Background: PTH regulates HDAC4 to control its dissociation from Runx2.
Results: PTH regulates phosphorylation and dephosphorylation of HDAC4 through PKA and partial degradation of HDAC4.
Conclusion: PTH controls MMP-13 transcription through PKA-dependent phosphorylation and dephosphorylation of HDAC4.
Significance: The results provide insight into how PTH participates in modification of HDAC4, which is crucial for understanding hormonal regulation of gene expression.
Keywords: Histone Deacetylase (HDAC), Osteoblast, Parathyroid Hormone, Phosphorylation, Protein Kinase A (PKA)
Abstract
Histone deacetylases (HDACs) are crucial regulators of gene expression in transcriptional co-repressor complexes. Previously, we reported that HDAC4 was a basal repressor of matrix metalloproteinase-13 (MMP-13) transcription and parathyroid hormone (PTH) regulates HDAC4 to control MMP-13 promoter activity through dissociation from Runx2. Here, we show that PTH induces the protein kinase A (PKA)-dependent phosphorylation of HDAC4 in the nucleus of the rat osteoblastic cell line, UMR 106–01. We demonstrate that PKA-dependent phosphorylated HDAC4 is released from Runx2 bound to the MMP-13 promoter in these cells. Point mutation of Ser-740 in rHDAC4 prevents the release of HDAC4 from Runx2 on the MMP-13 promoter and also prevents the PTH stimulation of MMP-13 transcription. Thus, PTH-induced phosphorylation of rHDAC4 at Ser-740 is crucial for regulating MMP-13 transcription in osteoblasts. PTH causes degradation of HDAC4, and this product appears in the cytoplasm. The cytoplasmic degradation of HDAC4 is blocked by PKA and lysosomal inhibitors, but is not affected by proteasome, caspase-3, or serine and aspartic protease inhibitors. In addition, the phosphatase inhibitor, okadaic acid, prevents degradation indicating that dephosphorylation is associated with degradation. These mechanisms regulating HDAC4 and their roles in such processes are crucial for bone and chondrocyte development. Our data support a link between PTH regulating HDAC4 phosphorylation by PKA, trafficking, partial degradation, and the control of MMP-13 transcription through association with Runx2.
Introduction
Epigenetic modification plays an important role in the control of cell fate during mammalian development. Among these, the most studied so far is DNA methylation, ATP-dependent chromatin remodeling, and post-translational modifications of histones, which includes the dynamic acetylation and deacetylation of epsilon-amino groups of lysine residues present in the tails of core histones. Histone deacetylases (HDACs)2 are crucial regulators of gene expression in transcriptional co-repressor complexes and control gene expression important for diverse cellular functions. The class I HDACs (HDAC1, -2, -3, and -8) have homology to the yeast global transcriptional regulator Rpd3 and are widely expressed. In contrast, the class II HDACs (HDAC4, -5, -6, -7, -9, and -10) show homology to yeast Hda1 and are expressed in cell type-restricted patterns. Among class II HDACs, HDAC4, -5, -7, and -9 form a subclass known as class IIa, whereas HDAC6 and 10 constitute class IIb. The class IIa histone deacetylases can be expressed in a tissue-specific fashion, and are regulated by nuclear-cytoplasmic shuttling (1).
HDAC3 binds class II HDACs (2, 3) and form large multi-protein complexes. These multi-protein complexes are recruited to specific DNA sequences and chromatin substrates by transcription factors (4). Genetic deletion of HDAC3 and HDAC4 revealed the roles of these proteins in chondrocyte maturation (5). Several class II HDACs appear to have a role in skeletal formation (6). Notably, Hdac4-null mice display premature ossification of developing bones due to constitutive Runx2 expression. Thus, HDAC4 regulates chondrocyte hypertrophy and endochondral bone formation by inhibiting the activity of Runx2 (7). In osteoblasts, HDAC4 and HDAC5 participate in TGFβ signaling pathways that suppress Runx2 activity (8). Moreover, HDAC4 and HDAC5 deacetylate Runx2 and lead to Smurf-mediated degradation of Runx2 (9). Thus, HDAC4 is considered an important target for osteoblast or chondrocyte differentiation.
Dephosphorylation by phosphatases and phosphorylation by serine/threonine kinases controls nuclear import and export of HDAC4 and influences HDAC4 repressive activity (10). 14-3-3 proteins shuttle class II HDACs to the cytoplasm (11). These proteins have a phosphoserine/threonine-binding motif through which they interact with phosphorylated HDACs (12). Subcellular localization of HDAC4, HDAC5, and HDAC7 is regulated by calcium-calmodulin-dependent kinase IV (CaMK IV) (13) or PKD phosphorylation and association with 14-3-3 proteins for export to the cytoplasm (14, 15). Backs et al. (16, 17) showed that CaMK II signals specifically to HDAC4 but not HDAC5 by binding to a unique kinase-docking site contained in HDAC4. HDAC4 can subsequently re-enter the nucleus after dephosphorylation and dissociation from 14-3-3 (11).
Parathyroid hormone (PTH) is an 84-amino acid peptide hormone, which functions as an essential regulator of calcium homeostasis and as a mediator of bone remodeling (18). PTH acts via the PTH/PTH-related protein 1 receptor (a G protein-coupled receptor) on osteoblast membranes (19), and both its anabolic and catabolic effects on bone appear to be primarily mediated by the cAMP/PKA pathways (20). Parathyroid hormone-related peptide (PTHrP) or forskolin are reported to cause dephosphorylation of hHDAC4 at Ser-246 by PP2A through PKA resulting in an increase in the nuclear localization of HDAC4, inhibition of myocyte enhancer factor 2 (MEF2) transcriptional activity, and suppression of collagen X expression in chondrocytes (21).
A further level of regulation of HDAC4 has been shown to be through its partial degradation. This has been previously shown to be due to cleavage by caspase (22), or through SUMOylation and proteasome degradation (23). Most recently, Backs et al. (24) showed that PKA induces cleavage of HDAC4 to produce an N-terminal fragment, which acts as a CaMKII-insensitive repressor that selectively inhibits MEF2. The cleavage of HDAC4 is associated with a PKA activated-serine protease.
We recently showed that HDAC4 repressed MMP-13 transcription under basal conditions and parathyroid hormone (PTH) regulates HDAC4 to control MMP-13 promoter activity through dissociation from Runx2 (25). Here, we report that PTH stimulates phosphorylation of rHDAC4 at Ser-740 in the nucleus of osteoblastic cells. Phosphorylated Ser-740 rHDAC4 is associated with release from Runx2 on the MMP-13 promoter and activation of the gene. HDAC4 is then partially degraded in the cytoplasm after PTH treatment, which is blocked by PKA, phosphatase, and lysosomal inhibitors. This is the first observation of this complete system of regulation of HDAC4.
EXPERIMENTAL PROCEDURES
Materials
Parathyroid hormone (rat PTH 1–34), prostaglandin E2, okadaic acid, and NH4Cl were purchased from Sigma-Aldrich. H89, GF109203, MG132, lactacystin, AcDEVDCHO, KN-62, KN-92, KN-93, Gö6976, 3.4 DCl, AEBSF, pepstatin A, and purified catalytic subunit of PKA were purchased from EMD Millipore.
Cell Culture
The UMR 106-01 cells were cultured in Eagle's minimal essential medium (EMEM) supplemented with 25 mm Hepes, pH 7.4, 1% nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, 5% fetal bovine serum. Saos-2 cells were cultured in α-MEM supplemented with 1% l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum.
Antibodies
Anti-HDAC4 (against 10 N-terminal amino acids), anti-GFP, and anti-β-actin were purchased from Cell Signaling Technology. Anti-HDAC4 (H92, against amino acids 530–631), anti-Runx2 (M-70), anti-Cdk2 (M2), and anti-tubulin (TU-2) were purchased from Santa Cruz Biotechnology.
Western Blot
UMR 106-01 cells were treated with or without rat PTH (1–34, 10−8 m) for the indicated times. The cells were washed twice in PBS, pH 7.4 and pelleted by centrifugation at 2000 rpm for 5 min at 4 °C. The pellets were resuspended in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm PMSF, 1 mm EDTA, 1% sodium deoxycholate, 0.1% SDS, and protease inhibitors) and incubated for 15 min at 4 °C. Amounts of total protein were determined by the Bradford dye binding (Bio-Rad) method. The preparation of cytoplasmic and nuclear extracts from cells was by the NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific). To examine the interaction between HDAC4 and Runx2 using immunoprecipitation, the GFP-HDAC4 or mutant HDAC4 expression plasmids were transfected into UMR 106-01 cells. The total lysates were precleared by incubating with Protein A/G-agarose beads (Santa Cruz Biotechnology). After the cleared supernatants had been incubated overnight with 2 μg/ml antibody at 4 °C, the agarose beads were washed three times with PBS. Proteins were separated using SDS-PAGE and were transferred to polyvinylidene difluoride membranes. Proteins were detected by SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) according to the manufacturer's instructions. Quantitation was obtained using ImageJ.
Dephosphorylating Proteins in Vitro with Calf Intestinal Alkaline Phosphatase (CIP)
CIP can be used to release phosphate groups from phosphorylated tyrosine, serine, and threonine residues in proteins. UMR 106-01 cells stimulated with or without PTH were washed twice by PBS. Nuclear extracts were divided into two aliquots (90 μg). One aliquot was treated for 60 min at 37 °C with 10 units of CIP, and one aliquot was mock-treated. The reaction was stopped with 50 mm EDTA. The samples were then resolved by SDS-PAGE.
In Vitro Phosphorylation Assay
The phosphorylation reaction was performed in 20 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 1 mm EDTA, with 50 μg of nuclear extracts from UMR 106–01 cells treated with or without PTH for 30, 120, 240 min, and purified catalytic subunit of PKA (1.0 or 5.0 μg) at 30 °C for 30 min. The samples were then resolved by SDS-PAGE.
Luciferase Activities
For transient transfections, UMR 106-01 cells were seeded in 12-well plates overnight and then transfected with indicated plasmids using GeneJammer (Stratagene) according to the manufacturer's protocol. After 2 days, the cells were treated with PTH (10−8 m) for 6 h. Lysates were analyzed immediately for luciferase activity using the luciferase assay reagent (Promega) and an OptiCompII luminometer (MGM Instruments, Inc., Hamden, CT). To make the mutation constructs of GFP-HDAC4, a site-directed mutagenesis kit (Agilent Technologies) was used, and detailed procedures were in accordance with the manufacturer's instructions.
Chromatin Immunoprecipitation Assays (ChIP Assays)
UMR 106-01 cells were incubated for 10 min at room temperature with medium containing 0.8% formaldehyde. Cells were then washed in ice-cold PBS containing protease inhibitors and 1 mm phenylmethylsulfonyl fluoride and resuspended in SDS lysis buffer (1% SDS, 10 mm EDTA, pH 8.0, 25 mm Tris-HCl, pH 8.1, containing protease inhibitors and 1 mm PMSF) for 10 min on ice. Samples were sonicated to reduce the DNA length to 0.5–1 kbp, cellular debris was removed by centrifugation, and the supernatant was diluted 10-fold in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, 167 mm NaCl supplemented with protease inhibitors). For PCR analysis, aliquots (1:100) of total chromatin DNA before immunoprecipitation were saved (input). Prior to chromatin immunoprecipitations, the samples were precleared with 80 μl of a 25% (v/v) suspension of DNA-coated protein A/G-agarose for 30 min at 4 °C. The supernatant was recovered and used directly for immunoprecipitation experiments with appropriate antibody overnight at 4 °C. Immune complexes were mixed with 60 μl of a 25% precoated protein A/G-agarose suspension followed by incubation for 1 h at 4 °C. Beads were collected and sequentially washed with 1 ml of each of the following buffers: low salt wash buffer, high salt wash buffer, and LiCl wash buffer. The beads were then washed twice using 1 ml of TE buffer. The immunocomplexes were eluted two times by adding elution buffer of 1% SDS, 0.1 m NaHCO3. For re-ChIP assays, the immunoprecipitated complexes obtained by ChIP with anti-Runx2 antibody were eluted in ChIP elution buffer and then diluted 5-fold and subjected to the ChIP procedure with anti-GFP. The cross-linking reaction was reversed with 5 m NaCl by 6 h incubation at 65 °C. The samples were digested with proteinase K (10 mg/ml), 40 mm Tris-HCl pH 6.5, 10 mm EDTA at 42 °C for 1 h, and DNA was recovered by phenol/chloroform extractions. The DNA was precipitated with two volumes of ethanol using glycogen as carrier. The input lysates were also processed as above. The DNA was resuspended in water and used for quantitative PCR. The sequences of the oligonucleotides of the rat MMP-13 promoter (the distal RD and proximal AP-1 sites, −204/−34) were as follows: forward primer 5′-CAGATGCGTTTTGATATGCC-3′, reverse primer 5′-AATAGTGATGAGTCACCACTT-3′. The PCR reactions and program are described in detail in a previous publication (25).
Statistical Analysis
All results are expressed as means ± S.E. of triplicate measurements with all experiments being repeated at least three times. Statistical analyses were carried out using Student's t test or one-way ANOVA using the Tukey HSD test.
RESULTS
PTH Affects the Modification of HDAC4 in the Nucleus through Protein Kinase A
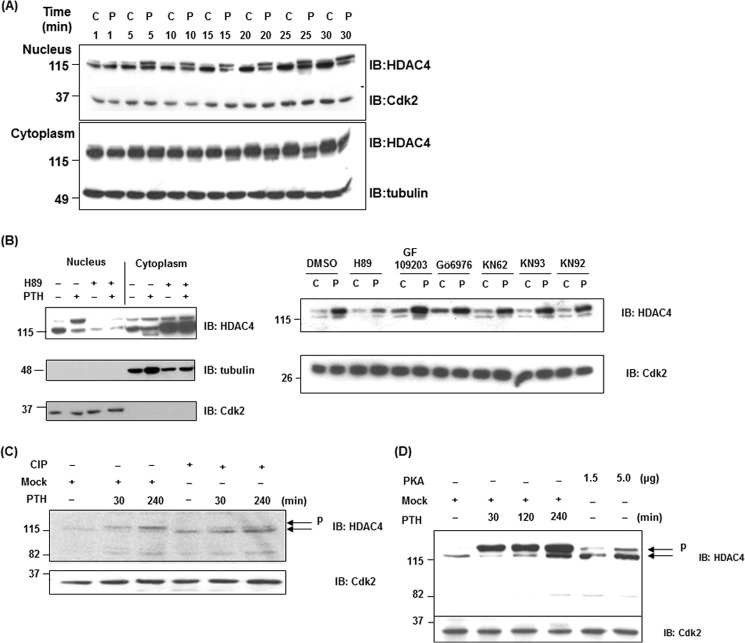
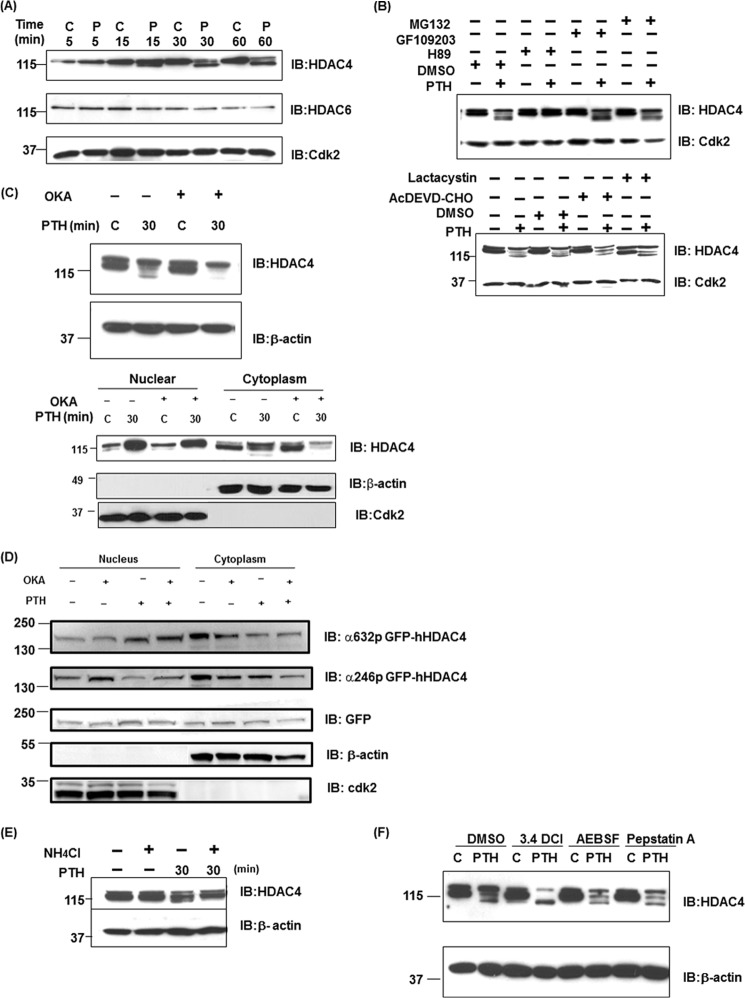
We focused on understanding the effects of PTH on phosphorylation of HDAC4 because it was previously shown that HDAC4 suppresses MMP-13 transcription through its association with Runx2; PTH regulates this repression in osteoblastic cells and this involves PKA (25). Phosphorylation and de-phosphorylation control nuclear export and import of HDAC4 (10). To begin deciphering the effects of PTH on HDAC4, we examined the nuclear fractions in UMR 106-01 cells after 5–30 min of exposure to PTH. A slower-migrating upper band was detected 5 min after PTH treatment only in the nucleus, which was apparent for up to 240 min (Fig. 1, A and C). A faster migrating band of HDAC4 was detectable in the cytoplasmic fraction after 15 min of PTH stimulation which we think is a partial degradation product (Fig. 1, A and B). We next examined whether PKA affected the appearance of a slower-migrating band of HDAC4 in the nucleus. We used the following inhibitors, protein kinase A (H89), protein kinase C (PKC) (GF109203), CaMK (KN62, 92, and 93) and PKD (Gö6976). As shown in Fig. 1B, H89 prevented the appearance of the slowly migrating band of HDAC4 in the nucleus in response to PTH but CaMK inhibitors had no effect, eliminating this pathway in PTH action in the nucleus. GF109203 and Gö6976 clearly increased the appearance of HDAC4 in the nucleus in the absence and presence of PTH treatment. H89 increased accumulation of HDAC4 in the cytoplasm with or without PTH. These data indicate that PKA activation may be required for import of HDAC4 to the nucleus as well as the phosphorylation in the nucleus after PTH treatment, whereas PKC and PKD activation may be related to export of HDAC4 to the cytoplasm. The faster-migrating band in the cytoplasm also appears to require PKA activation in response to PTH for its generation.
FIGURE 1.
PTH induces the phosphorylation of HDAC4 in the nucleus of osteoblastic cells. A, nuclear and cytoplasmic extracts from control or PTH treated (5, 10, 15, 20, 25, 30 min) UMR 106-01 cells were subjected to immunoblotting with anti-HDAC4, anti-Cdk, and anti-tubulin antibodies. Anti-Cdk2 was used for the nucleus as a loading control; anti-tubulin was used for the cytoplasm. B, UMR 106-01 cells were preincubated with protein kinase A inhibitor H89 (50 μm), CaMK inhibitors KN92 (5 μm), KN62 (5 μm), KN93 (5 μm), protein kinase C inhibitor GF109203 (5 μm), or PKD inhibitor Gö6976 (5 μm) for 30 min, and then treated with or without PTH (1–34, 10−8 m) for 30 min. Nuclear and cytoplasmic extracts were subjected to immunoblotting with anti-HDAC4, anti-Cdk, and anti-tubulin antibodies. Anti-Cdk2 was used for the nucleus and anti-tubulin was used for the cytoplasm as loading controls. C, nuclear extracts isolated from control and PTH-treated UMR 106-01 cells were incubated with buffer (Mock) or with 10 units CIP for 60 min at 37 °C. The samples were used for Western blot analysis using anti-HDAC4 and anti-Cdk antibodies. Anti-Cdk2 was used as a loading control. D, nuclear extracts isolated from control and PTH-treated UMR 106-01 cells were incubated with buffer (Mock) or with purified catalytic subunit of PKA (1.5 or 5 μg) for 30 min at 30 °C. The samples were used for Western blot analysis using anti-HDAC4 and anti-Cdk2 antibodies. Anti-Cdk2 was used as a loading control.
Protein Kinase A Induces the Nuclear Phosphorylation of HDAC4
PTH causes activation of protein kinase A, which phosphorylates proteins on serine/threonine residues in its specific recognition sequence in osteoblastic cells (26). We hypothesized that the slower migrating band of HDAC4 in the nucleus might be due to phosphorylation by protein kinase A. To investigate this hypothesis, we performed dephosphorylation assays using nuclear extracts from UMR 106-01 cells. The nuclear extracts were incubated with calf intestinal alkaline phosphatase (CIP) for 60 min at 37 °C. The samples were detected by Western blot analysis. The PTH-induced upper band of HDAC4 was decreased by CIP treatment in comparison with the mock treated samples to a molecular mass identical to the control cells (Fig. 1C). For a more direct confirmation of whether HDAC4 is phosphorylated by PKA, we performed in vitro phosphorylation assays. The nuclear extracts from control UMR 106–01 cells were incubated with the purified catalytic subunit of PKA for 30 min at 30 °C. After incubation, the samples were detected by Western blot analysis. The upper band of HDAC4 was increased by this enzymatic activity in comparison with the mock-treated cells. This upper band migrated identically to that from PTH-treated samples (Fig. 1D). These data indicate that PTH causes phosphorylation of HDAC4 by activation of PKA in the nucleus.
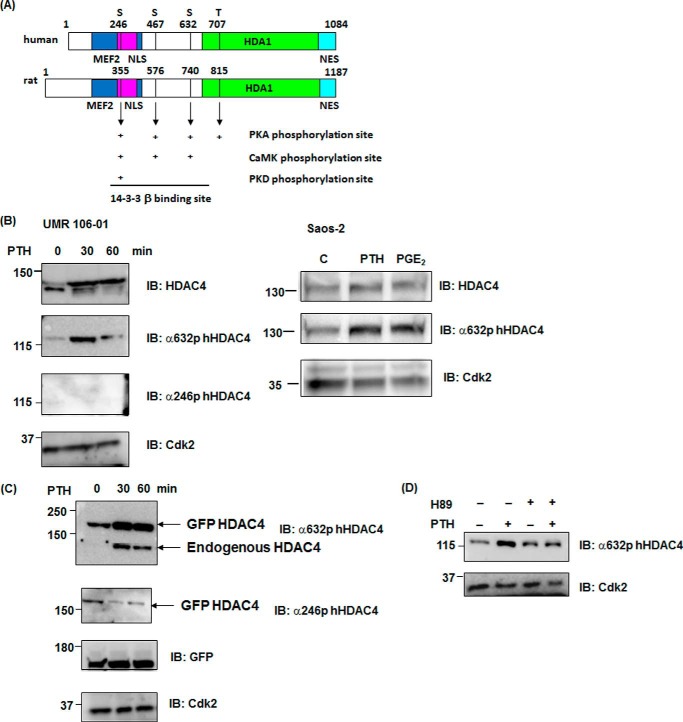
PTH Stimulates Phosphorylation at a Specific HDAC4 Site in the Nucleus
Next, we investigated which residues of HDAC4 are phosphorylated by PTH in the nucleus. Examination of the database of PKA-dependent phosphorylation sites (Phospho.ELM and KinasePhos), as shown in Fig. 2A, illustrated there were three serine (355, 576 and 740aa) and one threonine (815aa) residues in rat HDAC4. Interestingly, all sites in the rat HDAC4 were homologous to human sequences and r355 (h246), r576 (h467), and r740 (h632) were also CaMK-dependent phosphorylation sites (Fig. 2A). Ser-246, -467, and -632 in hHDAC4 are binding sites for 14-3-3 proteins and play a role in trafficking HDAC4 from nucleus to cytoplasm (13, 27). We examined whether phosphorylation of these sites of HDAC4 was stimulated by PTH using available HDAC4 phosphorylation antibodies. As shown in Fig. 2B, HDAC4 and its phosphorylation using anti phospho-Ser-632 (r740) HDAC4 were increased at 30 min after PTH stimulation, whereas phosphorylation using anti-phospho-Ser-246 (r355) was not detected in the nucleus of UMR 106-01 cells. Interestingly, phospho-Ser-632 HDAC4 was stimulated at 30 min with PTH or prostaglandin E2 (10−6 m) treatment in the human osteoblastic cell line, Saos-2 cells. Although total HDAC4 was increased for up to 240 min in UMR cells, phosphorylation at r740 HDAC4 was transiently increased at 30 min (Figs. 1B and 2, C and D). We have previously shown that PTH induces the transcription and total expression of HDAC4 at later times in these cells (25). Next, we overexpressed GFP-rHDAC4 constructs in UMR 106-01 cells. Phosphorylation using anti-phospho-Ser-632 was increased on GFP-HDAC4 and endogenous HDAC4 with PTH treatment whereas phosphorylation using anti-phospho-Ser-246 on GFP-HDAC4 was decreased (Fig. 2C). In addition the phosphorylation at Ser-740 was abolished in the presence of H89, indicating this is dependent on PKA. Thus, HDAC4 phosphorylation sites were differently modified by PTH stimulation. We hypothesized that a specific phosphorylation site of HDAC4 may be related to dissociation from Runx2 on the MMP-13 promoter.
FIGURE 2.
PTH causes phosphorylation of a specific site of HDAC4 in the nucleus. A, structure and PKA-dependent phosphorylation sites of human and rat HDAC4 proteins. MEF2 (MEF2C interaction domain): human (118–313), rat (227–422), NLS (nuclear localization signal): human (244–279), rat (353–388), HDA1 (HDA1-related domain): human (655–1084), rat (763–1187), and NES (nuclear export signal): human (1051–1084), rat (1154–1187). B, nuclear extracts from control or PTH-treated (0, 30, 60 min) UMR 106-01 cells were subjected to immunoblotting with anti-phosphoSer632 hHDAC4, anti-phosphoSer246 hHDAC4, and anti-Cdk antibodies. The nuclear extracts from control or PTH (human, 1–34, 10−8 m, 30 min) or prostaglandin E2 (10−6 m, 30 min) treated Saos-2 cells were subjected to immunoblotting with anti-HDAC4, anti-phosphoSer632 hHDAC4, and anti-Cdk antibodies. Anti-Cdk2 was used for the nucleus as a loading control. C, UMR 106-01 cells were transfected with GFP rHDAC4 construct. Cells were stimulated with PTH for 30 and 60 min. The nuclear extracts from control or PTH treated (0, 30, 60 min) UMR 106-01 cells were subjected to immunoblotting with anti-phosphoSer632 hHDAC4, anti-phosphoSer246 hHDAC4, GFP, and anti-Cdk antibodies. Anti-Cdk2 was used for the nucleus as a loading control. D, UMR 106–01 cells were preincubated with protein kinase A inhibitor H89 (50 μm) for 30 min, and then treated with or without PTH (1–34, 10−8 m) for 30 min. Nuclear extracts were subjected to immunoblotting with anti-phosphoSer632 hHDAC4 and anti-Cdk antibodies. Anti-Cdk2 was used as a loading control.
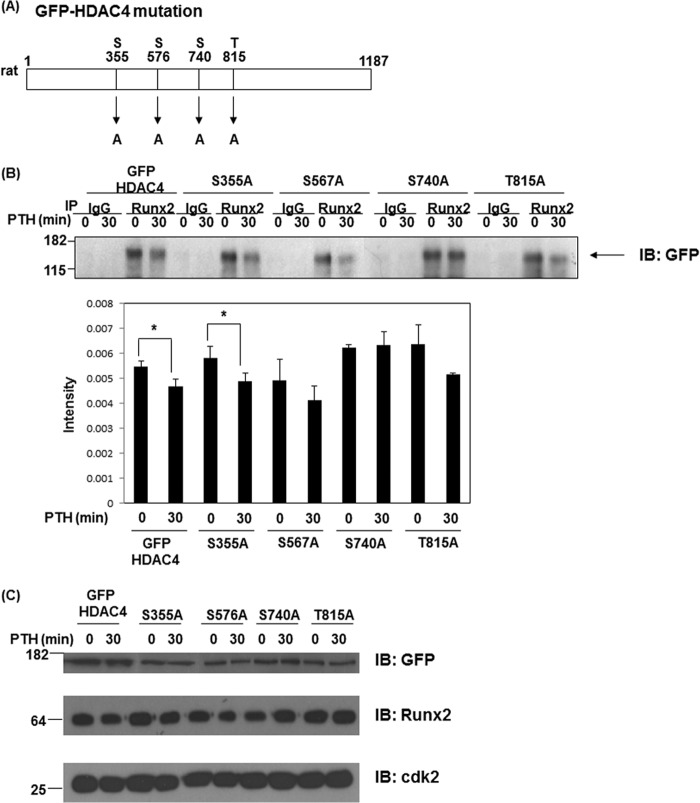
Mutation of Ser-740 in rHDAC4 Prevents Dissociation from Runx2
Next, to confirm our hypothesis, we performed immunoprecipitation assays of Runx2 using wild type and mutation constructs of rHDAC4. As shown in Fig. 3A, we made single point mutation constructs of rat HDAC4 (S355A, S576A, S740A, and T815A) which are activated by PKA, CaMK, or PKD-dependent pathways. Immunoprecipitated endogenous Runx2 bound to GFP-rHDAC4 (wild type) and four GFP-rHDAC4 mutant constructs under basal conditions. Runx2 binding with GFP-rHDAC4 (wild type) and GFP-rHDAC4 mutant constructs (S355A, S576A, and T815A) was decreased after PTH stimulation whereas the GFP-rHDAC4-S740A mutant remained associated with Runx2 (Fig. 3B). As shown in Fig. 3C, these protein levels were equally detected when we performed Western blots. This result suggests that Ser-740 in rat HDAC4 is a crucial phosphorylation site for dissociation from Runx2.
FIGURE 3.
Ser-740 site of HDAC4 is important for dissociation from Runx2. A, mutation sites of GFP-rHDAC4; Serine residues mutated to alanine. B, UMR 106–01 cells were transfected with GFP-rHDAC4 or mutant GFP-rHDAC4 constructs. Cells were stimulated with PTH (1–34, 10−8 m) for 30 min. Total cellular lysates isolated from UMR 106-01 cells were immunoprecipitated with anti-Runx2 or control rabbit IgG antibodies. Samples were immunoblotted with anti-GFP antibody. The quantitative intensity was obtained using ImageJ. *, p < 0.05 versus respective control. C, UMR 106–01 cells were transfected with GFP rHDAC4 or mutant GFP rHDAC4 constructs (S355A, S576A, S740A, and T815A). Total cell lysates were immunoblotted with anti-GFP, anti-Runx2, and anti-Cdk antibodies. Anti-Cdk2 was used as a loading control.
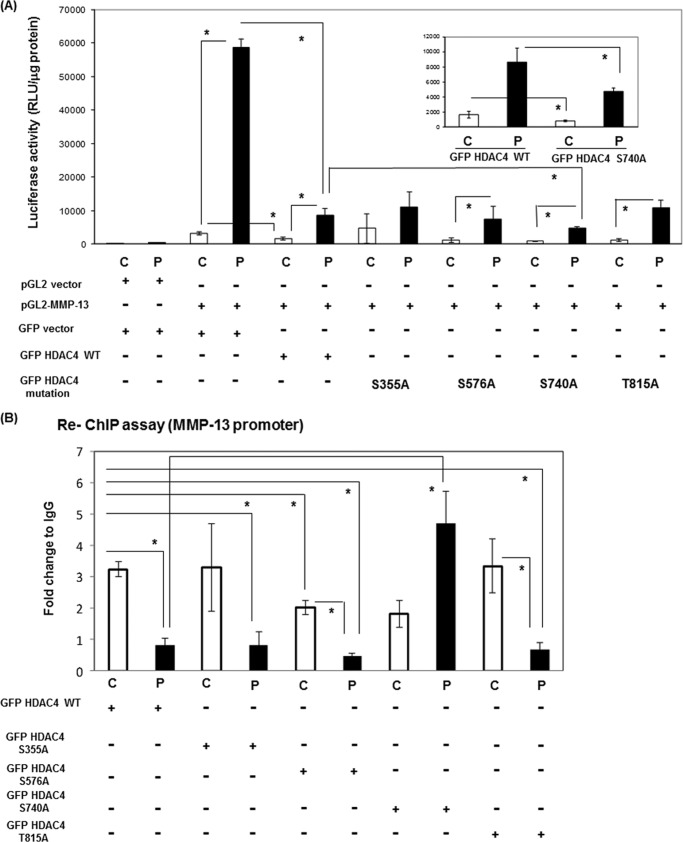
To investigate further whether Ser-740 in rHDAC4 is functionally related to dissociation from Runx2 on the runt domain of the MMP-13 promoter, we performed luciferase promoter assays using the GFP-rHDAC4 constructs. As shown in Fig. 4A, wild type GFP-rHDAC4 significantly inhibited MMP-13 transcriptional activity after PTH stimulation similarly to previous published data of ours (25). All phosphorylation site mutations, except the S740A mutation inhibited the PTH stimulation to a similar extent. The S740A mutation in rHDAC4 significantly inhibited the promoter activity more than the wild type rHDAC4. To further explore the potential role of HDAC4 interaction with Runx2, we investigated the presence of GFP-rHDAC4 or GFP-rHDAC4 mutant proteins with Runx2 at the Runx2 binding site of the MMP-13 promoter by ChIP re-ChIP assays of UMR 106–01 cells with or without PTH stimulation. As shown in Fig. 4B, the association of Runx2-GFP-rHDAC4 and Runx2-GFP-rHDAC4 mutations (S355A, S576A, and T815A) with the MMP-13 promoter was decreased after PTH stimulation using primers encompassing the RD and AP-1 sites. The association of Runx2-GFP-rHDAC4-S740A mutation actually increased with PTH treatment, indicating that the hormone-stimulated phosphorylation of this residue is required for dissociation of HDAC4 from Runx2 on the MMP-13 promoter.
FIGURE 4.
GFP-rHDAC4-S740A mutation abolished dissociation from Runx2 on the MMP-13 promoter. A, UMR 106–01 cells were transiently transfected with various vectors (100 ng of pGL2 vector, 100 ng of -148 rat MMP-13 promoter-Luc, 100 ng of GFP vector, 100 ng of GFP rHDAC4, and mutant GFP rHDAC4 constructs), and the luciferase activities were measured with (P) or without (C) 6 h of 10−8 m PTH (1–34) treatment. The luciferase activities were normalized to total protein. Error bars represent ± S.E. of three independent experiments. *, p < 0.05 versus respective control. B, UMR 106-01 cells were transfected with GFP rHDAC4 or mutant GFP rHDAC4 constructs (S355A, S576A, S740A, and T815A). The cells were treated with control or PTH (10−8 m) for 30 min, then prepared for ChIP assays. After immunoprecipitation of the cross-linked lysates with anti-Runx2 or with rabbit IgG as a negative control, the immune complexes were collected then diluted with ChIP dilution buffer and re-subjected to ChIP assays with anti-GFP or rabbit IgG. The DNA was subjected to PCR with primers that amplify the distal RD and proximal AP-1 sites of the endogenous rat MMP-13 promoter. Input DNA (1:100) is a positive control for the assay. Data are shown as -fold change to control after normalizing to input and IgG. Error bars represent ± S.E. of three independent experiments. *, p < 0.05 versus respective control.
Protein Kinase A and Lysosomal Inhibitors Prevent Partial HDAC4 Degradation
We had seen a faster-migrating band of HDAC4 in cytoplasmic and total cell lysates which appears to be a partial degradation product (Fig. 1, A and B) similar to that seen by Backs et al. (24). HDAC6 also slightly decreased after PTH treatment (Fig. 5A). We next examined whether PKA or other pathways affected the appearance of the faster migrating band and potential degradation of HDAC4 in the cytoplasm. We used protein kinase A (H89) and protein kinase C inhibitors (GF109203), proteasome inhibitors (MG132, lactacystin), and caspase-3 inhibitor (Ac-DEVD-CHO) (Fig. 5B). H89 was the only agent to block HDAC4 degradation (see also Fig. 1B), indicating that HDAC4 degradation occurs via non-proteosomal or non-caspase pathways and requires the PKA pathway in PTH action.
FIGURE 5.
PKA, phosphatase, and lysosomal inhibition blocks PTH-induced HDAC4 degradation. A, total cellular lysates (30 μg) isolated from 10−8 m PTH and control-treated UMR 106–01 cells for 5, 15, 30, and 60 min were used for Western blot analysis using anti-HDAC 4, 6, or Cdk2 antibodies. Anti-Cdk2 was used as the loading control. B, total cellular lysates isolated from UMR 106-01 cells, which had been preincubated with protein kinase A inhibitor H89 (50 μm) for 30 min, protein kinase C inhibitor GF109203 (5 μm) for 30 min, proteasome inhibitor MG132 (5 μm), lactacystin (5 μm), or caspase-3 inhibitor AcDEVD-CHO (100 μm) for 60 min, and then with or without PTH (10−8 m) stimulation for 30 min. Total cellular lysates were used for Western blot analysis using anti-HDAC4. Anti-Cdk2 was used as a loading control. C, total cellular lysates or nuclear/cytoplasmic extracts isolated from UMR 106-01 cells, which had been preincubated with phosphatase inhibitor, okadaic acid (OKA, 50 nm) for 60 min and then with or without PTH (10−8 m) stimulation for 30 min. Total cellular lysates or nuclear/cytoplasmic extracts were used for Western blot analysis using anti-HDAC4. Anti-β-actin was used for total cell lysates or cytoplasm as a loading control. Anti-Cdk2 was used as a nuclear loading control. D, nuclear and cytoplasmic extracts isolated from UMR 106-01 cells, which had been preincubated with phosphatase inhibitor, okadaic acid (OKA, 50 nm) for 60 min and then with or without PTH (10−8 m) stimulation for 30 min. The nuclear and cytoplasm extracts were subjected to immunoblotting with anti-phosphoSer632 hHDAC4, anti-phosphoSer246 hHDAC4, anti-GFP, anti-β-actin, and anti-Cdk antibodies. Anti-Cdk2 was used for the nucleus and anti-β-actin was used for cytoplasm as loading controls. E, total cellular lysates isolated from UMR 106-01 cells were preincubated with NH4Cl (20 mm) for 16 h and then with PTH (10−8 m) stimulation for 30 min. Total cellular lysates were used for Western blot analysis using anti-HDAC4. Anti-β-actin was used for loading control. F, UMR 106–01 cells were preincubated with vehicle, serine protease inhibitors, 3.4 DCl (50 μm) or AEBSF (200 μm), aspartic protease inhibitor pepstatin A (10 μm) for 90 min and then with or without PTH (10−8 m) stimulation for 30 min. Total cellular lysates were used for Western blot analysis using anti-HDAC4. Anti-β-actin was used as a loading control.
Since trafficking and partial degradation of HDAC4 are associated with phosphorylation, we used the phosphatase inhibitor, okadaic acid. The faster migrating band almost disappeared with PTH treatment in the presence of this inhibitor (Fig. 5C), indicating that HDAC4 degradation occurs through serine/threonine dephosphorylation. There was also less HDAC4 in the cytoplasm. Interestingly, the phosphatase inhibitor increased phospho-Ser-246 (r355) in the nucleus which had been seen to be reduced with PTH treatment (Fig. 5D). Phospho-Ser-632 (r740) HDAC4 was increased at 30 min after PTH stimulation as seen in Fig. 2, and okadaic acid additively increased phospho-Ser-632 (r740) with PTH treatment in the nucleus. In the cytoplasm, phosphorylation at both sites was decreased with PTH and okadaic acid. Thus, dephosphorylation likely by PP2A is also involved with partial degradation of HDAC4 and its trafficking from the nucleus in osteoblastic cells. PTH appears to cause the site-selective dephosphorylation or phosphorylation of HDAC4 in the nucleus.
The intracellular systems of protein degradation are classified as lysosomal and non-lysosomal. The lysosomal process involves uptake into lysosomes, and the non-lysosomal step generally involves tagging of proteins prior to degradation by the proteasome. Because proteasome inhibitors did not affect the degradation of HDAC4, we hypothesized that HDAC4 was degraded by the lysosome. To assess a relationship with the lysosome, we examined the effects of the lysosomal inhibitor, NH4Cl, on HDAC4 mobility in response to PTH. Preincubating cells with NH4Cl (Fig. 5E) blocked PTH-induced degradation of HDAC4. Chloroquine and bafilomycine A1 also blocked its degradation (data not shown). In addition, we used serine protease inhibitors (3.4 DCl, AEBSF) and an aspartic protease inhibitor (Pepstatin A). AEBSF and Pepstatin A had no effect on PTH-induced HDAC4 degradation whereas 3.4 DCl increases the faster migrating band (Fig. 5F). These results indicate that HDAC4 degradation is associated with lysosomal pathways but not serine or aspartic proteases.
DISCUSSION
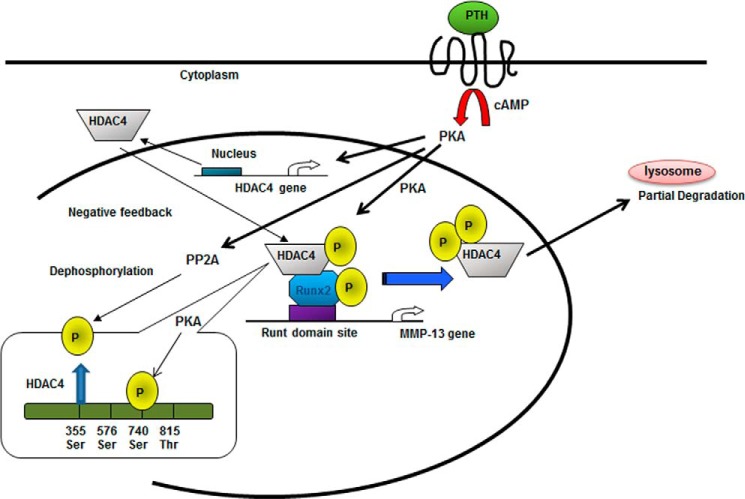
The results of this study show the effects of PTH on the phosphorylation and processing of HDAC4 in rat osteoblastic cells. We focused on HDAC4 function, because HDAC4 plays an important role to suppress chondrocyte and osteoblast differentiation through association with Runx2 and particularly suppresses MMP-13 gene expression in vitro and in vivo (7, 25). We conclude that PKA-dependent phosphorylated HDAC4, especially at Ser-740 in rHDAC4, is released from Runx2 bound to the MMP-13 promoter and exported to the cytoplasm and is then dephosphorylated in the cytoplasm and partially degraded through a lysosomal pathway (Fig. 6).
FIGURE 6.
Model of PKA-dependent phosphorylated regulation of HDAC4. PKA phosphorylates HDAC4 in the nucleus, especially inducing phosphorylation of HDAC4 at serine 740, which results in HDAC4 being released from Runx2 bound to the Runt domain site of the MMP-13 promoter. PKA phosphorylation also induces dephosphorylation of HDAC4 at serine 355 in the nucleus. After transport of HDAC4 into the cytoplasm, HDAC4 is partially degraded through the lysosomal pathway.
Work by several laboratories showed that HDAC4 phosphorylation, binding to 14-3-3 proteins and HDAC4 export to the cytoplasm is through phosphorylation by the CaMKII/IV pathway. HDAC4 is also required for hetero-oligomerization with HDAC5 and its regulation by CaMKII (16, 17). We demonstrate here that PTH induces HDAC4 phosphorylation, especially at Ser-740 in rHDAC4 through PKA activation in the nucleus, but not by CaMKII/IV as detected by use of inhibitors and antibodies. In addition, protein expression of HDAC5 is very low and PTH does not stimulate phosphorylation of HDAC5 and HDAC7 in UMR 106-01 cells and rat primary osteoblasts (data not shown). PTHrP or forskolin repress chick chondrocyte hypertrophy through PKA-dependent dephosphorylation of hHDAC4 at phospho-S246 by PP2A, which increases the nuclear localization of HDAC4 and inhibits MEF2 function (21). Consistent with their data, we found PTH decreases phosphorylation of Ser-355 in GFP-rHDAC4 (equivalent to hHDAC4 246S), but we could not detect endogenous phosphorylated Ser-355 rHDAC4 in UMR 106-01 cells, indicating that this requires overexpression of HDAC4. We showed that dephosphorylation of Ser-355 rHDAC4 is not related to dissociation from Runx2 or regulation of the transcriptional activity of MMP-13 (Fig. 4, A and B). However, the phosphatase inhibitor, okadaic acid, enhances phosphorylation of Ser-355 rHDAC4 in the nucleus and this phosphorylation was reduced after PTH stimulation whereas phosphorylation of Ser-740 rHDAC4 was increased by PTH and okadaic acid. Phosphorylation of Ser-355 rHDAC4 and 740 rHDAC4 may be regulated inversely by PTH through PP2A and PKA, controlling HDAC4 trafficking out of the nucleus and subsequent partial degradation. Therefore, we conclude that transient PTH-induced phosphorylation of Ser-740 rHDAC4 (Ser-632 in hHDAC4) via PKA activation plays a role to cause the induction of MMP-13 gene expression in osteoblasts. Moreover, PKA-activated phosphorylation and dephosphorylation of HDAC4 may regulate terminal chondrocyte differentiation through the MMP-13 gene in hypertrophic chondrocytes.
Prior work has shown that HDAC4 suppresses MMP-13 transcription and PTH controls HDAC4 functions via the dissociation from Runx2 and the regulated feedback of HDAC4 in osteoblasts (25). CaMK-activated phosphorylation of class II HDACs such as HDAC4, HDAC5 and HDAC7 causes binding to 14-3-3 proteins and results in dissociation from transcription factors such as MEF-2 and then HDAC4–14-3-3 complexes are exported to the cytoplasm (28, 29). We have shown that Runx2 is phosphorylated (Ser-28, Ser-347, and Thr-340), and PKA regulation of Ser-347 mRunx2 stimulates MMP-13 transcription (30). These phosphorylation sites of Runx2 may be related to binding to HDAC4 in the nucleus to regulate the MMP-13 gene via Runx2's association or dissociation with HDAC4. Berdeaux et al. (31) have shown that dephosphorylation of a Ser/Thr kinase, SIK1 at Ser-577 enhances MEF-2 activity through the phosphotransfer reaction of SIK1 to class II HDACs and PKA also inhibits SIK1 activity via phosphorylation. Although both activated SIK1 or CaMKI phosphorylate HDAC4 and induce nuclear exclusion, forskolin administration counters the ability of activated forms of either SIK1 or CAMKI to affect Ser-246 phosphorylation of HDAC4 by inducing the expression and/or activity of an HDAC4 phospho-Ser-246 phosphatase in chick chondrocytes (21). In contrast, we demonstrate that PKA activation by PTH in osteoblasts induces phosphorylation of HDAC4, especially Ser-740 on rHDAC4, which is related to dissociation from Runx2 on the MMP-13 promoter and regulates this gene transcription.
We have shown that HDAC4 is partially degraded in the cytoplasm after PTH stimulation in osteoblastic cells. The degradation of HDAC4 is inhibitable and induced immediately by phosphorylation via the PKA pathway. The partial degradation of HDAC4 has been previously shown to be due to cleavage by caspase-2 and caspase-3 (10, 22), or through SUMOylation-dependent proteasome degradation (23) or PKA-mediated HDAC4 cleavage through a serine protease (24). Our investigations, however, indicate that SUMO modification and alternative splicing of HDAC4 do not affect its degradation after PTH stimulation (data not shown). We have shown that HDAC4 is partially and selectively degraded and the degradation in the cytoplasm is associated with phosphorylation/dephosphorylation and the lysosomal pathway. We have shown that the degradation of HDAC4 is suppressed by lysosomal inhibitors.
In conclusion, we demonstrate here that PTH regulates numerous steps in the life cycle of HDAC4 in osteoblastic cells. PKA-dependent phosphorylation of rHDAC4 at Ser-740 is related to dissociation from Runx2 on the MMP-13 promoter and results in stimulation of gene transcription. PTH induces HDAC4 export to the cytoplasm. The enzyme is then partially degraded in steps which require PKA, phosphatase and lysosomal activity. These are the first findings of this system of regulation of HDAC4. It demonstrates a role for HDAC4 in hormonal regulation of gene expression in osteoblasts.
This work was supported, in whole or in part, by National Institutes of Health Grant DK47420 (to N. C. P.).
- HDAC
- histone deacetylase
- PTH
- parathyroid hormone
- PTHrP
- parathyroid hormone-related peptide
- MEF
- myocyte enhancer factor
- CIP
- calf intestinal alkaline phosphatase.
REFERENCES
- 1. Verdin E., Dequiedt F., Kasler H. G. (2003) Class II histone deacetylases: versatile regulators. Trends Genet. 19, 286–293 [DOI] [PubMed] [Google Scholar]
- 2. Fischle W., Dequiedt F., Hendzel M. J., Guenther M. G., Lazar M. A., Voelter W., Verdin E. (2002) Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45–57 [DOI] [PubMed] [Google Scholar]
- 3. Fischle W., Dequiedt F., Fillion M., Hendzel M. J., Voelter W., Verdin E. (2001) Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J. Biol. Chem. 276, 35826–35835 [DOI] [PubMed] [Google Scholar]
- 4. Razidlo D. F., Whitney T. J., Casper M. E., McGee-Lawrence M. E., Stensgard B. A., Li X., Secreto F. J., Knutson S. K., Hiebert S. W., Westendorf J. J. (2010) Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLoS One 5, e11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradley E. W., McGee-Lawrence M. E., Westendorf J. J. (2011) Hdac-mediated control of endochondral and intramembranous ossification. Crit. Rev. Eukaryot. Gene. Expr. 21, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Westendorf J. J. (2007) Histone deacetylases in control of skeletogenesis. J. Cell Biochem. 102, 332–340 [DOI] [PubMed] [Google Scholar]
- 7. Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. (2004) Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119, 555–566 [DOI] [PubMed] [Google Scholar]
- 8. Kang J. S., Alliston T., Delston R., Derynck R. (2005) Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J. 24, 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jeon E. J., Lee K. Y., Choi N. S., Lee M. H., Kim H. N., Jin Y. H., Ryoo H. M., Choi J. Y., Yoshida M., Nishino N., Oh B. C., Lee K. S., Lee Y. H., Bae S. C. (2006) Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 281, 16502–16511 [DOI] [PubMed] [Google Scholar]
- 10. Paroni G., Cernotta N., Dello Russo C., Gallinari P., Pallaoro M., Foti C., Talamo F., Orsatti L., Steinkühler C., Brancolini C. (2008) PP2A regulates HDAC4 nuclear import. Mol. Biol. Cell 19, 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grozinger C. M., Schreiber S. L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. U.S.A. 97, 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897 [DOI] [PubMed] [Google Scholar]
- 13. Zhao X., Ito A., Kane C. D., Liao T. S., Bolger T. A., Lemrow S. M., Means A. R., Yao T. P. (2001) The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 276, 35042–35048 [DOI] [PubMed] [Google Scholar]
- 14. Ha C. H., Wang W., Jhun B. S., Wong C., Hausser A., Pfizenmaier K., McKinsey T. A., Olson E. N., Jin Z. G. (2008) Protein kinase D-dependent phosphorylation and nuclear export of histone deacetylase 5 mediates vascular endothelial growth factor-induced gene expression and angiogenesis. J. Biol. Chem. 283, 14590–14599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vega R. B., Harrison B. C., Meadows E., Roberts C. R., Papst P. J., Olson E. N., McKinsey T. A. (2004) Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24, 8374–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Backs J., Backs T., Bezprozvannaya S., McKinsey T. A., Olson E. N. (2008) Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4. Mol. Cell. Biol. 28, 3437–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Backs J., Song K., Bezprozvannaya S., Chang S., Olson E. N. (2006) CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 116, 1853–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strewler G. J., Stern P. H., Jacobs J. W., Eveloff J., Klein R. F., Leung S. C., Rosenblatt M., Nissenson R. A. (1987) Parathyroid hormonelike protein from human renal carcinoma cells. Structural and functional homology with parathyroid hormone. J. Clin. Invest. 80, 1803–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jüppner H., Abou-Samra A. B., Freeman M., Kong X. F., Schipani E., Richards J., Kolakowski L. F., Jr., Hock J., Potts J. T., Jr., Kronenberg H. M., Segre G. V. (1991) A G protein-linked receptor for parathyroid hormone and parathyroid hormone-related peptide. Science 254, 1024–1026 [DOI] [PubMed] [Google Scholar]
- 20. Li X., Liu H., Qin L., Tamasi J., Bergenstock M., Shapses S., Feyen J. H., Notterman D. A., Partridge N. C. (2007) Determination of dual effects of parathyroid hormone on skeletal gene expression in vivo by microarray and network analysis. J. Biol. Chem. 282, 33086–33097 [DOI] [PubMed] [Google Scholar]
- 21. Kozhemyakina E., Cohen T., Yao T. P., Lassar A. B. (2009) Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol. Cell. Biol. 29, 5751–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu F., Dowling M., Yang X. J., Kao G. D. (2004) Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 279, 34537–34546 [DOI] [PubMed] [Google Scholar]
- 23. Kirsh O., Seeler J. S., Pichler A., Gast A., Müller S., Miska E., Mathieu M., Harel-Bellan A., Kouzarides T., Melchior F., Dejean A. (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21, 2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Backs J., Worst B. C., Lehmann L. H., Patrick D. M., Jebessa Z., Kreusser M. M., Sun Q., Chen L., Heft C., Katus H. A., Olson E. N. (2011) Selective repression of MEF2 activity by PKA-dependent proteolysis of HDAC4. J. Cell Biol. 195, 403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shimizu E., Selvamurugan N., Westendorf J. J., Olson E. N., Partridge N. C. (2010) HDAC4 represses matrix metalloproteinase-13 transcription in osteoblastic cells, and parathyroid hormone controls this repression. J. Biol. Chem. 285, 9616–9626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Swarthout J. T., D'Alonzo R. C., Selvamurugan N., Partridge N. C. (2002) Parathyroid hormone-dependent signaling pathways regulating genes in bone cells. Gene. 282, 1–17 [DOI] [PubMed] [Google Scholar]
- 27. Wang A. H., Kruhlak M. J., Wu J., Bertos N. R., Vezmar M., Posner B. I., Bazett-Jones D. P., Yang X. J. (2000) Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20, 6904–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gao C., Li X., Lam M., Liu Y., Chakraborty S., Kao H. Y. (2006) CRM1 mediates nuclear export of HDAC7 independently of HDAC7 phosphorylation and association with 14-3-3s. FEBS Lett. 580, 5096–5104 [DOI] [PubMed] [Google Scholar]
- 29. McKinsey T. A., Zhang C. L., Olson E. N. (2001) Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21, 6312–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Selvamurugan N., Shimizu E., Lee M., Liu T., Li H., Partridge N. C. (2009) Identification and characterization of Runx2 phosphorylation sites involved in matrix metalloproteinase-13 promoter activation. FEBS Lett. 583, 1141–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berdeaux R., Goebel N., Banaszynski L., Takemori H., Wandless T., Shelton G. D., Montminy M. (2007) SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 13, 597–603 [DOI] [PubMed] [Google Scholar]