Background: Rfu1 localizes to endosomes and has a role in ubiquitin homeostasis.
Results: Bro1 recruits Rfu1 to endosomes by binding through the Bro1 V domain and the YPEL motif of Rfu1, and Bro1 overexpression prevents Rfu1 degradation in heat-shocked cells.
Conclusion: Bro1 regulates both deuibiquitinating enzymes Doa4 and Rfu1, which inhibits Doa4.
Significance: The system that maintains ubiquitin homeostasis is elaborately regulated.
Keywords: Cell Biology, Deubiquitination, Endosome, Protein Degradation, Stress Response, Ubiquitin, Ubiquitin Ligase, Heat Shock
Abstract
Yeast Rfu1 (regulator for free ubiquitin chain 1) localizes to endosomes and plays a role in ubiquitin homeostasis by inhibiting the activity of Doa4. We show that Bro1, a member of the class E vacuolar protein sorting proteins that recruits Doa4 to endosomes and stimulates Doa4 deubiquitinating activity, also recruits Rfu1 to endosomes for involvement in ubiquitin homeostasis. This recruitment was mediated by the direct interaction between a region containing the YPEL motif in Rfu1 and the V domain in Bro1, which could be analogous to the interaction between the mammalian Alix V domain and YPXnL motifs of viral and cellular proteins. Furthermore, overexpression of Bro1, particularly the V domain, prevented Rfu1 degradation in response to heat shock. Thus, Bro1, a Doa4 positive regulator, regulated Rfu1, a negative regulator of Doa4. Rfu1 degradation partly involved the proteasome and a ubiquitin ligase Rsp5, suggesting that Rfu1 stability was regulated by ubiquitin-proteasome pathways.
Introduction
Ubiquitin is a highly conserved 76-amino acid protein that is covalently attached to lysine residues within target proteins via a carboxyl-terminal glycine (1). The ubiquitin modification plays a critical role in regulating various cellular processes, such as protein degradation, protein trafficking, chromosome surveillance, apoptosis, and signal transduction. To perform such a remarkable array of cellular tasks, many ubiquitins are required in a cell. Ubiquitins are expressed abundantly from several ubiquitin-encoding genes, and ubiquitin expression appears to be regulated because either an excess or shortage of ubiquitin is detrimental to cells (2). Indeed, a cell has several different mechanisms to control and maintain ubiquitin homeostasis. For example, in response to heat stress, which induces protein ubiquitination, the transcription of polyubiquitin genes is up-regulated to produce more ubiquitins (3). In addition, some deubiquitinating enzymes (Dubs)2 play important roles in ubiquitin homeostasis. For example, mice with a mutation in the gene encoding UCH-L1, an abundant brain-specific Dub that causes gracile axonal dystrophy, exhibit reduced ubiquitin levels (4). Similarly, the ataxia (axJ) mutation caused by reduced expression of Usp14, a proteasome-associated mammalian Dub, causes reduced ubiquitin levels (5). In yeast, mutations in Ubp6, an Usp14 homolog, cause reduced levels of monomer ubiquitin (6).
Endosomes facilitate the transport of membrane proteins between the plasma membrane and lysosomes/vacuoles (7). Many ubiquitins are used to regulate trafficking processes at endosomes (8). In yeast, Doa4 is an endosome-associated Dub that recovers ubiquitin from ubiquitinated cargos when they are incorporated into multivesicular bodies (9–13). In addition, Doa4 contributes to ubiquitin recycling and is important for maintaining the monomeric ubiquitin pool within a cell. In the absence of Doa4, the level of monomer ubiquitin decreases and small ubiquitin species or free ubiquitin chains increase (12–15). Previously, we identified Rfu1 (regulator for free ubiquitin chains 1) as an inhibitor of Doa4 (16). In the absence of Rfu1, the level of free ubiquitin chains decreased, and ubiquitin monomers increased. Moreover, Rfu1 inhibited the Doa4-mediated deubiquitination of a ubiquitinated cargo in vivo, and the deubiquitinating activity of Doa4 for ubiquitin chains in vitro. During heat shock, Rfu1 was degraded presumably to allow Doa4 to produce ubiquitin monomers to meet the need for more ubiquitin within the cell.
Interestingly, Doa4 is also regulated by another factor, Bro1. Bro1 is an ESCRT-III adaptor protein that binds to Snf7, one of the ESCRT-III subunits, to enhance the stability of ESCRT-III (17, 18). Bro1 recruits Doa4 to endosomes and activates Doa4 deubiquitinating activity (19–21). A mutation in Bro1 was reported to have a similar ubiquitin profile as the doa4 mutation (22). Notably, recent studies showed that that the V domains of mammalian Alix, a homolog of Bro1, and of various yeast Bro1 were bound to K63-linked ubiquitin chains (23–25). In this study, we examined the localization and degradation mechanisms of Rfu1 and revealed that both mechanisms are largely dependent on Bro1.
EXPERIMENTAL PROCEDURES
Media
Yeast strains were grown in YPAD medium (1% yeast extract, 2% Bacto-peptone, 2% glucose, and 0.002% adenine), in synthetic complete medium (SD: 0.67% yeast nitrogen base and 2% glucose supplemented with amino acids) or synthetic casamino medium (SC: 0.67% yeast nitrogen base, 2% glucose, and 0.5% casamino acids; if necessary, tryptophan, uracil, or adenine was added). For microscopy studies, 0.02% adenine was added.
Yeast Strains
A list of the yeast strains used in this study is provided in supplemental Table S1. To delete BRO1 with HIS3, a PCR-generated EcoRV fragment carrying HIS3 was inserted into the blunted HindIII sites (+446, +2339) of BRO1 (nucleotide −434 to +2585) in BSII to create E766. Using the E766 plasmid, a fragment covering −150 to +2535 of BRO1 with HIS3 inserted into BRO1, was generated by PCR and then transformed.
Plasmids
A list of the plasmids used in this study is provided in supplemental Table S2. The Rfu1-GFP and Rfu1 mutant-GFP plasmids (pRfu1(1–200)-GFP, pRfu1(1–124)-GFP, and pRfu1(61–200)-GFP), in which the fusion proteins are expressed under the RFU1 promoter, were created as follows. SpeI and EcoRI fragments of RFU1 with the RFU1 promoter (−740 to −1) were PCR amplified using genomic DNA as a template. These PCR fragments were cut with SpeI, and EcoRI was inserted into the SpeI and EcoRI sites of pGCU10 (26) to create pRfu1(1–200)-GFP and pRfu1(1–124)-GFP. For pRfu1(60–200)-GFP, two PCR fragments were obtained. The two fragments were cut with SpeI-BamHI and BamHI-EcoRI, respectively, and inserted into the SpeI and EcoRI sites of pGCU10.
MBP-Rfu1(1–200), -(1–140), -(1–172), -(61–200) (E382, E609, E392, and E393, respectively) were created as follows. The PCR fragments were cut with EcoRI and XhoI. The resultant fragments were ligated into the EcoRI-SalI fragment of pMAL-p2X (New England Biolabs, Inc.).
Plasmids expressing HA-tagged Bro1-N, Bro1-C, and Bro1-V under the GPD promoter (E710, E711, and E772, respectively), were created as follows. PCR fragments were generated using a genomic library, cut with KpnI-SalI, and ligated with the EcoRI-SalI fragment of pRS426 and KpnI-EcoRI fragment of the 3HA-GPD promoter from E276.
Plasmids expressing GST-Bro1, GST-Bro1-N, GST-Bro1-C, GST-Bro1-V, GST-Bro1-Vcomp, and GST-Bro1-D were created as follows. PCR fragments were generated using E548 as a template, digested with BamHI and SalI, and ligated with the BamHI-SalI fragment of pGEX4T-3.
Antibodies
For Western blotting, blots were incubated with a mouse anti-GFP monoclonal antibody (Roche Applied Science), anti-HA antibody (HA.11, COVANCE, Princeton, NJ), or anti-yeast PGK antibody (Molecular Probes, Eugene, OR), followed by horseradish peroxidase (HRP)-conjugated anti-mouse IgG (NA931V, Amersham Biosciences), and then visualized using ECL-plus reagent (Amersham Biosciences). To detect GST, an HRP-conjugated anti-GST antibody (Wako Chemicals) was used. A rabbit anti-yeast Bro1 antibody was generated by immunizing with purified GST-Bro1 V. To see ubiquitin profiles, blots were incubated with mouse anti-ubiquitin monoclonal antibody (P4D1-HRP, Santa Cruz Biotechnology).
Immunoblotting
Preparation of whole cell extracts and immunoblot analysis were performed as previously described (27) except cells were harvested in the early log phase. To analyze the overall ubiquitin profiles, total cell proteins were separated by 10–20% gradient gels (Biocraft Inc.) using Tricine-based buffer, followed by transfer to Immobilon-P membranes (Millipore). Blots were incubated with mouse anti-ubiquitin monoclonal antibody (P4D1-HRP, Santa Cruz). Alternatively, the blots were incubated with a mouse anti-GFP monoclonal antibody (Roche Applied Science), anti-HA antibody (HA.11, COVANCE), or anti-yeast PGK antibody (Molecular Probes), followed by HRP-conjugated anti-mouse IgG (NA931V, Amersham Biosciences), and then visualized using ECL-plus reagent (Amersham Biosciences). To detect GST, an HRP-conjugated anti-GST antibody (Wako Chemicals) was used. A rabbit anti-yeast Bro1 antibody was generated by immunizing with purified GST-Bro1 V.
Recombinant Protein Purification
MBP, MBP-Rfu1, and MBP fusions of the Rfu1 deletion mutants were purified as previously described (16). Recombinant GST, GST-Bro1, or the various GST-Bro1 deletion mutants were purified using glutathione chromatography as recommended by the manufacturer (Pharmacia). Recombinant proteins were eluted with 20 mm glutathione, 50 mm Tris-HCl (pH 8.0), and 2 mm DTT, dialyzed against 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 10% glycerol, and then stored −80 °C.
In Vitro Binding between Various MBP-fused Rfu1 and GST-fused Bro1 Proteins
MBP or various MBP-fused Rfu1 derivatives (3 μg each) were mixed with 18 μg of GST or various GST-Bro1 fusion proteins in buffer B (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10% glycerol) in a 200-μl reaction volume for 1 h at 25 °C, after which amylose resin was added. After 30 min, the resin was washed with buffer B and eluted with buffer B containing 10 mm maltose. The eluted samples were analyzed by immunoblot analysis using anti-MBP and anti-GST antibodies.
Microscopy
FM4-64 (Molecular Probes, Inc.) staining was performed as described (28). Cells were imaged at room temperature using a confocal microscope (LSM780; Carl Zeiss) equipped with an αPlan-Apochromat ×100 oil objective lens. Images were processed using the LSM image browser, and the brightness and contrast were adjusted using Adobe Photoshop CS4.
RESULTS
Bro1 Mediates the Endosomal Localization of Rfu1
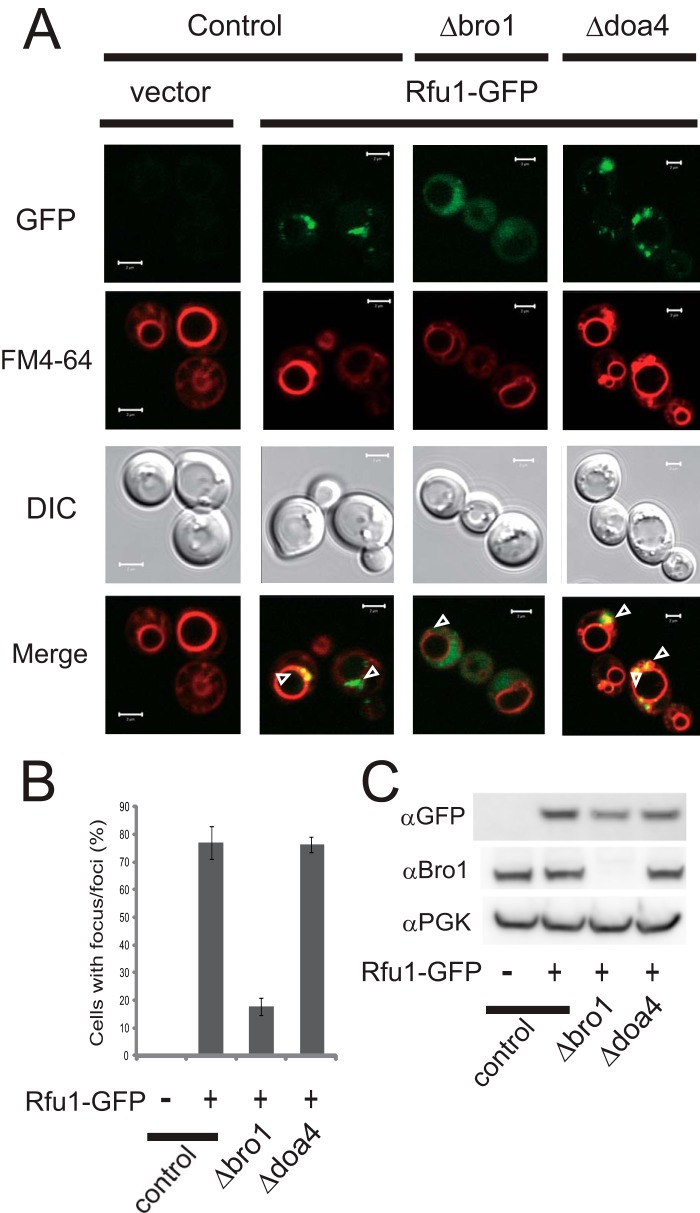
Both Rfu1 and Doa4 localize to endosomes (9, 29). Based on co-immunoprecipitation analyses, Rfu1 was noted to bind to Bro1 as well as Doa4 (16). Because the endosomal localization of Doa4 is largely dependent upon Bro1 (19–21), we tested whether Rfu1 localization was also affected by the presence of Bro1. The localization of exogenous Rfu1-GFP expressed under the RFU1 promoter was examined in Δrfu1Δvps4 cells. In Δvps4 cells, endosome-localized ESCRT factors are concentrated in the class E compartment, an aberrant late endosomal structure that is adjacent to the vacuole (30), and FM4-64 specifically labels class E compartments and vacuolar membranes. As expected, Rfu1-GFP fluorescence was present at foci that overlapped with FM4-64-stained class E compartments in Δvps4Δrfu1 cells (Fig. 1, A and B). In BRO1-deleted Δbro1Δvps4 Δrfu1 cells, however, the localization of Rfu1-GFP at class E compartments was mainly lost. Many Rfu1-GFP-expressing cells showed only highly diffuse cytosolic GFP fluorescence. Some Rfu1-GFP-expressing cells showed much lower levels of punctuate GFP fluorescence at class E compartments against highly diffuse cytosolic GFP fluorescence. An anti-GFP immunoblot analysis showed that Rfu1-GFP was expressed at 10–40% lower levels in Δbro1Δvps4Δrfu1 cells than in Δvps4Δrfu1 cells, suggesting that Bro1 might also affect the stability of Rfu1 (Fig. 1C). We confirmed that the localization of Rfu1-GFP was unaffected in Δdoa4Δvps4Δrfu1cells, suggesting that Doa4 was not involved in Rfu1 localization (Fig. 1, A and B). These results indicated that the endosomal localization of Rfu1 was mainly dependent on Bro1.
FIGURE 1.
Bro1-dependent endosomal localization of Rfu1-GFP. A, GFP and FM4-64 fluorescence and DIC microscopy of Rfu1-GFP in control (Δvps4Δrfu1), Δbro1 (Δbro Δvps4Δrfu1), and Δdoa4(Δdoa4Δvps4Δrfu1) cells. Arrowheads indicate the class E compartments. Scale bar, 2 μm. B, quantification of Rfu1-GFP foci in A. Cells containing GFP foci around the vacuolar membrane were counted among a total of 50 cells in each experiment, and mean values of three independent experiments are shown. S.E. are shown as bars. C, Rfu1-GFP expression as determined by anti-GFP immunoblot analysis. Top, anti-GFP immunoblot. Middle, anti-Bro1 immunoblot. Bottom, anti-phosphoglycerate kinase (PGK) immunoblot, a control for protein loading. DIC, differential interference contrast.
Bro1 and Rfu1 Directly Interact
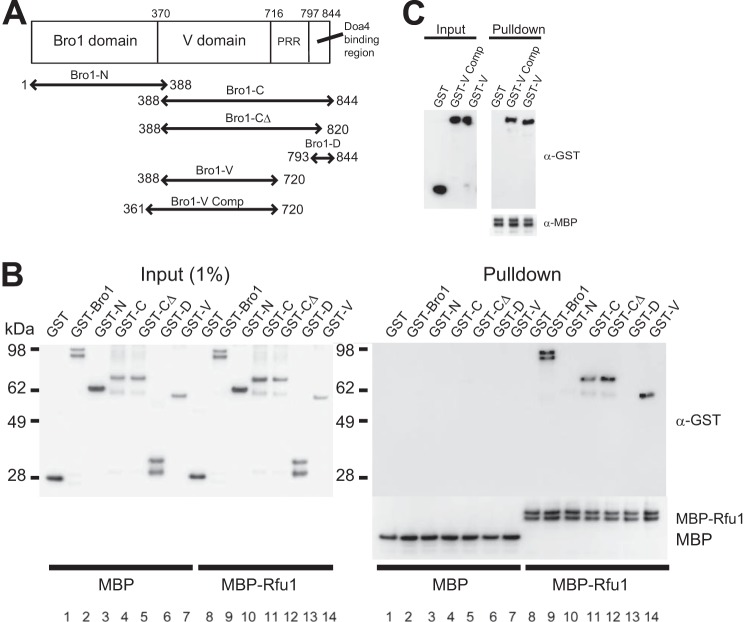
The association between Bro1 and Rfu1 was detected even in the Δdoa4 mutant (16). These results suggested that Doa4 was not required for the interaction between Bro1 and Rfu1, but rather that Bro1 and Rfu1 likely formed a direct interaction. We tested this possibility using purified recombinant proteins. MBP-Rfu1, but not MBP, specifically bound to GST-Bro1, but not GST (Fig. 2B, lanes 8 and 9). We observed that purified recombinant GST-Bro1 was always in a doublet form, and partial degradation might occur during the expression and purification of GST-Bro1. We subsequently examined which region of Bro1 bound to Rfu1 (Fig. 2A). We first divided Bro1 into the following two domains: the N-terminal Bro1 domain (Bro1-N, aa 1–388), which binds to Snf7 and stabilizes ESCRT-III (18, 31) and the C-terminal region (Bro1-C, aa 388–844). We found that MBP-Rfu1 specifically bound to GST-Bro1-C, but not GST-Bro1-N (Fig. 2B, lanes 10 and 11). The Bro1-C region could be divided into three regions. A search of the Pfam database revealed a domain called the V domain from aa 475–704 (32, 33). Secondary structure alignments of Bro1 and Alix suggested that a region within Bro1 from approximately aa 370–706 contains a complete V domain (33). It was previously reported that the Bro1 region from aa 793–844 was sufficient to mediate the interaction with Doa4 and that the PSVF sequence within aa 831–834 of Bro1 was also important for binding (20). Another study reported that Bro1 with a frameshift at aa 732 had impaired Doa4 binding (19). Thus, Bro1-C contains the V domain, a proline-rich region, and a binding region for Doa4 (20). An interaction was observed between MBP-Rfu1 and GST-Bro1-CΔ (aa 388–820, Fig. 2B, lane 12), which was defective in the Doa4-binding region. In contrast, MBP-Rfu1 did not interact with the Doa4-binding region of Bro1, GST-Bro1-D (aa 793–844; Fig. 2B, lane 13). We also observed an interaction between MBP-Rfu1 and GST-Bro1-V (aa 388–720) in which the proline-rich region was mostly deleted (Fig. 1B, lane 14). We determined that GST-Bro1-V (aa 388–720) bound to MBP-Rfu1 as efficiently as GST fusion of a region containing a predicted complete V domain, Bro1 (aa 361–720; Fig. 2C). These results indicated that a region containing the Bro1 V domain is a binding site for Rfu1.
FIGURE 2.
Direct interaction between Rfu1 and Bro1. A, Bro1 organization and constructs. B, analysis of MBP-Rfu1 binding to wild type Bro1 and various Bro1 deletion mutants in vitro. MBP or MBP-Rfu1 was mixed with GST, GST-Bro1, or the noted GST-Bro1 deletion mutants, and the proteins were isolated with amylase resin. Samples were examined by immunoblot analysis using anti-MBP and anti-GST antibodies. C, analysis of MBP-Rfu1 binding to Bro1-V and Bro1-V Comp.
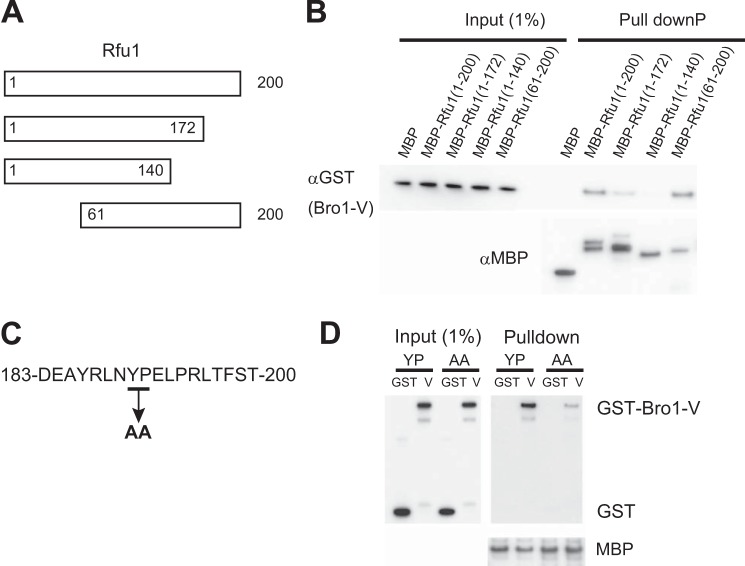
We subsequently generated MBP fusions for Rfu1 deletion mutants to determine which region of Rfu1 was responsible for Bro1-V binding (Fig. 3A). Rfu1 is a 200-amino acid protein, and aside from an N-terminal region with weak homology to the MIT (microtubule interacting and trafficking) domains, Rfu1 does not have distinct domains. Therefore, we generated various deletion mutants and tested their ability to bind to GST-Bro1-V (Fig. 3, A and B). We determined that Bro1-V bound to MBP-Rfu1(61–200), but exhibited significantly reduced binding to MBP-Rfu1(1–172) and MBP-Rfu1(1–140). These results indicated that the C-terminal region of Rfu1 encompassing aa 61–200, particularly the most C-terminal part of Rfu1, was responsible for Bro1-V binding.
FIGURE 3.
Importance of YPEL motif of Rfu1 for binding to Bro1 in vitro. A, Rfu1 organization and constructs. B, analysis of the interaction between various MBP-Rfu1 deletion mutants and Bro1-V(388–720). C, amino acid sequence of the Rfu1 carboxyl terminus. D, impaired binding of MBP-Rfu1(61–200,Y190A,P191A) with Bro1-V.
In mammalian cells, the V domain of ALIX, a homolog of Bro1, binds to YPXnL motifs (where X varies in sequence and length), which are found in the viral Gag proteins and cellular proteins (32–35). We searched for the YPXnL motif within Rfu1. A YPEL sequence was detected at aa 190–193, which is the most carboxyl-terminal region of Rfu1, and was within the region for Bro1 V binding (Fig. 3C). We therefore tested whether the YPEL sequence might be a binding motif for the Bro1 V domain. Indeed, the binding activity for GST-Bro1-V was reduced in a mutant in which YPEL was changed to AAEL in MBP-Rfu1(aa 61–200), indicating that the YPEL motif was important for binding to the Bro1 V domain (Fig. 3D).
Importance of the YPEL Motif of Rfu1 for the Localization of Endosome and Ubiquitin Homeostasis
The importance of the C-terminal region and the YPEL motif in Rfu1 for Bro1 binding was verified by examining the localization of various Rfu1-GFP fusions expressed in yeast (Fig. 4). When an N-terminal-truncated form of Rfu1(60–200)-GFP was expressed in Δvps4Δrfu1 cells, GFP foci co-localized with the class E compartments and exhibited a similar fluorescence pattern as Rfu1(1–200)-GFP, although its expression was lower than that of Rfu1(1–200)-GFP. In contrast, Rfu1(1–124)-GFP fluorescence localization was different. A significant decrease in specific localization on class E compartments were observed, and many Rfu1(1–124)-GFP expressing cells showed higher diffusive fluorescence; these fluorescence patterns were similar to those of Rfu1(1–200)-GFP expressed in BRO1-deleted cells (Fig. 1A). These results suggested that the carboxyl region was important for the endosomal localization of Rfu1-GFP.
FIGURE 4.
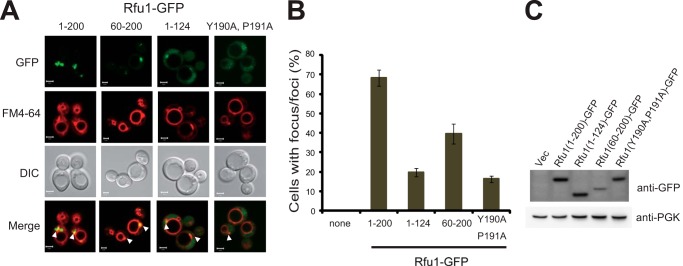
Localization of various Rfu1-GFP mutants. A, GFP and FM4-64 fluorescence and DIC microscopy of Rfu1-GFP, Rfu1(60–200)-GFP, Rfu1(1–124)-GFP, or Rfu1(Y190A,P191A) in Δvps4Δrfu1 cells. Arrowheads indicate the class E compartments. Scale bar, 2 μm. B, quantification of Rfu1-GFP foci in A. Cells containing GFP foci around the vacuolar membrane were counted among a total of 50 cells in each experiment, and mean values of at least three independent experiments are shown. S.E. are shown as bars. C, expression of Rfu1-GFP or various Rfu1-GFP mutants in Δvps4Δrfu1 cells. DIC, differential interference contrast.
We also examined the localization of a YPEL-mutated form of Rfu1(Y190A,P191A)-GFP. Its endosomal localization was mainly lost and there were considerably smaller and fewer foci at class E compartments with a more highly diffuse background (Fig. 4A). We observed that the expression level of Rfu1(Y190A,P191A)-GFP was slightly lower than that of Rfu1(1–200)-GFP, but much higher than that of Rfu1(60–200)-GFP, which showed foci at the class E compartments in Δvps4Δrfu1 cells (Fig. 4B). These results suggested that the YPEL motif was important for the endosomal localization of Rfu1.
We subsequently determined whether the YPEL motif of Rfu1 was important for ubiquitin homeostasis by examining cellular ubiquitin profiles. Rfu1-GFP or Rfu1(Y190A,P191A)-GFP was expressed in Δrfu1 cells, and cellular ubiquitin profiles were examined (Fig. 5A). Wild type cells grown in early growth phase showed a bulk ubiquitin profile with monomeric ubiquitin, free ubiquitin chains, and slowly migrated high-molecular weight forms (16, 36–38). As reported previously, the amount of free ubiquitin chains was decreased, whereas the level of monomeric ubiquitin was increased in Δrfu1 cells (16). We found that the expression of Rfu1-GFP, but not Rfu1(Y190A,P191A)-GFP restored a normal ubiquitin profile, indicating that the YPEL motif of Ruf1 was critical for maintaining ubiquitin homeostasis by Rfu1.
FIGURE 5.
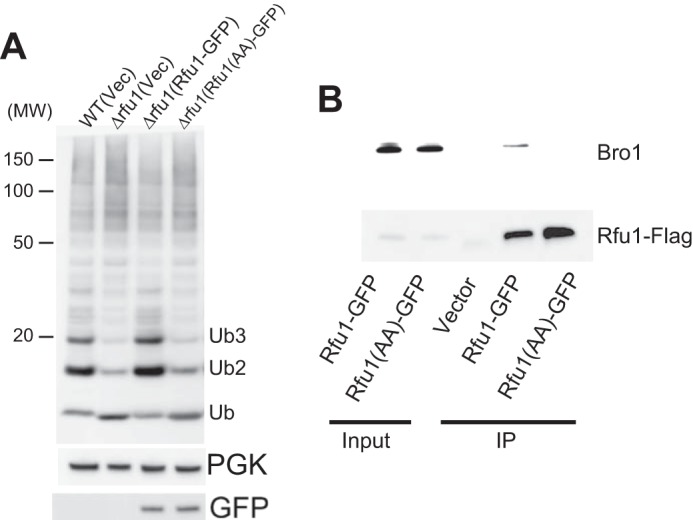
Requirement of the YPEL motif of Rfu1 for ubiquitin homeostasis. A, immunoblot analyses of wild-type, or Δrfu1 cells harboring a vector or a plasmid expressing Rfu1-GFP or Rfu1(Y190A,P191A)-GFP. Rfu1(Y190A,P191A)-GFP is indicated as Rfu1(AA)-GFP. Top, anti-ubiquitin immunoblot analysis. The position of size standard in kilodaltons is indicated on the left. Monomeric ubiquitin and ubiquitin chain positions are marked. Middle, phosphoglycerate kinase (PGK) immunoblot. Bottom, anti-GFP immunoblot. B, impaired association of Rfu1(Y190A,P191A)-GFP with endogenous Bro1 in vivo. Lysates of Δrfu1 cells harboring a vector or plasmids expressing Rfu1-GFP or Rfu1(Y190A,P191A)-GFP were immunoprecipitated (IP) with anti-GFP. The resulting immunocomplexes were analyzed by immunoblot using anti-GFP and anti-Bro1.
We investigated whether the YPEL motif of Rfu1 functioned in the interaction with Bro1 in vivo (Fig. 5B). Immunoprecipitation analysis using lysates from Δrfu1 cells expressing Rfu1-GFP or Rfu1(Y190A,P191A)-GFP was performed. Endogenous Bro1 was specifically immunoprecipitated by Rfu1-GFP, but not Rfu1(Y190A,P191A)-GFP. These results indicated that the YPEL motif of Rfu1 was important for ubiquitin homeostasis and Bro1 binding in vivo.
Rsp5 Contributes to Rfu1-3F Degradation
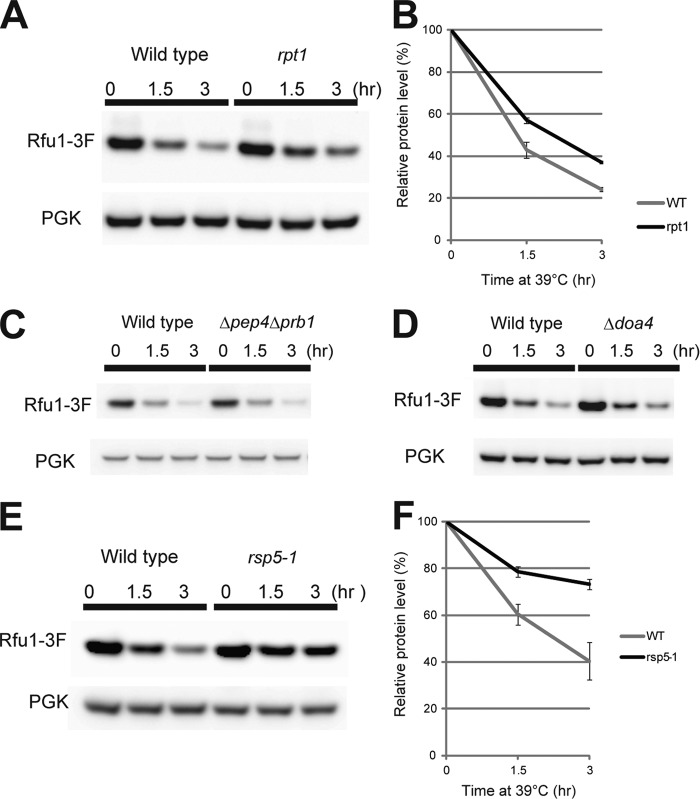
Rfu1–3xFLAG (Rfu1-3F) is degraded upon heat shock (16), possibly allowing more Rfu1-free Doa4 deubiquitination to produce ubiquitin monomers. Concurrently with the interaction analysis of Bro1 and Rfu1, we investigated the mechanism of Rfu1 degradation during heat shock. We found that degradation of exogenously expressed Rfu1-3F was partially inhibited in the rpt1 mutant, which has a temperature-sensitive mutation in one of the 26 S proteasome subunits (Fig. 6, A and B), suggesting that proteasomal degradation was involved. In contrast, Rfu1-3F degradation was not impaired in the Δpep4Δpbr1 mutant in which many vacuolar degradation events were inhibited (Fig. 6C). In addition, we found that Rfu1-3F degradation was not impaired by the doa4 mutation (Fig. 6D).
FIGURE 6.
Degradation of Rfu1-3xFLAG during heat shock is prevented in rpt1 and rsp5-1 mutant cells. A, Rfu1-3xFLAG and phosphoglycerate kinase (PGK) protein levels in rpt1 mutant cells. Cells expressing Rfu1-3xFLAG under the RFU1 promoter (E508) were grown in SC-Ura and then heat shocked at the indicated times in the presence of cycloheximide. B, protein levels were quantified from the bands in A. Data are the mean ± S.E. of three independent experiments. C, Rfu1-3xFLAG and PGK protein levels in Δpep4Δprb1 mutant cells. D, Rfu1-3xFLAG and PGK protein levels in control (Y860) and Δdoa4 cells (Y862). E, Rfu1-3xFLAG and PGK levels in rsp5-1. F, quantification of E.
We investigated which ubiquitin ligase (E3) was involved in Rfu1-3F degradation. We expressed Rfu1-3F exogenously in 54 nonessential deletion mutants of E3 ligase or E3 ligase candidates (supplemental Table S1), and examined the degradation of Rfu1-3F in these mutants. However, none of the mutants impaired Rfu1 degradation.3 Because Rsp5, an essential ubiquitin ligase, is recruited to endosomes (39), we tested a temperature-sensitive mutant of Rsp5, rsp5-1 (40). Rfu1-3F degradation was observed to be inhibited (Fig. 6, E and F), indicating that Rsp5 was involved in Rfu1-3F degradation.
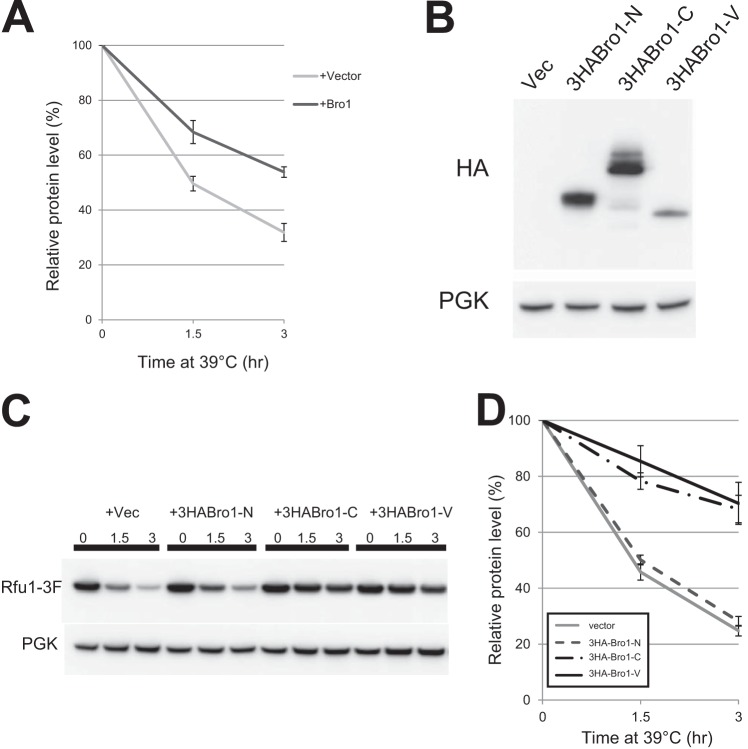
Bro1 Overexpression Prevents Rfu1-3xFLAG Degradation Upon Heat Shock
Bro1 recruits Doa4 to endosomes and activates the deubiquitinating activity of Doa4 (20). Therefore, it is possible that Bro1 has other effects on Rfu1 in addition to recruiting Rfu1 to endosomes. We therefore tested the effects of Bro1 overexpression on Rfu1-3F degradation. Cycloheximide was added when the temperature was shifted to 39 °C to prevent further production of Rfu1-3F. We observed that overexpression of full-length Bro1 inhibited Rfu1-3F degradation (Fig. 7A). We then examined the effects of overexpressing HA-tagged Bro1-N, Bro1-C, and Bro1-V. We observed that Bro1-C as well as Bro1-V inhibited Rfu1-3F degradation (Fig. 7, C and D). Bro1-N expression had no effect, although it was expressed at a significant level (Fig. 7B). These results indicate that the Bro1 V domain expression inhibited Rfu1 degradation after heat shock.
FIGURE 7.
Bro1 overexpression prevents Rfu1-3xFLAG degradation during heat shock. A, quantification of Rfu1-3xFLAG and phosphoglycerate kinase (PGK) protein levels. Cells (Y861) with or without full-length Bro1 overexpression were heat shocked at 39 °C at the indicated times in SC-Ura containing 50 μg/ml of cycloheximide. Data are the mean ± S.E. of four experiments. B, expression levels of HA-tagged Bro1 deletion mutants as determined by anti-HA immunoblot analysis. 3HA-Bro1-N, 3HA-Bro-C, and 3HA-Bro1-V indicate that 3HA-tagged Bro1 fragments of Bro1(1–388), Bro1(388–844), and Bro1(388–720), respectively. C, Rfu1-3xFLAG and PGK protein levels in cells (Y1007) with or without the overexpression of HA-tagged Bro1 deletion mutants during heat shock in the presence of cycloheximide. D, quantification of Rfu1-3xFLAG levels in C. Protein levels were quantified from the bands shown in C. Data are the mean ± S.E. of three independent experiments.
DISCUSSION
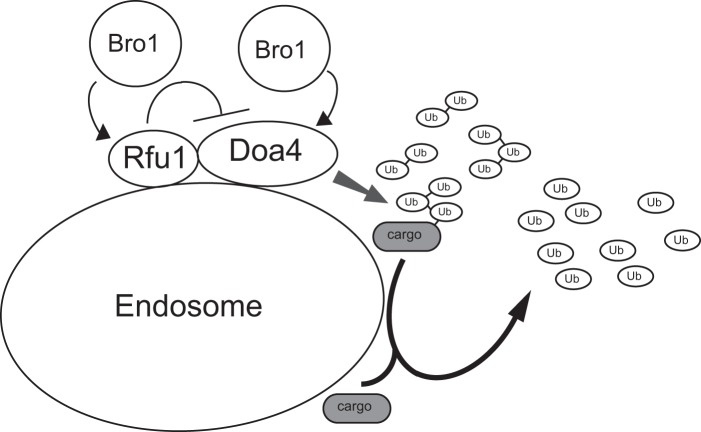
In addition to trafficking and sorting various cargos, endosomes seem to have an important role in ubiquitin homeostasis. In this study, we showed that the endosome localization of Rfu1 was important for ubiquitin homeostasis, and this was achieved by an Rfu1 interaction with Bro1. It was reasonable to assume that Rfu1 must be present on endosomes to elicit its function, because Doa4, a target of Rfu1, is localized at endosomes. Many ubiquitins are utilized on endosomes, and Bro1 recruits both Doa4 and Rfu1 for them to function. Thus, Bro1, a positive regulator of Doa4, also regulates a negative regulator of Doa4, Rfu1 (Fig. 8). Although the detailed regulation of Bro1 on Doa4 and Rfu1 is still unclear, from previous and present analysis, Bro1 was observed to bind to both Rfu1 and Doa4, but different regions within Bro1 mediate the binding to Doa4 and Rfu1; the C-terminal region is required for the interaction with Doa4, and the V domain mediate binding to Rfu1. Therefore, one Bro1 molecule may simultaneously interact with Doa4 and Rfu1, and regulate them. Certainly, it will be important to determine how the three proteins interact with each other.
FIGURE 8.
Model for relationship between Doa4, Rfu1, and Bro1.
Mammalian Alix V binds to the YPXnL motif of viral and cellular proteins, and these interactions are important for virus budding, multivesicular body sorting, or generation of exosomes (32–35). Our analysis revealed for the first time that the yeast Bro1 V domain also binds the YPXnL motif (YPEL in Rfu1) and has important functions as well, notably recruiting Rfu1 through the YPEL motif and preventing Rfu1 degradation when it was overexpressed. Therefore, although amino acid sequences are not entirely highly conserved between the V domains of Alix and Bro1, the specific binding mode for a YPXnL motif may be similar between them.
Recently, it was reported that the Bro1 V domain binds to K63-linked ubiquitin chains (25). Thus, it is tempting to speculate that the Bro1 V domain binds to both Rfu1 and K63-linked ubiquitin chains. In a cell, binding of the V domain to Rfu1 may affect the efficiency of the V domain binding to ubiquitin chains, or vice versa, and the regulated binding of Bro1 V domain to Rfu1 and ubiquitin chains may result in the regulation of ubiquitin homeostasis. Further detailed analysis on the V domain is required to clarify this point.
We also addressed the mechanism of Rfu1 degradation in response to heat shock. Our results showed that Rfu1 degradation was partially inhibited in the rpt1 mutant and that degradation was not inhibited by Δpep4Δprb1, suggesting that at least part, if not all, of Rfu1 is degraded by the proteasome. Furthermore, we found that Rsp5, a ubiquitin ligase, was required for Rfu1 degradation. Thus, a regulator of ubiquitin homeostasis may be regulated by a ubiquitin-related system; however, the detailed mechanisms for Rfu1 degradation along with the role of Bro1 in Rfu1 degradation remains to be investigated. Moreover, further analysis is required to clarify whether Rsp5 is directly or indirectly involved in Rfu1 degradation. Although we have not been able to detect the ubiquitinated form of Rfu1 by immunoprecipitation analysis in MG132-treated heat-shocked cells,3 Rfu1 may be ubiquitinated by Rsp5 during heat shock, and Bro1 may prevent Rfu1 ubiquitination. It is noteworthy that Rsp5 is also related to ubiquitin homeostasis (41).
In conclusion, we showed that Bro1 regulates the localization and the activity of Rfu1. Our data suggests that the system that maintains ubiquitin homeostasis would be elaborately regulated.
Supplementary Material
Acknowledgments
We thank Drs. K. Kohno, Y. Kimata, J. M. Huibregtse, and Y. Saeki for materials, and members of the Tanaka lab for helpful discussions.
This work was supported by Grant-in-Aids for Scientific Research (C) (to Y. K.), Specially Promoted Research, Scientific Research on Innovative Areas (to Y. K.), and the Takeda Science Foundation (to K. T.).

This article contains supplemental Tables S1 and S2.
Y. Kimura, J. Kawawaki, and K. Tanaka, unpublished observation.
- Dub
- deubiquitinating enzyme
- MBP
- maltose-binding protein
- Rfu1
- regulator for free ubiquitin chain 1
- Rfu1-3F
- Rfu1-3xFLAG
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- aa
- amino acid.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Kimura Y., Tanaka K. (2010) Regulatory mechanisms involved in the control of ubiquitin homeostasis. J. Biochem. 147, 793–798 [DOI] [PubMed] [Google Scholar]
- 3. Finley D., Ozkaynak E., Varshavsky A. (1987) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48, 1035–1046 [DOI] [PubMed] [Google Scholar]
- 4. Osaka H., Wang Y. L., Takada K., Takizawa S., Setsuie R., Li H., Sato Y., Nishikawa K., Sun Y. J., Sakurai M., Harada T., Hara Y., Kimura I., Chiba S., Namikawa K., Kiyama H., Noda M., Aoki S., Wada K. (2003) Ubiquitin carboxyl-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum. Mol. Genet. 12, 1945–1958 [DOI] [PubMed] [Google Scholar]
- 5. Anderson C., Crimmins S., Wilson J. A., Korbel G. A., Ploegh H. L., Wilson S. M. (2005) Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J. Neurochem. 95, 724–731 [DOI] [PubMed] [Google Scholar]
- 6. Chernova T. A., Allen K. D., Wesoloski L. M., Shanks J. R., Chernoff Y. O., Wilkinson K. D. (2003) Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J. Biol. Chem. 278, 52102–52115 [DOI] [PubMed] [Google Scholar]
- 7. Henne W. M., Buchkovich N. J., Emr S. D. (2011) The ESCRT pathway. Dev. Cell 21, 77–91 [DOI] [PubMed] [Google Scholar]
- 8. Clague M. J., Liu H., Urbé S. (2012) Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev. Cell 23, 457–467 [DOI] [PubMed] [Google Scholar]
- 9. Amerik A. Y., Nowak J., Swaminathan S., Hochstrasser M. (2000) The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11, 3365–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amerik A. Y., Hochstrasser M. (2004) Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695, 189–207 [DOI] [PubMed] [Google Scholar]
- 11. Katzmann D. J., Babst M., Emr S. D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 12. Dupré S., Haguenauer-Tsapis R. (2001) Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of Doa4p ubiquitin isopeptidase. Mol. Cell Biol. 21, 4482–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nikko E., André B. (2007) Evidence for a direct role of the Doa4 deubiquitinating enzyme in protein sorting into the MVB pathway. Traffic 8, 566–581 [DOI] [PubMed] [Google Scholar]
- 14. Papa F. R., Hochstrasser M. (1993) The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature 366, 313–319 [DOI] [PubMed] [Google Scholar]
- 15. Swaminathan S., Amerik A. Y., Hochstrasser M. (1999) The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10, 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kimura Y., Yashiroda H., Kudo T., Koitabashi S., Murata S., Kakizuka A., Tanaka K. (2009) An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell 137, 549–559 [DOI] [PubMed] [Google Scholar]
- 17. Odorizzi G., Katzmann D. J., Babst M., Audhya A., Emr S. D. (2003) Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116, 1893–1903 [DOI] [PubMed] [Google Scholar]
- 18. Wemmer M., Azmi I., West M., Davies B., Katzmann D., Odorizzi G. (2011) Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J. Cell Biol. 192, 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nikko E., André B. (2007) Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot. Cell 6, 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richter C., West M., Odorizzi G. (2007) Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 26, 2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luhtala N., Odorizzi G. (2004) Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amerik A., Sindhi N., Hochstrasser M. (2006) A conserved late endosome-targeting signal required for Doa4 deubiquitylating enzyme function. J. Cell Biol. 175, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dowlatshahi D. P., Sandrin V., Vivona S., Shaler T. A., Kaiser S. E., Melandri F., Sundquist W. I., Kopito R. R. (2012) ALIX is a Lys-63-specific polyubiquitin binding protein that functions in retrovirus budding. Dev. Cell 23, 1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keren-Kaplan T., Attali I., Estrin M., Kuo L. S., Farkash E., Jerabek-Willemsen M., Blutraich N., Artzi S., Peri A., Freed E. O., Wolfson H. J., Prag G. (2013) Structure-based in silico identification of ubiquitin-binding domains provides insights into the ALIX-V:ubiquitin complex and retrovirus budding. EMBO J. 32, 538–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pashkova N., Gakhar L., Winistorfer S. C., Sunshine A. B., Rich M., Dunham M. J., Yu L., Piper R. C. (2013) The yeast alix homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev. Cell 25, 520–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kimata Y., Iwaki M., Lim C. R., Kohno K. (1997) A novel mutation which enhances the fluorescence of green fluorescent protein at high temperatures. Biochem. Biophys. Res. Commun. 232, 69–73 [DOI] [PubMed] [Google Scholar]
- 27. Kimura Y., Koitabashi S., Kakizuka A., Fujita T. (2001) Initial process of polyglutamine aggregate formation in vivo. Genes Cells 6, 887–897 [DOI] [PubMed] [Google Scholar]
- 28. Vida T. A., Emr S. D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 30. Babst M., Wendland B., Estepa E. J., Emr S. D. (1998) The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim J., Sitaraman S., Hierro A., Beach B. M., Odorizzi G., Hurley J. H. (2005) Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell 8, 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S., Joshi A., Nagashima K., Freed E. O., Hurley J. H. (2007) Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 14, 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fisher R. D., Chung H. Y., Zhai Q., Robinson H., Sundquist W. I., Hill C. P. (2007) Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128, 841–852 [DOI] [PubMed] [Google Scholar]
- 34. Baietti M. F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., Zimmermann P., David G. (2012) Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell. Biol. 14, 677–685 [DOI] [PubMed] [Google Scholar]
- 35. Dores M. R., Chen B., Lin H., Soh U. J., Paing M. M., Montagne W. A., Meerloo T., Trejo J. (2012) ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J. Cell Biol. 197, 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Nocker S., Vierstra R. D. (1993) Multiubiquitin chains linked through lysine 48 are abundant in vivo and are competent intermediates in the ubiquitin proteolytic pathway. J. Biol. Chem. 268, 24766–24773 [PubMed] [Google Scholar]
- 37. Amerik A. Yu, Swaminathan S., Krantz B. A., Wilkinson K. D., Hochstrasser M. (1997) In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 16, 4826–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Amerik A. Y., Li S. J., Hochstrasser M. (2000) Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381, 981–992 [DOI] [PubMed] [Google Scholar]
- 39. Wang G., McCaffery J. M., Wendland B., Dupré S., Haguenauer-Tsapis R., Huibregtse J. M. (2001) Localization of the Rsp5p ubiquitin-protein ligase at multiple sites within the endocytic pathway. Mol. Cell Biol. 21, 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huibregtse J. M., Yang J. C., Beaudenon S. L. (1997) The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 94, 3656–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krsmanovi T., Kölling R. (2004) The HECT E3 ubiquitin ligase Rsp5 is important for ubiquitin homeostasis in yeast. FEBS Lett. 577, 215–219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.