase PhoQ binds to Mg2+ ions with an affinity of roughly 300 μM [32], the citrate sensing CitA binds to its ligand with a Kd of 5.5 μM [33], and NarX binds to nitrate with a Kd of 35 μM [34].
In the CusSs samples, trace amounts of copper were also detected by ICP-MS, though no copper ions were added to the samples and despite dialysis in preparation of the protein samples. ICP-MS of the buffers without protein indicates that they do not contain copper, suggesting the copper is bound to the protein. An explanation for this trace copper could be the presence of a high-affinity metal binding site in the protein, and that the copper may have been acquired from the bacterial growth medium and then retained through protein purification and dialysis. Based on the ICP-MS measurements of the CusSs samples, the copper is likely to be partially occupying one of the four metal ions binding sites, since the amount of silver plus copper does not exceed 4:1 metal ion to protein (Table 1). Ag(I) and Cu(I) are likely able to occupy the metal ion sites interchangeably because of their similar properties. However, Cu(I) binding data have not been reported here because of the difficulties of maintaining strict anaerobic conditions needed for experiments with Cu(I).
The ICP-MS and equilibrium dialysis data suggests the presence of a metal site in CusSs that can retain copper through extensive dialysis. The presence of a high-affinity metal binding site, in addition to the other metal binding sites, could potentially allow the formation of a response gradient in the signaling process. This idea has been suggested in the ‘rheostat’ model by Inouye et al, where depending on ligand concentration, the histidine kinase can be in an intermediate conformational state fluctuating between the on and off states [35]. Possibly, with one metal ion bound CusS may be minimally activated to allow basal level of expression of the CusCFBA efflux system. If metal concentrations increase, further metal binding to CusS may increase kinase activity, and ultimately increase the cellular response to counter the growing challenge of higher metal concentrations.
Both NMR (Figure 2) and CD spectra (Figure 3) show that the sensor domain of CusS is structured. Spectral changes suggest that additional structural changes take place upon metal binding. Increased dispersion of the Ag(I)-CusS NMR spectrum may indicate a stabilization of the secondary and tertiary structure due to metal binding. Detailed NMR analyses to interpret the structural effects could not be accomplished on these samples due to low concentrations, however, efforts are being made to optimize sample quality for future studies. In other metal-binding systems, such as CopZ and azurin, metal binding loops that are disordered in the apo state adopt structure in the metal-bound state [36, 37]. The bacterial metallochaperone CopZ coordinates Cu(I) through two cysteine residues in a MXCXXC motif located in a solvent exposed loop that transitions from an unstructured to structured state upon metal binding [38]. The spectral changes observed in CusSs upon metal binding are consistent with this type of structural alteration. Taken together these data provide strong evidence that the periplasmic domain of CusS acts as a specific Ag(I) receptor.
Bacterial histidine kinases are known to function as dimers [39] and signal transmission from the extracellular sensor domain to the kinase regions occurs through a dimeric interface [40]. There are numerous examples of sensor domains that in isolation are monomeric in the absence of ligand, but dimerize in the ligand – bound state (reviewed in [13]). Understanding how quaternary changes are induced by ligand binding is key to understanding how kinase activity is controlled. In the case of the aspartate receptor Tar, binding of aspartate to the periplasmic domain causes stabilization of the dimeric form of the protein and causes the inhibition of kinase activity of the histidine kinase CheA [41]. The periplasmic sensor domain of NarX is monomeric in the apo-state, but dimerizes in the presence of its ligand, nitrate. In this case, dimerization is thought to result in piston-like movements of the transmembrane helices that are propagated to the intracellular catalytic domains and alter activity [14]. Similarly, citrate binding to the periplasmic sensor domain of CitA induces dimerization and changes the position of the N and C-terminal helices of the domain. These sensor domain conformational changes induced by ligand binding are coupled to kinase activity.
Based on our analytical ultracentrifugation and crosslinking studies on the histidine kinase CusS, significant dimerization is observed in the periplasmic sensor domain of the protein only in the presence of Ag(I). The crosslinking data also show minor amounts of dimerization in the absence of silver, which may be caused due to residual copper bound to CusSs as seen in the ICP-MS data (Table 1). Minor amounts of higher molecular weight bands are also seen in the presence of silver, which may have formed due to an excess of crosslinker (Figure 5). Overall our data points to a model where the sensor domain of CusS dimerizes upon binding metal. However, it remains to be tested how ligand-induced dimerization of the sensor domain contributes to the kinase activation of full-length CusS.
Based on our study we can envision a model where the periplasmic domain of CusS acts as a receptor to sense the presence of elevated levels of Ag(I) (or Cu(I)) by direct binding from solution. Ag(I) binding causes local conformational changes in the periplasmic domain of CusS and enhances dimer formation between the sensor domains. These protein-protein interactions mediated by elevated periplasmic levels of silver or copper ions may be an important step, if not the only step, in activation of downstream signaling events that trigger the upregulation of Ag(I)-responsive genes. Further insight into ligand binding and conformational change leading to kinase activation will require knowledge of metal coordination by the periplasmic domain of CusS and its three dimensional structure.
Supplementary Material
Acknowledgments
The authors would like to thank Chad Park for assistance with analytical ultracentrifugation experiments. This work was supported by the National Institutes of Health grant GM079192 to M.M.M.
Abbreviations
- HK
histidine kinase
- TCS
two-component system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bragg PD, Rainnie DJ. The effect of silver ions on the respiratory chain of Escherichia coli. Can J Microbiol. 1974;20(6):883–9. doi: 10.1139/m74-135. [DOI] [PubMed] [Google Scholar]
- 2.Jung WK, Koo HK, Kim KW, Shin S, Kim SH, Park YH. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl Environ Microbiol. 2008;74(7):2171–8. doi: 10.1128/AEM.02001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson NJ, Winge DR. Copper metallochaperones. Ann Rev Biochem. 2010;79:537–62. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Nat Acad Sci U SA. 2009;106(20):8344–9. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27(2–3):313–39. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 6.Rademacher C, Masepohl B. Copper-responsive gene regulation in bacteria. Microbiol. 2012;158(Pt 10):2451–64. doi: 10.1099/mic.0.058487-0. [DOI] [PubMed] [Google Scholar]
- 7.Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Ann Rev Microbiol. 2010;64:539–59. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 8.Franke S, Grass G, Nies DH. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiol. 2001;147(Pt 4):965–72. doi: 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 9.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bact. 2003;185(13):3804–12. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munson GP, Lam DL, Outten FW, O’Halloran TV. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bact. 2000;182(20):5864–71. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudipaty SA, Larsen AS, Rensing C, McEvoy MM. Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol Lett. 2012;330(1):30–7. doi: 10.1111/j.1574-6968.2012.02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external zinc. J Bact. 2005;187(18):6333–40. doi: 10.1128/JB.187.18.6333-6340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung J, Hendrickson WA. Sensor domains of two-component regulatory systems. Curr Opin Microbiol. 2010;13(2):116–23. doi: 10.1016/j.mib.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17(2):190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kneuper H, Janausch IG, Vijayan V, Zweckstetter M, Bock V, Griesinger C, Unden G. The nature of the stimulus and of the fumarate binding site of the fumarate sensor DcuS of Escherichia coli. J Biol Chem. 2005;280(21):20596–603. doi: 10.1074/jbc.M502015200. [DOI] [PubMed] [Google Scholar]
- 16.Kramer J, Fischer JD, Zientz E, Vijayan V, Griesinger C, Lupas A, Unden G. Citrate sensing by the C4-dicarboxylate/citrate sensor kinase DcuS of Escherichia coli: binding site and conversion of DcuS to a C4-dicarboxylate- or citrate-specific sensor. J Bact. 2007;189(11):4290–8. doi: 10.1128/JB.00168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pappalardo L, Janausch IG, Vijayan V, Zientz E, Junker J, Peti W, Zweckstetter M, Unden G, Griesinger C. The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J Biol Chem. 2003;278(40):39185–8. doi: 10.1074/jbc.C300344200. [DOI] [PubMed] [Google Scholar]
- 18.Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit R, Miller SI. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26(2):165–74. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Sevvana M, Vijayan V, Zweckstetter M, Reinelt S, Madden DR, Herbst-Irmer R, Sheldrick GM, Bott M, Griesinger C, Becker S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J Mol Biol. 2008;377(2):512–23. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 20.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 21.Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283(2):489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 22.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen CA, Pelletier JJ, Paulus H, Xu MQ. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192(2):271–81. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 23.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4(5):603–14. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 25.Swillens S. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol Pharmacol. 1995;47(6):1197–203. [PubMed] [Google Scholar]
- 26.Dam J, Schuck P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Meth Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]
- 27.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 30.Dibrov P, Dzioba J, Gosink KK, Hase CC. Chemiosmotic mechanism of antimicrobial activity of Ag(+) in Vibrio cholerae. Antimicrob Agents Chemother. 2002;46(8):2668–70. doi: 10.1128/AAC.46.8.2668-2670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arguello JM, Raimunda D, Padilla-Benavides T. Mechanisms of copper homeostasis in bacteria. Front Cell Infec Microbiol. 2013;3:73. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesley JA, Waldburger CD. Comparison of the Pseudomonas aeruginosa and Escherichia coli PhoQ sensor domains: evidence for distinct mechanisms of signal detection. J Biol Chem. 2001;276(33):30827–33. doi: 10.1074/jbc.M104262200. [DOI] [PubMed] [Google Scholar]
- 33.Kaspar S, Perozzo R, Reinelt S, Meyer M, Pfister K, Scapozza L, Bott M. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol Microbiol. 1999;33(4):858–72. doi: 10.1046/j.1365-2958.1999.01536.x. [DOI] [PubMed] [Google Scholar]
- 34.Lee AI, Delgado A, Gunsalus RP. Signal-dependent phosphorylation of the membrane-bound NarX two-component sensor-transmitter protein of Escherichia coli: nitrate elicits a superior anion ligand response compared to nitrite. J Bact. 1999;181(17):5309–16. doi: 10.1128/jb.181.17.5309-5316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inouye M, Dutta R. Histidine Kinases in Signal Transduction 2002. Academic Press; [Google Scholar]
- 36.Hussain F, Wittung-Stafshede P. Impact of cofactor on stability of bacterial (CopZ) and human (Atox1) copper chaperones. Biochim Biophys Acta. 2007;1774(10):1316–22. doi: 10.1016/j.bbapap.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Granillo A, Wittung-Stafshede P. Tuning of copper-loop flexibility in Bacillus subtilis CopZ copper chaperone: role of conserved residues. J Phys Chem B. 2009;113(7):1919–32. doi: 10.1021/jp807594q. [DOI] [PubMed] [Google Scholar]
- 38.Winkler JR, Wittung-Stafshede P, Leckner J, Malmstrom BG, Gray HB. Effects of folding on metalloprotein active sites. Proc Nat Acad Sci USA. 1997;94(9):4246–9. doi: 10.1073/pnas.94.9.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao R, Stock AM. Biological insights from structures of two-component proteins. Ann Rev Microbiol. 2009;63:133–54. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldberg SD, Soto CS, Waldburger CD, Degrado WF. Determination of the physiological dimer interface of the PhoQ sensor domain. J Mol Biol. 2008;379(4):656–65. doi: 10.1016/j.jmb.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends in biochemical sciences. 2001;26(4):257–65. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.