Abstract
Nuclear paraspeckles are built co-transcriptionally around a long non-coding RNA, NEAT1. Here we report that transfected 20-mer phosphorothioate-modified (PS) antisense oligonucleotides (ASOs) can recruit paraspeckle proteins to form morphologically normal and apparently functional paraspeckle-like structures containing no NEAT1 RNA. PS-ASOs can associate with paraspeckle proteins, including P54nrb, PSF, PSPC1 and hnRNPK. NEAT1 RNA can be displaced by transfected PS-ASO from paraspeckles and rapidly degraded. Co-localization of PS-ASOs with P54nrb was observed in canonical NEAT1-containing paraspeckles, in perinucleolar caps upon transcriptional inhibition, and importantly, in paraspeckle-like or filament structures lacking NEAT1 RNA. The induced formation of paraspeckle-like and filament structures occurred in mouse embryonic stem cells expressing little or no NEAT1 RNA, suggesting that PS-ASOs can serve as seeding molecules to assemble paraspeckle-like foci in the absence of NEAT1 RNA. Moreover, CTN, an RNA reported to be functionally retained in paraspeckles, was also observed to localize to paraspeckle-like structures, implying that paraspeckle-like structures assembled on PS-ASOs are functional. Together, our results indicate that functional paraspeckles can form with short nucleic acids other than NEAT1 RNA.
INTRODUCTION
The mammalian cell nucleus is compartmentalized into discrete protein- and RNA-containing bodies that organize distinct nuclear processes from DNA replication and repair to RNA transcription and processing (1). Intriguingly, nuclear bodies maintain their structural integrity despite lacking defining membranes and exchange components dynamically with surrounding nucleoplasm. How these distinct structures assemble and are maintained is not fully understood. A critical step in nuclear body biogenesis is the initial seeding event that provides a scaffold for subsequent recruitment of additional components. RNA is an essential architectural element in nuclear body formation. This is not surprising since many nuclear bodies assemble around the sites of transcription. For example, histone H2B pre-mRNAs tethered to a specific chromatin region in HeLa cells recruit and retain NPAT, a histone locus body protein (2). Both coding and non-coding RNA can serve as scaffolds for the assembly of nuclear bodies (2). Many long non-coding RNAs (lncRNAs), such as Xist, Gomafu, Malat1, satIII, TUG1 and GRC, have been reported to localize to specific nuclear bodies (2–12). Relevant to this study is nuclear-enriched abundant transcript 1 (NEAT1), a lncRNA that localizes to and serves as a requisite structural component of a recently defined nuclear body, paraspeckle (6–9).
Nuclear paraspeckles are mammalian-specific ribonucleoprotein bodies (13,14). Core paraspeckle-localized proteins, including P54nrb, PSF and PSPC1, belong to the Drosophila behavior/human splicing (DBHS) family. These proteins contain two N-terminal tandem RNA recognition motifs, a conserved protein–protein interaction NONA/paraspeckle domain and a C-terminal coiled-coil domain (15). DBHS proteins have been shown to intertwine extensively as homo- and heterodimers via these core domains in an RNA-independent manner (15). Indeed, self-association of DBHS dimers appears to be essential for building the higher-ordered paraspeckle structures, since depletion of either P54nrb or PSF resulted in a loss of paraspeckles (16). However, tethering either DBHS protein to an engineered chromatin region in C2C12 myoblast genome failed to recruit RNA components including NEAT1 RNA (2,8), arguing that paraspeckles do not form via stochastic interactions of individual protein components.
Paraspeckles were found to be assembled hierarchically through recruitment of their components to the central scaffolding RNA, NEAT1 (2,8). NEAT1 is an abundant lncRNA transcribed by RNA pol II. Two variant transcripts of NEAT1, NEAT1_1 and NEAT1_2, share the same 5′-end but are processed alternatively at the 3′-termini. The architectural role of NEAT1 in paraspeckle formation and maintenance was proposed based on the observation that depletion of NEAT1 lncRNA resulted in the loss of paraspeckles (6,9,17). Moreover, paraspeckles are not present in human embryonic stem cells lacking NEAT1 RNA expression, supporting the indispensable role of NEAT1 in paraspeckle formation (18). Both isoforms of NEAT1 are present in paraspeckles (19). Interestingly, although overexpression of mouse NEAT1_1 in NIH3T3 cells can increase paraspeckle numbers (6), paraspeckle formation was only rescued by exogenous expression of mouse NEAT1_2, but not NEAT1_1, in NEAT1 knockout mouse embryonic fibroblast (MEF) cells, suggesting that NEAT1_1 alone is insufficient to maintain paraspeckle integrity (16). Finally, the hypothesis that NEAT1 RNA is the seeding molecule of paraspeckles was validated by the visualization of co-transcriptional assembly of paraspeckles on nascent NEAT1 transcripts (6,9), suggesting that NEAT1 is both sufficient and necessary to recruit protein components from soluble nucleoplasmic pool, triggering de novo paraspeckle formation.
Although the formation of paraspeckles has been characterized, the functions of paraspeckles are not fully understood. NEAT1−/− mice, despite losing paraspeckles completely, failed to exhibit any physiological defects under normal conditions (20). However, potential roles of NEAT1/paraspeckles in response to cellular stress were suggested. For example, NEAT1 RNA was induced upon virus infection (21,22) or by immune stimuli such as poly I:C (23). In addition, paraspeckle/NEAT1 RNA has been proposed to be involved in attenuating the cell death pathway, since increased sensitivity was observed in NEAT1−/− fibroblasts in the presence of a proteasome inhibitor, MG132 (24). Moreover, paraspeckles were implicated in nuclear RNA retention. Mouse CTN-RNA, an isoform of mCAT2 mRNA, is retained under normal conditions in nuclei and enriched in paraspeckles, through preferential association of P54nrb/PSF with the 3′-UTR of CTN-RNA containing hyper-A-to-I-edited, inverted SINE repeats (25). Intriguingly, 3′-UTR of CTN-RNA can be cleaved upon stress to release protein-coding mCAT2 mRNA into the cytoplasm (25). Since the inverted SINE elements are commonly located in the 3′-UTRs of many human and mouse RNAs (26), nuclear RNA retention by P54nrb/PSF may be considered as a general function of paraspeckles. It has also been reported recently that NEAT1 RNA played a role in cellular transcriptional control through sequestration of paraspeckle proteins, some of which are transcriptional factors (23,24). Although paraspeckles may contain additional RNA components, it is currently unknown whether the structures can be seeded by endogenous or exogenous nucleic acids other than NEAT1 RNA. Formation of nuclear bodies on foreign nucleic acids has been reported previously. For example, PML/PML-like nuclear bodies can be assembled in association with HSV-1 genome upon viral infection (27).
Phosphorothioate antisense oligonucleotides (PS-ASOs) with various 2′-chemical modifications have been shown to base pair with RNA and alter RNA functions by several mechanisms including RNase H1-mediated RNA cleavage (28,29). When transfected or micro-injected into mammalian cells, PS-ASOs, especially at high concentrations, are enriched in the nucleus and can induce the formation of bright spherical nuclear foci designated phosphorothioate bodies (PS bodies). PS bodies are stable electron-dense structures of 0.15–2.0 μm in diameter, whose formation is independent of the sequence or antisense activity of PS-ASOs (30). Although no relationship of PS-ASO foci to known nuclear structures, including nuclear speckles, PML bodies, Cajal bodies, centromere, replication and transcription sites, has been reported, occasional touching of PS-ASO-enriched nuclear bodies with SC35 was observed (30). Since paraspeckles are also frequently located adjacent to SC35-rich domains (13), we hypothesized that PS-ASOs may interact and co-localize with paraspeckle proteins in nuclear bodies.
In this report, we describe the co-localization of PS-ASOs with P54nrb/PSF to canonical NEAT1-containing paraspeckles as well as perinucleolar caps upon RNA pol II transcriptional inhibition. More importantly, we present that the transfected 20-mer PS-ASOs can recruit paraspeckle proteins and scaffold the assembly of morphologically normal and apparently functional paraspeckle-like structures containing no NEAT1 RNA, demonstrating the nucleation of nuclear bodies by PS-ASOs.
MATERIALS AND METHODS
Cell culture and transfection
HeLa, MEF, mouse hepatocellular SV40 large T-antigen carcinoma (MHT) and A431 cells were grown at 37°C, 8% CO2 in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin. Primary mouse hepatocytes were isolated and cultured as previously described (31). Mouse embryonic stem cells (mESCs) (Life Technologies) were cultured at 37°C, 5% CO2 in DMEM with high glucose supplemented with 15% FBS, 1×L-glutamine, 1×penicillin-streptomycin, 1× Non-Essential Amino Acids, 1×sodium pyruvate, 55 μM 2-mercaptoethanol and 50 μl LIF/500 ml. For siRNA treatment, cells at 70% confluency were transfected with 3 nM siRNA using Lipofectamine RNAiMax at a 6 μg/ml final concentration, and harvested 24 h after transfection for subsequent analyses. For antisense activity assays (Figure 2B and D), cells plated in six-well dishes at 70% confluency were transfected with ASOs at specified concentrations using Lipofectamine 2000 at a 4 μg/ml final concentration. Cells were harvested 4 h post-transfection for subsequent analyses. For assays examining effects of non-NEAT1-targeting ASOs on NEAT1 lncRNA level (Figure 6B, C and D), cells were harvested 18 h after ASO transfection, unless otherwise specified. For actinomycin D treatment, cells were treated for 90 min at specified concentrations. siRNAs and ASOs used in this study are listed in Supplementary Data.
Figure 2.
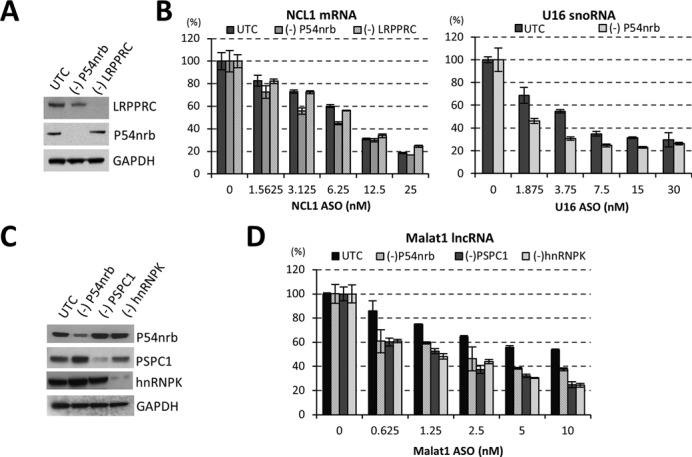
Reduction of paraspeckle proteins increases the activity of RNase H1-based ASOs. (A) siRNA-mediated reduction of P54nrb and LRPPRC proteins, as determined by western analysis. UTC, mock treated control cells. GAPDH served as a loading control. (B) Reduction of P54nrb, but not LRPPRC, increased the activity of ASO-mediated cleavage of NCL1 mRNA (left panel) or U16 snoRNA (right panel), as determined by qRT-PCR analysis. (C) siRNA-mediated reduction of paraspeckle proteins P54nrb, PSPC1 and hnRNPK, as determined by western analysis. GAPDH served as a loading control. (D) Reduction of other paraspeckle proteins can also increase ASO activity, as exemplified with an ASO targeting Malat1 lncRNA, as determined by qRT-PCR. The error bars represent standard deviation from three parallel experiments.
Figure 6.
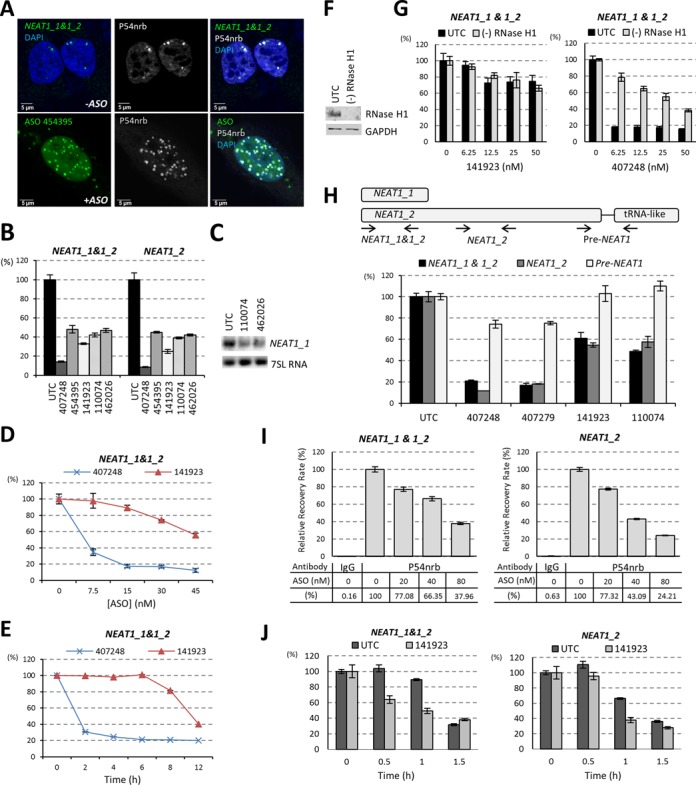
Transfected PS-ASOs can reduce NEAT1 RNA level. (A) PS-ASO transfection increased the number of “paraspeckles”, as indicated by P54nrb staining. In untreated HeLa cells (-ASO), paraspeckles were detected by staining of P54nrb and NEAT1 RNA-FISH, while in ASO transfected HeLa cells(+ASO), paraspeckle-like structures were indicated by P54nrb staining and co-localized ASOs. (B) NEAT1 RNA can be reduced by transfection of PS-ASOs that do not target NEAT1 (ISIS454395, ISIS141923, ISIS110074 and ISIS462026), as determined by qRT-PCR. A NEAT1-ASO ISIS407248 served as a positive control. (C) Northern analysis confirmed the reduction of NEAT1_1 RNA upon ASOs (ISIS110074 and ISIS462026) transfection. 7SL RNA served as a loading control. (D) Dose-dependent reduction of NEAT1_1&1_2 upon transfection of control ISIS141923. The levels of NEAT1 RNA were determined by qRT-PCR. A NEAT1-specific ASO ISIS407248 was included as a positive control. (E) Reduction of NEAT1 RNA by ISIS141923 occurred later than NEAT1-specific ISIS407248 directed RNase H1 cleavage in HeLa cells. The levels of NEAT1 RNA were determined by qRT-PCR. (F) siRNA-mediated reduction of RNase H1 in HeLa cells, as determined by western analysis. GAPDH served as a loading control. (G) Depletion of RNase H1 did not alter the reduction of NEAT1 RNA by a control ASO ISIS141923, but reduced NEAT1-specific ASO ISIS407248 activity, as determined by qRT-PCR. (H) The level of pre-NEAT1 RNA was not affected by transfection of non-NEAT1 PS-ASOs. Upper panel: schematic representation of relative positions of qRT-PCR primer-probe sets for NEAT1_1&1_2, NEAT1_2 and pre-NEAT1. Lower panel: qRT-PCR of HeLa cells transfected with either NEAT1-ASOs (ISIS407248 or ISIS407279) or control ASOs (ISIS141923 or ISIS110074) at a final concentration of 50 nM for overnight. (I) PS-ASO transfection reduced the level of P54nrb-associated NEAT1 RNA in HeLa cells. HeLa cells transfected with ISIS454395 for 8 h were subjected to RIP using anti-P54nrb or anti-mouse IgG (negative control) antibodies. NEAT1 RNA level was determined by qRT-PCR and the relative recovery rates were calculated and plotted. (J) Transfection of a control ASO ISIS141923 reduced the stability of both NEAT1_1&1_2 and NEAT1_2 RNAs in HeLa cells. The error bars in all panels represent standard deviation from three parallel experiments.
ASO pull-down
To isolate ASO-binding proteins (Figure 1A, B and C), neutravidin beads (50 μl) were incubated with 50 μl of 200 μM biotinylated 2′-MOE modified PS-ASO386652 at 4°C for 2 h in W-100 buffer (50 mM Tris, pH 7.5, 100 mM KCl, 5 mM Ethylenediaminetetraacetic acid (EDTA), 0.1% NP-40, 0.05% SDS) and blocked for 30 min with blocking buffer (10 mg/ml BSA, 1.2 mg/ml glycogen and 0.2 mg/ml tRNA in W-100 buffer). After washing three times with W-100 buffer, ASO-coated beads were incubated at 4°C for 3 h with 300 μg HeLa cell extracts prepared in buffer A (25 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 150 mM KCl, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM β-mercaptoethanol and one tablet of Protease Inhibitor Cocktail/50 ml (Roche)). After washing three times with 500 μl W-200 buffer (50 mM Tris, pH 7.5, 200 mM KCl, 5 mM EDTA, 0.1% NP-40, 0.05% SDS), beads were transferred to a 1 ml column and further washed seven times with W-200 buffer. Bound proteins were eluted by incubation with 100 μl of 50 μM 2′-MOE modified PS-ASO ISIS116847 in W-100 at room temperature for 30 min. The eluted materials were diluted with 100 μl W-100 supplemented with 100 units of RNaseOUT (Life Technologies) and incubated at 4°C for 2 h with 30 μl Neutravidin beads pre-incubated with 30 μl of 200 μM biotinylated RNA oligonucleotides complementary (XL180) or non-complementary (XL181) to ISIS116847. After washing seven times with W-200, the beads were treated with 100 μl Tris-EDTA buffer containing 5 units/μl RNase I at 30°C for 30 min, and the eluted proteins were ethanol precipitated and separated on 4–12% PAGE. Sliver-staining was carried out using ProteoSilver Plus Silver Stain Kit (Sigma) according to manufacturer's instructions. The protein bands were excised and identified through Alphalyse by mass spectrometry.
Figure 1.
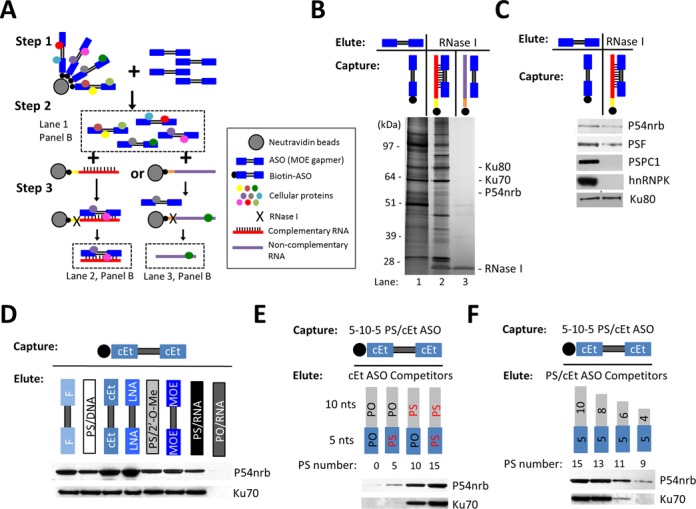
Paraspeckle proteins associate with PS-ASOs. (A) Schematic representation of experimental procedures of affinity selection of proteins bound to PS-ASO/RNA duplex. (B) Silver staining of co-selected proteins with ssPS-ASO (lane 1), PS-ASO/RNA duplex (lane 2) or control ssRNA (lane 3). (C) Western analysis followed by ASO pull-down confirmed the association of paraspeckle proteins with PS-ASO or PS-ASO/RNA duplex. (D) 2′-modifications of PS-ASOs can influence protein binding. The proteins captured with 5–10–5 cEt PS-ASO were eluted by competition with 5–10–5 ASOs bearing the same sequence but with distinct 2′-modifications, or with uniform ASOs with or without PS backbone modification, as indicated above lanes. The levels of P54nrb and Ku70 were determined by western analysis. (E and F) P54nrb/ASO interaction requires 10 or more PS backbone modified nts. ASO-associated proteins captured with 5–10–5 cEt PS-ASO were eluted with 15-mer ASOs containing different numbers of PS modified nts (panel E), or with PS-ASOs with varying length (panel F), and subjected to western analysis.
RNA preparation, northern hybridization and qRT-PCR
Total RNA was isolated using RNeasy Mini Kit (QIAGEN) or Tri-reagent (Sigma), according to manufacturer's instructions. For northern blotting, total RNA (10 μg) was resolved on the 1% agarose gel buffered with 1×MOPS and transferred onto a nylon membrane. Hybridization was performed at 58°C (for NEAT1 RNA) or 42°C (for 7SL RNA) using 5′-end [γ-32P] labeled oligonucleotide probes. The sequences for oligonucleotide probes are NEAT1 RNA, 5′- TTCAC AACAG CATAC CCGAG ACTAC TTCCC CATAC ATGCG TGACT AATAC -3′ and 7SL RNA, 5′- CTCAG CCTCC CGAGT AGCTG -3′. One-step quantitative real time-polymerase chain reaction (qRT-PCR) was performed using QuantiTect Probe RT-PCR Kit (QIAGEN), according to manufacturer's instructions. The target RNA expression level was normalized to total RNA measured by Quant-iT RiboGreen RNA Reagent (Life technologies). qRT-PCR primers and probes used in this study are listed in Supplementary Data.
Western blotting
Cell extracts were separated in 4–12% gradient sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Proteins were transferred to a nitrocellulose membrane using iBlot Gel Transfer Device (Life technologies). The membranes were blocked at room temperature for 30 min with blocking buffer containing 5% w/v nonfat dry milk in 1×PBS and incubated with primary antibodies in blocking buffer at 4°C for overnight. After washing three times with washing buffer (0.1% Tween-20 in 1×PBS) for 5 min each, membranes were incubated with secondary antibodies in blocking buffer at room temperature for 1 h. After washing three times with washing buffer for 5 min each, proteins were detected using ECL (Abcam). Antibodies for western blotting used in this study are listed in Supplementary Data.
RNA fluorescence in situ hybridization (RNA-FISH) and immunofluorescence
Cells seeded on glass-bottom culture dishes (MatTek) were transfected with fluorescently labeled ASOs for 6 h, unless otherwise specified. Cells were then fixed at room temperature for 30 min with 4% formaldehyde and permeabilized for 5 min with 0.1% Triton X-100 in 1× PBS. For NEAT1 RNA-FISH, fluorescently labeled RNA probes were generated using FISH Tag™ RNA Multicolor Kit (Life Technologies) according to the manufacturer's instructions. Primers for generating DNA templates for RNA probe synthesis are listed in Supplementary Data. After washing twice with 1×PBS and once with 2×saline-sodium citrate (SSC), permeabilized cells were incubated for overnight at 55°C with hybridization solution (50% formamide, 2XSSC, 5% dextran sulfate, 0.5 M EDTA, 0.01% Tween-20, 100 μg/ml yeast tRNA and 1X Denhardt's salt) containing fluorescently labeled RNA probes. The cells were washed twice with pre-warmed wash buffer (0.2× SSC and 50% formamide) at 55°C for 30 min each. After a post-fixation at room temperature for 30 min with 4% formaldehyde, indirect immunofluorescence (IF) staining was performed as described previously (32). Briefly, cells were blocked in blocking buffer (1 mg/ml BSA in 1×PBS) at room temperature for 30 min, and then incubated in blocking buffer with primary antibodies at room temperature for 2 h. After washing three times with washing buffer (0.1% Tween-20 in 1×PBS) for 5 min each, cells were incubated in blocking buffer with secondary antibodies at room temperature for 1 h. Finally, cells were washed three times in washing buffer for 5 min each, mounted using Prolong Gold anti-fade reagent with DAPI (Molecular Probes). Antibodies for IF used in this study are listed in Supplementary Data. Images were generated by Confocal Laser Scanning Biological Microscopy FV1000 Fluoview (Olympus) and analyzed using Fluoview Ver. 2.0b Viewer (Olympus). In Figure 7C, the construct for generating RNA-FISH probe for CTN-RNA was kindly provided by David L. Spector and Kannanganattu V. Prasanth, and the assay was carried out as described previously (25).
Figure 7.
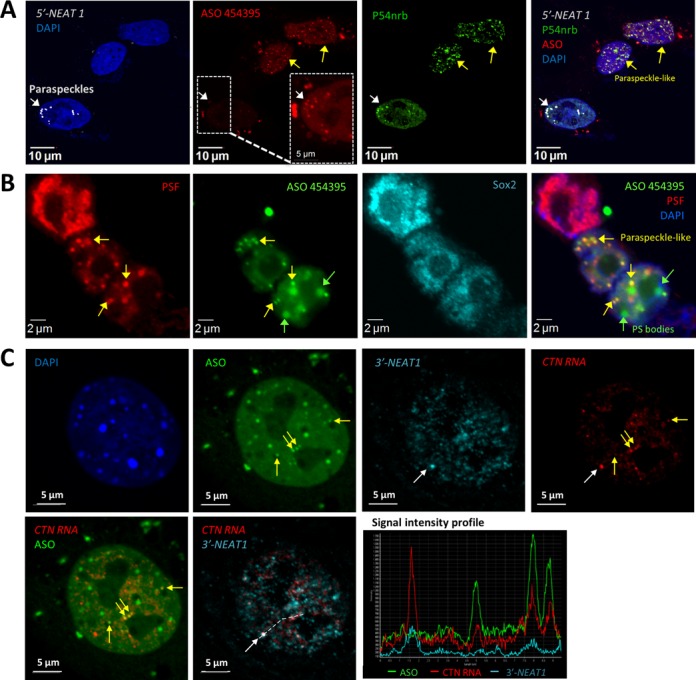
Transfected PS-ASOs can seed the formation of paraspeckle-like foci in the absence of NEAT1. (A) Co-localization of PS-ASOs with P54nrb in NEAT1 RNA-absent, nuclear paraspeckle-like foci in certain cells (yellow arrows), as indicated by IF staining of P54nrb and RNA-FISH of NEAT1 RNA in HeLa cells. The ASO signal was intensified for one cell (dashed box) to indicate the co-localization of PS-ASOs with P54nrb and NEAT1 RNA in a canonical paraspeckle (white arrow). (B) PS-ASOs can induce the formation of paraspeckle-like foci in mESCs, as indicated by IF staining of PSF and mESC marker Sox2. (C) Nuclear retention of mouse CTN-RNA by paraspeckle-like foci. RNA-FISH of mouse CTN-RNA and NEAT1 RNA in MEF cells transfected with ASO ISIS454395 indicated the localization of CTN-RNA in both canonical paraspeckles containing NEAT1 RNA (white arrow, and signal intensity profile for a region marked with a dash line) as well as in paraspeckle-like foci lacking NEAT1 RNA (yellow arrows, and signal intensity profile).
Non-disruptive RNA decay assay
Mock transfected HeLa cells (UTC) or HeLa cells transfected with 60 nM PS-ASO ISIS141923 for 5 h were pulse labeled with 0.5 mM 5′-ethynyl Uridine (EU) for 1 h to label nascent RNAs. At indicated time points after the removal of EU-containing medium, total RNA was isolated using Tri-reagent from different samples and 1 μg total RNA was subjected to nascent RNA capture using Click-iT® Nascent RNA Capture Kit (Life Technologies), according to manufacturer's instructions. Briefly, EU-RNAs were biotinylated by click reaction using biotin azide at a final concentration of 0.5 mM. Biotinylated-EU-RNAs (0.5 μg) were then incubate with 50 μl Dynabeads MyOne Streptavidin T1 magnetic beads at room temperature for 30 min. The first-strand cDNA synthesis was carried out immediately after binding and washing using the RNA captured on the beads as a template by the SuperScript VILO cDNA synthesis Kit (Life technologies), according to manufacturer's instructions. Reaction mixture was heated at 85°C for 5 min to terminate RT and release the cDNA from the beads. The kinetics of NEAT1 RNA decay was then determined by qRT-PCR.
RNA immunoprecipitation
Cell extracts were prepared in IP Lysis Buffer (Pierce) supplemented with Protease Inhibitor Cocktail (Sigma P8340) and 100 Unit/ml RNaseOUT Ribonuclease inhibitor (Life technologies). Protein A/G Magnetic beads (Millipore) were coated with 10 μg P54nrb antibody (Millipore 05–950) or negative control mouse IgG antibody. RNA immunoprecipitation (RIP) was carried out from equal amount of cell extracts using Magna RIP Kit (Millipore), according to manufacturer's instructions. The co-immunoprecipitated RNAs from beads and RNAs from 5% of input cell extracts were prepared using Tri-reagent (Sigma) and subjected to qRT-PCR analysis for NEAT1 RNA. The immunoprecipitation recovery rates of NEAT1 RNA from different samples were calculated relative to the recovery rate of UTC sample.
RESULTS
Nuclear paraspeckle proteins associate with PS-ASOs
To better understand the intracellular distribution of PS-ASOs, we sought to identify proteins with which PS-ASOs interact. An affinity selection approach was developed to isolate PS-ASO- and PS-ASO/RNA duplex-associated cellular proteins. Single-stranded (ss) ASO-binding proteins were first precipitated from HeLa cell extracts using biotinylated, PS-modified, 5–10–5 gapmer ASOs containing 10 deoxyribonucleotides in the middle flanked at both ends by five 2′-O-methoxyethyl (2′-MOE) modified ribonucleotides (Figure 1A). To increase the specificity, isolated proteins were eluted by competition using the PS-ASOs of the same sequence and modification but lacking 5′-biotin. ASO/RNA duplex-binding proteins were further isolated from the ASO-protein pool using a biotinylated RNA complementary to ASO competitors or a biotinylated non-complementary RNA to discount proteins captured by ssRNAs. The proteins that bound ssASO, ASO/RNA duplex and ssRNA were fractionated by SDS-PAGE and visualized by silver staining (Figure 1B). The interactions with other proteins and effects of other proteins on ASO activity and co-localization are the subject of other reports. ASO/RNA duplex-binding proteins identified by mass spectrometry include Ku70/K80 complex and a paraspeckle protein P54nrb. The co-selection of P54nrb and Ku80 by both ssASO and ASO/RNA duplex was confirmed by western analysis (Figure 1C). Additional major paraspeckle proteins, including PSF, PSPC1 and hnRNPK, were also isolated with PS-ASOs (Figure 1C). P54nrb and PSF associated with both ssASOs and ASO/RNA duplex, whereas PSPC1 and hnRNPK were preferentially enriched by selection with ssASOs.
Association of paraspeckle protein P54nrb with PS-ASOs appeared to be independent of ASO sequence (data not shown) but was influenced by the presence of 2′-modifications of the ASOs. The ssASO-binding proteins were isolated using a 5–10–5 PS-ASO with flanking sequences modified with 2′-constrained ethyl (2′-cEt) and eluted with PS-ASOs containing various 2′-modifications (Figure 1D). 2′-cEt, locked nucleic acid, and 2′-fluoro (2′-F) modified PS-ASOs exhibited stronger binding to P54nrb than did those modified with 2′-MOE or 2′-O-methyl (2′-O-Me). In contrast, the binding of Ku70 appeared to be independent of types of 2′-modifications. The PS backbone modification was critical for P54nrb and Ku70 binding, as RNA oligonucleotides with a phosphodiester backbone (PO) failed to compete with the captured PS-cEt ASOs to elute these proteins (Figure 1D). The failure of PO-RNAs to elute these proteins was not due to unexpected degradation of the PO-RNAs, since the PO-RNAs survived the experimental process, as determined by a PAGE analysis (our unpublished data). The requirement for the PS modification in protein binding was further demonstrated by elution of P54nrb and Ku70 from 5–10–5 PS-cEt ASOs using 15-mer ASOs containing different numbers of PS modifications (Figure 1E) and using PS-ASOs of different lengths (Figure 1F). Binding of P54nrb and Ku70 to ASOs requires 10 or more PS-modified nucleotides, consistent with previous findings that PS-ASOs associate with more proteins than do PO-ASOs (33).
Paraspeckle proteins negatively affect the antisense activities of PS-ASOs
Association of PS-ASOs with paraspeckle proteins, such as P54nrb, may affect the activity of ASO-directed RNase H1 cleavage in cells. To evaluate this possibility, P54nrb or a control protein, LRPPRC, were reduced by siRNA treatment at a final concentration of 3 nM in HeLa cells for 24 h, followed by transfection of PS-ASOs targeting either a NCL1 mRNA or a nuclear retained snoRNA U16. LRPPRC is an ASO-binding protein that has no obvious effect on ASO activity. Western analysis confirmed that P54nrb and LRPPRC proteins were significantly reduced (Figure 2A). Reduction of P54nrb, but not LRPPRC, moderately increased ASO activity, as evidenced by the enhanced reduction of the target RNA levels in P54nrb-depleted cells, when compared with mock transfected cells, especially at low ASO concentrations (Figure 2B). Similar effects were observed when P54nrb was reduced by a different siRNA, and when ASOs targeting other cellular RNAs were tested (data not shown). These results suggest that the observed enhancement of ASO activity is specific to P54nrb reduction.
To further evaluate the effects of additional ASO-associated paraspeckle proteins on PS-ASO activity, P54nrb, PSPC1 and hnRNPK levels were reduced individually in HeLa cells by corresponding siRNAs. Significant protein reductions were confirmed by western analysis (Figure 2C). Upon reduction of these proteins, the activity of a tested ASO targeting Malat1 lncRNA was significantly enhanced (Figure 2D), indicating that all these tested paraspeckle proteins negatively affect the antisense activity of PS-ASOs. As a control, no effect on activity of PS-ASOs was observed upon depletion of multiple other ASO-associated nuclear proteins, including NCL1 and KHSRP (Supplementary Figure S1), suggesting that the effects of these paraspeckle proteins on ASO activity, though moderate, were specific. These experiments were repeated multiple times over many months and the small but significant effects on ASO activity were reproducible.
PS-ASOs co-localize with nuclear paraspeckles
The association of PS-ASOs with paraspeckle proteins prompted us to investigate the potential relationship of PS-ASOs to nuclear paraspeckles. For this purpose, HeLa cells were transfected with 60 nM Cy3-labeled 5–10–5 PS/MOE-ASOs (ISIS446654). Fixed cells were stained by indirect immunofluorescence for P54nrb and PSPC1 (Figure 3A). Consistent with a previous report, the transfected PS-ASOs were predominantly localized in nucleus (30), while the cytoplasmic aggregates represented ASO-containing liposomes due to transfection of high amounts of ASOs by Lipofectamine, as well as some lysosome-localized ASOs (data not shown). Strikingly, all detected nuclear foci containing both P54nrb and PSPC1, presumably paraspeckles, overlapped with PS-ASO-enriched foci. However, not all bright PS-ASO foci contained paraspeckle marker proteins, indicating the existence of additional PS-ASO-containing nuclear bodies. In addition, co-localization of PS-ASOs with P54nrb in distinct nuclear foci was also confirmed by transient expression of GFP-tagged P54nrb in HeLa cells (Supplementary Figure S2). Finally, the localization of PS-ASOs to paraspeckles was validated by co-localizing PS-ASOs with NEAT1 RNA, the lncRNA delineating paraspeckles (Figure 3B) (14). NEAT1 RNA was detected by RNA-FISH using RNA probes against either the 5′ region shared by isoforms NEAT1_1 and NEAT1_2 (5′-probe) or a probe specific to the longer isoform NEAT1_2 (3′-probe). Signal from both probes co-localized with that from the transfected PS-ASOs in HeLa cells, indicating that PS-ASOs localize to canonical paraspeckles containing both paraspeckle proteins and the structural NEAT1 RNA. The co-localization was independent of PS-ASO-sequence and, at the concentrations studied (60 nM), was unaffected by different types of 2′-modification (Supplementary Figure S3A), consistent with the binding properties described above (Figure 1D). Moreover, similar co-localization was observed in multiple cell lines (Supplementary Figure S3B). Last but not least, PS-ASOs were transfected here at a concentration (60 nM) significantly higher than pharmacological concentrations to allow the visualization of ASO localization by the confocal microscope. However, under these conditions, neither significant cell death nor effects on cell proliferation rate was observed in ASO transfected cells (data not shown).
Figure 3.
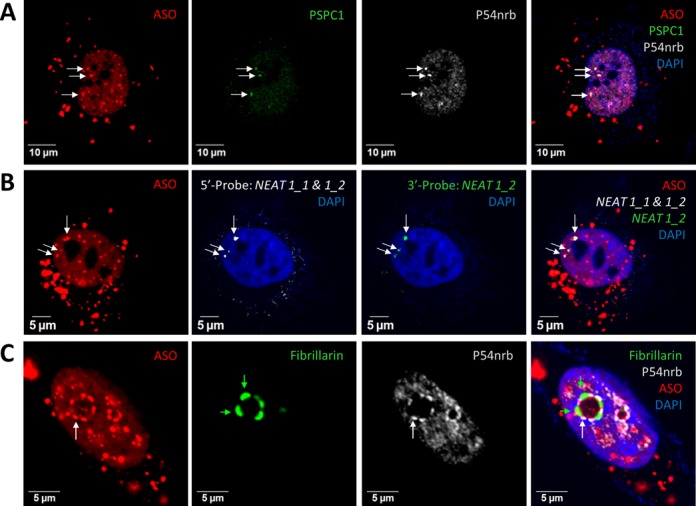
PS-ASOs can localize to bona fide paraspeckles. (A) Co-localization of transfected PS-ASOs with paraspeckle marker proteins PSPC1 and P54nrb in HeLa cells, as indicated by arrows. The nucleus was stained with DAPI. (B) Co-localization of PS-ASOs with structural RNA NEAT1 in paraspeckles, as indicated by arrows. NEAT1 RNA was detected by RNA-FISH using two RNA-probes specific to either NEAT1_1&1_2 (5′-probe) overlapping region or the 3′-end of NEAT1_2 (3′-probe). (C) PS-ASOs relocate with P54nrb to dark perinucleolar caps upon actinomycin D treatment. The light perinucleolar caps were marked by Fibrillarin staining. Green and white arrows indicate light and dark perinucleolar caps, respectively.
Paraspeckle proteins relocate to the perinucleolar region to form distinct cap structures upon transcriptional inhibition (34). When transcription is inhibited, NEAT1 RNAs dissociate from paraspeckles and are either degraded or relocated to other nuclear domains (6,9,17). To examine whether PS-ASOs relocate with paraspeckle proteins to the perinucleolar caps upon transcriptional inhibition, we treated HeLa cells pre-transfected with 60 nM ASO (ISIS446654) with actinomycin D (1.5 μg/ml, 1.5 h) to arrest transcription, and then stained the cells for P54nrb and fibrillarin, protein markers for previously defined dark and light perinucleolar caps, respectively (34). In contrast to the NEAT1 lncRNA, which dissociates from the paraspeckle proteins during this dramatic reorganization, PS-ASOs relocated with the paraspeckle proteins to the dark perinucleolar caps containing P54nrb (Figure 3C). Similar relocation to the perinucleolar caps was observed with PS-ASOs of different sequences and 2′-modifications (Supplementary Figure S4A) and in multiple human and mouse cell lines, including mESCs (Supplementary Figure S4B). Importantly, co-localization of PS-ASOs and P54nrb to the perinucleolar caps occurred in the absence of NEAT1 lncRNA, suggesting that the association of PS-ASOs with paraspeckle proteins is independent of NEAT1 lncRNA.
Paraspeckles and PS bodies are distinct nuclear foci containing PS-ASOs
Transfected PS-ASOs accumulate in 20–30 bright foci in the nucleus. An average of 3–10 of these PS-ASO foci were co-localized with P54nrb in paraspeckles, whereas the others contained TCP1-β, a protein we recently found to localize to PS bodies (Figure 4A) (35). Signal intensity profile analyses indicated the enrichment of transfected PS-ASOs in both nuclear bodies; however, no overlap between P54nrb and TCP1-β was observed, suggesting that PS bodies and paraspeckles are distinct. This view is further supported by the observations that PS bodies and paraspeckles differ slightly in shape (PS bodies are round; paraspeckles are irregular) and that, in contrast to the dynamic nature of paraspeckles, PS bodies are stable structures (30). It has been reported that neither the formation nor the maintenance of PS bodies requires ongoing RNA transcription (30). As a result, actinomycin D treatment had no effect on PS body integrity (Supplementary Figure S5).
Figure 4.
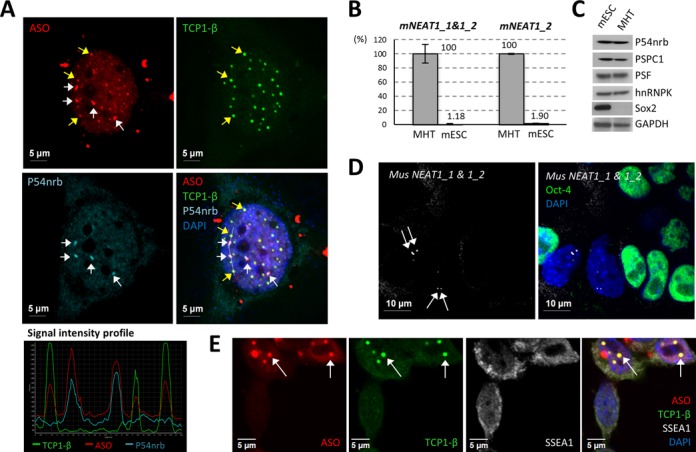
PS bodies and paraspeckles are distinct ASO-containing subnuclear bodies. (A) Transfected ASOs can co-localize with either PS body protein TCP1-β or with paraspeckle protein P54nrb in HeLa cells. White or yellow arrows indicate the co-localization of PS-ASOs with P54nrb in paraspeckles or TCP1-β in PS bodies, respectively. The signal intensity profile of several ASO-containing structures (marked with a dash line in the merge panel) confirmed the co-localization of the ASOs with either TCP1-β or P54nrb. (B) NEAT1 RNA was barely detectable by qRT-PCR in mESCs, as compared with the expression level in control MHT cells. Relative levels of NEAT1_1&1_2 and NEAT1_2 in MHT cells and mESCs are indicated. The error bars represent standard deviation from three parallel experiments. (C) Major paraspeckle proteins were expressed comparably between mESCs and MHT cells. Sox2 served as a mESC marker. GAPDH served as a loading control. (D) RNA-FISH of NEAT1 RNA showed no paraspeckle formation in mESCs. Oct-4 protein staining served as a mESC marker. Arrows indicate paraspeckles in differentiated cells. (E) PS bodies can form in mESCs lacking paraspeckles. Arrows indicate the co-localization of TCP1-β and ASOs in PS bodies. SSEA1 served as a mESC marker.
Paraspeckles and PS bodies are defined as distinct PS-ASO-containing nuclear foci through differences in morphology, dynamics and protein components. We found that PS-body formation is a separate event from localization of PS-ASOs to paraspeckles by analyzing ASO localization in mESCs, in which both isoforms of NEAT1 lncRNA are expressed at a barely detectable level compared with that in mouse MHT cells (Figure 4B) (36). Paraspeckle proteins examined were expressed at comparable levels in mESCs and in MHT cells (Figure 4C). In analyses combining NEAT1 RNA-FISH and IF staining of mESC marker Oct-4, paraspeckles, demarked by the enriched signal of NEAT1, were observed in some differentiated cells losing Oct-4 staining but not in mESCs (Figure 4D). The lack of paraspeckles in mESCs may be due to the poor expression of the structural RNA NEAT1. Importantly, neither the formation of PS bodies nor the co-localization of PS-ASOs with TCP1-β was impaired in mESCs lacking paraspeckles (Figure 4E). Together, these results indicate that PS bodies and ASO-paraspeckles are distinct nuclear foci and that paraspeckle localization of PS-ASOs is not a prerequisite for PS-body formation.
PS-ASOs attract paraspeckle proteins to ASO-induced nuclear filament structures
PS-ASOs, delivered to cells by either transfection or electroporation, were observed to form nuclear filament structures in a subset of cells (Figure 5A and Supplementary Figure S6A). ASO-filaments were usually observed around 4–8 h after ASO transfection in ∼10–15% of transfected cells in an ASO concentration- and sequence-dependent manner (data not shown). Although the formation and the functions of these ASO-filaments are yet to be elucidated, we found that some paraspeckle proteins, such as P54nrb, PSF and PSPC1, co-localized precisely with PS-ASOs in these filament structures (Figure 5A, Supplementary Figure S6B, and S6C). PS-ASOs with different sequences and distinct 2′-modifications were able to induce the formation of these nuclear filaments (Supplementary Figure S6B), and these structures were observed in other human and mouse cell lines after treatment with PS-ASOs (Supplementary Figure S6C). Since paraspeckle protein P54nrb was not detected in nuclear filament structures in cells not treated with ASOs, our data suggest that P54nrb is brought to these structures through its interaction with PS-ASOs.
Figure 5.
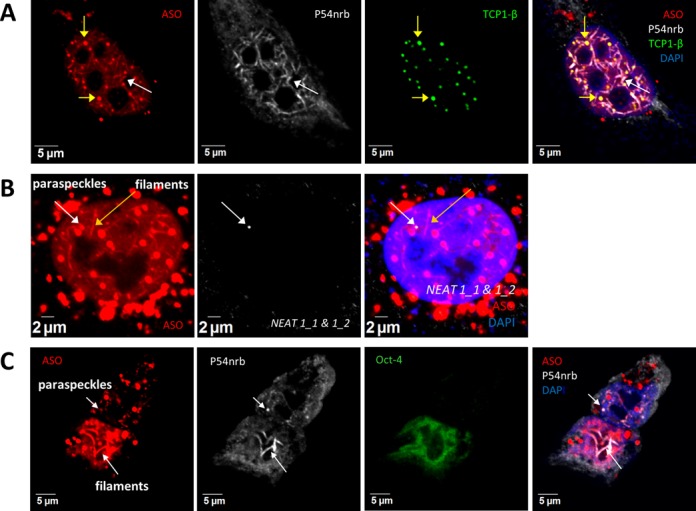
NEAT1 lncRNA is dispensable for the formation of ASO-induced nuclear filaments containing paraspeckle proteins. (A) Co-localization of paraspeckle protein P54nrb with transfected PS-ASOs in nuclear filaments in HeLa cells. Yellow arrows indicate the co-localized PS-ASOs and TCP1-β in PS bodies. The white arrow indicates nuclear filaments containing PS-ASOs and P54nrb. (B) NEAT1 RNA does not co-localize with ASO-filaments, as determined by RNA-FISH using a NEAT1 RNA probe targeting both isoforms in HeLa cells. The white or yellow arrows indicate paraspeckles and filaments, respectively. (C) P54nrb co-localizes with ASOs in nuclear filaments in mESCs. Co-localization of PS-ASOs with P54nrb in nuclear paraspeckles was observed in the differentiated cells that lose mESC marker Oct-4 but not in mESCs. Arrows indicate the co-localization of PS-ASOs with P54nrb.
The presence of paraspeckle proteins in the ASO-induced nuclear filaments raises the possibility that NEAT1, which scaffolds paraspeckles, is also present in and required for the integrity of ASO-induced nuclear filaments. In HeLa cells transfected with 60 nM ISIS446654, NEAT1 RNA was not observed in ASO-induced nuclear filaments (Figure 5B), even when fluorescence signals were enhanced. In addition, formation of P54nrb-containing nuclear filament structures was observed in a subpopulation of HeLa cells transfected with a NEAT1-specific ASO ISIS407248 (Supplementary Figure S6D). Importantly, despite low levels of expression of NEAT1 RNA and a lack of detectable paraspeckles, ASO-induced nuclear filaments containing P54nrb were observed in mESCs (Figure 5C). Together, these results indicate that NEAT1 lncRNA is not required for the formation of ASO-induced nuclear filaments.
PS-ASOs reduce NEAT1 RNA levels in transfected cells
An average of 5–20 paraspeckles per HeLa nucleus were documented previously (14). Interestingly, we noticed that the number of P54nrb-enriched subnuclear foci increased dramatically in some cells containing high concentration of PS-ASOs. In ∼10% of HeLa cells transfected with 60 nM ISIS454395, more than 40 P54nrb-containing PS-ASO foci per cell were observed (Figure 6A), whereas in mock-transfected HeLa cells, only a few paraspeckles (∼3–10 paraspeckles per cell), judged by the co-localization of NEAT1 RNA and P54nrb, were detected (Figure 6A and Supplementary Figure S7). We note that the level of P54nrb was not significantly changed upon transfection of cells with PS-ASOs (data not shown), so the increased number of P54nrb foci most likely stems from recruiting diffuse P54nrb from the nucleoplasm.
Since NEAT1 RNA is essential for initiating paraspeckle assembly (2,8), it is possible that ASO transfection induces up-regulation of NEAT1 RNA levels to increase paraspeckle numbers. However, our results do not support this hypothesis and to our surprise, NEAT1 RNA levels were moderately reduced upon ASO treatment. Transfection of HeLa cells with PS-ASOs (exemplified by ISIS454395, ISIS141923, ISIS110074 and ISIS462026) with no complementarity to NEAT1 RNA at a 60 nM concentration resulted in a 40–60% reduction of NEAT1 RNA (Figure 6B). As a positive control, transfection with 30 nM NEAT1-specific PS-ASO (ISIS407248), an ASO designed to reduce NEAT1 levels by directing RNase H1 cleavage, reduced the NEAT1 RNA levels by more than 85%. Consistently, reduction of NEAT1_1 RNA by transfection of 60 nM PS-ASOs ISIS110074 or ISIS462026 was confirmed by northern hybridization (Figure 6C).
ASO-induced NEAT1 RNA reduction appears not to be a secondary effect of transfection. Dose-dependent reduction of NEAT1 was observed in HeLa cells upon transfection of a control ASO ISIS141923 lacking a cellular target and has no significant complementarity to NEAT1 RNA (Figure 6D). While ISIS141923 caused moderate NEAT1 RNA reduction, a NEAT1-specific ASO ISIS407248 exhibited a more potent dose-dependent reduction of NEAT1 RNA (Figure 6D). It is unlikely that the ASO-induced NEAT1 RNA reduction was due to sequence-dependent off-target cleavage by RNase H1, since the NEAT1 reduction by the non-NEAT1-targeting ASO occurred between 6 and 8 h after transfection, whereas RNase H1-mediated cleavage by the specific ASO took place as early as 2 h after transfection (Figure 6E). Importantly, depletion of RNase H1 by siRNA treatment (Figure 6F) in HeLa cells did not affect NEAT1 reduction induced by control ASO (ISIS141923), but significantly decreased the antisense activity of the NEAT1-specific ASO (ISIS407248) as determined by qRT-PCR (Figure 6G). These results suggest that PS-ASOs can reduce NEAT1 RNA levels through mechanisms other than RNase H1-mediated cleavage.
To elucidate mechanisms of PS-ASO-induced NEAT1 degradation, we first evaluated whether the transcription of NEAT1 was affected in the presence of PS-ASOs by measuring the level of pre-NEAT1 RNA by qRT-PCR, using primers spanning the 3′-end of NEAT1_2 and the small tRNA-like domain, which is cleaved during the processing of NEAT1_2 RNA (Figure 6H) (16). Since the pre-NEAT1 RNA was not affected in the presence of 60 nM control PS-ASOs ISIS141923 or ISIS110074 (Figure 6H), the observed reduction in mature NEAT1 RNA levels appears not to occur at the transcriptional level. Given that P54nrb bound with higher affinity to PS-ASOs than to PO-RNAs (Figure 1D), it is very likely that the transfected PS-ASOs reduce NEAT1 RNA levels through competition for paraspeckle protein binding. NEAT1 RNAs, when dissociated from paraspeckle proteins, might be more susceptible to degradation, as the depletion of some paraspeckle proteins, including P54nrb and PSF, resulted in the reduction of NEAT1 RNA levels (16). To test this hypothesis, we first performed RIP with anti-P54nrb antibody from cell extracts prepared from HeLa cells transfected with increasing amount of PS-ASOs ISIS454395. The co-immunoprecipitated RNAs were analyzed by qRT-PCR (Figure 6I). The relative recovery rates of P54nrb-associated NEAT1 RNAs, detected by primer probe sets either specific to the long isoform (NEAT1_2) or to both isoforms (NEAT1_1 & 1_2) and corrected for mock transfected control, reduced in an ASO dose-dependent manner. Since it is true that transfected PS-ASOs can displace NEAT1 RNA from paraspeckles, we then evaluated the possibility that non-protein-bound NEAT1 RNAs can be rapidly degraded. To measure the decay rates of NEAT1 RNA in the presence or absence of PS-ASOs in a non-disruptive way, HeLa cells were either mock-transfected or transfected with 60 nM control PS-ASO ISIS141923 for 5 h before they were pulse-labeled with 0.5 mM 5′-ethynyl Uridine (EU) for 1 h to label nascent RNAs. At indicated time points after the removal of EU-containing medium, nascent EU-labeled RNAs were captured from total RNA by biotinylation of EU through click chemistry, followed by purification on streptavidin beads. NEAT1 RNA level was quantified by qRT-PCR. Indeed, a more rapid decay of NEAT1 RNA was observed upon control PS-ASO transfection (Figure 6J), implying that non-protein bound NEAT1 is less stable. These results suggest that PS-ASOs that bind to paraspeckle proteins can compete with NEAT1 RNA for protein binding, displacing NEAT1, and leading to a more rapid degradation of the RNA.
PS-ASOs seed the formation paraspeckle-like foci
Our observations imply that PS-ASOs initiate the assembly of paraspeckle-like foci in the absence of NEAT1 RNA. To test this possibility, combined NEAT1 RNA-FISH and IF staining of P54nrb was performed in fixed HeLa cells transfected with 60 nM PS-ASO ISIS454395 (Figure 7A). Strikingly, in ∼10–15% of cells that took up higher amount of PS-ASOs, higher numbers of NEAT1-absent ASO-P54nrb foci (paraspeckle-like structures) were observed (yellow arrows), while in cells that took up less PS-ASOs (∼85–90% of total transfected cells), NEAT1 RNA was observed to co-localize with PS-ASOs and P54nrb in the canonical paraspeckles (white arrow), suggesting that PS-ASOs can localize to both NEAT1-present paraspeckle structures and NEAT1-absent paraspeckle-like structures. In addition, co-localization of paraspeckle proteins with a NEAT1-specific ASO ISIS652721 (Cy3-conjugated ISIS407248) in nuclear foci was also observed in some HeLa cells (Supplementary Figure S8). More importantly, we found the co-localization of paraspeckle protein PSF with PS-ASO ISIS454395 in mESCs that expressing little or no NEAT1 RNA (Figure 7B). Together, these results suggest that PS-ASO-induced paraspeckle-like foci contain no NEAT1 RNA.
It has been reported that some paraspeckle proteins, such as P54nrb and PSF, are essential for building higher-ordered paraspeckle structures, since the depletion of either protein resulted in a significant reduction of long isoform NEAT1_2 RNA level, and therefore, the loss of paraspeckle structures (Supplementary Figure S9) (16,37). Different from canonical paraspeckles, the induced formation of paraspeckle-like foci, judged by the co-localization of PSF and PS-ASOs, was not abolished in P54nrb-depleted cells (Supplementary Figure S9), suggesting that the assembly of paraspeckle-like structure is different from that of canonical paraspeckles, and the architectural components of paraspeckle-like foci are, rather than NEAT1_2 RNA, PS-ASOs.
Paraspeckle proteins P54nrb and PSF associate preferentially with hyper-A-to-I-edited RNAs, and therefore, paraspeckles were proposed to be involved in nuclear retention of edited RNAs (18,25). To investigate the function of ASO-induced paraspeckle-like foci, we performed RNA-FISH of both NEAT1 RNA and a previously identified paraspeckle-retained RNA, CTN, in fixed MEF cells transfected with 60 nM PS-ASO (ISIS454395). CTN was observed in both canonical paraspeckles containing NEAT1 RNA and in ASO-induced paraspeckle-like structures lacking NEAT1 RNA (Figure 7C), indicating that the PS-ASO-seeded paraspeckle-like foci are functional.
DISCUSSION
In this study, we demonstrate that paraspeckle-like structures can be scaffolded on nucleic acids other than NEAT1 lncRNA. Association of PS-ASOs with paraspeckle proteins results in their co-localization in different nuclear structures including canonical NEAT1 RNA-containing paraspeckles, perinucleolar caps (upon transcriptional inhibition), nuclear filaments and NEAT1 RNA-absent paraspeckle-like structures. Our observations suggest that short 20-mer PS-ASOs can serve as seeding molecules to assemble distinct nuclear foci.
The major determinant of the observed co-localization of PS-ASOs with paraspeckle proteins and the inhibitory effects of paraspeckle proteins, such as P54nrb, PSPC1 and hnRNPK, on ASO antisense activity (Figure 2) is the binding affinity of PS-ASOs with paraspeckle proteins. Consistent with a previous study showing stronger binding affinity of PS-ASOs than PO-ASOs to many proteins (33), P54nrb bound more tightly to PS-ASOs than to PO-ASOs (Figure 1D). Since P54nrb bound both ssPS-ASO and PS-ASO/RNA duplexes (Figure 1C), it is possible that binding to P54nrb might reduce the amount of ssASOs that available to bind target mRNAs or might limit the accessibility of the ASO/mRNA duplex to RNase H1, resulting in the observed negative effect of P54nrb on PS-ASO-mediated target RNA reduction. These potential possibilities are currently under investigation. Although depletion of paraspeckle proteins caused only moderate effects on antisense activity of PS-ASOs, this is not surprising, since PS-ASOs associate with many cellular proteins and each protein may bind a portion of ASOs, leading to moderate influence of individual proteins on ASO activities.
PS-ASOs localized to canonical paraspeckles containing NEAT1 RNA or paraspeckle-like foci lacking NEAT1 RNA. Although it is possible that the PS-ASOs hybridize to NEAT1 RNA, this seems unlikely as there was no correlation between PS-ASO-paraspeckle localization and ASO-NEAT1 RNA complementarity. It is more likely that PS-ASOs can localize to paraspeckles through interactions with the paraspeckle proteins. Several evidences supporting this conclusion include that (i) PS-ASOs can pull down paraspeckles proteins from total cell lysate in the affinity selection assay, (ii) the association of PS-ASOs with paraspeckle proteins in the nuclear filaments or paraspeckle-like structures was not impaired in mESCs expressing little or no NEAT1 RNA, (iii) NEAT1-specific ASOs were also observed to co-localize with paraspeckle proteins in distinct nuclear foci and (iv) co-localization of PS-ASOs with paraspeckle proteins to the perinucleolar caps was observed upon actinomycin D treatment which dissociates NEAT1 RNA from paraspeckle proteins. One of the most important observations of this study is that PS-ASOs triggered the formation of P54nrb-containing nuclear structures morphologically similar to paraspeckles that do not contain NEAT1 RNA. This conclusion is supported by the findings that (i) the number of ASO-containing paraspeckle-like foci increased dramatically in a subset of ASO transfected cells, accompanied by a reduction in NEAT1 RNA, (ii) NEAT1 RNA was not detected in paraspeckle-like foci and (iii) formation of paraspeckle-like foci was observed in mESCs that express little or no NEAT1 RNA. However, it has been reported previously that chromatin immobilization of PSF recruits other paraspeckle proteins but not RNA components like NEAT1 and CTN (2,8). Therefore, enrichment of paraspeckle proteins in paraspeckle-like structures may not be a faithful reflection of bona fide formation of functional paraspeckles in vivo. By RNA-FISH we found that paraspeckle-like foci lacking NEAT1 retained CTN-RNA as did conical paraspeckles, implying that paraspeckle-like structures may not only be morphologically normal but also functional (Figure 7C). It is currently unknown how a short 20-mer PS-ASOs can induce the formation of paraspeckle-like structure, which is normally built on long NEAT1 RNAs. It is possible that PS-ASOs can serve as seeding molecules to dock certain paraspeckle proteins, such as P54nrb/PSF/PSPC1, leading to the subsequent assembly of other paraspeckle components. This view is supported by previous studies showing that paraspeckle formation is a seeded event by NEAT1 RNA (2,8). Paraspeckles may contain multiple NEAT1 RNA molecules that interact to form a lattice on which paraspeckles are built (6). Similarly, An ASO-induced paraspeckle-like structure may contain multiple PS-ASO molecules, since it is unlikely that a single PS-ASO molecule would result in sufficient fluorescent signal to allow the detection of a distinct structure under confocal microscopy.
Canonical paraspeckles and paraspeckle-like structures differ in their architectural components. Canonical paraspeckles contain and are built on NEAT1 RNA, while paraspeckle-like structures contain no NEAT1 RNA and are possibly built on PS-ASOs. In addition, PS-ASOs localize to canonical paraspeckles as soon as 2–3 h after transfection of 60 nM PS-ASO ISIS454395 in most well-transfected cells, while formation of paraspeckle-like structures was usually observed at a later time point, ∼5–8 h after transfection, coinciding with the reduction of NEAT1 RNA, in an ASO concentration-dependent manner in a subset of cells (∼10–15% of total transfected cells). Moreover, canonical paraspeckles often cluster around the NEAT1 gene locus through co-transcriptional assembly (8). In contrast, ASO-induced paraspeckle-like foci were scattered throughout the nucleus, supporting the view that paraspeckle-like foci do not require NEAT1 RNA. It is possible that PS-ASOs join pre-existing paraspeckles and replace NEAT1 RNA and that these structures then move to other subnuclear sites. It is also likely that PS-ASOs trigger the de novo on-site formation of paraspeckle-like foci independently of NEAT1 RNA by recruiting paraspeckle proteins from nucleoplasmic pool.
Transfected PS-ASOs moderately reduced both the cellular and P54nrb-associated NEAT1 RNA levels (Figure 6). This reduction in NEAT1 RNA levels does not appear to be due to RNase H1-mediated cleavage caused by imperfect base pairing of PS-ASOs with NEAT1 RNA since (i) PS-ASOs of different sequences caused similar effects, (ii) the onset of NEAT1 reduction occurred much later with non-NEAT1-targeting PS-ASOs than with the NEAT1-specific PS-ASO (Figure 6E) and (iii) the depletion of RNase H1 did not affect NEAT1 reduction by the non-NEAT1-targeting ASOs (Figure 6G). Given the short half-lives of NEAT1 RNAs (2 h for the short and 4 h for the long isoforms) (38), it is possible that the transfected PS-ASOs compete paraspeckle proteins from NEAT1 RNA, and that non-protein-bound NEAT1 is rapidly degraded. Previous studies have already demonstrated that reduction of paraspeckle proteins can reduce the level of NEAT1 RNA (9,16), implying protective roles of paraspeckle proteins in NEAT1 RNA stability. Indeed, transfection of PS-ASOs affected the stability rather than the transcription of NEAT1 RNA since pre-NEAT1 RNA level was not affected.
The function of PS-ASO-P54nrb/PSF containing foci has yet to be defined. On one hand, these nuclear bodies may serve as a reservoir to deposit excess PS-ASOs or to prevent hybridization of PS-ASO with target RNA to render it unable to exert antisense activity. On the other hand, protein sequestration by PS-ASOs into paraspeckle-like or filament structures may restrict functional available proteins. A recent study reported up-regulation of NEAT1 RNA upon viral infection or poly (I:C) stimulation as a result of facilitating transcription of antiviral genes such as IL8 through protein sequestration of PSF, a transcriptional repressor of IL8 (23). It was shown in that study that siRNA-mediated reduction of NEAT1 RNA reduced the up-regulation of IL8 mRNA by poly (I:C) stimulation, most likely due to the release of PSF from paraspeckles to repress the expression of IL8 mRNA. In our study, however, treatment of HeLa cells with NEAT1-specific PS-ASOs had no significant effect on Poly (I:C) induced IL8 mRNA up-regulation (data not shown). One explanation may be that PS-ASOs bind paraspeckle proteins including PSF thus limiting their function. Nonetheless, it is possible that the interaction between PS-ASO and paraspeckle proteins (e.g. PSF) may signal host immune response and help cells cope with excess foreign ASO molecules.
In conclusion, our results indicate that formation of paraspeckle-like structures is triggered by short PS-modified oligonucleotides in the absence of NEAT1 RNA. This is very likely due to high affinity between PS-ASOs and the paraspeckle proteins, most of which are RNA-binding proteins. These findings provide a possible explanation for the driving force of the formation of other endogenous nuclear bodies that contain RNAs and the ASO-induced formation of paraspeckle-like structures may provide a useful tool to study the biological function of paraspeckles.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Timothy A. Vickers, Walt F. Lima, Hongjiang Wu and C. Frank Bennett for insightful discussions; Thazha P. Prakash for ASO synthesis; Hong Sun for technical assistance. The CTN-RNA plasmid was kindly provided by David L. Spector and Kannanganattu V. Prasanth.
FUNDING
Source of Open Access funding: Internal funding from ISIS pharmaceuticals.
Conflict of interest statement. None declared.
REFERENCES
- 1.Spector D.L. Nuclear domains. J. Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- 2.Shevtsov S.P., Dundr M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 3.Clemson C.M., McNeil J.A., Willard H.F., Lawrence J.B. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sone M., Hayashi T., Tarui H., Agata K., Takeichi M., Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 6.Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., Lawrence J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawaguchi T., Hirose T. Architectural roles of long noncoding RNAs in the intranuclear formation of functional paraspeckles. Front. Biosci. 2012;17:1729–1746. doi: 10.2741/4015. [DOI] [PubMed] [Google Scholar]
- 8.Mao Y.S., Sunwoo H., Zhang B., Spector D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki Y.T., Ideue T., Sano M., Mituyama T., Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl Acad. Sci. U.S.A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng R., Shen Z., Tripathi V., Xuan Z., Freier S.M., Bennett C.F., Prasanth S.G., Prasanth K.V. Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells. J. Cell Sci. 2010;123:3734–3744. doi: 10.1242/jcs.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., Lin C., Liu W., Zhang J., Ohgi K.A., Grinstein J.D., Dorrestein P.C., Rosenfeld M.G. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valgardsdottir R., Chiodi I., Giordano M., Cobianchi F., Riva S., Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox A.H., Lam Y.W., Leung A.K., Lyon C.E., Andersen J., Mann M., Lamond A.I. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 14.Fox A.H., Lamond A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Passon D.M., Lee M., Rackham O., Stanley W.A., Sadowska A., Filipovska A., Fox A.H., Bond C.S. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl Acad. Sci. U.S.A. 2012;109:4846–4850. doi: 10.1073/pnas.1120792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L.L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Souquere S., Beauclair G., Harper F., Fox A., Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell. 2010;21:4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa S., Naganuma T., Shioi G., Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saha S., Murthy S., Rangarajan P.N. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J. Gen. Virol. 2006;87:1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q., Chen C.Y., Yedavalli V.S., Jeang K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio. 2013;4:e00596–00512. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Benard M., Fox A.H., et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 26.Chen L.L., Carmichael G.G. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 27.Everett R.D., Murray J. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 2005;79:5078–5089. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crooke S.T.V.T., Lima W., Wu H.-J. In: Antisense Drug Technology—Principles, Strategies, and Application. In: Crooke S.T., editor. Boca Raton, FL, USA: CRC Press; 2006. [Google Scholar]
- 29.Raal F.J., Santos R.D., Blom D.J., Marais A.D., Charng M.J., Cromwell W.C., Lachmann R.H., Gaudet D., Tan J.L., Chasan-Taber S., et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 30.Lorenz P., Baker B.F., Bennett C.F., Spector D.L. Phosphorothioate antisense oligonucleotides induce the formation of nuclear bodies. Mol. Biol. Cell. 1998;9:1007–1023. doi: 10.1091/mbc.9.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lima W.F., Prakash T.P., Murray H.M., Kinberger G.A., Li W., Chappell A.E., Li C.S., Murray S.F., Gaus H., Seth P.P., et al. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Liang X.H., Crooke S.T. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011;39:4875–4889. doi: 10.1093/nar/gkr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown D.A., Kang S.H., Gryaznov S.M., DeDionisio L., Heidenreich O., Sullivan S., Xu X., Nerenberg M.I. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem. 1994;269:26801–26805. [PubMed] [Google Scholar]
- 34.Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 35.Liang X.H., Shen W., Sun H., Prakash T.P., Crooke S.T. TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku484. doi:10.1093/nar/gku484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naganuma T., Hirose T. Paraspeckle formation during the biogenesis of long noncoding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tani H., Mizutani R., Salam K.A., Tano K., Ijiri K., Wakamatsu A., Isogai T., Suzuki Y., Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


