Abstract
During eukaryotic ribosome biogenesis, members of the conserved atypical serine/threonine protein kinase family, the RIO kinases (Rio1, Rio2 and Rio3) function in small ribosomal subunit biogenesis. Structural analysis of Rio2 indicated a role as a conformation-sensing ATPase rather than a kinase to regulate its dynamic association with the pre-40S subunit. However, it remained elusive at which step and by which mechanism the other RIO kinase members act. Here, we have determined the crystal structure of the human Rio1–ATP–Mg2+ complex carrying a phosphoaspartate in the active site indicative of ATPase activity. Structure-based mutations in yeast showed that Rio1's catalytic activity regulates its pre-40S association. Furthermore, we provide evidence that Rio1 associates with a very late pre-40S via its conserved C-terminal domain. Moreover, a rio1 dominant-negative mutant defective in ATP hydrolysis induced trapping of late biogenesis factors in pre-ribosomal particles, which turned out not to be pre-40S but 80S-like ribosomes. Thus, the RIO kinase fold generates a versatile ATPase enzyme, which in the case of Rio1 is activated following the Rio2 step to regulate one of the final 40S maturation events, at which time the 60S subunit is recruited for final quality control check.
INTRODUCTION
Ribosome assembly is a crucial energy-demanding task in every cell that ensures an adequate production of proteins to meet the overall cellular needs. In eukaryotes, ribosome synthesis, which begins in the nucleolus, is a very complex coordinated process involving several hundred assembly factors that ensure the synthesis of mature 18S ribosomal ribonucleic acid (rRNA) with around 33 ribosomal proteins (r-proteins) and mature 25S, 5.8S, 5S with ∼46 r-proteins, into 40S and 60S ribosomal subunits, respectively (1–3). Although it is clear that the function of the ribosome biogenesis factors is to facilitate proper ribosomal subunits assembly, the precise molecular mechanism by which they perform their in vivo function remains for most of them unresolved. Among the myriad of ribosome biogenesis factors, a few factors house enzymatic activities (e.g. NTPases, methyl transferases, etc.) and accordingly have been the focus in many investigations [see for review (1–3)].
Among these biogenesis factors with enzymatic activity are the RIO kinases, which are members of a protein family that is evolutionary conserved and present in all three domains of life. These RIO kinases exhibit an atypical kinase domain, the RIO domain, which is a trimmed version of the canonical eukaryotic protein kinases (ePKs) lacking the activation loop and the substrate recognition domain (4–7). In most Archaea and in lower eukaryotes (e.g. the yeast Saccharomyces cerevisiae), the RIO family is composed of two subfamilies: Rio1 and Rio2. In higher eukaryotes including human another subfamily member, RioK3 is present. In the past years, loss of function analyses have revealed that all eukaryotic members of the RIO family are involved in the late steps of small subunit (SSU) biogenesis, cell-cycle progression and tumorigenesis (4,8–15). Moreover, studies in human cells suggested that all three RIO kinases bind to late pre-40S particles and that both Rio1 and Rio2 act as kinases to trigger the release of late SSU biogenesis factors from the evolving 40S particles (13,14,16). Hence, models were proposed that assumed Rio1 and Rio2 phosphorylate ribosome biogenesis factors, thereby triggering their release. However, to date no physiological substrates of the RIO kinases participating in ribosome biogenesis have been identified [see also discussions in (13,14,16)].
Recently, the view that RIO kinases function exclusively as enzymes with kinase activity has changed, due to the finding that the crystal structure of the Rio2–ATP–Mg2+ complex carries a phosphoaspartate (pAsp) intermediate in its active site. This finding suggested an adenosine triphosphatase (ATPase) activity for Rio2, which could be experimentally verified by in vitro activity measurements (17). Moreover, the fit of the Rio2 X-ray structure into the cryo-electron microscopy density map of the late pre-40S particle (18) indicated that the catalytic site of Rio2 is occluded, making phosphorylation of a nearby protein substrate very difficult. Due to these and other findings, we have suggested that Rio2's catalytic ATPase activity could control the dynamic association of Rio2 with the evolving 40S subunit (17), thereby coordinating pre-40S maturation and ribosome biogenesis factors release.
In this study, we provide further insight into the conserved role of RIO kinases in ribosome synthesis by reporting the first X-ray crystal structure of a human RIO kinase, hsRio1, with bound adenosine triphosphate (ATP) and Mg2+ at 2.8-Å resolution. Similar to Rio2, hsRio1 carries a pAsp intermediate in the catalytic center, which is typically seen in P-type ATPases (19) and indicative of ATP hydrolysis. Indeed, in vitro assays showed that Rio1 acts predominantly as an ATPase, but also exhibits a weaker phosphorylation activity. In vivo analyses in yeast demonstrated that Rio1's ATPase activity is required for late 40S biogenesis to regulate its dynamic association with pre-40S particles. However, in contrast to Rio2, a rio1 dominant negative mutant mapping in the active site caused a block in pre-40S formation, but these precursor particles were trapped together with a number of late 40S biogenesis factors in association with 60S subunits. Thus, Rio1's ATPase activity could play a role in the recently described translation-like cycle between nascent 40S and 60S subunits, which has been suggested to be a final quality control checkpoint for 40S biogenesis.
MATERIALS AND METHODS
Yeast strains and yeast genetic methods
The S. cerevisiae strains used in this study are listed in Supplementary Table S1. All strains, unless otherwise specified, are derivatives of BY4741 (Euroscarf). Preparation of media, yeast transformation and genetic manipulations were done according to standard procedures.
Plasmid constructs
All recombinant deoxyribonucleic acid (DNA) techniques were performed according to standard procedures using Escherichia coli DH5α for cloning and plasmid propagation. Site-directed mutagenesis was performed by overlap extension polymerase chain reaction (PCR). All cloned DNA fragments generated by PCR amplification were verified by sequencing. Plasmids used in this study are listed in Supplementary Table S2. ctRio1 was amplified from Chaetomium thermophilum complementary DNA (20) and cloned into appropriate E. coli expression vector.
Purification and crystallization of hsRio1
A fragment of the hsRio1, residues 143-494, was cloned into the expression vector, pDEST527 with a Tobacco Etch Virus (TEV) protease cleavage site preceded by an N-terminal 6X His tag. The fragment was identified as a degradation product during previous attempts to purify the full-length protein. The hsRio1 (143-494) expression vector was transformed to RosettaTM (DE3) pLysS E. coli cells (Novagen) and expression carried out in both selenomethionine and Luria-Berti (LB) media. Expression in selenomethionine media (Shanghai Medicilon Inc.) was carried out according to the manufacturer's instructions. For expression in LB media, large cultures were started from dilution (1:100) of single colony overnight cultures in LB media with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. These cultures were incubated at 37°C until mid-log phase at which point the cells were induced with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and expression carried out at 20°C for 18 h. Harvested cell pellets were resuspended in lysis buffer containing 50 mM Tris pH 8.0, 200 mM NaCl, 2 mM MgCl2, 1 mM ATP, 0.2% β-mercaptoethanol, 0.1 mg/ml DNAse and 0.3 X BugbusterTM (Novagen). Lysis occurred with gentle stirring on ice for 45 min, and was followed by centrifugation at 18 000 rpm at 4°C for 30 min. The supernatant was filtered and loaded onto a pre-equilibrated 5 ml HisTrapTM column (GE Healthcare) attached to a fast protein liquid chromatography system at a flow rate of 2.5 ml/min. The HisTrapTM column was washed with 10 column volumes of lysis buffer (sans bugbuster and DNase) to remove any unbound protein. Bound hsRio1 (143-494) protein was subjected to an imidazole gradient of 100 mM to 1 M and the protein eluted between 150 mM and 300 mM. Partially purified fractions were pooled together and transferred to a dialysis membrane to which 1 mg/ml TEV protease, needed to cleave the histidine tag, was added. The protein was dialyzed overnight at 4°C into 50 mM Tris pH 8.0, 200 mM NaCl, 1 mM ATP, 2 mM MgCl2 and 0.2% β-mercaptoethanol and then loaded onto a 5 ml HisTrap column pre-equilibrated with lysis buffer. The HisTrap column was washed with lysis buffer to elute cleaved hsRio1 (143-494) that was further purified by size exclusion chromatography using Superdex 200 (GE Healthcare) pre-equilibrated in 50-mM Tris pH 8.0, 200 mM NaCl, 1 mM ATP, 2 mM MgCl2, 10% glycerol and 0.2% β-mercaptoethanol. Purified hsRio1 (143-494) with ATP/Mg2+ bound was concentrated to 20 mg/ml and subjected to robotic sparse matrix screening using Phoenix Liquid Handling System (Art Robbins). Rod-shaped crystals were observed in Wizard III screen in both the 1:1 and the 2:1 protein to well solution ratios. These crystals grew in 28% PEG 400, 0.1 M HEPES pH 7.5, 0.2 M CaCl2 and were optimized by varying the concentration of precipitant, PEG 400, and pH. Optimized crystals were flash frozen in their well solution with 20% (v/v) ethylene glycol as a cryoprotectant and shipped to Argonne National Labs (ANL) for data collection and processing.
Structure determination
Data were collected at 100K at the NE-CAT beamline (24-ID) at the Advanced Photon Source, ANL, Argonne, IL, USA. All data were indexed, integrated and scaled using HKL2000 program (21). The crystals belonged to the P3221space group with unit cell dimensions of a = b = 78.76, c = 110.57, and one molecule per asymmetric unit. The structure was solved by a combination of molecular replacement using the structure of Archaeoglobus fulgidus Rio1 (PDB: 1ZP9) and single-wavelength anomalous dispersion using seleno-methionine-modified crystals, using PHASER in PHENIX (21–23). The structure was rebuilt using COOT and refined in PHENIX (21,24). Data collection and refinement statistics are provided in Table 1.
Table 1. Data collection and refinement statistics.
| hsRio1/ATP/Mg2+ | |
|---|---|
| Data collection | |
| Space group | P3221 |
| Cell dimensions | |
| a = b, c (Å) | 78.76, 110.57 |
| α = β, γ (°) | 90.00, 120.00 |
| Resolution (Å) | 43.0–2.7 (2.80–2.70)a |
| Rsym or Rmerge | 0.066 (0.215)1 |
| I/σI | 12.7 (3.40)1 |
| Completeness (%) | 100 (100)1 |
| Redundancy | 2.0 (2.0)1 |
| Refinement | |
| Resolution (Å) | 43.0–2.7 (2.80–2.70) |
| No. reflections | 11330 (1101) |
| Rwork / Rfree | 20.4/24.3 (31.1/35.7)1 |
| No. atoms | 1924 |
| Protein | 1889 |
| Ligand/ion | 28 |
| Water | 7 |
| B-factors (average; Å2) | 58.8 |
| rms deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.35 |
aOne crystal was used for each data set. Values in parentheses are for highest-resolution shell.
pAsp hydroxylamine sensitivity and single turnover experiments
ctRio2 and ctHrr25 were purified as described earlier (17). ctRio1 was purified using the same conditions as described for ctRio2 (17). pAsp hydroxylamine sensitivity assays were essentially performed as described previously (17), except that buffer K200 (10% glycerol, 20 mM Tris-HCl pH 7.5, 200 mM KCl, 10 mM imidazole, 5 mM MgCl2, 2 mM β-mercaptoethanol with or without 0.5 M NaCl or 0.5 M Hydroxylamine-HCl) was used. Single turnover experiments using 1 μM of purified proteins, thin layer chromatography, phosphorylation assay and quantitation were performed as described previously (17).
Western blot analysis
The following primary antibodies were used in this study: anti-Rio2 (Santa-Cruz, Rio2 Y-220, cat. no. sc-98828; 1:500), anti-ProtA (Sigma, cat. no.P1291; 1:10 000), anti-Rps3 (1:5000), anti-rpS8 (1:8000), anti-eIF5b [1:2000 (25)], anti-Rpl25 (1:5000), anti-rpP0 [1:1000 (26)], anti-Rps26 (1:2000, kindly provided by Dr. Vikram Panse, unpublished data), anti-Tsr1, anti-Ltv1, anti-Enp1, anti-Nob1, anti-Dim1 and anti-Pno1 [all 1:2000 (27)]. Horseradish peroxidase conjugated goat anti-rabbit (Roche, cat. no. 170-6515; 1:8,000) and goat anti-mouse (Bio-Rad, cat. no. 170-6516; 1:3000) were used as secondary antibodies.
RNA work and northern blot analysis
RNA extraction and northern blotting experiments were performed as previously described (28). Probes used were previously described (29). Signals were acquired using a Phosphorimager screen and quantified with ImageJ.
Purification of ribosomal subunits from C. thermophilum and in vitro binding experiments
C. thermophilum mycelium was grown as described earlier (20). Mycelium was disrupted using a Retsch MM400 cryomill and resuspended in lysis buffer (20 mM HEPES pH 7.5, 100 mM KAc, 5 mM Mg(OAc)2, 0.1% NP40, 1 mM Dithiothreitol (DTT), FY protease inhibitors). Extract was clarified in a bench-top centrifuge followed by a spin in a JA25.5 rotor for 20 min at 16 000 rpm. Fifteen to twenty milliliters of extract was loaded on a high salt sucrose cushion (3 ml of 2 M sucrose solution and 3 ml of 1.5 M sucrose containing 20 mM HEPES pH 7.5, 0.5 M KOAc, 5 mM Mg(OAc)2, 1 mM DTT) and centrifuged in a Ti70 rotor at 53 500 rpm for 18 h at 4°C. The pellet, containing ribosomes, was resuspended in resuspension buffer (20 mM HEPES pH 7.5, 50 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT). To separate into subunits the 80S ribosomes were incubated in 1X subunit separation buffer (20 mM HEPES pH 7.5, 500 mM KCl, 1-mM DTT, 0.01% RNasin, 1 mM puromycin, 1.5 mM Mg(OAc)2) for 30 min on ice and then 5 min at room temperature. Five OD (OD260) of ribosomes were loaded on a 10–30% sucrose gradient (20 mM HEPES pH7.5, 500 mM KCl, 1 mM DTT, 1.5 mM Mg(OAc)2 and centrifuged using SW40 rotor at 23 000 rpm for 18 h. The 60S/40S peaks were harvested and buffer exchanged and concentrated in to 1 mM HEPES pH7.5, 2 mM Mg(OAc)2 and 1 mM DTT.
ctRio1 was purified using the same conditions as described for ctRio2 (17). 1:1 to 1:10 molar ratios of ct40S::ctRio1 or ct60S::ctRio1 were incubated for 10 min at room temperature and then separated on a 10–30% sucrose gradient. Collected fractions were TCA precipitated and proteins were separated on a NuPAGE sodium dodecyl sulphate (SDS) 4–12% gradient polyacrylamide gels (Invitrogen) and stained with colloidal Coomassie (Sigma-Aldrich).
Miscellaneous
Fluorescence in situ hybridization analysis was performed as described previously (28) using the following Cy3-conjugated probe 5′Cy3-ATG CTC TTG CCA AAA CAA AAA AAT CCA TTT TCA AAA TTA TTA AAT TTC TT-3′) for detection of ITS1-containing pre-rRNAs. Cells were examined by fluorescence microscopy using an Imager Z1 microscope (Carl Zeiss) with an X100, numerical aperture 1.4 Plan-Apo-Chromat oil immersion lens and a DICIII, 4,6-diamidino-2-phenylindole, or HECy3 filter, respectively. Snapshots were all taken with identical exposure time and further processed in batch. Polysome profiles were analyzed by sucrose gradient centrifugation as previously described in buffer containing 50 mM Tris-HCl, pH 7.5, 50 mM (NH4)2SO4, 12 mM MgCl2 (17). Affinity purifications of Tandem Affinity Purification (TAP)-tagged bait proteins were, unless otherwise indicated, performed in a buffer containing 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 5% glycerol and 0.1% NP40 essentially as described previously (17). For TEV protease cleavage, DTT and RNasin (Fermentas) were added to the buffer to a final concentration of 1 mM and 200 U/ml, respectively. Elutions from Sepharose Flag-M2 beads were performed in the presence of an excess of Flag peptide for 40 min at 4°C. The Flag eluates were precipitated by the addition of TCA and dissolved in SDS sample buffer before separation on NuPAGE SDS 4–12% gradient polyacrylamide gels (Invitrogen) and staining with colloidal Coomassie (Sigma-Aldrich). Mass spectrometry identification of the proteins contained in Coomassie-stained bands was performed as described previously (17).
RESULTS
Structure of the human Rio1 kinase
The human Rio1 protein (NP_113668.2) was cloned and expressed in E. coli. Diffraction-quality crystals were obtained from a purified fragment of hsRio1 consisting of residues 143-494 (see Table 1 for statistics), containing the entire RIO domain (aa 164-387) and 106 residues of the extended C-terminus. However, in the electron density, we could only observe residues 164-430, excluding residues 212-243 that are known to correspond to the flexible loop of the RIO kinases. Like canonical ePKs, the RIO domain of hsRio1 consists of an N-terminal lobe consisting of 5-stranded anti-parallel β-sheet (β1-5) and a single α-helix (αC) between β3 and β4 (Figure 1a). The entire subdomain is connected via a loop called the ‘hinge region’ to a C-terminal lobe containing three α-helices (αE, αF and αG) and a β-hairpin (β6-7). HsRio1, like other RIO kinases, contains a RIO kinase-specific α-helix preceding the β-sheet, called αR, and the Rio1 subfamily-specific ‘flexible loop’ located between β3 and αC. In this structure, the entire flexible loop is not visible in the electron density map, although it was observed in previous structures of the adenosine-bound archaeal Rio1 (30). Surprisingly, 64 amino acids of the C-terminus of this construct were not observed in the electron density map. This may be due to intrinsic disorder or a separate, randomly oriented subdomain. However, the data allowed positioning of 44 residues past the end of the conserved RIO domain, as shown in comparison with the structure of A. fulgidus Rio1 (afRio1; Figure 1b) (30). These amino acids form two α-helices (αH and αI) that interact via both hydrophobic and hydrogen-bonded interactions with the RIO domain, effectively extending the domain. The residues of these helices are moderately conserved in eukaryotic Rio1 sequences (Supplementary Figure S1).
Figure 1.
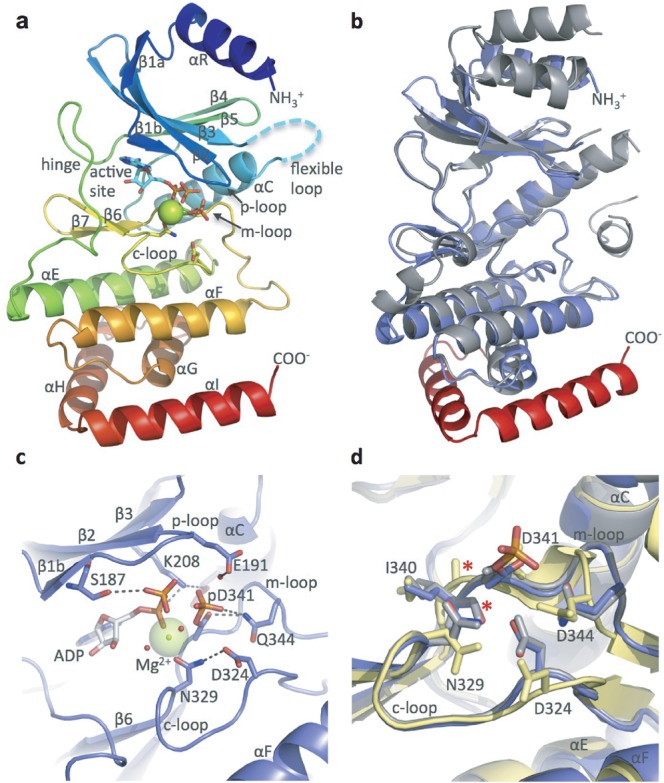
hsRio1 structure. (a) Overall structure of hsRio1 residues 140-430. The entire conserved RIO domain (αR-αG) is observed in the hsRio1 structure (colored blue to red N-terminal to C-terminal. In addition, two helices, αH and αI, C-terminal to the RIO domain are observed. Density is not observed for 32 residues of the flexible region (represented by the cyan dashed line). (b) Alignment of the archaeal Rio1 (gray) with the hsRio1 structure (blue in the RIO domain, red in the extension). (c) Detailed view of the active site and interactions with ADP, Mg2+ and the pAsp (pD341). Invariant residues from the p-, c- and m-loops are shown in sticks. (d) Alignment of the c- and m-loops from the A. fulgidus Rio1/ATP complex (active state; gray); the adenosine complex (inactive state; yellow) and hsRio1/ADP/pAsp (this structure; blue). The red asterisk shows the backbone carbonyl that flips when ATP binds to produce the active conformation. The residues are labeled according to numbering in hsRio1.
pAsp in the active site of human Rio1 enzyme
HsRio1 was crystallized with 1 mM ATP and 2 mM MgCl2 present in the crystallization solution. Upon fitting of a molecule of ATP in the active site, we observed additional unexplained positive density. Given that the recently solved structure of eukaryotic Rio2 from C. thermophilum (ctRio2) incubated with ATP and Mg2+ contained ADP-Mg2+ and a pAsp intermediate rather than ATP in the active site (17), we sought to fit the latter into the electron density. Refinement of the intermediate resulted in a better fit to the observed electron density suggesting the presence of a pAsp intermediate in the catalytic center. Supplementary Figure 2A and B shows composite omit map density calculated for the entire model but contoured around the ADP ligand and the pAsp and difference density calculated when the phosphate of the pAsp is omitted. The RIO kinase active site contains invariant aspartate and asparagine residues in the catalytic loop (c-loop), D324 (C. thermophilum ctRio1 D281; S. cerevisiae scRio1 D244) and N329 in hsRio1, and the invariant D341 (ctRio1 D298; scRio1 D261) in the metal binding loop (m-loop). D341 is the residue that is modified by the addition of a phosphate group. This phosphate group is within range for a water-mediated interaction with the invariant glutamate residue, E191, in the Rio1 phosphate-binding loop (p-loop) (Figure 1c). The conformation of the m-loop of hsRio1 and the position of the pAsp closely align with that determined for ctRio2 (Supplementary Figure S2c) (17).
Figure 2.
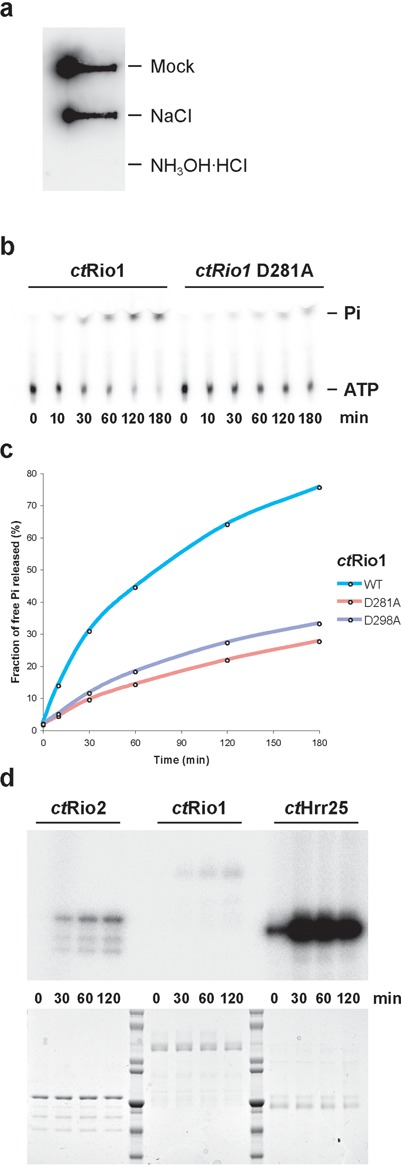
Rio1 has ATPase activity in vitro. (a) Hydroxylamine sensitivity of acyl-phosphate analysis. Phosphorimage of filter blot of 32P-labeled ctRio1 after treatment with assay buffer, 0.5 M NaCl or 0.5 M hydroxylamine (b–d) ctRio1 ATPase (% of γ-phosphate release) and kinase (autophosphorylation) activities were tested in single turnover experiments using 1 μM of the indicated purified recombinant protein (load is shown in (d), lower panel). (b–c) Time-dependent free phosphate released (%Pi) as obtained from thin layer chromatography experiments of single turnover assays. (d) Comparative analysis of time-dependent autophosphorylation activity as obtained by SDS-PAGE and detected by autoradiography from single turnover experiments performed with 1 μM of the indicated purified recombinant protein. Note that the amount of sample loaded in (d) is 100 times more than in (b).
Comparisons of several RIO kinase structures have led to the identification of at least two states (labeled ‘active’ and ‘inactive’) of the enzyme that are characterized primarily by the conformation of the m-loop, with additional altered positioning of the N-lobe relative to the C-lobe of the RIO domain and slight shifts in the m-loop. In this structure of hsRio1, the backbone of the m-loop aligns perfectly with the m-loop of the considered ‘active’ state of afRio1 (30) bound to ATP (RMSD 0.771 for overall alignment of 152 Cα atoms), and is therefore considered to be in the ‘active’ conformation (Figure 1d).
Rio1 can act as an ATPase in vitro
Due to the presence of a pAsp in the hsRio1 crystal structure, we wanted to gain further insight into Rio1's putative ATPase activity. For these in vitro studies we switched to C. thermophilum Rio1 (ctRio1), which exhibits better biochemical properties than the human ortholog (for sequence alignment see Supplementary Figure S1). Full-length recombinant ctRio1 protein was expressed and purified from E. coli and autophosphorylated in the presence of γ-32P-labeled ATP. The presence of a pAsp intermediate in solution was tested by taking advantage of the specific sensitivity of acyl-phosphate to hydroxylamine as described earlier [see (17) and references therein, and the Materials and Methods section]. This analysis showed that most of the autophosphorylation signal seen in ctRio1 (mock or 0.5M NaCl) is sensitive to hydroxylamine treatment, suggesting that ctRio1 contains a pAsp intermediate in solution (Figure 2a). Since pAsp intermediates are typically found in P-loop ATPases (19), we assayed purified ctRio1 catalytic activity in single turnover measurements using γ-32P-labeled ATP and compared the amounts of released radioactive γ-32P-labeled phosphate by thin-layer chromatography versus autophosphorylation analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. ctRio1 can hydrolyze ATP and most of the hydrolyzed γ-32P-labeled phosphate is released as free Pi, whereas Rio1's autophosphorylation activity remains very low (less than 0.1% of total ATP hydrolysis events) (Figure 2b–d and data not shown). Finally, mutation of ctRio1 at the predicted catalytic residues D281 (catalytic aspartate; scRio1 D244; hsRio1 D324) or D298 (pAsp, Mg2+ coordination; scRio1 D261; hsRio1 D341) to alanine resulted in a decrease of the overall ATPase activity (see Figure 2c), thus demonstrating ctRio1-dependent ATPase activity in vitro. Altogether, our previous study (17) and the results shown in this work support the model that the RIO protein family acts as ATPases.
Rio1's ATPase activity regulates pre-40S association
To gain insight into the in vivo role of Rio1's ATPase activity, we used the yeast (S. cerevisiae) system to genetically manipulate scRio1, which is essential for growth and its depletion causes accumulation of pre-40S particles containing immature 20S pre-rRNA (12). Moreover, catalytic dead mutants of both scRio1 and hsRio1 have been analyzed in vivo (8,13). Based on the crystal structure of human Rio1, we have generated active site mutants, rio1 D244A (catalytic aspartate) and rio1 D261A (phosphoapartate and Mg2+ coordination), which are predicted to block ATP hydrolysis. In both cases, yeast cells expressing these mutant Rio1 proteins are not viable [Figure 3a and (8)]. Subsequent phenotypic analysis of these mutants showed that Rio1's catalytic activity is required for the synthesis of the 40S subunits (Figure 3b) and efficient cytoplasmic 20S>18S rRNA processing (Figure 3c and d).
Figure 3.
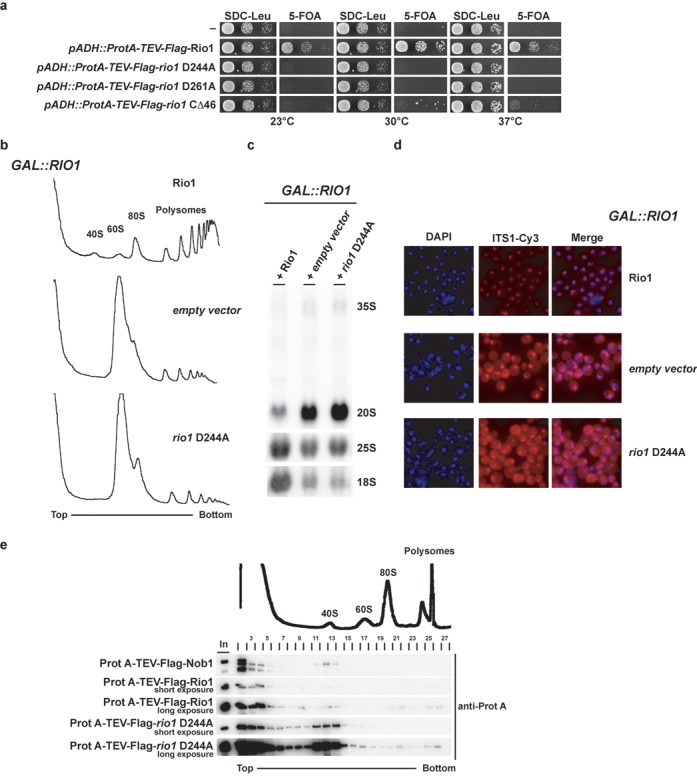
Rio1 catalytic activity is required for late 40S cytoplasmic maturation and regulates its pre-40S dynamic association. (a) Rio1 catalytic activity is essential for yeast growth. Viability of mutations affecting Rio1 catalytic residues (rio1 D244A catalytic Asp and rio1 D281A Mg2+ coordination and pAsp) was tested in a RIO knockout strain background complemented by a RIO1 gene carried on a plasmid (CEN3, URA3). For plasmid shuffling experiments serial dilutions of the indicated strains were spotted on SDC-Leu (loading control) and 5-FOA (shuffling) plates for 3 days at the indicated temperature. (b) Rio1 catalytic activity is required for 40S biogenesis. Polysomes profile analysis obtained from whole cell lysates from Rio1-depleted cells (16 h in SDC-Leu) either complemented by an empty vector or a vector (CEN3, LEU2) carrying a wild-type RIO1 or a catalytically inactive allele (rio1 D244A) is shown. The A254nm profiles of the derived sucrose gradients fractions are depicted. (c) Rio1 catalytic activity is required for 20S rRNA processing. Northern blot analysis of steady-state (pre-) rRNA species on Rio1-depleted cells (see above) using probes complementary to the ITS1, 18S and 25S mature region of the rRNA. (d) Rio1 catalytic activity is required for cytoplasmic processing at site D. Steady-state sub-cellular localization of ITS1 containing rRNA species was monitored by fluorescent in situ hybridization (FISH) on para-formalhehyde-fixed Rio1-depleted cells (see above) using a Cy3-conjugated probe complementary to the ITS1 region of the rRNA (see the Materials and Methods section). (e) Rio1 catalytic activity regulates its dynamic association with pre-40S. The co-sedimentation behavior of the indicated constructs was assayed by western blotting analysis using the indicated antibody from 5 to 40% sucrose gradient fractions of whole cell extract from wild-type cells co-expressing the indicated constructs. The A254nm profile of an exemplary derived sucrose gradients fraction is depicted.
Next, we analyzed whether the association of Rio1 with pre-40S subunits is regulated by its catalytic activity. For this purpose, we co-expressed N-terminally tagged wild-type and rio1 D244A mutant proteins in yeast and detected Rio1's binding to pre-ribosomal particles by sucrose gradient centrifugation. N-terminally tagged Nob1, which served as control, sedimented both as free and in a pre-40S-associated pool (Figure 3e). In contrast, wild-type Rio1 was only marginally associated with pre-40S particles, but the rio1 D244A ATPase mutant was significantly shifted into the 40S pool (Figure 3e). These results indicate that Rio1's catalytic activity regulates its dynamic association with the nascent 40S subunit.
Relocation of late pre-ribosome biogenesis factors into 80S-like particles upon overexpression of catalytic inactive Rio1
Due to the observation that rio1 D244A becomes stuck on pre-40S particles, we wondered whether Rio1 defective in ATP-hydrolysis could exert a dominant negative effect in vivo. As anticipated, GAL-promoter-induced overexpression of rio1 D244A in wild-type yeast harboring endogenous Rio1 caused a strong growth inhibition at all tested temperatures (Figure 4a). If this dominant negative effect were due to endogenous chromosomal Rio1 being unable to replace overproduced mutant rio1 D244A from pre-40S particles, we may neutralize this dominant effect by generating a second site mutation in rio1 D244A that diminishes its affinity to pre-40S subunits. We considered the C-terminal domain of Rio1 as a target for such a second site mutation, as previous studies indicated that C-terminal tagging or C-terminal mutations impair growth without affecting the in vitro enzymatic activity [see (8,31), Figure 3a and data not shown]. Based on these considerations, we deleted approximately half of Rio1's conserved C-terminal domain (rio1CΔ46; Supplementary Figure S1). Importantly, overexpressed rio1CΔ46 did not cause a dominant growth defect. However, when rio1CΔ46 and rio1 D244A mutations were combined intragenetically, the dominant negative phenotype was abolished (Figure 4a). To test whether suppression was caused by a diminished binding of Rio1 to pre-40S subunits due to the C-terminal truncation (see above), we affinity-purified TAP-tagged rio1CΔ46 and rio1CΔ80, respectively, from yeast lysates. Apparently, full-length Rio1 was co-enriched with 20S rRNA containing ribosomal subunits, but both carboxy-terminal truncations had lost their binding capacity to pre-ribosomal subunits (Figure 4b, left panel). Moreover, we investigated the binding of recombinant ctRio1 and ctRio1CΔ80 to isolated ct40S subunits. This in vitro assay recapitulated the previous in vivo findings that Rio1's carboxy-terminal domain is crucially involved in targeting the enzyme to pre-ribosomal particles (Figure 4b, right panel). Thus, the intragenic combination of the catalytic ATPase mutant (exhibiting enhanced pre-ribosome binding) with Rio1 mutations that decrease pre-ribosome binding suppresses the dominant phenotype.
Figure 4.
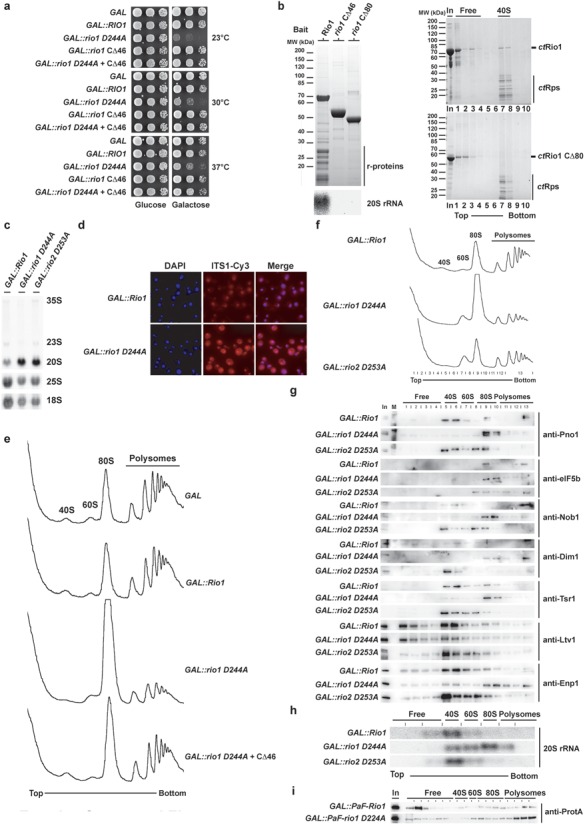
Rio1 dominant negative mutant shows a translation-like initiation defect and delocalization of late ribosome biogenesis factors. (a) Overexpression of rio1 D244A induces a dominant negative growth defect. Serial dilutions of wild-type cells carrying the multi-copy GAL empty vector or the same plasmid with indicated galactose-inducible Rio1 alleles were spotted on SDC-Ura (repressed) and SGC-Ura (induced) plates for 2 or 3 days at 30°C–37°C and 23°C, respectively. (b) Rio1 C-terminal part is required for (pre-) 40S binding. Wild-type yeast cells (BY4741-Euroscarf) transformed with a plasmid carrying the indicated bait protein fused with N-terminal ProtA-TEV-Flag under the control of the ADH1 promoter were grown in SDC-Leu up to OD600 = 1.5. Indicated bait proteins were purified by standard tandem affinity purification (see the Materials and Methods section). Co-immunoprecipitated proteins from Flag eluates were precipitated by addition of TCA and resolved on a 4–12% Nupage gel (Invitrogen) and visualized by coomassie staining. RNA was extracted by hot-phenol extraction and resolved on a 1.2% agarose formaldehyde/MOPS (3-(N-morpholino)propanesulfonic acid) gel, and transferred to a positively charged nylon membrane. For northern blot analysis membrane was hybridized with the indicated probe (see the Materials and Methods section) and exposed to a Phosphorimager screen. For in vitro binding (right panel) purified ct40S (10 pmol) and the indicated ctRio1 constructs (100 pmol) were mixed in binding buffer and incubated at Room Temperature (RT) for 10 min prior to loading on sucrose gradient. Fractions were collected, proteins were TCA precipitated, resolved by SDS-PAGE and visualized by coomassie staining. (c–d) Overexpression of catalytically inactive Rio1 induces cytoplasmic 20S rRNA processing defect. Northern blot analysis of steady-state (pre-) rRNA species using probes complementary to the ITS1, 18S and 25S mature region of the rRNA and steady-state subcellular localization of ITS1 containing rRNA species was monitored by FISH using a Cy3-conjugated probe complementary to the ITS1 region of the rRNA on yeast cells after 6 h induction of the dominant negative phenotype (see the Materials and Methods section). (e) Overexpression of rio1 D244A induces a translation initiation-like defect. Polysomes profile analysis obtained from whole cell lysates from yeast cells overexpressing the indicated constructs for 6 h is shown. The A254nm profiles of the derived sucrose gradients fractions are depicted. (f) Same as in (e) except that the indicated fractions were collected, TCA precipitated and analyzed by western blotting [shown in (g)]. (g) Overexpression of rio1 D244A induces relocalization of several late 40S biogenesis factors in the 80S fraction. TCA-precipitated fractions obtained from the sucrose gradient shown in (e) were analyzed by SDS-PAGE and western blotting using the indicated antibody (see the Materials and Methods section). (h) Overexpression of rio1 D244A induces relocalization of 20S rRNA in the 80S fraction. Similar to (g), RNA from sucrose gradient fractions was extracted and analyzed by northern using a probe complementary to the ITS1 region. (i) Overexpressed rio1 D244A mostly localized in 80S and polysomes fractions. Polysomes profile analysis obtained from whole cell lysates from yeast cells overexpressing Gal::ProtA-TEV-Flag-Rio1 or rio1 D244A was induced for 6 h prior to cycloheximide treatment. The indicated fractions were collected, TCA precipitated and analyzed by western blotting.
Next, we wanted to find out whether and where the 40S biogenesis pathway is inhibited upon expression of dominant rio1 D244A. As shown by northern blot analysis and fluorescence in situ hybridization using a probe for ITS1, 20S>18S rRNA processing (D-site cleavage), which occurs in the cytoplasm, was inhibited upon overexpression of the catalytic rio1 mutant (Figure 4c and d). Moreover, overexpression of rio1 D244A also impaired formation of 40S subunits, which is seen as a reduced 40S peak on sucrose gradients when compared to the 60S peak (Figure 4e). Unexpectedly, we observed the appearance of a huge 80S peak in the dominant mutant and reduced polysomes (Figure 4e). Such a ribosomal profile on sucrose gradients is reminiscent of mutants with defects in translation initiation (32–35). However, when the intragenic rio1 D244A + CΔ46 suppressor allele was analyzed in a similar way, 20S>18S rRNA processing and a normal polysome profile were restored (Figure 4e and data not shown). To test whether this peculiar phenotype of polysome loss and appearance of 80S ribosomes is unique for rio1 D244A, we tested another dominant biogenesis factor mutant, rio2 D253A (17). Although rio2 D253A induced a clear-cut 40S subunit biogenesis defect (compare 40/60S ratio), a strong increase in the 80S peak as evident in the rio1 D244A was not observed upon rio2 D253A overexpression (Figure 4f).
In light of previous findings showing that the final SSU maturation requires association of nascent 40S with 60S subunits to form 80S-like ribosomes (27,36) and that depletion of Fap7 induced a redistribution of other late 40S biogenesis factors and 20S rRNA into 80S-like particles (27), we compared how pre-40S biogenesis factors and 20S rRNA behave in rio1 and rio2 dominant negative ATPase mutants. Strikingly, we found for dominant rio1 D244A an almost complete relocation of both late 40S biogenesis factors (e.g. Pno1, Nob1, Tsr1, Dim1, Enp1) and 20S rRNA mostly into the 80S fraction of the sucrose gradient (Figure 4g and h). In contrast, in the case of dominant rio2 D253A these late pre-40S factors and the 20S rRNA co-sediment to a larger extent with the 40S particles (Figure 4g and h). Finally, we have analyzed the sedimentation behavior of the overexpressed Rio1 in sucrose gradient (Figure 4i). Interestingly, rio1 D244A relocates to the 80S and polysomes fractions (Figure 4i). We conclude from these studies that Rio1's catalytic activity is required for its own recycling and for the recycling of pre-40S biogenesis factors from nascent 40S subunits, and if defective these factors become stuck on 80S-like particles containing immature 20S rRNA (see also the Discussion section).
Rio1 associates with very late nascent pre-40S particles that also contain 60S subunits
To find out to which nascent pre-40S subunits in vivo Rio1 binds, we affinity-purified Rio1-containing pre-ribosomal particles from yeast and analyzed their protein and rRNA composition. Unexpectedly, particles pulled down by either Rio1 or mutant rio1 D244A not only contained 20S rRNA but also significant amounts of mature 25S rRNA together with 60S r-proteins (Figure 5). Similarly, the pre-40S factor and endonuclease Nob1 co-precipitated substantial amounts of 25S rRNA (36), but Rio2-associated particles were rather devoid of 25S rRNA and Rpl proteins (Figure 5).
Figure 5.
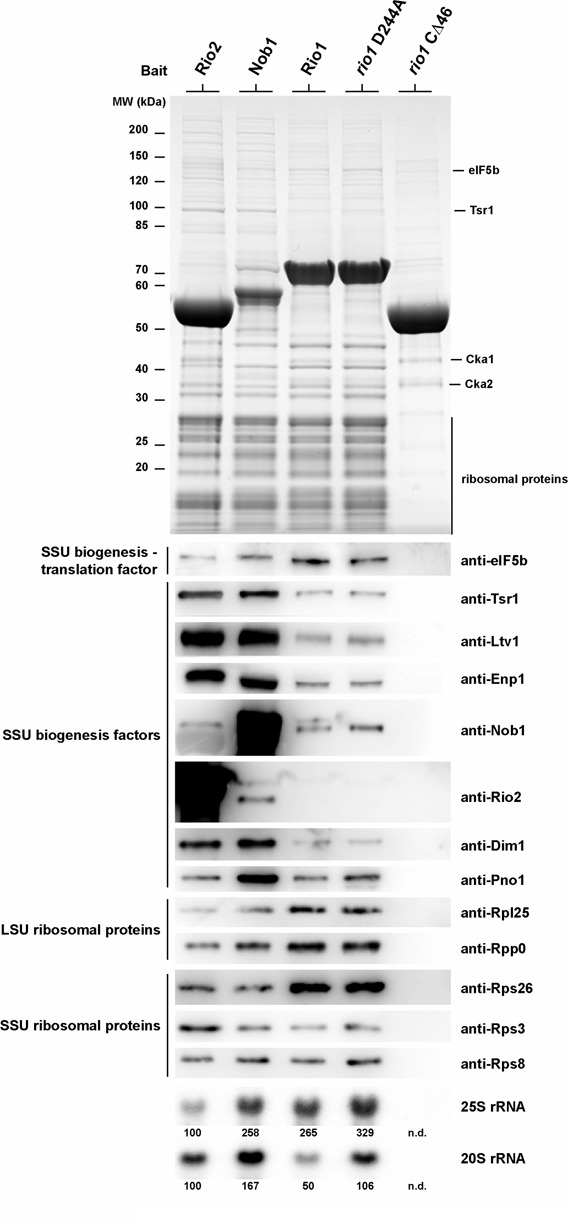
Rio1 associates at a very late step of pre-40S biogenesis via its conserved C-terminal region. Rio1 associates with a very late pre-40S particle lacking most late SSU ribosome biogenesis factors. Wild-type yeast cells (BY4741-Euroscarf) transformed with a plasmid carrying the indicated bait protein fused either with N-terminal ProtA-TEV-Flag (Nob1 and Rio1 constructs) or C-terminal Flag-TEV-ProtA (Rio2) under the control of the ADH1 promoter were grown in SDC-Leu up to OD600 = 1.5. Bait proteins were purified by standard tandem affinity purification (see the Materials and Methods section). Co-immunoprecipitated proteins from Flag eluates were precipitated by addition of TCA and resolved on a 4–12% Nupage gel (Invitrogen) and analyzed either by coomassie staining and mass-spectrometry or by western blotting using the indicated antibody. RNA was extracted by hot-phenol extraction and resolved on a 1.2% agarose Formaldehyde/MOPS gel, and transferred to a positively charged Nylon membrane. For northern blot analysis membrane was hybridized with the indicated probes (see the Materials and Methods section) and exposed to a Phosphorimager screen. Signals were quantified with ImageJ.
Although 20S rRNA is present in the Rio1-purified particle, late pre-40S biogenesis factors were absent or only weakly co-enriched (e.g. Rio2, Tsr1, Dim1, Nob1, Ltv1, Enp1) (Figure 5). This is in contrast to Rio2 and Nob1, which significantly co-purified these late pre-40S biogenesis factors (Figure 5). When we tested for Rps26, a well-known late assembling 40S r-protein (18,37), it was strongly co-enriched in the Rio1-purified, but not in the Rio2- or Nob1-purified particle. Taken together, these data suggest that Rio1 is present on a very late pre-40S particle that apparently follows the Rio2 particle. This Rio1-containing particle could correspond to one of the final nascent 40S subunit intermediates, which were postulated to be engaged in binding to the 60S subunit for a translation-like check-point cycle (27).
DISCUSSION
Using a combination of X-ray crystallography, enzymatic assays and in vivo functional analyses, we have unraveled common and different mechanistic aspects by which Rio1 (this study) and Rio2 (17) are involved in late 40S biogenesis. The atomic structures of eukaryotic hsRio1 and ctRio2 revealed an unusual pAsp intermediate, known to exist in the active site of P-type ATPases [see (17) and references therein]. Our subsequent enzymatic characterization indicated that both Rio1 and Rio2 predominantly act as ATPases in vitro. This finding allows the generalization of the concept that RIO protein kinases rather act as conformation-sensing ATPases.
The catalytic activities of Rio1 and Rio2 are required for maturation of the 40S subunit, and the common theme is that via their ATP hydrolysis activity they can control not only their dissociation from the pre-40S particles, but also affect recycling of other SSU biogenesis factors (13,14). Notably, the pre-60S maturation pathway lacks ATPases of the RIO-type, but instead a number of GTPases and ATPases are involved in the large subunit assembly where they participate in checkpoint control or assembly factor recycling (2,3,38,39).
Our study has provided evidence that Rio1 is associated with late pre-40S particles that succeeds earlier particles on which Rio2 is acting as a conformation-sensing ATPase, mediating its own release and concomitant recycling of several SSU assembly factors (e.g. Ltv1, Enp1). Since Rio1 is not a stoichiometric component of Nob1-associated or Pno1-associated particles, we suggest that Rio1, Nob1 and Pno1 are part of a discrete, probably short-lived pre-40S particle. Additionally, Rio2 is absent from the Rio1-purified particle, which is in good agreement with previous work in human cells (13). Thus, we conclude that Rio1's association with the pre-40S particle might occur after the release of Rio2, during which most of the other late SSU biogenesis factors dissociate.
Our findings concerning the function of Rio1 are related to a recently postulated model of final 40S maturation events, in which a translation-like cycle between nascent 40S and 60S subunits is thought to occur as a final quality control checkpoint for 40S biogenesis (27,36). Although a role of Rio1 was not considered in this quality control checkpoint, our data showed that overexpression of a Rio1 catalytic inactive mutant leads to cell growth inhibition, a translation initiation-like defect, and trapping of many late SSU biogenesis factors within the 80S fraction on sucrose gradients. These findings imply that Rio1's ATPase activity could play a so far unrecognized role in controlling this final maturation step, which is thought to correspond to a pioneer round of translation. Importantly, another study has demonstrated the involvement of an as yet unknown ATP-consuming factor in Nob1-dependent 20S rRNA processing (D-site cleavage) in vitro (36). Thus, it is tempting to speculate that Rio1 could be this unkown ATP-consuming enzyme on the very late pre-40S particle that is involved in ATP-dependent D-site cleavage.
How Rio1 and Rio2 performed their functions on the nascent pre-40S and how they control the dynamic associations of several late ribosome biogenesis factors is still unclear. We have previously suggested a Rio2-heterophosphorylation-independent mechanism associated with factor recycling (17). At this stage, we cannot rule out a Rio1-heterophosphorylation-independent mechanism enabling the release of a subset of late ribosome biogenesis factors. However, our structural and functional analysis shows very strong similarities in the mode of action of both Rio1 and Rio2. In addition, Rio1 associates with pre-40S particle largely devoid of most of the late SSU biogenesis factors. Moreover, despite the fact that Rio1 structural localization on a pre-40S particle is unknown, it is also unlikely that all these factors that are distributed all over the nascent SSU head domain (18,40) are all direct target of Rio1. Finally, from a structural and enzymatic point of view all RIO proteins lack the substrate recognition domain, a typical feature found in ePKs, and have poor kinase activity in vitro (4–7,17).
In conclusion, the various members of the conserved RIO protein family use an apparent kinase fold to act as ATPases, perhaps in a stepwise ATP-hydrolysis cycle as described for P-type ATPases (19), to monitor and regulate conformational progression and final maturation of the nascent 40S subunit. It is tempting to speculate that the ATPase activity of the RIO protein might be activated in a regulated fashion, probably depending on the proper assembly status of the nascent pre-40S particle. As soon as the pre-40S particle has reached an assembly intermediate competent for the RIO ATPase activity, ATP hydrolysis could occur leading to a RIO phosphoaspartate intermediate. Such an intermediate might shift the pre-40S particle into a different conformational state, which in turn triggers the release of a subset of ribosome biogenesis factors. The subsequent hydrolysis of the pAsp and/or release of the generated ADP from the active site may finally trigger the final release of these regulatory RIO ATPases from the pre-ribosome.
ACCESSION NUMBER
Atomic coordinates and structure factors for the crystal structure have been deposited in the Protein Data Bank under accession number 4OTP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [1].
Acknowledgments
The authors thank Dr. Panse for sharing unpublished Rps26 antibody; Dr. Karbstein, Dr. Ballesta and Dr. Dever for providing antibodies; Dr. Lechner and his team for performing mass-spectrometry analysis; Sabine Griesel for providing ctRio1 clone; Daniela Strauß for providing reagents, scientific discussion and support; and Drs. Tschochner, Milkereit and Griesenbeck and ‘the House of the Ribosome’ (University of Regensburg) for their generous support provided in the last stage of this study to S.F.-C.
Authors contributions: S.F.-C. performed all the in vivo yeast experiments, hydroxylamine sensitivity assay, single turnover analysis and Chaetomium thermophilum recombinant protein purifications. I.K. purified, crystallized and solved the structure of hsRio1. E.T. purified Chaetomium thermophilum ribosomal subunits and performed in vitro binding experiments. S.F.-C., N.L. and E.H. designed the experiments and wrote the manuscript. All authors discussed the experiments and commented on the manuscript.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
FUNDING
National Institute of General Medical Sciences, National Institutes of Health [P41 GM103403]; US DOE [DE-AC02-06CH11357]; National Institutes of Health National Cancer Institute [K22CA123152 to N.L.]; German Research Council [DFG Hu363/10-4 to E.H.]; Post-doctoral program of the Medical Faculty of the University of Heidelberg [to S.F.-C.]; Funding for open access charge: DFG via the open access publishing program [to S.F.-C./University of Regensburg]. This work was supported by the German Research Foundation (DFG) within the funding program Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kressler D., Hurt E., Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2009;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Thomson E., Ferreira-Cerca S., Hurt E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013;126:4815–4821. doi: 10.1242/jcs.111948. [DOI] [PubMed] [Google Scholar]
- 3.Woolford J.L., Jr, Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esser D., Siebers B. Atypical protein kinases of the RIO family in archaea. Biochem. Soc. Trans. 2013;41:399–404. doi: 10.1042/BST20120317. [DOI] [PubMed] [Google Scholar]
- 5.LaRonde-LeBlanc N., Wlodawer A. A family portrait of the RIO kinases. J. Biol. Chem. 2005;280:37297–37300. doi: 10.1074/jbc.R500013200. [DOI] [PubMed] [Google Scholar]
- 6.LaRonde-LeBlanc N., Wlodawer A. The RIO kinases: an atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim. Biophys. Acta. 2005;1754:14–24. doi: 10.1016/j.bbapap.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 7.Laronde N.A. The Ancient Microbial RIO Kinases. J. Biol. Chem. 2014;289:9488–9492. doi: 10.1074/jbc.R113.538090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angermayr M., Roidl A., Bandlow W. Yeast Rio1p is the founding member of a novel subfamily of protein serine kinases involved in the control of cell cycle progression. Mol. Microbiol. 2002;44:309–324. doi: 10.1046/j.1365-2958.2002.02881.x. [DOI] [PubMed] [Google Scholar]
- 9.Baumas K., Soudet J., Caizergues-Ferrer M., Faubladier M., Henry Y., Mougin A. Human RioK3 is a novel component of cytoplasmic pre-40S pre-ribosomal particles. RNA Biol. 2012;9:162–174. doi: 10.4161/rna.18810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geerlings T.H., Faber A.W., Bister M.D., Vos J.C., Raue H.A. Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S Pre-rRNA in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:22537–22545. doi: 10.1074/jbc.M300759200. [DOI] [PubMed] [Google Scholar]
- 11.Vanrobays E., Gelugne J.P., Gleizes P.E., Caizergues-Ferrer M. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:2083–2095. doi: 10.1128/MCB.23.6.2083-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanrobays E., Gleizes P.E., Bousquet-Antonelli C., Noaillac-Depeyre J., Caizergues-Ferrer M., Gelugne J.P. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 2001;20:4204–4213. doi: 10.1093/emboj/20.15.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widmann B., Wandrey F., Badertscher L., Wyler E., Pfannstiel J., Zemp I., Kutay U. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell. 2011;23:22–35. doi: 10.1091/mbc.E11-07-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zemp I., Wild T., O'Donohue M.F., Wandrey F., Widmann B., Gleizes P.E., Kutay U. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 2009;185:1167–1180. doi: 10.1083/jcb.200904048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Read R.D., Fenton T.R., Gomez G.G., Wykosky J., Vandenberg S.R., Babic I., Iwanami A., Yang H., Cavenee W.K., Mischel P.S., et al. A kinome-wide RNAi screen in Drosophila Glia reveals that the RIO kinases mediate cell proliferation and survival through TORC2-Akt signaling in glioblastoma. PLoS Genet. 2013;9:e1003253. doi: 10.1371/journal.pgen.1003253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyler E., Zimmermann M., Widmann B., Gstaiger M., Pfannstiel J., Kutay U., Zemp I. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA. 2010;17:189–200. doi: 10.1261/rna.2325911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira-Cerca S., Sagar V., Schafer T., Diop M., Wesseling A.M., Lu H., Chai E., Hurt E., LaRonde-LeBlanc N. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 2012;19:1316–1323. doi: 10.1038/nsmb.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strunk B.S., Loucks C.R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C.L., III, Karbstein K., Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 20.Amlacher S., Sarges P., Flemming D., van Noort V., Kunze R., Devos D.P., Arumugam M., Bork P., Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.McCoy A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2006;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Choi S.K., Olsen D.S., Roll-Mecak A., Martung A., Remo K.L., Burley S.K., Hinnebusch A.G., Dever T.E. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol. 2000;20:7183–7191. doi: 10.1128/mcb.20.19.7183-7191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilella M.D., Remacha M., Ortiz B.L., Mendez E., Ballesta J.P. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44’ have different functional roles. Eur. J. Biochem. 1991;196:407–414. doi: 10.1111/j.1432-1033.1991.tb15831.x. [DOI] [PubMed] [Google Scholar]
- 27.Strunk B.S., Novak M.N., Young C.L., Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira-Cerca S., Poll G., Gleizes P.E., Tschochner H., Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Pertschy B., Schneider C., Gnadig M., Schafer T., Tollervey D., Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 2009;284:35079–35091. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laronde-Leblanc N., Guszczynski T., Copeland T., Wlodawer A. Structure and activity of the atypical serine kinase Rio1. FEBS J. 2005;272:3698–3713. doi: 10.1111/j.1742-4658.2005.04796.x. [DOI] [PubMed] [Google Scholar]
- 31.Soudet J., Gelugne J.P., Belhabich-Baumas K., Caizergues-Ferrer M., Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2009;29:80–92. doi: 10.1038/emboj.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de la Cruz J., Iost I., Kressler D., Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deloche O., de la Cruz J., Kressler D., Doere M., Linder P. A membrane transport defect leads to a rapid attenuation of translation initiation in Saccharomyces cerevisiae. Mol. Cell. 2004;13:357–366. doi: 10.1016/s1097-2765(04)00008-5. [DOI] [PubMed] [Google Scholar]
- 34.Dong J., Lai R., Nielsen K., Fekete C.A., Qiu H., Hinnebusch A.G. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- 35.Valasek L., Nielsen K.H., Hinnebusch A.G. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lebaron S., Schneider C., van Nues R.W., Swiatkowska A., Walsh D., Bottcher B., Granneman S., Watkins N.J., Tollervey D. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 2012;19:744–753. doi: 10.1038/nsmb.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira-Cerca S., Poll G., Kuhn H., Neueder A., Jakob S., Tschochner H., Milkereit P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 38.Kappel L., Loibl M., Zisser G., Klein I., Fruhmann G., Gruber C., Unterweger S., Rechberger G., Pertschy B., Bergler H. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J. Cell Biol. 2012;199:771–782. doi: 10.1083/jcb.201205021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo Y., Granneman S., Thoms M., Manikas R.G., Tollervey D., Hurt E. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature. 2013;505:112–116. doi: 10.1038/nature12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Granneman S., Petfalski E., Swiatkowska A., Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 2010;29:2026–2036. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


