Abstract
Dual specificity phosphatase 6 (DUSP6) functions as a feedback attenuator of Fibroblast Growth Factor signaling during development. In vitro high throughput chemical screening attempts to discover DUSP6 inhibitors have yielded limited success. Yet, in vivo whole organism screens using zebrafish identified 1 (BCI) as an allosteric inhibitor of DUSP6. Here we designed and synthesized a panel of analogs to define structure-activity relationship (SAR) of DUSP6 inhibition. In vivo, high-content analysis in transgenic zebrafish coupled with cell-based chemical complementation assays identified structural features of the 1 pharmacophore that were essential for biological activity. In vitro assays of DUSP hyperactivation corroborated the results from in vivo and cellular SAR. The results reinforce the notion that DUSPs are druggable through allosteric mechanisms, and illustrate the utility of zebrafish as a model organism for in vivo SAR analyses.
Keywords: Cognition Network Technology, FGF signaling, BCI, high-content screening, zebrafish
Introduction
Dual specificity phosphatases (DUSPs) represent a large family of enzymes that catalyze the dephosphorylation of proteins on both phosphotyrosine and phosphoserine/phosphothreonine residues within the same substrate. DUSP6, also known as mitogen-activated protein kinase phosphatase 3 (MKP3), belongs to a subgroup of eleven dual specificity phosphatases that dephosphorylate and inactivate mitogen-activated protein kinases (MAPKs).[1–3] During development, Dusp6 functions as a negative feedback regulator of Fibroblast Growth Factor (FGF) signaling.[4–6] The discovery of potent and selective inhibitors of dual specificity phosphatases has been hindered by a high degree of conservation between the DUSP active sites and their shallow and feature-poor topology.[1] In addition, the presence of a reactive, active site cysteine, which is critical for enzymatic activity but displays high nucleophilicity and sensitivity to oxidation due to a low pKa sulfhydryl moiety, has hampered drug discovery efforts.[7] Perhaps not too surprisingly, in vitro screens for DUSP inhibitors have yielded hit compounds that were redox reactive,[8] lacked in vivo activity[9] or had activities not readily reconciled with DUSP inhibition.[10]
It was the advent of a whole organism live reporter for FGF activity (Tg(dusp6:EGFP)pt6)[11, 12] that enabled the discovery of a biologically active inhibitor of zebrafish Dusp6, (E)-2-benzylidene-3-(cyclohexylamino)-2,3-dihydro-1H-inden-1-one (BCI), shortly designated as 1.[13] Chemical complementation assays revealed that 1 specifically inhibited DUSP6 and DUSP1, but not the related Dusp5.[13] Interestingly, 1 lacked antiphosphatase activity in a traditional biochemical assay using bacterially produced recombinant protein, and therefore had been missed in prior in vitro screens for DUSP inhibitors.[14] Instead, 1 selectively inhibited DUSP activity in the presence of ERK, which activates DUSP6 through a conformational change that brings a general acid residue in close proximity to the active site cysteine, enhancing its nucleophilicity.[15] The zebrafish system therefore captured the inhibitory activity of 1 against the DUSPs’ biologically relevant phosphatase activity and provided a useful chemical probe to study the role of Dusp6 in embryonic development and in adult immunity[13, 16–21].
To explore structure-activity relationship (SAR) of 1 and DUSP6 inhibition, we synthesized a series of 29 analogs with modifications in four functional groups of the 1 pharmacophore. SAR was evaluated for FGF hyperactivation in vivo using transgenic zebrafish that report on FGF activity,[11] and for DUSP6 and DUSP1 inhibition in cell-based chemical complementation assays.[13] The SAR studies revealed a strong correlation between in vivo FGF hyperactivation and inhibition of cellular DUSPs and a lack of correlation between biological activity and whole organism toxicity. Biochemical assays for substrate-induced Dusp6 hyperactivation corroborated the inhibitory activities of the new analogs.
The selective inhibition of substrate-induced DUSP6 activation by 1 was reconciled by molecular modeling studies of the 1-DUSP6 interaction. Unbiased docking simulations supported that 1 would bind to the low-activity form of DUSP6, occupying a novel allosteric binding site adjacent to the phosphatase active site.[13] Refined scoring of potential docking modes using the Poisson-Boltzmann Surface Area (PBSA) binding free energy calculations indicated a preferred binding orientation for 1 where its cyclohexylamino-side chain and α,β-unsaturated ketone moiety form hydrogen bonds with DUSP6. Collectively, these results support the hypothesis that DUSPs can be targeted through allosteric mechanisms.
Results
Chemical synthesis of BCI analogs
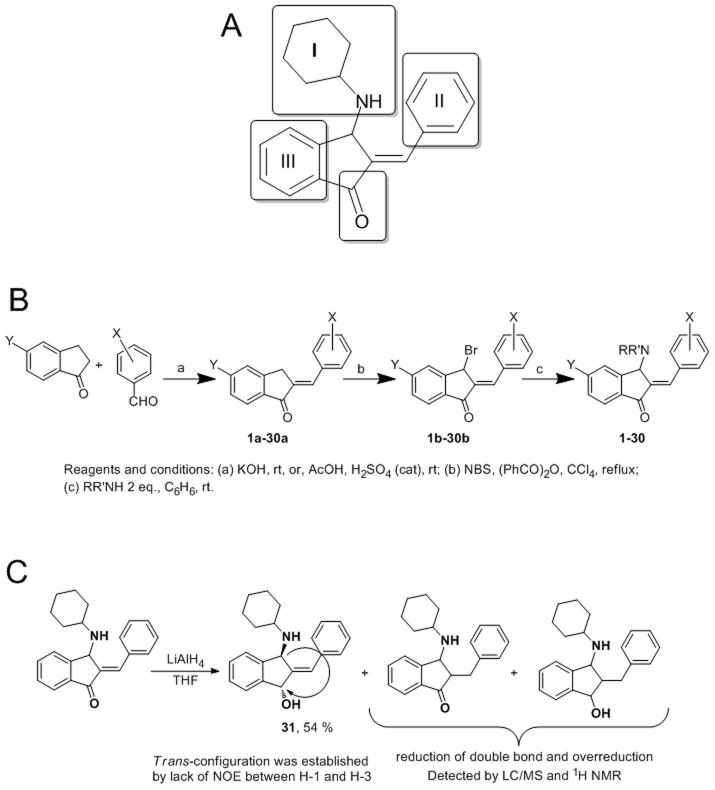
Compound 1 has four distinct, potential sites of modification (amine I, aromatic rings II and III, and the carbonyl group, Scheme 1A). We created a small library of 29 analogs, designed to probe spatial and hydrogen-bonding requirements of the aminoalkyl (I ring) system (Supplemental Materials, Table S1, compounds 2–11), to modulate the electrophilicity and hydrogen acceptor properties of the α,β-unsaturated ketone through electron-donating or electron-withdrawing substituents in the II and III rings (Supplemental Materials, Table S1, compounds 12–20), and to investigate whether combinations of multiple structural modifications would reach maximal potency (Supplemental Materials, Table S1, compounds 21–30).
Scheme 1. Design and generation of 1 and analogs.
A. Sites of modification of the scaffold. B. Synthesis of 1 and its analogs. C. Synthesis of aminoalcohol, 31.
A convenient and flexible route to 1 and its analogs is shown in Scheme 1B. This route allowed synthesis of the parent compound and a series of analogs with modified fragment I in Scheme 1A. The condensation of 5-substituted 1indanones with appropriate benzaldehydes afforded corresponding 2-benzylidene-1-indanones in quantitative yield. The bromination of 2-benzylidene-1-indanones with N-bromosuccinimde (NBS) provided 3-bromo-2-benzylidene-1-indanones.[22] The final step in synthesis is reaction of 3-bromo-2-benzylidene-1-indanones with amines as developed by Cromwell.[23] Reaction of the unsubstituted 3-bromo-2-benzylidene-1-indanone (1b, X, Y = H) with amines proceeds in two steps, giving initially a 2-[α-(alkylamino)benzyl]-1-indenone, which rearranges to the more thermodynamically stable 2-benzylidene-3-(alkylamino)-2,3-dihydro-1H-inden-1-one.[23]
The amine in position I was modified to examine steric effects, hydrogen-donating properties, effects of relative basicity of the amino group, and effects of introduction of additional H-donors and H-acceptors. 3-Bromo-2-benzylidene-1-indanone 1b was converted to analogs 1–11 by treatment with two equivalents of the corresponding primary or secondary amines in benzene at room temperature (Supplemental Materials, Table S2 & S3). The reaction provided the desired compounds, generally in high yield; one notable exception was the 4-piperazine analog 9 (BCI-187), which was afforded in 42% yield using 1.9 equivalents of piperazine. Using two equivalents of piperazine resulted in formation of a mixture of desired compound and isomerization products with migration of the piperazine group. Maury et al. reported that 1 and analogs are labile compounds that can undergo allylic rearrangements with migration of the amino group or prototropic isomerization of the double bond into an endocyclic alkene.[23] The rate of this isomerization is dependent on nature of the amine, but formation of the desired product was complete in our hands within 24 h; the one exception was reaction of 1b with aniline, which required 120 h and provided compound 4 (BCI-11) in 69% yield.
This method was found to be suitable for the synthesis of analogs containing additional substituents in rings II or III, as well as analogs containing combinations of different amines in position I. Acid- or base-catalyzed condensation using substituted benzaldehydes or indanones in the first step provided corresponding 2arylidene-1-indanones 12a-30a in good yield.[24] Following bromination with NBS, treatment of the resulting 3-bromo-2-arylidene-1-indanones 12b-30b with cyclohexylamine in benzene provided final products bearing substituents in rings II (12–17), C (18–20) and B and C (21, 22). Compounds 23–30 were synthesized as described above using morpholine.
To probe the importance of the carbonyl group, we also synthesized 2-benzylidene-3-cyclohexylaminoindan-1-ol 31 (BCI-10) with a trans relationship between the amine and hydroxyl groups (as documented by the absence of Nuclear Overhauser Effect (NOE) between H-1 and H-3) via reduction of 1 with LiAlH4 (Scheme 1C). The moderate yield of alcohol 31 was due to over-reduction of both the carbonyl group and the carbon-carbon double bond.
Structure-activity studies in zebrafish embryos
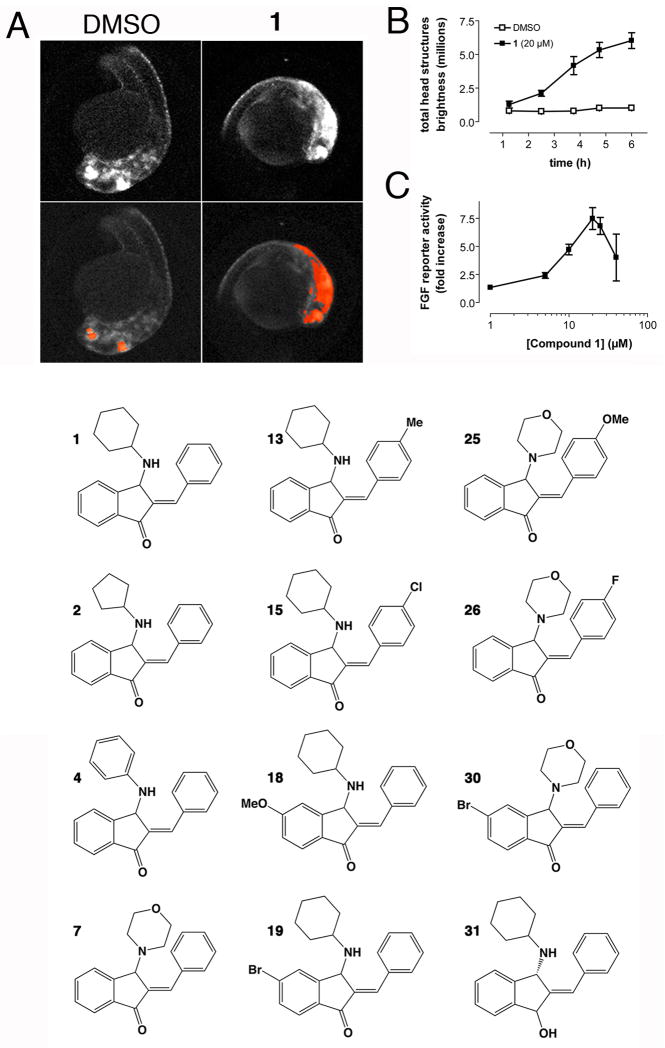
We first analyzed all analogs for hyperactivation of FGF signaling in the Tg(dusp6:EGFP)pt6 transgenic zebrafish model. In this assay, we previously established transgenic zebrafish that expresses destabilized GFP under the control of active FGF signaling. [11, 12] These transgenic embryos respond to FGF activation with GFP expression in specific areas of the brain, which can be quantified by automated image analysis using Cognition Network Technology (CNT) [25]. To determine optimal conditions for FGF reporter activation and to test whether the response was saturable, we performed time- and concentration-dependence experiments. 24 hours post-fertilization (hpf) embryos were treated with 20 μM of 1 and images were acquired as described previously.[25, 26] Figure 1A shows representative fluorescence micrographs of vehicle- or 1-treated embryos before and after CNT analysis.[26] Time course experiments in embryos treated with 1 showed that after 5 hours, GFP expression reached a maximum and remained stable for an additional hour (Fig. 1B). A concentration-dependence experiment at the 5 hour time point documented FGF reporter activation was maximal at 20 μM and declined at higher concentrations (Fig. 1C). We tested all 29 analogs to determine if any of the new compounds induced GFP expression similar to 1 (Supplemental Table S4). Dose-response curves were obtained showing nine compounds were equipotent to 1, and one agent, 7 (BCI-9) that had significantly higher activity than 1 (EC50 4.5 μM) (Table 1). In total, in vivo SAR identified 11 new compounds that showed concentration-dependent hyperactivation of FGF signaling in Tg(dusp6:EGFP)pt6 embryos (Table 1, relevant structures shown in Fig. 1D). Structural elements essential for activity were an aliphatic amino-alkyl side chain at C-3, and the α,β-unsaturated ketone moiety. Changes that were tolerated without loss of activity were moderately electron-donating or –withdrawing substitutents in the II and II rings. A planar aromatic amine in ring I and a strongly electron-withdrawing cyano substituent in rings II or III were not tolerated (See Discussion for more details). These data document that specific structural modifications affected biological activity, suggesting that 1 is a bona fide pharmacophore of in vivo FGF signaling.
Figure 1. Quantitation of in vivo FGF hyperactivation by automated image analysis.
A. Upper panel. Representative fluorescence micrographs of 24 hpf Tg(dusp6:EGFP)pt6 embryos treated for 5 hours with vehicle (1% DMSO) or 20 μM 1. The major bright head structures are eye and retina, mid-hindbrain boundary, and trigeminal ganglia. Lower panel. Archived scan images with CNT algorithm applied. Areas in red are regions of GFP expression in the head that exceeded a threshold relative to yolk sac fluorescence. B. Time course of FGF activation. 24 hpf Tg(dusp6:EGFP)pt6 embryos were exposed to vehicle (DMSO) or 20 μM 1 in 96 well plates, imaged every hour for 6 hours, and analyzed by the CNT ruleset. Data show total GFP intensity in the head from 8 embryos per condition ± SEM. C. Dose-response of FGF activation by 1 at 5 hours after treatment. D. Chemical structures of important BCI analogs and numbering scheme used in this study.
Table 1.
Summary of BCI analogs.
| Compound | Alias | FGF reporter activation in zebrafish | cellular inhibition of | ||
|---|---|---|---|---|---|
| EC50(μM) | p-value | DUSP6 | DUSP1 | ||
| 1 | BCI | 10.6 ± 0.8 (14) | 13.3 ± 1.8 (12) | 8.0 ± 0.6 (11) | |
| 2 | VNK164 | 13.3 ± 3.3 (2) | 0.56 | 15.8 ± 3.0 (3) | 8.3 ± 0.8 (3) |
| 3 | VNK165 | 12.3 ± 0.8 (2) | 0.21 | 25.0 ± 7.0(3) | 14.0 ± 2.8 (3) |
| 4 | BCI-11 | ≫25 | ND | ND | |
| 5 | BCI-8 | 13.6 (1) | ND | ND | |
| 7 | BCI-9 | 4.5 ± 0.6 (7) | ≪ 0.01 | 50.8 ± 4.1 (7) | 28.4 ± 1.8 (6) |
| 12 | VNK211 | 22.6 ± 2.6 (3) | 0.04 | ND | ND |
| 13 | VNK212 | 12.0 ± 1.5 (5) | 0.47 | 64.7 ± 15.7 (3) | 25.5 ± 6.1 (3) |
| 14 | VNK303 | 8.9 ± 2.5 (2) | 0.6 | ND | ND |
| 15 | VNK183 | 10.0 ± 1.1 (4) | 0.66 | 37.8 ± 11.8 (3) | 14.7 ± 4.4 (3) |
| 16 | VNK297 | ~10.0(1) | ND | ND | |
| 18 | VNK216 | 10.4 ± 0.6 (3) | 0.81 | 67.8 ± 14.2 (3) | 28.0 ± 5.0 (3) |
| 19 | VNK215 | 12.0 ± 3.0 (6) | 0.67 | 55.9 ± 8.6 (9) | 28.6 ± 3.8 (11) |
| 24 | VNK256 | ≫25 | ≫ 100 (2) | ≫100 (2) | |
| 25 | VNK269 | 7.1 ± 0.7 (2) | 0.02 | ND | ND |
| 26 | VNK304 | 10.3 (2) | ND | ND | |
| 28 | VNK296 | ≫25 | ≫ 100 (2) | ≫ 100 (2) | |
| 30 | VNK299 | ≫25 | ND | ND | |
| 31 | BCI-10 | ≫25 | ND | ND | |
average EC50 ± S.E. of (n) experiments.
p-value: compared with BCI by Student's t-test (two tailed, unequal variances)
Identification of a non-toxic analog of 1 with cellular and in vivo activity
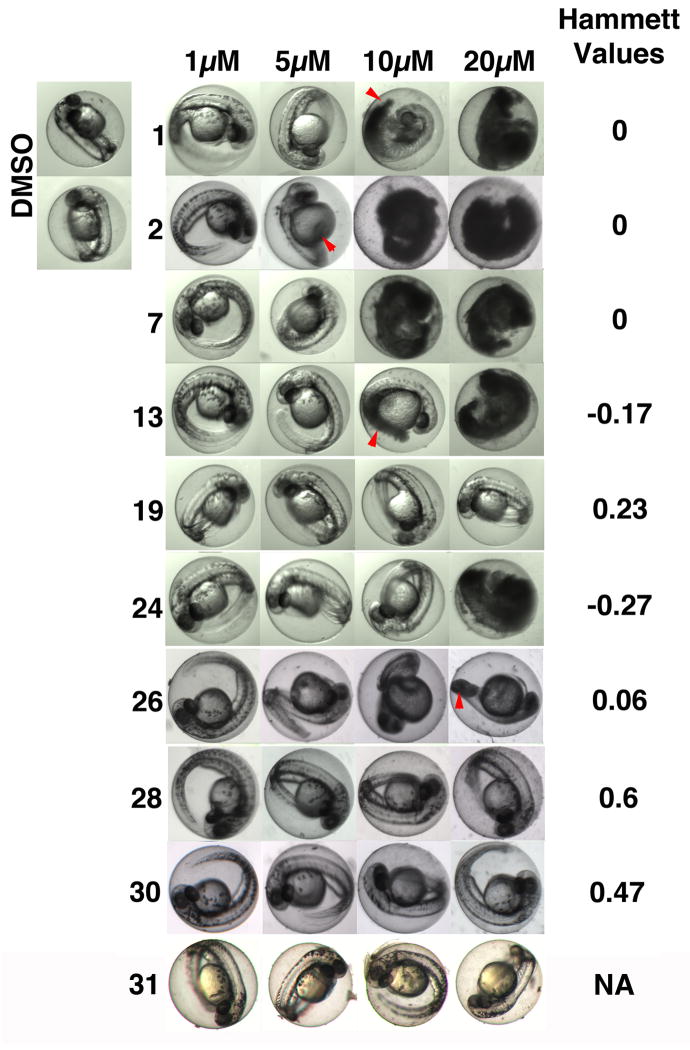
Because the primary assay for biological activity involved the use of a living vertebrate animal, we were able to observe whole organism toxicities upon compound treatment, and to relate toxicity to in vivo target activity and chemical reactivity. The 1 pharmacophore contains an electrophilic α,β-unsaturated ketone moiety, and although many drugs on the market are electrophilic, the presence of such elements is often viewed as a liability in drug development due to possible non-selective modification of cellular nucleophiles, leading to off-target effects and toxicity. Indeed, many of the active agents at the highest doses started to show whole organism toxicity at the time of imaging as manifested by gross morphological changes (data not shown). We therefore assessed toxicity upon prolonged exposure to agents (24 hours) by visual inspection of larvae for morphological changes such as a bent tail phenotype and the appearance of opaque, necrotic cells (Fig. 2 & Supplemental Table S4 & S5). Cellular toxicity was confirmed by staining with acridine orange (a vital dye that labels cells with damaged cell membranes), revealing the presence of dead cells in the tail (data not shown).
Figure 2. Compound 19 lacks whole organism toxicity.
Images show transmitted light micrographs of embryos in the presence of vehicle (DMSO), or the indicated concentrations of test agents after 24 hours of treatment. Toxicity manifests itself by the appearance of opaque cells, indicating necrosis (red arrows demarcate examples of necrotic cells). Compound 19, 27, and 29, 30 exhibited minimal toxicity. Hammett Sigma values indicate the degree of electrophilicity of the α,β-unsaturated ketone moiety.
Several agents exhibited little or no toxicity at concentrations that hyperactivated FGF signaling in vivo (Supplemental Table S4 and S5). Conversely, we found some agents that did not hyperactivate FGF signaling but caused developmental toxicity, and several that were devoid of toxicity and activity, including the alcohol 31 (Supplemental Table S4 and S5 and Fig. 2). The latter observation suggests that while the α,β-unsaturated ketone moiety is important for FGF hyperactivation, it also may also be a contributing factor in embryo toxicity. To further explore this hypothesis, we examined whether there was a correlation between toxicity and electrophilicity by calculating Hammett s constants for 1 and nine analogs with or without in vivo FGF enhancing activity. Hammett s constants ranged from −0.54 (low electrophilicity) to 0.60 (high electrophilicity), covering the entire spectrum of chemical reactivities in the series (Fig. 2).
At concentrations at or above their EC50 (5–10 μM), analogs with negative Hammett s values (lower electrophilicity) showed toxicity (Fig. 2). In contrast, compounds with positive Hammett values appeared to be less toxic. Thus, surprisingly, agents with predicted high electrophilicity were generally better tolerated. There seemed to be no correlation between toxicity and in vivo target activity, as two inactive analogs (21 (BCI-266) and 24 (BCI-256)) were also toxic. Collectively, the data document that electrophilicity does not significantly contribute to toxicity. This suggests that, while the unsaturated ketone is required for activity, it does not indiscriminately modify essential cellular constituents, and that the untoward effects of compounds are due to off-target effects unrelated to chemical reactivity. One agent in particular, compound 19 (BCI-215), did not show any toxicity at concentrations two times the EC50 for FGF activation (Fig. 2). To probe for possible developmental delays or defects, we performed a hatching study and found that larvae treated with 19 developed normally. After a 6 hour exposure followed by washout, 100% of vehicle- or compound 19-treated embryos, at two times the EC50 for FGF activation (20 μM), had hatched by 56 hpf (data not shown). In contrast, no embryos hatched after exposure to 1 or 7 at their EC50 (10 or 5 μM, respectively). Thus, 19 lacked whole organism toxicity at concentrations that activated FGF signaling. To corroborate the results from the zebrafish developmental toxicity studies, we performed a cytotoxicity experiment in EA.hy926 cells, a hybridoma cell line that retains many properties of normal endothelial cells [27–29]. While both agents were relatively non-toxic under the conditions of the assay, BCI showed signs of cell loss, nuclear condensation, and necrosis (PI staining) at concentrations above 25 μM (Supplementary Fig. S1). In contrast, BCI-215 was devoid of cellular toxicity at concentrations up to 50 μM (Supplementary Fig. S1). Therefore, the cellular assay recapitulated differences in developmental toxicity.
Analogs of 1 inhibit DUSPs in a chemical complementation assay
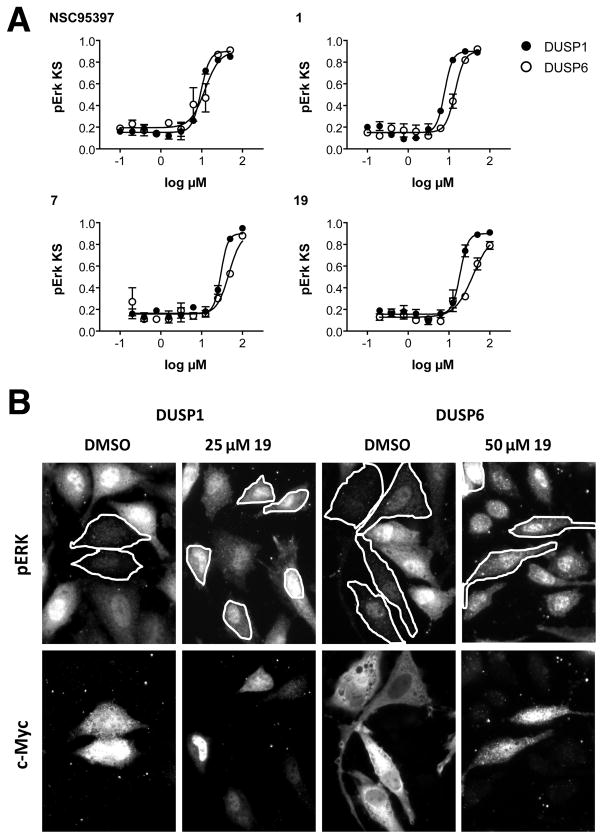
To test whether the FGF hyperactivating activities of the new analogs were due to DUSP inhibition, we tested seven agents that had shown robust activity in the zebrafish and that were available in sufficient quantities, for DUSP6 and DUSP1 inhibitory activity in our mammalian cell-based chemical complementation assay (Table 1).[30] In these assays, HeLa cells were transfected with Myc-tagged DUSP1 or DUSP6, and stimulated with phorbol ester (TPA) to activate the ERK pathway. In this assay, the expression of active phosphatases decreases TPA-induced ERK phosphorylation. Thus, compounds that inhibit DUSP activity restore pERK levels in the DUSP overexpressing cells. Restoration of ERK phosphorylation can be quantified by comparing pERK distributions of treated and untreated cell populations by Kolmogorov-Smirnov (KS) statistics. Figure 3A shows that 1, 7, and 19 concentration-dependently increased pERK levels in DUSP-overexpressing cells with IC50 values in the micromolar range. Figure 3B shows representative images for 19 that illustrate restoration of ERK phosphorylation in DUSP-overexpressing cells. Table 1 shows that all agents with activity in the zebrafish also inhibited DUSP6 and DUSP1 in mammalian cells, whereas two inactive compounds (24 (BCI-256) and 28 (BCI-296)) lacked antiphosphatase activity. Hence, the data support the hypothesis that in vivo activity of the analogs is due to DUSP inactivation, and validate the core structure of 1 as a pharmacophore for DUSP inhibition.
Figure 3. Inhibition of DUSP6 and DUSP1 by analogs of compound 1 in mammalian cells.
A. Concentration-dependent inhibition of DUSP1 (closed symbols) and DUSP6 (open symbols) by 1 and analogs and a previously described multi-targeted DUSP inhibitor (NSC95397). B. Representative fluorescence micrographs of ERK phosphorylation (upper panel) and DUSP expression (lower panel) in the presence or absence of compound 19. Restoration of ERK phosphorylation in DUSP-expressing cells is observed after treatment with 19.
Analogs of 1 suppress ERK-stimulated activation of DUSP6
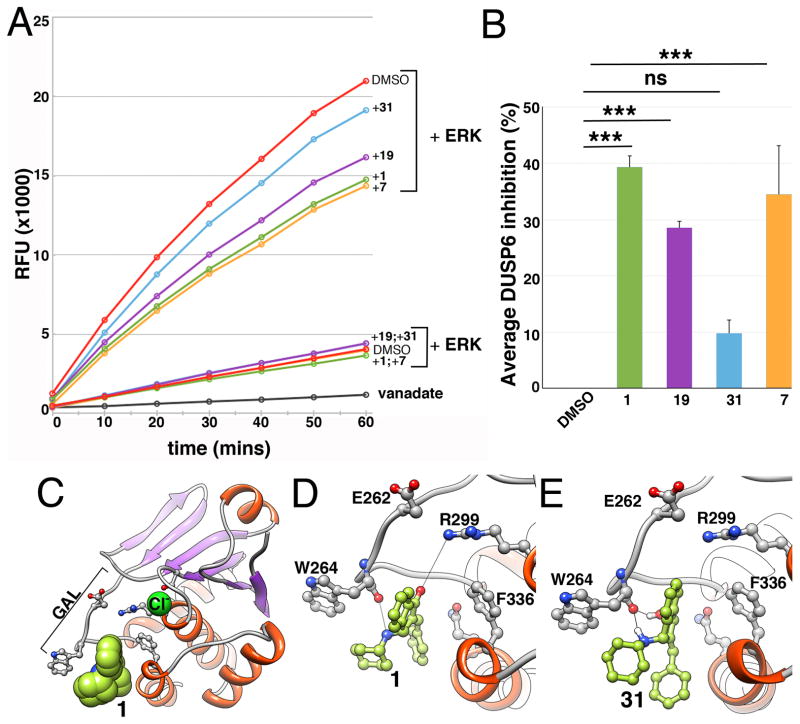
DUSP6 activity is stimulated upon substrate binding. In the absence of ERK, DUSP6 has low basal catalytic activity, which upon interaction with ERK is significantly enhanced.[15] In vitro studies with 1 suggested that its mechanism of action was through suppressing this ERK-stimulated activation of DUSP6.[13] We confirmed that analogs of 1 also inhibited the activation of DUSP6 by ERK binding (Fig. 4A, B). 1, 7 and 19 significantly suppressed activation of DUSP6 with similar magnitudes (Fig. 4A, B). Consistent with previous data,[13] none of the agents tested inhibited basal phosphatase activity (Fig 4A). In contrast, suppression of DUSP6 activation by compound 31, which failed to hyperactivate FGF signaling in the transgenic embryos, was insignificant (Fig. 4A, B). Thus in vitro inhibition of DUSP6 by analogs of 1 correlated with in vivo activity.
Figure 4. Compound 1, 7, and 19 inhibit ERK2 stimulated activation of DUSP6 catalytic activity in vitro.
A. DUSP6 phosphatase activity in the presence and absence of ERK2. Inclusion of 1, 7 and 19 partially suppressed DUSP6 activation by ERK2, but not basal phosphatase activity. In contrast, compound 31 showed only a minor effect. Data are from a single experiment that has been repeated twice with identical results. Vanadate was used as a positive control. Note that in assays in the absence of ERK, results from treatment with 1 overlapped with 7 and 19 with 31. B. Graph showing average percent inhibition of DUSP6 hyperactivation ± SD from three independent experiments by 1, 7, 31 and 19. Percent inhibition was graphed from results at 10 minutes when the assay was within the linear range. ***, p<0.001 by one-way ANOVA compared with vehicle control. C. Overview of putative allosteric binding site of 1 and docking orientation (chloride ion highlights catalytic site). D. Close-up view of 1’s interactions with allosteric binding site residues. Two hydrogen bonds are observed (black dotted lines), between the ketone oxygen of 1 and Arg299, and the amine of 1 and Trp264’s backbone oxygen. E. Close-up view of 31’s interactions with allosteric binding site residues. Hydrogen bonding of amine in 31 and Trp264’s backbone oxygen is present. In contrast, hydrogen bonding with Arg299 as noted in 1 is absent.
Refinement of the DUSP6-1 binding model by computational modeling and simulation
We previously established the necessity of both the cyclohexylamino substituent and the benzylidene substituent for 1’s biological activity.[13] These experimental findings were consistent with a computational model of 1 binding to an allosteric site on DUSP6 in an orientation where both of these rings make contact with the interior of the pocket through hydrophobic interactions. Occupation of this site by 1 is thought to prevent a conformational change in DUSP6, which otherwise would accommodate ERK binding upon positioning Asp262 (in the general acid loop, GAL) close to Arg299 of the phosphatase catalytic site (Fig. 4C and Movie S1).[31] Ensemble docking and clustering approach that we previously utilized identified multiple potential orientations for 1 in the allosteric pocket with comparable interaction scores.[13] The scoring incorporated only pairwise atomic interactions as a weighted sum of van der Waals interactions and hydrogen bond formation energies. The allosteric pocket of DUSP6, however, features charged Asp262 and Arg299 side-chains and hydrophobic patches, whose interactions are not accounted for by these interactions. Hence, we rigorously evaluated electrostatic and solvation effects to gain insights that would complement the SAR studies. To this end, we refined the structural model for DUSP6-1 interaction by rescoring docking modes with the help of PBSA calculations implemented in the FRED application (OpenEye, Santa Fe, NM).[32] PB and SA components of this scoring method describe electrostatic interactions and solvation effects, respectively. The method is computationally more expensive than force-field based scoring, hence is generally applied for refinement in post-processing of docking poses.[33]
Application of this method permitted us to prioritize the binding pose of 1 shown in Figures 4C and 4D. Consistent with the existing model and the fact that deletion of the benzylidene moiety abolishes biological activity[13] the refined model showed the indanone ring buried deep in the binding pocket. Two previously undetected hydrogen bonds were observed using molecular dynamics simulations starting from this docking pose (Movie S2): the first is between the ketone oxygen of 1 and the Arg299 side-chain, and the second between 1’s secondary amine of the cyclohexylamino side chain and the Trp264 backbone oxygen. In addition, Trp264 makes hydrophobic contacts with the aminocyclohexane moiety. To confirm these computational predictions, we analyzed docking of 31 to DUSP6 using the same parameters as DUSP6-1 (Fig. 4E). No hydrogen bond formation was observed between Arg299 and α,β-unsaturated alcohol moiety in 31, in support of a model where the α,β-unsaturated ketone moiety in 1 is required for DUSP6 inhibition through hydrogen bonding but not covalent modification, consistent with experimental observations.
Discussion and Conclusions
Despite the evidence that DUSPs play important roles in a variety of maladies, including cancer[34], inflammation[35], and immunity[16], they have largely eluded attempts to discover biologically active small molecule inhibitors. The reasons for this are numerous and include overlapping substrate specificity, shallow and feature-poor active sites, redox sensitivity, and the use of in vitro assays that do not recapitulate the activity of these enzymes their biological context. We recently identified a small molecule, compound 1, that inhibits the activation of DUSP6 by ERK, and presumably binds to a novel allosteric site on DUSP6. In this report, we designed a series of novel 1-analogs to explore their SARs through in vivo studies using transgenic zebrafish, and confirmation by in vitro phosphatase assay.
In vivo SAR supports the importance of: Ring I
Changing the size of the aliphatic I-ring in analogs 2 (BCI-164) and 3 (BCI-165) preserved activity, while replacement of the cyclohexyl ring with a phenyl ring in analog 4 (BCI-11) abolished activity (Table 1). Shortening the side chain by incorporating the nitrogen into the aliphatic ring preserved activity (compound 5 (BCI-8)). Introduction of heteroatoms into Ring I abolished activity (analogs 8 –11). A notable exception appeared to be the morpholine-substituted compound 7, which in vivo had the maximal activity of all the BCI analogs.
Rings II and III
Substituents at the para position in Ring II with electron-donating (OMe: 12 (BCI-211), Me: 13 (BCI-212)) or electron-withdrawing groups (F: 14 (BCI-303), Cl: 15 (BCI-183), 3,4-Cl2: 16 (BCI-297)) did not substantially change activity compared to 1 (Table 1). The strongest electron-withdrawing cyano group (compound 17 (BCI-7)), however, was not tolerated. Similar results were obtained for compounds with substitutions in ring C (compounds 18–20), where methoxy- (compound 18 (BCI-216)) and bromo (compound 19 (BCI-215)) substituents were tolerated, but a cyano group was not (compound 20 (BCI-169)). Finally, reduction of the α,β-unsaturated ketone to the α,β-unsaturated alcohol (compound 31, Scheme 1C) abolished activity. These results are consistent with the prediction by the model that the carbonyl group of the indanone system engaged in a hydrogen bond with Arg299, and with a minor contribution of an additional hydrogen bond between the cyclohexylamino moiety and Trp264.
Double substitutions and combination with a morpholino substitutent in Ring I
Simultaneous substitutions in both the II and III-ring with methoxy groups or halogens rendered the scaffold inactive (21, 22 (BCI-283)). Because of the unique ability of the morpholino stubstituent in Ring I to increase potency (analog 7), we asked if inclusion of a morpholino moiety in Ring I position along with substituents in Rings II and III would improve the activity; this hypothesis was not supported. While the activity of a fluoro-substituted compound was preserved (14 vs. 26 (BCI-304)), the replacement of cyclohexylamine with morpholine did not render previously inactive compounds active (21 vs. 23 (BCI-267), 22 vs. 29 (BCI-282)). Furthermore, the morpholino substituent abolished the activity of three active agents (analogs 24, 27 (BCI-271), 28 (BCI-296)). Thus, the ability of the morpholino group to preserve or improve potency was limited to unsubstituted (7) and para-fluoro substituted (26) analogs.
Activity in vivo and DUSP inhibition
There is considerable debate about whether potent and selective inhibitors of DUSPs can be obtained. Multiple attempts at discovering DUSP inhibitors have failed, and because adequate probes are lacking, the question of what degree of specificity is required to elicit desired biological responses has been intractable. The dearth of chemical probes for DUSP activity has also prevented proof-of-principle studies in mammals. Our studies demonstrate that DUSPs may be druggable through exploitation of allosteric mechanisms. First, 1 and seven analogs that activated FGF signaling in vivo also inhibited mammalian DUSP1 and DUSP6 in cultured cells, whereas agents that lacked in vivo activity were devoid of antiphosphatase activity. This is a significant finding as prior to our current studies, only three agents had ever shown confirmed activity in the mammalian cell-based chemical complementation assay, namely the thiol poison, phenyl arsine oxide[36], the glutathione-depleting alkaloid, sanguinarine[30], and the para-quinone, NSC95397.[37] Second, experimental data were consistent with computational modeling predictions based on the DUSP6-1 interactions at the allosteric site, lending credence to the proposed mechanism of inhibition. Taken together, the results validate the 1 scaffold as a bona fide pharmacophore for allosteric DUSP6 inhibition.
All agents with in vivo activity were inhibitors of both DUSP6 and DUSP1. Because there is no published X-ray crystal structure of DUSP1, we created a homology model and showed that the allosteric site also exists on DUSP1 (data not shown). Therefore, the 1 pharmacophore might not be expected to show selectivity for either DUSP.
Although we observed a correlation between in vivo and cellular activity, there were differences in potency between these assays. The most likely reasons for this are solubility, uptake, and/or protein binding as zebrafish assays are performed in an unbuffered aqueous solution, whereas cellular assays are conducted in complete growth medium with serum. The most striking difference between assays was observed in the in vitro DUSP hyperactivation assay, where compounds showed only partial activity at high concentrations (100 μM). While we have observed this phenomenon before[13], a definitive explanation is lacking. One reason for the lackluster in vitro activity could be that multiple binding processes and enzymatic reactions with different affinities and kinetics occur concurrently, possibly affecting the enzyme-inhibitor interaction. Alternative reasons is that in vitro assays do not faithfully recapitulate biological conditions due to lack of a proper microenvironment and accessory/scaffolding proteins.
Neither the dual inhibitory nature of 1 and analogs, nor the presence of an electrophilic α,b-unsaturated ketone appeared to influence FGF hyperactivation. More importantly, the fact that embryos treated with compound 19 developed normally suggests that neither a lack of selectivity nor the presence of a potentially electrophilic moiety were causes for toxicity. These features make compound 19 not only an attractive candidate for further evaluation in mammals, but also provide the research community with a much cleaner probe than 1 to investigate the biological functions of DUSP1 and DUSP6. The data demonstrate the rich potential of zebrafish in early drug discovery and identify compound 19 as a candidate for proof-of-principle studies to investigate the role of Dusp6 in embryonic development and in disease models.
DUSPs have long eluded drug discovery efforts using the contemporary single-target, biochemical assay-based discovery paradigm. Their active sites are shallow, and their catalytic activity depends on a highly reactive, redox-sensitive cysteine. Prior discovery efforts, therefore, have been exceedingly good at discovering redox-active, nonselective inhibitors with lack of or promiscuous cellular activity. Our findings suggest that targeting DUSPs by allosteric mechanisms can circumvent many of the problems caused by the nature of the DUSPs’ catalytic cavity. The zebrafish in particular has been indispensable in discovering such inhibitors, and the present data create continued enthusiasm for the further identification of DUSP inhibitors by phenotypic discovery in transgenic zebrafish.
Experimental Sections
Chemical Synthesis
The synthesis of and analytical data for all compounds are described in the Supplementary Materials.
Zebrafish maintenance and compound treatment
All procedures involving zebrafish were reviewed and approved by the University of Pittsburgh Institutional Animal Care and Use Committee. Tg (dusp6:eGFP) pt6 embryos were obtained by natural mating and incubated at 28.5 °C.[26] One transgenic embryo was placed into every well of a 96-well plate in 200 μL of E3 (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4). Compounds were dissolved as 100X stock solutions in DMSO and 2 μL added directly to octuplicate wells. For the SAR studies, a negative control (8 wells of DMSO (1%)) was included on every plate. A full dose-response of 1 was run on each day of experiments.
Automated imaging and analysis
At the end of compound treatment, embryos were anesthesized with 40 μg/ml MS222 (tricaine methanesulfonate, Sigma) in E3. Plates were loaded into an ImageXpress Ultra high-content reader (Molecular Devices) and imaged using a 4X objective at excitation/emission wavelengths of 488/525 nm (GFP).[26] Archived scan images were uploaded into Developer (Definiens AG) and analyzed for GFP expression in the head using a simplified version of our previously described CNT ruleset.[26] A GFP threshold was set based on well background fluorescence and regions within the zebrafish larva were classified as positive for GFP expression if their fluorescence intensity exceeded this threshold. GFP expressing areas were merged and the four largest objects selected for quantitation. [Total head structures brightness = (mean GFP intensity) * (area of the four head structures)]. EC50 values were determined from dose-response curves by a four-parameter logistic equation where the bottom and top were defined as the magnitude of FGF activation by 1% DMSO and by the maximum response elicited by the positive plate control (usually seen with 20 μM 1), respectively. EC50 values in Table 1 are the averages ± SEM of (n) independent experiments.
Developmental toxicity assessments
After drug treatment and GFP quantitation, embryos in microplates were returned to the incubator overnight in the continued presence of test agents. After a total of 24 hours treatment, wells were examined visually for signs of toxicity such as changes in gross morphology, necrosis, heart-beat, and circulation to tail. Selected larvae were photographed on a transmitted light microscope to document toxicity.
Cell Culture
HeLa cells were obtained from ATCC (Manassas, VA), and were maintained in Dulbecco’s minimum essential medium containing 10% fetal bovine serum (HyClone, Logan, UT), and 1% penicillin-streptomycin (Invitrogen) in a humidified atmosphere of 5% CO2 at 37 °C. EA.hy926 cells (ATCC CRL-2922), a hybridoma cell line that retains many properties of normal endothelial cells [27–29] were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 μg/ml Penicillin-streptomycin, and 2 mM glutamine.
Cytotoxicity of BCI analogs in cultured cells
EA.hy926 cells (10,000/well) were plated in the wells of a 384 well microplate, allowed to attach overnight, and treated with five-point, 1/3 dose reduction gradients of 1 and 19. The non-selective thiol poison phenylarsine oxide (PAO), which induces apoptosis in normal cells [38] was included as a positive control. After six hours of continuous exposure, cells were stained with 1 μg/ml propidium iodide (PI) and 10 μg/ml Hoechst 33342 to visualize necrotic cells and nuclei, respectively. Cells were imaged live on an ArrayScan II high-content reader. Numbers of nuclei per imaging field, nuclear condensation, and the percentage of PI positive cells determined by the Target Activation Bioapplication as described [38].
Antibodies and plasmids
Rabbit polyclonal phospho-ERK was from Cell Signaling Technology (Beverly, MA). Mouse monoclonal anti-c-myc (9E10) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Secondary antibodies were AlexaFluor 488-conjugated goat anti-mouse and AlexaFluor 594-conjugated goat anti-rabbit (Invitrogen). c-Myc-DUSP6/MKP3 (PYST1) was in pSG5 [39, 40]. c-Myc-DUSP1-pcDNA3.1 was subcloned from a pET15b vector encoding DUSP1/MKP1/CL100 into pcDNA3.1 (both original plasmids were gifts from Steve Keyse, CRUK, Dundee).
Chemical complementation assay for DUSP6 and DUSP1
Compounds were analyzed for inhibition of DUSP1 and DUSP6 in intact cells as described. [13] Briefly, HeLa cells were transfected in 384 well plates with human c-Myc-DUSP6 or c-Myc-DUSP1 using Fugene HD (Roche). After 48h in culture, cells were treated in quadruplicate wells for 15 min with ten two-fold concentration gradients of 1 and analogs or the nonselective DUSP inhibitor, NSC95397 (positive control), and stimulated for 15 min with phorbol ester (TPA, 500 ng/ml). Cells were immunostained with a mixture of anti-pERK (1:200 dilution) and anti-c-Myc (1:100 dilution) antibodies. Positive pERK and c-Myc-DUSP signals were visualized with AlexaFluor-594 (pERK) and Alexa-488 (c-Myc) conjugated secondary antibodies, respectively. Plates were analyzed by three-channel multiparametric analysis for pERK and c-Myc-DUSP intensities in an area defined by nuclear staining using the Target Activation Bioapplication on the ArrayScan II (Thermo Fisher Cellomics, Pittsburgh, PA). DUSP transfected cells were classified as expressors if their average c-Myc staining intensity exceeded a threshold defined as the mean intensity + 2 SD of untransfected cells. pERK levels were quantified in the DUSP-expressing subpopulation by Kolmogorov-Smirnov statistics, comparing the cumulative pERK distribution of each test well to a reference distribution from 14 DUSP-transfected and vehicle-treated wells. High KS values denote large differences in ERK phosphorylation levels compared with vehicle control and indicate suppression of DUSP activity. KS values were plotted against compound concentration and IC50 values calculated by fitting curves to a four parameter logistic equation, with the top defined by the maximum KS value obtained in the presence of the highest concentration of the positive control (1 or NSC95397).
In vitro Phosphatase Assays
3-O-Methylfluorescein Phosphate (OMFP) based ERK2 induced activation of DUSP6 assays were performed as described.[13] Recombinant His-tagged Dusp6 was expressed from a bacterial expression vector and 250 ng were incubated with 100μM 1, 7, 19 and 31. To assay activated DUSP6, 210 ng of recombinant ERK2 (Cell Signaling Technology, Danvers, MA) was added to DUSP6/compound mixtures before the addition of OMFP (100 μM). The final reaction volume was 15 μl. OMF fluorescence was measured on an M5 multimode reader (Molecular Devices) at excitation/emission wavelengths of 485/525nm at ten minute intervals for 1 hour at room temperature.
Statistical significance
In vivo and cell-based assay data were analyzed by two-tailed Student’s t-test assuming unequal variances. For in vitro phosphatase activity assays, one-way ANOVA was performed between all groups, followed by post hoc comparison of means using Tukeys multiple range test. P-values of italic>0.05 and bold>0.001 were considered statistically significant for in vitro assays.
Molecular modeling
The scientific application FRED (OpenEye, Santa Fe, NM) was used for rescoring docking poses of 1 at the putative binding site in DUSP6 (PDB: 1MKP). AM1-BCC partial charges for 1 were calculated using QUACPAC[41] and conformers were generated using OMEGA[42] with default options. Conformational changes in DUSP6 upon ERK binding (60 ns; Movie S1) and of the 1-DUSP6 complex (1.4 ns; Movie S2) were performed using NAMD[43] and CHARMM force field using water simulations.[44]
Supplementary Material
Movie S1: Dusp6 General Acid Loop Motions
The movie shows DUSP6 general acid loop (GAL) motions from an unbiased molecular dynamics simulation. Simulation was performed in the presence of water (not shown) using PDB structure 1MKP. The GAL contains the catalytic residue Asp262, which is required for formation of the “catalytic triad” (with Cys293 and Arg299). Upon formation of the catalytic triad, DUSP6 performs its dephosphorylation function at a higher catalytic rate. This conformational shift is also referred to as the catalytic activation of DUSP6. The GAL in its active form from DUSP10 structure (PDB id: 1ZZW) is shown in yellow for comparison. Upon formation of the catalytic triad, Asp262 is shown in sphere representation.
Movie S2: BCI and DUSP6 Interactions
The movie shows interactions of 1 in the allosteric pocket of DUSP6. Simulation was performed in the presence of water (not shown) using PDB structure 1MKP. The bound conformation of 1 was obtained using molecular docking. 1 buries its hydrophobic surface in the allosteric binding pocket and forms hydrogen bonds with Arg299 side-chain and Trp264 backbone oxygen. Hydrogen bond formation is indicated by blue dashed lines. 1 binding in the allosteric pocket prevents a general acid loop (GAL) movement that brings Asp262 into proximity with the catalytic cysteine (Cys293), thereby inhibiting catalytic activation of DUSP6.
Acknowledgments
We thank Nina Senutovitch and Richard DeBiasio for technical assistance with the EA.hy926 cell line and Donna Huryn for critically reading the manuscript. Grants from the National Institutes of Health supported this work [Grants R01 GM099738 and P41 GM103712 to I.B., CA78039 to A.V., HD053287 to M.T and A.V, and HL088016 to M.T.]. This project used the UPCI Chemical Biology Facility supported in part by award NIH P30CA047904. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health (NIH).
Abbreviations
- BCI
(E)-2-benzylidene-3-(cyclohexylamino)-2,3-dihydro-1H-inden-1-one
- CNT
cognition network technology
- DUSP
dual specificity phosphatase
- EC50
Half maximal effective concentration
- FGF
fibroblast growth factor
- FITC
fluorescein isothiocyanate
- GAL
General Acid Loop
- HBSS
Hank’s Balanced Salt Solution
- HCS
high-content screening
- hpf
hours post fertilization
- MAPK
mitogen-activated protein kinase or MAP kinase
- MKP3
MAP kinase phosphatase 3
- NBS
N-bromosuccinimide
- PBSA
Poisson-Boltzmann Surface Area
- PDB
Protein Data Bank
- PTP
protein tyrosine phosphatase
- SAR
structure-activity relationship
References
- 1.Farooq A, Zhou MM. Cell Signal. 2004;16(7):769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Keyse SM. Curr Opin Cell Biol. 2000;12(2):186–192. doi: 10.1016/s0955-0674(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 3.Patterson KI, Brummer T, O'Brien PM, Daly RJ. Biochem J. 2009;418(3):475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 4.Kawakami Y, Rodriguez-Leon J, Koth CM, Buscher D, Itoh T, Raya A, Ng JK, Esteban CR, Takahashi S, Henrique D, Schwarz MF, Asahara H, Izpisua Belmonte JC. Nat Cell Biol. 2003;5(6):513–519. doi: 10.1038/ncb989. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Scott DA, Hatch E, Tian X, Mansour SL. Development. 2007;134(1):167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsang M, Maegawa S, Kiang A, Habas R, Weinberg E, Dawid IB. Development. 2004;131(12):2769–2779. doi: 10.1242/dev.01157. [DOI] [PubMed] [Google Scholar]
- 7.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Cell. 2005;120(5):649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 8.Brisson M, Nguyen T, Vogt A, Yalowich J, Giorgianni A, Tobi D, Bahar I, Stephenson CR, Wipf P, Lazo JS. Mol Pharmacol. 2004;66(4):824–833. doi: 10.1124/mol.104.001784. [DOI] [PubMed] [Google Scholar]
- 9.Johnston PA, Foster CA, Shun TY, Skoko JJ, Shinde S, Wipf P, Lazo JS. Assay Drug Dev Technol. 2007;5(3):319–332. doi: 10.1089/adt.2007.066. [DOI] [PubMed] [Google Scholar]
- 10.Nemoto K, Vogt A, Oguri T, Lazo JS. Prostate. 2004;58(1):95–102. doi: 10.1002/pros.10292. [DOI] [PubMed] [Google Scholar]
- 11.Molina GA, Watkins SC, Tsang M. BMC Dev Biol. 2007;7:62. doi: 10.1186/1471-213X-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsang M. Birth Defects Res C Embryo Today. 2010;90(3):185–192. doi: 10.1002/bdrc.20183. [DOI] [PubMed] [Google Scholar]
- 13.Molina G, Vogt A, Bakan A, Dai W, Queiroz de Oliveira P, Znosko W, Smithgall TE, Bahar I, Lazo JS, Day BW, Tsang M. Nat Chem Biol. 2009;5(9):680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazo JS, Aslan DC, Southwick EC, Cooley KA, Ducruet AP, Joo B, Vogt A, Wipf P. J Med Chem. 2001;44(24):4042–4049. doi: 10.1021/jm0102046. [DOI] [PubMed] [Google Scholar]
- 15.Camps M, Nichols A, Gillieron C, Antonsson B, Muda M, Chabert C, Boschert U, Arkinstall S. Science. 1998;280(5367):1262–1265. doi: 10.1126/science.280.5367.1262. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Yu M, Lee WW, Tsang M, Krishnan E, Weyand CM, Goronzy JJ. Nat Med. 2012;18(10):1518–1524. doi: 10.1038/nm.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maillet M, Purcell NH, Sargent MA, York AJ, Bueno OF, Molkentin JD. J Biol Chem. 2008;283(45):31246–31255. doi: 10.1074/jbc.M806085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuda M, Nogare DD, Somers K, Martin K, Wang C, Chitnis AB. Development. 2013;140(11):2387–2397. doi: 10.1242/dev.091348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niwa Y, Shimojo H, Isomura A, Gonzalez A, Miyachi H, Kageyama R. Genes Dev. 2011;25(11):1115–1120. doi: 10.1101/gad.2035311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simoes FC, Peterkin T, Patient R. Development. 2011;138(15):3235–3245. doi: 10.1242/dev.059634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers JR, Planamento J, Ebrom P, Krulewitz N, Wade E, Pownall ME. Dev Biol. 2013;378(2):107–121. doi: 10.1016/j.ydbio.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Pearson B, Ayer R, Cromwell N. J Org Chem. 1962;27:3038–3044. [Google Scholar]
- 23.Maury G, Wu E, Cromwell N. J Org Chem. 1968;33(5):1900–1907. [Google Scholar]
- 24.Murray R, Cromwell N. J Org Chem. 1976;41(22):3540–3545. [Google Scholar]
- 25.Saydmohammed M, Vollmer LL, Onuoha EO, Vogt A, Tsang M. Birth Defects Res C Embryo Today. 2011;93(3):281–287. doi: 10.1002/bdrc.20216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogt A, Codore H, Day BW, Hukriede NA, Tsang M. J Vis Exp. 2010;(40) doi: 10.3791/1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgell CJ, McDonald CC, Graham JB. Proc Natl Acad Sci U S A. 1983;80(12):3734–3737. doi: 10.1073/pnas.80.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgell CJ, Haizlip JE, Bagnell CR, Packenham JP, Harrison P, Wilbourn B, Madden VJ. In Vitro Cell Dev Biol. 1990;26(12):1167–1172. doi: 10.1007/BF02623694. [DOI] [PubMed] [Google Scholar]
- 29.Bauer J, Margolis M, Schreiner C, Edgell CJ, Azizkhan J, Lazarowski E, Juliano RL. J Cell Physiol. 1992;153(3):437–449. doi: 10.1002/jcp.1041530302. [DOI] [PubMed] [Google Scholar]
- 30.Vogt A, Lazo JS. Pharmacol Ther. 2005;107(2):212–221. doi: 10.1016/j.pharmthera.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Rigas JD, Hoff RH, Rice AE, Hengge AC, Denu JM. Biochemistry. 2001;40(14):4398–4406. doi: 10.1021/bi002951v. [DOI] [PubMed] [Google Scholar]
- 32.Grant J, Pickup B, Nicholls A. J Comput Chem. 2001;22:608–640. [Google Scholar]
- 33.Kuhn B, Gerber P, Schulz-Gasch T, Stahl M. Journal of medicinal chemistry. 2005;48(12):4040–4048. doi: 10.1021/jm049081q. [DOI] [PubMed] [Google Scholar]
- 34.Keyse SM. Cancer Metastasis Rev. 2008;27(2):253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Shepherd EG, Nelin LD. Nat Rev Immunol. 2007;7(3):202–212. doi: 10.1038/nri2035. [DOI] [PubMed] [Google Scholar]
- 36.Vogt A, Cooley KA, Brisson M, Tarpley MG, Wipf P, Lazo JS. Chem Biol. 2003;10(8):733–742. doi: 10.1016/s1074-5521(03)00170-4. [DOI] [PubMed] [Google Scholar]
- 37.Vogt A, McDonald PR, Tamewitz A, Sikorski RP, Wipf P, Skoko JJ, 3rd, Lazo JS. Mol Cancer Ther. 2008;7(2):330–340. doi: 10.1158/1535-7163.MCT-07-2165. [DOI] [PubMed] [Google Scholar]
- 38.Vogt A, McPherson PA, Shen X, Balachandran R, Zhu G, Raccor BS, Nelson SG, Tsang M, Day BW. Chem Biol Drug Des. 2009;74(4):358–368. doi: 10.1111/j.1747-0285.2009.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dowd S, Sneddon AA, Keyse SM. J Cell Sci. 1998;111:3389–3399. doi: 10.1242/jcs.111.22.3389. [DOI] [PubMed] [Google Scholar]
- 40.Groom LA, Sneddon AA, Alessi DR, Dowd S, Keyse SM. Embo J. 1996;15(14):3621–3632. [PMC free article] [PubMed] [Google Scholar]
- 41.Jakalian A, Jack DB, Bayly CI. Journal of computational chemistry. 2002;23(16):1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 42.Bostrom J, Greenwood JR, Gottfries J. J Mol Graph Model. 2003;21(5):449–462. doi: 10.1016/s1093-3263(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 43.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD., Jr Journal of computational chemistry. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1: Dusp6 General Acid Loop Motions
The movie shows DUSP6 general acid loop (GAL) motions from an unbiased molecular dynamics simulation. Simulation was performed in the presence of water (not shown) using PDB structure 1MKP. The GAL contains the catalytic residue Asp262, which is required for formation of the “catalytic triad” (with Cys293 and Arg299). Upon formation of the catalytic triad, DUSP6 performs its dephosphorylation function at a higher catalytic rate. This conformational shift is also referred to as the catalytic activation of DUSP6. The GAL in its active form from DUSP10 structure (PDB id: 1ZZW) is shown in yellow for comparison. Upon formation of the catalytic triad, Asp262 is shown in sphere representation.
Movie S2: BCI and DUSP6 Interactions
The movie shows interactions of 1 in the allosteric pocket of DUSP6. Simulation was performed in the presence of water (not shown) using PDB structure 1MKP. The bound conformation of 1 was obtained using molecular docking. 1 buries its hydrophobic surface in the allosteric binding pocket and forms hydrogen bonds with Arg299 side-chain and Trp264 backbone oxygen. Hydrogen bond formation is indicated by blue dashed lines. 1 binding in the allosteric pocket prevents a general acid loop (GAL) movement that brings Asp262 into proximity with the catalytic cysteine (Cys293), thereby inhibiting catalytic activation of DUSP6.