Abstract
Constitutive expression of interferons (IFNs) and activation of their signaling pathways have pivotal roles in host responses to malignant cells in the tumor microenvironment. IFNs are induced from the innate immune system and in tumors through stimulation of Toll-like receptors (TLRs) and through other signaling pathways in response to specific cytokines. Although in the oncologic context IFNs have been thought of more as exogenous pharmaceuticals, the autocrine and paracrine actions of endogenous IFNs probably have even more critical effects on neoplastic disease outcomes. Through high-affinity cell surface receptors, IFNs modulate transcriptional signaling, leading to regulation of over 2000 genes with varying patterns of temporal expression. Induction of the gene products by both unphosphorylated and phosphorylated STAT1 after ligand binding, results in alterations in tumor cell survival, inhibition of angiogenesis, and augmentation of actions of T, natural killer (NK), and dendritic cells. The interferon-stimulated gene (ISG) signature can be a favorable biomarker of immune response but, in a seemingly paradoxical finding, a specific subset of the full ISG signature indicates an unfavorable response to DNA damaging interventions such as radiation. IFNs in the tumor microenvironment thus can alter the emergence, progression, and regression of malignancies.
Although in an oncologic context IFNs have been often thought of more as exogenous pharmaceuticals, the autocrine and paracrine actions of endogenous IFNs probably have even more critical effects in contributing to tumor outcomes in patients. Constitutive expression of interferons (IFNs) and activation of their signaling pathways have pivotal roles in host responses to malignant cells in the tumor microenvironment. Induction of IFNs in immune effector cells, together with sustained effects of STAT1, can result in direct alterations in tumor cell survival, inhibition of angiogenesis, and augmentation of actions of T, NK cells and dendritic cells. These effects derive from immune cell recognition of tumors, endothelial cell proliferation, and response of tumors to exogenous DNA damage. With receptors present on almost every cell type, IFNs through their cellular actions can alter the emergence, progression, and regression of malignancies (Table 1). The interferon-stimulated gene (ISG) signature can be a favorable biomarker of immune response but, in a seemingly paradoxical finding, a specific subset of the ISG signature indicates an unfavorable response to DNA damaging interventions such as radiation.
Table 1.
Receptors and Signaling Molecules in IFN Pathways
| Receptors responding to pathogenic molecules |
| - Toll-like receptors (TLRs) |
| TLR3: dsRNA (poly I:C) |
| TLR4: LPS (paclitaxel) |
| TLR7/8: ssRNA (imiquimod) |
| TLR9: CpG DNA |
| - RIG-I, MDA5: dsRNA |
| - cGAS: cytoplasmic ss and dsDNA |
| Signaling molecules involved in IFN production |
| - TBK1, IKK |
| - IRF1, IRF2, IRF3, IRF5, IRF7, IRF8 |
| - NF-κB, AP-1 |
| - STING |
| Receptors responding to IFNs |
| - IFNAR1/ IFNAR2: type I IFNs |
| - IFNGR1/ IFNGR2: type II IFN |
| - IFNLR1/ IL-10R2: type III IFNs |
| Signaling molecules responding to IFNs |
| - JAK1, JAK2, TYK2 |
| - STAT1, STAT2 (PY- and U-STATs) |
| - IRF9 |
| - PI3K, MAPKs |
IFNs, a family of secreted α-helical cytokines, are induced by the innate immune system through stimulation of Toll-like receptors (TLRs) and other signaling pathways in response to specific extracellular biomolecules (pathogen- or damage-associated molecular patterns, PAMPs or DAMPs). Through high-affinity cell surface receptors, IFNs activate kinase-driven signaling, leading to the induction of over 2000 transcriptionally regulated ISGs with varying patterns of temporal expression after ligand binding. Although most genes (>1500) are stimulated, some are suppressed (~300).1–7 These ISGs, stimulated by exogenous IFNs at the RNA level up to 100 fold include structural proteins, transcription factors, adaptors, enzymes, and secreted proteins.5
Expression arrays and cytogenetic analyses have identified somatic, homozygous deletions of the chromosomal locus for IFNs-α and IFN-β and germline mutations of ISGs in colon, lung, prostate, breast, head and neck, and pancreatic carcinomas, melanoma, and hematologic malignancies.8–17 Epigenetic and genetic silencing of signaling pathways stimulated by IFNs is also likely to influence tumor development.18–21 Although we will draw on insights from studies of actions of exogenously added IFNs, our focus is to illustrate how endogenous host IFNs can potently influence early regression or later either stability or progression of the neoplastic process. Since tenets regarding their protein structure, receptors, and intracytoplasmic signaling have been the basis for new insights concerning endogenous IFNs and their activation, we will begin with a short overview of canonical findings and understandings.
GENES, RECEPTORS, PROTEINS, AND CANONICAL SIGNALING
Classification of the several types and families of IFNs comes from commonality in both primary structures and their influence on three dimeric target receptors. Based on similarities and differences, there are three major classes of IFNs.22–25 Type I IFNs include the IFN-α family with its many isoforms, IFN-β, and other IFNs of less studied significance in humans, IFN-ω, IFN-τ, IFN-κ, and IFN-ε.23, 26 The sole type II IFN is IFN-γ.27 A more recently discovered family, type III IFNs or IFNs-λ (IL-28/29) and its isoforms are produced by mucosal epithelial cells.24 Type III IFNs share structural homology and induction pathways with type I IFNs but with cell lineage distribution of its unique receptor restricted to mucosal epithelial cells and plasmacytoid dendritic cells (pDC).24, 28
The genes for the human type I IFNs including those encoding 14 subspecies of IFN-α, are clustered at 9p21.22, 26 As proteins, the human IFN-α subspecies have about 50% sequence identity; IFN-β is about 20% identical to IFN-α2. IFNs-α and IFN-β have 186–190 amino acids and have a cleavable signal peptide resulting in secreted proteins of 165 or 166 amino acids. Structure–function analyses have shown that the NH2 terminus of type I IFNs is important for biological activity.26 The gene encoding IFN-γ, located on human chromosome 12, has three introns, encodes a protein of 146 amino acids, functions as a dimer, and has minimal homology with type I IFNs.27 NK and T cells are major sources of IFN-γ. Type I IFNs are produced predominantly by dendritic cells but can be induced in all cell types including T cells, monocytes, fibroblasts, and epithelial cells.
Virus or microbial gene products, such as dsRNA, ssRNA, dsDNA, ssDNA, or cell wall constituents (PAMPs) bind to specific membrane proteins (TLRs) to trigger type I IFN synthesis.29–34 dsRNA can be recognized by TLR3 and also by two cytosolic RNA-helicases, RIG-I and MDA5, both of which are ISGs.33–35 Viral single-stranded RNAs are recognized by TLR7 and TLR8 and viral and cellular DNAs (DAMPs) by TLR9 and STING, all present in endosomal membranes.29, 30, 32, 33 Adaptor proteins connect TLRs to specific protein kinases such as TBK1 and IKK to activate transcription factors NFκB, IRF3, IRF7, and AP-1.32, 33
For IFN-β induction, NFκB, the AP-1 complex composed of ATF2/c-jun and IRF1, and IRF3 or IRF7 are required.35, 36 IFN-β induces further IRF7 synthesis, which, in turn, induces transcription of IFN-α1 and other IFN-α genes.31 Inhibition of IFN signaling blocks robust production of IFNs-α. Synthesis of various IFNs is, therefore, intimately linked and further influenced by interferon regulatory factors (IRFs), a family of nine transcription factors that have common DNA binding domains in their N-terminus. IRFs were first identified through the role of IRF1 in inducing IFN-β.37–39 IRF1 is expressed constitutively and also in response to IFN-γ as is IRF8.40 Although IRF3 and IRF7 are important in inducing Type I IFNs, IRF1 and possibly IRF5 may determine which species of IFNs are induced by TLR activation. IRF7 amplifies the phosphorylated activation of constitutive IRF3 leading to additional synthesis of IFNs and probably also induction of specific ISGs such as CXCL10.37–39
IFNs themselves, once secreted, bind to glycosylated, species-specific, heterodimeric cell surface transmembrane proteins that trigger signaling through their cytoplasmic domains.22, 25, 27, 41–43 To elicit responses, IFNs-α and IFN-β bind to two receptor subunits, IFNAR-1 and IFNAR-2. IFN-β interacts with the receptor heterodimer differently than does IFN-α2; specific anchor points for IFN-β induce a different conformational change in IFNAR1 with greater induction of a subset of ISGs.43, 44 IFNAR1 is at least in part degraded by an E3 ubiquitin ligase, which when decreased in activity increases IFNAR1 and suppresses tumorigenicity of a human melanoma xenograft.45 IFN-γ binds to a different heterodimeric receptor consisting of two subunits, IFNGR-1 and IFNGR-2.27 The critical event in signaling by IFNs is this ligand-initiated dimerization of the subunits of the receptors that results in cascades of downstream phosphorylation. The activated cytoplasmic domains of receptors for IFNs signal by binding to JAKs (Janus-activated kinases) and STATs (signal transducers and activators of transcription), which in turn phosphorylate STAT1 and STAT2.25, 46–48
After phosphorylation by JAKs, STATs form active homomeric or heteromeric dimers that bind to cis-acting sequences in the promoters of ISGs. For full activation of STATs, further phosphorylation at specific serine residues may be required. ISGF3, the canonical transcription factor activated by type I IFNs, is composed of STAT1, STAT2 and IRF9. It binds to ISREs (IFN-Stimulated Response Elements) in ISG promoters.49 In the IFN-γ signaling pathway, the two receptor subunits, JAK1, and JAK2, activate Gamma-Activated Factor (GAF), a homodimer of tyrosine-phosphorylated STAT1, which binds to the Gamma-Activated Sites (GAS) in ISG promoters.28 In addition to the canonical STAT pathways, IFNs can additionally activate transcription through other pathways.23, 47, 50 For example, type I IFNs can also trigger GAF formation and gene induction by through GAS elements. Furthermore, STAT1 can form heterodimers with other STATs.50 Other kinases, such as PI3K and p38 MAPK, can be activated by type I IFNs and can influence specific patterns of induction of ISGs.47, 50, 51 Protein tyrosine phosphatases (PTP) have a regulatory role in suppressing signaling by IFNs. Inhibition of PTPs can prolong signaling and potentiate antitumor activities of IFNs.52–54 Inhibition of the PTP SHP-1 can be accomplished by inhibitors with potentiation of effects of IFNs.55–57 NK cells in which SHP-2 has been silenced have elevated cytolytic activity with increased IFN-γ consistent with immune activation.56 Also vitally important in negatively regulating activity of IFNs in immune and other cell types are the ISG SOCS proteins, especially SOCS1.58–60
CONSTITUTIVE ENDOGENOUS IFNs
Constitutive production of type I and II IFNs was suggested to have potentially important roles in the absence of viral infection over 30 years ago.61–63 In contrast to “emergency” production of IFNs induced by virus infection, “physiological” low levels of IFNs may be produced continuously and remain localized with no systemic release or effects.61, 63–65 Transcriptional regulation of the constitutive expression of IFNs has not been studied as extensively as that of virus-induced IFN expression but in part may result from different induction mechanisms.66 Constitutive expression of IFN-β may be maintained in part by IRF7 but also of importance may be non-canonical binding by c-Jun and RelA of the IFN-β gene promoter.66–68 The expression of IFNs-α and IFN-β may additionally be regulated by repressors, IRF2 and a homodimer of the p50 NF-κB subunit.69–72 Constitutive low levels of IFNs achieve various physiological functions not only through downstream events but also by maintaining signaling of other cytokines. A small amount of IFNs-α and IFN-β has proven to be important for efficient response to cytokines including IFN-γ and IL-6 through signaling crosstalk.73–76 In addition to signaling crosstalk, adequate expression of signaling components, such as IRF7 and STAT1 and STAT2, can be constitutively maintained by low levels of type I and II IFNs.66, 77, 78 Constitutive expression of endogenous IFNs may thus maintain physiological functions, such as homeostasis of immune cell activities, hematopoietic stem cell niches, and bone remodeling.63, 66, 68, 79–81
ANTICANCER EFFECTS OF ENDOGENOUS IFNS
Endogenous IFN-γ is the basis of a tumor surveillance system for both chemically induced and spontaneously arising tumors in mice. IFN-γ receptor (ifngr1)-deficient mice or stat1-deficient mice developed tumors more rapidly compared to wild-type mice.82 Endogenous type I IFNs were also required to inhibit the growth of primary carcinogen-induced and transplantable tumors.83 The responsiveness to endogenous IFNs was critical in preventing tumorigenesis through tumor surveillance; loss of sensitivity to endogenous IFNs can contributed to escape of tumor cells from host immune surveillance.
Among aberrant IFN responses in cancer have been decreased responses in some human lymphoblastoid cells.84 Furthermore, about 30% of melanoma and lung carcinoma cell lines have inactivated IFN-γ pathway components, including inactive JAK2 and the lack of JAK1 or IFN-γR.82 Lack of STAT1, STAT2, and IRF9 was observed in melanoma cell lines and primary melanocytes; expression of STAT1 was also missing in some chronic myeloid leukemia (CML) cells obtained from patient tissues.85–87 Thus, loss of responsiveness to IFNs may be a critical component of the host response to tumors.
Additionally suggesting a critical role of induction and signaling by IFNs in cancer pathogenesis has been the identification of genotypic variants that affect tumor initiation and progression. Gene expression arrays and cytogenetic analyses have identified somatic homozygous deletions in the 9p21 chromosomal locus for IFNs; furthermore mutations of ISGs have been found in melanoma, colon, lung, and hematologic malignancies.8, 11–14, 17 IFN-β-deficient or IFNAR1− deficient mouse embryo fibroblasts underwent spontaneous transformation in vitro.88 Oncogenes such as RAS or the human papillomavirus protein E6E7 have downregulated TLR signaling.88, 89 Other viral oncoproteins can block signaling by interfering with functions of ISGs or ISGF3.90–92 Alterations in TLR signaling have been suggested to be important in oncogenesis because of gain of function mutations in the major adaptor proteins, MYD88, in a subset of aggressive diffuse B cell lymphomas.93 In chronic lymphocytic leukemia, heterozygous mutations in the DNA binding domain of IRF4, both constitutively expressed and induced through TLR ligation, have been identified.94 Other genetic alterations of IRF4 have been identified in a subset of B cell lymphomas (a translocation) and cutaneous carcinomas (a germ line SNP).95, 96 Overexpression of IRF5 in breast carcinoma cells inhibited in vitro and in vivo cell growth and furthermore IRF5 was downregulated in human breast carcinomas.97
A controlled study of over 4000 individuals has correlated SNPs in IRFs, IFN-γ, and IFN-γR2 with colorectal carcinoma risk and progression.17 Further confirming the potential importance of IFN signaling pathways in the development of colorectal carcinoma has been correlation of SNPs in JAKs and STAT1 with increased risk.98 Overexpression of IRF8 in transformed cells with the CML Bcr-Abl translocation inhibited leukemogenesis overcame resistance to the tyrosine kinase inhibitor, imatinib, and was correlated with reduced BCL-2 expression.99 IRF4 inhibited granulocytic differentiation; its loss enhanced progression of a CML-like disease in IRF8−/− mice.100 An additional role for the IFN system in CML was suggested by identification of a genomic SNP in the promoter for IFN-γ that resulted in a better response to imatinib.101 Germ line mutations in the ISG, RNASEL, have been correlated with increased risk for prostate, breast, head and neck, and pancreatic carcinomas.9, 16 Indeed GWAS analyses have identified at STAT1 and IRF1 as among the transcription factors overall most commonly containing SNPs in cancer.102 Thus, aberrant and augmented IFN signaling can result in significant modulation of early events of tumor pathogenesis.
INCREASED ENDOGENOUS IFNS IN PATHOLOGICAL CONDITIONS
Although endogenous IFNs may be important for cellular homeostasis and resistance to cancer growth, production of endogenous IFNs can also contribute to pathology. Aberrantly regulated constitutive production of IFNs can also influence development of autoimmune diseases. In patients with autoimmune diseases, including type I diabetes mellitus (DM), systemic lupus erythematosus (SLE), or Sjögren’s syndrome, type I IFN expression or an ISG signature has been identified.66 The IFN-α pathway wss activated in a subgroup of SLE patients with increased disease severity and more double stranded DNA (dsDNA).103 Accelerated IFNs-α and IFN-β responses has led to a psoriasis-like skin inflammation.70, 104 SLE symptoms were ameliorated in IFNAR-deficient mice.105 A monoclonal neutralizing antibody against IFN-α has thus been in clinical trials to treat SLE.106
Increased expression of ISGs has been identified in cancer cells compared to corresponding normal primary cells or normal tissues with correlations to the degree of tumor invasion.107–110 Expression of some IFN-induced genes was higher in metastatic cancer cells than non-metastatic cells.111, 112 Depletion of IRF2, a repressor of IFN-α/β expression, significantly increased tumor growth in a xenograft model with increased proliferation and decreased apoptotic cell death113. Recurrent alteration in IRF2 and its inactivation has been identified in HBV-related hepatocellular carcinoma113.
CAUSES OF INCREASED ENDOGENOUS IFN PRODUCTION
Oncoviruses
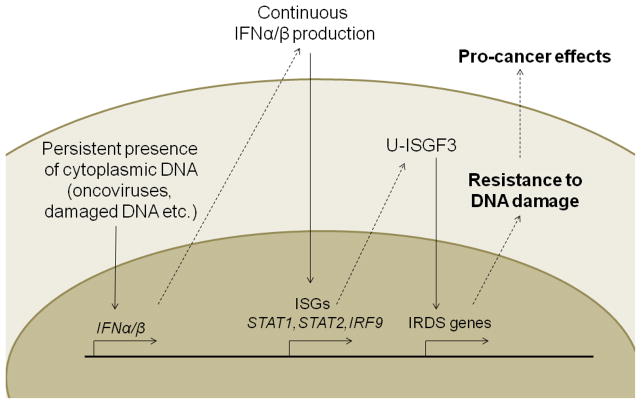
About 18% of human cancers can be caused by infection with about 12% being caused by one of seven different pathogens including Helicobacter pylori (approximately 5% of all cancers), human papilloma viruses (HPV, 5%), hepatitis B and C viruses (HBV and HCV, 5%), Epstein-Barr virus (EBV, 1%), human immunodeficiency virus (HIV, 1%), and human herpes virus 8 (HHV, 1%).114 Tumor development in carriers with latent or persistent infections often occurs decades after initial exposure to the virus.115 Although RNA viruses are potent inducers of IFN production, DNA viruses are known to be relatively poor.116 Therefore, consistent exposure to a low level of IFNs-α and IFN-β, induced by persistent infection with DNA- or retro-viruses, might contribute to the increase of resistance to DNA damage through the expression of IFN-related DNA damage resistant gene products over a long period117 (Figure 1).
Figure 1. Causes and consequences of increased endogenous IFN production.
Persistent infection with oncoviruses (DNA- or retroviruses) or exposure to DNA damaging agents (UV light, DNA damaging chemicals, etc.) leads to cytoplasmic DNA, which induces IFN-α/β production. Chronic exposure of cells to type I IFN increases the expression of ISGs, including STAT1, STAT2, and IRF9. U-ISGF3, formed by increased levels of unphosphorylated STAT1 and STAT2, and IRF9. U-ISGF3 selectively induces a subset of ISGs, the IRDS genes, leading to resistance to DNA damage. U-ISGF3; unphosphorylated IFN stimulated gene factor 3, IRDS; IFN-related DNA damage resistance signature
Cytosolic dsDNA from DNA viruses can also induce IFNs-α and/or IFN-β in a TLR-independent fashion.30, 118 Host cytosolic DNA as well as virus DNA has also induced expression of type I IFNs, which may be linked to autoimmune diseases in humans. Failure to clear self-DNA leads to an inappropriate activation of type I IFN production through TLR-independent signaling.119, 120 Both self and non-self cytosolic DNAs may trigger production of type I IFNs through STING (stimulator of interferon genes)-dependent cytosolic DNA sensing pathways.30 Cytoplasmic DNA triggers this signaling pathway by binding to a the DNA-sensing protein, cyclic GMP-AMP synthase (cGAS).121 The DNA-bound activated cGAS synthesizes cyclic guanosine monophosphate-adenosine monophosphate (cGMP-AMP or cGAMP), a second messenger that activates an adaptor protein and traffics rapidly to peri-nuclear endosomes with tank binding kinase 1 (TBK1), through the autophagosome in response to cytoplasmic DNA.122 TBK1 activates IRF3 and IRF7, which translocate into the nucleus to stimulate the expression of type I IFNs. In addition to cGAS, many other proteins, including meiotic recombination 11 homolog A (MRE11), DEAD-box helicase family 41 (DDX41), and IFI16, also recognize cytoplasmic DNA and facilitate STING activity30.
DNA damaging agents
In addition to virus infection, DNA damaging agents including ultraviolet (UV) light and constituents of cigarette smoke can be causative factors in cancer. UV irradiation initiated melanoma genesis with a persistent ISG response signature.123 Damaged DNA leaking into the cytoplasm may also trigger STING-dependent signaling with induction of type I IFNs. MRE11-RAD50, a DNA damage-sensing complex, physically interacted with damaged DNA in the cytoplasm, activated STING and IRF3, and induced the production of type I IFNs.124 Aberrant self DNA was normally eliminated by cellular DNAses such as DNAse II and DNAse III (3′ repair exonuclease I, TREX1). Defects in these DNAses caused inefficient removal of cytosolic DNA, leading to increased type I IFNs.125 When dsDNA was oxidized by reactive oxygen species (ROS) or superoxide induced by UV irradiation or activated neutrophils, it became more resistant to DNAse III (TREX1), increasing the expression of type I IFNs.126 Thus, through the STING and/or other nucleotide recognition proteins, damaged self-DNA fragments may be causative in constitutive production of IFNs.
Loss of p53 function
The tumor suppressor p53, frequently mutated in tumors, functions as transcription factor to promote or repress transcription of many protein-encoding genes and noncoding RNAs.127 Wild-type p53 is inactivated in tumors by various means including overexpression of the p53 inhibitor MDM2.128 Loss of p53 function has an important role in cancer progression and resistance to therapy. DNA demethylation in p53-null cells resulted in a lethal induction of IFN-β and ISGs through the IFNAR type I receptor.20 These strong responses were triggered by dsRNAs expressed from multiple noncoding genes, including major classes of short, interspersed nuclear elements (SINEs) B1/B2. The transcription of noncoding RNA, normally repressed by the combination of p53 and DNA methylation, was increased >100 fold in the absence of both p53 and DNA methylation. Resulting high levels of IFN-β and ISGs led to caspase activation and apoptosis in p53 null mouse embryo fibroblasts. Cells that escaped the suicidal IFN responses survived to develop into tumors. 20
Loss of responsiveness to IFNs or the inability of cells to produce IFNs in cells with aberrant p53 function may be an important major mechanism of tumor progression. Loss of p53 function has been correlated with resistance to DNA damaging agents independent of other risk factors.129 However, while p53 deficiency increased apoptosis in the presence of DNA demethylating agent, decitabine, it decreased apoptosis induced by a DNA damaging agent, doxorubicin.20 Whether loss of p53 increases in resistance to DNA damage through induction of an IFN-response induced by low levels of IFNs has not been examined. However, higher levels of STAT1 proteins have been identified in cells expressing mutant p53 or no p53.117, 130 Increased levels of STAT1 protein had pro-tumor and metastatic effects,112, 131 and also are able to induce high levels of the IRDS gene products,132, 133 suggesting that p53 deficiency induces DNA damage resistance, at least partially, through the induction of the IRDS genes.
PROCANCER EFFECTS OF INCREASED ENDOGENOUS IFNS
Although the production of endogenous IFNs is increased under pathological conditions, the amount is still much lower compared to therapeutically applied IFNs. The pattern of ISG expression correlated to the concentration of IFNs present in the cancer microenvironment. Constitutive low levels of IFNs were able to induce steady-state upregulation of ISGs in cancer cells, including IFI27, ISG15, and BST2, with no increase of pro-apoptotic and anti-proliferative ISGs, which were induced by the higher doses of IFNs used therapeutically.133, 134
Some studies have identified expression of ISGs in tumors from patients with various different types of cancers resistant to chemotherapy and radiation therapy.135–137 Interestingly, only a minor subset of ISGs (about 30 out of >100 total ISGs) were consistently overexpressed in therapy-resistant cancer cells. This subgroup of ISGs included IFI27 (ISG12), ISG15 (G1P2), BST2, OAS1, OAS3, OASL, and others. These genes, comprising an IFN-related DNA damage-resistant signature (IRDS), were also upregulated in cancers compared to normal tissues.107–110, 137 High expression of IRDS genes promoted tumor growth and metastasis as well as resistance to chemotherapy and radiation.112, 138–140
Selective induction of IRDS genes can be mediated by unphosphorylated STAT1 (U-STAT1).132, 133 STAT1 overexpression in mice conferred protection from radiation and mediated amplification of metastatic potential.112, 131 U-STAT1, together with U-STAT2 and IRF9, formed un-phosphorylated IFN-stimulated gene factor 3 (U-ISGF3).117 High levels of the component proteins, STAT1, STAT2, and IRF9 phosphorylated and unphosphorylated, were induced in response to IFNs (Figure 1). Chronic exposure to low levels of IFN-β and increased U-ISGF3 levels thus may lead to enhanced expression of IRDS genes with no increase of other ISGs that are anti-proliferative and/or pro-apoptotic with resulting resistance to DNA damage.117
The unresolved puzzles in understanding actions of IFNs in tumor cell survival and apoptosis can be, however, exemplified by two ISGs, ISG12 (IFI27) and ISG 6–16 (G1P3), commonly induced in IFN-responsive cells.141–143 ISG 6–16 gene encodes a low-molecular-weight mitochondrial protein that stabilized mitochondrial function and opposed apoptosis. ISG 6–16 antagonized TRAIL-mediated apoptosis and cytotoxicity in both cell lines and fresh myeloma cells through antagonism of TRAIL-mediated mitochondrial potential loss and cytochrome c release. In contrast, ISG12 expression has sensitized cells to apoptotic stimuli through mitochondrial membrane destabilization.
THERAPEUTIC APPLICATION OF IFNS
While it has been known for more than 40 years that IFNs could influence outcomes of both transplantable and viral-induced experimental murine tumors,23 a precursor to studies considering a mechanistic role of IFNs and STATs in the microenvironment have studies evaluating whether IFNs could influence an induced, non-viral (carcinogen) malignancy. The first such study used an unpurified mouse IFN preparation and identified inhibition of development of subcutaneous fibrosarcomas after 3-methylcholanthrene.144 A subsequent study with purified IFN-β after the bladder-specific carcinogen N-[4-(5-nitro-2-furyl)-2-thiazolyl]formamide (FANFT) introduced into the diet identified complete inhibition of transitional carcinoma development by IFN-β.145 Similar effectiveness occurred with an oral TLR inducer of IFNs. 145
Subsequent studies have further confirmed the role of IFNs-α, IFN-β, and IFN-γ in inhibiting carcinogen-induced malignancy. IFN-γ inhibited hepatic carcinogenesis in mice in response to diethylnitrosamine with an increase in infiltration of CD8 T cells and NK cells but additionally an accumulation of p53 in hepatocytes sensitizing them to apoptosis.146 In rats treated with IFN-α for carcinogenesis induced by diethylnitrosamine both preneoplastic foci and tumors in the liver were reduced, an effect that was increased with more sustained administration of IFN-α.147 Furthermore, IFN-α seemed to reduce frequency of hepatocellular carcinomas in hepatitis B virus-infected patients with cirrhosis after surgical resection.148
Activation of anti-tumor immunity
Induced IFNs and ISG proteins have potent influences on lytic activity and ISG products from immune effector cells, on induction of angiogenesis, and on tumor cell invasiveness. Unraveling mechanisms of anti-tumor effects of IFNs and aberrations in their signaling pathways has been a complex and a still evolving, uncompleted research challenge. Further complexities but also increased clarity has begun to emerge in elucidating cellular effects of IFNs and the protein products of ISGs on host cellular mechanisms, particularly new understandings of influences on immune effector cells (Figure 2), of resistance to malignant development and progression.
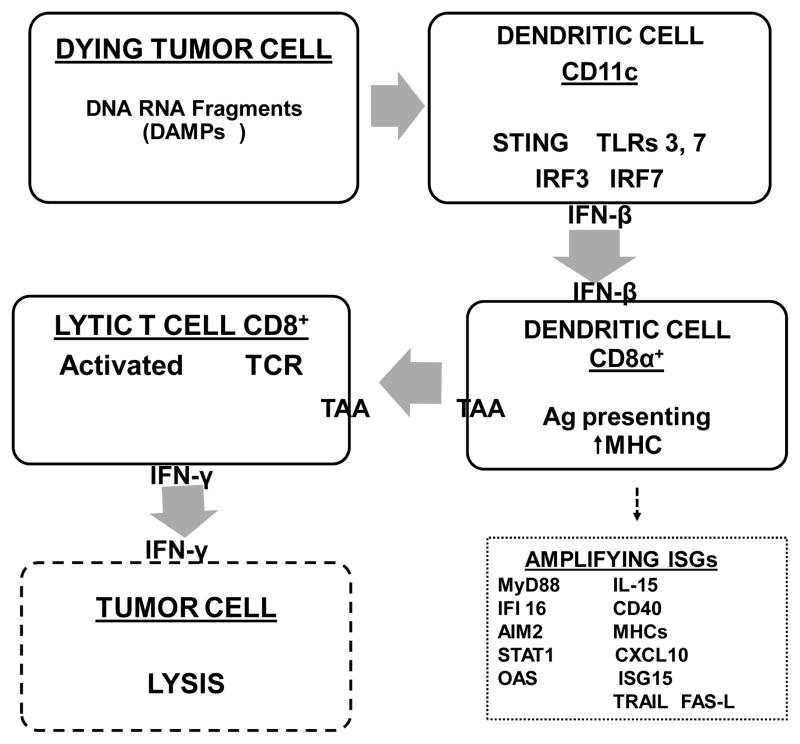
Figure 2. Assumed cascade of events in T cell activation resulting from triggering of IFN synthesis and secretion by release of nucleotide fragments into the microenvironment from dying tumor cells.
Consistent with existing evidence detailed in the text (although not at all steps are yet completely confirmed), fragments of RNA and DNA are released into the microenvironment and processed through endosomal pathways of CD11c+ plasmacytoid dendritic cells for secretion of IFN-β. Subsequent steps are activation of CD8a+ dendritic cells, with presentation through MHC pathways of tumor-associated antigens (TAA) through the CD8+ T cell receptor (TCR), and secretion of IFN-γ. 2,3 Various steps in the complete cascade, probably similar to but not necessarily identical to those resulting in constitutive IFN secretion and autoimmune diseases, are amplified by protein products of ISGs, A few examples are given.
Amplification of innate and specific immune responses has resulted from both Type I and Type II IFNs with influences on antigen recognition and processing and cytolytic activity.149–154 Plasmacytoid dendritic cells have been found to secrete high levels of IFNs-α and IFN-β in response have been identified in the T cell zone of lymphatic tissue in tumors of various types.150, 155–157 Activated NK and T cells have a critical role in production and action of IFNs for protection from oncogenesis and in controlling growth of syngeneic and transplanted tumors. Maturation of dendritic cells, themselves a primary source in the innate immune system for production of IFNs-α and IFN-β, was also influenced by IFNs.155–157
IFNs have stimulated activity of cytotoxic and helper T cells, natural killer (NK) cells, macrophages, and dendritic cells. One of the first immune effectors identified as augmented in activity by IFNs were NK cells whose cytotoxic activity in both mouse and man could be augmented. Activation of NK cells and monocytes was identified both in vitro and in vivo.158–164 Effects of IFNs on NK cells can be further amplified by production of the ISG IL-15 by dendritic cells resulting in their initial priming for cytotoxicity.165
Molecular and cellular effects of IFNs and ISG proteins provide a basis for understanding effects on innate and acquired host immune responses in the tumor microenvironment. Equivalent antitumor effectiveness of IFNs in vivo for syngeneic tumor cells occurred regardless of whether tumor cells were sensitive or resistant to antiproliferative effects of IFNs in vitro.166, 167 Subsequent studies, using genetically mutant mice, have more rigorously suggested the critical importance of immune cell regulation in antitumor effects of IFNs. STAT1 knockout mice implanted with IFN-sensitive tumors and treated with exogenous IFN-α did not survive longer than control mice.168 Conversely, wild-type (STAT1+/+) mice implanted with STAT1-null tumor cells were able to mount an effective antitumor response following IFNs, suggesting the importance of host cells.169 Extension of these studies to either IFN-γR or STAT1 deficient mice identified development of both spontaneous tumors of diverse histologies and methylcholanthrene-induced fibrosarcomas at significantly greater frequency than in wild-type littermates, emphasizing further the critical role of endogenous IFN in host response to primary tumor.82, 170 In addition to IFN-γ, endogenous type I IFNs were required to prevent carcinogen-induced and transplantable tumors.83 In contrast to IFN-γ for which tumor cells were important targets, host hematopoietic cells (NK cells, dendritic cells) were critical targets for the protective antitumor responses leading to tumor elimination by IFNs-α and IFN-β.83, 171
This solid initial groundwork in understanding on immune effectors cells has been now followed by additional by excellent insights into T cell and dendritic cell interactions as modified by IFNs.2, 3, 150–153 These findings in the last few years emphasize importance of the CD8a+ DC subset as critical for CD8+ T cell priming and tumor rejection.; 2, 3, 172 Histologic identification of a tumor-infiltrating lymphocyte that has been correlated with a favorable clinical outcome in diverse malignancies including melanoma has identified activated CD8+ T cells that had with a ISG expression signature.153 Effects of IFNs on innate immune responses can then be subsequently translated into longer lived memory of not only response of T but also of B cells.173, 174
Following binding of IFNs to immune effector cells, upregulated ISGs include major histocompatibility (MHC) components leading to eventual activation of CD8+ cytotoxic T cells.93, 175, 176 IFNs increased transcription of MHC class I genes and induced expression of additional proteins required for surface expression of the mature MHC class I complex. MHC Class I proteins associate with β2-microglobulin, also an ISG, in the endoplasmic reticulum for transport to the cell surface; augmentation of β2-microglobulin has been identified in patients after IFN-α2 or IFN-β.177, 178 Processing of potential antigenic peptides for loading onto class I molecules occurs in the proteasome whose three subunits (LMP-2, LMP-7, and LMP-10) together with Transporter for Processing (TAP) are all ISGs that can also be induced by IRF7.179–182 Findings of increases in MHC Class II and other ISGs in vitro led to assessment of changes in monocyte cell-surface markers using treatment of patients with IFN-β with significantly increased monocytes positive for HLA-DQ.183, 184 Additionally, levels of OASs were significantly increased in patient peripheral blood mononuclear cells.184 These translational results of increases in ISGs in cells from treated patients were founded and initiated based upon the findings a few years prior of increases in these stimulated RNAs in IFN-treated cells.183 Additional translational studies subsequently confirmed increases in patients of the in vitro demonstrations of MHC Class I and Class II components by IFN-γ.185
IFNs have also increased tumor-associated antigens such as carcinoembryonic antigen and TAG-72 on the membranes of tumor cells both in vitro and in vivo.186 potentially thus facilitating antigen-specific T cell immunity as suggested by studies combining IFN-α with a CEA poxvirus vaccine.187 In addition to MHC and tumor-associated antigens as ISGs that can influence T cell function, other ISGs such as SECTM1 can serve as a co-stimulatory ligand for T cells after TCR activation.188
A number of ISGs function as chemoattractant to both lymphocytes and monocytes, recruiting these cells into tissues with induction by IFNs both in vitro and in patients.189–191 These IFN-induced chemokines have included CCL5 (RANTES), CXCL10 (IP-10), CCL2 (MCP-1), CCL3 (MIP-1α), CXCL9 (MIG), and CXCL11(I-TAC).23 STAT1 induction of chemokines CXCL9, CXCL10, CXCL11, and CXCL16 controlled recruitment of protective, antigen-specific Th1 cells into peripheral tissues.192 Other ISG protein products, such as phospholipid scramblase 1 (PLSCR1), can provide macrophages with a signal for engulfment after tumor cell apoptosis.193–195 PLSCR1 itself also may increase expression of other ISGs influencing apoptosis and other effects of IFNs.193–195
ISG15, a secreted protein induced by IFNs-α and IFN-β, induced IFN-γ synthesis by T cells and proliferation of NK and lymphokine-activated killer (LAK) cells.196 A recently identified mutation in ISG15 secretion resulted in decreased production of IFN-γ by NK and T cells, impaired protective immunity to mycobacteria, and has resulted in new insights into the immunomodulatory functions of secreted ISG15.197, 198 Four of the top 36 expressed genes in an expression array of melanoma cells (ISG15, USP18, UBE1L, UBE2L6) constitute part of a cascade of ubiquitin-related ISGs that can influence both innate immunity for malignant cells and cellular signaling.199 Melanoma cells producing high levels of ISG15 were those able to induce e-cadherin expression on dendritic cells.200 Conjugation of ISG15 to STAT1 and JAK1 and components of other signaling pathways (ERK1, phospholipase C, and heat shock proteins) suggest that regulation of ISG15 family molecules may have profound effects in the tumor microenvironment.201–204 Consistent with this, an siRNA to the ISG, USP18, that deconjugates ISG15 from target proteins, increased expression of TRAIL and thus promoted apoptosis.205 The functional effects of the ubiquitin-like ISGs may be explained by ISGylation and deISGylation stabilizing or altering binding of a target protein to its typical interaction partner at the protein-protein or protein-RNA interface.198, 202, 206
In contrast to immune augmenting effects, type I IFNs can also suppress T cell function through inhibition of secretion of IL-17.207, 208 Reduced responsiveness in T cells to IFN-α and IFN-γ with decreases in pSTAT1 can result from myeloid-derived suppressor cells.209 Possibly most important in suppressing T cell function in the tumor microenvironment is the protein, Programmed-cell Death-1 (PD-1), an inhibitory receptor on T-cells, activated in the tumor microenvironment by its ligands PD-L1 and PD-L2. PD-L1 secretion can be upregulated on tumor cells and tumor-associated stromal cells by IFNs-α and IFN-γ.210–213 Upregulation of PD-L1, a ligand detrimental to host response, by IFN-γ was mediated at least in part by IRF1 transcription. Thus, while PD-L1, an ISG detrimental to host T cell response, other ISGs such as HLA components and CXCL10, have been essential mediators—again illustrating the research challenges for dissecting the importance of individual products of ISGs in the tumor microenvironment.
Inhibition of angiogenesis
IFNs inhibit angiogenesis both by inhibiting endothelial cells through increases and decreases of proteins mediating the angiogenic process. Inhibition of angiogenesis by IFNs occurred before antiproliferative effects on tumor cells and was identified in vivo within 24h of tumor cell inoculation.214 IFN-sensitive and –resistant bladder carcinoma cells in mice after IFN-α had decreased tumor cell growth with reduced vascularization by suppression of the angiogenic cytokine, basic fibroblast growth factor (bFGF).215 Knockout studies confirmed signaling through STAT1 as necessary for reduction by IFNs of bFGF signaling. 215 Compared with wild-type mice, IFNAR receptor knockout mice had increased angiogenesis and tumorigenesis.216 In addition to bFGF, IFNs can inhibit other angiogenesis mediators such as VEGF by regulating VEGF promoter activity, in part at least mediated by a reduction in its promoter, HIF-1α, that was reduced by IFN-γ.217, 218 Contributing to the reduction in VEGFA by IFN-γ may be a conformational change in VEGF-A RNA 3′ untranslated region to reduce VEGF-A translation through integration of hypoxia and STAT signaling.219 In addition to IFN-γ, reductions in VEGF-A can also result from IFN-α2 in treated patients.220 Furthermore, IL-8, a mediator of angiogenesis, was inhibited in vitro and in vivo by IFN-α2b and IFN-β.221, 222
Another ISG product family, the guanylate binding proteins (GBPs), a family seven human GTPases of the dynamin superfamily that are potently induced in part by IFNs, modulated proliferation and spreading of endothelial cells through matrix metalloproteases in vitro.223–225 hGBP1 also inhibited proliferation of endothelial cells stimulated by VEGF or bFGF through its C-terminal alpha helices.223, 226 Thus, induction of ISGs, such as GBP1 and others such as IFI16 and PML that function as angiostatic inhibitors, coupled with downregulation of other factors such as bFGF, VEGF, and IL8, may all contribute to inhibition of angiogenesis by IFNs.225–228
Endogenous Type I IFN signaling has a role as a negative regulator of angiogenesis, keeping the “angiogenic switch” in the off position. Indeed, mice lacking IFN-β (IFN-β−/−) had faster growing melanomas and sarcomas with better developed blood vessels than did wild-type littermates.229 These tumors from the IFN-β−/− mice had enhanced infiltration by a neutrophilic cell population consistent with myeloid-derived suppressor cells (MDSCs) and increased expressing of VEGF and matrix metalloproteinase 9. These finding thus further suggest that endogenous IFN-β may play an important role in regulating tumor-induced angiogenesis. Further supporting this was the finding that in vitro treatment of tumor-infiltrating neutrophils with low levels of IFN-β reduced expression of pro-angiogenic factors to normal levels.229
An essential part of the malignant process, the motility of transformed cells and their invasion of normal tissues, can be inhibited by several ISGs. One of these genes, highly induced by IFNs-α and IFN-β in most tissues, is MX1, the gene product of which is a GTPase that suppressed motility and invasiveness of prostate carcinoma and melanoma in both in vitro and in vivo assessments.230 Another ISG product, schlafen 5, decreased melanoma cell invasiveness into three-dimensional collagen.231 Also, the ISG protein, IFIT2 (p54), inhibited migration of oral squamous carcinomas in vitro, possibly through interaction with cytokeratins.232 Thus, both the necessary steps for regional and distant metastases, invasion and angiogenesis, are suppressed by IFNs and its ISG protein products.
Activation of apoptosis of cancer cells
At higher doses exogenous IFNs, however, can result in pro-apoptotic effects either directly or through induction of secreted ISGs such as TNF-alpha related apoptosis inducing ligand (TRAIL/Apo2L) or Fas ligand (FasL) by immune effector cells.23, 233, 234 While expression may vary between cell types, pro-apoptotic, specific ISGs may be induced however only by a high dose of IFNs and/or last for short times because of IFN-induced negative regulators.117 These pro-apoptotic ISGs include TRAIL, Fas/FasL, XIAP associated factor-1 (XAF-1), PKR, 2′ 5′ A oligoadenylate synthetase (OAS), ISG12 (IFI27) and ISG 6–16 (G1P3), death activating protein kinases (DAP kinase), phospholipid scramblase (PLSCR1), IRFs, and promyelocytic leukemia gene (PML). Among them, OAS, IFI27, and PLSCR1 have been identified as induced by U-ISGF3, suggesting that they might have dual role in regulating apoptosis.117 While influences of concentration of IFNs and the kinetics of specific ISGs on apoptosis in cells of varied histologies are beyond the scope of this review, these are detailed in other reviews.233, 234
PERSPECTIVES AND CLINICAL IMPLICATIONS
The effects of IFNs in cancer biology are different depending on the levels of IFNs and the time of exposure to them (Figure 3). Either promoting or blocking IFN responses can be a good clinical strategy to cure cancers, depending on which effects of IFNs we need to target.
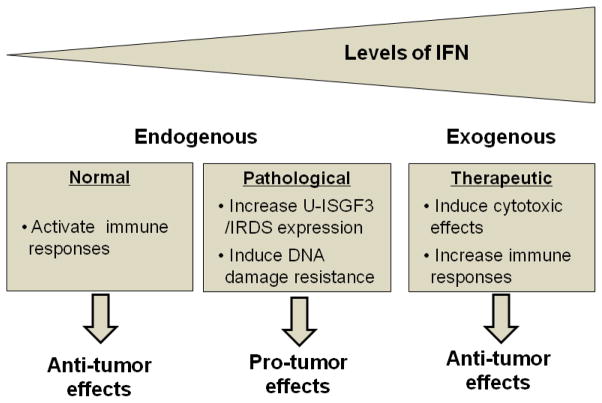
Figure 3. Effects of IFNs in tumor biology.
The effects of IFNs are determined by the levels of IFNs and the duration of exposure to them. In normal conditions, very low levels of endogenous type I IFNs are constitutively expressed and activate immune responses, inhibiting tumor progression (left panel). In pathological conditions, the production of endogenous IFNs is increased, and chronic exposure to IFNs enhances the expression of the IRDS genes through U-ISGF3, resulting in enhanced resistance to DNA damage (middle panel). For therapeutic purposes, high levels of exogenous IFNs are used to increase the expression of cytotoxic genes as well as to stimulate anti-cancer immunity (right panel). IRDS; IFN-related DNA damage resistance signature
Good or bad IFNs
The ability of type I IFNs to induce pro-apoptotic and anti-proliferative responses in a variety of cell types has led logically to exploration of their potential as antitumor therapeutic agents. However, more recent studies have shown that constitutive exposure of cells to a low level of type I IFN leads to steady-state expression of the IRDS subset of ISGs and that, via detailed mechanisms that are still obscure, the IRDS proteins mediate phenotypes that are favorable to the tumor, including facilitation of EMT and metastasis, suppression of T cell toxicity and resistance to therapies that damage DNA136 (Figure 4).
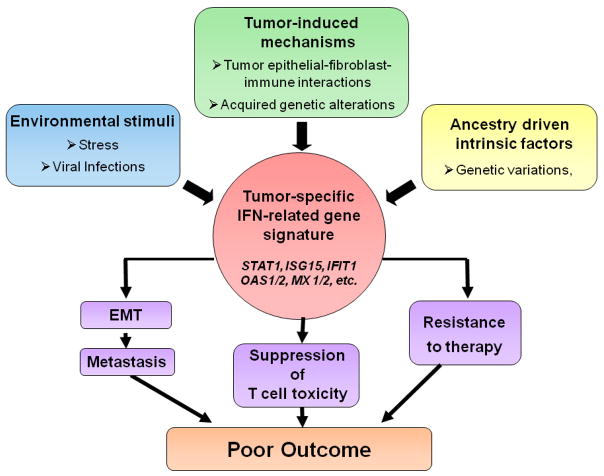
Figure 4. Potential origin and effects of the IRDS in tumor biology.
Tumor-induced mechanisms, environmental stimuli, or ancestry-driven intrinsic factors may induce the IRDS in tumors. This signature may lead to poor outcomes by mechanisms that include EMT and consequently increased metastatic potential, suppression of T-cell toxicity, and resistance to therapy. IRDS; IFN-related DNA damage resistance signature, EMT; epithelial to mesenchymal transition
This figure and legend are modified from the original in the paper by Wallace et al136 with the permission of Dr. Ambs.
Endogenous interferons
IFNs might be produced constitutively in the tumor microenvironment by the tumor cells themselves, by the surrounding stroma, or by invading immune cells, especially macrophages and dendritic cells. It is also possible that the IRDS signature may be generated by additional mechanisms still to be discovered, including genetic factors (Figure 4). If IFNs in the tumor microenvironment are indeed responsible for the IRDS signature, a reasonable therapeutic strategy may be to inhibit transiently the ability of the tumor cells to respond to IFNs in advance of ionizing radiation therapy or chemotherapy. Such inhibition might be achieved by using antibodies against the type I IFN receptor, which has been successful in mice235 or by using tyrosine kinase inhibitor of JAK1 and JAK2, which block responses to all known IFNs. JAK1/JAK2 inhibitors are already approved for clinical use in rheumatoid arthritis, psoriasis and other diseases.236 In treating common cancers such as prostate or breast, for example, it could be of major benefit to patients if down-regulation of the expression of the IRDS proteins in the tumor allowed a given clinical effect to be achieved with a lower dose of radiation or DNA damaging agents, thus reducing collateral damage. In coming years assessing these and other new insights into action of endogenous IFNs and ISGs in the tumor microenvironment should yield further improvements in patient outcomes.
Exogenous interferons
Although there have been some successes, except as a prototype for immune modulation and recombinant DNA pharmaceuticals, IFNs have not had much success as therapeutic agents in cancer. Our current realization that chronic exposure to type I IFN might be beneficial to tumors should be factored into the design of future trials. It may not be a good idea to prolong the exposure of tumors to exogenous IFNs, by using long-acting derivatives; a better strategy may be to use repeated pulses of short-acting IFNs in a protocol that minimizes down regulation and maximizes expression of the full set of ISGs, which encode many pro-apoptotic and anti-proliferative proteins. In other words, design the protocol to allow expression to subside of IFN-induced proteins, especially SOCS1, that suppress the expression of the full set of ISGs, but not the IRDS subset, following the first administration of IFN before giving a second dose. Additionally, when exogenous IFNs are to be combined with therapeutic regimens that cause DNA damage (ionizing radiation and many chemotherapeutic drugs), it is likely to be best to administer the drugs or radiation when the IRDS pattern of gene expression is minimal in the tumors, either immediately after administration of IFNs, when the full set of ISGs are still expressed, or long after, when expression of the IRDS proteins has subsided.
Acknowledgments
Supported in part by NIH P30 CA043703
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Borden EC, Williams BR. Interferon-stimulated genes and their protein products: what and how? J Interferon Cytokine Res. 2011;31:1–4. doi: 10.1089/jir.2010.0129. [DOI] [PubMed] [Google Scholar]
- 2.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hertzog P, Forster S, Samarajiwa S. Systems biology of interferon responses. J Interferon Cytokine Res. 2011;31:5–11. doi: 10.1089/jir.2010.0126. [DOI] [PubMed] [Google Scholar]
- 6.Rusinova I, Forster S, Yu S, Kannan A, Masse M, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waddell SJ, Popper SJ, Rubins KH, Griffiths MJ, Brown PO, et al. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS One. 2010;5:e9753. doi: 10.1371/journal.pone.0009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns P, Tokino K, Eby Y, Sidransky D. Homozygous deletions of 9p21 in primary human bladder tumors detected by comparative multiplex polymerase chain reaction. Cancer Res. 1994;54:1422–1424. [PubMed] [Google Scholar]
- 9.Casey G, Neville PJ, Plummer SJ, Xiang Y, Krumroy LM, et al. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet. 2002;32:581–583. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz MO, Rubin CM, Harden A, Ziemin S, Larson RA, et al. Deletions of interferon genes in acute lymphoblastic leukemia. N Engl J Med. 1990;322:77–82. doi: 10.1056/NEJM199001113220202. [DOI] [PubMed] [Google Scholar]
- 12.Green WB, Slovak ML, Chen IM, Pallavicini M, Hecht JL, et al. Lack of IRF-1 expression in acute promyelocytic leukemia and in a subset of acute myeloid leukemias with del(5)(q31) Leukemia. 1999;13:1960–1971. doi: 10.1038/sj.leu.2401596. [DOI] [PubMed] [Google Scholar]
- 13.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 14.Olopade OI, Buchhagen DL, Malik K, Sherman J, Nobori T, et al. Homozygous loss of the interferon genes defines the critical region on 9p that is deleted in lung cancers. Cancer Res. 1993;53:2410–2415. [PubMed] [Google Scholar]
- 15.Rennert H, Bercovich D, Hubert A, Abeliovich D, Rozovsky U, et al. A novel founder mutation in the RNASEL gene, 471 delAAAG, is associated with prostate cancer in Ashkenazi Jews. Am J Hum Genet. 2002;71:981–984. doi: 10.1086/342775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rokman A, Ikonen T, Seppala EH, Nupponen N, Autio V, et al. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am J Hum Genet. 2002;70:1299–1304. doi: 10.1086/340450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slattery ML, Lundgreen A, Bondurant KL, Wolff RK. Interferon-signaling pathway: associations with colon and rectal cancer risk and subsequent survival. Carcinogenesis. 2011;32:1660–1667. doi: 10.1093/carcin/bgr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulaeva OI, Draghici S, Tang L, Kraniak JM, Land SJ, et al. Epigenetic silencing of multiple interferon pathway genes after cellular immortalization. Oncogene. 2003;22:4118–4127. doi: 10.1038/sj.onc.1206594. [DOI] [PubMed] [Google Scholar]
- 19.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2013;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110:E89–98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reu FJ, Bae SI, Cherkassky L, Leaman DW, Lindner D, et al. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24:3771–3779. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- 22.Bekisz J, Schmeisser H, Hernandez J, Goldman ND, Zoon KC. Human interferons alpha, beta and omega. Growth Factors. 2004;22:243–251. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- 23.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotenko SV. IFN-lambdas. Curr Opin Immunol. 2011;23:583–590. doi: 10.1016/j.coi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 26.Pestka S. The human interferon-alpha species and hybrid proteins. Semin Oncol. 1997;24:S9-4–S9-17. [PubMed] [Google Scholar]
- 27.Farrar MA, Schreiber RD. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 28.Durbin RK, Kotenko SV, Durbin JE. Interferon induction and function at the mucosal surface. Immunol Rev. 2013;255:25–39. doi: 10.1111/imr.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhat N, Fitzgerald KA. Recognition of Cytosolic DNA by cGAS and other STING-dependent sensors. Eur J Immunol. 2013 doi: 10.1002/eji.201344127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2013 doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 33.Khoo JJ, Forster S, Mansell A. Toll-like receptors as interferon-regulated genes and their role in disease. J Interferon Cytokine Res. 2011;31:13–25. doi: 10.1089/jir.2010.0095. [DOI] [PubMed] [Google Scholar]
- 34.Sen GC, Sarkar SN. Transcriptional signaling by double-stranded RNA: role of TLR3. Cytokine Growth Factor Rev. 2005;16:1–14. doi: 10.1016/j.cytogfr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, et al. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 36.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 38.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 39.Taniguchi T. Aimez-vous Brahms? A story capriccioso from the discovery of a cytokine family and its regulators. Nat Immunol. 2009;10:447–449. doi: 10.1038/ni0509-447. [DOI] [PubMed] [Google Scholar]
- 40.Tailor P, Tamura T, Ozato K. IRF family proteins and type I interferon induction in dendritic cells. Cell Res. 2006;16:134–140. doi: 10.1038/sj.cr.7310018. [DOI] [PubMed] [Google Scholar]
- 41.Novick D, Orchansky P, Revel M, Rubinstein M. The human interferon-gamma receptor. Purification, characterization, and preparation of antibodies. J Biol Chem. 1987;262:8483–8487. [PubMed] [Google Scholar]
- 42.Ruzicka FJ, Jach ME, Borden EC. Binding of recombinant-produced interferon beta ser to human lymphoblastoid cells. Evidence for two binding domains. J Biol Chem. 1987;262:16142–16149. [PubMed] [Google Scholar]
- 43.Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, et al. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 2011;146:621–632. doi: 10.1016/j.cell.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piehler J, Thomas C, Garcia KC, Schreiber G. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol Rev. 2012;250:317–334. doi: 10.1111/imr.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar KG, Liu J, Li Y, Yu D, Thomas-Tikhonenko A, et al. Raf inhibitor stabilizes receptor for the type I interferon but inhibits its anti-proliferative effects in human malignant melanoma cells. Cancer Biol Ther. 2007;6:1437–1441. doi: 10.4161/cbt.6.9.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brierley MM, Fish EN. Review: IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J Interferon Cytokine Res. 2002;22:835–845. doi: 10.1089/107999002760274845. [DOI] [PubMed] [Google Scholar]
- 47.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 48.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 49.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 51.Ezell SA, Polytarchou C, Hatziapostolou M, Guo A, Sanidas I, et al. The protein kinase Akt1 regulates the interferon response through phosphorylation of the transcriptional repressor EMSY. Proc Natl Acad Sci U S A. 2012;109:E613–621. doi: 10.1073/pnas.1115029109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 53.Wu TR, Hong YK, Wang XD, Ling MY, Dragoi AM, et al. SHP-2 is a dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J Biol Chem. 2002;277:47572–47580. doi: 10.1074/jbc.M207536200. [DOI] [PubMed] [Google Scholar]
- 54.Yi T, Pathak MK, Lindner DJ, Ketterer ME, Farver C, et al. Anticancer activity of sodium stibogluconate in synergy with IFNs. J Immunol. 2002;169:5978–5985. doi: 10.4049/jimmunol.169.10.5978. [DOI] [PubMed] [Google Scholar]
- 55.Kundu S, Fan K, Cao M, Lindner DJ, Zhao ZJ, et al. Novel SHP-1 inhibitors tyrosine phosphatase inhibitor-1 and analogs with preclinical anti-tumor activities as tolerated oral agents. J Immunol. 2010;184:6529–6536. doi: 10.4049/jimmunol.0903562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purdy AK, Campbell KS. SHP-2 expression negatively regulates NK cell function. J Immunol. 2009;183:7234–7243. doi: 10.4049/jimmunol.0900088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Win-Piazza H, Schneeberger VE, Chen L, Pernazza D, Lawrence HR, et al. Enhanced anti-melanoma efficacy of interferon alfa-2b via inhibition of Shp2. Cancer Lett. 2012;320:81–85. doi: 10.1016/j.canlet.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev Immunol. 2002;2:410–416. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- 59.Krebs DL, Hilton DJ. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- 60.Starr R, Fuchsberger M, Lau LS, Uldrich AP, Goradia A, et al. SOCS-1 binding to tyrosine 441 of IFN-gamma receptor subunit 1 contributes to the attenuation of IFN-gamma signaling in vivo. J Immunol. 2009;183:4537–4544. doi: 10.4049/jimmunol.0901010. [DOI] [PubMed] [Google Scholar]
- 61.Bocci V. Is interferon produced in physiologic conditions? Medical hypotheses. 1980;6:735–745. doi: 10.1016/0306-9877(80)90091-2. [DOI] [PubMed] [Google Scholar]
- 62.Bocci V. Roles of interferon produced in physiological conditions. A speculative review. Immunology. 1988;64:1–9. [PMC free article] [PubMed] [Google Scholar]
- 63.Gresser I. Biologic effects of interferons. The Journal of investigative dermatology. 1990;95:66S–71S. doi: 10.1111/1523-1747.ep12874776. [DOI] [PubMed] [Google Scholar]
- 64.Tovey MG, Streuli M, Gresser I, Gugenheim J, Blanchard B, et al. Interferon messenger RNA is produced constitutively in the organs of normal individuals. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:5038–5042. doi: 10.1073/pnas.84.14.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yaar M, Palleroni AV, Gilchrest BA. Normal human epidermis contains an interferon-like protein. The Journal of cell biology. 1986;103:1349–1354. doi: 10.1083/jcb.103.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balachandran S, Beg AA. Defining emerging roles for NF-kappaB in antivirus responses: revisiting the interferon-beta enhanceosome paradigm. PLoS pathogens. 2011;7:e1002165. doi: 10.1371/journal.ppat.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hata N, Sato M, Takaoka A, Asagiri M, Tanaka N, et al. Constitutive IFN-alpha/beta signal for efficient IFN-alpha/beta gene induction by virus. Biochemical and biophysical research communications. 2001;285:518–525. doi: 10.1006/bbrc.2001.5159. [DOI] [PubMed] [Google Scholar]
- 69.Cheng CS, Feldman KE, Lee J, Verma S, Huang DB, et al. The specificity of innate immune responses is enforced by repression of interferon response elements by NF-kappaB p50. Science signaling. 2011;4:ra11. doi: 10.1126/scisignal.2001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hida S, Ogasawara K, Sato K, Abe M, Takayanagi H, et al. CD8(+) T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-alpha/beta signaling. Immunity. 2000;13:643–655. doi: 10.1016/s1074-7613(00)00064-9. [DOI] [PubMed] [Google Scholar]
- 71.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 72.Senger K, Merika M, Agalioti T, Yie J, Escalante CR, et al. Gene repression by coactivator repulsion. Molecular cell. 2000;6:931–937. doi: 10.1016/s1097-2765(05)00081-x. [DOI] [PubMed] [Google Scholar]
- 73.Mitani Y, Takaoka A, Kim SH, Kato Y, Yokochi T, et al. Cross talk of the interferon-alpha/beta signalling complex with gp130 for effective interleukin-6 signalling. Genes to cells: devoted to molecular & cellular mechanisms. 2001;6:631–640. doi: 10.1046/j.1365-2443.2001.00448.x. [DOI] [PubMed] [Google Scholar]
- 74.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, et al. Functional role of type I and type II interferons in antiviral defense. Science (New York, NY ) 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 75.Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, et al. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science (New York, NY ) 2000;288:2357–2360. doi: 10.1126/science.288.5475.2357. [DOI] [PubMed] [Google Scholar]
- 76.Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-alpha/beta revisited. Nature reviews. Molecular cell biology. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 77.Fleetwood AJ, Dinh H, Cook AD, Hertzog PJ, Hamilton JA. GM-CSF- and M-CSF-dependent macrophage phenotypes display differential dependence on type I interferon signaling. Journal of leukocyte biology. 2009;86:411–421. doi: 10.1189/jlb.1108702. [DOI] [PubMed] [Google Scholar]
- 78.Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of interferon-beta to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. The Journal of biological chemistry. 2006;281:31119–31130. doi: 10.1074/jbc.M604958200. [DOI] [PubMed] [Google Scholar]
- 79.Hamilton JA, Whitty GA, Kola I, Hertzog PJ. Endogenous IFN-alpha beta suppresses colony-stimulating factor (CSF)-1-stimulated macrophage DNA synthesis and mediates inhibitory effects of lipopolysaccharide and TNF-alpha. Journal of immunology (Baltimore, Md : 1950) 1996;156:2553–2557. [PubMed] [Google Scholar]
- 80.Sato T, Onai N, Yoshihara H, Arai F, Suda T, et al. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nature medicine. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- 81.Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, et al. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- 82.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunn GP, Bruce AT, Sheehan KC, Shankaran V, Uppaluri R, et al. A critical function for type I interferons in cancer immunoediting. Nat Immunol. 2005;6:722–729. doi: 10.1038/ni1213. [DOI] [PubMed] [Google Scholar]
- 84.Pfeffer LM, Donner DB. The down-regulation of alpha-interferon receptors in human lymphoblastoid cells: relation to cellular responsiveness to the antiproliferative action of alpha-interferon. Cancer research. 1990;50:2654–2657. [PubMed] [Google Scholar]
- 85.Wong LH, Krauer KG, Hatzinisiriou I, Estcourt MJ, Hersey P, et al. Interferon-resistant human melanoma cells are deficient in ISGF3 components, STAT1, STAT2, and p48-ISGF3 gamma. The Journal of biological chemistry. 1997;272:28779–28785. doi: 10.1074/jbc.272.45.28779. [DOI] [PubMed] [Google Scholar]
- 86.Heyman M, Grander D, Brondum-Nielsen K, Cederblad B, Liu Y, et al. Interferon system defects in malignant T-cells. Leukemia. 1994;8:425–434. [PubMed] [Google Scholar]
- 87.Landolfo S, Guarini A, Riera L, Gariglio M, Gribaudo G, et al. Chronic myeloid leukemia cells resistant to interferon-alpha lack STAT1 expression. The hematology journal: the official journal of the European Haematology Association/EHA. 2000;1:7–14. doi: 10.1038/sj.thj.6200004. [DOI] [PubMed] [Google Scholar]
- 88.Chen HM, Tanaka N, Mitani Y, Oda E, Nozawa H, et al. Critical role for constitutive type I interferon signaling in the prevention of cellular transformation. Cancer Sci. 2009;100:449–456. doi: 10.1111/j.1349-7006.2008.01051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shmulevitz M, Pan LZ, Garant K, Pan D, Lee PW. Oncogenic Ras promotes reovirus spread by suppressing IFN-beta production through negative regulation of RIG-I signaling. Cancer Res. 2010;70:4912–4921. doi: 10.1158/0008-5472.CAN-09-4676. [DOI] [PubMed] [Google Scholar]
- 90.Katze MG, He Y, Gale M., Jr Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 91.O’Brien PM, Saveria Campo M. Evasion of host immunity directed by papillomavirus-encoded proteins. Virus Res. 2002;88:103–117. doi: 10.1016/s0168-1702(02)00123-5. [DOI] [PubMed] [Google Scholar]
- 92.Rincon-Orozco B, Halec G, Rosenberger S, Muschik D, Nindl I, et al. Epigenetic silencing of interferon-kappa in human papillomavirus type 16-positive cells. Cancer Res. 2009;69:8718–8725. doi: 10.1158/0008-5472.CAN-09-0550. [DOI] [PubMed] [Google Scholar]
- 93.Schiavoni G, Mattei F, Gabriele L. Type I Interferons as Stimulators of DC-Mediated Cross-Priming: Impact on Anti-Tumor Response. Front Immunol. 2013;4:483. doi: 10.3389/fimmu.2013.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Havelange V, Pekarsky Y, Nakamura T, Palamarchuk A, Alder H, et al. IRF4 mutations in chronic lymphocytic leukemia. Blood. 2011;118:2827–2829. doi: 10.1182/blood-2011-04-350579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han J, Qureshi AA, Nan H, Zhang J, Song Y, et al. A germline variant in the interferon regulatory factor 4 gene as a novel skin cancer risk locus. Cancer Res. 2011;71:1533–1539. doi: 10.1158/0008-5472.CAN-10-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Salaverria I, Philipp C, Oschlies I, Kohler CW, Kreuz M, et al. Translocations activating IRF4 identify a subtype of germinal center-derived B-cell lymphoma affecting predominantly children and young adults. Blood. 2011;118:139–147. doi: 10.1182/blood-2011-01-330795. [DOI] [PubMed] [Google Scholar]
- 97.Bi X, Hameed M, Mirani N, Pimenta EM, Anari J, et al. Loss of interferon regulatory factor 5 (IRF5) expression in human ductal carcinoma correlates with disease stage and contributes to metastasis. Breast Cancer Res. 2011;13:R111. doi: 10.1186/bcr3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Slattery ML, Lundgreen A, Kadlubar SA, Bondurant KL, Wolff RK. JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol Carcinog. 2013;52:155–166. doi: 10.1002/mc.21841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burchert A, Cai D, Hofbauer LC, Samuelsson MK, Slater EP, et al. Interferon consensus sequence binding protein (ICSBP; IRF-8) antagonizes BCR/ABL and down-regulates bcl-2. Blood. 2004;103:3480–3489. doi: 10.1182/blood-2003-08-2970. [DOI] [PubMed] [Google Scholar]
- 100.Yamamoto M, Kato T, Hotta C, Nishiyama A, Kurotaki D, et al. Shared and distinct functions of the transcription factors IRF4 and IRF8 in myeloid cell development. PLoS One. 2011;6:e25812. doi: 10.1371/journal.pone.0025812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim DH, Kong JH, Byeun JY, Jung CW, Xu W, et al. The IFNG (IFN-gamma) genotype predicts cytogenetic and molecular response to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res. 2010;16:5339–5350. doi: 10.1158/1078-0432.CCR-10-1638. [DOI] [PubMed] [Google Scholar]
- 102.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kirou KA, Lee C, George S, Louca K, Peterson MG, et al. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis and Rheumatism. 2005;52:1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 104.Arakura F, Hida S, Ichikawa E, Yajima C, Nakajima S, et al. Genetic control directed toward spontaneous IFN-alpha/IFN-beta responses and downstream IFN-gamma expression influences the pathogenesis of a murine psoriasis-like skin disease. Journal of immunology (Baltimore, Md : 1950) 2007;179:3249–3257. doi: 10.4049/jimmunol.179.5.3249. [DOI] [PubMed] [Google Scholar]
- 105.Nacionales DC, Kelly-Scumpia KM, Lee PY, Weinstein JS, Lyons R, et al. Deficiency of the type I interferon receptor protects mice from experimental lupus. Arthritis and Rheumatism. 2007;56:3770–3783. doi: 10.1002/art.23023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yao Y, Richman L, Higgs BW, Morehouse CA, de los Reyes M, et al. Neutralization of interferon-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis and Rheumatism. 2009;60:1785–1796. doi: 10.1002/art.24557. [DOI] [PubMed] [Google Scholar]
- 107.Andersen JB, Hassel BA. The interferon regulated ubiquitin-like protein, ISG15, in tumorigenesis: friend or foe? Cytokine & growth factor reviews. 2006;17:411–421. doi: 10.1016/j.cytogfr.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 108.Hatano H, Kudo Y, Ogawa I, Tsunematsu T, Kikuchi A, et al. IFN-induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:6097–6105. doi: 10.1158/1078-0432.CCR-07-4761. [DOI] [PubMed] [Google Scholar]
- 109.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Suomela S, Cao L, Bowcock A, Saarialho-Kere U. Interferon alpha-inducible protein 27 (IFI27) is upregulated in psoriatic skin and certain epithelial cancers. The Journal of investigative dermatology. 2004;122:717–721. doi: 10.1111/j.0022-202X.2004.22322.x. [DOI] [PubMed] [Google Scholar]
- 111.Cai D, Cao J, Li Z, Zheng X, Yao Y, et al. Up-regulation of bone marrow stromal protein 2 (BST2) in breast cancer with bone metastasis. BMC cancer. 2009;9:102. doi: 10.1186/1471-2407-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khodarev NN, Roach P, Pitroda SP, Golden DW, Bhayani M, et al. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One. 2009;4:e5821. doi: 10.1371/journal.pone.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nature genetics. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Parkin DM. The global health burden of infection-associated cancers in the year 2002. International journal of cancer. Journal international du cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 115.Avanzi S, Alvisi G, Ripalti A. How virus persistence can initiate the tumorigenesis process. World journal of virology. 2013;2:102–109. doi: 10.5501/wjv.v2.i2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cann A. Principles of Molecular Virology. 5. Philadelphia: Elsevier; 2012. [Google Scholar]
- 117.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, et al. IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. The EMBO journal. 2013;32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goubau D, Deddouche S, Reis E, Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. The Journal of experimental medicine. 2005;202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nature immunology. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 121.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (New York, NY ) 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zaidi MR, Davis S, Noonan FP, Graff-Cherry C, Hawley TS, et al. Interferon-gamma links ultraviolet radiation to melanomagenesis in mice. Nature. 2011;469:548–553. doi: 10.1038/nature09666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, et al. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Atianand MK, Fitzgerald KA. Molecular basis of DNA recognition in the immune system. Journal of immunology (Baltimore, Md : 1950) 2013;190:1911–1918. doi: 10.4049/jimmunol.1203162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gehrke N, Mertens C, Zillinger T, Wenzel J, Bald T, et al. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity. 2013;39:482–495. doi: 10.1016/j.immuni.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 127.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nature reviews. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 128.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 129.Aas T, Borresen AL, Geisler S, Smith-Sorensen B, Johnsen H, et al. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nature medicine. 1996;2:811–814. doi: 10.1038/nm0796-811. [DOI] [PubMed] [Google Scholar]
- 130.Townsend PA, Cragg MS, Davidson SM, McCormick J, Barry S, et al. STAT-1 facilitates the ATM activated checkpoint pathway following DNA damage. J Cell Sci. 2005;118:1629–1639. doi: 10.1242/jcs.01728. [DOI] [PubMed] [Google Scholar]
- 131.Khodarev NN, Minn AJ, Efimova EV, Darga TE, Labay E, et al. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer research. 2007;67:9214–9220. doi: 10.1158/0008-5472.CAN-07-1019. [DOI] [PubMed] [Google Scholar]
- 132.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cheon H, Yang J, Stark GR. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research. 2011;31:33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Levin D, Harari D, Schreiber G. Stochastic receptor expression determines cell fate upon interferon treatment. Molecular and cellular biology. 2011;31:3252–3266. doi: 10.1128/MCB.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Duarte CW, Willey CD, Zhi D, Cui X, Harris JJ, et al. Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype-specific manner. PLoS One. 2012;7:e29653. doi: 10.1371/journal.pone.0029653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wallace TA, Martin DN, Ambs S. Interactions among genes, tumor biology and the environment in cancer health disparities: examining the evidence on a national and global scale. Carcinogenesis. 2011;32:1107–1121. doi: 10.1093/carcin/bgr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Khodarev NN, Beckett M, Labay E, Darga T, Roizman B, et al. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Luszczek W, Cheriyath V, Mekhail TM, Borden EC. Combinations of DNA methyltransferase and histone deacetylase inhibitors induce DNA damage in small cell lung cancer cells: correlation of resistance with IFN-stimulated gene expression. Molecular cancer therapeutics. 2010;9:2309–2321. doi: 10.1158/1535-7163.MCT-10-0309. [DOI] [PubMed] [Google Scholar]
- 140.Rickardson L, Fryknas M, Dhar S, Lovborg H, Gullbo J, et al. Identification of molecular mechanisms for cellular drug resistance by combining drug activity and gene expression profiles. British journal of cancer. 2005;93:483–492. doi: 10.1038/sj.bjc.6602699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cheriyath V, Glaser KB, Waring JF, Baz R, Hussein MA, et al. G1P3, an IFN-induced survival factor, antagonizes TRAIL-induced apoptosis in human myeloma cells. J Clin Invest. 2007;117:3107–3117. doi: 10.1172/JCI31122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cheriyath V, Leaman DW, Borden EC. Emerging roles of FAM14 family members (G1P3/ISG 6–16 and ISG12/IFI27) in innate immunity and cancer. J Interferon Cytokine Res. 2011;31:173–181. doi: 10.1089/jir.2010.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rosebeck S, Leaman DW. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis. 2008;13:562–572. doi: 10.1007/s10495-008-0190-0. [DOI] [PubMed] [Google Scholar]
- 144.Salerno RA, Whitmire CE, Garcia IM, Huebner RJ. Chemical carcinogenesis in mice inhibited by interferon. Nat New Biol. 1972;239:31–32. doi: 10.1038/newbio239031a0. [DOI] [PubMed] [Google Scholar]
- 145.Borden EC, Sidky YA, Erturk E, Wierenga W, Bryan GT. Protection from carcinogen-induced murine bladder carcinoma by interferons and an oral interferon-inducing pyrimidinone, bropirimine. Cancer Res. 1990;50:1071–1074. [PubMed] [Google Scholar]
- 146.Luth S, Schrader J, Zander S, Carambia A, Buchkremer J, et al. Chronic inflammatory IFN-gamma signaling suppresses hepatocarcinogenesis in mice by sensitizing hepatocytes for apoptosis. Cancer Res. 2011;71:3763–3771. doi: 10.1158/0008-5472.CAN-10-3232. [DOI] [PubMed] [Google Scholar]
- 147.Nakaji M, Yano Y, Ninomiya T, Seo Y, Hamano K, et al. IFN-alpha prevents the growth of pre-neoplastic lesions and inhibits the development of hepatocellular carcinoma in the rat. Carcinogenesis. 2004;25:389–397. doi: 10.1093/carcin/bgh028. [DOI] [PubMed] [Google Scholar]
- 148.Oon CJ, Chen WN. Lymphoblastoid alpha-interferon in the prevention of hepatocellular carcinoma (HCC) in high-risk HbsAg-positive resected cirrhotic HCC cases: a 14-year follow-up. Cancer Invest. 2003;21:394–399. doi: 10.1081/cnv-120018231. [DOI] [PubMed] [Google Scholar]
- 149.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 150.Gajewski TF, Fuertes MB, Woo SR. Innate immune sensing of cancer: clues from an identified role for type I IFNs. Cancer Immunol Immunother. 2012;61:1343–1347. doi: 10.1007/s00262-012-1305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hervas-Stubbs S, Perez-Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A, et al. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res. 2011;17:2619–2627. doi: 10.1158/1078-0432.CCR-10-1114. [DOI] [PubMed] [Google Scholar]
- 153.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 154.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Asselin-Paturel C, Trinchieri G. Production of type I interferons: plasmacytoid dendritic cells and beyond. J Exp Med. 2005;202:461–465. doi: 10.1084/jem.20051395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Radvanyi LG, Banerjee A, Weir M, Messner H. Low levels of interferon-alpha induce CD86 (B7.2) expression and accelerates dendritic cell maturation from human peripheral blood mononuclear cells. Scand J Immunol. 1999;50:499–509. doi: 10.1046/j.1365-3083.1999.00625.x. [DOI] [PubMed] [Google Scholar]
- 157.Santini SM, Lapenta C, Logozzi M, Parlato S, Spada M, et al. Type I interferon as a powerful adjuvant for monocyte-derived dendritic cell development and activity in vitro and in Hu-PBL-SCID mice. J Exp Med. 2000;191:1777–1788. doi: 10.1084/jem.191.10.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Edwards BS, Merritt JA, Fuhlbrigge RC, Borden EC. Low doses of interferon alpha result in more effective clinical natural killer cell activation. J Clin Invest. 1985;75:1908–1913. doi: 10.1172/JCI111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Herberman RB, Djeu JY, Ortaldo JR, Holden HT, West WH, et al. Role of interferon in augmentation of natural and antibody-dependent cell-mediated cytotoxicity. Cancer Treat Rep. 1978;62:1893–1896. [PubMed] [Google Scholar]
- 160.Kirkwood JM, Ernstoff MS, Trautman T, Hebert G, Nishida Y, et al. In vivo biological response to recombinant interferon-gamma during a phase I dose-response trial in patients with metastatic melanoma. J Clin Oncol. 1990;8:1070–1082. doi: 10.1200/JCO.1990.8.6.1070. [DOI] [PubMed] [Google Scholar]
- 161.Maluish AE, Leavitt R, Sherwin SA, Oldham RK, Herberman RB. Effects of recombinant interferon-alpha on immune function in cancer patients. J Biol Response Mod. 1983;2:470–481. [PubMed] [Google Scholar]
- 162.Schiller JH, Storer B, Paulnock DM, Brown RR, Datta SP, et al. A direct comparison of biological response modulation and clinical side effects by interferon-beta ser, interferon-gamma, or the combination of interferons beta ser and gamma in humans. J Clin Invest. 1990;86:1211–1221. doi: 10.1172/JCI114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Silver HK, Connors JM, Karim KA, Kong S, Spinelli JJ, et al. Effect of lymphoblastoid interferon on lymphocyte subsets in cancer patients. J Biol Response Mod. 1983;2:428–440. [PubMed] [Google Scholar]
- 164.Trinchieri G, Santoli D, Koprowski H. Spontaneous cell-mediated cytotoxicity in humans: role of interferon and immunoglobulins. J Immunol. 1978;120:1849–1855. [PubMed] [Google Scholar]
- 165.Lucas M, Schachterle W, Oberle K, Aichele P, Diefenbach A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 2007;26:503–517. doi: 10.1016/j.immuni.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Belardelli F, Gresser I, Maury C, Maunoury MT. Antitumor effects of interferon in mice injected with interferon-sensitive and interferon-resistant Friend leukemia cells. II. Role of host mechanisms. Int J Cancer. 1982;30:821–825. doi: 10.1002/ijc.2910300622. [DOI] [PubMed] [Google Scholar]
- 167.Reid TR, Race ER, Wolff BH, Friedman RM, Merigan TC, et al. Enhanced in vivo therapeutic response to interferon in mice with an in vitro interferon-resistant B-cell lymphoma. Cancer Res. 1989;49:4163–4169. [PubMed] [Google Scholar]
- 168.Lesinski GB, Anghelina M, Zimmerer J, Bakalakos T, Badgwell B, et al. The antitumor effects of IFN-alpha are abrogated in a STAT1-deficient mouse. J Clin Invest. 2003;112:170–180. doi: 10.1172/JCI16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Badgwell B, Lesinski GB, Magro C, Abood G, Skaf A, et al. The antitumor effects of interferon-alpha are maintained in mice challenged with a STAT1-deficient murine melanoma cell line. J Surg Res. 2004;116:129–136. doi: 10.1016/j.jss.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 170.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 171.Swann JB, Hayakawa Y, Zerafa N, Sheehan KC, Scott B, et al. Type I IFN contributes to NK cell homeostasis, activation, and antitumor function. J Immunol. 2007;178:7540–7549. doi: 10.4049/jimmunol.178.12.7540. [DOI] [PubMed] [Google Scholar]
- 172.Mescher MF, Curtsinger JM, Agarwal P, Casey KA, Gerner M, et al. Signals required for programming effector and memory development by CD8+ T cells. Immunol Rev. 2006;211:81–92. doi: 10.1111/j.0105-2896.2006.00382.x. [DOI] [PubMed] [Google Scholar]
- 173.Coro ES, Chang WL, Baumgarth N. Type I IFN receptor signals directly stimulate local B cells early following influenza virus infection. J Immunol. 2006;176:4343–4351. doi: 10.4049/jimmunol.176.7.4343. [DOI] [PubMed] [Google Scholar]
- 174.Le Bon A, Thompson C, Kamphuis E, Durand V, Rossmann C, et al. Cutting edge: enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J Immunol. 2006;176:2074–2078. doi: 10.4049/jimmunol.176.4.2074. [DOI] [PubMed] [Google Scholar]
- 175.Pamer E, Cresswell P. Mechanisms of MHC class I--restricted antigen processing. Annu Rev Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 176.Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 177.Liberati AM, Horisberger MA, Garofani P, De Angelis V, Ferrajoli A, et al. Interferon-alpha-induced biologic modifications in patients with chronic myelogenous leukemia. J Interferon Res. 1994;14:349–355. doi: 10.1089/jir.1994.14.349. [DOI] [PubMed] [Google Scholar]
- 178.Witt PL, Storer BE, Bryan GT, Brown RR, Flashner M, et al. Pharmacodynamics of biological response in vivo after single and multiple doses of interferon-beta. J Immunother Emphasis Tumor Immunol. 1993;13:191–200. doi: 10.1097/00002371-199304000-00006. [DOI] [PubMed] [Google Scholar]
- 179.Cramer LA, Nelson SL, Klemsz MJ. Synergistic induction of the Tap-1 gene by IFN-gamma and lipopolysaccharide in macrophages is regulated by STAT1. J Immunol. 2000;165:3190–3197. doi: 10.4049/jimmunol.165.6.3190. [DOI] [PubMed] [Google Scholar]
- 180.El Hage F, Durgeau A, Mami-Chouaib F. TAP expression level in tumor cells defines the nature and processing of MHC class I peptides for recognition by tumor-specific cytotoxic T lymphocytes. Ann N Y Acad Sci. 2013;1283:75–80. doi: 10.1111/j.1749-6632.2012.06777.x. [DOI] [PubMed] [Google Scholar]
- 181.Johnsen A, France J, Sy MS, Harding CV. Down-regulation of the transporter for antigen presentation, proteasome subunits, and class I major histocompatibility complex in tumor cell lines. Cancer Res. 1998;58:3660–3667. [PubMed] [Google Scholar]
- 182.Zhang L, Pagano JS. Interferon regulatory factor 7 mediates activation of Tap-2 by Epstein-Barr virus latent membrane protein 1. J Virol. 2001;75:341–350. doi: 10.1128/JVI.75.1.341-350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Friedman RL, Manly SP, McMahon M, Kerr IM, Stark GR. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell. 1984;38:745–755. doi: 10.1016/0092-8674(84)90270-8. [DOI] [PubMed] [Google Scholar]
- 184.Spear GT, Paulnock DM, Jordan RL, Meltzer DM, Merritt JA, et al. Enhancement of monocyte class I and II histocompatibility antigen expression in man by in vivo beta-interferon. Clin Exp Immunol. 1987;69:107–115. [PMC free article] [PubMed] [Google Scholar]
- 185.Borden EC, Rinehart JJ, Storer BE, Trump DL, Paulnock DM, et al. Biological and clinical effects of interferon-beta ser at two doses. J Interferon Res. 1990;10:559–570. doi: 10.1089/jir.1990.10.559. [DOI] [PubMed] [Google Scholar]
- 186.Guadagni F, Schlom J, Johnston WW, Szpak CA, Goldstein D, et al. Selective interferon-induced enhancement of tumor-associated antigens on a spectrum of freshly isolated human adenocarcinoma cells. J Natl Cancer Inst. 1989;81:502–512. doi: 10.1093/jnci/81.7.502. [DOI] [PubMed] [Google Scholar]
- 187.Hance KW, Rogers CJ, Zaharoff DA, Canter D, Schlom J, et al. The antitumor and immunoadjuvant effects of IFN-alpha in combination with recombinant poxvirus vaccines. Clin Cancer Res. 2009;15:2387–2396. doi: 10.1158/1078-0432.CCR-08-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang T, Huang C, Lopez-Coral A, Slentz-Kesler KA, Xiao M, et al. K12/SECTM1, an interferon-gamma regulated molecule, synergizes with CD28 to costimulate human T cell proliferation. J Leukoc Biol. 2012;91:449–459. doi: 10.1189/jlb.1011498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cole KE, Strick CA, Paradis TJ, Ogborne KT, Loetscher M, et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med. 1998;187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89:207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Marfaing-Koka A, Devergne O, Gorgone G, Portier A, Schall TJ, et al. Regulation of the production of the RANTES chemokine by endothelial cells. Synergistic induction by IFN-gamma plus TNF-alpha and inhibition by IL-4 and IL-13. J Immunol. 1995;154:1870–1878. [PubMed] [Google Scholar]
- 192.Mikhak Z, Fleming CM, Medoff BD, Thomas SY, Tager AM, et al. STAT1 in peripheral tissue differentially regulates homing of antigen-specific Th1 and Th2 cells. J Immunol. 2006;176:4959–4967. doi: 10.4049/jimmunol.176.8.4959. [DOI] [PubMed] [Google Scholar]
- 193.Frasch SC, Henson PM, Kailey JM, Richter DA, Janes MS, et al. Regulation of phospholipid scramblase activity during apoptosis and cell activation by protein kinase Cdelta. J Biol Chem. 2000;275:23065–23073. doi: 10.1074/jbc.M003116200. [DOI] [PubMed] [Google Scholar]
- 194.Silverman RH, Halloum A, Zhou A, Dong B, Al-Zoghaibi F, et al. Suppression of ovarian carcinoma cell growth in vivo by the interferon-inducible plasma membrane protein, phospholipid scramblase 1. Cancer Res. 2002;62:397–402. [PubMed] [Google Scholar]
- 195.Zhou Q, Zhao J, Al-Zoghaibi F, Zhou A, Wiedmer T, et al. Transcriptional control of the human plasma membrane phospholipid scramblase 1 gene is mediated by interferon-alpha. Blood. 2000;95:2593–2599. [PubMed] [Google Scholar]
- 196.D’Cunha J, Knight E, Jr, Haas AL, Truitt RL, Borden EC. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci U S A. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Campbell JA, Lenschow DJ. Emerging roles for immunomodulatory functions of free ISG15. J Interferon Cytokine Res. 2013;33:728–738. doi: 10.1089/jir.2013.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Giannakopoulos NV, Luo JK, Papov V, Zou W, Lenschow DJ, et al. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem Biophys Res Commun. 2005;336:496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- 200.Padovan E, Terracciano L, Certa U, Jacobs B, Reschner A, et al. Interferon stimulated gene 15 constitutively produced by melanoma cells induces e-cadherin expression on human dendritic cells. Cancer Res. 2002;62:3453–3458. [PubMed] [Google Scholar]
- 201.Cong XL, Lo MC, Reuter BA, Yan M, Fan JB, et al. Usp18 promotes conventional CD11b+ dendritic cell development. J Immunol. 2012;188:4776–4781. doi: 10.4049/jimmunol.1101609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ritchie KJ, Hahn CS, Kim KI, Yan M, Rosario D, et al. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10:1374–1378. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- 203.Yan M, Luo JK, Ritchie KJ, Sakai I, Takeuchi K, et al. Ubp43 regulates BCR-ABL leukemogenesis via the type 1 interferon receptor signaling. Blood. 2007;110:305–312. doi: 10.1182/blood-2006-07-033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang D, Zhang DE. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res. 2011;31:119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Potu H, Sgorbissa A, Brancolini C. Identification of USP18 as an important regulator of the susceptibility to IFN-alpha and drug-induced apoptosis. Cancer Res. 2010;70:655–665. doi: 10.1158/0008-5472.CAN-09-1942. [DOI] [PubMed] [Google Scholar]
- 206.Skaug B, Chen ZJ. Emerging role of ISG15 in antiviral immunity. Cell. 2010;143:187–190. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Guo B, Chang EY, Cheng G. The type I IFN induction pathway constrains Th17-mediated autoimmune inflammation in mice. J Clin Invest. 2008;118:1680–1690. doi: 10.1172/JCI33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Henry T, Kirimanjeswara GS, Ruby T, Jones JW, Peng K, et al. Type I IFN signaling constrains IL-17A/F secretion by gammadelta T cells during bacterial infections. J Immunol. 2010;184:3755–3767. doi: 10.4049/jimmunol.0902065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mundy-Bosse BL, Lesinski GB, Jaime-Ramirez AC, Benninger K, Khan M, et al. Myeloid-derived suppressor cell inhibition of the IFN response in tumor-bearing mice. Cancer Res. 2011;71:5101–5110. doi: 10.1158/0008-5472.CAN-10-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Cho HY, Lee SW, Seo SK, Choi IW, Choi I. Interferon-sensitive response element (ISRE) is mainly responsible for IFN-alpha-induced upregulation of programmed death-1 (PD-1) in macrophages. Biochim Biophys Acta. 2008;1779:811–819. doi: 10.1016/j.bbagrm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 211.Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274) FEBS Lett. 2006;580:755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 212.Muhlbauer M, Fleck M, Schutz C, Weiss T, Froh M, et al. PD-L1 is induced in hepatocytes by viral infection and by interferon-alpha and -gamma and mediates T cell apoptosis. J Hepatol. 2006;45:520–528. doi: 10.1016/j.jhep.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 213.Peng W, Liu C, Xu C, Lou Y, Chen J, et al. PD-1 blockade enhances T-cell migration to tumors by elevating IFN-gamma inducible chemokines. Cancer Res. 2012;72:5209–5218. doi: 10.1158/0008-5472.CAN-12-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sidky YA, Borden EC. Inhibition of angiogenesis by interferons: effects on tumor- and lymphocyte-induced vascular responses. Cancer Res. 1987;47:5155–5161. [PubMed] [Google Scholar]
- 215.Dinney CP, Bielenberg DR, Perrotte P, Reich R, Eve BY, et al. Inhibition of basic fibroblast growth factor expression, angiogenesis, and growth of human bladder carcinoma in mice by systemic interferon-alpha administration. Cancer Res. 1998;58:808–814. [PubMed] [Google Scholar]
- 216.McCarty MF, Bielenberg D, Donawho C, Bucana CD, Fidler IJ. Evidence for the causal role of endogenous interferon-alpha/beta in the regulation of angiogenesis, tumorigenicity, and metastasis of cutaneous neoplasms. Clin Exp Metastasis. 2002;19:609–615. doi: 10.1023/a:1020923326441. [DOI] [PubMed] [Google Scholar]
- 217.Hiroi M, Mori K, Sakaeda Y, Shimada J, Ohmori Y. STAT1 represses hypoxia-inducible factor-1-mediated transcription. Biochem Biophys Res Commun. 2009;387:806–810. doi: 10.1016/j.bbrc.2009.07.138. [DOI] [PubMed] [Google Scholar]
- 218.von Marschall Z, Scholz A, Cramer T, Schafer G, Schirner M, et al. Effects of interferon alpha on vascular endothelial growth factor gene transcription and tumor angiogenesis. J Natl Cancer Inst. 2003;95:437–448. doi: 10.1093/jnci/95.6.437. [DOI] [PubMed] [Google Scholar]
- 219.Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, et al. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–919. doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yurkovetsky ZR, Kirkwood JM, Edington HD, Marrangoni AM, Velikokhatnaya L, et al. Multiplex analysis of serum cytokines in melanoma patients treated with interferon-alpha2b. Clin Cancer Res. 2007;13:2422–2428. doi: 10.1158/1078-0432.CCR-06-1805. [DOI] [PubMed] [Google Scholar]
- 221.Reznikov LL, Puren AJ, Fantuzzi G, Muhl H, Shapiro L, et al. Spontaneous and inducible cytokine responses in healthy humans receiving a single dose of IFN-alpha2b: increased production of interleukin-1 receptor antagonist and suppression of IL-1-induced IL-8. J Interferon Cytokine Res. 1998;18:897–903. doi: 10.1089/jir.1998.18.897. [DOI] [PubMed] [Google Scholar]
- 222.Singh RK, Gutman M, Llansa N, Fidler IJ. Interferon-beta prevents the upregulation of interleukin-8 expression in human melanoma cells. J Interferon Cytokine Res. 1996;16:577–584. doi: 10.1089/jir.1996.16.577. [DOI] [PubMed] [Google Scholar]
- 223.Guenzi E, Topolt K, Cornali E, Lubeseder-Martellato C, Jorg A, et al. The helical domain of GBP-1 mediates the inhibition of endothelial cell proliferation by inflammatory cytokines. EMBO J. 2001;20:5568–5577. doi: 10.1093/emboj/20.20.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Guenzi E, Topolt K, Lubeseder-Martellato C, Jorg A, Naschberger E, et al. The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J. 2003;22:3772–3782. doi: 10.1093/emboj/cdg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Vestal DJ, Jeyaratnam JA. The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J Interferon Cytokine Res. 2011;31:89–97. doi: 10.1089/jir.2010.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Raffaella R, Gioia D, De Andrea M, Cappello P, Giovarelli M, et al. The interferon-inducible IFI16 gene inhibits tube morphogenesis and proliferation of primary, but not HPV16 E6/E7-immortalized human endothelial cells. Exp Cell Res. 2004;293:331–345. doi: 10.1016/j.yexcr.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 227.Cheng X, Liu Y, Chu H, Kao HY. Promyelocytic leukemia protein (PML) regulates endothelial cell network formation and migration in response to tumor necrosis factor alpha (TNFalpha) and interferon alpha (IFNalpha) J Biol Chem. 2012;287:23356–23367. doi: 10.1074/jbc.M112.340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Taylor KL, Leaman DW, Grane R, Mechti N, Borden EC, et al. Identification of interferon-beta-stimulated genes that inhibit angiogenesis in vitro. J Interferon Cytokine Res. 2008;28:733–740. doi: 10.1089/jir.2008.0030. [DOI] [PubMed] [Google Scholar]
- 229.Jablonska J, Leschner S, Westphal K, Lienenklaus S, Weiss S. Neutrophils responsive to endogenous IFN-beta regulate tumor angiogenesis and growth in a mouse tumor model. J Clin Invest. 2010;120:1151–1164. doi: 10.1172/JCI37223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Mushinski JF, Nguyen P, Stevens LM, Khanna C, Lee S, et al. Inhibition of tumor cell motility by the interferon-inducible GTPase MxA. J Biol Chem. 2009;284:15206–15214. doi: 10.1074/jbc.M806324200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Katsoulidis E, Mavrommatis E, Woodard J, Shields MA, Sassano A, et al. Role of interferon {alpha} (IFN{alpha})-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J Biol Chem. 2010;285:40333–40341. doi: 10.1074/jbc.M110.151076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lai KC, Liu CJ, Chang KW, Lee TC. Depleting IFIT2 mediates atypical PKC signaling to enhance the migration and metastatic activity of oral squamous cell carcinoma cells. Oncogene. 2013;32:3686–3697. doi: 10.1038/onc.2012.384. [DOI] [PubMed] [Google Scholar]
- 233.Chawla-Sarkar M, Lindner DJ, Liu YF, Williams BR, Sen GC, et al. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003;8:237–249. doi: 10.1023/a:1023668705040. [DOI] [PubMed] [Google Scholar]
- 234.Kotredes KP, Gamero AM. Interferons as inducers of apoptosis in malignant cells. J Interferon Cytokine Res. 2013;33:162–170. doi: 10.1089/jir.2012.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Miller JC, Ma Y, Bian J, Sheehan KC, Zachary JF, et al. A critical role for type I IFN in arthritis development following Borrelia burgdorferi infection of mice. J Immunol. 2008;181:8492–8503. doi: 10.4049/jimmunol.181.12.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mesa RA, Yasothan U, Kirkpatrick P. Ruxolitinib. Nat Rev Drug Discov. 2012;11:103–104. doi: 10.1038/nrd3652. [DOI] [PubMed] [Google Scholar]