Abstract
Genome-wide association studies (GWAS) have begun to identify the common genetic component to ischaemic stroke (IS). However, IS has considerable phenotypic heterogeneity. Where clinical covariates explain a large fraction of disease risk, covariate informed designs can increase power to detect associations. As prevalence rates in IS are markedly affected by age, and younger onset cases may have higher genetic predisposition, we investigated whether an age-at-onset informed approach could detect novel associations with IS and its subtypes; cardioembolic (CE), large artery atherosclerosis (LAA) and small vessel disease (SVD) in 6,778 cases of European ancestry and 12,095 ancestry-matched controls. Regression analysis to identify SNP associations was performed on posterior liabilities after conditioning on age-at-onset and affection status. We sought further evidence of an association with LAA in 1,881 cases and 50,817 controls, and examined mRNA expression levels of the nearby genes in atherosclerotic carotid artery plaques. Secondly, we performed permutation analyses to evaluate the extent to which age-at-onset informed analysis improves significance for novel loci. We identified a novel association with an MMP12 locus in LAA (rs660599; p = 2.5×10−7), with independent replication in a second population (p = 0.0048, OR(95% CI) = 1.18(1.05–1.32); meta-analysis p = 2.6×10−8). The nearby gene, MMP12, was significantly overexpressed in carotid plaques compared to atherosclerosis-free control arteries (p = 1.2×10−15; fold change = 335.6). Permutation analyses demonstrated improved significance for associations when accounting for age-at-onset in all four stroke phenotypes (p<0.001). Our results show that a covariate-informed design, by adjusting for age-at-onset of stroke, can detect variants not identified by conventional GWAS.
Author Summary
Ischaemic stroke places an enormous burden on global healthcare. However, the disease processes that lead to stroke are not fully understood. Genome-wide association studies have recently established that common genetic variants can increase risk of ischaemic stroke and its subtypes. In this study, we aimed to identify novel genetic associations with ischaemic stroke and its subtypes by addressing the fact that younger onset cases may have a stronger genetic component, and using this information in our analyses. We identify a novel genetic variant on chromosome 11 (rs660599), which is associated with increased risk of large artery stroke. We also show that mRNA expression of the nearest gene (MMP12) is higher in arteries with the disease process underlying large artery stroke (atherosclerosis). Finally, we evaluate our novel analysis approach, and show that our method is likely to identify further associations with ischaemic stroke.
Introduction
Genome-wide association studies (GWAS) in ischaemic stroke have begun to identify the common genetic variants that confer risk of the disease. However, there is considerable heterogeneity present in stroke phenotypes: GWAS analyses have primarily looked at the three main subtypes; cardioembolic (CE), large artery atherosclerosis (LAA) and small vessel disease stroke (SVD). Within these subtype analyses, numbers of cases are smaller, but the expectation is that the effects of SNPs identified within the subtypes will be considerably larger. Indeed, all validated GWAS SNPs for ischaemic stroke to date have been stroke subtype-specific [1], [2], [3], [4], [5], indicating the importance of subtyping of cases.
Clinical risk factors are important in stroke; as many as 77% of first-ever stroke patients are hypertensive [6], and other factors such as diabetes mellitus and elevated serum cholesterol confer a considerable proportion of disease risk [7]. These risk factors increase in prevalence in older age groups, suggesting older stroke patients may have a reduced stroke-specific genetic contribution. Indeed, IS is uncommon in individuals below middle age, but increases greatly in prevalence beyond the age of 65 [8], with a lifetime risk of 1 in 5 for women and 1 in 6 for men [9].
Under the assumptions of the liability threshold model, the low prevalence of IS in younger age ranges suggests that individuals who do suffer strokes in this age group are likely to have an increased genetic predisposition. This is supported by family history data; with stronger family history seen in younger onset cases [10], [11], [12], and twin studies [13], which suggest that early onset cases may have higher heritability. We recently showed stronger effects for all stroke-associated SNPs in younger age groups, found evidence genome-wide that a significant number of SNPs show stronger association p-values when the oldest cases are removed, and showed increased pseudoheritability estimates for younger onset cases in certain stroke subtypes, thereby supporting this hypothesis [14]. However, the question of how best to integrate this information into GWAS analyses of ischaemic stroke remains unanswered. Previous GWAS have analysed younger subsets of ischaemic stroke cases [1], [15], but this approach may not be optimal for existing GWAS datasets if the increase in odds ratios for SNPs in younger cases are not sufficient to justify discarding a large proportion of the ascertained cases. All previous young onset analyses have been restricted to all ischaemic stroke cases versus controls; this may be particularly relevant given that all known loci for ischaemic stroke to date are for stroke subtypes [16].
A recent publication [17], outlined a novel method of informing genetic association analyses on important clinical covariates. Using the liability threshold model in conjunction with estimates of disease prevalence for individuals with specific clinical covariates, the method estimates posterior disease liabilities for each individual in a GWAS, and uses these liabilities in regression analyses to test for association with genome-wide SNPs. This approach avoids issues due to multiple testing across age-at-onset thresholds, and provides a simple solution that is rooted is previous epidemiological research. In the present study, we extend the clinical covariate informed analysis approach to imputed genotypes, informing our analyses on the age-at-onset to identify novel variants associated with IS. We perform a genome-wide analysis with four stroke phenotypes (IS, CE, LAA, SVD), and then determine the utility of the approach in ischaemic stroke GWAS, testing whether SNPs increase in significance.
Results
Association analysis
We performed age-at-onset informed association analysis for a total of 6,778 ischaemic stroke cases and 12,095 controls across four ischaemic stroke phenotypes; all IS and the three major subtypes: CE, LAA, and SVD (Table 1); with 1,637, 1,316, and 1,108 cases in the CE, LAA and SVD analyses respectively. With the exception of the young Milanese cohort, the age-at-onset distributions were similar in all cohorts (Table S3).
Table 1. Sample size of discovery populations.
| Study Population | IS | CE | LAA | SVD | Controls |
| Belgium – Immunochip | 396 | 147 | 57 | 49 | 319 |
| Germany-Immunochip | 421 | 127 | 101 | - | 2,355 |
| Krakow – Immunochip | 384 | 119 | 33 | 28 | 255 |
| Sweden – Immunochip | 796 | 246 | 56 | 183 | 997 |
| UK – Immunochip | 867 | 130 | 152 | 257 | 1,790 |
| Germany – WTCCC2 | 1,174 | 330 | 346 | 106 | 797 |
| UK – WTCCC2 | 2,374 | 474 | 498 | 460 | 5,175 |
| Milano | 366 | 64 | 73 | 25 | 407 |
| Total (Discovery) | 6,778 | 1,637 | 1,316 | 1,108 | 12,095 |
IS, all ischaemic stroke; CE, cardioembolic stroke; LAA, large artery stroke; SVD, small vessel disease.
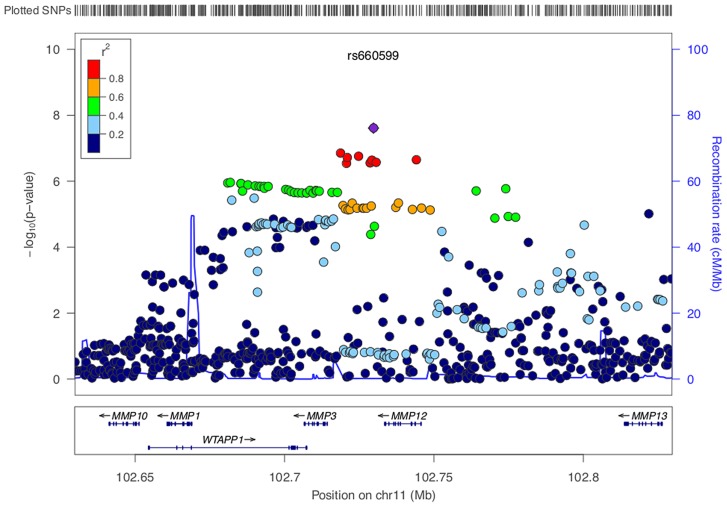
We identified a group of twenty SNPs proximal to MMP3 and MMP12 on chromosome 11 in the LAA subtype that met our criteria for replication. The strongest associated of these was rs662558 (p = 1.4×10−7), a SNP that is in 1000 Genomes, but not HapMap II. Therefore, to enable replication in existing METASTROKE datasets, which were imputed to HapMap II, we selected the most strongly associated SNP from the HapMap II panel, which was in perfect LD with the lead SNP in our discovery meta-analysis (rs660599: uninformed, p = 1.6×10−6; informed, p = 2.5×10−7; Figure 1) [16]. We found no evidence of between-study heterogeneity at either SNP (Cochran's Q p = 0.22 and p = 0.19 for rs662558 and rs660599, respectively). The evidence of an age-at-onset effect at rs660599 was p = 0.011 (from permutations). We calculated age-at-onset quartiles for all large artery stroke cases from the discovery cohorts, and used these to evaluate this region at different age-at-onset thresholds. The median age-at-onset was 71 years, and the interquartile range was between 61 and 78 years. Post-hoc analyses of rs660599 in the discovery cohorts using logistic regression (full details in Text S2) showed considerably stronger associations in younger age-at-onset quantiles (Q1; OR(95% CI) = 1.83 (1.46–2.30), Q1–Q2; 1.56 (1.33–1.83), Q1–Q3; 1.30 (1.14–1.49), Q1–Q4; 1.30 (1.15–1.46)). No other regions met our criteria for replication.
Figure 1. LocusZoom plot of MMP12 association using age-at-onset informed approach.
SNPs are colored based on their correlation (r2) with the labeled top SNP, which has the smallest P value in the region. The fine-scale recombination rates estimated from 1000 Genomes (EUR) data are marked in light blue, with genes marked below by horizontal blue lines. Arrows on the horizontal blue lines show the direction of transcription, and rectangles are exons. SNP p-values are from the discovery meta-analysis only with the exception of rs660599, for which the given p-value indicates the overall evidence for association from the discovery and replication cohorts.
Replication analysis
The associated locus was evaluated in a further 1,881 large artery stroke cases and ancestry matched controls in 9 cohorts from METASTROKE (Table 2). We found evidence for replication of the SNP (rs660599) in all large artery stroke cases of European Ancestry (p = 0.0048, OR(95% CI) = 1.18(1.05–1.32)). Combining this result with the discovery p-value gave a genome-wide significant p-value of 2.6×10−8 (Table 3). Secondly, we used the Han and Eskin random effects meta-analysis approach to evaluate the association [18] after including a further 355 cases and 1,390 controls of Pakistani ancestry. The evidence for replication in this sample was p = 0.0063, giving an overall p-value of 3.4×10−8. Age-at-onset information was available across all age-at-onset quantiles for a subset of the replication studies (1,240 cases, 9,238 controls; ASGC, HVH, ISGS/SWISS, MGH-GASROS, Utrecht). We evaluated the SNP (rs660599) in these studies at different age-at-onset quantiles using logistic regression, meta-analysing as previously. We again found the strongest effects in the youngest age quantile, consistent with a stronger effect in younger onset cases (Q1; OR(95% CI) = 1.27(1.02–1.57), Q1–Q2; 1.18(1.00–1.39), Q1–Q3; 1.22(1.05–1.40), Q1–Q4; 1.22(1.07–1.41)).
Table 2. Sample size of replication populations.
| Study Population | LAA (age<61) | LAA (age<71) | LAA (age<78) | LAA (all ages) | IS | Controls |
| ARIC | - | - | - | 31 | 385 | 8,803 |
| ASGC | 81 | 179 | 277 | 421 | 1,162 | 1,244 |
| deCODE | - | - | - | 255 | 2,391 | 26,970 |
| GEOS | - | - | - | 37 | 448 | 498 |
| HVH | 18 | 39 | 63 | 71 | 566 | 2,072 |
| ISGS/SWISS | 84 | 130 | 179 | 217 | 1,070 | 1,370 |
| MGH-GASROS (Affymetrix) | 31 | 60 | 79 | 102 | 485 | 3,030 |
| MGH-GASROS (Illumina) | 22 | 47 | 59 | 68 | 296 | 377 |
| PROMISe | 134 | 230 | 301 | 324 | 556 | 1,145 |
| RACE | - | - | - | 355 | 1,390 | 5,308 |
| Total (Replication) | 370 | 685 | 958 | 1,881 | 8,749 | 50,817 |
LAA, large artery stroke; IS, all ischaemic stroke; ARIC, the Atherosclerosis Risk in communities study; ASGC, the Australian Stroke Genetics collaboration; deCODE, deCODE genetics; GEOS, the Genetics of early onset stroke study; HVH, the heart and vascular health study; ISGS/SWISS, the Ischaemic stroke genetics study/Siblings with Ischaemic stroke study; MGH-GASROS, Massachusetts General Hospital – Genetics affecting stroke risk and outcome; PROMISe, Prognostic modeling in ischaemic stroke study [55]; RACE, Risk Assessment of Cerebrovascular Events study. For further details of these populations please see the original METASTROKE publication [16].
Table 3. Evidence for association of A allele of rs660599 (chromosome 11; Base position 102,234,967) with large artery atherosclerotic stroke and all ischaemic stroke.
| Subtype | SNP | RAF | p-value (discovery) | OR (95% CI) (EUR replication) | p-value (EUR replication, overall) | p-value (ALL replication, overall) |
| LAA | rs660599 | 0.19 | 2.5.×10−7 | 1.18 (1.05–1.32) | 0.0048, 2.6×10−8 | 0.0063, 3.4×10−8 |
| IS | “ | “ | 3.2×10−4 | 1.05 (1.00–1.11) | 0.050, 1.9×10−4 | 0.098, 3.6×10−4 |
| CE | “ | “ | 0.13 | - | - | - |
| SVD | “ | “ | 0.30 | - | - | - |
LAA, large artery stroke; IS, all ischaemic stroke; SNP, single nucleotide polymorphism; RAF, risk allele frequency; OR, odds ratio; 95% CI, 95% confidence interval; EUR, meta-analysis in individuals of European ancestry alone; ALL, trans-ethnic meta-analysis of all individuals. Forest plots of effect sizes and standard errors for each replication centre are given in Figures S3, S4.
mRNA expression in carotid plaques
mRNA expression of the two proximal genes, MMP3 and MMP12 was analysed from 29 carotid, 15 abdominal aorta, 24 femoral plaques, and 28 atherosclerosis free left internal thoracic artery controls. MMP12 expression was upregulated in carotid plaques compared with left internal thoracic artery controls (P = 1.2×10−15; fold change [FC] = 335.6). It was also upregulated in femoral plaques (P = 3.2×10−14; FC = 306.0) and abdominal plaques (P = 5.0×10−11; FC = 399.3) compared with controls. Conversely, MMP3 was not significantly overexpressed in carotid, femoral or abdominal plaques versus controls (p>0.05).
Regulatory information from ENCODE
Eight SNPs were identified that were perfect proxies (r2 = 1) with the associated SNP (rs660599) in the region. Seven of the SNPs were in an intergenic region between MMP3 and MMP12, while one fell within an intron of MMP12. We investigated the evidence that any of these SNPs are functional variants using RegulomeDB [19]. Of the eight SNPs, we found strong evidence that one of these SNPs (rs586701) affects binding. The SNP overlaps both CHIP-seq and DNA-seq peaks from ENCODE analyses, indicating that there is open chromatin in the region, and therefore that the SNP is likely to be functional. There is also evidence from a separate CHIP-seq analysis that the SNP affects protein binding [20], and evidence from multiple sources that the SNP overlaps a predicted motif [21], [22], [23]. Histone modifications were observed in CHIP-seq experiments from ENCODE in a number of cells types, including Human umbilical vein endothelial (Huvec) cells. Two other SNPs (rs17368582, rs2276109) in moderate LD with the associated SNP (r2 = 0.64) have been previously shown to directly influence MMP12 expression by affecting the affinity of an AP-1 binding site in the MMP12 promoter region [24], [25]. Using RegulomeDB, we found further evidence from ENCODE that one of these SNPs (rs2276109) is indeed functional, giving evidence that the associated locus in this analysis is likely to affect MMP12 expression through altered transcription. Detailed results for all analysed SNPs are given in Table S1. Additionally, we investigated if these SNPs (rs17368582, rs2276109, rs586701) were associated with MMP12 expression in tissues from the GTEx project [26]. However, we could not confirm an association with MMP12 expression in any relevant tissues (p>0.4 in whole blood, tibial artery, aortic artery).
Evaluation of age-at-onset informed approach
Finally, we evaluated the overall utility of the age-at-onset informed approach in permutation analyses for SNPs that met p-value thresholds in the case control discovery data set. We generated 1000 permutations of age-at-onset within each centre, and performed age-at-onset informed analysis and subsequent meta-analysis for these SNPs, in the relevant stroke subtype.
We compared the sum of the meta-analysis Z scores from all SNPs with p<0.05 in the observed age at onset informed meta-analysis with those from permutations. At this p-value selection threshold, we found strong evidence (p<0.001) for genome-wide age-at-onset effects in each of the stroke phenotypes, with consistently increased summed Z scores in the observed age-at-onset informed meta-analysis compared to the permutations (Figure 2, red points, right hand axis). These results suggest that many of the risk variants for each stroke subphenotype have a higher frequency in younger onset cases. As the p-value selection threshold decreased, the summed Z score statistic became less significant in each stroke type, possibly reflecting lower overall power when fewer SNPs are included, even as these SNPs may have larger average effects. Further details are seen from the median proportion of SNPs more significant in the age-at-onset informed analysis than in the permutations (Figure 2, blue points, left hand axis). For CE and LAA stroke, the proportions increased with more stringent p-value thresholds (from 52.1% to 56.3% for p<0.05 and p<0.00005 thresholds in CE, and from 51.4% to 56.0% for p<0.05 and p<0.00005 thresholds in LAA). Interestingly, in the all ischaemic stroke analysis the median proportion of SNPs more significant in the observed results than permutations dropped from 55.1% for SNPs with p<0.05 to 49.2% for only SNPs with p<0.00005. This result may indicate a reduced proportion of true associations at stricter p-value thresholds for all ischaemic stroke compared to the subtypes, which is consistent with the observation that all common variants associated with stroke are for stroke subtypes, rather than for the phenotype of all ischaemic stroke [16].
Figure 2. Evaluation of evidence genome-wide for SNPs exhibiting greater significance using the age-at-onset informed approach compared to permutations.
-log10(p value) from permutations for evidence of age-at-onset effect at given SNP p-value selection threshold shown in red; median proportion of SNPs (with IQR) more significant in observed age-at-onset informed meta-analysis compared to permutations shown in blue; horizontal line at p = 0.05 in red; horizontal line at median proportion of SNP = 0.5 in blue; IS, all ischaemic stroke; CE, cardioembolic stroke; LAA, large artery atherosclerotic stroke; SVD, small vessel disease. See Table S5 for number of SNPs included at each p-value selection threshold.
The previously reported GWAS associations from a recent ischaemic stroke meta-analysis (9p21, HDAC9, PITX2, ZFHX3) were all found to be more significant using the age-at-onset informed approach than the uninformed analysis (Figure 3). The increase in significance ranged from over half an order of magnitude (7.9×10−9 to 1.5×10−9 for rs879324 in ZFHX3, CE), to under half an order of magnitude (5.7×10−9 to 2.5×10−9 for rs2107595 in HDAC9, LVD). To ensure these analysis methods were comparable, we calculated genomic inflation factors and plotted QQ-plots. These were similar in the standard and the age-at-onset informed approach (Table S4, Figure S1, S2). For these four associated SNPs, we further used the permuted data sets to assess the observation of increased significance in the age-at-onset informed analysis. We compared the observed meta-analysis p-value to those from the permutations, generating an empirical p-value by dividing the number of permutations more significant than the observed results by the number of permutations. In LAA stroke, we observed a significant age-at-onset effect (p = 0.018, 0.011 and 0.002 for the HDAC9, MMP12 and 9p21-associated SNPs in Figure 3, respectively). Similarly, for CE, we observed a significant age-at-onset effect for rs879324 (ZFHX3, p = 0.026), and a near-significant effect in rs6843082 (PITX2, p = 0.081). This result provides further evidence that risk variants associated with ischaemic stroke subtypes have a stronger role in younger onset cases, and suggests that the age-at-onset informed approach will produce improved significance when the magnitude of genetic effects are stronger in younger onset cases.
Figure 3. Meta-analysis p-values of known loci for ischaemic stroke subtypes using age-at-onset informed approach compared to uninformed approach.
-log10 of p-values derived from meta-analysis of all discovery cohorts using age-at-onset informed approach (red) and uninformed approach (blue). 9p21 (rs1004638), MMP12 (rs660599) and HDAC9 (rs2107595) p-values calculated within large artery atherosclerosis subtype of stroke, PITX2 (rs6843082) and ZFHX3 (rs879324) p-values calculated with cardioembolic stroke subtype.
Discussion
We used a large GWAS dataset to evaluate the utility of an age-at-onset informed analysis approach to ischaemic stroke, and to identify novel variants associated with ischaemic stroke phenotypes. We identified a novel MMP12 locus that is associated with large artery atherosclerotic stroke, and verified that the age-at-onset informed approach produces improved significance for loci associated with each of the stroke phenotypes studied, as well as demonstrating that it increased the significance of four previous GWAS associations with ischemic stroke, all without systematic inflation of the test statistic. Importantly, the novel associated SNP would not have been identified using a standard logistic regression framework.
We identified a group of SNPs proximal to Matrix Metalloproteinase 12 (MMP12) that showed increased significance when using the age-at-onset informed approach. The increase in significance from the equivalent uninformed analysis was of almost an order of magnitude (from p = 1.6×10−6 to p = 2.5×10−7 for rs660599). We took a single SNP from this region forward for replication in an independent dataset, finding further evidence that the region is associated with large artery stroke. Two SNPs (rs17368582, rs2276109) in this LD-block have previously been shown to directly influence MMP12 expression by affecting the affinity of an AP-1 binding site in the MMP12 promoter region [24], [25], and another variant in this block (rs17361668) is associated with increased fibrinogen levels, leading to an increased risk of developing advanced carotid atherosclerotic lesions, and an increased risk of myocardial infarction. We identified a second functional candidate (rs586701), which falls within both CHIP-seq and DNA-seq peaks from ENCODE, and is in complete LD with the associated SNP in our analysis.
We investigated mRNA expression of MMP12 and MMP3 in carotid atherosclerotic plaques in individuals from the Tampere Vascular Study. MMP12 was overexpressed in diseased tissue compared to healthy controls, while no significant difference was found for the other nearby gene, MMP3. MMP12 is a member of the Matrix Metalloproteinase (MMP) family of proteases, which are capable of degrading extracellular matrix proteins, and have a prominent role in atherosclerosis. They are thought to promote macrophage invasion [27], [28], [29], promote angiogenesis [30], and show increased activity in atheromatous plaques [31]. MMP12 deletions are associated with smaller, more stable lesions in the brachiocephalic artery of rabbits [32], and reduced elastin degradation in the aortic arch [33], indicating that MMP12 may have a role in destabilising plaques. Studies in humans have found MMP12 is localized to the core of advanced plaques, in macrophages with decreased arginase-I expression [34], that MMP12 localizes selectively to macrophages at the borders of the lipid core [35], and that MMP12 is significantly overexpressed in ruptured plaques when compared with thick or thin cap plaques, or with plaques with pathological intimal thickening [36]. This indicates that MMP12 is likely be involved in late-stage plaque instability: our study suggests that genetic variation impacts on this process.
Secondly, we performed extensive permutation analyses to assess the utility of the age-at-onset informed approach genomewide. In each phenotype studied we found evidence that SNPs were more strongly associated using the approach than would be expected by chance, indicating that multiple risk variants are likely to be more common in younger onset cases. The significance was strongest when more SNPs were included in the analysis, which likely reflects the cumulative impact of age-at-onset effects on many SNPs. An alternative explanation might be that the increased significance for lower p-value thresholds is the result of the cumulative effects of subtle confounding. However, this is unlikely because any subtle biases will also be present in the permutations, and should therefore not affect the significance of the results. This result supports observations from family history and prospective cohort studies, which have observed stronger effects in younger onset cases [6], [11]. Furthermore, all known associations with stroke were more significant using the age-at-onset informed approach. The increase in significance was around half an order of magnitude (e.g from p = 7.9×10−9 to 1.5×10−9 for ZFHX3, Figure 2), and was significant in all but one locus, as assessed by permutation. Taken together, these results indicate that age-at-onset is an important measure to stratify stroke cases, and show that, as expected by theory [17], integrating this information into association studies is likely to increase power to identify novel loci when the relative contribution of genetic is dependent on age-at-onset.
Our study has limitations. We used imputed data from the Immunochip platform, meaning we only had access to ∼40% of the genome across all centres. Secondly, cases were drawn from a number of international centres, meaning that despite efforts to standardize phenotyping, we cannot rule out differences in screening and clinical ascertainment.
Of complex diseases, IS has a particularly large degree of heterogeneity, exemplified by the fact that all validated associations identified to date have been within subtypes defined by clinical and radiological information. Further heterogeneity by risk factor and clinical covariate profiles is likely to exist, but the optimal method of incorporating this information into analyses remains an unanswered question. Our results indicate that a covariate-informed design, conditioning on age-at-onset of stroke, can unearth further associated variants. We provide evidence for this by identifying an association with a novel MMP12 locus in large artery stroke, supported by increased mRNA expression of the implicated gene in carotid plaques. GWAS in ischaemic stroke have begun to identify the genetic component of the disease, but these results are not yet clinically useful. Our study suggests that a more refined approach to analysis of genetic data, incorporating covariate information, is an important step in this process, and will help to ensure success in future GWAS.
Materials and Methods
Ethics statement
All studies were approved by their local ethics committees; all patients gave informed consent.
Description of datasets
The initial dataset consisted of 6,778 ischaemic stroke cases of European ancestry and 12,095 ancestry-matched controls from the Wellcome Trust Case-Control Consortium II project in ischaemic stroke [1], as well as a cohort from Milan, Italy [16]. These included 2,858 cases and 5,716 matched controls genotyped using the Immunochip platform; and 3,940 cases genotyped using either the Illumina 610 k or 660 k platforms matched with 6,379 controls genotyped on the Illumina Human 1.2M Duo (UK), Illumina Human 550 k (German) and Illumina 610 k platforms (Italian) (Table 1). The Immunochip cases were described in the previous WTCCC2 ischaemic study, where they formed the replication effort [1], as well as in a recent paper [37]. Genotyping of the five Immunochip case cohorts on the commercially available Immunochip array (Illumina, San Diego, CA, USA) was performed at the Sanger Centre, Hinxton, Cambridge UK. Swedish controls were provided and genotyped by the Swedish SLE network, Uppsala, Sweden. Belgian control samples were provided through the efforts of the International Multiple Sclerosis Genetics Consortium (IMSGC). German controls were derived from the PopGen biobank, [38]. UK controls were derived from the 1958 Birth cohort. Any of the 1958 Birth controls overlapping with those from the WTCCC2 datasets, as assessed by IBD estimates, were removed prior to analysis. Standard quality control procedures were undertaken on all centres, before centre-wise imputation to the 1000 Genomes phase 1 integrated variant set (March 2012), using IMPUTE v2.2.0 [39], [40]. SNPs with poor imputation quality (info<0.3) or low minor allele frequency (MAF<0.01) were discarded.
Ischemic stroke was defined as a typical clinical syndrome with radiological confirmation; ascertained cases were classified into individual stroke subtypes using the Trial of Org 10172 in acute stroke (TOAST) criteria in all centres [41]. Age-at-onset was defined as age at first hospital admission for stroke; where this information was unavailable, age at blood draw was used (7.3% of cases). The age-at-onset and gender distributions of the populations are given in Table S3. Age-at-onset quantiles were calculated from all the cases from the discovery datasets in the four stroke phenotypes (all IS and the three stroke subtypes: CE, LAA, SVD) and these were used to evaluate associated loci at different age-at-onset thresholds.
Association analysis
The prevalence of ischaemic stroke by age was obtained from a recent publication [9]; gender-specific estimates were averaged, and prevalences within each of the stroke subtypes were assumed to be approximately 20% of the overall total, similar to proportions seen in population-based studies [42]. We modeled phenotype data using a continuous unobserved quantitative trait called the disease liability, which we used to approximate the effect of age-at-onset on the liability scale, based on estimates of ischaemic stroke prevalence by age from epidemiological data (full details in Text S2). We developed two models for our analysis; one based on the prevalence rates for all ischaemic stroke cases, and secondly for the three stroke subtypes. We used these models to calculate posterior mean liabilities after conditioning on age-at-onset for the four stroke phenotypes separately. Controls were modeled in the same way, but were assumed to take the posterior mean from the lower (unaffected) portion of the distribution in the liability threshold model. Where age data was missing, individuals were assigned the median age value. Full descriptions of the models used and the formulae used to calculate posterior mean liabilities are given in Text S2. Regression was then performed on posterior liabilities by multiplying the number of samples by the squared correlation between the expected genotype dosage and posterior mean liabilities for each of the discovery cohorts in the four ischaemic stroke phenotypes (CE, LAA, SVD, IS), following a previous approach [17]. Ancestry-informative principal components were included where appropriate (6 of 8 centres), using the EIGENSTRAT procedure [43]. All analysis was performed using the R statistical software.
The results from each centre were meta-analysed for each of the four phenotypes using Stouffer's Z-score weighted approach, as implemented in METAL [44]. Genomic control was used to correct for any residual inflation due to population stratification [45]. Between-study heterogeneity was assessed using Cochran's Q statistic. We considered only SNPs present in at least 75% of the cases, and with no evidence of heterogeneity (Cochran's Q p-value>0.001). All SNPs analysed were either genotyped or imputed in both the Immunochip and the genome-wide datasets. After meta-analysis, the resulting p-values were compared with the equivalent values from an unconditioned analysis. For SNPs more significant in the age-at-onset informed analysis and with p<5×10−6, we determined the evidence of a true age-at-onset effect by generating 1000 permutations of age-at-onset and rerunning the age-at-onset informed analysis, meta-analysing as previously. We calculated an empirical p-value by dividing the number of permuted observations showing greater significance in the meta-analysis than the observed results by the number of permutations. Any novel SNP with a meta-analysis p<5×10−6 and evidence of an age-at-onset effect at p<0.05 were taken forward for replication. We set the experiment-wide significance threshold at p<5×10−8.
Replication analysis
Replication of an associated variant was performed in a further 10 cohorts from METASTROKE. Nine of the centres used a cross-sectional design, while one was a large prospective, population based cohort (ARIC). Nine of the centres were of European ancestry, while one consisted of individuals of Pakistani ancestry (RACE) (Table 2). All centres used a case-control methodology; centres with a cross sectional design used logistic regression to model the association of genotype dosages from imputation with the dichotomous outcome of ischaemic stroke and prospective cohorts used Cox proportional-hazards models to evaluate time to first stroke, fitting an additive model relating genotype dose to the stroke outcome. European ancestry replication centres were meta-analysed using a fixed effects inverse-variance weighted method. To assess the evidence for association of the SNP for replication samples of all ancestries, we performed a trans-ethnic meta-analysis using a random-effects model to control for any resulting heterogeneity [18]. To evaluate the overall evidence for association, the results of the discovery and replication analyses were combined using Fisher's Method.
mRNA expression in carotid atherosclerotic plaques
Expression of the two genes proximal to the associated variant was tested in atherosclerotic plaques from the Tampere Vascular study [27], [46], [47], [48], [49]. Carotid, femoral, and aortic atherosclerotic plaques constituting the intima and inner media were prospectively obtained between 2005 and 2009 from patients fulfilling the following inclusion criteria: (1) carotid endarterectomy attributable to asymptomatic or symptomatic >70% carotid stenosis, or (2) femoral or (3) aortic endarterectomy with aortoiliac or aortobifemoral bypass attributable to symptomatic peripheral arterial disease. Whole thickness left internal thoracic artery samples obtained during coronary artery bypass surgery and identified as being microscopically atherosclerosis free were used as controls. The patients were consecutively recruited and stratified according to indication for surgery. All open vascular surgical procedures were performed at the Division of Vascular Surgery and Heart Center, Tampere University Hospital.
Fresh tissue samples were immediately soaked in RNALater solution (Ambion Inc) and homogenized using an Ultra-Turrax T80 homogenizer (IKA). RNA was extracted with the Trizol reagent (Invitrogen) and miRNEasy Mini-Kit (Qiagen) with the RNase-Free DNase Set (Qiagen) according to manufacturer instructions. The RNA isolation protocol was validated by analyzing the integrity of the RNA with the RNA 6000 Nano Chip Kit (Agilent). The expression levels were analyzed with an Illumina HumanHT-12 v3 Expression BeadChip (Illumina). In brief, 300–500 ng of RNA was reverse transcribed in cRNA and biotin-UTP labeled using the IlluminaTotalPrep RNA Amplification Kit (Ambion), and 1500 ng of cRNA was then hybridized to the Illumina HumanHT-12 v3 Expression BeadChip.
The BeadChips were scanned with the Illumina iScan system. After background subtraction, raw intensity data were exported using the Illumina Genome Studio software. Further data processing was conducted by means of R language and appropriate Bioconductor modules. Data were log2-transformed, and robust multichip average and robust spline normalization (rma_rsn) were used. Accuracy of the expression array was validated with qRT-PCR [50]. mRNA Expression levels in the tissues were determined; a fold change statistic was estimated between the two tissues, and significance was calculated using a t test.
Regulatory information using RegulomeDB
Recent evidence indicates that a significant proportion of GWAS SNPs fall within regions that are likely to affect binding of nearby proteins, such as transcription factor binding sites [51], [52]. We used the RegulomeDB database to access regulatory information from ENCODE and other existing publications [19], investigating the evidence that the SNPs in the associated locus have a regulatory function. First, the linkage-disequilibrium (LD) patterns amongst the most strongly associated SNPs were determined. We then used PLINK to determine the LD structure of the associated region, using LD-patterns from the 85 Utah residents from the 1000 Genomes project [53], [54]. All SNPs with r2>0.6 were identified within a 2,000 kb window from the index SNP. All of the SNPs identified were then investigated using RegulomeDB to determine the evidence that any of the SNPs have a regulatory function.
Evaluation of age-at-onset informed approach
Permutation analysis was performed to evaluate the age-at-onset informed approach, to show that including age at onset information directly led to the increased significance, due solely to inclusion of age-at-onset information at tested SNPs. First, we identified a set of SNPs enriched for true association in the case control analysis of ischaemic stroke and subtypes. An expanded set of discovery and METASTROKE studies were analysed using standard case control methods and subsequent meta-analysis (see Table S2). SNPs with p<0.05 and no evidence of heterogeneity (p>0.0001) were extracted and pruned for LD (300 kb window, r2<0.25), leaving a set of almost independent SNPs for further analysis. Each retained SNP represented the most significant association in each LD block, as determined by the “clump” procedure in PLINK, based on LD patterns from the CEU individuals from 1000 Genomes. The number of SNPs used in each analysis is given in Table S5. These SNP subsets were derived for ischaemic stroke, and for each stroke subset and then used in the age-at-onset informed analysis. Analysis was performed as previously for each stroke subtype using the age-at-onset informed method within studies and meta-analysis across studies (giving observed results, as obtained above). We then performed a permutation study to obtain the expected distribution of p-values at these SNPs. Age at onset for cases was permuted within stroke subtypes within each study, and then the data were re-analysed, for 1000 permutations. Two summary statistics were constructed: (1) within permutations, we compared p-values from analysis of permuted age at onset with p-values from the observed data, and tabulated the proportion of SNPs with increased significance in the observed data set than in the permuted data set; across permutations, we calculated the median proportion of SNPs with increased significance in the observed data; (2) Within permutations, we converted each SNP p-value to a Z score and summed the absolute value of the Z score across SNPs (sumZ). An empirical p-value for the age-informed analysis was calculated from the proportion of simulated data sets where sumZ exceeded the value in the observed analysis. This analysis was performed at SNP subsets defined from four SNP p-value thresholds in the discovery and METASTROKE studies: p<0.05, p<0.005, p<0.0005, and p<0.00005.
Finally, we assessed the evidence of an age-at-onset effect at the four stroke loci identified in the METASTROKE ischaemic stroke collaboration (9p21, HDAC9, PITX2, ZFHX3) [16]. For each SNP, we generated an empirical p-value from the proportion of permutations showing stronger association than in the observed age-at-onset informed analysis.
Supporting Information
QQ-plots for cardioembolic stroke and all ischaemic stroke analyses. QQ-plots of expected p-values (x-axis) against observed p-values (y-axis) for analyses of (clockwise from top left) cardioembolic stroke (age-at-onset informed), cardioembolic stroke (uninformed), all ischaemic stroke (uninformed), all ischaemic stroke (age-at-onset informed). Lambda values for each plot are given in Table S4.
(DOCX)
QQ-plots for large artery atherosclerotic stroke and small vessel disease stroke analyses. QQ-plots of expected p-values (x-axis) against observed p-values (y-axis) for analyses of (clockwise from top left) large artery stroke (age-at-onset informed), large artery stroke (uninformed), small vessel stroke (uninformed), small vessel stroke (age-at-onset informed). Lambda values for each plot are given in Table S4.
(DOCX)
Forest plot of SNP effects for rs660599 in the large artery atherosclerotic stroke replication populations. ASGC, the Australian Stroke Genetics collaboration; deCODE, deCODE genetics; GEOS, the Genetics of early onset stroke study; HVH, the heart and vascular health study; ISGS/SWISS, the Ischaemic stroke genetics study/Siblings with Ischaemic stroke study; MGH-GASROS, Massachusetts General Hospital – Genetics affecting stroke risk and outcome. PROMISe, Prognostic modeling in ischaemic stroke study; RACE, Risk Assessment of Cerebrovascular Events study.
(DOCX)
Forest plot of SNP effects for rs660599 in the large artery atherosclerotic stroke replication populations for cases with age <61 years. ASGC, the Australian Stroke Genetics collaboration; HVH, the heart and vascular health study; ISGS/SWISS, the Ischaemic stroke genetics study/Siblings with Ischaemic stroke study; MGH-GASROS, Massachusetts General Hospital – Genetics affecting stroke risk and outcome. PROMISe, Prognostic modeling in ischaemic stroke study.
(DOCX)
Results from RegulomeDB, showing the evidence that SNPs in the associated MMP12 region have a regulatory function. Scores indicate the following degrees of evidence: Score 2b, TF binding + any motif + DNase Footprint + DNase peak; Score 4, TF binding + DNase peak; Score 5, TF binding or DNase peak; Score 6, other; “No data” indicates that RegulomeDB holds no information about the given SNP, meaning there currently exists no evidence to suggest that the SNP has a regulatory function. In some cases this may indicate that the SNP falls within a protein-coding region. SNP, single nucleotide polymorphism.
(DOCX)
Expanded set of populations used to generate SNPs with p<0.05 to evaluate the age-at-onset informed approach. ARIC, The Atherosclerosis Risk in Communities study; ASGC, Australian Stroke Genetics Collaborative; CHS, Cardiovascular Health Study; FHS, Framingham Heart Study; HPS, Heart Protection Study; HVH, The Heart and Vascular Health Study; ISGS/SWISS, The Ischemic Stroke Genetics Study/Sibling with Ischaemic Stroke Study; MGH-GASROS, The MGH Genes Affecting Stroke Risk and Outcome Study; WTCCC2-Germany, The Wellcome Trust Case-Consortium II Munich; WTCCC2-UK, The Wellcome Trust Case-Consortium II UK; RACE, Risk Assessment of Cerebrovascular Events Study, Pakistan.
(DOCX)
Age and gender distributions of populations. ARIC, The Atherosclerosis Risk in Communities study; ASGC, Australian Stroke Genetics Collaborative; CHS, Cardiovascular Health Study; FHS, Framingham Heart Study; HPS, Heart Protection Study; HVH, The Heart and Vascular Health Study; ISGS/SWISS, The Ischemic Stroke Genetics Study/Sibling with Ischaemic Stroke Study; MGH-GASROS, The MGH Genes Affecting Stroke Risk and Outcome Study; WTCCC2-Germany, The Wellcome Trust Case-Consortium II Munich; WTCCC2-UK, The Wellcome Trust Case-Consortium II UK; RACE, Risk Assessment of Cerebrovascular Events Study, Pakistan. IS, all ischaemic stroke; CE, cardioembolic stroke; LAA, large artery stroke; SVD, small vessel disease.
(DOCX)
Genomic inflation (λ) rates for discovery populations for age-at-onset informed and uninformed approaches. IS, all ischaemic stroke; CE, cardioembolic stroke; LAA, large artery stroke; SVD, small vessel disease.
(DOCX)
Number of SNPs used in evaluation of age-at-onset informed approach. IS, all ischaemic stroke; CE, cardioembolic stroke; LAA, large artery stroke; SVD, small vessel disease.
(DOCX)
Membership of Wellcome Trust Case Control Consortium 2 (WTCCC2).
(DOCX)
Liability threshold models.
(DOCX)
Acknowledgments
The authors thank all study staff and participants for their important contributions, and METASTROKE for granting access to study data.
Funding Statement
The principal funding for this study was provided by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z and WT084724MA). HSM is supported by an NIHR Senior Investigator award and the NIHR Biomedical Research Centre at Cambridge. We acknowledge support from the National Institutes of Health Research Biomedical Research Centre at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. The PROMISe Study (The Netherlands) was made possible, in part, by a Complementation Grant to PIWdB from the Biobanking and Biomolecular Resources Research Infrastructure in the Netherlands (BBMRI-NL). SA is supported in part by a grant from the Netherlands Heart Foundation (grant no. 2005B031) and a grant from the Dutch Brain Foundation (project 2008(1).10). The Australian Stroke Genetics Collaboration (ASGC) Australian population control data was derived from the Hunter Community Study. We also thank the University of Newcastle for funding and the men and women of the Hunter region who participated in this study. This research was funded by grants from the Australian National and Medical Health Research Council (NHMRC Project Grant ID: 569257), the Australian National Heart Foundation (NHF Project Grant ID: G 04S 1623), the University of Newcastle, the Gladys M Brawn Fellowship scheme and the Vincent Fairfax Family Foundation in Australia. EGH is supported by the Australian NHMRC Fellowship scheme. The Genetics of Early Onset Stroke (GEOS) Study, Baltimore, USA was supported by the National Institutes of Health Genes, Environment and Health Initiative (GEI) Grant U01 HG004436, as part of the GENEVA consortium under GEI, with additional support provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK072488); and the Office of Research and Development, Medical Research Service, and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs. Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the National Institutes of Health to the Johns Hopkins University (contract number HHSN268200782096C). Assistance with data cleaning was provided by the GENEVA Coordinating Center (U01 HG 004446; PI Bruce S Weir). Study recruitment and assembly of datasets were supported by a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control and by grants from the National Institute of Neurological Disorders and Stroke (NINDS) and the NIH Office of Research on Women's Health (R01 NS45012, U01 NS069208-01). The Heart and Vascular Health Study (HVH) research reported in this article was funded by NHLBI grants R01 HL085251 and R01 HL073410 The Ischemic Stroke Genetics Study (ISGS)/Siblings With Ischemic Stroke Study (SWISS) study was supported in part by the Intramural Research Program of the National Institute on Aging, NIH project Z01 AG-000954-06. ISGS/SWISS used samples and clinical data from the NIH-NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds), human subjects protocol numbers 2003-081 and 2004-147. ISGS/SWISS used stroke-free participants from the Baltimore Longitudinal Study of Aging (BLSA) as controls. The inclusion of BLSA samples was supported in part by the Intramural Research Program of the National Institute on Aging, NIH project Z01 AG-000015-50, human subjects protocol number 2003-078.The ISGS study was funded by NIH-NINDS Grant R01 NS-42733 (JFM, P.I.). The SWISS study was funded by NIH-NINDS Grant R01 NS-39987 (JFM, P.I.). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (http://biowulf.nih.gov). The MGH Genes Affecting Stroke Risk and Outcome Study (MGH-GASROS) GASROS was supported by The National Institute of Neurological Disorders and Stroke (U01 NS069208), the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research 0775010N, the National Institutes of Health and National Heart, Lung, and Blood Institute's STAMPEED genomics research program (R01 HL087676) and a grant from the National Center for Research Resources. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research resources. Milano - Besta Stroke Register Collection and genotyping of the Milan cases within CEDIR were supported by Annual Research Funding of the Italian Ministry of Health (Grant Numbers: RC 2007/LR6, RC 2008/LR6; RC 2009/LR8; RC 2010/LR8). FP6 LSHM-CT-2007-037273 for the PROCARDIS control samples. The Wellcome Trust Case-Control Consortium 2 (WTCCC2) The principal funding for the WTCCC2 stroke study was provided by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z and WT084724MA). The Stroke Association provided additional support for collection of some of the St George's, London cases. The Oxford cases were collected as part of the Oxford Vascular Study which is funded by the Medical Research Council, Stroke Association, Dunhill Medical Trust, National Institute of Health Research (NIHR) and the NIHR Biomedical Research Centre, Oxford. PMR has a Wellcome Trust Senior Investigator Award and an NIHR Senior Investigator Award. The Edinburgh Stroke Study was supported by the Wellcome Trust (clinician scientist award to CS), and the Binks Trust. Sample processing occurred in the Genetics Core Laboratory of the Wellcome Trust Clinical Research Facility, Western General Hospital, Edinburgh. This work was supported by grants received from the German Federal Ministry of Education and Research (BMBF) in the context of the e:Med program (e:AtheroSysMed) and the FP7 European Union project CVgenes@target (261123) to Martin Dichgans. Much of the neuroimaging occurred in the Scottish Funding Council Brain Imaging Research Centre (www.sbirc.ed.ac.uk), Division of Clinical Neurosciences, University of Edinburgh, a core area of the Wellcome Trust Clinical Research Facility and part of the SINAPSE (Scottish Imaging Network – A Platform for Scientific Excellence) collaboration (www.sinapse.ac.uk), funded by the Scottish Funding Council and the Chief Scientist Office. Collection of the Munich cases and data analysis was supported by the Vascular Dementia Research Foundation. This work made use of data and samples generated by the 1958 Birth Cohort (NCDS). Access to these resources was enabled via the 58READIE Project funded by Wellcome Trust and Medical Research Council (grant numbers WT095219MA and G1001799). A full list of the financial, institutional and personal contributions to the development of the 1958 Birth Cohort Biomedical resource is available at http://www2.le.ac.uk/projects/birthcohort. Tampere Vascular Study (TVS) was supported by the European Union 7th Framework Programme funding for the AtheroRemo project [201668], the Finnish Foundation of Cardiovascular Research (TL), the Finnish Cultural Foundation, the Tampere Tuberculosis Foundation, the Emil Aaltonen Foundation (NO & TL), the Tampere University Hospital Medical Fund (grant 9M048 and 9N035) and the the Foundation of Clinical Chemistry. Immunochip case data for UK, Polish, Belgian, German and Swedish cohorts was genotyped by the Sanger Centre, Cambridge, UK as part of the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z and WT084724MA), as were the UK and Polish control cohorts. German Control Genotypes was provided through the POPGEN Consortium. Swedish Immunochip Control Samples wereprovided by the Swedish SLE network and the Uppsala Bioresource. Belgian Immunochip Control Sample Genotypes were provided through the efforts of the International Multiple Sclerosis Genetics Consortium (IMSGC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, et al. (2012) Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet 44: 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holliday EG, Maguire JM, Evans TJ, Koblar SA, Jannes J, et al. (2012) Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet 44: 1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gschwendtner A, Bevan S, Cole JW, Plourde A, Matarin M, et al. (2009) Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol 65: 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, et al. (2009) A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet 41: 876–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, et al. (2008) Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol 64: 402–409. [DOI] [PubMed] [Google Scholar]
- 6. Seshadri S, Beiser A, Kelly-Hayes M, Kase CS, Au R, et al. (2006) The lifetime risk of stroke: estimates from the Framingham Study. Stroke 37: 345–350. [DOI] [PubMed] [Google Scholar]
- 7. O'Donnell MJ, Xavier D, Liu L, Zhang H, Chin SL, et al. (2010) Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. Lancet 376: 112–123. [DOI] [PubMed] [Google Scholar]
- 8. Rothwell PM, Coull AJ, Silver LE, Fairhead JF, Giles MF, et al. (2005) Population-based study of event-rate, incidence, case fatality, and mortality for all acute vascular events in all arterial territories (Oxford Vascular Study). Lancet 366: 1773–1783. [DOI] [PubMed] [Google Scholar]
- 9. Seshadri S, Wolf PA (2007) Lifetime risk of stroke and dementia: current concepts, and estimates from the Framingham Study. Lancet Neurol 6: 1106–1114. [DOI] [PubMed] [Google Scholar]
- 10. Seshadri S, Beiser A, Pikula A, Himali JJ, Kelly-Hayes M, et al. (2010) Parental occurrence of stroke and risk of stroke in their children: the Framingham study. Circulation 121: 1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jerrard-Dunne P, Cloud G, Hassan A, Markus HS (2003) Evaluating the genetic component of ischemic stroke subtypes: a family history study. Stroke 34: 1364–1369. [DOI] [PubMed] [Google Scholar]
- 12. MacClellan LR, Mitchell BD, Cole JW, Wozniak MA, Stern BJ, et al. (2006) Familial aggregation of ischemic stroke in young women: the Stroke Prevention in Young Women Study. Genet Epidemiol 30: 602–608. [DOI] [PubMed] [Google Scholar]
- 13. Brass LM, Isaacsohn JL, Merikangas KR, Robinette CD (1992) A study of twins and stroke. Stroke 23: 221–223. [DOI] [PubMed] [Google Scholar]
- 14. Traylor M, Bevan S, Rothwell PM, Sudlow C, Dichgans M, et al. (2013) Using phenotypic heterogeneity to increase the power of genome-wide association studies: application to age at onset of ischaemic stroke subphenotypes. Genetic Epidemiology 37: 495–503. [DOI] [PubMed] [Google Scholar]
- 15. Cheng YC, O'Connell JR, Cole JW, Stine OC, Dueker N, et al. (2011) Genome-wide association analysis of ischemic stroke in young adults. G3 (Bethesda) 1: 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, et al. (2012) Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol 11: 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zaitlen N, Lindstrom S, Pasaniuc B, Cornelis M, Genovese G, et al. (2012) Informed conditioning on clinical covariates increases power in case-control association studies. PLoS Genet 8: e1003032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han B, Eskin E (2011) Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet 88: 586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22: 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, et al. (2010) Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell 19: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, et al. (2009) Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, et al. (2008) JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res 36: D102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, et al. (2011) Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 21: 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jormsjo S, Ye S, Moritz J, Walter DH, Dimmeler S, et al. (2000) Allele-specific regulation of matrix metalloproteinase-12 gene activity is associated with coronary artery luminal dimensions in diabetic patients with manifest coronary artery disease. Circ Res 86: 998–1003. [DOI] [PubMed] [Google Scholar]
- 25. Motterle A, Xiao Q, Kiechl S, Pender SL, Morris GE, et al. (2012) Influence of matrix metalloproteinase-12 on fibrinogen level. Atherosclerosis 220: 351–354. [DOI] [PubMed] [Google Scholar]
- 26. The Genotype-Tissue Expression (GTEx) project. Nature Genetics 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oksala N, Levula M, Airla N, Pelto-Huikko M, Ortiz RM, et al. (2009) ADAM-9, ADAM-15, and ADAM-17 are upregulated in macrophages in advanced human atherosclerotic plaques in aorta and carotid and femoral arteries–Tampere vascular study. Ann Med 41: 279–290. [DOI] [PubMed] [Google Scholar]
- 28. Shipley JM, Wesselschmidt RL, Kobayashi DK, Ley TJ, Shapiro SD (1996) Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc Natl Acad Sci U S A 93: 3942–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson JL, Baker AH, Oka K, Chan L, Newby AC, et al. (2006) Suppression of atherosclerotic plaque progression and instability by tissue inhibitor of metalloproteinase-2: involvement of macrophage migration and apoptosis. Circulation 113: 2435–2444. [DOI] [PubMed] [Google Scholar]
- 30. Pepper MS (2001) Role of the matrix metalloproteinase and plasminogen activator-plasmin systems in angiogenesis. Arterioscler Thromb Vasc Biol 21: 1104–1117. [DOI] [PubMed] [Google Scholar]
- 31. Choudhary S, Higgins CL, Chen IY, Reardon M, Lawrie G, et al. (2006) Quantitation and localization of matrix metalloproteinases and their inhibitors in human carotid endarterectomy tissues. Arterioscler Thromb Vasc Biol 26: 2351–2358. [DOI] [PubMed] [Google Scholar]
- 32. Johnson JL, George SJ, Newby AC, Jackson CL (2005) Divergent effects of matrix metalloproteinases 3, 7, 9, and 12 on atherosclerotic plaque stability in mouse brachiocephalic arteries. Proc Natl Acad Sci U S A 102: 15575–15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luttun A, Lutgens E, Manderveld A, Maris K, Collen D, et al. (2004) Loss of matrix metalloproteinase-9 or matrix metalloproteinase-12 protects apolipoprotein E-deficient mice against atherosclerotic media destruction but differentially affects plaque growth. Circulation 109: 1408–1414. [DOI] [PubMed] [Google Scholar]
- 34. Thomas AC, Sala-Newby GB, Ismail Y, Johnson JL, Pasterkamp G, et al. (2007) Genomics of foam cells and nonfoamy macrophages from rabbits identifies arginase-I as a differential regulator of nitric oxide production. Arterioscler Thromb Vasc Biol 27: 571–577. [DOI] [PubMed] [Google Scholar]
- 35. Halpert I, Sires UI, Roby JD, Potter-Perigo S, Wight TN, et al. (1996) Matrilysin is expressed by lipid-laden macrophages at sites of potential rupture in atherosclerotic lesions and localizes to areas of versican deposition, a proteoglycan substrate for the enzyme. Proc Natl Acad Sci U S A 93: 9748–9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morgan AR, Rerkasem K, Gallagher PJ, Zhang B, Morris GE, et al. (2004) Differences in matrix metalloproteinase-1 and matrix metalloproteinase-12 transcript levels among carotid atherosclerotic plaques with different histopathological characteristics. Stroke 35: 1310–1315. [DOI] [PubMed] [Google Scholar]
- 37. Kilarski L, Achterberg S, Devan WJ, Traylor M, Malik R, et al. (2014) Meta-analysis in over 17,900 cases of ischaemic stroke reveals a novel association at 12q24.12. Neurology (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, et al. (2006) PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet 9: 55–61. [DOI] [PubMed] [Google Scholar]
- 39. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marchini J, Howie B (2010) Genotype imputation for genome-wide association studies. Nat Rev Genet 11: 499–511. [DOI] [PubMed] [Google Scholar]
- 41. Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, et al. (1993) Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 24: 35–41. [DOI] [PubMed] [Google Scholar]
- 42. Lovett JK, Coull AJ, Rothwell PM (2004) Early risk of recurrence by subtype of ischemic stroke in population-based incidence studies. Neurology 62: 569–573. [DOI] [PubMed] [Google Scholar]
- 43. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 44. Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- 46. Levula M, Oksala N, Airla N, Zeitlin R, Salenius JP, et al. (2012) Genes involved in systemic and arterial bed dependent atherosclerosis–Tampere Vascular study. PLoS One 7: e33787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Niinisalo P, Oksala N, Levula M, Pelto-Huikko M, Jarvinen O, et al. (2010) Activation of indoleamine 2,3-dioxygenase-induced tryptophan degradation in advanced atherosclerotic plaques: Tampere vascular study. Ann Med 42: 55–63. [DOI] [PubMed] [Google Scholar]
- 48. Oksala N, Levula M, Pelto-Huikko M, Kytomaki L, Soini JT, et al. (2010) Carbonic anhydrases II and XII are up-regulated in osteoclast-like cells in advanced human atherosclerotic plaques-Tampere Vascular Study. Ann Med 42: 360–370. [DOI] [PubMed] [Google Scholar]
- 49. Oksala N, Parssinen J, Seppala I, Raitoharju E, Kholova I, et al. (2013) Association of Neuroimmune Guidance Cue Netrin-1 and its Chemorepulsive Receptor UNC5B with Atherosclerotic Plaque Expression Signatures and Stability in Human(s) - Tampere Vascular Study. Circ Cardiovasc Genet 6: 579–87. [DOI] [PubMed] [Google Scholar]
- 50. Raitoharju E, Seppala I, Lyytikainen LP, Levula M, Oksala N, et al. (2013) A comparison of the accuracy of Illumina HumanHT-12 v3 Expression BeadChip and TaqMan qRT-PCR gene expression results in patient samples from the Tampere Vascular Study. Atherosclerosis 226: 149–152. [DOI] [PubMed] [Google Scholar]
- 51. Schork AJ, Thompson WK, Pham P, Torkamani A, Roddey JC, et al. (2013) All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet 9: e1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Achterberg S, Kappelle LJ, Algra A (2008) Prognostic modelling in ischaemic stroke study, additional value of genetic characteristics. Rationale and design. Eur Neurol 59: 243–252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
QQ-plots for cardioembolic stroke and all ischaemic stroke analyses. QQ-plots of expected p-values (x-axis) against observed p-values (y-axis) for analyses of (clockwise from top left) cardioembolic stroke (age-at-onset informed), cardioembolic stroke (uninformed), all ischaemic stroke (uninformed), all ischaemic stroke (age-at-onset informed). Lambda values for each plot are given in Table S4.
(DOCX)
QQ-plots for large artery atherosclerotic stroke and small vessel disease stroke analyses. QQ-plots of expected p-values (x-axis) against observed p-values (y-axis) for analyses of (clockwise from top left) large artery stroke (age-at-onset informed), large artery stroke (uninformed), small vessel stroke (uninformed), small vessel stroke (age-at-onset informed). Lambda values for each plot are given in Table S4.
(DOCX)
Forest plot of SNP effects for rs660599 in the large artery atherosclerotic stroke replication populations. ASGC, the Australian Stroke Genetics collaboration; deCODE, deCODE genetics; GEOS, the Genetics of early onset stroke study; HVH, the heart and vascular health study; ISGS/SWISS, the Ischaemic stroke genetics study/Siblings with Ischaemic stroke study; MGH-GASROS, Massachusetts General Hospital – Genetics affecting stroke risk and outcome. PROMISe, Prognostic modeling in ischaemic stroke study; RACE, Risk Assessment of Cerebrovascular Events study.
(DOCX)
Forest plot of SNP effects for rs660599 in the large artery atherosclerotic stroke replication populations for cases with age <61 years. ASGC, the Australian Stroke Genetics collaboration; HVH, the heart and vascular health study; ISGS/SWISS, the Ischaemic stroke genetics study/Siblings with Ischaemic stroke study; MGH-GASROS, Massachusetts General Hospital – Genetics affecting stroke risk and outcome. PROMISe, Prognostic modeling in ischaemic stroke study.
(DOCX)
Results from RegulomeDB, showing the evidence that SNPs in the associated MMP12 region have a regulatory function. Scores indicate the following degrees of evidence: Score 2b, TF binding + any motif + DNase Footprint + DNase peak; Score 4, TF binding + DNase peak; Score 5, TF binding or DNase peak; Score 6, other; “No data” indicates that RegulomeDB holds no information about the given SNP, meaning there currently exists no evidence to suggest that the SNP has a regulatory function. In some cases this may indicate that the SNP falls within a protein-coding region. SNP, single nucleotide polymorphism.
(DOCX)
Expanded set of populations used to generate SNPs with p<0.05 to evaluate the age-at-onset informed approach. ARIC, The Atherosclerosis Risk in Communities study; ASGC, Australian Stroke Genetics Collaborative; CHS, Cardiovascular Health Study; FHS, Framingham Heart Study; HPS, Heart Protection Study; HVH, The Heart and Vascular Health Study; ISGS/SWISS, The Ischemic Stroke Genetics Study/Sibling with Ischaemic Stroke Study; MGH-GASROS, The MGH Genes Affecting Stroke Risk and Outcome Study; WTCCC2-Germany, The Wellcome Trust Case-Consortium II Munich; WTCCC2-UK, The Wellcome Trust Case-Consortium II UK; RACE, Risk Assessment of Cerebrovascular Events Study, Pakistan.
(DOCX)
Age and gender distributions of populations. ARIC, The Atherosclerosis Risk in Communities study; ASGC, Australian Stroke Genetics Collaborative; CHS, Cardiovascular Health Study; FHS, Framingham Heart Study; HPS, Heart Protection Study; HVH, The Heart and Vascular Health Study; ISGS/SWISS, The Ischemic Stroke Genetics Study/Sibling with Ischaemic Stroke Study; MGH-GASROS, The MGH Genes Affecting Stroke Risk and Outcome Study; WTCCC2-Germany, The Wellcome Trust Case-Consortium II Munich; WTCCC2-UK, The Wellcome Trust Case-Consortium II UK; RACE, Risk Assessment of Cerebrovascular Events Study, Pakistan. IS, all ischaemic stroke; CE, cardioembolic stroke; LAA, large artery stroke; SVD, small vessel disease.
(DOCX)
Genomic inflation (λ) rates for discovery populations for age-at-onset informed and uninformed approaches. IS, all ischaemic stroke; CE, cardioembolic stroke; LAA, large artery stroke; SVD, small vessel disease.
(DOCX)
Number of SNPs used in evaluation of age-at-onset informed approach. IS, all ischaemic stroke; CE, cardioembolic stroke; LAA, large artery stroke; SVD, small vessel disease.
(DOCX)
Membership of Wellcome Trust Case Control Consortium 2 (WTCCC2).
(DOCX)
Liability threshold models.
(DOCX)