Background: Particulate methane monooxygenase (pMMO) is a membrane-bound metalloenzyme that oxidizes methane to methanol.
Results: Metal binding data and crystal structures reveal that zinc inhibits pMMO at two sites.
Conclusion: Zinc does not inhibit pMMO by binding at the active site but may hinder another function such as proton transfer.
Significance: New insight into the function of an environmentally and industrially important enzyme has been obtained.
Keywords: Copper, Crystal Structure, Enzyme Inhibitor, Membrane Enzyme, Metalloprotein, Zinc, Methane Monooxygenase, Methanotroph
Abstract
Particulate methane monooxygenase (pMMO) is a membrane-bound metalloenzyme that oxidizes methane to methanol in methanotrophic bacteria. Zinc is a known inhibitor of pMMO, but the details of zinc binding and the mechanism of inhibition are not understood. Metal binding and activity assays on membrane-bound pMMO from Methylococcus capsulatus (Bath) reveal that zinc inhibits pMMO at two sites that are distinct from the copper active site. The 2.6 Å resolution crystal structure of Methylocystis species strain Rockwell pMMO reveals two previously undetected bound lipids, and metal soaking experiments identify likely locations for the two zinc inhibition sites. The first is the crystallographic zinc site in the pmoC subunit, and zinc binding here leads to the ordering of 10 previously unobserved residues. A second zinc site is present on the cytoplasmic side of the pmoC subunit. Parallels between these results and zinc inhibition studies of several respiratory complexes suggest that zinc might inhibit proton transfer in pMMO.
Introduction
Methanotrophic bacteria, organisms that oxidize methane to methanol in the first step of their metabolism (1), have become increasingly important in the quest for efficient conversion of abundant natural gas to useable fuels and chemicals (2). Methane oxidation is catalyzed by methane monooxygenase enzymes (MMOs).3 The majority of methanotrophs produces a membrane-bound or particulate MMO (pMMO) (3), and a few strains can also express a soluble MMO (sMMO) (4) under conditions of copper limitation (5). Whereas sMMO has a single well characterized diiron catalytic center (6), pMMO contains several distinct metal-binding sites (7). pMMO is composed of three subunits, pmoB, pmoA, and pmoC, arranged in an α3β3γ3 trimer (8). The pmoB subunit comprises two periplasmic domains connected by two transmembrane helices. In Methylococcus capsulatus (Bath) pMMO, these soluble domains house two copper sites, modeled as monocopper and dicopper (8). The monocopper site is not conserved in other pMMOs (9, 10). Biochemical and mutagenesis data from our laboratory indicate that the active site is located at the dicopper site (11, 12). The pmoA and pmoC subunits are composed entirely of transmembrane helices. An additional metal-binding site is present in the pmoC subunit, exposed to the center of the pMMO trimer.
In the structures of pMMO from M. capsulatus (Bath) and Methylocystis species strain M, this pmoC metal-binding site is occupied by zinc (10). The zinc ion is coordinated by two histidines, an aspartic acid, and a fourth ligand assigned as a solvent molecule (10). Because pMMO does not contain any zinc upon isolation, this zinc ion is derived from the millimolar zinc concentrations required for crystallization. By contrast, this site is partially occupied by copper in Methylosinus trichosporium OB3b pMMO, which can be crystallized in the absence of zinc (9). In this case, the copper might originate from excess copper added during cell lysis. The pmoC metal-binding site has attracted much attention due to a proposal that it houses a diiron active site similar to that in sMMO (13). However, we have clearly demonstrated that pMMO activity requires copper and not iron. We have also shown that the activity derives from the pmoB subunit (11, 12).
Several observations suggest that the pmoC zinc/copper site may still be functionally relevant. First, all the ligands are strictly conserved (see below and Ref. 14). Second, the best enzymatic activity is obtained from intact pMMO containing all three subunits, suggesting that the transmembrane regions have an important function (11). Third, previous studies have shown that zinc inhibits pMMO activity in M. capsulatus (Bath) and M. trichosporium OB3b membrane extracts (15–17). Up to seven zinc ions have been suggested to bind to M. capsulatus (Bath) pMMO and have been proposed to inhibit by either replacing active site copper ions or another copper site that is involved in reducing the active site (15–17). Based on the crystal structures, which were determined after these studies, it is also possible that inhibition is due to zinc occupation of the pmoC intramembrane metal-binding site.
Understanding the molecular basis for zinc inhibition requires a systematic analysis. Previous studies used membrane extracts with undefined metal concentrations or EDTA-treated samples in which pMMO is already partially inhibited (16–18). Here, we have used pMMOs from M. capsulatus (Bath) and Methylocystis species strain Rockwell to rigorously address the unresolved questions of how many zinc ions are required to inhibit pMMO, where zinc binds to inhibit pMMO, and whether zinc replaces copper in pMMO.
EXPERIMENTAL PROCEDURES
Cell Growth
M. capsulatus (Bath) cells were grown as described previously (19) in 12–15-liter fermentations using nitrate mineral salts medium (0.2% w/v KNO3, 0.1% w/v MgSO4·7H2O, and 0.001% w/v CaCl2·2H2O) and 3.9 mm phosphate buffer, pH 6.8, supplemented with 50 μm CuSO4·5H2O, 80 μm NaFe(III)EDTA, 1 μm Na2MoO4·2H2O, and trace metals solution. Cells were cultivated with a 4:1 air/methane ratio at 45 °C and 300 rpm agitation. Methylocystis sp. str. Rockwell was grown in 12–15-liter fermentations using salts solution (0.08% w/v KNO3, 0.02% w/v K2SO4, 0.003% w/v MgSO4·7H2O, and 0.0007% w/v CaCl2·2H2O) and 3.9 mm phosphate buffer, pH 7.0, supplemented with 50 μm CuSO4·5H2O, 40 μm FeSO4·7H2O, and trace elements solution. Methylocystis sp. str. Rockwell was grown with a 3:1 air/methane ratio at 30 °C and 300 rpm. Cells were harvested when they reached an A600 of 5.0–8.0 by centrifugation at 5000–7000 × g for 10 min. Cells were washed 1–3 times with 25 mm PIPES, pH 6.8–7.0, before freezing in liquid nitrogen and storing at −80 °C.
Membrane Isolation and Metal Removal
Frozen M. capsulatus (Bath) cells were thawed in 25 mm PIPES, pH 7.0–7.2, 250 mm NaCl and lysed by sonicator or microfluidizer. Methylocystis sp. str. Rockwell cells were lysed similarly but with the addition of 500 μm CuSO4·5H2O and using degassed buffers. Cell debris was removed by centrifugation at 20,000–24,000 × g for 1–2 h. Membranes were pelleted by centrifugation at 125,000 × g and washed 2–3 times in 25 mm PIPES, pH 7.0–7.2, 250 mm NaCl before freezing in liquid nitrogen and storing at −80 °C. Preparation of apo membranes (membrane-bound pMMO with all metal ions removed) was carried out as described previously (11, 18) using 50 mm ascorbate and 50 mm potassium cyanide. Metal removal efficiency was assessed by ICP-AES (Varian Vista MPX) at the Integrated Molecular Structure Education and Research Center of Northwestern University. Protein concentrations were measured using the Bio-Rad DC assay.
Metal Reconstitution and Activity Assays
For copper reconstitutions, apo membranes were diluted to 2.5 mg/ml and incubated with various equivalents of CuSO4·5H2O at room temperature for 30 min. Reconstituted samples were either assayed for activity immediately or the excess copper was removed by washing membranes 3–4 times by repeated ultracentrifugation (125,000 × g for 10 min) and resuspension with Dounce homogenizers in metal-free lysis buffer. For some experiments, zinc was added directly to these copper-reconstituted membranes and incubated for 30 min at room temperature prior to activity analysis. For other experiments, membranes were only washed once after copper loading and then incubated with different stoichiometric amounts of ZnSO4·7H2O for 30 min at room temperature. Excess metals were then washed out three times using ultracentrifugation and resuspension. For all metal reconstitutions, the concentrations of metal stock solutions were routinely measured by ICP-AES. The stoichiometries of copper, iron, and zinc binding were measured by ICP-AES and the Bio-Rad DC assay for protein concentrations. Activity assays were carried out as described previously (11). In brief, membranes (3–4 mg/ml) were reduced with either duroquinol or 5.4 mm NADH in a 2-ml vial, and 1 ml of headspace gas was replaced with methane gas. Samples were incubated at 45 °C for 3 min at 200 rpm and then heat inactivated at 80 °C for 10 min. Samples were then cooled on ice and centrifuged to remove insoluble membranes and reductants. The supernatant (3 μl) was injected onto an HP-PLOT Q column (Agilent). Calibration curves were generated from methanol standards (Sigma, spectrophotometric grade >99%). For calculation of IC50 values from inhibition curves, a modified dose-response curve was fit to the form y = a + (b/(1 + (x/IC50)∧d)).
Protein Purification of pMMO from Methylocystis sp. str. Rockwell
Methylocystis sp. str. Rockwell membranes were solubilized with 1.5 mg of n-dodecyl β-d-maltopyranoside/mg of crude protein. Solubilized membranes were diluted with column equilibration buffer (50 mm Tris, pH 8.5, 0.05% n-dodecyl β-d-maltopyranoside) before loading onto a Source 15Q column (GE Healthcare). pMMO was eluted using a 0–0.8 m NaCl gradient, concentrated using a 50-kDa molecular mass cutoff Amicon concentrator, and then frozen in liquid nitrogen. All purification buffers were degassed and kept under argon balloons throughout purification. The identity of all three pMMO subunits was confirmed by mass spectrometry. Purified protein was run on a 4–16% blue native-polyacrylamide gel (Invitrogen), and a band at ∼450 kDa was cut out for mass spectrometry. Some blue native-polyacrylamide gel slices were alkylated with iodoacetamide and run on a 15% SDS-polyacrylamide gel, and all bands were cut out for mass spectrometry.
Nano-LC-ESI-MS/MS Analysis for Protein Identification of Purified Methylocystis sp. str. Rockwell pMMO
Prior to mass spectrometry, Coomassie Blue was removed from the gel bands, and for optimal identification of proteins, membrane proteins were cleaved by CNBr and trypsin as described previously (20). For nano-LC-ESI-MS/MS experiments, 200 ng of peptides were separated on a reverse-phase LC system (UltiMate 3000; Dionex, Idstein, Germany) on-line coupled to an ESI-mass spectrometer (LTQ Velos ETD or Orbitrap Elite, both from Thermo Fisher Scientific). Peptide ions were analyzed in a data-dependent mode using abundant ions for additional tandem-MS experiments by collision-induced dissociation to obtain amino acid sequence information.
Protein identification was performed with ProteomDiscoverer Version 1.4 software (Thermo Fisher Scientific) using the Mascot Version 2.3 search algorithm (Matrixscience, London, UK) with tolerances for peptide masses and fragment masses of 0.4 Da for the LTQ Velos ETD mass spectrometer or 10 ppm and 0.4 Da for the Orbitrap Elite system. Searches were performed allowing two missed cleavage sites after CNBr and tryptic cleavage. Homoserine lactone modification of methionine was considered as variable modification. All data were searched against a database created by DecoyDatabaseBuilder (21) containing UniProt/Tremble entries (released October, 2013) with one additional shuffled decoy of each protein for false discovery rate calculations. Additionally, a taxonomy filter for proteobacteria (35,857,698 sequences) was used.
Crystallization and Structure Determination of pMMO from Methylocystis sp. str. Rockwell
For crystallization, purified Methylocystis sp. str. Rockwell pMMO protein was thawed and exchanged into degassed 50 mm Tris, pH 8.5, 0.03% n-undecyl β-d-maltopyranoside. Crystals grew in the sitting drop geometry at room temperature within 1–4 weeks. Drops contained 2.5 μl of 20 mg/ml pMMO protein, 0.5 μl of 85 mm n-octyl β-d-thiomaltoside, and 2 μl of crystallization solution (10% PEG 3000, 200 mm magnesium formate dihydrate, 100 mm sodium cacodylate trihydrate, pH 6.5). Crystals were flash-frozen using 30% PEG 3000, 200 mm magnesium formate dihydrate, 100 mm sodium cacodylate trihydrate, pH 6.5, as a cryoprotectant. For soaking experiments, crystals were soaked in mother liquor containing either 2 mm CuSO4·5H2O or 2 mm ZnSO4·7H2O for 10–24 h prior to flash freezing. Iron soaks were carried out as described previously (22).
Data sets were collected at the GM/CA-CAT and LS-CAT beamlines at the Advanced Photon Source at Argonne National Laboratory. Data were integrated and scaled using HKL2000 (23) and further processed anisotropically using the UCLA anisotropy server, which performs ellipsoidal truncation of the data including only reflections with I/σI >3.0 (24). The anisotropic resolution cutoff resulted in a lower completeness. Rmeas and CC1/2 were calculated using XDS. The structure was solved by molecular replacement with PHASER (25) using the coordinates of Methylocystis species strain M pMMO (PDB code 3RFR) as a starting model. Methylocystis sp. str. Rockwell pMMO has 94:97.4, 96:97.6, and 96.9:97.7% identity/similarity to the Methylocystis sp. str. M pmoB, pmoA, and pmoC subunits, respectively. Modeling and refinement were performed using Coot (26) and Refmac5 (27). Tight noncrystallographic symmetry was used during refinement for each structure. In the purified as-isolated structure, the restraints included residues 29–416 (pmoB), 11–252 (pmoA), 16–196 (pmoC), and 226–256 (pmoC). For the copper-soaked structure, the restrained residues were 29–416 (pmoB), 11–252 (pmoA), 16–195 (pmoC), and 226–256 (pmoC). For the zinc-soaked structure, the restrained residues were 29–417 (pmoB), 11–252 (pmoA), 16–210 (pmoC), and 224–256 (pmoC). The final model for the purified as-isolated pMMO structure included the following: pmoB residues 29–418, pmoA residues 9–252, pmoC residues 16–198 and 226–256, and three polyalanine helices consisting of up to 25 residues, six copper ions, four lipid molecules, and 117 water molecules. The final model for the copper-soaked structure included pmoB residues 29–418, pmoA residues 9–252, and pmoC residues 16–199 and 224–256, three polyalanine helices consisting of up to 24 residues, and six copper ions. The final model for the zinc-soaked structure included pmoB residues 29–418, pmoA residues 9–252, and pmoC residues 16–210 and 224–256, three polyalanine helices consisting of up to 25 residues, five zinc ions, three copper ions, and one cacodylate molecule. Simulated annealing composite omit maps were calculated for each structure using Phenix (28).
X-ray Absorption Spectroscopy
As-isolated and zinc-loaded M. capsulatus (Bath) crude membranes were resuspended in 30% glycerol, loaded into Lucite cells wrapped with Kapton tape, flash-frozen in liquid nitrogen, and stored at −80 °C until data collection. Copper XAS data were collected at the Stanford Synchrotron Radiation Light Source (SSRL) on beamline 9-3, and zinc XAS data were collected at the National Synchrotron Light Source (NSLS) on beamline X3A. Both beamlines were equipped with Si(220) double crystal monochromators and in-line harmonic rejection mirrors. During data collection, samples were maintained at 10 K using an Oxford Instruments continuous-flow liquid helium cryostat at SSRL and at 25 K using a liquid helium Displex cryostat at NSLS. Protein fluorescence excitation spectra at SSRL were collected using a 100-element Canberra germanium solid-state detector. At NSLS, spectra were collected on a 13-element Canberra germanium solid-state detector. For copper data collection at SSRL, a 6-μm nickel filter and solar slits were placed between the cryostat and detector for background noise filtration; x-ray energies were internally calibrated by collecting copper foil absorption spectra simultaneously with protein data, and the monochromator was calibrated for each spectrum based on the first inflection point of the copper foil spectrum (assigned to 8980.3 eV). XAS spectra were measured in 5-eV steps in the pre-edge region (8750–8960 eV), 0.25 eV steps in the edge region (8986–9050 eV), and 0.05 Å−1 increments in the EXAFS region (to k = 13 Å−1 for copper), integrating from 1 to 20 s in a k3 weighted manner for a total scan length of ∼40 min. For zinc data collection at NSLS, a 3-μm copper filter was placed between the cryostat and detector for background noise filtration; x-ray energies were internally calibrated by collecting zinc foil absorption spectra simultaneously with protein data, and the monochromator was calibrated for each spectrum based on the first inflection point of the zinc foil spectrum (assigned to 9659 eV). XAS spectra were measured in 5-eV steps in the pre-edge region (9350–9640 eV), 0.25 eV steps in the edge region (9640–9690 eV), and 0.05 Å−1 increments in the EXAFS region (to k = 13 Å−1 for zinc), integrating from 1 to 14 s in a k3 weighted manner for a total scan length of ∼45 min. For all samples, 4–5 (SSRL) and 10–12 (NSLS) spectra were collected on multiple independent versions of each sample to ensure spectral reproducibility.
During data processing, each fluorescence channel of each scan was examined for spectral anomalies prior to averaging, and for copper spectra, each fluorescence channel was closely monitored for photoreduction. XAS data were processed using the Macintosh OS X version of the EXAFSPAK program suite (29) integrated with the Feff Version 8 software (30) for theoretical model generation. Data reduction utilized a polynomial function in the pre-edge region and a three-region cubic spline throughout the EXAFS region. Data were converted to k-space using E0 values of 9000 eV for copper and 9680 eV for zinc. The k-cubed weighted EXAFS was truncated at 1.0 and 12.85 Å−1 for copper and at 1.0 and 12.87 Å−1 for zinc, for filtering purposes. These k ranges correspond to a spectral resolution of ∼0.13 Å for all metal-ligand interactions under consideration; therefore, only independent scattering environments outside 0.13 Å were considered resolvable in the EXAFS fitting analysis (31). EXAFS fitting analysis was performed on raw/unfiltered data using single scattering Feff theoretical models calculated for oxygen, sulfur, carbon, and metal coordination to simulate the nearest neighbor ligand environments. During copper data simulation, a scale factor of 1.0 and threshold energies of −12.0, −14.0, and −16.0 eV were utilized for Cu-O/N/C, Cu-S, and Cu-Cu scattering, respectively. During zinc data simulation, a scale factor of 1.0 and a threshold energy of −15.25 eV were utilized for Zn-O/N/C and S scattering. Scale factors and E0 values used during the simulations were calibrated by fitting crystallographically characterized models at different metal oxidation states, as described previously (32, 33). Criteria for judging the best fit simulation utilized both the lowest mean square deviation between data and fit corrected for the number of degrees of freedom (F′) (34) and a reasonable Debye-Waller factor (σ2 <0.006 Å2).
RESULTS
Zinc Inhibition of M. capsulatus (Bath) pMMO
To study the effects of zinc binding on pMMO, membrane extracts were used instead of purified protein because of the low activity of purified preparations (35). As-isolated membranes typically have activities of 25–40 nmol of methanol/min·mg (duroquinol reductant) or 80–130 nmol of methanol/min·mg (NADH reductant) and contain 5–9 copper ions per 100-kDa αβγ protomer (Table 1). To allow for the possibility that zinc inhibition might be affected by adventitiously bound copper, copper-reconstituted membranes were also prepared for zinc titration experiments. After loading apo membranes (membrane-bound pMMO with all metal ions removed) (Table 1) with copper, both duroquinol and NADH-driven methane oxidation activities were recovered. The highest activity was observed after adding 2 eq of copper per 100-kDa protomer (Fig. 1A), consistent with previous results (11). In a similar experiment, apo membranes were loaded with copper, and the excess copper ions were removed by ultracentrifugation and resuspension in metal-free buffer three times before activity analysis. Under these conditions, the highest activity for both duroquinol and NADH reductants was also observed with 2 eq of copper bound per 100-kDa protomer (Fig. 1B), which resulted from loading 3 eq of copper before washing. Although both reductants yield activity in copper-reconstituted membranes, the NADH activity recovered was usually ∼25% of the as-isolated activity, whereas duroquinol usually resulted in ∼50% of the as-isolated activity (Fig. 1). Previous EXAFS data indicate that the dicopper center is reassembled using this reconstitution procedure (11). Because only two copper ions are tightly bound after removal of excess metals, the dicopper site is probably not fully loaded, with some copper occupying the mononuclear site resulting in a lower percentage of activity recovered.
TABLE 1.
Metal content of M. capsulatus (Bath) crude membranes before (as-isolated) and after (apo) cyanide treatment (average of 3–5 samples)
| Copper/100-kDa protomer | Iron/100-kDa protomer | Zinc/100-kDa protomer | |
|---|---|---|---|
| As-isolated membranes | 7 ± 2 | 0.29 ± 0.07 | 0.1 ± 0.3 |
| Apo membranes | 0.04 ± 0.03 | 0.06 ± 0.02 | 0.01 ± 0.02 |
FIGURE 1.
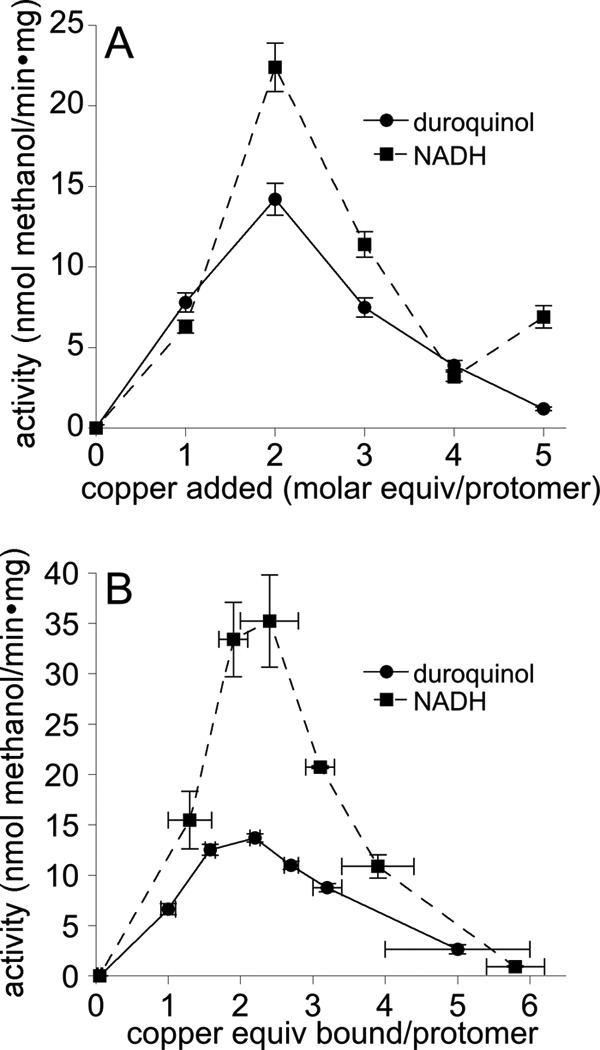
Methane oxidation activity of apo membrane-bound M. capsulatus (Bath) pMMO after metal loading using two copper reconstitution methods. A, apo M. capsulatus (Bath) membranes were incubated with 1–5 eq of CuSO4 per 100-kDa protomer. As-isolated crude membranes had an activity of 28 ± 2 nmol of methanol/min·mg (duroquinol) or 94 ± 7 nmol of methanol/min·mg (NADH). The maximum activity for both reductants occurred after addition of 2 eq of copper, and the activity recovered was 51% for duroquinol and 24% for NADH. B, apo membranes were incubated with 1, 2, 3, 4, 5 or 10 eq of copper per 100-kDa protomer as in A, but then was washed three times to remove excess copper prior to activity measurements. Copper ions bound per 100-kDa pMMO protomer after excess metal removal are reported on the x axis. As-isolated crude membranes had an activity of 27 ± 1 nmol of methanol/min·mg (duroquinol) and 139 ± 5 nmol of methanol/min·mg (NADH). The maximum activity recovered was 50% for duroquinol and 25% for NADH.
As observed previously, addition of excess copper inhibits pMMO activity (Fig. 1, A and B). This excess copper does not wash out readily as 4–6 copper ions per 100-kDa protomer remain bound after incubating apo membranes with 10 eq of copper and then removing excess copper by washing (Fig. 1B). Inhibition by excess copper has been attributed to peroxide formation (11), and these extra copper ions may be binding to lipids (36). Desalting columns were tested as a second method of metal removal, and 2 eq of copper per 100-kDa protomer remained bound after adding 10 eq. The desalting method clearly removes more adventitiously bound metal, but this proved impractical because the membranes adhere to the column. Thus, the washing method was used to remove excess metal ions for all subsequent experiments. Adventitious metal binding only occurs when 4 or more equivalents of copper are loaded before washing and can be avoided by adding less copper (1–3 eq). Unless noted otherwise, for all experiments reported here involving copper-reconstituted, membrane-bound pMMO, 3 eq of copper are loaded initially, which results in ∼2 eq of copper after washing.
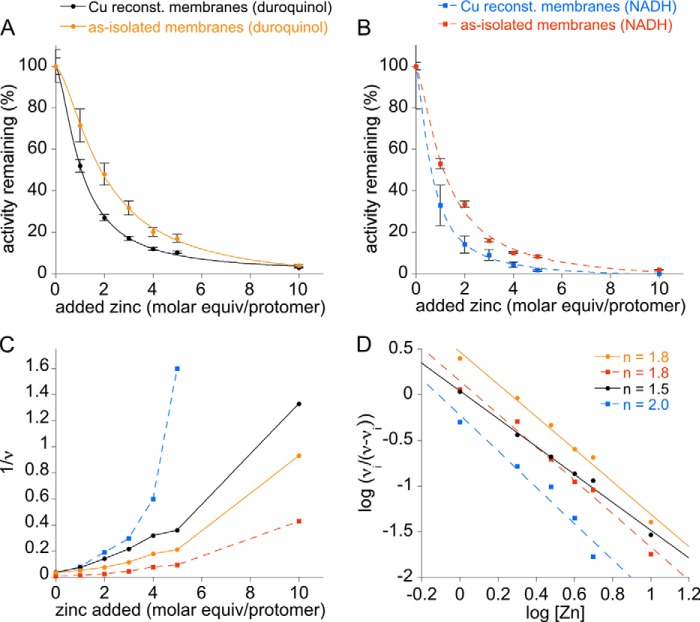
Both as-isolated and copper-reconstituted membrane-bound pMMO were then treated with increasing amounts of zinc. Complete inhibition is observed after 10 eq of zinc per protomer are added, with a dramatic decrease occurring after the addition of 2 eq of zinc (Fig. 2, A and B). With both duroquinol and NADH reductants, the as-isolated membranes have slightly higher IC50 values for zinc inhibition (59 and 36 μm, respectively) than those measured for copper-reconstituted membranes (36 and 22 μm, respectively). Dixon plots reveal a parabolic pattern, consistent with multisite inhibition (Fig. 2C) (37). Hill plots yield slopes ranging from −1.5 to −2.0, indicative of two zinc-binding sites (Fig. 2D). To probe the nature of these zinc-binding sites, copper-reconstituted membranes treated with up to 30 eq of zinc were repeatedly ultracentrifuged and resuspended in metal-free buffer to remove excess zinc. After this procedure, 0.5–0.8 zinc ions per protomer remain, indicating that zinc can only bind tightly to one inhibitory site under these conditions (Fig. 3A). Importantly, the copper stoichiometry remains unchanged upon zinc binding. Thus, the more tightly bound zinc does not displace copper ions in the active site, and the zinc-binding site is not occupied by copper in copper-reconstituted pMMO. This result suggests that pMMO activity does not require copper binding at this site and that this site is likely apo in vivo. Binding of zinc at this site reduces both duroquinol and NADH-driven membrane-bound pMMO activity by 40–60% (Fig. 3B).
FIGURE 2.
Zinc inhibition of methane oxidation activity of as-isolated and copper-reconstituted membrane-bound M. capsulatus (Bath) pMMO. A, duroquinol was used as the reductant. The as-isolated activity was 28 ± 2 nmol of methanol/min·mg, and the copper-reconstituted activity was 27 ± 2 nmol of methanol/min·mg. The IC50 values for as-isolated and copper-reconstituted membranes were 59 and 36 μm, respectively. B, NADH was used as the reductant. The as-isolated activity was 130 ± 4 nmol of methanol/min·mg, and the copper reconstituted activity was 37 ± 8 nmol of methanol/min·mg. The IC50 values for the as-isolated and copper-reconstituted membranes were 36 and 22 μm, respectively. C, Dixon plots (1/ν versus [I]) for the inhibition data in A and B reveal a parabolic shape indicative of multisite inhibition. D, Hill plots (log (νi/ν − νi) versus log [I]) were used to determine the number of zinc-binding sites. Linear regression lines fit to data for as-isolated membranes were y = 0.46812 − 1.7731x (duroquinol, R = 0.99383) and y = 0.15082 − 1.8165x (NADH, R = 0.99214). For copper-reconstituted membranes, fits were y = 0.043844 − 1.524x (duroquinol, R = 0.99611) and y = −0.22384 − 1.9747x (NADH, R = 0.97396).
FIGURE 3.
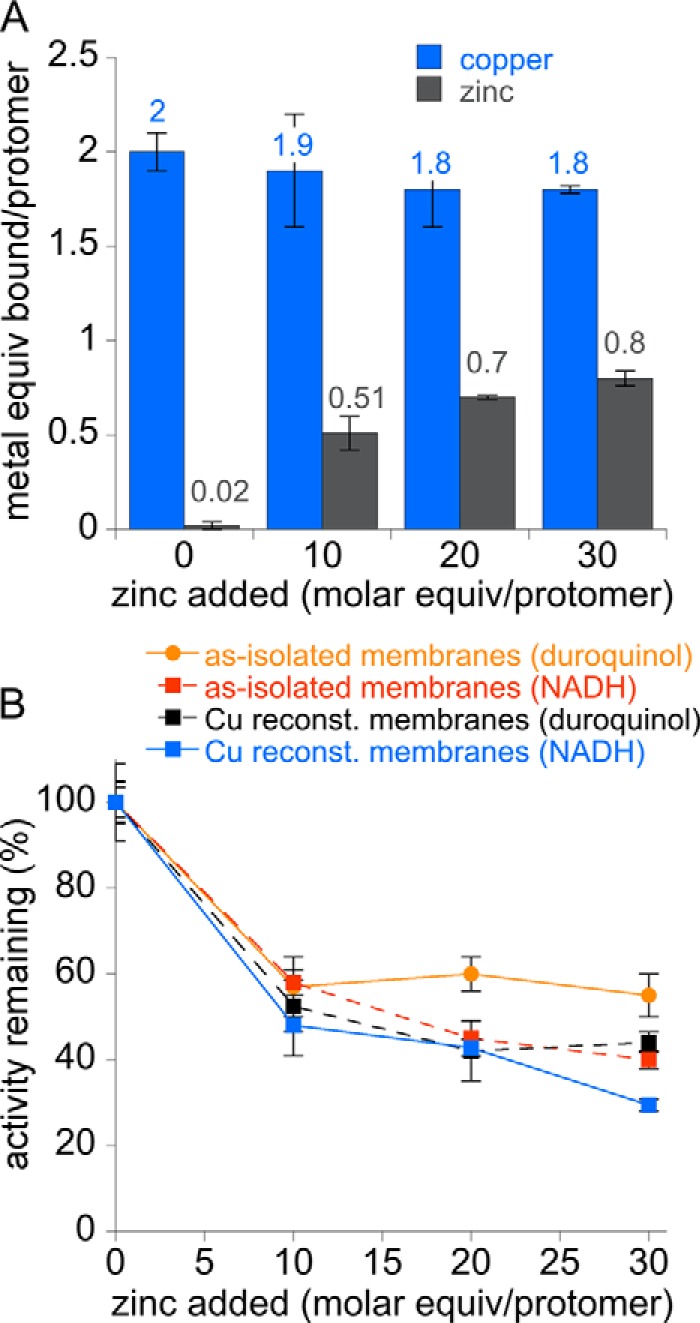
Metal content and methane oxidation activity after zinc-loading copper-reconstituted and as-isolated membrane-bound M. capsulatus (Bath) pMMO. A, metal content of copper-reconstituted membranes after addition of 0–30 m eq of ZnSO4 followed by removal of excess metals. B, activity of copper-reconstituted and as-isolated membranes after addition of zinc and removal of excess metals.
We then used the same procedure to load apo membrane-bound pMMO with zinc. Up to 1.6 zinc ions are measured (Fig. 4A), which is twice as much zinc as in the copper- and zinc-loaded protein (Fig. 3A). Therefore, two binding sites may be present and could be the active site in pmoB as well as the pmoC site. Addition of copper to these zinc-loaded membranes results in loss of half the zinc ions (Fig. 4B). From these data, it appears that when zinc is loaded first, copper can replace one zinc site, which is likely the more accessible pmoC site. The activity of the zinc- and copper-loaded membrane-bound pMMO is 11–18% of the copper-reconstituted membrane-bound pMMO activity (Table 2). This activity is lower than the 40–60% observed for copper- and zinc-loaded pMMO (Fig. 3B), even though the metal stoichiometries are similar (Fig. 3A and 4B), which would be consistent with zinc occupying the active site when loaded first.
FIGURE 4.
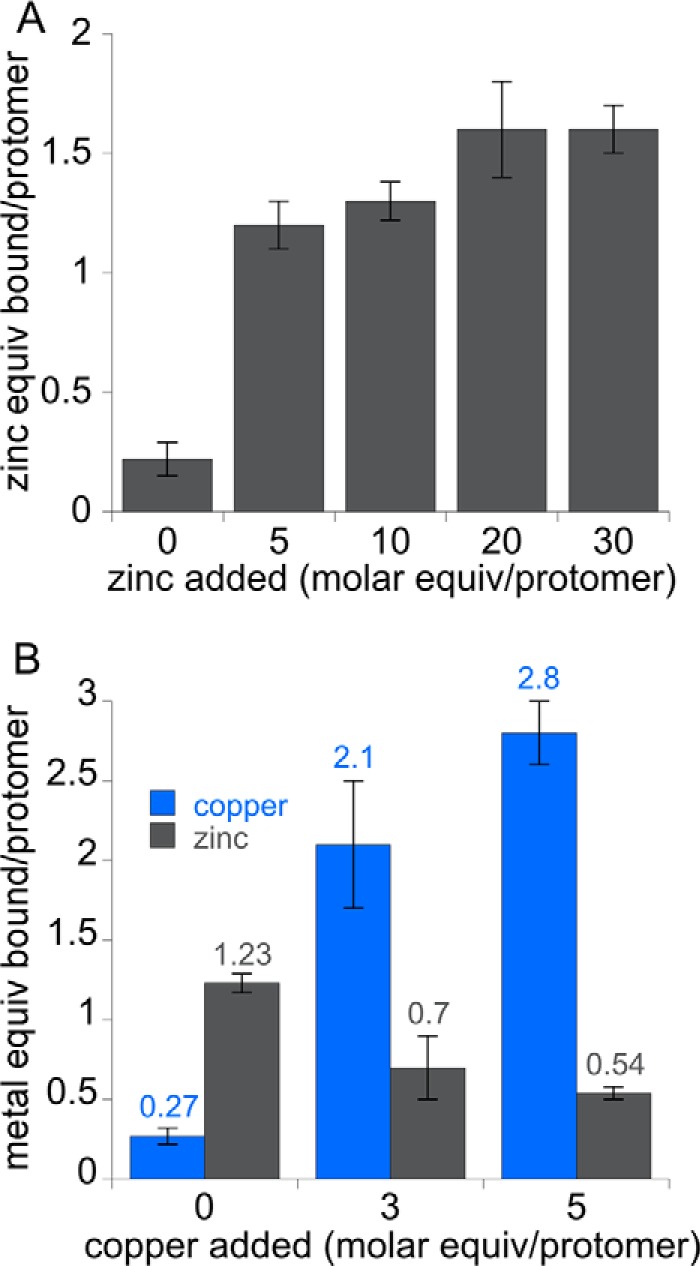
Metal content of apo M. capsulatus (Bath) membranes after zinc and zinc/copper loading. A, apo membranes loaded with 0–30 m eq of zinc followed by removal of excess metal. B, apo membranes loaded with 10 m eq of zinc followed by 0–5 m eq of copper and then removal of excess metals.
TABLE 2.
Specific methane oxidation activities of apo M. capsulatus (Bath) pMMO when loaded with zinc first and then copper
| Equivalent of zinc/copper added | Reductant | Activity | Percent of copper reconstituteda |
|---|---|---|---|
| nmol methanol/min·mg | |||
| 10 zinc/3 copper | Duroquinol | 5.1 ± 0.9 | 17 ± 3 |
| NADH | 7.7 ± 0.9 | 18 ± 2 | |
| 10 zinc/5 copper | duroquinol | 4.1 ± 0.2 | 14 ± 1 |
| NADH | 4.8 ± 0.6 | 11 ± 2 |
a The activities of the copper-reconstituted membrane-bound pMMO were 30 ± 2 (duroquinol) and 43 ± 3 (NADH).
EXAFS Analysis of Zinc-loaded M. capsulatus (Bath) pMMO
Analysis of the copper K-edge EXAFS for as-isolated membrane-bound M. capsulatus (Bath) pMMO indicates that copper coordination is structurally unperturbed by zinc binding. The copper EXAFS in the absence and presence of zinc (Fig. 5, A and C, respectively) appear nearly identical and are consistent with spectra previously reported for as-isolated and oxidized M. capsulatus (Bath) pMMO (32). Fourier transforms of the copper EXAFS in the absence and presence of zinc (Fig. 5, B and D, respectively) both show broad first coordination shell features at a phase-shifted bond length (R + Δ) of <2.0 Å, a well resolved feature at R + Δ <2.5 Å, and long range scattering signals at R + Δ >2.5 Å. Again, these spectra match data previously reported for M. capsulatus (Bath) pMMO. The close agreement of the copper EXAFS among samples suggests a common coordination geometry and copper redox state, and the current spectra are in close agreement with XANES spectra previously reported for M. capsulatus (Bath) pMMO (data not shown). Metrical parameters obtained from simulations of the copper EXAFS for pMMO in the absence and presence of zinc are virtually identical, with the copper-first shell ligand coordination environment constructed of ∼1 Cu-(O/N) ligand at 1.92 Å, a second set of ∼2.5 Cu-(O/N) ligands centered at 2.10 Å, and a well defined Cu-Cu vector at 2.48 Å (Table 3). Long range Cu···C scattering, in the range of 2.7 to 4.0 Å, is also observed in both samples. The copper-first shell ligand bond lengths are within technique error limits of ±0.02 Å of values we reported previously (32), but there are subtle differences in coordination numbers. The vector at 2.48 Å in the current data is best fit by metal-metal coordination as opposed to metal-carbon at a similar distance, with a significant improvement (15%) in the quality of fit for a Cu-Cu vector as opposed to a Cu···C vector (Table 3).
FIGURE 5.
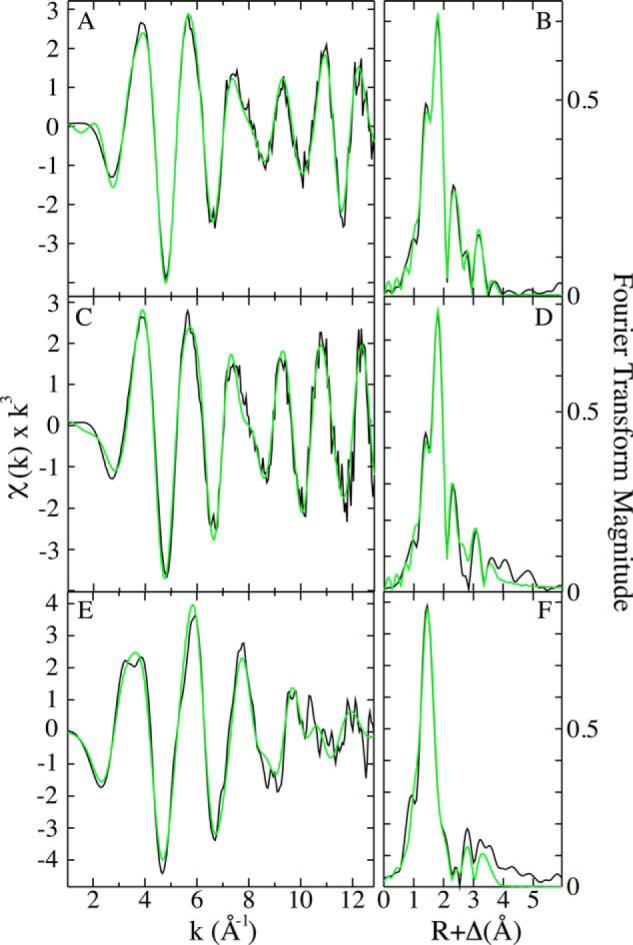
Copper and zinc EXAFS data and simulations for as-isolated membrane-bound M. capsulatus (Bath) pMMO. Raw k3-weighted copper EXAFS data and phase-shifted Fourier transforms of EXAFS are shown, respectively, for as-isolated membrane-bound M. capsulatus (Bath) pMMO without zinc (A and B) and with zinc (C and D). Raw k3-weighted zinc EXAFS data and phase-shifted Fourier transforms are shown for as-isolated membrane-bound M. capsulatus (Bath) pMMO with zinc (E and F). Raw unfiltered data are shown in black, and best-fit simulations are shown in green.
TABLE 3.
Summary of M. capsulatus (Bath) pMMO copper and zinc EXAFS simulation coordination parameters for progressive inclusion of scattering environments
Values obtained are from fitting spectra averaged from the 2–3 independent data sets for each sample. Copper spectra labeled under number .6 fits compare simulations in which the copper-copper vector is substituted by a Cu · · · C fit. Best fit parameters for all samples are in boldface.
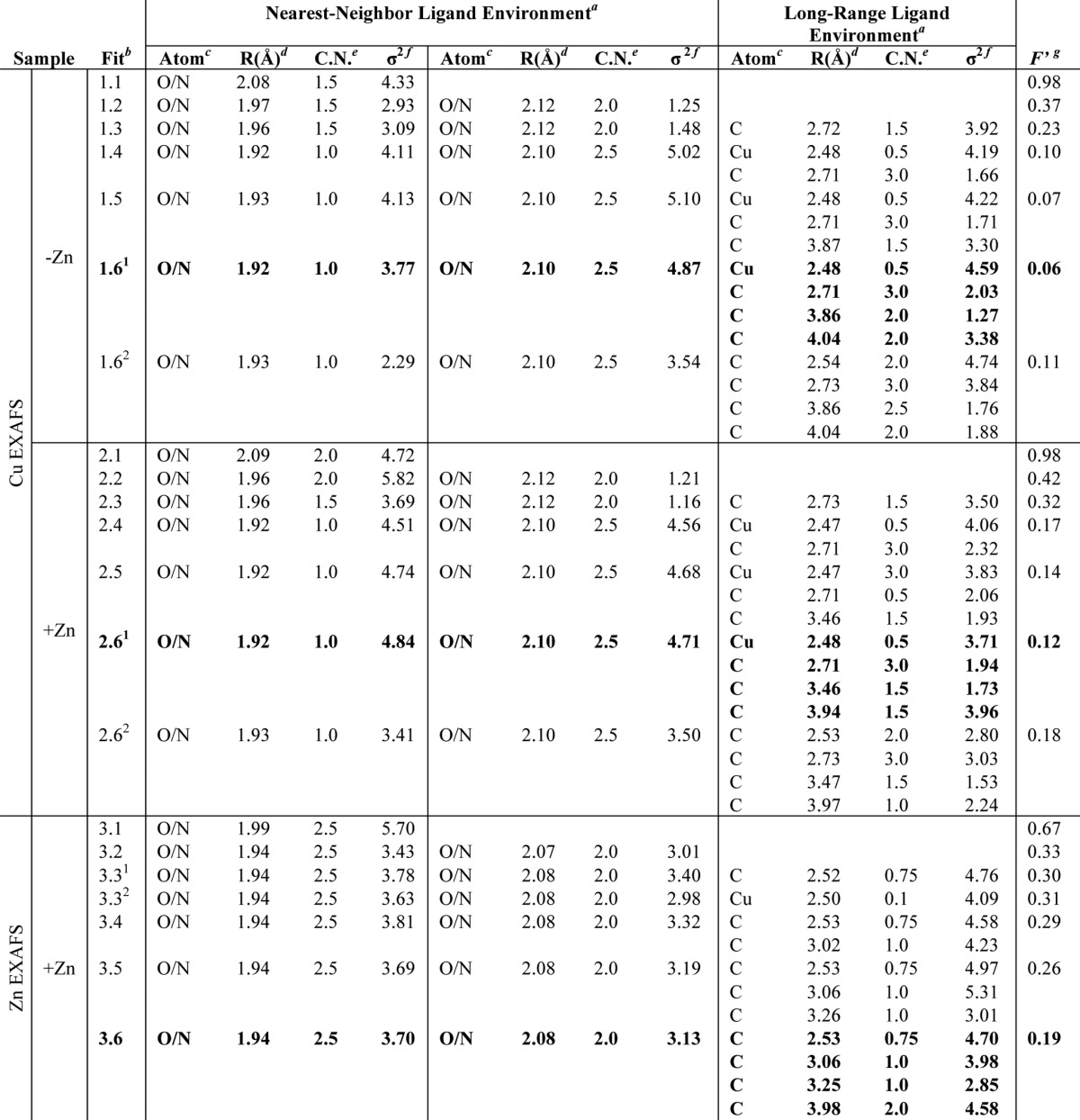
a Independent metal-ligand scattering environment is shown.
b Initial number corresponds to sample type, and the extension number corresponds to number of shells included in the fit.
c Scattering atoms are as follows: O is oxygen; N is nitrogen; Cu is copper, and C is carbon.
d Average metal-ligand bond length from two independent samples.
e Average metal-ligand coordination number is from two independent samples.
f Average Debye-Waller factor in Å2 × 103 is from two independent samples.
g Number of degrees of freedom weighted mean square deviation between data and fit is shown.
Analysis of the zinc K-edge EXAFS for as-isolated membrane-bound M. capsulatus (Bath) pMMO in the presence of zinc indicates a four coordinate ligand environment with no observable long range zinc-metal interaction. The zinc EXAFS and Fourier transforms for pMMO (Fig. 5, E and F) are consistent with a zinc first shell ligand coordination geometry that is tetrahedral and relatively symmetric. The camelback beat pattern observed in the zinc EXAFS between k of 3–4 is consistent with spectral anomalies observed from metal-imidazole scattering from metal coordinated to histidine residues (38). Long range (R + Δ > 2.5 Å) metal-ligand scattering is observed in the Fourier transform of the zinc EXAFS (Fig. 5F). Simulations of the zinc EXAFS indicate a zinc first shell coordination environment constructed of two independent Zn-(O/N) environments consisting of ∼4 total ligands, divided equally, and centered at 1.94 and 2.08 Å (Table 3). Zinc-oxygen and zinc-nitrogen bond lengths in the range of 1.94–2.08 Å most closely match tetrahedral Zn-(O/N) model compounds published in the Cambridge Structural database (39). Long range scattering can be fit for only Zn···C interactions at 2.53, 3.06, 3.25, and 3.98 Å. Attempts to fit the 2.53 Å feature with a metal were unsuccessful as were attempts to fit the 3.06-, 3.25-, and 3.98-Å features as a multiple scattering imidazole unit (40).
Activity and Crystal Structure of Methylocystis sp. str. Rockwell pMMO
To investigate the effects of zinc addition on the pMMO structure, we used pMMO from Methylocystis sp. str. Rockwell because it crystallizes more reproducibly than M. capsulatus (Bath) pMMO and is amenable to in crystallo metal binding studies. Methylocystis sp. str. Rockwell pMMO is 46.5:62.4, 57.9:76.2, and 57.8:70% identical/similar to the M. capsulatus (Bath) pmoB, pmoA, and pmoC subunits, respectively. The ligands to the pmoB dicopper site and the pmoC metal-binding site are conserved in Methylocystis sp. str. Rockwell pMMO, but the pmoB monocopper site is only present in M. capsulatus (Bath) pMMO. Membrane-bound Methylocystis sp. str. Rockwell pMMO contains 4.2 ± 2 copper ions per 100-kDa protomer, which decreases to 2.2 ± 0.4 upon purification. After protein concentration, only 0.9 ± 0.1 copper per 100-kDa protomer remains bound (Table 4). The activity of membrane-bound Methylocystis sp. str. Rockwell pMMO at 30 °C is 20 ± 3 nmol of methanol/min·mg using NADH as a reductant and 4.5 ± 0.9 nmol of methanol/min·mg using duroquinol as a reductant. After solubilization with n-dodecyl β-d-maltopyranoside, activities of 0.5 ± 0.1 nmol of methanol/min·mg (NADH reductant) and 0.54 ± 0.09 nmol of methanol/min·mg (duroquinol reductant) were measured. The purified protein does not exhibit measurable activity, likely due to the low copper stoichiometry.
TABLE 4.
Metal content of Methylocystis sp. str. Rockwell pMMO (average of 3–4 samples)
| Copper/100-Da protomer | Iron/100-Da protomer | Zinc/100-Da protomer | |
|---|---|---|---|
| Membranes | 4.2 ± 2 | 0.4 ± 0.2 | 0.1 ± 0.1 |
| Purified pMMO (dilute) | 2.2 ± 0.4 | 0.06 ± 0.03 | 0.1 ± 0.1 |
| Purified pMMO (concentrated) | 0.9 ± 0.1 | 0.06 ± 0.04 | 0.13 ± 0.08 |
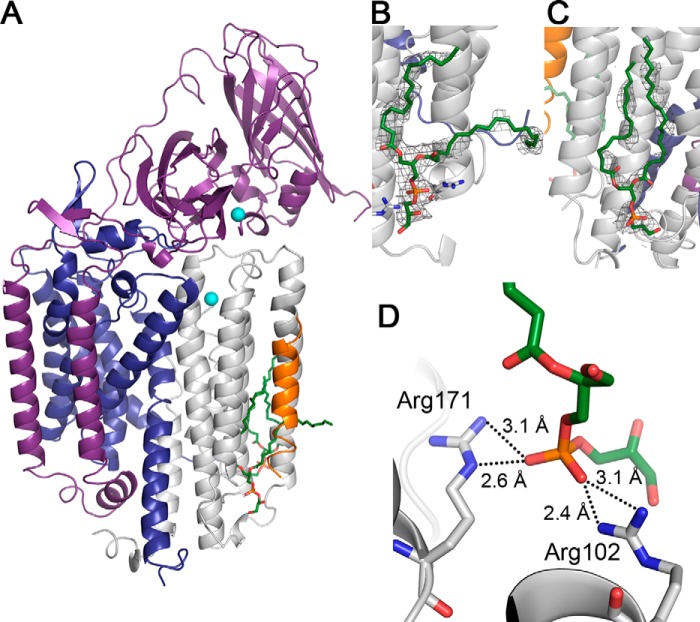
The crystal structure of Methylocystis sp. str. Rockwell pMMO was solved to 2.6 Å resolution using anisotropic data (Table 5). The anisotropy resulted in a low completeness in the higher resolution shells, as typically observed for ellipsoidally truncated data (9, 41–44). It has the same architecture as the three previous pMMO structures (8–10) and contains one pMMO trimer in the asymmetric unit. The Methylocystis sp. str. Rockwell pMMO crystals are particularly useful for metal binding experiments because the crystallization condition does not contain zinc, and the protein loses most of the metal upon concentration. The Methylocystis sp. str. Rockwell pMMO structure is very similar to those from M. trichosporium OB3b and Methylocystis sp. str. M (9, 10) and also contains a transmembrane helix of unknown identity closely associated with the pmoC subunit of each protomer (Fig. 6A). Extensive efforts to identify this mystery helix via mass spectrometry were not successful. Additional transmembrane helices are also present in the structures of Pseudomonas stutzeri cbb3 cytochrome oxidase (45) and Escherichia coli AcrB (46). The cbb3 cytochrome oxidase helix could not be identified by mass spectrometry, and the AcrB helix was assigned as YajC (46). Interestingly, we also detected YajC in our samples by mass spectrometry.
TABLE 5.
Data collection and refinement statistics for Methylocystis sp. str. Rockwell pMMO
Values in parentheses refer to the highest resolution shell.
| As isolated (copper anomalous) | Copper soaked (copper anomalous) | Zinc soaked (zinc anomalous) | Zinc soaked (copper anomalous) | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a, b, and c | 117.5, 118.5, 189.4 Å | 116.4, 184.7, and 188.6 Å | 120.9, 185.5, and 192.7 Å | 121.1, 185.7, and 193.2 Å |
| Resolution | 50–2.59 Å (2.62–2.59 Å) | 50–3.15 Å (3.2–3.15 Å) | 50–3.33 Å (3.39–3.33 Å) | 50–3.59 Å (3.65–3.59 Å) |
| Anisotropic truncation limit | 3.1, 2.6, 3.0 Å | 3.7, 3.1, 3.4 Å | 3.7, 3.3, 3.7 Å | 3.9, 3.6, 3.8 Å |
| Wavelength | 1.37765 Å | 1.37765 Å | 1.27816 Å | 1.37765 Å |
| Rsym or Rmerge | 0.145 | 0.174 (0.619) | 0.164 (0.394) | 0.166 (0.468) |
| Rmeas | 0.176 (1.16) | 0.153 (0.591) | 0.144 (0.481) | 0.152 (0.879) |
| CC1/2 | 0.998 (0.684) | 0.998 (0.840) | 0.999 (0.925) | 0.999 (0.841) |
| I/σI | 18.9 (2.9) | 21.5 (4.4) | 23.6 (8.9) | 23.7 (8.0) |
| Completeness | 72.6% (2.4%) | 77.8% (4.9%) | 80.4% (4.6%) | 87.7% (6.7%) |
| Redundancy | 14.4 (9.2) | 13.7 (11.5) | 14.9 (14.5) | 14.8 (14.0) |
| Refinement | ||||
| No. of reflections | 94,045 | 55,888 | 51,695 | |
| Rwork/Rfree | 0.237/0.289 | 0.221/0.280 | 0.224/0.281 | |
| Average B-factor | 42.2 Å2 | 57.5 Å2 | 54.7 Å2 | |
| Root mean square deviations | ||||
| Bond lengths | 0.0119 Å | 0.0125 Å | 0.0134 Å | |
| Bond angles | 1.666° | 1.6825° | 1.7454° | |
| Residues in favored region of Ramachandran plot | 89.3% | 84.6% | 82.6% | |
| Residues in allowed region of Ramachandran plot | 8.6% | 11.5% | 13.5% | |
FIGURE 6.
Crystal structure of Methylocystis sp. str. Rockwell pMMO. A, one protomer shown with the pmoB, pmoA, pmoC, and mystery helix polypeptides in purple, blue, gray, and orange, respectively. Copper is depicted as cyan spheres, and lipids are shown as green sticks. B, zoomed-in view of the lipid-binding site located between the pmoC subunit and the mystery helix. The mystery helix and a pmoC helix consisting of amino acids 66–92 are omitted for clarity. C, zoomed-in view of the lipid-binding site located at the pmoC/pmoA interprotomer interface. The 2mFo − DFc composite omit map calculated with simulated annealing is superimposed over the lipids in both B and C in gray (1σ). D, hydrogen bonding interactions between the pmoC subunit and the lipid located at the site between pmoC and the mystery helix. The mystery helix is not shown in this panel. Nitrogen, phosphorus, and oxygen are colored blue, orange, and red, respectively.
The three mystery helices are associated with hairpin-shaped electron densities that were modeled as phospholipids (Fig. 6, B and C). The identity of the lipids could not be determined from the electron density, but they were modeled as a 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-rac-(1-glycerol)). This lipid was chosen because type II methanotrophs predominantly contain C18 lipids with either a phosphatidylglycerol or phosphatidylethanolamine headgroup (47, 48). The phosphate group of the lipid interacts with Arg-102 and Arg-171 from the pmoC subunit (Fig. 6D), which are both highly conserved residues. A second lipid-binding site located at the inter-protomer pmoC/pmoA interface is only present in one protomer. The phosphate group of this second lipid interacts with the peptide backbone via a water molecule. These are the first lipid-binding sites observed in a pMMO structure.
Metal-binding sites in Methylocystis sp. str. Rockwell pMMO
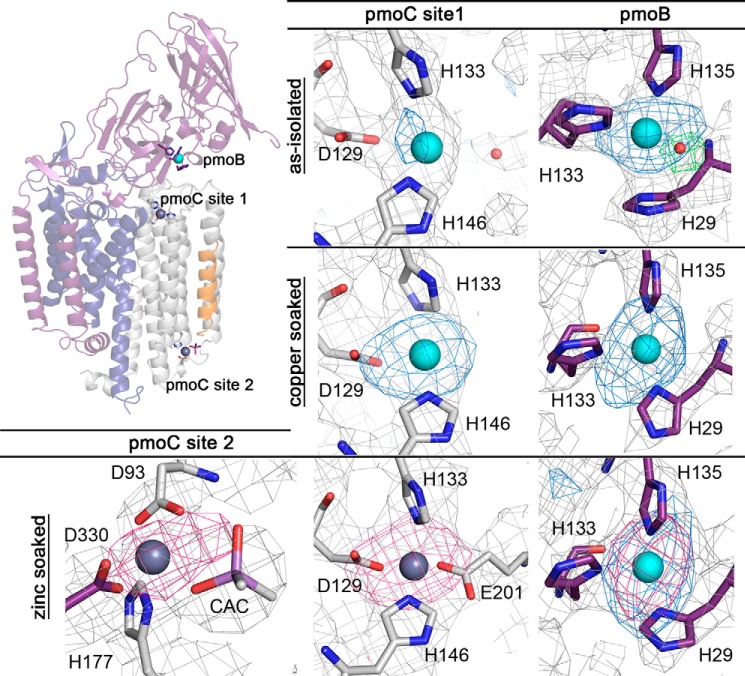
In the structure of purified as-isolated Methylocystis sp. str. Rockwell pMMO, copper anomalous difference Fourier maps reveal two copper-binding sites. One copper ion was modeled at the pmoB metal-binding site coordinated by His-29, His-133, His-135, and a solvent molecule (Fig. 7). This solvent molecule was observed for the first time in a pMMO structure, likely due to the improved resolution of 2.6 Å and is within hydrogen bonding distance of conserved residue Glu-31. Although a dicopper center is modeled at this site in the M. capsulatus (Bath) pMMO structure (8), only one copper ion could be modeled in the Methylocystis sp. str. Rockwell site, consistent with the low copper stoichiometry in the purified protein (Table 4). Similarly, only one protomer in the asymmetric unit of the Methylocystis sp. strain M pMMO structure could be modeled as a dicopper center, with the other two protomer sites modeled as monocopper, also due to low copper stoichiometry (10). This loss of copper could not be prevented by performing concentration steps anaerobically or by using nonchelating buffers. The second site, at the pmoC metal-binding site, had weaker anomalous difference density but could be modeled as a monocopper center with 0.7–0.8 occupancy. This copper ion is coordinated by Asp-129, His-133, His-146, and a solvent molecule (Fig. 7). Copper also weakly occupies this site in the M. trichosporium OB3b pMMO structure (9).
FIGURE 7.
Metal centers of purified Methylocystis sp. str. Rockwell pMMO. The pmoC metal sites for purified as-isolated, copper-soaked, and zinc-soaked crystals (left and center) and the pmoB metal sites for as-isolated, copper-soaked, and zinc-soaked crystals (right) are shown. Ligands from the pmoC and pmoB subunits are shown in gray and purple, respectively. Copper and zinc ions are shown as cyan and dark gray spheres. Simulated annealing composite omit maps are shown in gray and contoured at 1σ. Anomalous difference Fourier maps calculated using data collected at the copper and zinc absorption edges are superimposed in blue and magenta, respectively, and are contoured at 3σ. The Fo − Fc map for the solvent ligand in the upper right panel is superimposed in green and contoured at 3σ.
After soaking Methylocystis sp. str. Rockwell pMMO crystals in 2 mm CuSO4, the occupancy of the pmoC metal site increases as shown by a significant increase in the intensity of the copper anomalous peak from 4–5 to 9–10σ. The solvent molecule is not observed in the copper-soaked structure due to the lower resolution. The anomalous density at the pmoB site remains unchanged, however (10–14σ in both data sets, Fig. 7). Soaking pMMO crystals in 2 mm ZnSO4 yields zinc anomalous density in the pmoC metal-binding site (8–10σ) (Fig. 7). No signal is observed at this site in copper anomalous maps (Fig. 7), suggesting that zinc replaces any copper that is initially present. Notably, a second zinc anomalous peak (∼6σ) is observed on the cytoplasmic side of two of the three protomers. This zinc ion is coordinated by pmoC residues His-177 and Asp-93 (Fig. 7). Residue Asp-330 from a symmetry related pmoB subunit and a cacodylate molecule from the crystallization buffer are adjacent to one of these sites but not the other. The same two residues coordinate a zinc site in the Methylocystis sp. str. M pMMO structure cocrystallized with zinc (10). These two residues are not conserved in M. capsulatus (Bath) pMMO, but a cytoplasmic zinc site was also present in that structure, coordinated by pmoA His-11 and pmoC Glu-284. Zinc anomalous density was also observed at the pmoB copper site, but this signal was likely due to copper absorption because the peak intensities were very similar for both metals at this site, and the absorption edge of zinc is only slightly higher than that of copper. The low resolution of the copper- and zinc-soaked data sets (3.15–3.6 Å) preclude more detailed comparisons and conclusions regarding relative occupancies.
Thus, copper can occupy the pmoB metal site, and both copper and zinc can occupy the pmoC metal site. These findings agree with the M. capsulatus (Bath) metal loading data (Figs. 4B, 3A) and support the hypothesis that the pmoC subunit can be loaded with either copper or zinc and that the copper in the active site is not readily displaced by zinc. Iron(II) was also soaked into pMMO crystals using an in crystallo method for metal loading ribonucleotide reductase (22), but no iron was detected in the crystals either by fluorescence scans or analysis of anomalous data.
Interestingly, zinc binding at the pmoC site in the zinc-soaked structure stabilizes pmoC residues 200–210, which are not visible in any other pMMO structures, including the current as-isolated and copper-soaked structures. As a result, residue Glu-201 replaces the previously modeled water molecule as the fourth ligand to the zinc ion (Fig. 7). These 10 residues interact with the pmoA subunit from the neighboring protomer. It remains unclear whether they connect to pmoC residue 224 of the same or the neighboring protomer, opening the possibility of a domain swap within the pMMO trimer (Fig. 8).
FIGURE 8.
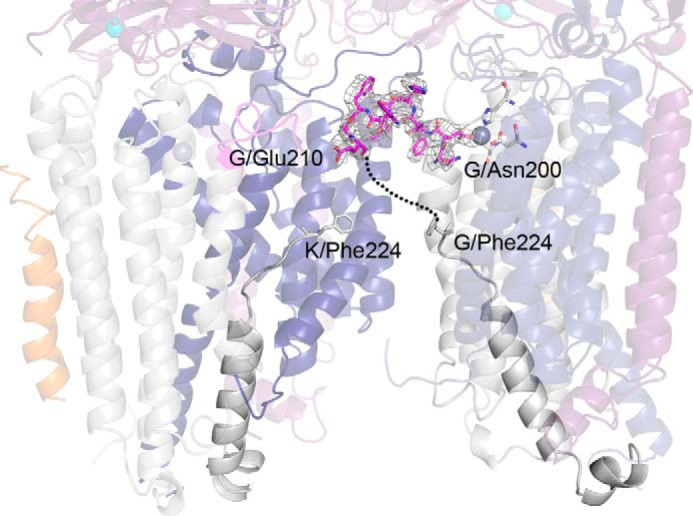
Previously unobserved residues modeled in the Methylocystis sp. str. Rockwell pMMO pmoC subunit. Residues 200–210 (magenta) are ordered in the zinc-soaked structure and provide a fourth amino acid ligand, Glu-201, to the zinc site. The simulated annealing composite omit map is superimposed in gray (1σ). The C-terminal residue of this stretch, Glu-210, may connect to either the C terminus of pmoC within the same protomer (chain G, intervening disordered residues denoted with a dotted line) or to the C terminus of pmoC within the neighboring protomer (chain K).
DISCUSSION
The effects of zinc on activity of membrane-bound pMMO from M. capsulatus (Bath) were assessed using both as-isolated and copper-reconstituted samples with NADH and duroquinol as reductants. Under these conditions, complete inhibition is observed upon addition of 10 eq of zinc per 100-kDa protomer (Fig. 2, A and B), and the data are consistent with two zinc-binding sites (Fig. 2, C and D). If excess zinc is removed, 40–60% inhibition is observed, and the amount of bound zinc is consistent with a single zinc-binding site (Fig. 3). Addition of zinc does not affect the copper stoichiometry, and EXAFS data show that zinc binding does not disrupt the dicopper center of pMMO (Fig. 3A and Table 3). By contrast, treatment of apo membranes with zinc prior to copper loading reduces activity by ∼80–90% (Table 2). These findings suggest that zinc inhibition of copper-loaded pMMO occurs at a site distinct from the copper-active site, but that preloading with zinc leads to some occupancy of the active site and thus a higher level of inhibition.
The crystal structures of Methylocystis sp. str. Rockwell support this model and strongly suggest that the higher affinity zinc site is the pmoC metal-binding site. This site is partially occupied by copper upon isolation, and it can be loaded with copper or zinc by soaking (Fig. 7). Zinc is coordinated tetrahedrally by conserved pmoC residues Asp-129, His-133, His-146, and Glu-201 (Fig. 7), consistent with the 4.5 (± 0.5) first shell coordination environment determined by EXAFS (Table 3). Because copper inhibition of pMMO does not occur in the presence of catalase, copper binding to the periplasmic pmoC metal site does not appear to inhibit pMMO (11). This may be due to the absence of the Glu-201 ligand when copper is bound. The apparent lability of this site combined with the effects of zinc addition on activity are consistent with this site being unoccupied by metal ions under physiological conditions. The identity of the second zinc-binding site is less clear, but it could correspond to the solvent-exposed site on the cytoplasmic side of pmoC (Fig. 7). Consistent with a lower affinity and a less specific site, the exact location and ligation of this site differs between the Methylocystis pMMO structures and the M. capsulatus (Bath) structure.
These findings indicate that the molecular basis for zinc inhibition is more complex than simply disrupting the active site. One possibility is that zinc inhibits proton transfer steps. Zinc is a well known inhibitor of proton translocation and has been used to trace proton pathways in many membrane-bound enzymes, including the cytochrome bc1 complex, cytochrome c oxidase, and bacterial photosynthetic reaction centers. In these systems, zinc can inhibit proton uptake from the cytoplasm (N side) or proton release to the periplasm/matrix (P side). The cytochrome bc1 complex and reaction center each have one inhibitory zinc-binding site, whereas zinc inhibition in cytochrome c oxidase involves multiple sites. On the P side of the bc1 complex, zinc prevents proton release from a bound quinol (49), and on the P side of cytochrome c oxidase, a proton pumping exit pathway is blocked (50–53). In cytochrome c oxidase, zinc also prevents proton pumping by binding at the entrance of the D and K proton translocation pathways on the N side (50, 54, 55). In the reaction center, zinc binds to the N side, blocking proton transfer from the cytoplasm to a bound quinone (56–58).
There are several parallels between these studies and what is observed for pMMO. The coordination of the pmoC zinc site is similar to the sites observed in the other systems. In all these enzymes, zinc is coordinated by histidine and carboxylate residues, usually in a tetrahedral geometry (55, 56, 59, 60). The pmoC zinc site is quite similar to a site in the Rhodobacter sphaeroides reaction center that is ligated by two histidines, an aspartate, and a water molecule (56). The pmoC zinc site is solvent-accessible and is located 20 Å from the dicopper-active site in the pmoB subunit. Similarly, the zinc-binding sites in cytochrome c oxidase and the reaction center are at least 20 Å from the proton start or destination site and are housed in solvent-accessible regions of the membrane domains. In cytochrome c oxidase, zinc only binds tightly to the P side in the presence of membrane potential or enzyme turnover (51–53, 55). It is possible that the second zinc-binding site in pMMO is also low affinity in the absence of turnover, and this site may be the cytoplasmic zinc-binding site. The zinc inhibitory sites on the N sides of both cytochrome c oxidase (55) and the reaction center (56) are situated similarly to the second zinc-binding site in the pMMO structures. Finally, zinc only partially inhibits pMMO activity, similar to the 50–90% inhibition observed for cytochrome c oxidase (51) and the 80–90% inhibition in the cytochrome bc1 complex (49).
By analogy to these systems, it may be that zinc binding to pmoC on the P side of pMMO (pmoC site 1, Fig. 7) prevents an important proton release step from a bound reductant such as a quinol. Quinols have long been proposed to be the physiological reductant of pMMO. Only quinols yield activity for solubilized pMMO (61), and improved activity has been reported for pMMO in the presence of a type 2 NADH:quinone oxidoreductase (62–64), which may provide reduced quinones directly to pMMO. In one possible model, a quinol could bind near the zinc site and supply electrons to the active site. Of potential relevance to this idea is the observation that pmoC residues 200–210 become ordered in the zinc-soaked Methylocystis sp. str. Rockwell pMMO structure (Fig. 8). Zinc binding could preclude conformational changes in these residues required for quinol binding and subsequent fast release of protons. Because zinc binding does not completely abolish pMMO activity, this scenario would require the existence of several proton transfer pathways. It is also significant that lipids are found associated with the pmoC subunit (Fig. 6, B and C); lipids exert stabilizing lateral pressure on membrane proteins and can affect proton transport (65). Efforts to test these hypotheses in a proteoliposome system are underway.
Acknowledgments
Use of the Advanced Photon Source, an Office of Science User Facility operated for the United States Department of Energy Office of Science by Argonne National Laboratory, was supported by the United States Department of Energy under Contract DE-AC02–06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corp. and Michigan Technology Tri-Corridor Grant 085P1000817. GM/CA-CAT has been funded in whole or in part with Federal funds from the National Institutes of Health Grant Y1-CO-1020 from NCI and Grant Y1-GM-1104 from NIGMS. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource and at the National Synchrotron Light Source. SSRL is a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by National Institutes of Health, NCRR, Biomedical Technology Program. NSLS, located at Brookhaven National Laboratory, is supported by the United States Department of Energy, Division of Materials Sciences and Division of Chemical Sciences, under Contract DE-AC02-98CH10886.
This work was supported, in whole or in part, by National Institutes of Health Grants GM070473 (to A. C. R.) and DK068139 (to T. L. S.).
The atomic coordinates and structure factors (codes 4PHZ, 4PI0, and 4PI2) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MMO
- methane monooxygenase
- pMMO
- particulate methane monooxygenase
- sMMO
- soluble methane monooxygenase
- SSRL
- Stanford Synchrotron Radiation Lightsource
- NSLS
- National Synchrotron Light Source
- EXAFS
- extended x-ray absorption fine structure
- ESI
- electrospray ionization
- str.
- strain
- ICP-AES
- inductively coupled plasma-atomic emission spectroscopy.
REFERENCES
- 1. Semrau J. D., Dispirito A. A., Yoon S. (2010) Methanotrophs and copper. FEMS Microbiol. Lett. 34, 496–531 [DOI] [PubMed] [Google Scholar]
- 2. Conrado R. J., Gonzalez R. (2014) Envisioning the bioconversion of methane to liquid fuels. Science 343, 621–623 [DOI] [PubMed] [Google Scholar]
- 3. Culpepper M. A., Rosenzweig A. C. (2012) Architecture and active site of particulate methane monooxygenase. Crit. Rev. Biochem. Mol. Biol. 47, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tinberg C. E., Lippard S. J. (2011) Dioxygen activation in soluble methane monooxygenase. Acc. Chem. Res. 44, 280–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murrell J. C., McDonald I. R., Gilbert B. (2000) Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 8, 221–225 [DOI] [PubMed] [Google Scholar]
- 6. Rosenzweig A. C., Frederick C. A., Lippard S. J., Nordlund P. (1993) Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 366, 537–543 [DOI] [PubMed] [Google Scholar]
- 7. Rosenzweig A. C. (2008) The metal centres of particulate methane monooxygenase. Biochem. Soc. Trans. 36, 1134–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lieberman R. L., Rosenzweig A. C. (2005) Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 434, 177–182 [DOI] [PubMed] [Google Scholar]
- 9. Hakemian A. S., Kondapalli K. C., Telser J., Hoffman B. M., Stemmler T. L., Rosenzweig A. C. (2008) The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry 47, 6793–6801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith S. M., Rawat S., Telser J., Hoffman B. M., Stemmler T. L., Rosenzweig A. C. (2011) Crystal structure and characterization of particulate methane monooxygenase from Methylocystis species strain M. Biochemistry 50, 10231–10240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Balasubramanian R., Smith S. M., Rawat S., Yatsunyk L. A., Stemmler T. L., Rosenzweig A. C. (2010) Oxidation of methane by a biological dicopper centre. Nature 465, 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Culpepper M. A., Cutsail G. E., 3rd, Hoffman B. M., Rosenzweig A. C. (2012) Evidence for oxygen binding at the active site of particulate methane monooxygenase. J. Am. Chem. Soc. 134, 7640–7643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martinho M., Choi D. W., Dispirito A. A., Antholine W. E., Semrau J. D., Münck E. (2007) Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: evidence for a diiron center. J. Am. Chem. Soc. 129, 15783–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hakemian A. S., Rosenzweig A. C. (2007) The biochemistry of methane oxidation. Annu. Rev. Biochem. 76, 223–241 [DOI] [PubMed] [Google Scholar]
- 15. Chen C. L., Chen K. H., Ke S. C., Yu S. S., Chan S. I. (2004) Preparation and characterization of a (Cu, Zn)-pMMO from Methylococcus capsulatus (Bath). J. Inorg. Biochem. 98, 2125–2130 [DOI] [PubMed] [Google Scholar]
- 16. Cook S. A., Shiemke A. K. (1996) Evidence that copper is a required cofactor for the membrane-bound form of methane monooxygenase. J. Inorg. Biochem. 63, 273–284 [Google Scholar]
- 17. Takeguchi M., Miyakawa K., Okura I. (1999) The role of copper in particulate methane monooxygenase from Methylosinus trichosporium OB3b. J. Mol. Catal. A 137, 161–168 [DOI] [PubMed] [Google Scholar]
- 18. Smith S. M., Balasubramanian R., Rosenzweig A. C. (2011) Metal reconstitution of particulate methane monooxygenase and heterologous expression of the pmoB subunit. Methods Enzymol. 495, 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Whittenbury R., Phillips K. C., Wilkinson J. F. (1970) Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 61, 205–218 [DOI] [PubMed] [Google Scholar]
- 20. Helling S., Hüttemann M., Kadenbach B., Ramzan R., Vogt S., Marcus K. (2012) Discovering the phosphoproteome of the hydrophobic cytochrome c oxidase membrane protein complex. Methods Mol. Biol. 893, 345–358 [DOI] [PubMed] [Google Scholar]
- 21. Reidegeld K. A., Eisenacher M., Kohl M., Chamrad D., Körting G., Blüggel M., Meyer H. E., Stephan C. (2008) An easy-to-use Decoy Database Builder software tool, implementing different decoy strategies for false discovery rate calculation in automated MS/MS protein ientifications. Proteomics 8, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 22. Voegtli W. C., Sommerhalter M., Saleh L., Baldwin J., Bollinger J. M., Jr., Rosenzweig A. C. (2003) Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2. J. Am. Chem. Soc. 125, 15822–15830 [DOI] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006) Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams P. D., Afonine P. V., Bunkczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. George G., Pickering I. (1995) EXAFSPAK: a suite of computer programs for analysis of x-ray absorption spectra. SSRL, Stanford [Google Scholar]
- 30. Ankudinov A. L., Rehr J. J. (1997) Relativistic calculations of spin-dependent X-ray absorption spectra. Phys. Rev. B 56, R1712–R1715 [Google Scholar]
- 31. Lee P. A., Citrin P. H., Eisenberger P., Kincaid B. M. (1981) Extended x-ray absorption fine structure- its strengths and limitations as a structural tool. Rev. Mod. Phys. 53, 769–806 [Google Scholar]
- 32. Lieberman R. L., Kondapalli K. C., Shrestha D. B., Hakemian A. S., Smith S. M., Telser J., Kuzelka J., Gupta R., Borovik A. S., Lippard S. J., Hoffman B. M., Rosenzweig A. C., Stemmler T. L. (2006) Characterization of the particulate methane monooxygenase metal centers in multiple redox states by X-ray absorption spectroscopy. Inorg. Chem. 45, 8372–8381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang B., Alam S. L., Meyer H. H., Payne M., Stemmler T. L., Davis D. R., Sundquist W. I. (2003) Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J. Biol. Chem. 278, 20225–20234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bencze K. Z., Kondapalli K. C., Stemmler T. L. (2007) in Applications of Physical Methods to Inorganic and Bioinorganic Chemistry: Handbook, Encyolopedia of Inorganic Chemistry (Scott R. A., Lukehart C. M., eds) 2nd Ed., pp. 513–528, John Wiley & Sons, Ltd, Chichester, UK [Google Scholar]
- 35. Lieberman R. L., Shrestha D. B., Doan P. E., Hoffman B. M., Stemmler T. L., Rosenzweig A. C. (2003) Purified particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a dimer with both mononuclear copper and a copper-containing cluster. Proc. Natl. Acad. Sci. U.S.A. 100, 3820–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monson C. F., Cong X., Robison A. D., Pace H. P., Liu C., Poyton M. F., Cremer P. S. (2012) Phosphatidylserine reversibly binds Cu2+ with extremely high affinity. J. Am. Chem. Soc. 134, 7773–7779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Segel I. H. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, pp.465–473, John Wiley & Sons, Inc., New York [Google Scholar]
- 38. Wang S., Lee M. H., Hausinger R. P., Clark P. A., Wilcox D. E., Scott R. A. (1994) Structure of the dinuclear active site of urease. X-ray absorption spectroscopic study of native and 2-mercaptoethanol-inhibited bacterial and plant enzymes. Inorg. Chem. 33, 1589–1593 [Google Scholar]
- 39. Allen F. H. (2002) The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Crystallogr. Sect. B Struct. Sci. 58, 380–388 [DOI] [PubMed] [Google Scholar]
- 40. Stemmler T. L., Sossong T. M., Jr., Goldstein J. I., Ash D. E., Elgren T. E., Kurtz D. M., Jr., Penner-Hahn J. E. (1997) EXAFS comparison of the dimanganese core structures of manganese catalase, arginase and manganese-substituted ribonucleotide reductase and hemerythrin. Biochemistry 36, 9847–9858 [DOI] [PubMed] [Google Scholar]
- 41. Reiter N. J., Osterman A., Torres-Larios A., Swinger K. K., Pan T., Mondragón A. (2010) Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468, 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McLellan J. S., Yang Y., Graham B. S., Kwong P. D. (2011) Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 85, 7788–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brohawn S. G., del Mármol J., MacKinnon R. (2012) Crystal structure of the human K2P TRAAK, a lipid- and mechano-sensitive K+ Ion Channel. Science 335, 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamashita S., Takeshita D., Tomita K. (2014) Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase. Structure 22, 315–325 [DOI] [PubMed] [Google Scholar]
- 45. Buschmann S., Warkentin E., Xie H., Langer J. D., Ermler U., Michel H. (2010) The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science 329, 327–330 [DOI] [PubMed] [Google Scholar]
- 46. Törnroth-Horsefield S., Gourdon P., Horsefield R., Brive L., Yamamoto N., Mori H., Snijder A., Neutze R. (2007) Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure 15, 1663–1673 [DOI] [PubMed] [Google Scholar]
- 47. Bowman J. P., Skerratt J. H., Nichols P. D., Sly L. I. (1991) Phospholipid fatty acid and lipopolysaccharide fatty acid signature lipids in methane utilizing bacteria. FEMS Microbiol. Lett. 85, 15–22 [Google Scholar]
- 48. Fang J., Barcelona M. J., Semrau J. D. (2000) Characterization of methanotrophic bacteria on the basis of intact phospholipid profiles. FEMS Microbiol. Lett. 189, 67–72 [DOI] [PubMed] [Google Scholar]
- 49. Lee D. W., El Khoury Y., Francia F., Zambelli B., Ciurli S., Venturoli G., Hellwig P., Daldal F. (2011) Zinc inhibition of bacterial cytochrome bc1 reveals the role of cytochrome b E295 in proton release at the Qo site. Biochemistry 50, 4263–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Faxén K., Salomonsson L., Adelroth P., Brzezinski P. (2006) Inhibition of proton pumping by zinc ions during specific reaction steps in cytochrome c oxidase. Biochim. Biophys. Acta 1757, 388–394 [DOI] [PubMed] [Google Scholar]
- 51. Kuznetsova S. S., Azarkina N. V., Vygodina T. V., Siletsky S. A., Konstantinov A. A. (2005) Zinc ions as cytochrome c oxidase inhibitors: Two sites of action. Biochemistry 70, 128–136 [DOI] [PubMed] [Google Scholar]
- 52. Mills D. A., Schmidt B., Hiser C., Westley E., Ferguson-Miller S. (2002) Membrane potential-controlled inhibition of cytochrome c oxidase by zinc. J. Biol. Chem. 277, 14894–14901 [DOI] [PubMed] [Google Scholar]
- 53. Vygodina T. V., Zakirzianova W., Konstantinov A. A. (2008) Inhibition of membrane-bound cytochrome c oxidase by zinc ions: High-affinity Zn2+-binding site at the P-side of the membrane. FEBS Lett. 582, 4158–4162 [DOI] [PubMed] [Google Scholar]
- 54. Muramoto K., Hirata K., Shinzawa-Itoh K., Yoko-o S., Yamashita E., Aoyama H., Tsukihara T., Yoshikawa S. (2007) A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 104, 7881–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qin L., Mills D. A., Hiser C., Murphree A., Garavito R. M., Ferguson-Miller S., Hosler J. (2007) Crystallographic location and mutational analysis of Zn and Cd inhibitory sites and role of lipidic carboxylates in rescuing proton path mutants in cytochrome c oxidase. Biochemistry 46, 6239–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Axelrod H. L., Abresch E. C., Paddock M. L., Okamura M. Y., Feher G. (2000) Determination of the binding sites of the proton transfer inhibitors Cd2+ and Zn2+ in bacterial reaction centers. Proc. Natl. Acad. Sci. U.S.A. 97, 1542–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Paddock M. L., Graige M. S., Feher G., Okamura M. Y. (1999) Identification of the proton pathway in bacterial reaction centers: Inhibition of proton transfer by binding of Zn2+ or Cd2+. Proc. Natl. Acad. Sci. U.S.A. 96, 6183–6188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Utschig L. M., Ohigashi Y., Thurnauer M. C., Tiede D. M. (1998) A new metal-binding site in photosynthetic bacterial reaction centers that modulates QA to QB electron transfer. Biochemistry 37, 8278–8281 [DOI] [PubMed] [Google Scholar]
- 59. Berry E. A., Zhang Z., Bellamy H. D., Huang L. (2000) Crystallographic location of two Zn2+-binding sites in the avian cytochrome bc1 complex. Biochim. Biophys. Acta 1459, 440–448 [DOI] [PubMed] [Google Scholar]
- 60. Giachini L., Francia F., Veronesi G., Lee D. W., Daldal F., Huang L. S., Berry E. A., Cocco T., Papa S., Boscherini F., Venturoli G. (2007) X-ray absorption studies of Zn2+ binding sites in bacterial, avian, and bovine cytochrome bc1 complexes. Biophys. J. 93, 2934–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shiemke A. K., Cook S. A., Miley T., Singleton P. (1995) Detergent solubilization of membrane-bound methane monooxygenase requires plastoquinol analogs as electron donors. Arch. Biochem. Biophys. 321, 421–428 [DOI] [PubMed] [Google Scholar]
- 62. Choi D. W., Kunz R. C., Boyd E. S., Semrau J. D., Antholine W. E., Han J. I., Zahn J. A., Boyd J. M., de la Mora A. M., DiSpirito A. A. (2003) The membrane-associated methane monooxygenase pMMO and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J. Bacteriol. 185, 5755–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cook S. A., Shiemke A. K. (2002) Evidence that a type-2 NADH:quinone oxidoreductase mediates electron transfer to particulate methane monooxygenase in Methylococcus capsulatus. Arch. Biochem. Biophys. 398, 32–40 [DOI] [PubMed] [Google Scholar]
- 64. Shiemke A. K., Arp D. J., Sayavedra-Soto L. A. (2004) Inhibition of membrane-bound methane monooxygenase and ammonia monooxygenase by diphenyliodonium: implications for electron transfer. J. Bacteriol. 186, 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Palsdottir H., Hunte C. (2004) Lipids in membrane protein structures. Biochim. Biophys. Acta 1666, 2–18 [DOI] [PubMed] [Google Scholar]