Abstract
Chlorhexidine is one of the most widely used biocides in health and agricultural settings as well as in the modern food industry. It is a cationic biocide of the biguanide class. Details of its mechanism of action are largely unknown. The frequent use of chlorhexidine has been questioned recently, amidst concerns that an overuse of this compound may select for bacteria displaying an altered susceptibility to antimicrobials, including clinically important anti-bacterial agents. We generated a Salmonella enterica serovar Typhimurium isolate (ST24CHX) that exhibited a high-level tolerant phenotype to chlorhexidine, following several rounds of in vitro selection, using sub-lethal concentrations of the biocide. This mutant showed altered suceptibility to a panel of clinically important antimicrobial compounds. Here we describe a genomic, transcriptomic, proteomic, and phenotypic analysis of the chlorhexidine tolerant S. Typhimurium compared with its isogenic sensitive progenitor. Results from this study describe a chlorhexidine defense network that functions in both the reference chlorhexidine sensitive isolate and the tolerant mutant. The defense network involved multiple cell targets including those associated with the synthesis and modification of the cell wall, the SOS response, virulence, and a shift in cellular metabolism toward anoxic pathways, some of which were regulated by CreB and Fur. In addition, results indicated that chlorhexidine tolerance was associated with more extensive modifications of the same cellular processes involved in this proposed network, as well as a divergent defense response involving the up-regulation of additional targets such as the flagellar apparatus and an altered cellular phosphate metabolism. These data show that sub-lethal concentrations of chlorhexidine induce distinct changes in exposed Salmonella, and our findings provide insights into the mechanisms of action and tolerance to this biocidal agent.
Keywords: Salmonella, biocide tolerance, chlorhexidine, proteomics, transcriptomics, whole genome sequencing, SNP typing
Introduction
Chlorhexidine (1,6 bis(4′-chlorophenylbiguanide)hexane) is a cationic biocide of the biguanide class. It has a broad spectrum of action and is effective against Gram-negative and Gram-positive bacteria as well as yeasts, protozoa and some lipophilic viruses (Jones, 2000). This biocide is used as a disinfectant, an antiseptic and a preservative. The application of chlorhexidine in both medical and agricultural settings, along with its use as an active component in domestic cleaning agents, is increasing (Nde et al., 2009).
Chlorhexidine has been in use for over 50 years (Block and Furman, 2002; Gilbert and Moore, 2005). Despite its broad use, precise details of its mode of action are lacking (Jones, 2000; Galice et al., 2006). Nonetheless the general mechanism of action for chlorhexidine, like other cationic compounds, involves the bacterial cell membrane (Gilbert and Moore, 2005). In Gram-negative bacteria, chlorhexidine can bind to divalent cations such as Ca2+ and Mg2+ normally associated with lipopolysaccharide, and to negatively charged proteins and acidic phospholipids (Jones, 2000). Chlorhexidine is composed of a six carbon chain, a structure that is smaller compared to other cationic compounds. In addition it contains an inflexible hydrophobic region. Its size and molecular inflexibility are thought to limit its ability to fold sufficiently to allow its insertion into the membrane (Gilbert and Moore, 2005). It has been hypothesized that chlorhexidine can form a molecular bridge between adjacent phospholipid head groups thereby displacing any divalent cations. Consequently, this binding event can alter membrane fluidity and the activity of membrane-bound enzymes and osmoregulation mechanisms (Gilbert and Moore, 2005). Chlorhexidine treatment has been associated with the loss of membrane integrity, causing widespread membrane damage, and at high concentrations, the formation of large phosphate containing moieties, which may lead to the precipitation of cytoplasmic content, including the coagulation and precipitation of nucleic acids, proteins, and the leakage of cellular materials (Jones, 2000; Hope and Wilson, 2004; Cheung et al., 2012). It is thought that chlorhexidine may also inhibit oxygen utilization (Barrett-Bee et al., 1994) and energy production in bacteria (Nde et al., 2009). Other effects caused by this biocide include DNA damage (Yeung et al., 2007; Nde et al., 2009) and the inhibition of protein synthesis (Nde et al., 2009).
Like other biocides, the wide-spread use of chlorhexidine has been questioned recently, due to the possibility that the overuse of such agents may select for mutants displaying a reduced susceptibility to it and other related and non-related antimicrobials, including clinically important agents (Gilbert and Moore, 2005). Bacteria demonstrating a reduced susceptibility to chlorhexidine have been documented (Thomas and Stickler, 1979; Kropinski et al., 1982). Chlorhexidine tolerance has been selected in vitro in a number of different bacterial species including Salmonella (Braoudaki and Hilton, 2004; Condell et al., 2012a), Escherichia coli (Braoudaki and Hilton, 2004) and Staphylococcus aureus (Kaatz et al., 2005; Huet et al., 2008). Chlorhexidine tolerance has been reported to result from modification/(s) in cell wall structure and/or composition (Russell and Furr, 1986; Russell and Path, 1986; Tattawasart et al., 2000; Denyer and Maillard, 2002), and has also been associated with an over-expression of efflux pumps (Levy, 2000; Poole, 2004; Huet et al., 2008). In some instances the tolerant phenotype that develops following chlorhexidine exposure is associated with a reduced susceptibility to clinically important antimicrobial agents (Braoudaki and Hilton, 2004; Condell et al., 2012a).
Salmonella is an important food-borne pathogen and the causative agent of typhoid fever and gastroenteritis in humans (Coburn et al., 2007). It can survive in non-host environments, including a range of food matrices (Gast and Holt, 1998; Shi et al., 2007) and the food processing environment itself (Nesse et al., 2003; Vestby et al., 2009). The elimination of Salmonella through effective control measures, including adequate hygienic cleaning and sanitation, is essential to reduce its transmission and impact on public health.
Following several rounds of in vitro selection using sub-lethal concentrations of chlorhexidine, a S. Typhimurium strain was recovered, which had developed a high-level chlorhexidine tolerance phenotype (ST24CHX); a >50-fold increase in the minimum inhibitory concentration (MIC) (Condell et al., 2012a). In this study, we compared the susceptible Salmonella Typhimurium (ST24WT) with ST24CHX its isogenic chlorhexidine tolerant counterpart using a variety of molecular techniques, facilitating a study of the classical biochemical pathway. The alterations identified based on these approaches were subsequently validated. Using these techniques, several cellular signals involved in the bacterial response to chlorhexidine exposure were identified. Our findings provide new insights into the mechanisms of action of this biocidal agent.
Materials and methods
Bacterial culture, in vitro mutant selection, DNA, RNA, and protein preparation
All Salmonella isolates were stored on beads in cryopreservation fluid at −80°C (Technical Service Consultants Ltd, Lancashire, England). Isolates were streaked onto Mueller-Hinton agar (Oxoid, Cambridge, UK). A single colony was picked and subsequently used to inoculate Mueller-Hinton (MH) broth (Oxoid, Cambridge, UK), then incubated for 16–18 h at 37°C shaking at 250 rpm. The culture was used to inoculate fresh aliquots of MH broth at a dilution of 1:100. Cultures were harvested at the mid-logarithmic growth phase (OD610 nm = 0.6) for the assays described below.
Based on earlier work, a Salmonella enterica serovar Typhimurium isolate, denoted as ST24WT, was chosen for further study (Condell et al., 2012a); this isolate demonstrated a reduced susceptibility to three food-grade biocide formulations that were significantly above the mean MIC for a collection of 189 Salmonella isolates. When tested against chlorhexidine, ST24WT displayed an MIC of 1.96 μg/ml. Following several rounds of in vitro selection using sub-lethal concentrations of chlorhexidine (Condell et al., 2012a), a high-level tolerance phenotype was developed with an increase in the MIC to >50 μg/ml being noted. The isogenic, chlorhexidine tolerant, mutant was designated ST24CHX (Condell et al., 2012a). Chlorhexidine is commercially available at concentrations from 0.5 through 4% and in a variety of formulations for healthcare applications (Milstone et al., 2008).
For purification of DNA, a 5 ml aliquot of the 16–18 h culture in MH broth (as outlined above) was removed. For purification of temporal RNA and protein 50 ml of the mid-logarithmic growth phase (outlined above) was divided in two. One-half was untreated whilst the second-half was exposed to chlorhexidine. Both ST24WT and ST24CHX were exposed to 0.5 × MIC of the reference strain ST24WT, equivalent to 1 μg/ml chlorhexidine, and incubated for 30 min at 37°C. Following the incubation period 25 ml volumes from each sample were removed for proteomic analysis (see below). Additionally 2 ml from each culture was removed for RNA purification (see below). All samples were prepared in triplicate on independent occasions.
Genomic DNA isolation and single nucleotide polymorphism (SNP) analysis
Genomic DNA was isolated, for both ST24WT and ST24CHX, using the DNeasy blood and tissue kit (Qiagen, Inc., Valencia, California), following the manufacturers' instructions. The isolated DNA was submitted for commercial whole genome sequencing. Illumina 100 bp paired end sequences for each sample were generated from the HiSeq platform (Source BioScience, Nottingham, UK), with a mean of 3405 Mbases. Only those reads ≥Q30 were used for downstream analysis. MAQ (Li et al., 2008) version 0.4.7 was used to map reads to, and to generate SNPs from, the reference genome Salmonella enterica subsp. enterica serovar Typhimurium str. SL1344 (FQ_312003). In-house perl scripts were used to annotate the resulting SNP and indel files. Locations of SNPs within genes were confirmed using the Salmonella Typhimurium SL1344 transcriptional map provided by Kröger et al. (2012). A summary of the detected SNPs is provided in Table 1.
Table 1.
Single nucleotide polymorphisms (SNPs) that differentiate the reference, chlorhexidine susceptible, Salmonella Typhimurium ST24WT and the isogenic, chlorhexidine tolerant, mutant ST24CHX detected (A) within coding genes and (B) in intergenic regions.
| (A) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Nucleotide* | Gene name | Reference ID | Gene locations | Strand | ST24WT aa* (codon) | ST24CHX aa* (codon) | Gene function | Functional category | ||
| SL1344 | ST24WT | ST24CHX | |||||||||
| NON-SYNONYMOUS | |||||||||||
| 319066 | G | G | C | sciK1 | SL1344_0274 | 318655..319140 | + | V(GTG) | Q(CAG) | SPI-6 associated, Hcp like protein, component of type VI secretory system | Virulence |
| 319067 | T | T | A | sciK1 | SL1344_0274 | 318655..319140 | + | V(GTG) | |||
| 3504469 | T | T | G | lptC | SL1344_3289 | 3503949..3504524 | + | L(CTG) | R(CGG) | Lipopolysaccharide export system protein, forms ABC transporter with LptBFG | Transport and permeability |
| 638182 | C | C | T | ramR | SL1344_0568 | 637634..638215 | − | A(GCA) | T(ACA) | TetR family transcriptional regulator, repressor of ramA | Drug transport/ metabolism |
| 3781244 | G | G | A | pitA | SL1344_3554 | 3779941..3781314 | + | W(TGG) |
 (TAG) (TAG) |
Low-affinity inorganic phosphate transporter, PiT family | Inorganic ion transport and metabolism |
| 2502787 | G | A | G | yfdC | SL1344_2362 | 2502523..2503464 | + | G(GGC) | S(AGC) | Putative formate/nitrate transporter | Anaerobic metabolism, transport and permeability |
| 3542329 | C | T | C | oadA2 | SL1344_3324 | 3541466..3543241 | − | G(GGC) | S(AGC) | Oxaloacetate decarboxylase subunit alpha, involved in pyruvate metabolism, conversion of oxaloacetate to pyruvate and carbon dioxide | Amino acid metabolism, energy production |
| 4182738 | A | A | T | − | SL1344_4465 | 4812650..4815415 | + | Y(TAT) | F(TTT) | NtrC family of transcriptional regulator, involved in activation of nitrogen metabolism related promoters and sigma 54 | Regulation |
| 4812741 | T | C | T | L(CTG) | P(CCG) | ||||||
| 2043305 | A | C | A | − | SL1344_1929 | 2041795..2042394 | V(GTT) | A(GCT) | Bacteriophage SLP203, putative bacteriophage tail protein | Bacteriophage | |
| 2043305 | A | C | A | − | SL1344_1930 | 2042364..2043959 | − | S(TCT) | A(GCT) | Bacteriophage SLP203, putative bacteriophage tail fiber protein | Bacteriophage |
| 2042368 | A | A | G | − | V(GTA) | G(GGA) | |||||
| 2045590 | T | T | C | − | SL1344_1934 | 2045576..2045989 | − | K(AAA) | Q(CAA) | Bacteriophage SLP203, bacteriophage protein | Bacteriophage |
| 2066438 | C | A | C | − | SL1344_1960 | 2065512..2066507 | − | A(GCG) | S(TCG) | Bacteriophage SLP203, bacteriophage protein | Bacteriophage |
| 2766226 | A | A | G | gpP | SL1344_2590 | 2766099..2766791 | − | V(GTT) | A(GCT) | Bacteriophage SLP272, replication protein P | Bacteriophage |
| DEGENERATIVE | |||||||||||
| 334377 | C | S | C | − | SL1344_0286 | 332521..336123 | + | G(GGC) | SPI 6 associated. Rhs family protein- membrane protein of unknown function | Virulence | |
| 3032926 | G | R | G | avrA | SL1344_2845 | 3032477..3033382 | − | L(CTG) | SPI 1 associated. Type III secretion system effector protein- regulator | Virulence | |
| 2509924 | C | S | C | pgtP | SL1344_2367 | 2509846..2511237 | + | R(CGT) | Phosphoglycerate transporter protein, induced by pgtBC | Inorganic ion transport and metabolism | |
| 4404698 | T | K | T | nudC | SL1344_4105 | 4404662..4405435 | + | W(TGG) | Hypothetical NADH pyrophosphatase | Inorganic ion transport and metabolism | |
| 699483 | A | M | A | rlpA | SL1344_0626 | 699240..700307 | − | A(GCT) | Rare lipoprotein A, role in cell division, contains SPOR domain, may bind peptidoglycan | ||
| 2733943 | T | Y | T | SL1344_2552 | 2732413..2734785 | − | A(GCA) | Bacteriophage SLP272, tail fiber- like protein | Bacteriophage | ||
| NON-CODING | |||||||||||
| 4372550 | A | T | A | − | rRNA | 3588585.. 3591592 | − | 23S ribosomal RNA | Transcription, ribosomal structure and translation | ||
| 4415836 | G | A | G | − | rRNA | 4372578..4374122 | + | 16S ribosomal RNA | Transcription, ribosomal structure and translation | ||
| 1282541 | A | R | A | STnc150 | SL1344_ncRNA_25 | 1282521..1282678 | − | Experimentally verified small RNA (Sittka et al., 2008) | Regulation | ||
| 3099732 | G | T | G | ||||||||
Nucleotide denoted by S is either C or G; R is either G or A; K is either T or G; M is either A of C; Y is either T or C.
| (B) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Nucleotide | Gene name | Reference ID | Location | Stand | Description | Function | Functional category | |||
| SL1344 | ST24WT | ST24CHX | |||||||||
| INTERGENIC | |||||||||||
| 274042 | C | C | M | − | SL1344_0700 | Intergenic, pseudogene (783428..784579) | + | Pseudogene | UDP-galactopyranose mutase, involved in outer membrane biogenesis | ||
| 3708204 | C | S | C | glpR | SL1344_3489 | Intergenic, pseudogene (3707770..3708527) | − | Pseudogene | Hypothetical repressor of the glpD, glpFK, glpTQ, and glpACB operons involved in glycerol-3-phosphate metabolism | Anaerobic metabolism, lipid metabolism | |
| 4217516 | A | G | A | − | Intergenic | 182 bp up-stream of 16S ribosomal RNA (4416018..4417560) | |||||
| 3485325 | A | A | M | − | Intergenic | 98 bp up-stream of 16S ribosomal RNA (4217614..4219158) | |||||
| 3485327 | A | A | W | − | Intergenic | 35 bp up-stream of SL1344_3266 (folP), dihydropteroate synthase, involved in folate biosynthesis (- strand). | |||||
| 783643 | T | T | K | − | Intergenic | 37 bp up-stream of SL1344_3266 (folP), dihydropteroate synthase, involved in folate biosynthesis (- strand) | |||||
| 3848707 | A | W | A | − | Intergenic | 28 bp up-stream of SL1344_3605 (lpfB), fimbrial chaperone protein (- strand) | |||||
| 1549186 | A | W | A | − | Intergenic | 1000 bp up-stream of SL1344_1445 (ydeI), hypothetical protein of unknown function | |||||
| 1180820 | A | R | A | − | Intergenic | 385 bp up-stream from SL1344_1072 (ycdW), hypothetical 2-hydroxyacid dehydrogenase | |||||
| 2073115 | T | T | G | − | Intergenic | Bacteriophage SL203. Within a DNA invertase fragment | Bacteriophage | ||||
| 2045028 | G | A | G | − | Intergenic | Bacteriophage SLP203 | Bacteriophage | ||||
| 2762013 | T | C | T | − | Intergenic | Bacteriophage SLP203 | Bacteriophage | ||||
| 2762182 | A | G | A | − | Intergenic | Bacteriophage SLP272 | Bacteriophage | ||||
| 1282534 | T | W | T | − | Intergenic | Bacteriophage SLP272 | Bacteriophage | ||||
| 3117665 | T | A | T | − | SL1344 repeat region 2 | Intergenic, repeat region (3099171..3100233) | + | Repeat region | CRISPR repeat region | Bacteriophage | |
| 3117692 | T | A | T | − | SL1344 repeat region 3 | Intergenic, repeat region (3116271..3117792) | + | Repeat region | CRISPR repeat region | Bacteriophage | |
| 3435062 | G | G | R | − | SL1344 repeat region 3 | 3116271..3117792 | + | Repeat region | CRISPR repeat region | Bacteriophage | |
| 3590829 | G | A | G | − | SL1344 repeat region 10 | 3435027..3435149 | + | Repeat region | CRISPR repeat region | Bacteriophage | |
*Nucleotide denoted by S is either C or G; R is either G or A; K is either T or G; M is either A of C; Y is either T or C; W is either T or A.
RNA purification and quantification
RNA was purified from 2 ml of each mid-logarithmic phase culture (OD610 nm = 0.6), without chlorhexidine treatment and following exposure to 0.5 × MIC of the reference strain, equivalent to 1 μg/ml chlorhexidine, for 30 min using an Ambion RiboPure Bacteria Kit (Life Technologies Corporation, Carlsbad, CA). RNA was purified in accordance with the manufacturer's instructions, with the exception that the final DNase treatment step was carried out twice for each sample. Purified RNA was quantified using a ND-1000 spectrophotometer (Nanodrop, Thermo Scientific, Waltham, Massachusetts), and the quality of the RNA was assessed using a 2100 Bioanalyser (Agilent, Waldbronn, Germany).
Transcriptomic analysis of purified RNA
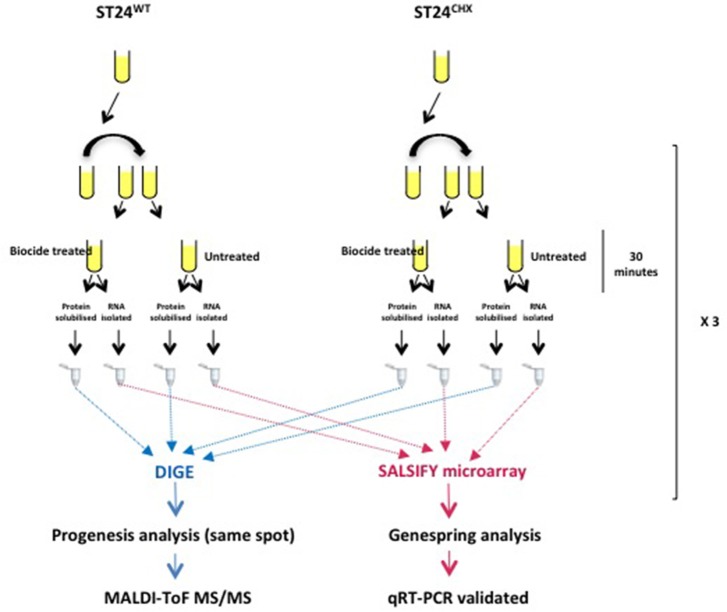
Conversion to and labeling of cDNA and hybridizations using SALSIFY microarrays (Design ID 026881) (Agilent technologies, Santa Clara, California), were carried out as described previously (Ygberg et al., 2006; Finn et al., 2013). In brief, RNA was converted to cDNA and fluorescently labeled with Cy3-dCTP using random priming. Genomic DNA was isolated from ST24WT as described above. The genomic DNA (gDNA) was then labeled with Cy5-dCTP as a reference source. The labeled cDNA and gDNA were combined, denatured and hybridized to the microarray. Subsequently, the hybridization was carried out for 16–18 h at 65°C. The microarray slides were then washed according to manufacturer's instructions (http://www.chem.agilent.com/Library/usermanuals/Public/G2534-90004_HybridizationChamber_User.pdf). Microarray slides were cleaned with inert gas to remove any debris before scanning with the Agilent Microarray scanner system (Agilent technologies, Santa Clara, California). Scans were carried out at 5 μm resolution with Green and Red PMT values set to 100 % and an XDR value of 0.1. Images generated were saved as multi-image.tiff files. Feature extraction software (Agilent Technologies, Santa Clara, California) was used to extract the data. Hybridizations were repeated on three biological replicates (Figure 1).
Figure 1.
Proteomic and transcriptomic experimental design for the analysis of differentially expressed genes and proteins comparing the reference chlorhexidine susceptible Salmonella Typhimurium, ST24WT and its, isogenic, chlorhexidine tolerant mutant, ST24CHX.
The microarray transcriptional data were analyzed by using both GeneSpring GX software version 7.3 (Agilent Technologies, Santa Clara, California) and using R 2.14.2 (R Core Team, 2011), with base package statistics and the attached package Limma (version 3.10.2) (Smyth and Speed, 2003; Smyth, 2005; Ritchie et al., 2007). Genes with a fold change >2 and p < 0.05 were considered to be differentially expressed. Only those genes confirmed as differentially expressed by both the GeneSpring and R analytical methods, and detected by more than one oligonucleotide probe, were considered as significant, and are discussed further below. A summary of the statistical and expression data for all individual differentially expressed genes in provided in Table S1. Data from this study has been deposited in NCBI's Gene Expression Omnibus (GEO accession number GSE59566).
qRT-PCR
Quantitative reverse transcriptase (RT)-mediated PCR (qRT-PCR) was performed on five selected gene targets, using specific primer pairs for three biological replicates (Table S2). Targets for validations were chosen as representative of the functional categories determined as differentially expressed from the microarray results, as well as one target non-differentially expressed. Bacterial cells were cultured to mid-logarithmic phase and total RNA was purified and quantified as outlined above. The levels of each RNA transcript were determined using a real-time One-Step RT-PCR assay (Eppendorf, Hamburg, Germany) with a QuantiTect SYBR Green RT-PCR kit (Qiagen, Hilden, Germany). The RT-PCR reaction was carried out in 25 μl volumes consisting of 12.5 μl 2 X QuantiTect SYBR Green RT-PCR Master Mix, 10 pmol of each primer pair, 32 ng of the purified RNA sample and 0.25 μl of QuantiTect RT Mix. RT-negative controls were included for each RNA sample to confirm that each was devoid of contaminating gDNA. A “no template control” was also included to ensure the reaction was free from contaminating RNA. Each qRT-PCR cycle consisted of an initial reverse transcription step of 30 min at 50°C followed by incubation at 95°C for 15 min. This latter step was immediately followed by 40 cycles of amplification consisting of; 15 s at 94°C, 30 s at 60°C and 30 s at 72°C. At the end of the 40 cycles the temperature was raised from 72 to 95°C, slowly, over 20 min. A final melt step, of 15 s at 95°C, ended the amplification programme. Relative gene expression was determined using the ΔΔCT method. Two housekeeping genes were selected for this purpose and these included the DNA-directed RNA polymerase subunit B gene (rpoB) and the DNA gyrase subunit A gene (gyrA). A summary of all qRT-PCR data is provided in Table S3.
Fluorescent differential gel electrophoresis (DIGE) analysis of proteins
For proteomic analysis, cell pellets were prepared from the 25 ml volumes of the mid-logarithmic phase cultures (OD610 nm = 0.6) by centrifugation at 3220 × g for 10 min. The resulting cell pellets were maintained on ice and washed in 1 ml ice-cold 10 mM Tris-Cl [pH 8], 1 mM EDTA (TE buffer) three times. The washed cell pellet was then resuspended in 200 μl solubilization buffer (7 M urea, 2 M thiourea, 1% ASB-14) for 18 h at room temperature. Proteins were quantified using the RCDC Protein Assay (Bio-Rad, Hercules, California) according to the manufacturer's instructions.
All samples were diluted to a final protein concentration of 5 μg/ml and adjusted to pH 8.5 using 10 mM NaOH. A pooled internal standard comprising 33.33 μg of each sample was labeled with Cy2 dye (GE Healthcare, Buckinghamshire, UK) following the manufacturer's instructions. For each sample, 60 μg of protein was labeled with Cy3 or Cy5 (GE Healthcare, Buckinghamshire, UK), to control for dye swap (Figure 1).
Samples were then pooled as follows; 50 μg each of the reference strain ST24WT samples (either Cy3 or Cy5 labeled) were combined with 50 μg of the corresponding mutant ST24CHX sample (labeled with the alternative dye). This was followed by the addition of 50 μg of internal standard. Pooled protein samples were then diluted to 450 μl in solubilization buffer (30 mM DTT, 0.5% IPG buffer [GE healthcare, Buckinghamshire, UK] and 0.01% bromophenol blue) for first dimension separation.
First dimension separation was carried out using Immobiline DryStrip gels (IPG strips) (GE healthcare, Buckinghamshire, UK) as previously described (Nally et al., 2005, 2007). In brief, 24 cm pH 4–7 IPG strips were rehydrated with protein samples overnight, as per the manufacturer's instructions. Isoelectric focusing was carried out using the Ettan IPGphor II Isoelectric focusing system (GE Healthcare, Buckinghamshire, UK) with the following parameters: Step and hold (Sth) 3500 V for 75,000 V/h, Gradient 8000 V for 10 min, Sth 8000 V for 1 h, Sth 100 V for 5 h. At the end of the isoeletric focusing step, strips were equilibrated for 10 min in equilibration buffer (6M urea, 0.75 M Tris-Cl pH 8.8, 29.3% (w/w) glycerol, 1% (w/v) SDS) containing 1% (w/v) DTT with gentle shaking at 80 rpm, followed by washing for 10 min in equilibration buffer containing 2.5% (w/v) iodoacetamide with shaking at 80 rpm. Strips were then loaded on a 12% (w/v) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel for the second dimension separation. Gels were electrophoresed at 2 W (per gel) for 1 h, followed by 4 W (per gel) for 18 h and, finally, at 12 W (per gel) until the dye front reached the bottom of the gel on visual inspection using the Ettan DALTsix Electrophoresis system. The entire process was carried out using low fluorescent glass plates and in the dark. Gels were subsequently imaged on a Typhoon™ variable mode imager (GE Healthcare, Buckinghamshire, UK) according to the manufacturer's instructions, and the output analyzed using Progenesis SameSpots software (©Nonlinear Dynamics Ltd., Newcastle, UK). Only t-test analysis spots with a p < 0.05, power >80% and q < 0.05 were considered differentially expressed. The experimental design is summarized in Figure 1. A summary of the statistical and expression data for all individual differentially expressed protein spots in provided in Table S4.
Excision and digestion of spots of interest for identification by mass spectrometry (MS)
A 2-D SDS-PAGE master gel was electrophoresed on 24 cm IPG strips as described above, with the exception that only 400 μg of un-labeled internal standard was loaded. The second dimension separation was carried out as before, with the exception that glass plates were pre-treated for 1.5 h with a bind silane solution (80% ethanol, 2% glacial acetic acid, 0.1% bind saline) (GE Healthcare) and reference markers were attached as outlined in the manufacturer's instructions (GE Healthcare). The resolved proteins were visualized using SYPRO-Ruby stain (Sigma, Saint Louis, Missouri) as per manufacturer's instructions. The master gel was scanned using the Typhoon™ variable mode imager (GE Healthcare), and the output analyzed using Progenesis SameSpots software. A pick list was generated, which included the coordinates of both differentially expressed and non-differentially expressed spots on the master gel. The protein spots on the pick list were excised and digested using the Ettan Spot Handling Workstation (GE Healthcare), and the resulting peptide mixtures used for MS-based protein identification.
All MS and MS/MS analyses were performed using a 4800 MALDI TOF/TOF (Applied Biosystems, Foster City, CA, USA). For each sample one MS spectrum was acquired, and the eight most intense precursors were subsequently selected for MS/MS analysis (Sergeant et al., 2011). An Applied Biosystems GPS-server was used for database searches with an in-house MASCOT platform (Matrix Science, www.matrixscience.com, London, UK). The spectra from one spot (combined MS and eight MS/MS spectra) were submitted and compared against the NCBInr database limited to bacterial proteins only (downloaded from the NCBI server on 26/09/2011 containing 8,874,873 bacterial sequences). A mass window of 100 ppm for the precursor and 0.75 Da for the fragments was tolerated. During the database searches the following parameters were defined: two missed cleavages, fixed carbamidomethylation of cysteine, variable oxidation of methionine and tryptophan to kynurenine or double oxidation to N-formylkynurenine. All the identification data are included in Table S5. All identifications were manually validated, and extra precursors were selected for fragmentation if the data obtained was judged to be insufficient. The MS-spectra of spots wherein the same protein was identified were compared, and extra precursors (unique to one of the spectra) were fragmented to distinguish the molecular forms present in the individual spots (Carpentier et al., 2011).
Phenotypic microarray assay
The chlorhexidine susceptible reference strain ST24WT and chlorhexidine tolerant mutant ST24CHX were examined for phenotypic divergence using Omnilog™ phenotypic microarrays (using all plates denoted PM 1 through 20) (BioLog Inc., Hayward, California). Bacterial cell suspensions were prepared and PM plates were inoculated following manufacturers' instructions. These plates were incubated at 37°C for 48 h in an Omnilog™ microplate reader (BioLog Inc., Hayward, California).
The digital imagery of this instrument tracks changes in the respiration of bacterial cultures growing in individual wells over time. The Omnilog™ output for a given plate consists of an optical density (OD) reading for each well, recorded every 15 min over the 48 h period. The data output for the reference and mutant strains were analyzed using Omnilog™ PM software, and negative controls (wells containing the inoculated Omnilog™ growth medium, but without any substrate, used to normalize differences in inoculums and redox dye oxidation between samples) were subtracted from each reading for each plate. The resulting kinetic profiles for ST24WT and ST24CHX were compared using an integration function. A divergent phenotype was identified when a difference in Omnilog™ units of 20,000 or greater between the strains was obtained following integrative function analysis. A summary of the differential phenotypes is provided in Table S6.
Results and discussion
Chlorhexidine tolerance has been documented in several bacterial genera including Pseudomonas, Klebsiella, and Serratia (Nde et al., 2009). Bacteria demonstrating a reduced susceptibility to chlorhexidine can be selected in vitro, as in the case of Salmonella (Braoudaki and Hilton, 2004; Condell et al., 2012b), Escherichia coli (Braoudaki and Hilton, 2004) and Staphylococcus aureus (Kaatz et al., 2005; Huet et al., 2008). Based on our current understanding of the modes of action related to various biocides, tolerance to these compounds, including chlorhexidine, typically does not develop following mutation to a particular target gene but rather involves broader cellular changes, such as up-regulated efflux pump activity or alterations in cell wall permeability (Poole, 2002). In some instances, the tolerant phenotype that develops may correlate with a reduced susceptibility to other antimicrobial agents, for example, chlorhexidine exposure has been associated with a reduced susceptibility to clinically important antimicrobial agents (Braoudaki and Hilton, 2004; Condell et al., 2012a). In this study, our aim was to describe the genotype and phenotype(s) exhibited by S. Typhimurium following sub-lethal exposure to chlorhexidine. Two S. Typhimurium isolates, a chlorhexidine susceptible reference strain, denoted as ST24WT and its isogenic chlorhexidine tolerant counterpart ST24CHX, were studied in detail. We explored whether or not tolerance to chlorhexidine was a multi-factorial process, and if such a phenotype evolves following one or more mutations in a target gene, as occurs with triclosan (Heath and Rock, 2000; Condell et al., 2012b; Sheridan et al., 2012).
To assist with a logical approach to describing the data from this study, the results and discussion are outlined as follows; firstly, the bacterial response of ST24WT to chlorhexidine exposure is described. This section is followed by a description of the response of the chlorhexidine tolerant ST24CHX following exposure to the biocide. Finally, a comparison of ST24WT and ST24CHX is shown and a general chlorhexidine response network in Salmonella Typhimurium based on these data is proposed.
Response of Salmonella typhimurium ST24WT to sub-lethal chlorhexidine exposure
Bacterial responses comparing S. Typhimurium ST24WT in the absence of biocide and following a 30 min exposure to 0.5 × MIC of chlorhexidine was carried out. Figure 1 provides an overview of the experimental strategy used.
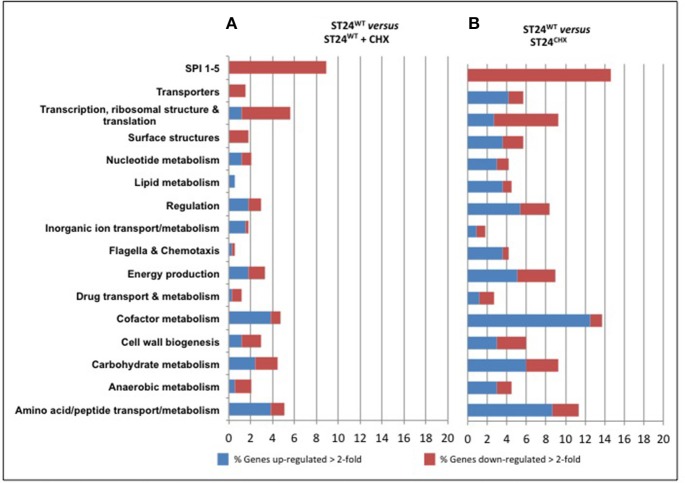
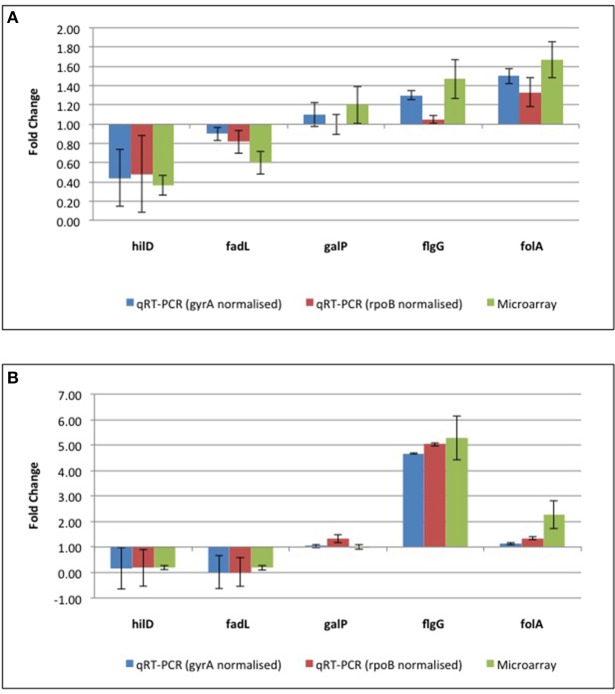
A total of 247 genes were significantly differentially expressed (2-fold-change, p < 0.05), following sub-lethal exposure to chlorhexidine (Table S1); 81 of these genes were up-regulated whilst 166 were down-regulated (Figure 2). Differentially expressed genes of known function were assigned to functional categories (Figure 3 and Table S1). Transcriptomic data were subsequently validated using qRT-PCR for 5 selected genes. Similar fold-change values were obtained from both independent analyses, for all five genes (Figure 4A, Table S3). Results from transcriptomic profiling revealed that the functional categories containing the largest number of up-regulated genes were involved in general cell metabolism; amino acid transport/metabolism, cofactor metabolism and carbohydrate metabolism. Conversely, the functional groups containing the largest number of down-regulated genes included virulence, transcription, translation and ribosomal structure, along with phage-associated genes. For each functional category, the numbers of differentially expressed genes are shown as a percentage of the total number of genes in that category (as determined by KEGG, Figure 3). Corresponding gene lists are provided in Table S1.
Figure 2.
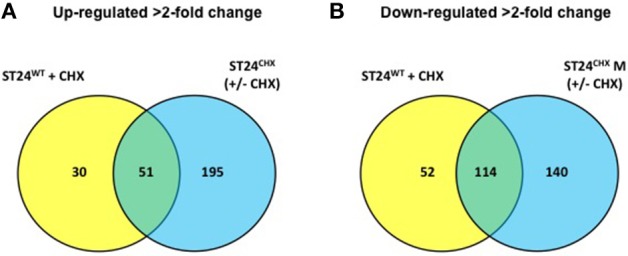
Number and distribution of genes (A) up-regulated in the sensitive ST24WT following chlorhexidine exposure and/or the tolerant mutant ST24CHX relative to the reference strain (ST24WT) without chlorhexidine exposure. (B) Down-regulated in the sensitive ST24WT following chlorhexidine exposure and/or the tolerant mutant ST24CHX. The figure shows the differentially expressed genes relative to the reference strain (ST24WT) without chlorhexidine exposure.
Figure 3.
Functional groups of genes differentially expressed between (A) the reference ST24WT in the absence of chlorhexidine exposure and the reference isolate following chlorhexidine exposure and (B) the mutant isolate without chlorhexidine exposure. The red and blue coloring indicates the percentage of those genes from each functional category that were differentially expressed. The numbers of genes included in the functional group analysis is given in Table S1. The lists of genes comprising the functional groups retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/).
Figure 4.
qRT-PCR confirmation of microarray data from (A) the reference ST24WT chlorhexidine treated, relative to the same isolate in the absence of chlorhexidine exposure and (B) the mutant isolate, ST24CHX, relative to the reference ST24WT. Details of protein results are provided in Table S4, with qRT-PCR and transcriptomic results shown in Table S3.
DIGE analysis compared the proteome of the susceptible ST24WT reference strain in the absence and presence of chlorhexidine. A total of 1431 protein spots were aligned by Progenesis SameSpots software. No differentially expressed proteins were detected when the proteomes of ST24WT was compared before and after chlorhexidine exposure for 30 min. This contrasts to the response of S. Typhimurium following the treatment to other biocidal agents. Numerous differentially expressed proteins were apparent in S. Typhimurium following 30 min exposure to the biocide triclosan, a similar response was also noted in E. coli following exposure to the same agent (Condell et al., 2012b; Sheridan et al., 2012).
General cell metabolism of ST24WT following sub-lethal exposure to chlorhexidine
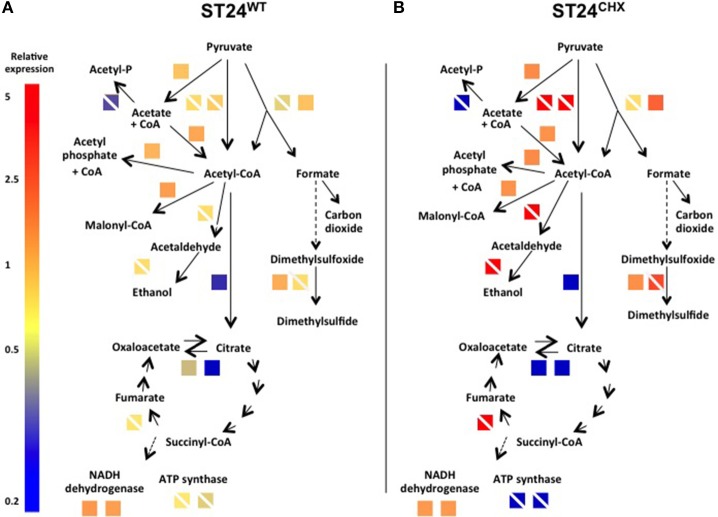
When analyzed using the transcriptomic platform, genes associated with central metabolism and energy production in ST24WT exhibited altered expression. Enzymes associated with glycolysis and the TCA cycle were down-regulated, whilst those of other alternative pathways, such as mixed acid fermentation, fatty acid biosynthesis and glyceroplipid metabolism were up-regulated (Figure 5A). The expression of formate dehydrogenase was increased, and this enzyme functions in the conversion of formate to CO2 in the absence of alternative electron acceptors. In addition, acetyl-CoA carboxylase, an enzyme that converts acetyl-CoA to malonyl-CoA, a regulatory step for the inititation of fatty acid biosynthesis, was also up-regulated. Genes associated with anaerobic glycerolipid metabolism, such as glyB, were up-regulated in ST24WT. Interestingly, genes encoded by the pdu operon, which is associated with the catabolism of 1,2-propanediol and linked with the formation of polyhedral bodies- large proteinaceous structures of unknown function (Bobik et al., 1999; Havemann et al., 2002), were up-regulated. This metabolic pathway is known to be involved with anoxic metabolism in Salmonella (Bobik et al., 1999; Havemann et al., 2002), and it has previously been proposed that chlorhexidine blocks oxygen utilization in bacteria (Barrett-Bee et al., 1994). ST24WT appeared to up-regulate alternative metabolic pathways for anoxic energy production to mitigate this inhibitory action of chlorhexidine (Figure 5A).
Figure 5.
Expression of differentially expressed genes and/or proteins associated with anaerobic metabolism/energy production in (A) the reference isolate ST24WT following chlorhexidine exposure and (B) the mutant isolate ST24CHX compared to its isogenic ST24WT without chlorhexidine exposure. Enzyme names, descriptions, gene identifiers and fold-change values are given in Table 2, Tables S1, S4. Color blocks without a white diagonal line represent gene fold change values (Table S1), color blocks containing a white diagonal line represent protein fold change values (Table 2, Table S4).
Cofactor metabolism of ST24WT following sub-lethal exposure to chlorhexidine
In addition to alterations in the general cell metabolism, genes associated with cofactor synthesis in ST24WT were differentially-expressed. Six % of genes known to be associated with cofactor biosynthesis (as obtained from KEGG) were up-regulated in ST24WT, following a 30 min exposure to chlorhexidine. These markers coded for enzymes involved in the synthesis of thiamine, folate, molybdopterin and porphyrin (vitamin B12) amongst others (Figure 6). The altered expression of these cofactor associated genes may be linked with the changes in general cell metabolism outlined earlier. For example, the increased expression detected in enzymes involved in both the synthesis and transport of thiamine may be a bacterial response designed to satisfy the increased demand for thiamine as a cofactor for pyruvate dehydrogenase and 2-oxoglutarate decarboxylase. Similarly, vitamin B12 is required for the utilization of 1,2-propanediol as a carbon source by the pdu operon and molybopterin as a cofactor of formate dehydrogenase. All were up-regulated in ST24WT, following exposure to chlorhexidine (Gladyshev et al., 1994; Bobik et al., 1999).
Figure 6.
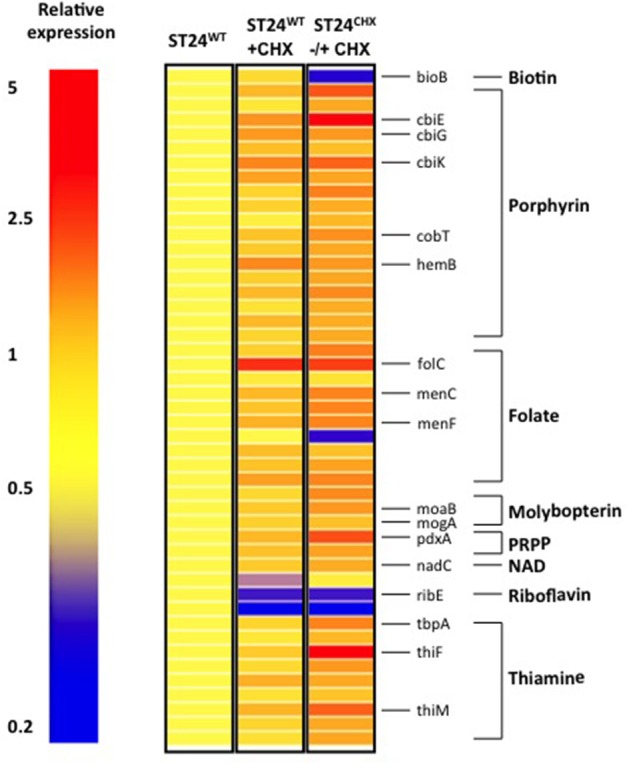
Chlorhexidine tolerance is associated with differentially expressed genes associated with cofactor metabolism. Relative expression refers to the fold-change difference in expression between the ST24WT, without chlorhexidine treatment relative to the conditions listed in the figure.
Modifications in membrane structural and transport systems of ST24WT following sub-lethal exposure to chlorhexidine
Reduced susceptibility to chlorhexidine associated with modifications in the cell wall/ cell membrane has been reported previously (Russell and Furr, 1986; Russell and Path, 1986; Tattawasart et al., 2000). As an initial step in forming contact with the bacterial cell, due to its positive charge, chlorhexidine is thought to associate with negatively-charged residues on the LPS structures of Gram-negative bacteria (Tattawasart et al., 2000). Modifications of the LPS structure, such as those that contribute to reducing its negative charge along with a reduction in the O-antigen polymer are associated with a decrease in susceptibility to cationic agents (Tattawasart et al., 2000).
In this study, several changes in the expression of genes associated with cellular permeability were noted in ST24WT following chlorhexidine exposure. An increase in the expression of genes associated with the biosynthesis of the peptidoglycan subunits (murG) and responsible for the cross-linking between these subunits (murD, murG) was noted. Up-regulation of these genes may provide a protective modification that contributes to an increase in thickness and/or the cross-linking of the peptidoglycan layer, changes that would affect the efficacy of antimicrobial agent action and/or transport (Gilbert and Brown, 1980; McDonnell and Russell, 1999). Additionally, ST24WT exhibited a down-regulation in genes of the rfb locus, involved in O-antigen synthesis; namely rfbU and rfbA. The O-antigen provides several anionic binding sites (Zorko and Jerala, 2008) and modification may result in altered chlorhexidine binding, thereby reducing the permeability of the cell to this agent. Modifications in O-antigen structure have previously been associated with an increased tolerance to clinically important cationic antimicrobials in Pseudomonas (Bryan et al., 1984; Hasegawa et al., 1997; Tattawasart et al., 2000).
Efflux pump activity of ST24WT following sub-lethal exposure to chlorhexidine
It has previously been reported that an increase in the expression of certain efflux systems is correlated with a reduction in the susceptibility to chlorhexidine in several bacterial species studied, including P. aeruginosa (mediated through MexCD–OprJ), S. aureus (QacAM MepA) and E. coli (AcrAB-TolC) (Brown and Skurray, 2001; Levy, 2002; Poole, 2004; Huet et al., 2008). However, in this study the reference isolate ST24WT showed no increase in the expression of any efflux systems following chlorhexidine exposure for 30 min. However, a decrease in the expression of AcrE, a component of the AcrEF-TolC efflux system and the multi-drug efflux associated gene ydhE, a member of the MATE super-family (Omote et al., 2006) were observed. From the study reported by Gilbert and Moore, the over-expression of efflux pumps do not reduce the antimicrobial effect of chlorhexidine, as the biocide does not become solubilized in the bacterial membrane core (Gilbert and Moore, 2005). Thus, the contribution of these efflux systems in terms of altered chlorhexidine susceptibility requires further investigation.
SOS response of ST24WT following sub-lethal exposure to chlorhexidine
Twelve genes encoding DNA binding proteins (other than transcriptional regulators) or associated with DNA damage/repair mechanisms were up-regulated (Table S1). Following exposure to chlorhexidine, ST24WT showed an increase, in four genes associated with the SOS response (Table S1). Recent work has suggested DNA damage as a means by which chlorhexidine inactivates periodontal bacteria (Yeung et al., 2007; Nde et al., 2009). It was reported that chlorhexidine can cause DNA-DNA cross-linking, thereby interfering with DNA metabolism and leading to DNA strand breaks (Yeung et al., 2007; Nde et al., 2009). If the same mechanism holds true for Salmonella, then the up-regulation in the DNA repair systems along with DNA binding proteins might represent the bacterial response to mitigate the DNA cross-linking effects of chlorhexidine. A similar feature was reported earlier in E. coli (Allen et al., 2006)
Transcription and translation in ST24WT following sub-lethal exposure to chlorhexidine
Several genes encoding proteins associated with transcription and translation were down-regulated following chlorhexidine exposure (Figure 3). A decrease in the expression of the α-sub-unit of DNA-directed RNA polymerase (rpoA) was detected. Furthermore, there was a down-regulation in 11 genes coding for ribosomal subunits, including multiple 30S and 50S ribosomal sub-unit components, at the RNA level. Chlorhexidine has previously been associated with a decrease in protein synthesis (Galice et al., 2006; Nde et al., 2009).
Virulence gene expression in ST24WT following sub-lethal exposure to chlorhexidine
Transcriptomic profiling of the chlorhexidine susceptible ST24WT revealed a correlation between chlorhexidine exposure and a reduction in the expression of virulence associated genes (Figure 7). A total of 8% of the genes encoded by SPI-1, SPI-2, SPI-3, SPI-4, and SPI-5 (obtained from KEGG) were down-regulated following chlorhexidine exposure in ST24WT. Studies examining the association between chlorhexidine tolerance and alterations in virulence phenotypes are lacking in the literature. It has been reported that reduced chlorhexidine susceptibility in a collection of β-haemolytic E. coli correlated with four virulence factor genotypes (Beier et al., 2005). However, a reduction in virulence factor expression in Streptococcus agalactiae exposed to sub-inhibitory concentrations of chlorhexidine was reported (Galice et al., 2006), while sub-lethal concentrations resulted in a significant reduction in infectivity of mice by E. coli and Klebsiella aerogenes (Holloway et al., 1986). This reduction in virulence following chlorhexidine exposure may contribute to its potency as a therapeutic antimicrobial agent.
Figure 7.
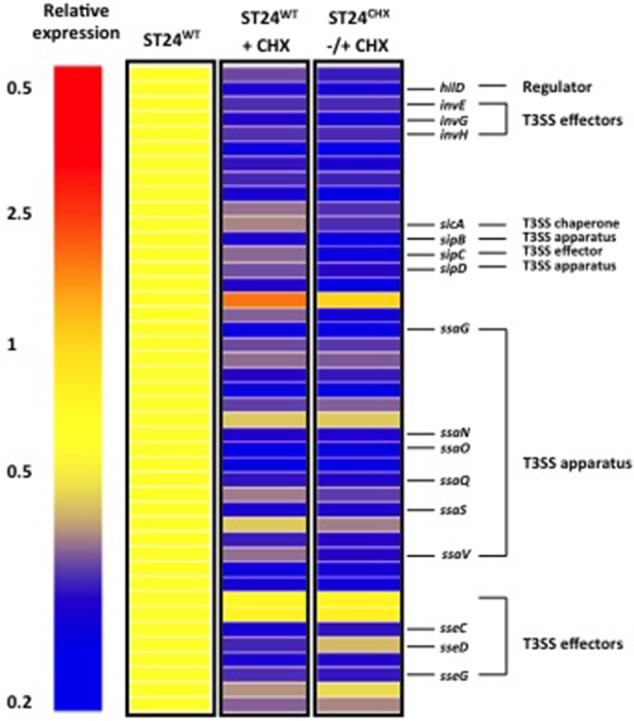
Chlorhexidine tolerance is associated with differential expression of SPI-1 and SPI-2 virulence genes. Relative expression refers to the fold-change in expression between the reference ST24WT, without chlorhexidine treatment relative to the conditions listed in the figure.
Response of Salmonella typhimurium ST24CHX to sub-lethal chlorhexidine exposure
The reference isolate, ST24WT was exposed to several rounds of in vitro selection using sub-lethal concentrations of chlorhexidine, as described previously by Condell et al. (2012a). A S. Typhimurium isolate denoted as ST24CHX was recovered which exhibited a high-level chlorhexidine tolerance phenotype, with a >50-fold increase in the MIC in comparison to ST24WT (Condell et al., 2012a). The transcriptomic and proteomic responses of ST24CHX were studied following a 30-min exposure to this biocide, in the same way as described for the wild-type isolate. Alterations in the bacterial response were compared to those detected for ST24CHX without biocide exposure.
Exposure of ST24CHX to chlorhexidine did not cause differential expression of any genes. Similarly, no differentially expressed proteins were detected. We conclude that exposure to 0.5 × MIC chlorhexidine for 30 min did not affect the transcription or translation of any genes in the tolerant mutant.
General comparisons of SNPs, gene and protein expression in ST24WT and ST24CHX
The mutant (ST24CHX) was studied in detail, using several analytical approaches to identify cellular mechanisms associated with the tolerance to chlorhexidine. Results from these analyses were compared to those from the reference susceptible isolate (ST24WT), described above, to identify any changes of interest.
Sixty SNPs that distinguished ST24CHX from its isogenic chlorhexidine susceptible reference ST24WT were identified. Of these, 18 SNPs were located in intergenic regions and 42 SNPs occurred within genes. The intergenic SNPs included 5 located within bacteriophage regions (annotated as L203 or SLP22), 4 located within clustered regularly interspaced short palindromic repeat (CRISPR) regions and 2 identified within pseudogenes. Of the SNPs that occurred within genes, 17 were synonymous, 15 were non-synonymous, 6 were degenerative and 4 were within non-coding genes. The non-synonymous and degenerative polymorphisms occurred within genes associated with bacteriophage, drug transport, virulence, transport/permeability and general cell metabolism, whilst the non-coding SNPs were within sRNA and ribosomal RNA regions. A summary of all SNPs identified between the reference and the mutant isolates are given in Table 1.
Differentially expressed transcripts were identified when comparing the transcriptomes of ST24WT and ST24CHX, without chlorhexidine exposure. A total of 500 genes were differentially expressed between both isolates (Table S1), with 246 of these being up-regulated and 254 down-regulated (Figure 2). Of these differentially expressed genes, those of known function were assigned into functional categories (Figure 3 and Table S1). As before, functional categories containing the largest number of up-regulated genes included general cell metabolism; amino acid, cofactor and carbohydrate transport/metabolism. Conversely, the functional groups containing the largest number of down-regulated genes included virulence, transcription, translation and ribosomal structure, along with phage-associated genes. Gene lists of the differentially expressed transcripts and their functional categories are provided in Table S1. A qRT-PCR assay was performed on 5 selected genes, and similar fold changes were obtained from both independent analyses for all of the chosen genes (Figure 4B). A summary of all qRT-PCR data are provided in Table S3.
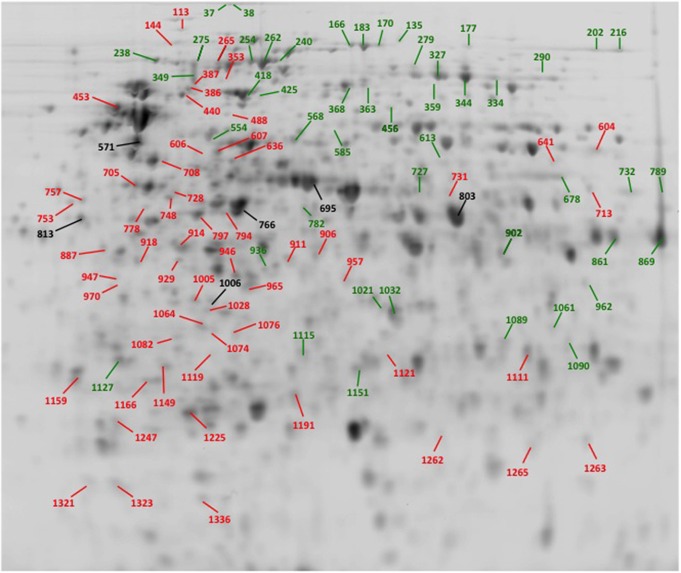
Proteomic analysis revealed a total of 470 protein spots differentially expressed, without chlorhexidine exposure, between ST24WT and ST24CHX (>2-fold change, p < 0.05). Among the differentially expressed proteins, 208 were up-regulated and 262 were down-regulated in ST24CHX relative to the reference isolate (Figure 8). Of the up-regulated proteins a total of 48 protein spots were identified with statistical significance and these corresponded to 34 individual proteins (Table 2, Tables S4, S5). These were divided into three functional categories; general cell metabolism (24 proteins), stress response (5 proteins) and transport and permeability (5 proteins). Up-regulated proteins are indicated in green in Figure 8, denoted by the green traces in Figure 9, and functionally categorized in Table 2.
Figure 8.
A 2-D proteome map of ST24WT and ST24CHX, without chlorhexidine treatment. The spot numbers corresponds to identified proteins as described in Table 2, Tables S4, S5. Those spots labeled in black represent non-differentially expressed proteins (see Table S5). Protein spots labeled green (and denoted as Group A, see Table 2, Table S4) were significantly up-regulated in ST24CHX relative to ST24WT, whilst spots labeled in red represent Group B and were significantly down-regulated in ST24CHX relative to ST24WT (Table 2, Table S4).
Table 2.
Identification and functional classification of proteins differentially expressed between the reference, chlorhexidine susceptible, Salmonella Typhimurium ST24WT, and the isogenic chlorhexidine tolerant mutant ST24CHX.
| Spot # | Protein name | Function | Functional category* | p-value | Fc† | GI number |
|---|---|---|---|---|---|---|
| PROTEINS UP-REGULATED | ||||||
| 290 | Anaerobic dimethyl sulfoxide reductase chain A (DmsA) | Terminal electron acceptor, reduction of DMSO to DMS | Anaerobic metabolism, Energy metabolism | 4.12E-05 | 3.40 | gi|323210416 |
| 202, 216 | Bifunctional acetaldehyde-CoA/alcohol dehydrogenase (AdhE) | Reduction of acetyl-CoA to acetaldehyde and then ethanol | Anaerobic metabolism, Energy metabolism | 1.48E-02 | 18.03 | gi|16760134, gi|16760134 |
| 135, 327, 334, 344, 359 | Formate acetyltransferase 1 (PflB) | Conversion of pyruvate and CoA to formate and acetyl-CoA | Anaerobic metabolism, Energy metabolism | 7.21E-04, 2.83E-05, 6.39E-05, 5.26E-05, 3.10E-04 | 6.53, 10.05, 7.45, 10.43, 3.71 | gi|213029313, gi|16759843 |
| 177 | Succinate dehydrogenase flavoprotein subunit (SdhA) | Fumarate and succinate interconversion, under anaerobic conditions | Anaerobic metabolism, Energy metabolism | 2.65E-05 | 8.43 | gi|261245997 |
| 861, 869 | Glyceraldehyde-3-phosphate dehydrogenase (GapA) | Conversion of glyceraldehydes-3-phosphate to D-glycerate 1,3-bisphosphate. Glycolysis | Energy metabolism | 5.74E-05, 7.95E-11 | 9.90, 15.34 | gi|16760605 |
| 554 | Phosphoglyceromutase (PmgI) | Conversion of 2-phospho-D-glycerate to 3-phospho-D-glycerate. Glycolysis | Energy metabolism | 5.18E-04 | 2.70 | gi|213023081 |
| 279 | Pyruvate kinase (PykF) | Conversion of phosphoenolpyruvate to pyruvate. Glycolysis | Energy metabolism | 4.21E-05 | 5.72 | gi|197362573 |
| 275, 349 | Dihydrolipoamide acetyltransferase (E2) (AceF) | Component of the pyruvate dehydrogenase complex | Energy metabolism | 2.03E-07, 9.76E-06 | 6.99, 9.91 | gi|62178722 |
| 363, 368 | Transketolase (TktA) | Transfer of a ketol group between several donors and acceptors, links glycolysis and the pentose phosphate pathway | Carbohydrate metabolism | 1.53E-04, 2.11E-04 | 4.33, 4.61 | gi|213850121, gi|301159632 |
| 936 | Ribose-phosphate pyrophosphokinase (PrsA) | Conversion of D-ribose-5-phosphate to 5-phospho-alpha-D-ribose 1-diphosphate (PRPP). Pentose-phosphate pathway | Carbohydrate metabolism, Co-factor metabolism | 1.11E-06 | 3.26 | gi|16760673 |
| 1089 | 2-deoxyribose-5-phosphate aldolase (DeoC) | Conversion of 2-deoxy-D-ribose 5-phosphate to D-glyceraldehyde 3-phosphate and acetaldehyde. Pentose-phosphate pathway | Carbohydrate metabolism, Nucleotide metabolism | 1.07E-04 | 4.22 | gi|320089000 |
| 568 | D-mannonate oxidoreductase (UxuB) | Conversion of D-fructuronate to D-mannonate. Pentose glucuronate interconversion | Carbohydrate metabolism | 4.17E-06 | 4.06 | gi|16761914 |
| 727 | Mannitol-1-phosphate 5-dehydrogenase (MtlD) | Converts mannose-1-phosphate to fructose-6-phosphate | Carbohydrate metabolism | 8.98E-05 | 3.65 | gi|16766971 |
| 613 | Alpha-galactosidase (MelA) | Hydrolysis of terminal, non-reducing alpha-D-galactose residues in alpha-D-galactosides; galactomannans, galactose oligosaccharides and galactolipids. Glycero/sphingolipid metabolism | Lipid metabolism, Carbohydrate metabolism, Anaerobic metabolism | 2.89E-04 | 3.65 | gi|16767548 |
| 585 | Glycerol kinase (GlpK) | Conversion of glycerol to glycerol-3-phosphate, synthesis of triglycerides and glycerophospholipids | Lipid metabolism, Anaerobic metabolism | 2.46E-06 | 9.03 | gi|16762349 |
| 782 | Galactonate dehydratase (DgoD) | Conversion of D-galactonate to 2-dehydro-3-deoxy-D-galactonate, glycero/sphingolipid metabolism | Carbohydrate metabolism | 2.32E-05 | 4.03 | gi|224585626 |
| 678 | Serine hydroxymethyltransferase (GlyA) | Conversion of 5,10-methylenetetrahydrofolate and glycine to tetrahydrofolate and L-serine | Cofactor metabolism | 3.92E-06 | 5.43 | gi|213163620 |
| 1061 | Pyridoxamine kinase (PdxY) | Conversion of pyridoxal (vitamin B6) to pyridoxal 5’-phosphate | Cofactor metabolism | 3.43E-04 | 2.76 | gi|16764798 |
| 1151 | Dihydropteridine reductase (NfnB) | Conversion of 5,6,7,8-tetrahydropteridine to 6,7-dihydropteridine. Folate biosynthesis | Cofactor metabolism | 1.28E-03 | 1.93 | gi|238911529 |
| 1032 | 2-hydroxy-3-oxopropionate reductase (GarR) | Conversion of glycerate to 2-hydroxy-3-oxopropanoate, produces of NADH | Other | 2.39E-03 | 2.43 | gi|25283646 |
| 789 | Glucose-1-phosphatase/inositol phosphatase (Agp) | Hydrolysis of phosphate from α-D-glucose-1-phosphate and 1-D-myo-inositol-hexakisphosphate. Glucose and inositol metabolism | Carbohydrate metabolism | 5.84E-07 | 8.81 | gi|323212400 |
| 37, 38, 240, 254, 262, | Elongation factor G (FusA) | Required for the catalysis of the GTP-dependent translocation step during translation elongation | Transcription, ribosomal structure and translation | 1.54E-05, 2.80E-05, 1.11E-06, 2.38E-04, 9.20E-07, | 6.45, 7.51, 9.05, 5.64, 9.26, | gi|16762837, gi|213586434 |
| 425 | Prolyl-tRNA synthetase (ProS) | Member of the aminoacyl-tRNA synthetases | Transcription, ribosomal structure and translation | 4.75E-05 | 4.79 | gi|162139614 |
| 1127 | Heat shock protein (GrpE) | Prevents aggregation of denatured proteins, in association with dnaK, dnaJ | Stress response | 8.48E-05 | 2.28 | gi|340000338 |
| 1115 | DNA-binding transcriptional regulator (PhoP) | Cytoplasmic regulator of phoP/phoQ, controls the transcription of genes involved in virulence, transport and LPS modification | Stress response, regulation | 8.28E-04 | 3.02 | gi|16764586 |
| 732 | Peptidyl-prolyl cis-trans isomerase (SurA) | Facilitate the proper folding of proteins in the periplasm | Stress response | 1.34E-03 | 3.72 | gi|16759087 |
| 238 | Outermembrane protein assembly factor (YeaT) | Forms a complex with YfgL, YfiO, and NlpB. Involved in outer membrane protein biosynthesis and assembly | Stress response | 2.57E-04 | 2.06 | gi|16763614 |
| 418 | Heat shock protein 90 (HtpG) | Molecular chaperone protein with ATPase activity | Stress response | 2.85E-05 | 4.44 | gi|161615315 |
| 1090 | Osmolarity response regulator (OmpR) | Part of the EnvZ-OmpR system, involved in the regulation of numerous genes including the outer membrane porin genes in response to osmolarity | Transport/Permeability, regulation | 8.48E-05 | 2.00 | gi|15803909 |
| 962 | Phosphate transporter ATP-binding protein (PstB) | Component of the phosphate starvation ABC transporter (pstABCS) | Transport/Permeability | 3.44E-05 | 3.33 | gi|16762475 |
| 1021 | Amine ABC transporter, periplasmic amine-binding protein (YehZ) | Uptake protein for the transport of quaternary amines | Transport/ Permeability | 1.11E-03 | 2.81 | gi|323231218 |
| 902 | Mannose-specific enzyme IIAB (ManX) | Mannose specific component of the phosphotransferase system | Transport/ Permeability, Carbohydrate metabolism | 3.16E-04 | 3.01 | gi|16765171 |
| 456 | Putative ABC transporter, ATP-binding protein (YjjK) | Hydrolyses ATP, coupled to the translocation of a substrate across the membrane. | Transport/Permeability | 3.99E-05 | 3.38 | gi|168262364 |
| PROTEINS DOWN-REGULATED | ||||||
| 606, 607 | Pyruvate kinase (PykF) | Conversion of phosphoenolpyruvate to pyruvate. Pyruvate metabolism | Energy metabolism | 6.86E-04, 2.85E-05 | 450, 534 | gi|213855848, gi|161503526 |
| 906 | Glyceraldehyde-3-phosphate dehydrogenase (GapA) | Conversion of glyceraldehydes-3-phosphate to D-glycerate 1,3-bisphosphate. Glycolysis | Energy metabolism | 7.84E-05, 1.85E-04 | 11.45, 3.84 | gi|16760605, |
| 914 | Phosphate acetyltransferase (Pta) | Conversion of acetyl-CoA and phosphate to CoA and acetyl phosphate. Glycolysis | Energy metabolism | 1.21E-04 | 18.71 | gi|322652083 |
| 965, 946 | Phosphopyruvate hydratase (Eno) | Conversion of 2-phospho-D-glycerate to phosphoenolpyruvate. Glycolysis | Energy metabolism | 1.65E-06, 3.16E-08 | 5.91, 16.11 | gi|297521596 |
| 911 | Phosphofructokinase (FruK) | Conversion of D-fructose-6-phosphate to D-fructose 1,6-bisphosphate | Energy metabolism | 1.63E-04 | 4.75 | gi|16761146 |
| 970, 1159 | Phosphoglycerate kinase (Pgk) | Conversion of 3-phospho-D-glycerate to 3-phsopho-D-glyceroyl phosphate. Glycolysis | Energy metabolism | 5.62E-06, 4.22E-08 | 17.35, 10.75 | gi|224584862 |
| 1265 | Acetate kinase (AckA) | Conversion of acetate to acetyl phosphate. Pyruvate metabolism | Energy metabolism | 6.88E-05 | 44.55 | gi|213581081 |
| 713 | Fumarate reductase, flavoprotein subunit (FrdA) | Conversion of succinate to fumarate | Anaerobic metabolism, Energy metabolism | 2.07E-06 | 9.62 | gi|326626034 |
| 641 | Bifunctional acetaldehyde CoA/alcohol dehydrogenase (AdhE) | Reduction of Acetyl-CoA to acetaldehyde and then to ethanol | Anaerobic metabolism, Energy metabolism | 2.43E-04 | 4.40 | gi|323270918 |
| 778 | F0F1 ATP synthase beta subunit (AtpD) | Beta subunit of membrane-bound ATP synthase | Energy metabolism | 5.11E-06 | 12.54 | gi|112791348 |
| 1076 | F0F1 ATP synthase subunit alpha (AtpA) | Alpha subunit of membrane-bound ATP synthase | Energy metabolism | 2.42E-06 | 10.14 | gi|213852796 |
| 1111, 1263 | Uridine phosphorylase (Udp) | Conversion of uridine and phosphate to uracil and alpha-D-ribose 1-phosphate | Nucleotide metabolism | 1.70E-06, 4.48E-05 | 6.40, 9.38 | gi|158428692, gi|213421695 |
| 1191 | Uracil phosphoribosyltransferase (Upp) | Conversion of UMP and diphosphate to uracil and 5-phospho-alpha-D-ribose 1-diphosphate | Nucleotide metabolism | 5.56E-05 | 2.33 | gi|213419931 |
| 731 | Inosine 5′-monophosphate dehydrogenase (GuaB) | Conversion of inosine monophosphate to xanthosine 5′-phosphate | Nucleotide metabolism | 1.98E-03 | 1.95 | gi|16765831 |
| 794 | Fructose 1, 6-bisphosphatase II (GlpX) | Conversion of D-fructose 1,6-bisphosphate to D-fructose 6-phosphate. Mannose/Fructose metabolism | Carbohydrate metabolism | 3.23E-05 | 13.86 | gi|16767351 |
| 929 | Transaldolase B (TalB) | Conversion of sedoheptulose 7-phosphate and D-glyceraldehyde 3-phosphate to D-erythrose 4-phosphate and D-fructose 6-phosphate | Carbohydrate metabolism | 2.46E-05 | 8.87 | gi|16759000 |
| 604 | Lysine decarboxylase (CadA) | Role in pH homeostasis. Conversion L-Lysine to cadaverine and carbon dioxide | Amino acid/peptide transport/metabolism | 3.90E-04 | 6.23 | gi|213053291 |
| 636, 1119 | Aspartate ammonia-lyase (AspA) | Conversion of L-aspartate to fumarate and ammonia | Amino acid/ peptide transport/ metabolism | 4.83E-04, 3.27E-08 | 7.71, 12.42 | gi|213582259, gi|167553587 |
| 1121 | Aminoacyl-histidine dipeptidase (PepD) | Metabolism of glutathione and amino acids | Amino acid/peptide transport/metabolism | 1.11E-03 | 2.81 | gi|16763698 |
| 1262 | Enoyl-(acyl carrier protein) reductase (FabI) | An NADH-dependent trans-2-enoyl-ACP reductase. A key regulator of fatty acid biosynthesis | Lipid metabolism | 5.25E-07 | 16.31 | gi|16765044 |
| 1225,1323 | Inorganic pyrophosphatase (Ppa) | Conversion of one molecule of pyrophosphate to two phosphate ions | Other | 1.46E-05, 3.49E-06 | 1.61, 10.38 | gi|16763234 |
| 113, 144 | DNA directed RNA polymerase subunit beta (RpoB) | Component of DNA-dependent RNA polymerase- catalyses the transcription of DNA into RNA | Transcription, ribosomal structure and translation | 5.49E-04, 4.81E-04 | 21.19, 10.59 | gi|213161404 |
| 797 | DNA-directed RNA polymerase subunit alpha (RpoA) | Component of DNA-dependent RNA polymerase- catalyses the transcription of DNA into RNA | Transcription, ribosomal structure and translation | 9.66E-05 | 2.42 | gi|293393287 |
| 957 | Methionine aminopeptidase (Map) | Removes the N-terminal methionine from nascent proteins, release of N-terminal amino-acids | Transcription, ribosomal structure and translation | 4.72E-05 | 2.19 | gi|16759205 |
| 748 | GTP-dependent nucleic acid-binding protein (YchF) | May act as a translation factor | Transcription, ribosomal structure and translation | 5.47E-05 | 2.00 | gi|16759205 |
| 705, 708, 918, 1166, | 30S ribosomal protein S1 (RpsA) | Structural ribosomal protein | Transcription, ribosomal structure and translation | 1.62E-08, 1.55E-07, 7.64E-05, 3.05E-05, | 15.82, 42.48, 8.12, 17.03 | gi|213418378, gi|213027002, gi|213021855 |
| 1321 | 30S ribosomal protein S2 (RpsB) | Structural ribosomal protein | Transcription, ribosomal structure and translation | 2.79E-07 | 40.92 | gi|16759206 |
| 488 | Translation initiation factor IF-2 (InfB) | Promotes the GTP-dependent binding of the initiator tRNA to the small subunit of the ribosome | Transcription, ribosomal structure and translation | 8.57E-06 | 11.34 | gi|213423445 |
| 1149, 1336 | Elongation factor Ts (Tsf) | Catalyses the release of guanosine diphosphate from EF-Tu | Transcription, ribosomal structure and translation | 1.69E-07, 6.07E-06 | 13.38, 25.60 | gi|213028021, gi|213618628 |
| 265, 353, 386, 387 | Elongation factor G (FusA) | Catalyses the translocation of the tRNA and mRNA down the ribosome at the end of each round of polypeptide elongation | Transcription, ribosomal structure and translation | 1.70E-04, 1.48E-04, 1.42E-05, 6.48E-05 | 6.59, 8.14, 6.37, 4.12 | gi|16762837, gi|213416464 |
| 1028, 1064 | Elongation protein Tu (TufB) | Mediates the entry of aminoacyl-tRNA into a free site on the ribosome | Transcription, ribosomal structure and translation | 1.96E-03, 5.49E-06 | 19.62, 7.71 | gi|315253052 |
| 1082 | Putative elongation factor (YeiP) | Facilitates translation | Transcription, ribosomal structure and translation | 2.78E-07 | 16.87 | gi|56127134 |
| 887 | Lysyl-tRNA synthetase (LysS) | Member of the aminoacyl-tRNA synthetases | Transcription, ribosomal structure and translation | 1.53E-04 | 6.25 | gi|16766341 |
| 453 | Molecular chaperone (DnaK) | Involved in DNA replication and in response to hyperosmotic shock | Stress response | 9.19E-06 | 8.07 | gi|161504855 |
| 440, 1005 | High temperature protein G (HtpG) | Molecular chaperone protein with ATPase activity | Stress response | 5.56E-05, 2.67E-04 | 21.70, 75.16 | gi|320084762, gi|213022746 |
| 1074 | ATP dependent protease binding subunit (ClpB) | Component of Clp protease | Stress response | 7.20E-04 | 7.17 | gi|4102206 |
| 728 | ATP-dependent protease ATP-binding subunit (HslU) | ATPase subunit of a protease degradation complex, ATP binding and hydrolysis component | Stress response | 1.33E-08 | 10.93 | gi|213051554 |
| 753, 757, 1247 | Phosphoenolpyruvate-protein phosphotransferase (PtsI) | Component of the PTS system. Transport and conversion of phosphoenolpyruvate to pyruvate | Transport/Permeability, energy metabolism | 1.73E-07, 1.16E-05, 6.10E-07 | 39.30, 29.51, 20.87 | gi|213051554, gi|213586857 |
| 947 | Flagellar hook-associated protein (FlgL) | Role in flagellar biosynthesis, hook associated protein | Flagella and chemotaxis | 3.48E-04 | 5.31 | gi|56413829 |
Functional categories assigned according to KEGG.
Fc, fold change. (see also Figure 8).
Figure 9.
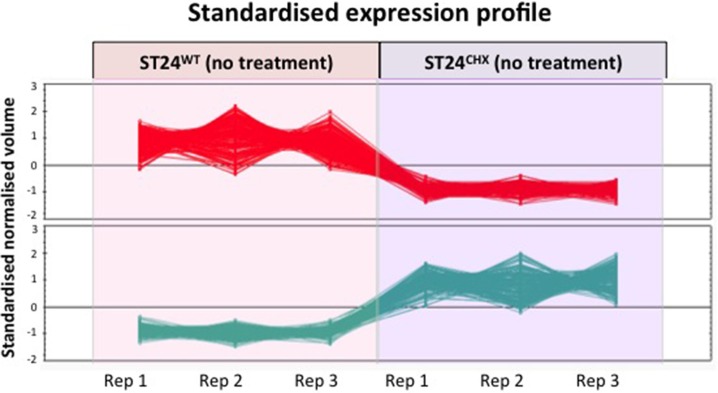
Differential expression of proteins between the reference strain ST24WT and mutant ST24CHX. Profiles indicated in red represent down-regulated proteins in ST24CHX relative to ST24WT, those in green are up-regulated. The data points represent the normalized expression level of individual protein spots (differentially expressed between the wild-type and the mutant), identified using Progenesis SameSpots software (© Nonlinear Dynamics Ltd., Newcastle, UK). The various replicates of the same protein spot, between the reference and the mutant are linked with lines.
Of the down-regulated proteins a total of 57 were identified with statistical significance and these corresponded to 39 proteins (Table 2, Tables S4, S5). The latter could be divided into four functional categories including; general cell metabolism (33 proteins), stress response (4 proteins), motility (1 protein) and transport and permeability (1 protein). Down-regulated proteins are indicated in red in Figure 8, denoted by red traces in Figure 9 and categorized, as before in Table 2.
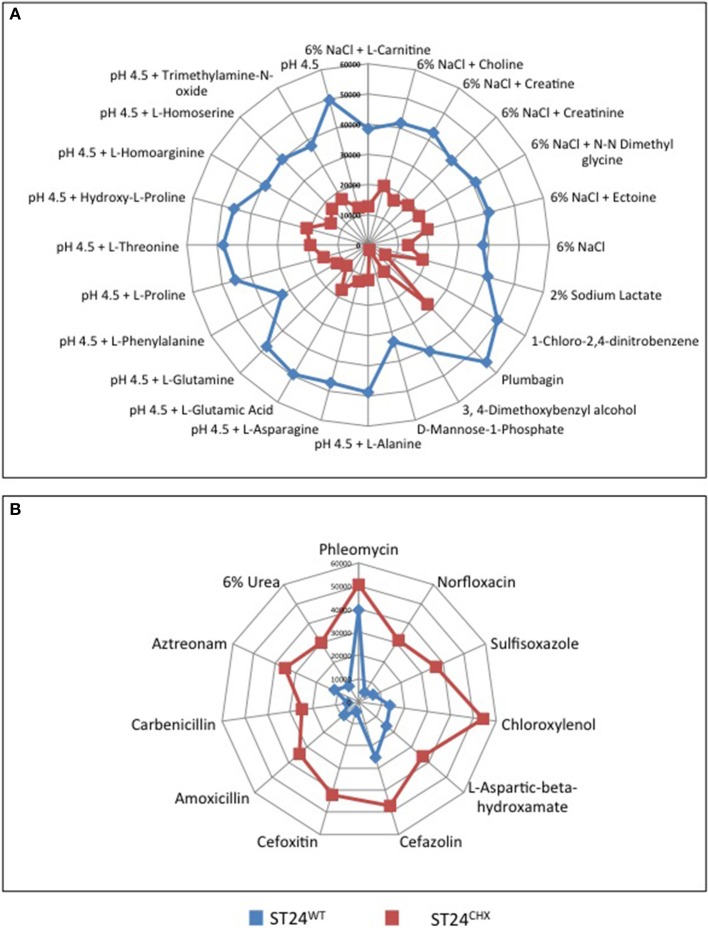
Phenotypes expressed by ST24WT and ST24CHX were compared, using the Omnilog™ phenotypic microarray platform. No significant differences in the growth rates of ST24WT and ST24CHX were detected in MH broth or in the Omnilog™ inoculation fluid (data not shown). Data from the phenotypic microarray showed, however, that alterations in the respiration of the chlorhexidine tolerant ST24CHX, relative to susceptible ST24WT, were apparent when grown on a variety of substrates and in the presence of antimicrobial compounds. All of these are listed in Table S6 and displayed as a series of heatmaps in Figures S1A and B. An increased respiration rate was also noted for ST24CHX, indicating enhanced tolerance, to 5 cell wall damaging agents, 2 DNA damaging agents, a folate synthesis inhibitor, and a glutamate up-take inhibitor (Figure 10). Conversely, ST24CHX was found to be more susceptible to 19 osmolytes (Figure 10), 16 cell wall/cell membrane damaging agents, 9 protein synthesis inhibitors, 6 respiration un-couplers, 3 oxidizing agents, and 11 agents causing damage to DNA or inhibiting DNA synthesis, replication or transcription, when compared to ST24WT.
Figure 10.
Metabolic differences that distinguish the sensitive reference ST24WT, from the chlorhexidine tolerant mutant ST24CHX on (A) osmolytes where enhanced respiration was recorded in the reference strain and (B) substrates where enhanced respiration was recorded in the mutant isolate. The blue lines represent the metabolism of ST24WT in Omnilog units, whilst the red lines represent the metabolism of ST24CHX. The center of the graph (0 Omnilog units) equates to no metabolic activity. The periphery of the graph (60,000 Omnilog units) equates to maximum metabolic activity. Each spoke of the graph represents a different compound with the box on each spoke indicating the metabolic activity of ST24WT/ST24CHX in the presence of that particular compound.
Modifications in general cell metabolism comparing ST24WT and ST24CHX
Relative to ST24WT the chlorhexidine tolerant mutant ST24CHX displayed multiple alterations in the general cell metabolism, at both the transcriptomic and proteomic levels (Encheva et al., 2009). The alterations were independent of chlorhexidine exposure (detected in ST24CHX with and without exposure to the same agent). These metabolic modifications were similar to those detected in ST24WT following chlorhexidine exposure, described above. However, metabolic changes in ST24CHX were more wide ranging (Figure 5B).
An increase was detected in ST24CHX in the level of pyruvate formate lyase. This enzyme catalyzes the non-oxidative fermentation of pyruvate to formate and acetyl-CoA, thus providing the main substrate for mixed acid fermentations. Additional up-regulation in multiple enzymes involved in mixed acid fermentation was observed in the mutant isolate including an alcohol/aldehyde dehydrogenase enzyme, which converts acetyl-coA to acetaldehyde and then subsequently to ethanol. Additionally, there was a mutation in a putative formate transporter (Table 1). Furthermore, the anaerobic dimethyl sulfoxide (DMSO) reductase (dmsA) was up-regulated, an enzyme, which works as part of an electron transport chain along with, formate dehydrogenase. An increase was also noted in the expression of succinate dehydrogenase, which catalyzes the conversion of fumarate to succinate under anaerobic conditions. The genomic comparison identified a non-synonymous polymorphism in the α-subunit of oxaloacetate decarboxylase, an enzyme involved in the anaerobic citrate fermentation, a pathway that converts citrate to acetate, formate and CO2 (Bott, 1997). Multiple genes associated with propanediol utilization were also up-regulated in the mutant isolate, with and without chlorhexidine treatment (Table S1).
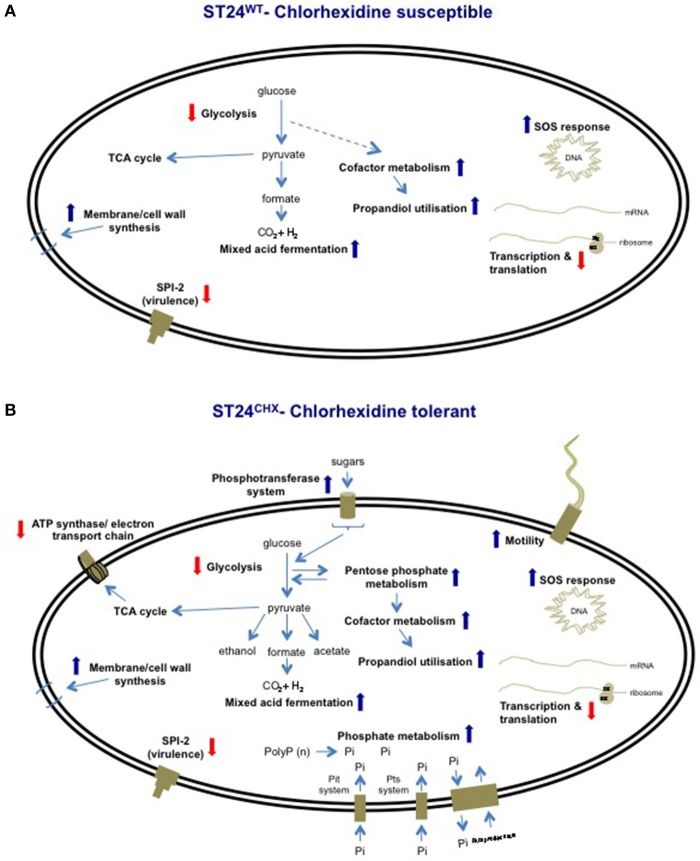
Given these observations, we hypothesize that chlorhexidine targets component/(s) of the respiratory chain. This phenotype could be expected to result in a reduction in oxygen consumption concomitant with a decrease in the proton motive force (PMF). This scenario would in turn result in reduced ATP production, which could be expected to have a pleiotropic effect leading, for example, to a reduction in nucleic acid and protein synthesis and ultimately the disruption of the structural integrity of the bacterial membrane. However, these detrimental effects may be circumvented should the bacterial cell divert the electron flow through the reduction of organic acids as described above. A similar situation has been previously reported where the oxidation of formate, catalyzed by formate dehydrogenase and succinate, by succinate dehydrogenase, afforded E. coli and Salmonella some protection from a cationic-based antimicrobial peptide (Barker et al., 2000). More recently, Cheung et al. reported that following chlorhexidine exposure, two enzymes involved in mixed acid fermentation, succinate dehydrogenase and lactate dehydrogenase, were both up-regulated in E. coli (Cheung et al., 2012), a feature that would appear to support our hypothesis. Nde et al. similarly found that chlorhexidine exposure in P. aeruginosa resulted in the down-regulation of several enzymes involved in oxidative phosphorylation and the up-regulation of enzymes involved in anaerobic metabolism, suggesting a dynamic shift in metabolic activity toward an anoxic profile. Nde and colleagues concluded that chlorhexidine suppressed energy metabolism via aerobic respiration.
In support of this hypothesis, an increase was observed in the expression of pyruvate dehydrogenase (quinone) at the transcriptional level in the chlorhexidine tolerant ST24CHX, with and without chlorhexidine treatment, relative to ST24WT. This enzyme catalyzes the conversion of pyruvate to acetate and, importantly, is coupled to the electron transport chain at the level of ubiquinone, in the terminal electron transport system. An increase in activity was similarly detected for the NADH dehydrogenase complex I, an enzyme involved in the anaerobic metabolism of fumarate. Several enzymes linked with glycerolipid metabolism, including the β-subunit of the anaerobic glycerol-3-phosphate dehydrogenase described above, were up-regulated in ST24CHX. This pathway shuttles electrons to the terminal electron transport chain at the level of succinate dehydrogenase. Enzymes in this pathway were up-regulated at both the transcriptomic and proteomic levels in ST24CHX (Figures S2A and B). These alterations in electron flow in ST24CHX were also reflected at the level of its metabolism when analyzed by the phenotypic microarray; wherein the mutant displayed an increased susceptibility to 6 respiration un-couplers (Figure S1 and Table S6).
Consistent with altered metabolism, the differential expression of genes involved in the utilization of other carbon sources were noted in ST24CHX relative to ST24WT, independent of chlorhexidine exposure (Table 2, Table S1). At both the RNA and protein levels, several transporters for different sugars, along with enzymes for their conversion to intermediates of the glycolytic pathway, were up-regulated in the mutant, including those for mannose, mannitol, 1-D-myo-inositol-4-phosphate, sorbitol and galactose (Table 2, Table S1). Carbon and electron flow within glycolysis and pyruvate metabolism was also altered with an increase in some enzymes and a decrease in others (Table 2, Table S1 and Figure S3). Interestingly, there was a marked increase in transketolase expression together with other enzymes of the pentose phosphate pathway (PPP) and pentose-glucuronate interconversions in ST24CHX. However, a decrease in transaldolase expression was noted (Table 2, Table S1). This feature suggested that the flow of carbon reducing power was away from glycolysis, being shuttled instead toward the PPP. Within this pathway, enzymes for the synthesis of the cofactor phosphoribosyl pyrophosphate (PRPP) were up-regulated (Table 2, Table S1 and Figure S4).
Based on the transcriptomic and proteomic data, several global regulators that were up-regulated in the mutant relative to ST24WT may account for at least some of the transcriptomic, proteomic and phenotypic changes observed. As an example, up-regulation of creB was noted in ST24CHX. CreB is thought to be a global regulator (Avison et al., 2001), and its expression is induced by the fermentation of carbon sources through glycolysis and anaerobic reactions, similar to those described above (Cariss et al., 2008). It has been demonstrated that CreB can control the expression of talA, an enzyme associated with the shuttling of glyceraldehyde-3-phophate away from glycolysis to the non-oxidative PPP (Avison et al., 2001). This is consistent with the up-regulation of enzymes associated with the PPP pathway noted in our study. Furthermore, CreB is also known to positively regulate phosphate acetyltransferase (pta), which was also up-regulated in ST24CHX at the transcriptomic and proteomic levels, along with acetate kinase (ackA), which was up-regulated at the protein level. Both of these enzymes are required for efficient carbon flux to support growth using sugars transported via the phosphotransferase system (PTS) (Avison et al., 2001), many of which were up-regulated in ST24CHX (Table 2).
The ferric up-take regulator (fur) was up-regulated in the mutant, compared with its wild-type progenitor. Fur is involved in metal-dependent transcriptional regulation in response to different metal concentrations and the redox state of the bacterial cell (Escolar et al., 1999; Troxell et al., 2011). Several cellular targets known to be positively regulated by Fur displayed an increased expression in ST24CHX, including those involved in anaerobic metabolism, such as dimethyl sulfoxide reductase (dmsA), which was up-regulated at both the RNA and protein levels, and those involved in glycerol/glycerolipid metabolism (glpAB), which were up-regulated at the RNA level. Decreased expression of cellular targets known to the negatively regulated by Fur, including those involved in glycolysis was also noted; for example, phosphofructokinase (pfkA), which was detected as down-regulated at the RNA and protein levels. It is reasonable to hypothesize that both Fur and CreB are important regulators of the chlorhexidine tolerant phenotype described in this study. Current work involving knock-out studies is on-going in order to further elucidate the possible role of these regulators in the chlorhexidine tolerance response.
Modifications in membrane structural and transport systems between ST24WT and ST24CHX
Relative to ST24WT the chlorhexidine tolerant mutant ST24CHX displayed multiple alterations in peptidoglycan synthesis/cross-linking, independent of chlorhexidine exposure. These alterations were similar to those detected in ST24WT following a 30 min sub-lethal chlorhexidine exposure, described earlier. However, modifications were more extensive in ST24CHX, with additional markers being detected. These included the up-regulation of the peptidoglycan synthetic gene murF and those involved in cross-linking; ppbC, pbpG. A reduction in expression was noted for loci encoding component parts of the LPS structure, such as rpfU, rfbF, and rfbH. Conversely, increased expression was observed in 2 enzymes associated with LPS biosynthesis, namely kdsA, which plays a key role in LPS biosynthesis and lpxK which catalyses the transfer of a phosphate from ATP to the 4′-position of the lipid A disaccharide (Table S1). A SNP was detected in the lptC-encoding gene, a component of a LPS export system (Tran et al., 2010). A mutation in this gene was previously found to impart a reduced susceptibility to the bile salt sodium deoxycholate in Salmonella (Hernández et al., 2012).
Based on the data obtained in this study, extensive modifications in the expression of genes/proteins associated with permeability were noted, suggesting modifications to the membrane function as important determinants of tolerance. These membrane alterations were similarly reflected in the data obtained from the phenotypic microarray. Some of the modifications noted in the membrane/cell wall structure of the mutant correlated with an increased sensitivity to 20 osmolytes (Figure 10A, Figure S1A and Table S6B). Similarly, the mutant exhibited an increased susceptibility to cell wall disrupting agents, including vancomycin and three β-lactam antimicrobials, along with 12 other membrane active agents, including colistin, a receptor for which was up-regulated (Figure S1B and Table S6B). Conversely, modifications in the cell wall also correlated with an increased tolerance to 5 clinically important, cell wall damaging agents, including 2 cephalosporins and 2 β-lactams (Figure 10B, Figure S1B and Table S6A). Earlier data reported the susceptible profile of ST24WT to b-lactam and cephalosporin antibiotics (Condell et al., 2012a). Based on these observations, it is tempting to speculate that the nature of the diverse phenotype reported here (Figure 10B and Tables S6A,B) may be reflective of changes occurring at the level of the bacterial membrane, in response to the stress imposed following exposure to chlorhexidine.
Efflux pump activity comparisons between ST24WT and ST24CHX
Similar efflux expression patterns were noted in ST24CHX and ST24WT, as described above (Table S1). In this case, a reduction in the expression of acrB and acrF, components of the AcrAB-TolC and AcrEF-TolC tripartite efflux systems (Zheng et al., 2009) was recorded. Interestingly, a mutation in ramR was also identified in ST24CHX (Table 1). RamR acts as a repressor of ramA (O'Regan et al., 2009), a known global transcriptional regulator of these latter efflux systems (Zheng et al., 2009). Mutations in ramR have previously been documented and these are associated with a de-repression of ramA and consequently the up-regulation of the aforementioned efflux systems (Abouzeed et al., 2008; O'Regan et al., 2009). However, in this case, it is possible that a mutation in ramR noted in ST24CHX, and which occurred within a helix-turn-helix motif, resulted in a reduction in ramA expression and, as a consequence, lowered the expression of both efflux systems. Moreover, in ST24CHX up-regulation in the expression of emrR, a negative regulator of the efflux system emrAB-tolC (Tanabe et al., 2009) was observed (Table S1) and this may lead to a more pronounced repression of this system. Similarly, there was also a decrease in the expression of the multi-drug efflux associated gene ydhE.
In respect of other efflux systems in ST24CHX, members of the major facilitator superfamily (MFS family) including bcr and smvA (Paulsen and Brown, 1996) and the ABC superfamily multi-drug transporter family; mdlB (Sulavik et al., 2001) were up-regulated at the transcriptional level (Table S1). Precisely how efflux systems might play a role in the susceptibility of bacteria toward chlorhexidine remains to be elucidated.
SOS response comparisons between ST24WT and ST24CHX
Relative to ST24WT the chlorhexidine tolerant mutant ST24CHX displayed multiple alterations in the SOS response, independent of chlorhexidine exposure. Adaptations noted were similar to those detected in ST24WT following chlorhexidine exposure. These modifications were more extensive in ST24CHX. An increased expression of 12 genes associated with DNA damage repair was observed in ST24CHX, relative to ST24WT (Table S1). The up-regulated genes included those associated with the SOS response-DNA damage repair system, consisting of recR, recO, recG, uvrA, uvrD, and ruvB (Janion, 2008), indicating chlorhexidine tolerance was associated with an induction of this repair system. This alteration in DNA metabolism in the mutant was further supported by data from the phenotypic microarray. Chlorhexidine tolerance correlated with a decrease in susceptibility to 2 DNA damaging agents (Figure S1B and Table S6). The hypothesis that an induction of the DNA repair systems along with DNA binding proteins might represent the bacterial response to mitigate the DNA cross-linking effects of chlorhexidine appears to hold with regards ST24CHX. However, the specific DNA defense system induced in response to chlorhexidine correlated with a reduced tolerance to other DNA damaging agents; an increase in susceptibility to 10 such agents was detected from the phenotypic microarray (Figure S1B and Table S6).
A comparison of transcription and translation between ST24WT and ST24CHX
As was discussed above, ST24WT in response to chlorhexidine exposure showed a decrease in the expression of transcription and translation associated genes and proteins. Similarly a decrease in the expression of genes associated with these cellular functions was noted in ST24CHX, relative to ST24WT, in this case the decrease was independent of chlorhexidine exposure (Table 2, Table S1). Reduced expression of rpoA at the RNA and protein levels were recorded, along with a reduction in the expression of the β-encoding sub-unit (rpoB) at the protein level. Furthermore, some 17 ribosomal structural genes were down-regulated, and 2 of these were also affected at the protein level. The ribosomal maturation factor (encoded by yfiA) and several translation factors including those for translation initiation and elongation were down-regulated (Table 2, Table S1). Two SNPs were detected in the mutant at 2 loci coding for ribosomal RNA (rRNA); 1 coding for a 23S rRNA and the other for a 16S rRNA. There were also 2 SNPs detected at 35 and 37 bp up-stream of a 16S rRNA, and these polymorphisms may have an effect on transcription (Table 1).
Mutations associated with changes in the expression of components of the cellular transcription and translation machinery, were reflected in the metabolism recorded from the phenotypic microarray data. ST24CHX displayed an increased tolerance to an agent affecting protein synthesis, namely L-aspartic-beta-hydroxamate and an increased sensitivity to rifampicin an inhibitor of RNA polymerase, along with seven inhibitors of protein synthesis, including drugs of the macrolide, polyketide and aminoglycoside classes (Figure S1B and Table S6). These results further support the hypothesis that chlorhexidine affects bacterial protein synthesis as a component of its mechanism of action, as previously described by Nde et al. (2009).
Comparison of the virulence features of ST24WT and ST24CHX
ST24WT following exposure to chlorhexidine exhibited a decrease in the expression of virulence associated genes. Similarly a decrease in the expression of virulence genes was noted in ST24CHX, relative to ST24WT, once more in this case the decrease was independent of chlorhexidine exposure. A down-regulation was noted in 12% of the virulence genes listed from KEGG, comprising SPI-1 through -5 in ST24CHX relative to ST24WT (Figures 2, 7). Two mutations in skiK1, a gene coding for a SPI-6 associated component of a type VI secretory system, were detected in the mutant isolate, along with a mutation in a second SPI-6 gene; coding for a protein of the Rhs family, and a mutation in a SPI-1 associated regulator protein (avrA). These results further indicate a relationship between chlorhexidine and a reduction in virulence gene expression. Further work confirmed a significantly attenuated virulence potential in ST24CHX, and a decrease in survival in THP-1 macrophages compared with ST24WT was also noted (data not shown). Previous work demonstrated a relationship between central metabolism, the TCA cycle and the intracellular survival of S. Typhimurium (Bowden et al., 2010). It is possible that the reduced survival of ST24CHX is linked with the metabolic modifications it displayed.
Altered phosphate metabolism in ST24CHX
Changes in phosphate metabolism were detected in ST24CHX, but not in ST24WT after sub-lethal chlorhexidine exposure. Inorganic phosphate is an essential element required for the phosphorylation of nucleic acids, lipids, sugars, proteins and for cell signaling. Salmonella species possess two major systems which function in the up-take of inorganic phosphate; the PTS and PIT systems. Both exhibited altered levels of expression in ST24CHX relative to the reference isolate. The ptsABCS system codes for a phosphate starvation ABC transporter that is induced under conditions where limited phosphate exists. In the mutant, this transporter was up-regulated based on the protein data (Table 2). Additionally, a SNP was detected in the PIT system in ST24CHX which introduced a premature stop codon (TGG-TAG) in the distal part of pitA (Table 1). PitA is a low affinity transporter thought to be constitutively expressed in Salmonella (Jackson et al., 2008). These features may indicate an increase in phosphate up-take in the mutant isolate. A mutation was also noted in a phosphate-linked antiporter (pgtP), which functions in the low affinity exchange of cytoplasmic inorganic phosphate for external phosphoglycerate (Varadhachary and Maloney, 1991). A down-regulation of two genes known to induce the expression of the latter antiporter, pgtC and pgtB, was also previously reported (Yang et al., 1988). The abundance of the inorganic pyrophosphatase enzyme (encoded by the ppa gene) was reduced in the mutant isolate (Table 2). These features suggest that an extensive change in phosphate metabolism had occurred in the mutant. Chlorhexidine is known to associate strongly with phosphate ions contained within the LPS (Tattawasart et al., 2000), and this biocide can form cytoplasmic complexes with phosphate containing moieties, which may lead to the precipitation of cytoplasmic contents (Cheung et al., 2012). Alterations in phosphate transport and its subsequent metabolism in the mutant appear to reflect these biocide-induced modifications following in vitro selection, wherein the cell may be acting to limit chlorhexidine adsorption to phosphate groups at the bacterial outer surface and consequently reduce the susceptibility of the bacterium to the biocide.
Altered flagella gene regulation in ST24CHX
Modifications in flagella-mediated motility are an important factor in environmental adaptation and in adherence to various surfaces. Up-regulation of a number of flagella structural genes were detected in the ST24CHX alone, independent of exposure to chlorhexidine, relative to ST24WT. These included flagellar hook-base body genes; flgA, flgC, flgF, flgG, flgH, flgI, fliF, fliH, fill, fliM, fliO (Table S1). The regulatory modification resulting in this up-regulation may be related to two SNPs detected in ST24CHX one of which was located in the SL1344_4465 gene, a hypothetical transcriptional regulator of the NtrC family that may activate RpoN (σ54) (Table 1). σ54 in turn acts as a positive regulator of σ28 (fliA) (Dong et al., 2011). The action of σ28 is counteracted by an anti-σ28 sigma-factor, FlgM, which was down-regulated in the ST24CHX mutant. This decreased expression may result in the overexpression of σ28, a phenomenon previously shown to result in increased expression of all flagellar genes of the temporally regulated middle gene promoters (Chilcott and Hughes, 2000). Based on our transcriptomic data, the genes identified that were up-regulated were all flagellar-middle genes. An increased swarm motility phenotype was also observed in the ST24CHX, compared to ST24WT, in the presence of low levels of chlorhexidine (data not shown). An up-regulation in the expression of the flagellar apparatus has previously been associated with a tolerance to antimicrobial agents in Salmonella (Kim et al., 2003; Kim and Surette, 2003; Butler et al., 2010) and may now be extended to chlorhexidine tolerance.
Metabolic defense network elicited by exposure of Salmonella to chlorhexidine
Based on the observations outlined above describing alterations in expression of genes and proteins involved in general cell metabolism, membrane structure, efflux, the SOS-response, transcription and translation and virulence, it may be reasonable to hypothesize that exposure of ST24WT to chlorhexidine elicits a distinct cellular response. Furthermore, these data point to the fact that chlorhexidine tolerance in ST24CHX was also associated with broad cellular alterations; the mutant isolate demonstrated changes in general cell metabolism, membrane structure, efflux, the SOS-response, transcription and translation, virulence, phosphate metabolism and motility. Overlaps were noted between the alterations seen in ST24WT following chlorhexidine exposure and changes in the geno-/phenotype in ST24CHX relative to ST24WT, independent of chlorhexidine exposure. These features were suggestive of a general chlorhexidine defense network (Figure 11). Based on the transcriptomic profiling a common response was observed for 165 genes; wherein 51 were up-regulated and 114 genes were down-regulated in both isolates (Figure 2). Furthermore, the distribution of chlorhexidine-regulated genes between the functional categories was similar for both (Figure 3). The proposed network involved multiple components, which produced changes in the activity of several metabolic pathways, shifting toward an anoxic profile, a feature designed to mitigate the oxygen inhibitory effects of chlorhexidine. Moreover a common up-regulation in the SOS-response was observed and this may be part of the bacterial response to the DNA damaging effects of this agent. Similar modifications in protein systhesis and bacterial cell wall synthesis were observed, all designed to reduce the antimicrobial effects of chlorhexidine. Finally a decrease in the expression of virulence associated cell targets along with those associated with transcription and translation was detected in both isolates (Figure 11).
Figure 11.
Overview of cellular process altered by chlorhexidine, relative to the chlorhexidine-sensitive ST24WT, in (A) the same isolate following chlorhexidine exposure and (B) in the chlorhexidine tolerant ST24CHX, with and without chlorhexidine exposure. Red arrowheads shown signify reduced expression and blue arrowheads signify increased expression.
Furthermore, data from this study showed that the increased chlorhexidine tolerance of ST24CHX was associated with more extensive alterations of the same components involved in this proposed defense network, when all of the functional categories were considered; relative to ST24WT(Figure 11). Interestingly, ST24CHX also elaborated some unique responses including modifications in phosphate metabolism and alterations in the expression of chemotaxis and mobility associated genes that may contribute to the increased chlorhexidine tolerance of the mutant.
Conclusions
This study investigated the bacterial response to chlorhexidine exposure at a number of biochemical levels in the bacterial cell. Results show that chlorhexidine elicits a broad range of effects on Salmonella, with an impact on central cellular processes including aerobic energy production and protein synthesis. This chlorhexidine defense network was conserved between the isogenic bacteria studied (Figure 11). Although ST24CHX elaborated additional modifications, which extended the proposed defense network, a divergent defense response involving the up-regulation of additional targets was noted which may be mediated by CreB and Fur (Figure 11).
Results from this study when compared to those of Condell et al. (2012b) suggest that the number of cellular alterations and the ease at which a tolerant phenotype to an active biocide develops can differ hugely depending on the mechanism of action of the agent(s). A tolerant phenotype to the biocidal agent triclosan developed at a faster rate compared with the emergence of chlorhexidine tolerance (Condell et al., 2012a). The numbers of alterations in the proteome of the triclosan tolerant isolate, compared with its susceptible progenitor were fewer, at 33, compared to the 470 proteins differentially expressed between ST24WT and ST24CHX. These differences in the number of alterations may reflect the mechanism of action of the two agents; triclosan has a cellular target, FabI, whereas chlorhexidine appears to have a broad spectrum mechanism.
Investigations of this nature are important in an effort to extend our understanding of how biocides act to eliminate bacteria, and the mechanisms by which a tolerance could develop. Such data are often lacking, even for the most commonly used biocides, and this information could assist the future refining and optimization of active formulations, to overcome potential tolerance mechanisms thereby improving biosecurity measures.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the financial support provided by the Food Institutional Research Measure (FIRM), administered by the Department of Agriculture, Food and the Marine (grant number: 08/RD/TAFRC/616) and Dr. Caitriona Scaife for her assistance with the Typhoon™ variable mode imager and Progenesis SameSpots software (Protein Separations Laboratory, Conway Institute, University College Dublin).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00373/abstract
(A) Comparative respiration of ST24WT, and the chlorhexidine tolerant mutant, ST24CHX, on PM plates 1–10, containing energy sources and osmolytes. Substrates where a significant difference in the respiration between ST24WT and ST24CHX was recorded are indicated with arrows and details are provided in Table S6. (B) Comparative respiration of ST24WT and the chlorhexidine tolerant mutant, ST24CHX, on PM plates 11–20, containing antimicrobial compounds. Compounds where a significant difference in the respiration between ST24WT and ST24CHX were recorded, indicating a possible alteration in susceptibility, are indicated with arrows and details are provided in Table S6.
Relative expression of enzymes that were determined to be differentially expressed and associated with (A) glycerolipid metabolism in the reference isolate ST24WT. (B) Glycerolipid metabolism in the mutant ST24CHX, (C) amino acid metabolism in the reference isolate ST24WT, and (D) amino acid metabolism in the mutant ST24CHX. Enzyme names, descriptions, gene identifiers and fold-change values are given in Table 2, Tables S1, S4. Color blocks without a white diagonal line represent gene fold change values (Table S1), color blocks containing a white diagonal line represent protein fold change values (Table 2, Table S4).
Relative expression of enzymes determined as differentially expressed and associated with carbohydrate metabolism in (A) the reference isolate ST24WT and (B) the isogenic mutant ST24CHX. Enzyme names, descriptions, gene identifiers and fold-change values are given in Table 2, Tables S1, S4. Color blocks without a white diagonal line represent gene fold change values (Table S1), color blocks containing a white diagonal line represent protein fold change values (Table 2, Table S4).
Relative expression of enzymes determined as differentially expressed and associated with carbohydrate metabolism; the pentose phosphate and pentose glucuronate pathways in (A) the reference isolate ST24WT and (B) the mutant isolate ST24CHX.
References
- Abouzeed Y. M., Baucheron S., Cloeckaert A. (2008). ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52, 2428–2434 10.1128/AAC.00084-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M. J., White G. F., Morby A. P. (2006). The response of Escherichia coli to exposure to the biocide polyhexamethylene biguanide. Microbiology 152, 989–1000 10.1099/mic.0.28643-0 [DOI] [PubMed] [Google Scholar]
- Avison M. B., Horton R. E., Walsh T. R., Bennett P. M. (2001). Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. Biochemistry Mosc. 276, 26955–26961 10.1074/jbc.M011186200 [DOI] [PubMed] [Google Scholar]
- Barker H., Kinsella N., Jaspe A., Thorster F., O'Connor C. D. (2000). Formate protects stationary-phase Escherichia coli and Salmonella cells from killing by a cationic antimicrobial peptide. Mol. Microbiol. 2000, 1518–1529 10.1046/j.1365-2958.2000.01820.x [DOI] [PubMed] [Google Scholar]
- Barrett-Bee K., Newboult L., Edwards S. (1994). The membrane destabilising action of the antibacterial agent chlorhexidine. FEMS Microbiol. Lett. 119, 249–253 10.1111/j.1574-6968.1994.tb06896.x [DOI] [PubMed] [Google Scholar]
- Beier R. C., Bischoff K. M., Ziprin R. L., Poole T. L., Nisbet D. J. (2005). Chlorhexidine susceptibility, virulence factors, and antibiotic resistance of beta-hemolytic Escherichia coli isolated from neonatal swine with diarrhea. Bull. Environ. Contam. Toxicol. 75, 835–844 10.1007/s00128-005-0826-5 [DOI] [PubMed] [Google Scholar]
- Block C., Furman M. (2002). Association between intensity of chlorhexidine use and micro-organisms of reduced susceptibility in a hospital environment. J. Hosp. Infect. 51, 201–206 10.1053/jhin.2002.1246 [DOI] [PubMed] [Google Scholar]
- Bobik T. A., Havemann G. D., Busch R. J., Williams D. S., Aldrich H. C. (1999). The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelle involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181, 5967–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott M. (1997). Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167, 78–88 10.1007/s002030050419 [DOI] [PubMed] [Google Scholar]
- Bowden S. D., Ramachandran V. K., Knudsen G. M., Hinton J. C., Thompson A. (2010). An incomplete TCA cycle increases survival of Salmonella Typhimurium during infection of resting and activated murine macrophages. PLoS ONE 5:e13871 10.1371/journal.pone.0013871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braoudaki M., Hilton A. C. (2004). Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J. Clin. Microbiol. 42, 73–78 10.1128/JCM.42.1.73-78.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. H., Skurray R. A. (2001). Staphylococcal multidrug efflux protein QacA. J. Mol. Microbiol. Biotechnol. 3, 163–170 [PubMed] [Google Scholar]
- Bryan L. E., O'Hara K., Wong S. (1984). Lipopolysaccharide changes in impermeability-type aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26, 250–255 10.1128/AAC.26.2.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M. T., Wang Q., Harshey R. M. (2010). Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. U.S.A. 107, 3776–3781 10.1073/pnas.0910934107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariss S. J. L., Tayler A. E., Avison M. B. (2008). Defining the growth conditions and promoter-proximal DNA sequences required for activation of gene expression by CreBC in Escherichia coli. J. Bacteriol. 190, 3930–3939 10.1128/JB.00108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier S. C., Panis B., Renaut B., Samyn B., Vertommen A., Vanhove A. C., et al. (2011). The use of 2D-electrophoresis and de novo sequencing to characterise inter- and intra-cultivar protein polymorphisms in an allopolyploid crop. Phytochemistry 72, 1243–1250 10.1016/j.phytochem.2010.10.016 [DOI] [PubMed] [Google Scholar]
- Cheung H.-Y., Wong M. M.-K., Cheung S.-H., Liang L. Y., Lam Y.-W., Chiu S.-K. (2012). Differential actions of chlorhexidine on the cell wall of Bacillus subtilis and Escherichia coli. PLoS ONE 7:e36659 10.1371/journal.pone.0036659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott G. S., Hughes K. T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708 10.1128/MMBR.64.4.694-708.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B., Grassl G. A., Finlay B. B. (2007). Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 85, 112–118 10.1038/sj.icb.7100007 [DOI] [PubMed] [Google Scholar]
- Condell O., Iversen C., Cooney S., Power K., Ciara W., Burgess C., et al. (2012a). Efficacy of biocides used in the modern food industry to control Salmonella—links between biocide tolerance and resistance to clinically relevant antimicrobial compounds. Appl. Environ. Microbiol. 78, 3087–3097 10.1128/AEM.07534-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condell O., Sheridan A., Power K. A., Bonilla-Santiago R., Sergeant K., Renaut J., et al. (2012b). Comparative proteomic analysis of Salmonella tolerance to the biocide active agent triclosan. J. Proteomics 75, 4505–4519 10.1016/j.jprot.2012.04.044 [DOI] [PubMed] [Google Scholar]
- Denyer S. P., Maillard J. Y. (2002). Cellular impermeability and uptake of biocides and antibiotics in Gram-negative bacteria. Soc. Appl. Microbiol. Symp. Ser. 92, S35–S45 10.1046/j.1365-2672.92.5s1.19.x [DOI] [PubMed] [Google Scholar]
- Dong T., Yu R., Schellhorn H. (2011). Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 79, 375–386 10.1111/j.1365-2958.2010.07449.x [DOI] [PubMed] [Google Scholar]
- Encheva V., Shah H. N., Gharbia S. E. (2009). Proteomic analysis of the adaptive response of Salmonella enterica serovar Typhimurium to growth under anaerobic conditions. Microbiology 155, 2429–2441 10.1099/mic.0.026138-0 [DOI] [PubMed] [Google Scholar]
- Escolar L., Pérez-Martín J., De Lorenzo V. (1999). Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181, 6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn S., Händler K., Condell O., Colgan A., Cooney C., McClure P., et al. (2013). ProP is required for the survival of desiccated Salmonella Typhimurium on a stainless steel surface. Appl. Environ. Microbiol. 79, 4376–4384 10.1128/AEM.00515-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galice D. M., Bonacorsi C., Soares V. C. G., Raddi M. S. G., Fonseca L. M. D. (2006). Effect of subinhibitory concentration of chlorhexidine on Streptococcus agalactiae virulence factor expression. Int. J. Antimicrob. Agents 28, 143–146 10.1016/j.ijantimicag.2006.03.024 [DOI] [PubMed] [Google Scholar]
- Gast R. K., Holt P. S. (1998). Persistence of Salmonella Enteritidis from one day of age until maturity in experimentally infected layer chickens. Poult. Sci. 77, 1759–1762 10.1093/ps/77.12.1759 [DOI] [PubMed] [Google Scholar]
- Gilbert P., Brown M. R. W. (1980). Cell wall-mediated changes in sensitivity of Bacillus megaterium to chlorhexidine and 2-phenoxyethanol, associated with growth rate and nutrient limitation. J. Appl. Bacteriol. 48, 223–230 10.1111/j.1365-2672.1980.tb01221.x [DOI] [PubMed] [Google Scholar]
- Gilbert P., Moore L. E. (2005). Cationic antiseptics: diversity of action under a common epithet. J. Appl. Microbiol. 99, 703–715 10.1111/j.1365-2672.2005.02664.x [DOI] [PubMed] [Google Scholar]
- Gladyshev V. N., Khangulov S. V., Milton J. A., Stadtman T. C. (1994). Coordination of selenium to molybdenum in formate dehydrogenase H from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 91, 7708–7711 10.1073/pnas.91.16.7708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kobayashi I., Saika T., Nishida M. (1997). Drug-resistance patterns of clinical isolates of Pseudomonas aeruginosa with reference to their lipopolysaccharide compositions. Chemotherapy 43, 323–331 10.1159/000239585 [DOI] [PubMed] [Google Scholar]
- Havemann G. D., Sampson E. M., Thomas A., Bobik T. A. (2002). PduA is a shell protein of polyhedral organelles involved in coenzyme B12 dependent degradation of 1, 2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 184, 1253–1261 10.1128/JB.184.5.1253-1261.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath R. J., Rock C. O. (2000). A triclosan-resistant bacterial enzyme. Nature 406, 145–146 10.1038/35022656 [DOI] [PubMed] [Google Scholar]
- Hernández S. B., Cota I., Ducret A., Aussel L., Casadesu J., Herna S. B. (2012). Adaptation and preadaptation of Salmonella enterica to bile. PLoS Genet. 8:e1002459 10.1371/journal.pgen.1002459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway P. M., Bucknall R. A., Denton G. W. (1986). The effects of sub-lethal concentrations of chlorhexidine on bacterial pathogenicity. J. Hosp. Infect. 8, 39–46 10.1016/0195-6701(86)90103-9 [DOI] [PubMed] [Google Scholar]
- Hope C. K., Wilson M. (2004). Analysis of the effects of chlorhexidine on oral biofilm vitality and structure based on viability profiling and an indicator of membrane integrity. Antimicrob. Agents Chemother. 48, 1461–1468 10.1128/AAC.48.5.1461-1468.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet A. A., Raygada J. L., Mendiratta K., Seo S. M., Kaatz G. W. (2008). Multidrug efflux pump overexpression in Staphylococcus aureus after single and multiple in vitro exposures to biocides and dyes. Microbiology 154, 3144–3153 10.1099/mic.0.2008/021188-0 [DOI] [PubMed] [Google Scholar]
- Jackson R. J., Binet M. R. B., Lee L. J., Ma R., Graham A. I., McLeod C. W., et al. (2008). Expression of the PitA phosphate/metal transporter of Escherichia coli is responsive to zinc and inorganic phosphate levels. FEMS Microbiol. Lett. 289, 219–224 10.1111/j.1574-6968.2008.01386.x [DOI] [PubMed] [Google Scholar]
- Janion C. (2008). Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int. J. Biol. Sci. 4, 338–344 10.7150/ijbs.4.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. (2000). Chlorhexidine: is it still the gold standard? Periodontology 2000, 55–62 10.1111/j.1600-0757.1997.tb00105.x [DOI] [PubMed] [Google Scholar]
- Kaatz G. W., McAleese F., Seo S. M. (2005). Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49, 1857–1864 10.1128/AAC.49.5.1857-1864.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Killam T., Sood V., Michael G., Surette M. G. (2003). Swarm-cell differentiation in Salmonella enterica serovar Typhimurium results in elevated resistance to multiple antibiotics. J. Bacteriol. 185, 3111–3117 10.1128/JB.185.10.3111-3117.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Surette M. G. (2003). Swarming populations of Salmonella represent a unique physiological state coupled to multiple mechanisms of antibiotic resistance. Biol. Proced. Online 5, 189–196 10.1251/bpo61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C., Dillon S. C., Cameron A. D. S., Papenfort K., Sivasankaran S. K., Hokamp K., et al. (2012). The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 109, E1277–E1286 10.1073/pnas.1201061109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A. M., Kuzio J., Angus B. L., Hancock R. E. (1982). Chemical and chromatographic analysis of lipopolysaccharide from an antibiotic-supersusceptible mutant of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 21, 310–319 10.1128/AAC.21.2.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. B. (2000). Antibiotic and antiseptic resistance: impact on public health. Pediatr. Infect. Dis. J. 19, S120–S122 10.1097/00006454-200010001-00008 [DOI] [PubMed] [Google Scholar]
- Levy S. B. (2002). Active efflux, a common mechanism for biocide and antibiotic resistance. Soc. Appl. Microbiol. Symp. Ser. 92, S65–S71 10.1046/j.1365-2672.92.5s1.4.x [DOI] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 10.1101/gr.078212.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell G., Russell A. D. (1999). Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12, 147–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstone A. M., Passaretti C. L., Perl T. M. (2008). Chlorhexidine: expanding the armamentarium for infection control and prevention. Clin. Infect. Dis. 46, 274–281 10.1086/524736 [DOI] [PubMed] [Google Scholar]
- Nally J. E., Whitelegge J. P., Aguilera R., Pereira M. M., Blanco D. R., Lovett M. A. (2005). Purification and proteomic analysis of outer membrane vesicles from a clinical isolate of Leptospira interrogans serovar Copenhageni. Proteomics 5, 144–152 10.1002/pmic.200400880 [DOI] [PubMed] [Google Scholar]
- Nally J. E., Whitelegge J. P., Bassilian S., Blanco R., Lovett M. A. (2007). Characterization of the outer membrane proteome of Leptospira interrogans expressed during acute lethal infection. Infect. Immun. 75, 766–773 10.1128/IAI.00741-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nde C. W., Jang H.-J., Toghrol F., Bentley W. E. (2009). Global transcriptomic response of Pseudomonas aeruginosa to chlorhexidine diacetate. Environ. Sci. Technol. 43, 8406–8415 10.1021/es9015475 [DOI] [PubMed] [Google Scholar]
- Nesse L. L., Nordby K., Heir E., Bergsjoe B., Vardund T., Nygaard H., et al. (2003). Molecular analyses of Salmonella enterica isolates from fish feed factories and fish feed ingredients. Appl. Environ. Microbiol. 69, 1075–1081 10.1128/AEM.69.2.1075-1081.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omote H., Hiasa M., Matsumoto T., Otsuka M., Moriyama Y. (2006). The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27, 587–593 10.1016/j.tips.2006.09.001 [DOI] [PubMed] [Google Scholar]
- O'Regan E., Quinn T., Pagès J.-M., McCusker M., Piddock L., Fanning S. (2009). Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar Enteritidis: involvement of ramA and other global regulators. Antimicrob. Agents Chemother. 53, 1080–1087 10.1128/AAC.01005-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I. A. T., Brown M. H. (1996). Proton-dependent multidrug efflux systems. Microbiology 60, 575–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2002). Mechanisms of bacterial biocide and antibiotic resistance. Soc. Appl. Microbiol. Symp. Ser. 92, 55S–65S 10.1046/j.1365-2672.92.5s1.8x [DOI] [PubMed] [Google Scholar]
- Poole K. (2004). Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10, 12–26 10.1111/j.1469-0691.2004.00763.x [DOI] [PubMed] [Google Scholar]
- R Core Team (2011). R: A Language And Environment For Statistical Computing. Available online at: http://www.r-core.org
- Ritchie M. E., Silver J., Oshlack A., Holmes M., Diyagama D., Holloway A., et al. (2007). Bioinformatics original paper two-colour microarrays. Bioinformatics 23, 2700–2707 10.1093/bioinformatics/btm412 [DOI] [PubMed] [Google Scholar]
- Russell A. D., Furr J. R. (1986). The effects of antiseptics, disinfectants and preservatives on smooth, rough and deep rough strains of Salmonella Typhimurium. Int. J. Pharm. 32, 115–123 10.1016/0378-5173(86)90017-7 [DOI] [Google Scholar]
- Russell A. D., Path F. R. C. (1986). Chlorhexidine: antibacterial action and bacterial resistance. Infection 14, 212–215 10.1007/BF01644264 [DOI] [PubMed] [Google Scholar]
- Sergeant K., Spiess N., Renaut J., Wilhelm E., Hausman J. F. (2011). One dry summer: a leaf proteome study on the response of oak to drought exposure. J. Proteomics 74, 1385–1395 10.1016/j.jprot.2011.03.011 [DOI] [PubMed] [Google Scholar]
- Sheridan A., Lenehan M., Condell O., Bonilla-Santiago R., Sergeant K., Renaut J., et al. (2012). Proteomic and phenotypic analysis of verocytotoxigenic Escherichia coli (VTEC) with tolerance to triclosan. J. Proteomics 80, 78–90 10.1016/j.jprot.2012.12.025 [DOI] [PubMed] [Google Scholar]
- Shi X., Namvar A., Kostrzynska M., Hora R., Warriner K. (2007). Persistence and growth of different Salmonella serovars on pre- and postharvest tomatoes. J. Food Prot. 70, 2725–2731 [DOI] [PubMed] [Google Scholar]
- Sittka A., Lucchini S., Papenfort K., Sharma C. M., Rolle K., Tim T. (2008). Deep sequencing analysis of small non-coding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4:e1000163 10.1371/journal.pgen.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. (2005). Limma: linear models for microarray data, in Bioinformatics and Computational Biology Solutions Using R and Bioconductors, eds Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W. (New York, NY: Springer; ), 397–420 [Google Scholar]
- Smyth G. K., Speed T. (2003). Normalization of cDNA microarray data. Methods 31, 265–273 10.1016/S1046-2023(03)00155-5 [DOI] [PubMed] [Google Scholar]
- Sulavik M. C., Houseweart C., Cramer C., Jiwani N., Murgolo N., Greene J., et al. (2001). Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45, 1126–1136 10.1128/AAC.45.4.1126-1136.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M., Szakonyi G., Brown K. A., Henderson P. J. F., Nield J., Byrne B. (2009). Biochemical and biophysical research communications the multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro. Biochem. Biophys. Res. Commun. 380, 338–342 10.1016/j.bbrc.2009.01.081 [DOI] [PubMed] [Google Scholar]
- Tattawasart U., Maillard J., Furr J. R., Russell A. D. (2000). Outer membrane changes in Pseudomonas stutzeri resistant to chlorhexidine diacetate and cetylpyridinium chloride. Int. J. Antimicrob. Agents 16, 233–238 10.1016/S0924-8579(00)00206-5 [DOI] [PubMed] [Google Scholar]
- Thomas B., Stickler D. J. (1979). Chlorhexidine resistance and the lipids of Providencia stuartii. Microbios 24, 141–150 [PubMed] [Google Scholar]
- Tran A. X., Dong C., Whitfield C. (2010). Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 10.1074/jbc.M110.144709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B., Fink R. C., Porwollik S., McClelland M., Hassan H. M. (2011). The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol. 11:236 10.1186/1471-2180-11-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadhachary A., Maloney P. C. (1991). Reconstitution of the phosphoglycerate transport protein of Salmonella Typhimurium. J. Biol. Chem. 266, 130–135 [PubMed] [Google Scholar]
- Vestby L. K., Møretrø T., Langsrud S., Heir E., Nesse L. L. (2009). Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet. Res. 5:20 10.1186/1746-6148-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.-L., Goldrick D., Hong J.-S. (1988). Identification of the products and nucleotide sequences of two regulatory genes involved in the exogenous induction of phosphoglycerate transport in Salmonella Typhimurium. J. Bacteriol. 170, 4299–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung S. Y., Huang C. S., Chan C. P., Lin C. P., Lim H. N., Lee P. H., et al. (2007). Antioxidant and pro-oxidant properties of chlorhexidine and its interaction with calcium hydroxide solutions. Int. Endod. J. 40, 837–844 10.1111/j.1365-2591.2007.01271.x [DOI] [PubMed] [Google Scholar]
- Ygberg S. E., Clements M. O., Rytkönen A., Thompson A., Holden D. W., Hinton J. C. D., et al. (2006). Polynucleotide phosphorylase negatively control spv virulence gene expression in Salmonella enterica. Infect. Immun. 74, 1243–1254 10.1128/IAI.74.2.1243-1254.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Cui S., Meng J. (2009). Effect of transcriptional activators RamA and SoxS on expression of multidrug efflux pumps AcrAB and AcrEF in fluoroquinolone-resistant Salmonella Typhimurium. J. Antimicrob. Chemother. 63, 95–102 10.1093/jac/dkn448 [DOI] [PubMed] [Google Scholar]
- Zorko M., Jerala R. (2008). Alexidine and chlorhexidine bind to lipopolysaccharide and lipoteichoic acid and prevent cell activation by antibiotics. J. Antimicrob. Chemother. 62, 730–737 10.1093/jac/dkn270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Comparative respiration of ST24WT, and the chlorhexidine tolerant mutant, ST24CHX, on PM plates 1–10, containing energy sources and osmolytes. Substrates where a significant difference in the respiration between ST24WT and ST24CHX was recorded are indicated with arrows and details are provided in Table S6. (B) Comparative respiration of ST24WT and the chlorhexidine tolerant mutant, ST24CHX, on PM plates 11–20, containing antimicrobial compounds. Compounds where a significant difference in the respiration between ST24WT and ST24CHX were recorded, indicating a possible alteration in susceptibility, are indicated with arrows and details are provided in Table S6.
Relative expression of enzymes that were determined to be differentially expressed and associated with (A) glycerolipid metabolism in the reference isolate ST24WT. (B) Glycerolipid metabolism in the mutant ST24CHX, (C) amino acid metabolism in the reference isolate ST24WT, and (D) amino acid metabolism in the mutant ST24CHX. Enzyme names, descriptions, gene identifiers and fold-change values are given in Table 2, Tables S1, S4. Color blocks without a white diagonal line represent gene fold change values (Table S1), color blocks containing a white diagonal line represent protein fold change values (Table 2, Table S4).
Relative expression of enzymes determined as differentially expressed and associated with carbohydrate metabolism in (A) the reference isolate ST24WT and (B) the isogenic mutant ST24CHX. Enzyme names, descriptions, gene identifiers and fold-change values are given in Table 2, Tables S1, S4. Color blocks without a white diagonal line represent gene fold change values (Table S1), color blocks containing a white diagonal line represent protein fold change values (Table 2, Table S4).
Relative expression of enzymes determined as differentially expressed and associated with carbohydrate metabolism; the pentose phosphate and pentose glucuronate pathways in (A) the reference isolate ST24WT and (B) the mutant isolate ST24CHX.