The regulated loading of helicase MCM2–7 onto origins of DNA replication is the prerequisite for replication fork establishment and genomic stability. However, the helicase loading process is not well understood. Samel et al. reveal that the MCM2–7 complex adopts a closed ring structure, suggesting that helicase loading requires active ring opening. A chemical biology approach demonstrates that regulated MCM2–7 ring opening at the Mcm2/5 interface is essential for prereplicative complex formation. This study establishes the existence of a unique DNA entry gate that is critical for helicase loading and DNA replication.
Keywords: DNA replication, replicative helicase, pre-RC, DNA licensing, cancer, genomic stability
Abstract
The regulated loading of the replicative helicase minichromosome maintenance proteins 2–7 (MCM2–7) onto replication origins is a prerequisite for replication fork establishment and genomic stability. Origin recognition complex (ORC), Cdc6, and Cdt1 assemble two MCM2–7 hexamers into one double hexamer around dsDNA. Although the MCM2–7 hexamer can adopt a ring shape with a gap between Mcm2 and Mcm5, it is unknown which Mcm interface functions as the DNA entry gate during regulated helicase loading. Here, we establish that the Saccharomyces cerevisiae MCM2–7 hexamer assumes a closed ring structure, suggesting that helicase loading requires active ring opening. Using a chemical biology approach, we show that ORC–Cdc6–Cdt1-dependent helicase loading occurs through a unique DNA entry gate comprised of the Mcm2 and Mcm5 subunits. Controlled inhibition of DNA insertion triggers ATPase-driven complex disassembly in vitro, while in vivo analysis establishes that Mcm2/Mcm5 gate opening is essential for both helicase loading onto chromatin and cell cycle progression. Importantly, we demonstrate that the MCM2–7 helicase becomes loaded onto DNA as a single hexamer during ORC/Cdc6/Cdt1/MCM2–7 complex formation prior to MCM2–7 double hexamer formation. Our study establishes the existence of a unique DNA entry gate for regulated helicase loading, revealing key mechanisms in helicase loading, which has important implications for helicase activation.
DNA replication is a carefully choreographed process that is central to genome integrity, while misregulation of DNA replication leads to genomic instability, disease, or cancer. A principal component of the DNA replication machinery is the replicative helicase that separates the two DNA strands at the replication fork. Minichromosome maintenance proteins 2–7 (MCM2–7) represent the core of the replicative helicase in eukaryotes. However, the MCM2–7 helicase does not bind to DNA by itself but instead is loaded at DNA replication origins by a number of factors. The origin recognition complex (ORC) and Cdc6 recognize the DNA replication origin (Bell and Stillman 1992; Speck et al. 2005; Speck and Stillman 2007) and recruit Cdt1 and MCM2–7 to DNA, which leads to formation of a prereplicative complex (pre-RC) (Bowers et al. 2004; Randell et al. 2006). During pre-RC assembly, two MCM2–7 hexamers are loaded into a head-to-head double hexamer around dsDNA (Evrin et al. 2009; Remus et al. 2009).
Helicase loading occurs from late M to early G1 phase of the cell cycle (Weinreich et al. 1999), although, at this point, the loaded double hexamer has yet to acquire helicase activity. At the onset of S phase, the MCM2–7 helicase becomes activated by two kinases (cyclin-dependent kinase [CDK] and Dbf4-dependent kinase [DDK]) (Tanaka et al. 2007; Zegerman and Diffley 2007; Sheu and Stillman 2010) and several replication factors (Tanaka and Araki 2010; Heller et al. 2011). Consequently, the MCM2–7 double hexamer separates into two single hexamers. Each single hexamer associates with Cdc45 and GINS (go-ichi-ni-san, Japanese for 5-1-2-3, named after Sld5, Psf1, Psf2, and Psf3, which make up GINS) to form a Cdc45/GINS/MCM2–7 complex (CMG) (Ilves et al. 2010). While the MCM2–7 double hexamer encircles dsDNA (Evrin et al. 2009; Remus et al. 2009), the CMG encircles ssDNA (Fu et al. 2011). How this dsDNA-to-ssDNA transition occurs remains unknown, but the process is thought to require opening of the MCM2–7 ring. The CMG complex represents the active replicative helicase that moves ahead of the replication fork to unwind the DNA (Gambus et al. 2006; Ilves et al. 2010; Fu et al. 2011).
Assembly of MCM2–7 double hexamer by ORC/Cdc6 and Cdt1 requires ATP hydrolysis (Bowers et al. 2004; Randell et al. 2006; Fernández-Cid et al. 2013). In the absence of ATP hydrolysis, only an initial ORC/Cdc6/Cdt1/MCM2–7 (OCCM) complex is formed, which contains a single MCM2–7 hexamer (Evrin et al. 2013) and multiple Cdt1 molecules (Takara and Bell 2011). ATP hydrolysis by Orc1 and Cdc6 leads to rapid Cdt1 release and formation of a pre-RC intermediate containing only one ORC, Cdc6, and the MCM2–7 hexamer, respectively, known as the ORC/Cdc6/MCM2–7 (OCM) complex (Fernández-Cid et al. 2013). The OCM complex can recruit a second MCM2–7 hexamer (Evrin et al. 2014) to facilitate MCM2–7 double hexamer formation (Evrin et al. 2009; Remus et al. 2009). Interestingly, the OCM complex is subject to quality control. Phosphorylation of ORC by CDK in S phase, which functions to inhibit pre-RC assembly, allows OCCM formation, but then ATP hydrolysis-driven Cdt1 release leads to disassembly of the OCCM complex, preventing OCM formation. Although we have gained insights into the mechanism of MCM2–7 double hexamer assembly (Riera et al. 2014; Yardimci and Walter 2014), it remains uncertain at which point during pre-RC formation the MCM2–7 ring is opened and dsDNA is inserted.
The six subunits of the MCM2–7 hexamer are assembled as a ring; their subunit arrangement is Mcm3–Mcm5–Mcm2–Mcm6–Mcm4–Mcm7 (Davey et al. 2003; Costa et al. 2011; Sun et al. 2013). Currently, it is unclear whether the six MCM2–7 subunits form a closed circular ring or whether the ring can transition between an open and a closed state by itself (Evrin et al. 2009; Remus et al. 2009; Costa et al. 2011; Sun et al. 2013). Clearly, for DNA loading, the ring needs to be in an open conformation so that dsDNA can slip inside. An electron microscopy (EM) analysis showed that Drosophila melanogaster MCM2–7 adopts a spiral shape with a gap between Mcm2 and Mcm5 (Costa et al. 2011). On the other hand, analysis of Mcm ATP-binding mutants in an ssDNA-binding assay showed that a weak Mcm2/5 interface can spring open in the absence of ATP and close in the presence of ATP. In addition, the analysis of ATPase mutants indicates that the Mcm2/6 ATPase pair modulates the activity of the putative Mcm2/5 gate during this process or could function as an alternative gate (Bochman and Schwacha 2007, 2010). Then again, MCM2–7 cannot bind to dsDNA on its own (Evrin et al. 2009; Remus et al. 2009) but is loaded by ORC, Cdc6, and Cdt1 onto DNA, suggesting that these factors could promote MCM2–7 ring opening (Coleman et al. 1996; Donovan et al. 1997; Weinreich et al. 1999; Evrin et al. 2009; Remus et al. 2009). If the MCM2–7 ring is constitutively closed or cycles through open and closed conformations, whether a specific DNA entry gate is used for helicase loading on dsDNA and how MCM2–7 double hexamer formation and helicase loading are coordinated remain important unresolved questions.
In this study, we examined the Saccharomyces cerevisiae (Sc) MCM2–7 structure and reveal that this complex adopts a topological closed conformation. We used a rapamycin-inducible linkage to systematically study the role of all Mcm2–7 interfaces during pre-RC formation and discovered that the MCM2–7 ring opening at the Mcm2/Mcm5 interface is essential for pre-RC formation. We show that DNA insertion into the MCM2–7 ring occurs prior to ATP hydrolysis at the stage of OCCM formation. Importantly, a linkage between Mcm2 and Mcm5 hinders the establishment of a stable OCM complex in vitro and blocks helicase loading in vivo, demonstrating that the Mcm2/5 gate is crucial for helicase loading and DNA replication.
Results
Structural analysis of the MCM2–7 complex by EM
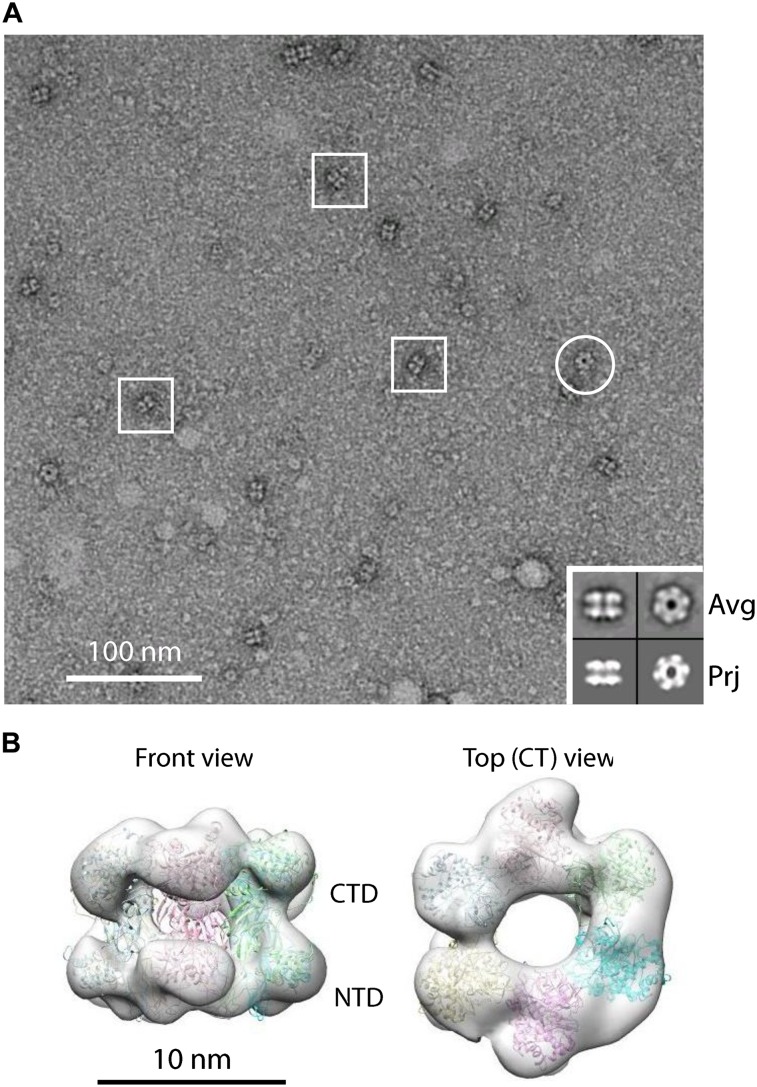
The MCM2–7 complex consists of six homologous proteins that are well conserved from yeast to humans (Bochman and Schwacha 2009; O’Donnell et al. 2013). These proteins associate into a stable hexamer with a mass of 605 kDa. The structure of the hexameric S. cerevisiae MCM2–7 complex has not yet been reported. Therefore, it is unknown whether the complex adopts an open ring conformation similar to Drosophila MCM2–7 (Costa et al. 2011) or whether the MCM2–7 hexamer is in a closed ring conformation, which would indicate that ORC/Cdc6 and Cdt1 need to open the MCM2–7 ring during helicase loading. We used EM to study the organization of the purified ScMCM2–7 complex and found that, in most particle images, MCM2–7 assumes a rectangular shape, corresponding to the hexamer in side views (Fig. 1A). Ring-shaped particles with six distinguishable densities corresponding to the top view were also observed, although at a lower frequency (Fig. 1A). Three-dimensional (3D) reconstruction of the budding yeast MCM2–7 shows that the complex is nearly sixfold symmetrical and adopts a closed ring conformation (Fig. 1B). Six copies of the near full-length crystal structure of an archaeal Sulfolobus solfataricus MCM (Protein Data Bank [PDB] ID 3F9V) can be fitted as rigid bodies into the 3D EM map, only with several small densities left unoccupied, which could correspond to the N-terminal or C-terminal extensions of eukaryotic MCM2–7 (Fig. 1B; Brewster et al. 2008). Hence, our EM analysis establishes that the budding yeast MCM2–7 complex assumes a closed circular conformation in solution.
Figure 1.
MCM2–7 is a topological closed ring. (A) An EM image of MCM2–7 in ATPγS. Three side views are marked with squares, and one top view is marked with a circle. A small inset (bottom right corner) shows reference-free class averages of the side and top views in the top row and a corresponding view reprojected from the 3D model in the bottom row. Each box is 27 nm in size. (B) Surface display of a front and a top view of 3D reconstruction of MCM2–7. Six copies of the crystal structure of the archaeal Sulfolobus solfataricus MCM (shown in cartoon view; Protein Data Bank [PDB] ID: 3F9V) (Brewster et al. 2008) are docked inside the EM map, which is shown as a semitransparent gray surface.
Design of conditional linkage between Mcm subunits
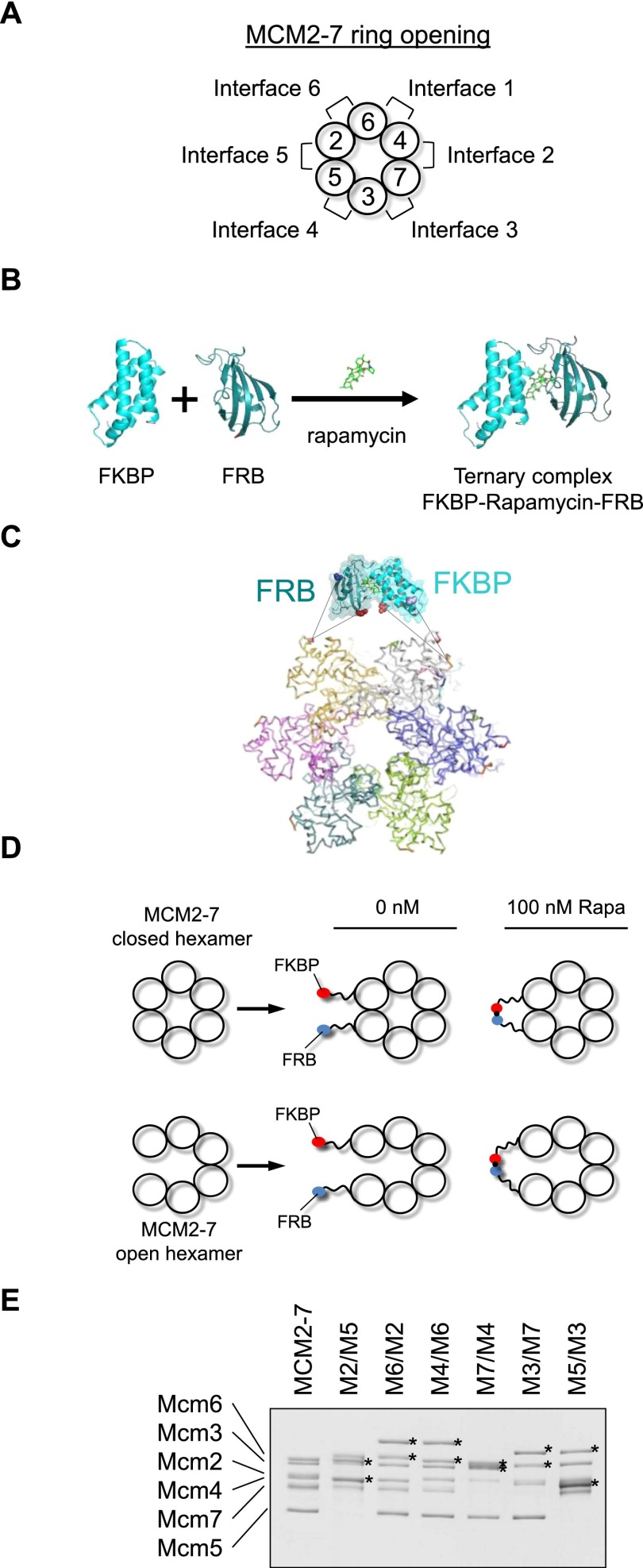
There are six interfaces at which the toroidal MCM2–7 ring could open up, but it is currently not known which one functions during pre-RC formation (Fig. 2A). As the helicase loading factors Orc1–6, Cdc6, and Cdt1 form a complex interface with MCM2–7 (Sun et al. 2013), we expect MCM2–7 ring opening to occur at one specific subunit interface: the DNA entry gate. This model makes the key prediction that artificial interconnection of the two Mcm subunits that make up the DNA entry gate should hinder DNA insertion into the MCM2–7 ring. Consequently, this connection should abolish helicase loading onto DNA. Here we used a chemical biology approach involving MCM2–7 variants that contain FK506- and rapamycin-binding protein (FKBP) and FKBP–rapamycin binding (FRB) protein–protein interaction domains attached to alternating Mcm subunits (Fig. 2B–E). As FKBP and FRB interact only in the presence of rapamycin (Fig. 2B), a conditional small molecule-dependent connection of neighboring Mcm subunits could be generated. MCM2–7 tolerates neither N-terminal nor C-terminal fusions on subunit Mcm5 (Hoang et al. 2007); therefore, we generated internal fusion proteins. The design of these internal Mcm–FKBP/FRB fusions took into account the following considerations: The six Mcm proteins are highly conserved but contain short stretches that are variable in amino acid sequence and length. Such regions form loops on the surface of the archaeal Methanobacterium thermoautotrophicum (Mt) Mcm crystal structure (Fletcher et al. 2003). This raises the possibility of inserting small globular domains of FKBP and FRB into these loops without altering the underlying structure of the Mcm proteins. Indeed, a similar internal fusion approach using an affinity tag has been shown to work for Mcm5 (Leon et al. 2008). To allow the FRB and FKBP domains to interact freely, we added 20-amino-acid long linkers on each end of the interaction domains (Fig. 2C–E). It is noteworthy that simultaneous insertion of FKBP and FRB into the N-terminal domains of two neighboring Mcm subunits did not compromise the ability to purify hexameric MCM2–7 complexes. The addition of FRB and FKBP to the Mcm subunits (Mx/My, where x and y indicate the two neighboring FRB and FKBP fused Mcm subunits) led to an obvious shift in molecular weight (Fig. 2E). Thus, we were in the position to investigate the involvement of each of the six possible MCM2–7 subunit interfaces for helicase loading.
Figure 2.
FKBP and FRB–MCM2–7. (A) MCM2–7 is arranged in a specific subunit order that creates six different subunit interfaces. (B) FKBP and FRB interact only in a rapamycin-dependent fashion. (C) The rapamycin-induced interaction between FKBP and FRB creates a linkage between Mcm subunits. (D) We designed 20-amino-acid-long linkers between FRB/FKBP and Mcm subunits to minimize any effect on MCM2–7 ring opening or closing. (E) Purified MCM2–7 and the six MCM2–7–FKBP/FRB protein complexes. Proteins are labeled with Mx/My, where x and y indicate the two neighboring FRB- and FKBP-fused Mcm subunits.
Characterization of the linked MCM2–7 complexes
To examine whether the conditional link between two neighboring Mcm subunits affected pre-RC formation we used an in vitro pre-RC assay (Evrin et al. 2009). We combined ORC, Cdc6, Cdt1, and either wild-type MCM2–7 or various FRB/FKBP MCM2–7 complexes and analyzed their binding to origin DNA coupled to magnetic beads in both the presence and absence of rapamycin (Fig. 3A). A low-salt wash removed unbound proteins, and, in turn, a mixture of pre-RC intermediates, including associated and loaded MCM2–7, could be observed (Randell et al. 2006; Tsakraklides and Bell 2010; Fernández-Cid et al. 2013; Frigola et al. 2013; Evrin et al. 2014). On the other hand, a high-salt wash removed ORC, Cdc6, and various MCM2–7 loading intermediates but not the MCM2–7 double hexamer, as this complex encircles DNA and is high-salt-resistant (Evrin et al. 2009; Remus et al. 2009).
Figure 3.
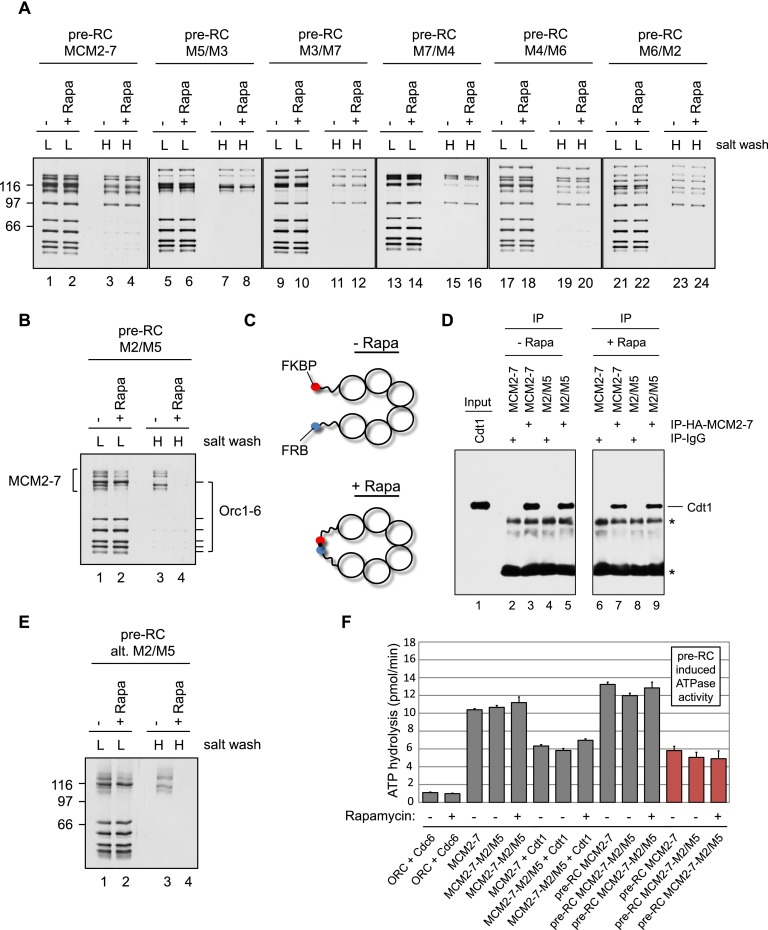
Only MCM2–7–M2/M5 in the presence of rapamycin displays a helicase loading defect. (A) Pre-RC reactions using MCM2–7 and five MCM2–7–FKBP/FRB protein complexes were assembled in the presence or absence of rapamycin and ATP, washed with low-salt (L) or high-salt (H) buffer, and analyzed by silver staining. The smallest subunit of the Orc1–6 complex stains only weakly by silver staining. (B) Pre-RC reactions using MCM2–7–M2/M5 were assembled in the presence or absence of rapamycin and ATP, washed with low salt (L) or high salt (H), and analyzed by silver staining. (C) Illustration showing the rapamycin-induced linkage between neighboring Mcm subunits. (D) The MCM2–7–Cdt1 interaction analysis used IgG control beads or MCM2–7 (HA-Mcm3) coupled to anti-HA beads. A 30% input is shown along with 100% of the immunoprecipitate. Asterisks mark nonspecific IgG-related bands. (E) Pre-RC reactions using an alternative MCM2–7–M2/M5 construct were assembled in the presence or absence of rapamycin and ATP, washed with low salt (L) or high salt (H), and analyzed by silver staining. (F) Analysis of ATPase activities during pre-RC assembly with wild-type (wt) MCM2–7 or MCM2–7–M2/M5 in the presence or absence of rapamycin. The ATP hydrolysis rates were determined for the indicated proteins in the presence of ARS1 origin DNA. Together, ORC, Cdc6, Cdt1, and MCM2–7 induced a strong ATPase activity. The pre-RC-induced ATPase activity is shown in red (ATPase activity of the full pre-RC reaction with the ATPase activity of the individual components [ORC/Cdc6 + Cdt1/MCM2–7] subtracted).
We found that wild-type MCM2–7 and five MCM2–7 gate fusions supported MCM2–7 association with ORC/Cdc6 and also salt-resistant MCM2–7 double hexamer formation. In the presence of rapamycin, we observed equal MCM2–7 association and loading, indicating that rapamycin addition has no influence on pre-RC formation. Interestingly, one specific fusion—the MCM2–7-M2/M5 construct—in the absence of rapamycin showed efficient MCM2–7 association and salt-resistant MCM2–7 loading (Fig. 3B, lanes 1,3). In contrast, in the presence of rapamycin, this construct displayed reduced MCM2–7 association and no high-salt-resistant loading. Thus, the rapamycin-induced link between Mcm2 and Mcm5 affected MCM2–7 association and blocked double hexamer formation (Fig. 3C).
It is conceivable that rapamycin-induced interaction between FRB and FKBP promotes intermolecular complex formation, linking two or more MCM2–7 hexamers together, which could explain the observed defect in pre-RC formation. To test this, we performed gel filtration and observed that MCM2–7–M2/M5 fractionated in the absence and presence of rapamycin at the size of a single hexamer (605 kDa) (Supplemental Fig. S1), demonstrating that rapamycin does not induce intermolecular interactions between hexamers.
To address whether the FKBP–FRB interaction affected the interactions between MCM2–7 and Cdt1, we incubated MCM2–7 or MCM2–7–M2/M5 in the absence and presence of rapamycin with Cdt1 and probed complex formation using an MCM2–7 immunoprecipitation assay. We observed coimmunoprecipitation (co-IP) of Cdt1 with MCM2–7 and MCM2–7–M2/M5 in a similar fashion (Fig. 3D). Another possibility is that the interaction between FRB and FKBP could alter a specific protein interaction during pre-RC formation. Thus, we generated an alternative Mcm5 construct: The linkage between Mcm2 and Mcm5 spanned a different region. This alternative MCM2–7–M2/M5 complex behaved similarly to the original construct, displaying a rapamycin-dependent defect in MCM2–7 association with ORC/Cdc6 and no MCM2–7 double hexamer formation (Fig. 3E).
The MCM2–7 helicase is an AAA+ ATPase. Notably, mutations in ATPase domains of individual Mcm subunits are affecting the ATPase activity of the whole complex (Bochman and Schwacha 2009); therefore, MCM2–7 ATPase measurements can identify structural defects in helicase mutants (Evrin et al. 2014). To identify whether MCM2–7-M2/M5 compared with MCM2–7 has normal or altered ATPase activity and study the influence of rapamycin on MCM2–7–M2/M5 ATP hydrolysis, we performed ATPase assays with the purified proteins (Fig. 3F). Interestingly, for MCM2–7, MCM2–7–M2/M5 without rapamycin, and MCM2–7–M2/M5 with rapamycin, we observed a very similar ATPase activity. Cdt1 has been shown to reduce the ATP hydrolysis of MCM2–7 (Fernández-Cid et al. 2013), which we also observed in all three conditions. Moreover, addition of ORC/Cdc6 and Cdt1 to MCM2–7 led to an induction of pre-RC ATPase activity. The “pre-RC-induced ATPase activity” is defined as the ATPase activity of the full pre-RC reaction minus the activity of the individual components (ORC/Cdc6 + Cdt1/MCM2–7) (Fernández-Cid et al. 2013). Indeed, MCM2–7 and MCM2–7–M2/M5 produced very similar pre-RC-induced ATPase activities regardless of the presence of rapamycin (Fig. 3F, marked in red). These results demonstrate that the MCM2–7–M2/M5 complex has a normal ATPase activity. Importantly, we found that the addition of rapamycin did not alter MCM2–7–M2/M5 ATPase activity, indicating that the rapamycin-induced FKBP–FRB interaction has no significant influence on the overall structure of the complex. Finally, MCM2–7–M2/M5 promoted normal pre-RC-induced ATPase activity, highlighting that the complex is functional for the activation of Orc1/Cdc6 ATP hydrolysis (Fernández-Cid et al. 2013).
The MCM2–7–M2/M5 strain displays rapamycin-dependent lethality
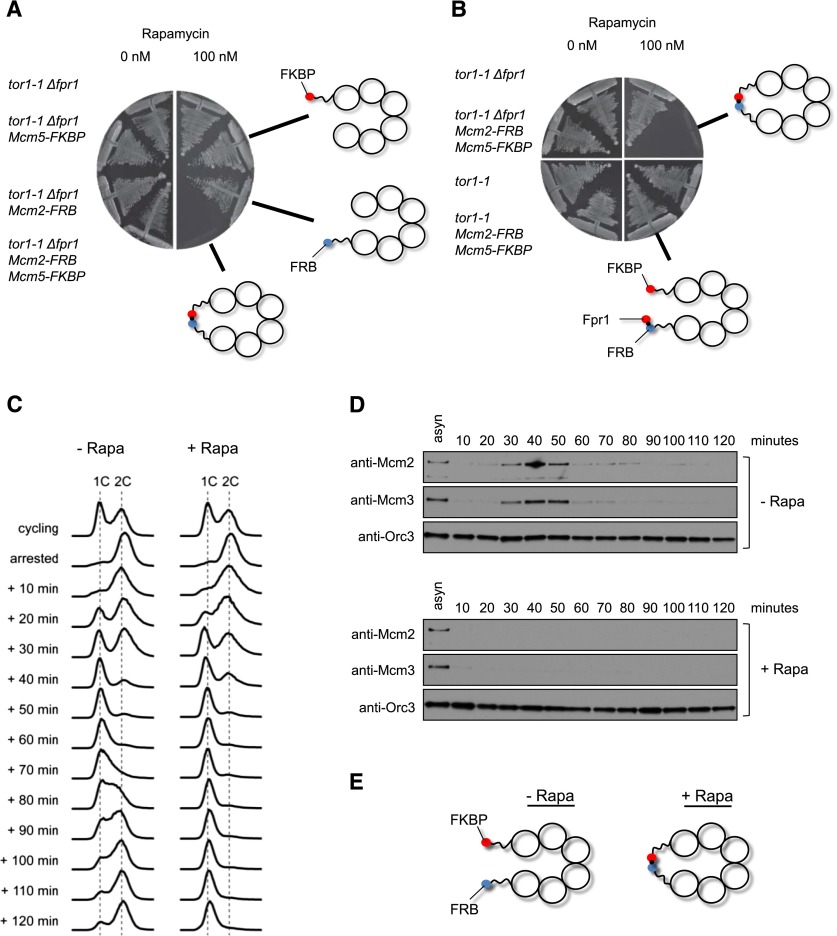
To address the in vivo functionality of the MCM2–7–M2/M5 complex, we replaced the wild-type copies of the Mcm2 and Mcm5 genes with Mcm2–FRB and Mcm5–FKBP fusions in S. cerevisiae. In order to test the sensitivity of this strain to rapamycin, we used the TOR1-1 mutation, which confers rapamycin resistance growth, and a deletion of the Fpr1 gene, which eliminates the most abundant FKBP-like protein and reduces binding of “free” yeast FKBP-like proteins to the FRB moiety fused to Mcm2 (Gruber et al. 2006). One-hundred nanomolar rapamycin inhibited the growth of Mcm5–FKBP Mcm2–FRB cells but not the control strain, Mcm5–FKBP strain, or Mcm2–FRB strain (Fig. 4A), while all strains grew equally well in the absence of rapamycin (Supplemental Fig. S2A), implying that interaction between Mcm5–FKBP and Mcm2–FRB indeed inactivates MCM2–7 function.
Figure 4.
A rapamycin-induced linkage between Mcm2 and Mcm5 affects cell viability, cell cycle progression, and MCM2–7 loading in vivo. (A) Linking Mcm2 with Mcm5 is lethal for the cell. The strains were plated on YPD containing 0 or 100 nM rapamycin. The plates were incubated at 30°C. (B) Fpr1 can bypass the lethal effect of the Mcm2–Mcm5 linkage. The strains were plated on YPD containing 0 or 100 nM rapamycin, and the plates were incubated for 2 d at 30°C. (C) Flow cytometry of the MCM2–7–M2/M5 yeast strain in the presence or absence of rapamycin. The cells were arrested in G2/M phase and then released into the cell cycle. In the presence of rapamycin, the strain progressed very slowly from G1 to S phase. (D) Chromatin-binding analysis for Mcm2, Mcm3, and Orc3 of samples shown in C. (E) Illustration showing the rapamycin-induced linkage between neighboring Mcm subunits.
It is possible that the linkage of Mcm2 and Mcm5 does not cause the rapamycin-dependent lethality per se, but rather it is caused by the formation of a bulky FRB–rapamycin–FKBP complex in their vicinity. To exclude this possibility, we tested whether the rapamycin-dependent lethality is suppressed by the presence of yeast Fpr1 protein, whose complexes with rapamycin should compete with rapamycin–FKBP complexes associated with Mcm2–FRB. Crucially, the presence of Fpr1 fully relieved the inhibition by rapamycin of Mcm5–FKBP Mcm2–FRB cell growth (Fig. 4B). As the linkage between Mcm2 and Mcm5 is at the N-terminal section, it could be argued that this location interferes with pre-RC formation. Therefore, we generated a C-terminal MCM2–7–M2/M5 construct and analyzed whether the overexpression of this mutant in the presence of wild-type MCM2–7 (not overexpressed) caused rapamycin-dependent dominant lethality. We observed that rapamycin produced dominant lethality in the case of MCM2–7–M2/M5 N-terminal and MCM2–7–M2/M5 C-terminal overexpression but not when wild-type MCM2–7 was overexpressed (Supplemental Fig. S2B). This indicates that both N-terminal- and C-terminal-attached FKBP–FRB linkages affect cell growth. To exclude the possibility that the linkage between neighboring Mcm subunits causes a general rapamycin-dependent growth defect, we overexpressed two different gate constructs, MCM2–7–M2/M6 (which is located next to M2/M5) and MCM2–7–M6/M4, along with wild-type MCM2–7 and MCM2–7–M2/M5. Next, we analyzed all strains for rapamycin-dependent dominant lethality. We found that rapamycin did not affect strains overexpressing MCM2–7, MCM2–7–M2/M6, or MCM2–7–M6/M4 but caused a severe growth defect with MCM2–7–M2/M5 (Supplemental Fig. S3). This result indicates that the rapamycin-dependent linkage has no effect on cell growth in general unless the linkage is placed between the Mcm2 and Mcm5 subunits.
Connection of Mcm2 and Mcm5 proteins hinders MCM2–7 association with chromosomes
To test whether rapamycin perturbs the cell cycle progression of Mcm5–FKBP Mcm2–FRB cells, we used a strain whose APC/C coactivator, Cdc20, was under the control of the methionine-repressible MET3 promoter, which, upon arrest and release, enables us to study the synchronous transition of M phase into the cell cycle (Uhlmann et al. 2000). Cells were grown asynchronously in medium lacking methionine and then arrested in M phase using methionine-containing medium. Rapamycin was added for the last 25 min of the M-phase arrest before the culture was washed and released in medium lacking methionine, which allowed synchronous transition through G1, S, and G2 phases (Fig. 4C). The addition of rapamycin did not affect the 2C-to-1C transition, but cell cycle progression slowed down significantly during the G1–S transition, while in the absence of rapamycin, the cells progressed through S phase and into G2 phase. This result indicates that the interaction between Mcm5–FKBP and Mcm2–FRB is affecting cell cycle progression into S phase. To identify the reason of this cell cycle defect, we determined the MCM2–7 chromatin association in these cells. We observed that the addition of rapamycin impaired the association of Mcm2 and Mcm3 with chromatin (Fig. 4D,E), indicating that interaction between Mcm5–FKBP and Mcm2–FRB abrogates the chromatin association of MCM2–7 in vivo.
Mcm2–Mcm5 linkage affects establishment of pre-RC complexes but not their maintenance
Helicase loading is a multistep reaction involving the formation of an initial OCCM complex followed by ATP hydrolysis and Cdt1 release, which in turn generates an OCM complex that is competent to recruit a second MCM2–7 hexamer for MCM2–7 double hexamer formation. It is not entirely clear how the rapamycin-dependent connection of Mcm2 and Mcm5 influences this assembly pathway. One possibility is that rapamycin affects the establishment of one specific pre-RC intermediate. Alternatively, it might alter complex stability, indicating a defect in complex maintenance. We first addressed the question of establishment and asked whether MCM2–7–M2/M5 supports OCCM formation in the absence and presence of rapamycin. To arrest pre-RC formation at the OCCM stage prior to Cdt1 release, we used ATPγS, an ATP analog that can only very slowly be hydrolyzed. We observed that rapamycin led to a slightly reduced association of Cdt1/MCM2–7–M2/M5 with ORC/Cdc6, when compared with the reaction in the absence of rapamycin (Fig. 5A, cf. lanes 12 and 15). Importantly, wild-type MCM2–7 was not affected by rapamycin (Fig. 5A, lanes 6,9), indicating that the connection between Mcm2 and Mcm5 affects, to some extent, the establishment of the OCCM complex. On the other hand, in the presence of ATP, hydrolysis of ATP occurs rapidly, generating an OCM complex. Rapamycin impeded stable OCM formation and resulted in Cdt1/MCM2–7–M2/M5 release (Fig. 5A, lanes 13,16), implying that a stable ORC/Cdc6–MCM2–7–M2/M5 interface was not established upon ATP hydrolysis. Importantly, this effect was not seen with wild-type MCM2–7 and rapamycin (Fig. 5A, lanes 7,10).
Figure 5.
A rapamycin-induced linkage between Mcm2 and Mcm5 affects pre-RC establishment but not maintenance. (A) Pre-RC reactions using wild-type (wt) MCM2–7 or MCM2–7–M2/M5 were assembled in the presence or absence of rapamycin and with either ATPγS or ATP, washed with low salt (L) or high salt (H), and analyzed by silver staining. (B) Pre-RC reactions using MCM2–7 or MCM2–7–M2/M5 were assembled in the absence of rapamycin and with ATPγS, incubated for 15 min, washed with low salt, incubated for the indicated times in the presence or absence of rapamycin, washed with low salt, and analyzed by silver staining. (C) Pre-RC reactions using wild-type MCM2–7 or MCM2–7–M2/M5 were assembled with ATP in the absence of rapamycin, incubated for 3 min, washed with low salt, incubated for the indicated time in the presence or absence of rapamycin, washed with low salt, and analyzed by silver staining. (D) Pre-RC reactions using wild-type MCM2–7 or MCM2–7–M2/M5 were assembled with ATP in the absence of rapamycin, incubated for 15 min, washed with high salt, incubated for the indicated time in the presence or absence of rapamycin, washed with low salt, and analyzed by silver staining.
Consistent with these findings, we did not observe high-salt stable MCM2–7 double hexamer formation in the presence of rapamycin and MCM2–7–M2/M5, while MCM2–7 supported high-salt stable complex formation in the presence of rapamycin (Fig. 5A, lanes 8,11,14,17). To address whether rapamycin also affects maintenance of the OCCM, OCM, or MCM2–7 double hexamer, we assembled the complexes in the absence of rapamycin and then added rapamycin afterward to assess their stability. In the case of the ATPγS-arrested OCCM complex, we observed that rapamycin had no influence on complex maintenance over a time frame of 20 min (Fig. 5B, cf. lanes 6–10 and 11–15). Importantly, even the OCM complex was stable in the presence of rapamycin (Fig. 5C, cf. lanes 6–10 and 11–15), and the MCM2–7–M2/M5 double hexamer was equally stable in the presence or absence of rapamycin over 20 min (Fig. 5D). These results show that the rapamycin-induced linkage affects only establishment of pre-RC complexes but does not affect complex maintenance. This is reminiscent of the CDK-dependent inhibition of pre-RC formation, which also blocks OCM establishment (Fernández-Cid et al. 2013; Frigola et al. 2013), suggesting that successful DNA insertion at the Mcm2/5 interface could be surveyed by a quality control mechanism.
Helicase loading onto DNA precedes ORC/Cdc6 ATP hydrolysis
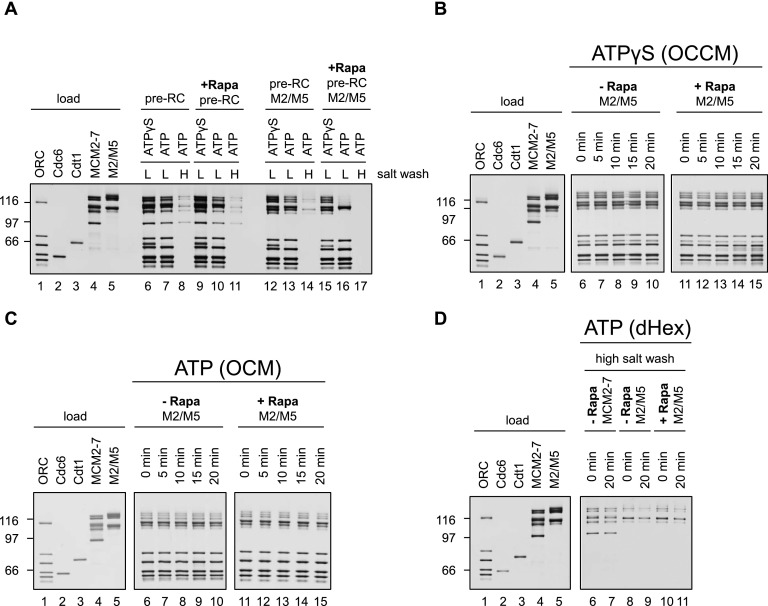
Currently, it is not clear at which stage of pre-RC formation DNA insertion occurs. The high-salt stability of the MCM2–7 double hexamer suggests that helicase loading and MCM2–7 double hexamer formation are coordinated (Evrin et al. 2009; Remus et al. 2009). On the other hand, recent EM data suggested that DNA loading could occur during OCCM formation (Sun et al. 2013). We wanted to resolve this question by investigating whether helicase loading prior to ATP hydrolysis can be observed at the stage of OCCM formation. To distinguish between associated and loaded MCM2–7, we used a differential salt wash with 300 or 500 mM NaCl in an attempt to remove ORC, Cdc6, and Cdt1 and probe for the presence of loaded MCM2–7.
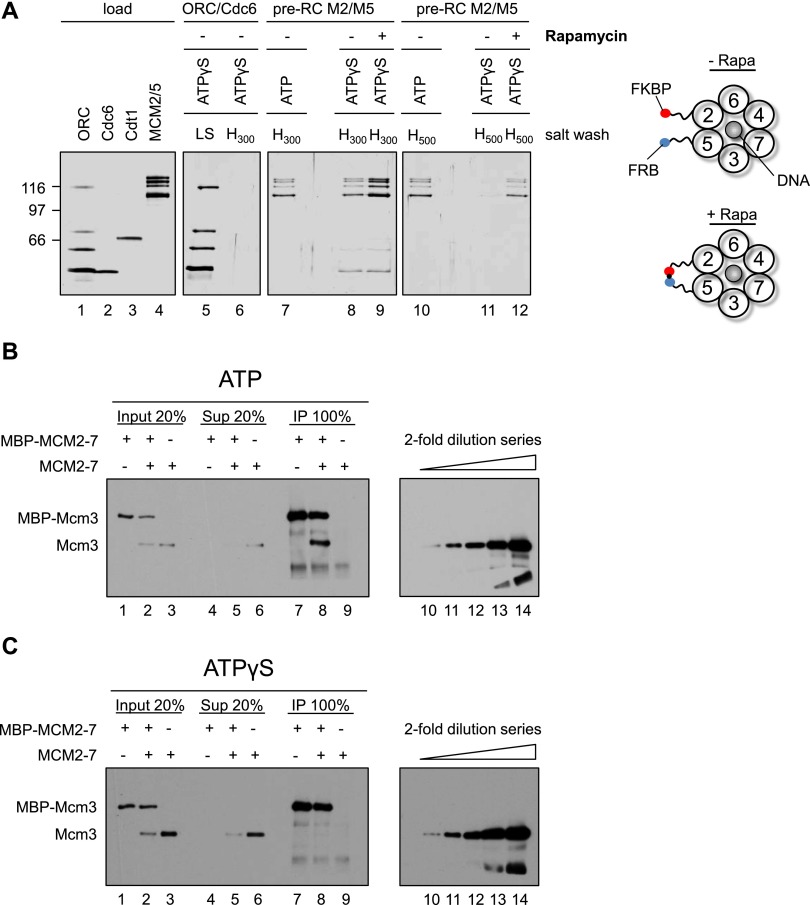
We reasoned that the Mcm2–Mcm5 interface could be particularly salt-sensitive (Bochman and Schwacha 2008). Thus, we assembled OCCM complexes with ATPγS and MCM2–7–M2/M5 and subsequently incubated the reactions in the absence or presence of rapamycin to identify whether rapamycin stabilized the Mcm2–Mcm5 interface. Next, we washed the complexes with either 300 mM NaCl (Fig. 6A, lanes 8,9) or 500 mM NaCl (Fig. 6A, lanes 11,12). As controls, we formed MCM2–7–M2/M5 complexes in ATP, a condition that allows double hexamer formation, and tested the salt stability with 300 or 500 mM NaCl salt washes (Fig. 6A, lanes 7,10). In addition, we assembled an ORC/Cdc6 complex in ATPγS, which was washed with 300 mM NaCl (Fig. 6A, lanes 5,6) to test the stability of the ORC/Cdc6/DNA complex. The 300 mM salt wash of the OCCM complex revealed that some MCM2–7–M2/M5 remained associated with the DNA, while even more MCM2–7–M2/M5 was observed in rapamycin-treated samples (Fig. 6A, lanes 8,9). However, the salt wash of the ORC/Cdc6 complex showed that 300 mM NaCl is sufficient to remove all ORC/Cdc6 from DNA (Fig. 6A, lanes 5,6), but in the case of the OCCM, some ORC/Cdc6 remained associated with DNA after the 300 mM salt wash (Fig. 6A, cf. lane 6 and 8,9). This suggests that an interaction between MCM2–7 and ORC/Cdc6 stabilizes the complex. Indeed, in the case of the OCCM complex (+rapamycin), we observed more salt-resistant MCM2–7–M2/M5 (Fig. 6A, lane 9) when compared with the double hexamer (Fig. 6A, lane 7). This suggests that the OCCM-to-MCM2–7 double hexamer transition limits pre-RC formation due to either quality control mechanisms or reaction time (Fernández-Cid et al. 2013; Frigola et al. 2013).
Figure 6.
Helicase loading occurs at the stage of OCCM formation. (A) Helicase loading occurs in the presence of ATPγS. Reactions using ORC/Cdc6 only (lanes 5,6) or MCM2–7–M2/M5 in the context of the pre-RC (lanes 7–12) were assembled without rapamycin using either ATPγS or ATP, incubated for 12 min, incubated 3 min more in the presence or absence of rapamycin, washed with high salt, ([H300− 300 mM NaCl] or [H500− 500 mM NaCl]), and analyzed by silver staining. This experiment shows that a rapamycin-dependent linkage at the Mcm2–Mcm5 gate hinders MCM2–7 release. (B) In the presence of ATP, MCM2–7 double hexamer formation is observed. Pre-RC assays were assembled in the presence of ATP and washed with low-salt buffer. When tagged and untagged MCM2–7 was used, equimolar amounts of each complex were combined in pre-RC reactions. Complexes were released from DNA via DNase I digestion, immunoprecipitated with anti-MBP beads (IP), and analyzed by Western blotting together with input and supernatant (Sup) using anti-Mcm3 antibodies. (C) In the presence of ATPγS, no MCM2–7 dimerization was observed. The experiment was performed as described in B, but with ATPγS. (B,C) Anti-Mcm3 Western blots (lanes 10–14) containing a twofold dilution series of MCM2–7 input for the respective experiments are shown (lane 14, 100% input; lane 13, 50% input; lane 12, 25% input; lane 11, 12.5% input, and lane 10, 6.75% input).
When the OCCM was washed with 500 mM salt, we detected very little MCM2–7–M2/M5 and no ORC, but rapamycin stabilized MCM2–7–M2/M5 significantly (Fig. 6A, lane 12). Interestingly, the level of loaded MCM2–7 within the rapamycin-treated OCCM was similar to the amount of high-salt-resistant MCM2–7 double hexamer (Fig. 6A, cf. lanes 10 and 12). However, when the same experiments were performed with wild-type MCM2–7, no rapamycin-dependent stabilization was observed with either 300 mM or 500 mM salt washes (Supplemental Fig. S4).
In summary, we were able to detect loaded MCM2–7 at the stage of OCCM formation. To rule out the possibility that ATPγS was hydrolyzed during the reaction, which would produce a high-salt-resistant MCM2–7 double hexamer, we analyzed the MCM2–7 dimerization state. We assembled pre-RC reactions with mixed populations of maltose-binding protein (MBP)-tagged and untagged MCM2–7 using ATP or ATPγS and then released the resulting complexes from the DNA and performed MBP immunoprecipitations. In the case of ATP, we observed co-IP of untagged MCM2–7 (Fig. 6B, lane 8), indicating the formation of the MCM2–7 double hexamer. However, in the case of ATPγS, we did not detect untagged MCM2–7 (Fig. 6C, lane 8), highlighting that no ATP hydrolysis-driven MCM2–7 double hexamer formation had occurred.
Thus, our data show that MCM2–7 loading has already taken place at the stage of OCCM formation, prior to ORC/Cdc6 ATP hydrolysis. The finding that, in the absence of rapamycin, only very small amounts of loaded MCM2–7 were detectable after a 500-mM salt wash indicates that the MCM2–7 hexamer encircling dsDNA is salt-sensitive (Bochman and Schwacha 2008). However, rapamycin-assisted Mcm2 and Mcm5 linking stabilized the complex significantly.
Discussion
MCM2–7 double hexamer assembly is a critical process, and misregulation of this reaction can have fatal consequences for the cell (Blow and Gillespie 2008). Here, we show that MCM2–7 forms a closed circular complex, consistent with the finding that MCM2–7 by itself cannot associate with DNA under physiological conditions (Coleman et al. 1996; Donovan et al. 1997; Evrin et al. 2009; Remus et al. 2009). ORC/Cdc6 and Cdt1 promote helicase loading onto DNA. Using a chemical biology approach, we identified the interface between Mcm2 and Mcm5 as the DNA entry gate during pre-RC formation, and five potential other gates cannot substitute for this. Surprisingly, our study reveals that helicase loading occurs at the stage of OCCM formation prior to ATP hydrolysis, demonstrating that helicase loading and double hexamer formation are separate events. Importantly, failed DNA loading leads to abortive OCM formation and release of MCM2–7 from the ORC/Cdc6/DNA complex. Thus, we identify the helicase loading event itself as an important quality control mechanism of pre-RC formation and DNA replication.
The MCM2–7 proteins form the core of the eukaryotic replicative helicase. In bacteria and archaea, the replicative helicase is arranged as a topologically closed ring (Pape et al. 2003; Itsathitphaisarn et al. 2012). In eukaryotes, several structural models have been observed, including a closed circular hexamer, as presented in this study (Fig. 1); open spiral-shaped hexamers (Costa et al. 2011; Lyubimov et al. 2012); and closed circular double hexamers (Evrin et al. 2009; Remus et al. 2009). MCM2–7, like all helicases, belongs to the AAA+ family of ATPases. In many cases, the structural organization of these hexameric helicases is strongly influenced by ATP binding. For example, archaeal MCM becomes stabilized upon addition of ATP (Sakakibara et al. 2009), while the SV40 large T antigen helicase requires ATP for its oligomerization into hexamers (Wang and Prives 1991), and S. cerevisiae MCM2–7 is stabilized by ATP as well (A Riera and C Speck, unpubl.). In fact, in the absence of ATP or the presence of chloride ions, the S. cerevisiae MCM2–7 ring becomes destabilized at the Mcm2/5 interface (Bochman and Schwacha 2008; Bochman et al. 2008).
In the same way, MCM2–7 from D. melanogaster or Encephalitozoon cuniculi assumes an open ring conformation (with an opening at the Mcm2/Mcm5 interface) when purified in the absence of ATP (Costa et al. 2011; Lyubimov et al. 2012). However, the ATP concentration in vivo is virtually constant, suggesting that an open MCM2–7 ring conformation is rare inside the living cell. Nevertheless, buffer conditions used during EM sample preparation may affect the MCM2–7 ring stability (Bochman and Schwacha 2008; Bochman et al. 2008), which may explain the observation of open MCM2–7 rings in electron micrographs. Another possibility is that Cdt1 interactions with MCM2–7 split open the MCM2–7 ring prior to helicase loading (Coleman et al. 1996; Blow and Gillespie 2008; Bochman and Schwacha 2009; Takara and Bell 2011; Arias-Palomo et al. 2013), although Cdt1 is not sufficient to promote origin binding of MCM2–7 (Evrin et al. 2013; Frigola et al. 2013). We therefore suggest that MCM2–7 is arranged in a closed circular conformation in vivo and that it needs to be actively opened by ORC/Cdc6 and Cdt1 during helicase loading. Opening of the MCM2–7 ring during helicase loading could occur at each of the six subunit interfaces (Fig. 2A), as is the case in bacteria (Arias-Palomo et al. 2013).
Here, we demonstrate that the FKBP–FRB linkage at five different interfaces (Mcm2/6, Mcm6/4, Mcm4/7, Mcm7/3, and Mcm3/5) does not affect MCM2–7 double hexamer formation, while an opening at the Mcm2/5 interface has been proved to be essential for MCM2–7 helicase loading. Since the rapamycin-induced linkage between Mcm2 and Mcm5 is sufficient to block helicase loading, it is clear that none of the five other potential gates can substitute for the Mcm2/5 interface. Thus, our data identified a unique DNA entry gate for the regulated loading of the eukaryotic replicative helicase onto DNA. This finding is consistent with studies that showed that a weak Mcm2/5 interface serves as an ATP-dependent discontinuity (“gate”) within the ring structure (Bochman and Schwacha 2007, 2010; Costa et al. 2011).
The mechanism of bacterial helicase ring opening has been recently described. It was found that DnaC, the helicase loader, adopts a spiral shape, interacts tightly with DnaB, and breaks the hexameric ring open. The cryo-EM structure of the S. cerevisiae OCCM complex has identified a spiral shape in ORC/Cdc6, but it is not yet clear whether this is the main mechanism that functions for MCM2–7 ring opening. It is interesting to note that ORC/Cdc6 and Cdt1 open the MCM2–7 ring at an interface that is inherently unstable (Bochman and Schwacha 2008; Bochman et al. 2008). This indicates that helicase opening at the Mcm2/5 interface is very specific—the other interfaces have evolved stronger protein interactions that cannot be broken (Davey et al. 2003)—and this result also suggests that even relatively weak forces could promote MCM2–7 ring opening at the Mcm2/5 interface during pre-RC formation. It is unknown whether the same gate functions during S phase, but the functional analysis is currently ongoing.
How helicase loading and MCM2–7 double hexamer assembly are coordinated has been under intense scrutiny for the past 10 years. Early models suggested that helicase loading occurs in an ATP hydrolysis-dependent fashion (Bowers et al. 2004; Randell et al. 2006; Costa et al. 2011). Later, after the discovery of the MCM2–7 double hexamer (Evrin et al. 2009; Remus et al. 2009; Gambus et al. 2011), it was suggested that MCM2–7 double hexamer formation and DNA loading are coordinated (Remus et al. 2009). Now, our data demonstrate that helicase loading occurs at the stage of OCCM formation prior to MCM2–7 double hexamer formation and ATP hydrolysis. This is analogous to the loading of the PCNA trimeric ring by replication factor C (RFC), where RFC loads the PCNA ring onto DNA in the absence of ATP hydrolysis, and only PCNA release from RFC requires ATP hydrolysis by RFC (Gomes and Burgers 2001; Johnson et al. 2006). Thus, our data show that ORC/Cdc6 and RFC act in a similar manner. Furthermore, our data are consistent with a recent EM study that proposed that DNA loading occurs at the stage of OCCM formation (Sun et al. 2013). Indeed, the OCCM structure showed that the MCM2–7 ring adopts a closed conformation, indicating, in the context of the data presented here, that loading has been completed. Nevertheless, we observed that the Mcm2/5 interface limits the salt stability of the OCCM complex (Fig. 6A), suggesting that protein interactions within the double hexamer seal the Mcm2/5 interface, resulting in higher salt stability.
Our results show that ATPase activity is not required for helicase loading; thus, the question remains: What is the role of ATP hydrolysis during pre-RC formation? We suggest that ATP hydrolysis alters the structure of MCM2–7 to promote MCM2–7 double hexamer formation. Indeed, the OCCM cannot recruit a second MCM2–7 hexamer, but upon ATP hydrolysis, the OCM is competent for MCM2–7 dimerization (Evrin et al. 2013, 2014; Fernández-Cid et al. 2013). It is clear that the recruitment of the second MCM2–7 hexamer requires Cdt1 (Fernández-Cid et al. 2013). However, whether the second MCM2–7 hexamer also requires ORC/Cdc6 for loading or becomes loaded in an entirely different way is still unknown (Riera et al. 2014; Yardimci and Walter 2014).
Moreover, DNA insertion may function for quality control. It has been shown that ORC phosphorylated by CDK or Orc1–5, missing Orc6, supports OCCM formation but then fails to establish a stable OCM complex upon ATP hydrolysis (Fernández-Cid et al. 2013; Frigola et al. 2013). It is interesting that rapamycin-induced linkage of Mcm2 and Mcm5 also affects the OCCM-to-OCM transition (Figs. 5A, 7A). We suggest that failed DNA insertion during MCM2–7 loading triggers ATP hydrolysis-dependent disassembly of the OCCM complex, which could be an important contributor to quality control during pre-RC formation. One could speculate that similar opening and quality control reactions also exist during helicase activation, when MCM2–7 ring opening occurs to extrude one strand of DNA for CMG assembly.
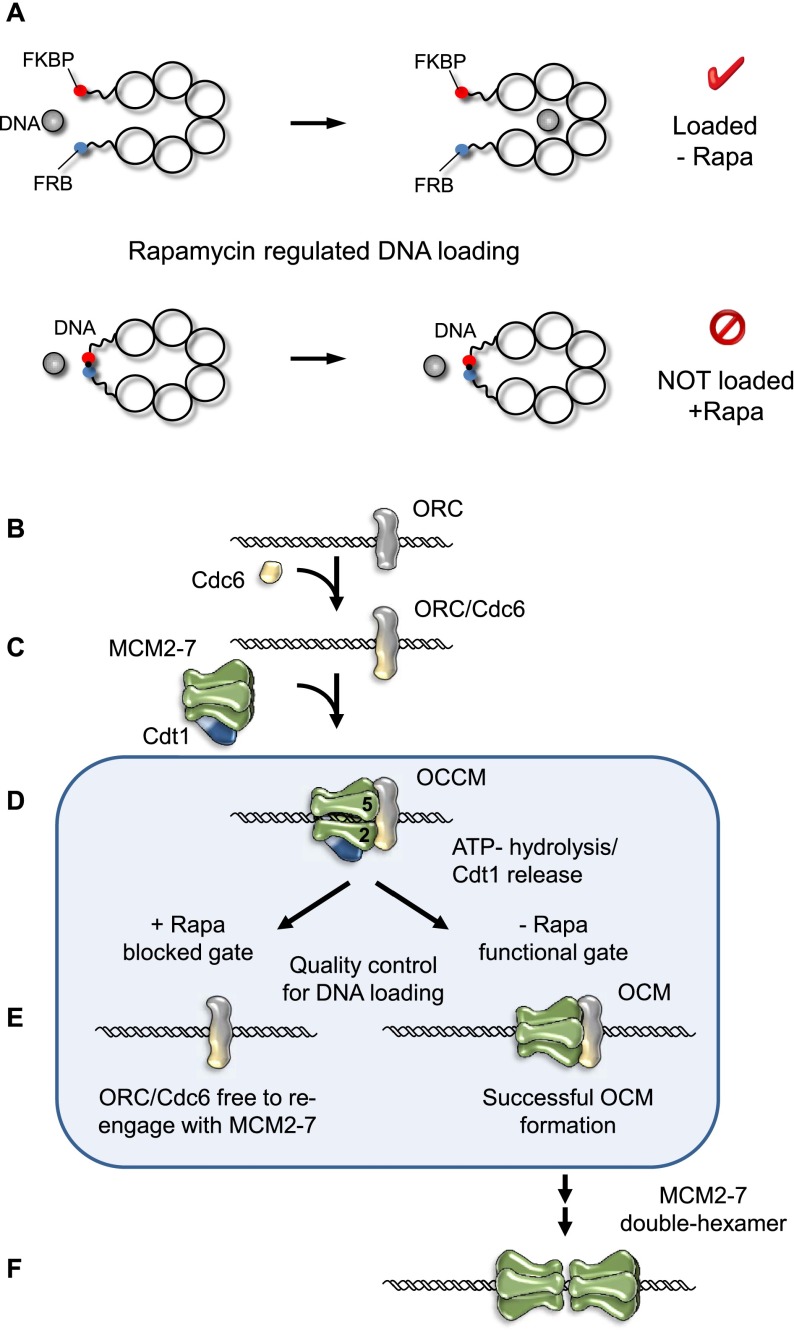
Figure 7.
A model illustrating regulated helicase loading during pre-RC assembly. (A) Illustration showing the rapamycin-induced linkage between neighboring Mcm subunits and the effect of this linkage on helicase loading during pre-RC assembly. (B–E) Proposed model of pre-RC formation. ORC recognizes the replication origin (B) and recruits Cdc6 to form the ORC/Cdc6 complex (C). (D) Cdt1/MCM2–7 binding to ORC/Cdc6 promotes helicase loading via the Mcm2/5 entry gate and OCCM complex formation. (E) ATP hydrolysis triggers Cdt1 release. If productive helicase loading occurs, the stable OCM can form. If helicase loading fails due to a rapamycin-induced linkage between Mcm2 and Mcm5, then MCM2–7–M2/M5 is released. This in turn makes ORC/Cdc6 available to re-engage with the next Cdt1/MCM2–7 complex. Failed helicase loading could represent an important quality control mechanism for pre-RC formation. (F) Successful OCM formation allows the recruitment of a second MCM2–7 hexamer to promote MCM2–7 double hexamer formation.
A model explaining how pre-RC formation is coordinated with helicase loading at a unique DNA entry gate
In this model (Fig. 7B–F), we explain how helicase loading is coordinated with MCM2–7 double hexamer formation. Pre-RC formation is initiated with the binding of ORC to the replication origin (Fig. 7B). In late M phase, Cdc6 joins ORC to form an ORC/Cdc6 complex (Fig. 7C; Speck et al. 2005; Speck and Stillman 2007; Sun et al. 2012). Under physiological conditions, MCM2–7 forms closed circular rings (Fig. 1). ORC/Cdc6 is competent to recruit a Cdt1/MCM2–7 heptamer (Fig. 7D; Takara and Bell 2011; Evrin et al. 2013; Sun et al. 2013). Our data demonstrate that a linkage between Mcm2 and Mcm5 does not prevent OCCM formation but blocks the ATP hydrolysis-dependent establishment of a stable OCM complex, which leads to disassembly of the complex and in turn makes ORC/Cdc6 available again to re-engage Cdt1/MCM2–7 (Fig. 7E). We suggest that the linkage interferes with DNA loading and that ATP hydrolysis-dependent MCM2–7 release serves as a quality control step during pre-RC formation. On the other hand, successful helicase loading onto DNA during OCCM formation, as we observed here, allows Cdt1 release and establishment of a stable OCM complex. The OCM is competent to recruit a second MCM2–7 hexamer, which functions for double hexamer formation (Fig. 7F). We propose that within the MCM2–7 double hexamer, a weak Mcm2/5 interface becomes stabilized, which could explain the salt stability of the complex.
Material and methods
Cloning of Mcm–FKBP/FRB
The FKBP/FRB insertion sites were chosen based on a structural model of the archaeal M. thermoautotrophicum MCM protein (Fletcher et al. 2003), the secondary structure and sequence alignments of the MtMCM sequence, and those of Mcm2 to Mcm7 from S. cerevisiae. The construction of the FKBP/FRB constructs was based on the plasmids pESC-Mcm2–Mcm7 (pCS14), pESC-Mcm6–Mcm7 (pCS15), and pESC-HA-Mcm3–Mcm5 (pCS232) (Evrin et al. 2009). Initially, restriction sites (AsiSI and AscI) were introduced at the sites of insertion using site-directed mutagenesis (Agilent). To maintain the reading frame, an additional nucleotide was inserted after the 8-base-pair (bp) restriction site (AsiSI: a; AscI: c). The FRB domain, including linkers, was fused into Mcm2, Mcm3, and Mcm4 via AsiSI restriction sites (AIAGANTCTPRGSGMLPSGMASR-FRB-TSYPYDVPDYAGANDGAAIA). The FKBP domain, including linkers, was fused into Mcm5, Mcm6, and Mcm7 via AscI (SSYPYDVPDYASLGGPSSPKKKRKVSR-FKBP-TSYISFLNSDLINSRTQRVDGQIGAP). The FKBP and FRB genes, including linkers, were codon-optimized for expression in S. cerevisiae (Genscript).
Cloning of Flag-Mcm3
We replaced the HA tag of Mcm3 in pCS232 with a Flag tag using a NotI digest and two complementary oligonucleotides that contain the NotI restriction site and encode the Flag tag sequence generating pCS502.
Generation of the Mcm2–FRB and Mcm5–FKBP strains
Integration of the Mcm2–FRB and Mcm5–FKBP genes into S. cerevisiae (YC475) was performed as previously described (Longtine et al. 1998). Transformed yeast clones were analyzed by PCR for the successful genomic integration of the FRB/FKBP insert. The strain used for the time course analysis was transformed with pMet-Cdc20-TRP (pCS604) followed by selection on tryptophan- and methionine-deficient medium as described (Uhlmann et al. 2000).
Expression and purification of proteins
ORC was expressed by using baculovirus-infected cells and purified as described (Klemm et al. 1997). Cdc6 and Cdt1 were expressed in bacteria and purified as described (Speck et al. 2005; Evrin et al. 2009). MCM2–7, FKBP/FRB–MCM2–7, and MBP-MCM2–7 were expressed in S. cerevisiae and purified as described (Evrin et al. 2009; Fernández-Cid et al. 2013).
In vitro pre-RC assembly assay
The pre-RC was assembled in a one-step reaction: 40 nM ORC, 80 nM Cdc6, 40 nM Cdt1, and 40 nM Mcm2–7 in buffer A (50 mM HEPES-KOH at pH 7.5, 100 mM KGlu, 10 mM MgAc, 50 mM ZnAc, 3 mM ATP or ATPγS, 5 mM DTT, 0.1% Triton X-100, 5% glycerol) were added to 6 nM pUC19-ARS1 plasmid beads (Evrin et al. 2009) for 15 min at 24°C or 3 min for OCM formation. The beads were washed three times with buffer A plus 1 mM EDTA or buffer B (50 mM HEPES-KOH at pH 7.5, 1 mM EDTA, 300 mM NaCl or “high salt” 500 mM NaCl, 5% glycerol, 0.1% Triton X-100, 5 mM DTT) before digestion with 1 U of DNase I in buffer A plus 5 mM CaCl2 for 6 min at 24°C. The samples were separated by PAGE and analyzed by silver staining. All samples marked with “high salt” were washed with 500 mM NaCl. For assays performed in the presence of rapamycin, rapamycin was added to all buffers at a final concentration of 100 nM.
HA-MCM2–7 pull-down reactions
One microgram of HA-MCM2–7 was immobilized on IgG control beads (anti-MBP [New England Biolabs] or anti-HA [Sigma]) on protein G beads [GE)] for 15 min at 24°C with mixing in 200 µL of buffer MB (50 mM Hepes-KOH at pH 7.5, 300 mM KGlu, 10 mM MgAc, 50 μM ZnAc, 3 mM ATP, 5 mM DTT, 0.1% Triton X-100, 10% glycerol, 0.1% BSA) followed by three washes with 200 μL of buffer MB. Purified Cdt1 (3.65 pmoles) were incubated with IgG control or HA-MCM2–7 beads for 10 min with mixing at 24°C in 200 µL of buffer MB. Afterward, the beads were washed three times with 200 µL of buffer MB. The bound proteins were analyzed by SDS-PAGE and Western blotting with anti-Cdt1 N-terminal (CS1411) antibody (Fernández-Cid et al. 2013).
MBP Immunoprecipitation assay
One standard size pre-RC reaction was prepared as described above (pre-RC assay) using MBP-tagged protein (20 nM), MBP-tagged and untagged protein (20 nM each), or only untagged protein (20 nM). After 4 min of incubation, the beads were washed twice with buffer A before DNA digestion with 0.03 U of DNase I in buffer A plus 5 mM CaCl2 for 7 min at 24°C. Reaction products were immunoprecipitated with anti-MBP (New England Biolabs) antibody coupled to protein G beads (Invitrogen) for 4 min at 24°C, washed three times with buffer A, and analyzed by Western blot.
Acknowledgments
We thank Bruce Stillman for Mcm2, Mcm3, Cdt1 (CS1411), and Orc3 (SB3) antibodies; Stefan Gruber and Kim Nasmyth for the rapamycin-resistant yeast strain; Luis Aragon for the Pmet-Cdc20-TRP plasmid; and the Medical Research Council for funding. S.A.S. was supported by a Fellowship from the German Research Foundation (DFG; grant SA 2181/1).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.242404.114.
References
- Arias-Palomo E, O’Shea VL, Hood IV, Berger JM. 2013. The bacterial DnaC helicase loader is a DnaB ring breaker. Cell 153: 438–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Stillman B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ. 2008. Replication licensing and cancer—a fatal entanglement? Nat Rev Cancer 8: 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. 2007. Differences in the single-stranded DNA binding activities of MCM2–7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J Biol Chem 282: 33795–33804 [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. 2008. The Mcm2–7 complex has in vitro helicase activity. Mol Cell 31: 287–293 [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. 2009. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 73: 652–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. 2010. The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2–7 ‘gate.’ Nucleic Acids Res 38: 6078–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Bell SP, Schwacha A. 2008. Subunit organization of Mcm2–7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol 28: 5865–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP. 2004. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol Cell 16: 967–978 [DOI] [PubMed] [Google Scholar]
- Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, Klein MG, Chen XS. 2008. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci 105: 20191–20196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. 1996. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell 87: 53–63 [DOI] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. 2011. The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18: 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, Indiani C, O’Donnell M. 2003. Reconstitution of the Mcm2–7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem 278: 4491–4499 [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. 1997. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci 94: 5611–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. 2009. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci 106: 20240–20245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Fernandez-Cid A, Zech J, Herrera MC, Riera A, Clarke P, Brill S, Lurz R, Speck C. 2013. In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2–7 hexamer dimerization. Nucleic Acids Res 41: 3162–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Fernandez-Cid A, Riera A, Zech J, Clarke P, Herrera MC, Tognetti S, Lurz R, Speck C. 2014. The ORC/Cdc6/MCM2–7 complex facilitates MCM2–7 dimerization during prereplicative complex formation. Nucleic Acids Res 42: 2257–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Cid A, Riera A, Tognetti S, Herrera MC, Samel S, Evrin C, Winkler C, Gardenal E, Uhle S, Speck C. 2013. An ORC/Cdc6/MCM2–7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell 50: 577–588 [DOI] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. 2003. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol 10: 160–167 [DOI] [PubMed] [Google Scholar]
- Frigola J, Remus D, Mehanna A, Diffley JF. 2013. ATPase-dependent quality control of DNA replication origin licensing. Nature 495: 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Gambus A, Khoudoli GA, Jones RC, Blow JJ. 2011. MCM2–7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem 286: 11855–11864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes XV, Burgers PM. 2001. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J Biol Chem 276: 34768–34775 [DOI] [PubMed] [Google Scholar]
- Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. 2006. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell 127: 523–537 [DOI] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. 2011. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang ML, Leon RP, Pessoa-Brandao L, Hunt S, Raghuraman MK, Fangman WL, Brewer BJ, Sclafani RA. 2007. Structural changes in Mcm5 protein bypass Cdc7–Dbf4 function and reduce replication origin efficiency in Saccharomyces cerevisiae. Mol Cell Biol 27: 7594–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. 2010. Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol Cell 37: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. 2012. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Yao NY, Bowman GD, Kuriyan J, O’Donnell M. 2006. The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem 281: 35531–35543 [DOI] [PubMed] [Google Scholar]
- Klemm RD, Austin RJ, Bell SP. 1997. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell 88: 493–502 [DOI] [PubMed] [Google Scholar]
- Leon RP, Tecklenburg M, Sclafani RA. 2008. Functional conservation of β-hairpin DNA binding domains in the Mcm protein of Methanobacterium thermoautotrophicum and the Mcm5 protein of Saccharomyces cerevisiae. Genetics 179: 1757–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lyubimov AY, Costa A, Bleichert F, Botchan MR, Berger JM. 2012. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc Natl Acad Sci 109: 11999–12004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell M, Langston L, Stillman B. 2013. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol 5: a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Meka H, Chen S, Vicentini G, van Heel M, Onesti S. 2003. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep 4: 1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP. 2006. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol Cell 21: 29–39 [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. 2009. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera A, Tognetti S, Speck C. 2014. Helicase loading: how to build a MCM2–7 double-hexamer. Semin Cell Dev Biol 30: 104–109 [DOI] [PubMed] [Google Scholar]
- Sakakibara N, Schwarz FP, Kelman Z. 2009. ATP hydrolysis and DNA binding confer thermostability on the MCM helicase. Biochemistry 48: 2330–2339 [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. 2010. The Dbf4–Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Stillman B. 2007. Cdc6 ATPase activity regulates ORC × Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem 282: 11705–11714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Chen Z, Li H, Stillman B. 2005. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol 12: 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kawakami H, Zech J, Speck C, Stillman B, Li H. 2012. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure 20: 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Evrin C, Samel SA, Fernandez-Cid A, Riera A, Kawakami H, Stillman B, Speck C, Li H. 2013. Cryo-EM structure of a helicase loading intermediate containing ORC–Cdc6–Cdt1–MCM2–7 bound to DNA. Nat Struct Mol Biol 20: 944–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takara TJ, Bell SP. 2011. Multiple Cdt1 molecules act at each origin to load replication-competent Mcm2–7 helicases. EMBO J 30: 4885–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H. 2010. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma 119: 565–574 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tsakraklides V, Bell SP. 2010. Dynamics of pre-replicative complex assembly. J Biol Chem 285: 9437–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Wang EH, Prives C. 1991. ATP induces the assembly of polyoma large tumor antigen into hexamers. Virology 184: 399–403 [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B. 1999. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc Natl Acad Sci 96: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Walter JC. 2014. Prereplication-complex formation: a molecular double take? Nat Struct Mol Biol 21: 20–25 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]