Cyclic electron flow is essential for survival under high light and anoxic conditions.
Abstract
To investigate the functional importance of Proton Gradient Regulation5-Like1 (PGRL1) for photosynthetic performances in the moss Physcomitrella patens, we generated a pgrl1 knockout mutant. Functional analysis revealed diminished nonphotochemical quenching (NPQ) as well as decreased capacity for cyclic electron flow (CEF) in pgrl1. Under anoxia, where CEF is induced, quantitative proteomics evidenced severe down-regulation of photosystems but up-regulation of the chloroplast NADH dehydrogenase complex, plastocyanin, and Ca2+ sensors in the mutant, indicating that the absence of PGRL1 triggered a mechanism compensatory for diminished CEF. On the other hand, proteins required for NPQ, such as light-harvesting complex stress-related protein1 (LHCSR1), violaxanthin de-epoxidase, and PSII subunit S, remained stable. To further investigate the interrelation between CEF and NPQ, we generated a pgrl1 npq4 double mutant in the green alga Chlamydomonas reinhardtii lacking both PGRL1 and LHCSR3 expression. Phenotypic comparative analyses of this double mutant, together with the single knockout strains and with the P. patens pgrl1, demonstrated that PGRL1 is crucial for acclimation to high light and anoxia in both organisms. Moreover, the data generated for the C. reinhardtii double mutant clearly showed a complementary role of PGRL1 and LHCSR3 in managing high light stress response. We conclude that both proteins are needed for photoprotection and for survival under low oxygen, underpinning a tight link between CEF and NPQ in oxygenic photosynthesis. Given the complementarity of the energy-dependent component of NPQ (qE) and PGRL1-mediated CEF, we suggest that PGRL1 is a capacitor linked to the evolution of the PSII subunit S-dependent qE in terrestrial plants.
The conversion of solar energy into chemical energy and building material by oxygenic photosynthesis, as performed by plants, green algae, and cyanobacteria, supports much of the life on our planet. The production of oxygen and the assimilation of carbon dioxide into organic matter determines, to a large extent, the composition of our atmosphere. Plant photosynthesis is achieved thanks to a series of reactions that occur mainly in the chloroplast, resulting in light-dependent water oxidation, NADP+ reduction, and ATP formation (Whatley et al., 1963). Two separate photosystems (PSI and PSII) and an ATP synthase (ATPase) embedded in the thylakoid membrane catalyze these reactions. The ATPase produces ATP at the expense of the proton motive force that is generated by the light reactions (Mitchell, 1961). The cytochrome (cyt) b6f complex assures the link between the two photosystems by transferring electrons from the membrane-bound plastoquinone to a soluble carrier, plastocyanin, or cyt c6 and functions in the pumping of protons. NADPH and ATP that are produced by linear electron flow from PSII to PSI are fueled into the Calvin Benson Bassham cycle (Bassham et al., 1950) to fix CO2. In parallel, cyclic electron flow (CEF) between the cyt b6f complex and PSI may occur, which would solely lead to the production of ATP. CEF around PSI has been first recognized by Arnon (1959) and is involved in the reequilibration of the ATP poise and prevention of overreduction of the PSI acceptor side (Alric, 2010; Peltier et al., 2010; Leister and Shikanai, 2013; Shikanai, 2014). In microalgae and vascular plants, CEF operates via an NAD(P)H dehydrogenase-like complex (NDH)-dependent and/or PROTON GRADIENT REGULATION5 (PGR5)-related pathway (Alric, 2010; Peltier et al., 2010; Leister and Shikanai, 2013; Shikanai, 2014). The thylakoid protein Proton Gradient Regulation5-Like1 (PGRL1; DalCorso et al., 2008) has been first discovered as a novel component for the PGR5-dependent CEF pathway in Arabidopsis (Arabidopsis thaliana), as its knockout causes a PGR5-like photosynthetic phenotype and is suggested to operate as a ferredoxin-plastoquinone reductase (Hertle et al., 2013). PGRL1 is also important for efficient CEF in the green alga Chlamydomonas reinhardtii, which becomes particularly evident under settings where CEF is induced, such as in acclimation to iron deficiency, high light (HL), or anaerobic growth conditions (Petroutsos et al., 2009; Iwai et al., 2010; Tolleter et al., 2011; Terashima et al., 2012). Remarkably, a CEF protein supercomplex composed of PSI-light-harvesting complex I (LHCI), LHCII, the cyt b6f complex, ferredoxin-NADPH oxidoreductase, and PGRL1 was isolated from state 2 conditions (Iwai et al., 2010). Under anaerobic conditions, the Ca2+ sensor CAS and Anaerobic response1 (ANR1) were shown to interact with PGRL1 in vivo (Terashima et al., 2012) and were found to be associated with the C. reinhardtii CEF supercomplex. Consistently, depletion of CAS and ANR1 by artificial microRNA expression in C. reinhardtii resulted in strong inhibition of CEF under anoxia, which could be partially rescued by an increase in the extracellular Ca2+ concentration, inferring that CEF is Ca2+ dependent (Terashima et al., 2012). Notably, the regulation of the proton motive force by a two-pore potassium channel in the thylakoid membrane of Arabidopsis (AtTPK3), is also Ca2+ dependent (Carraretto et al., 2013), suggesting that Ca2+-dependent activation of CEF and the channel may work hand in hand.
qE, the energy-dependent component of nonphotochemical quenching (NPQ) that occurs due to thermal dissipation of excess absorbed light energy (Li et al., 2000; Peers et al., 2009), is dependent on rapid luminal acidification upon photosynthetic electron transfer (Wraight and Crofts, 1970; Li et al., 2000). Thus, processes such as CEF that contribute to the pH gradient across the thylakoid membrane are interrelated to NPQ, as an acidified lumen is required for efficient qE (Joliot and Finazzi, 2010). In vascular plants, PSII subunit S (PSBS) is essential for efficient qE (Li et al., 2000), whereas qE induction in the green alga C. reinhardtii is mediated by light-harvesting complex stress-related protein3 (LHCSR3), an ancient light-harvesting protein that is missing in vascular plants (Peers et al., 2009). The moss Physcomitrella patens, which possesses genes encoding for PSBS and LHCSR proteins, utilizes both types of regulatory proteins to operate qE (Alboresi et al., 2010), suggesting that land plants evolved a novel PSBS-dependent qE mechanism before losing the ancestral LHCSR-dependent qE found in algae. This makes mosses a very interesting subject for investigating the interrelation and evolution of the CEF and NPQ molecular effectors.
Mosses diverged from vascular plants early after land colonization and are one of the oldest groups of land plants present on earth. This places the moss model system P. patens (Rensing et al., 2008) evolutionarily in the middle between algae and vascular plants and makes it an ideal model organism for the study of the evolution of photosynthetic organisms. Analysis of photosynthesis in P. patens can provide insights into the events leading to adaptation to the harsher physicochemical conditions of the terrestrial environment (Rensing et al., 2008), as evidenced by the presence and functional overlap of LHCSRs and PSBS (Alboresi et al., 2010).
To obtain insights into the interrelation and evolution of CEF and NPQ, we knocked out the PGRL1 gene from P. patens and analyzed functional phenotypic consequences. Moreover, we compared these phenotypes with phenotypic analyses of C. reinhardtii pgrl1, npq4, and pgrl1 npq4 single and double mutants lacking PGRL1, LHCSR3, and both PGRL1 and LHCSR3, respectively. The data provided strong evidence that the green cut protein PGRL1 (Karpowicz et al., 2011) is required for acclimation to anoxia both in algae and mosses. Moreover, an involvement of PGRL1 in the evolution of PSBS-dependent qE in terrestrial plants is implied.
RESULTS
Generation, Phenotype, and Photosynthetic Activity of P. patens pgrl1 Knockout Mutants
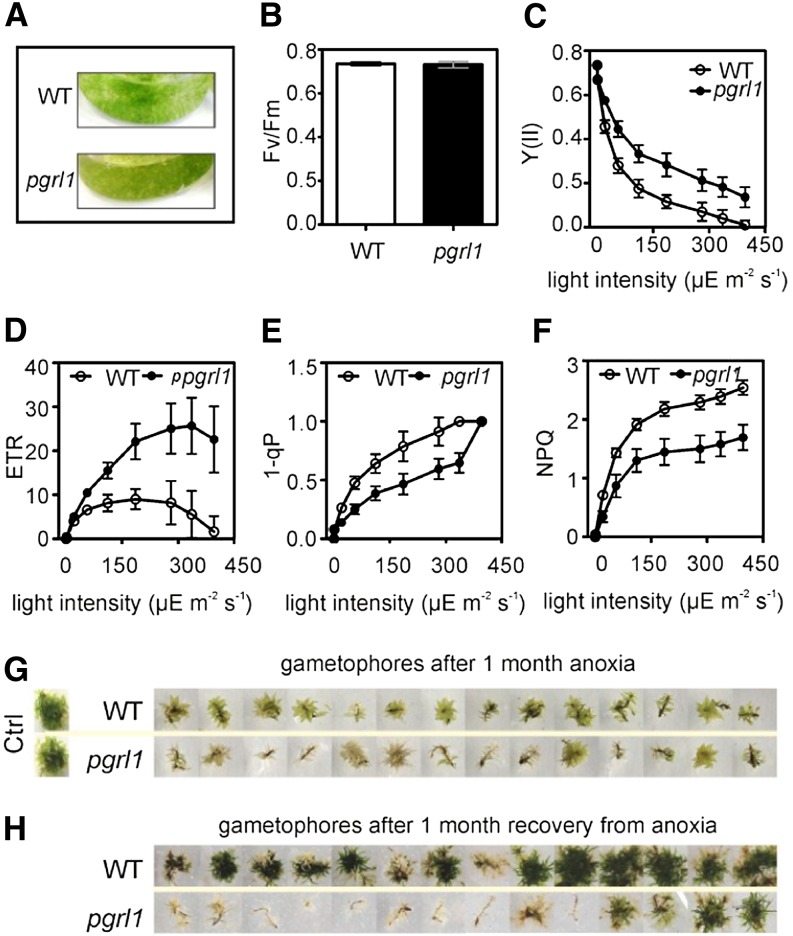
To investigate the functional properties of PGRL1 in P. patens, we took advantage of gene knockout by homologous recombination (Supplemental Fig. S1). In the model plant Arabidopsis, the lack of PGRL1 leads to slightly impaired growth and pale-green leaves (DalCorso et al., 2008). We compared the phenotype and growth rate of P. patens protonema and their emerging gametophores in the wild type and pgrl1. Moss growth under optimal conditions in liquid or solid basal KNOP media did not show major deviation in the morphology and growth rate of protonemal filament between the two genotypes (Supplemental Fig. S2). In pgrl1, chloronemal apical cells developed well into caulonemal cells and did not show differences in length and chlorosis compared with the wild type (Supplemental Fig. S2A). The side branches from some subapical cells of caulonemal filaments developed into leafy shoots, the gametophores (Supplemental Fig. S2B), with no major differences in leaf form and rhizoid length (Supplemental Fig. S2, C and D). Similarly, no differences in development of the protonema were observed between the two genotypes grown in liquid culture (Supplemental Fig. S2E). To assess functional consequences of the absence of PGRL1, chlorophyll fluorescence measurements were performed on protonema and gametophores of the P. patens wild type and pgrl1. Under optimal growth conditions, NPQ was induced to the same extent in both genotypes and life stages by increasing light intensity (Supplemental Fig. S3). However, when grown in basal KNOP media to a high-density culture for 1 month, pgrl1 showed browning of the tissue compared with the wild type (Fig. 1A). When measuring the key photosynthetic parameters in the P. patens pgrl1, maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) values did not differ significantly from those of the wild type (Fig. 1B). On the other hand, pgrl1 PSII quantum efficiency [Y(II)] values were higher than wild-type ones at increasing light intensities (Fig. 1C). Moreover, higher electron transfer rates (ETRs) at saturating light were observed in pgrl1 (Fig. 1, D–F), as already noticed in C. reinhardtii (Tolleter et al., 2011). The redox state of the primary electron-accepting plastoquinone of PSII fraction and NPQ increased with rising light intensities both in the wild type and pgrl1, although we observed lower absolute values for pgrl1 (Fig. 1, E and F). Taken together, these results suggest that PGRL1 modulates photosynthetic acclimation responses to high-density culture conditions in P. patens.
Figure 1.
Key photosynthetic parameters and phenotype of the P. patens wild type (WT) and pgrl1 grown in high-density cultures after 1-month anoxia and followed by 1-month recovery from anoxic treatment. A, Phenotype of P. patens protonema grown in liquid high-density culture. B, Fv/Fm measured in dark-adapted protonema tissue (C–F). Irradiance dependency of Y(II; C), ETR (D), 1-qP (E), and NPQ (F) in protonema. Each data set represents the mean of five independent measurements ± sd. Differences between pgrl1 and wild-type values (C–F) are statistically significant according to Student’s t test (P < 0.0001). G, Gametophore phenotypes after 1-month anaerobic growth. Control (Ctrl) plants were grown for 1 month under aerobic conditions. H, Gametophore phenotypes after 1-month recovery from the anaerobic treatment. The results shown are representative of three different biological replicates, each performed in technical triplicates.
The Anaerobic Response of the P. patens pgrl1 Is Compromised
Although cultures were subjected to shaking, high-density cultivation is known to lead to anoxia, a growth condition that induces CEF in green algae (Tolleter et al., 2011). Thus, we assumed that the phenotype observed in high-density cultures was an indirect response to hypoxic/anoxic conditions in the P. patens pgrl1.
To test whether the anaerobic growth response was altered in anaerobically treated plants, we grew P. patens gametophores on solid basal KNOP media in anaerobic bags for 1 month to induce long-term anaerobiosis (AN; Fig. 1G). After one month of anaerobic treatment, 85% of the pgrl1 and only 7% of the wild-type gametophores showed extensive bleaching (Fig. 1G). Right after 30 d of AN, wild-type and pgrl1 gametophores were shifted back to optimal aerobic growth conditions. Interestingly, 92% of the wild type but only 14% of the pgrl1 gametophores were able to recover (Fig. 1H). These values were somehow expected when considering the massive chlorosis observed for pgrl1 during the anaerobic treatment (Fig. 1G). All gametophores, which were whitish, did not recover from the stress conditions. During the recovery period, the survived mutants showed slower growth compared with the wild type. This clearly supports our hypothesis that PGRL1 plays a role in anaerobic responses in P. patens.
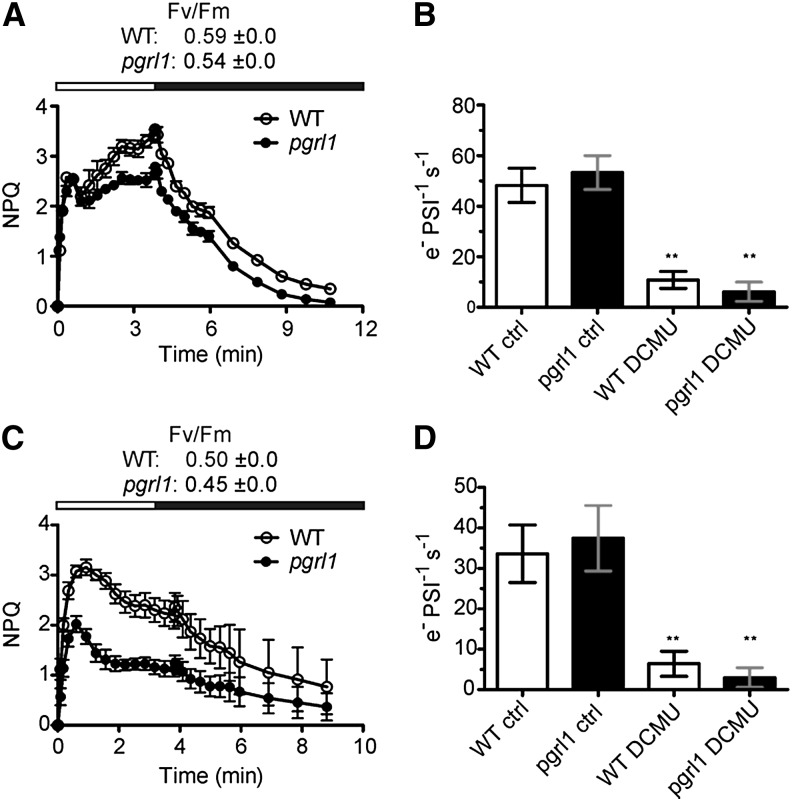
To further investigate the role of PGRL1 in photosynthetic acclimation to AN, short-term experiments were performed. These data showed that in P. patens pgrl1 protonema, NPQ measured during light-to-dark transition after short-term AN (7 h bubbling with argon in presence of light) is diminished compared with the wild type (Fig. 2A). Accordingly, measurement of the CEF activity by following the kinetics of the electrochromic carotenoid band shift relaxation in presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), a technique previously employed to assess this parameter in plants and algae (Joliot and Joliot, 2002; Terashima et al., 2012), revealed CEF capacity in the mutant was significantly reduced by almost 2-fold (Fig. 2B). At the same time, the overall electron flow rate (linear electron flow [LEF] and CEF) was not affected under these growth conditions in the mutant as measured in absence of DCMU (Fig. 2B).
Figure 2.
Time course of NPQ and CEF activity in P. patens wild-type (WT) and pgrl1 mutant protonema after short-term anoxia and HLC treatment. P. patens wild-type and pgrl1 protonema were bubbled with argon for 7 h in the light (A and B) or exposed to HLC conditions (400 µE m–2 s–1, 4°C) for 12 h (C and D). Time course of NPQ was measured in 40-min-dark-adapted tissue of P. patens (A and C). White bars indicate irradiation with light at 700 µE m–2 s–1, whereas black bars indicate darkness. CEF activity measurements were performed by following relaxation of the carotenoid electrochromic band shift at 520 nm in dark-adapted tissues without (ctrl) and after addition of DCMU to the samples (B and D). Each data set represents the mean of three independent measurements ± sd. Asterisks indicate statistically significant differences according to Student’s t test (**P < 0.05).
NPQ Kinetics and CEF Activity in Response to HL Cold in P. patenspgrl1 Protonema Compared with the Wild Type
To test whether PGRL1 is also required to cope with other stress conditions known to induce NPQ and CEF in vascular plants, the pgrl1 mutant and the wild type were challenged with HL cold (HLC) stress (400 µE m–2 s–1 at 4°C; Cornic et al., 2000; Savitch et al., 2011; Ivanov et al., 2012). Seven-day-old protonema liquid cultures of the two genotypes were transferred from optimal to HLC growth conditions. Already after 12 h, distinct photosynthetic stress responses were observed between the two genotypes (Fig. 2, C and D). Similar to what we reported for anaerobic conditions, a 12-h HLC treatment resulted in slightly lower PSII quantum efficiency in pgrl1 (Fv/Fm: 0.45 ± 0.0) compared with the wild type (Fv/Fm: 0.50 ± 0.0; Fig. 2C). As expected, NPQ was reduced in pgrl1, and at the same time, the efficiency of CEF was diminished more than 2-fold as assessed by the measurement of the carotenoid electrochromic band shift relaxation at 520 nm (Fig. 2D).
Proteome Analysis of the P. patens Wild Type and pgrl1 under Short-Term AN
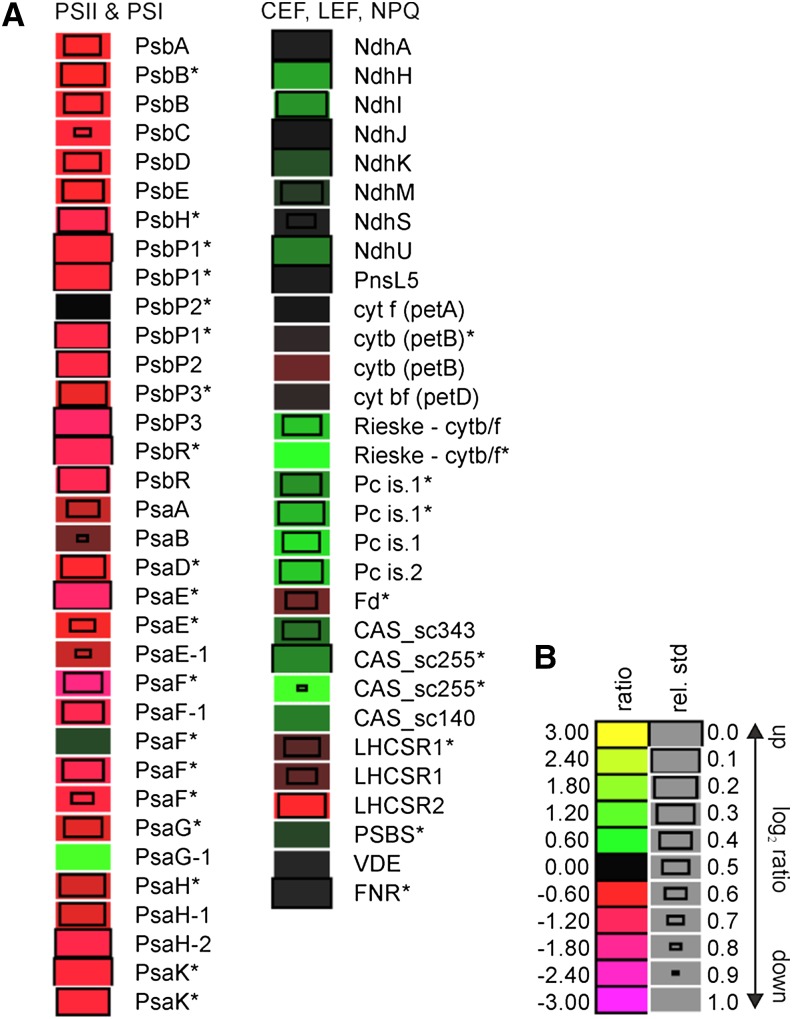
To further characterize the anaerobic response of the P. patens pgrl1 mutant, we took advantage of metabolic (14N/15N) labeling and performed quantitative proteomic analyses. To this end, wild-type protonema was labeled with Ca(15NO3)2, and pgrl1 protonema was labeled with Ca(14NO3)2. The differentially labeled protonema tissues of the wild type and pgrl1 were shifted to short-term anaerobic conditions. After the treatment, tissues from both genotypes were harvested, blotted dry, and quick frozen in liquid nitrogen. Total proteins were extracted from both genotypes, mixed based on equal protein amounts (40 µg from each strain), and fractionated by SDS-PAGE. After Coomassie Brilliant Blue staining, protein bands were excised and digested with trypsin, and peptides were subjected to liquid chromatography coupled with high-resolution mass spectrometry. Two independent biological replicates were analyzed, designated set1 and set2, allowing the identification of 9,461 and 5,893 peptides, respectively (Supplemental Table S1) using X! Tandem (Craig and Beavis, 2004) and Open Mass Spectrometry Search Algorithm (OMSSA; Geer et al., 2004). Posterior error probability of less than 0.05 was ensured using qvality (Käll et al., 2009). Peptide quantification was performed using pyQms (Höhner et al., 2013; Barth et al., 2014) with parameters described elsewhere (Höhner et al., 2013; Barth et al., 2014). Mass spectrometry data handling, storage, retention time alignment, and enhancement were done as described in Barth et al. (2014). Protein amounts of the pgrl1 knockout mutant (labeled with 14N) compared with the wild type (labeled with 15N) in set1 and set2 as such the ratios have been normalized to the median. Overall, more peptides could be quantified in set1 (7,593) compared with set2 (5,880; Supplemental Table S1). A ratio between pgrl1 and the wild type could be calculated for 1,946 proteins. Of those, 23 proteins (including groups) showed an induction of at least 100% (log2 ratio ≥ 1), and 35 proteins were down-regulated by at least 100% (log2 ratio ≤ –1). Please see Supplemental Table S1 for detailed information, sequence coverage, and annotations from PFAM, KEGG, and others. The mean ratios (14N-pgrl1 AN/15N-wild type AN) for PSII and PSI subunits and other photosynthetic proteins are listed in Figure 3. LHCII, LHCI, and ATPase protein are reported in Supplemental Figure S4. Proteins found in only one of the two replicates are marked with asterisks.
Figure 3.
Relative abundance of photosynthetic proteins in the P. patens pgrl1 compared with the wild type after short-term anoxia (AN). Protein expression levels were determined by comparative proteomics approach. After 7-h bubbling with argon, pgrl1 and wild-type protonema were harvested, and total protein was extracted. Equal protein amount (40 µg) from each sample was mixed and analyzed. In this graph, the ratios of the 14N labeled (pgrl1) versus 15N labeled (wild type) are presented. A, Protein ratios shown in the figure are based on two independent experiments (log2 transformed). Asterisks indicate proteins that have been quantified in a group of proteins. A log2 ratio greater than 0 (pgrl1/wild type) indicates an increase (green), a log2 ratio less than 0 indicates a decrease (red) in abundance in pgrl1, and a log2 ratio of zero indicates no changes in protein levels. PSI components were annotated as reported by Busch et al. (2013), whereas, for NDH complex, the annotation of Armbruster et al. (2013) was followed. B, Legend to heat map showing log2 values that correspond to different shades of red and green. Relative sd of a ratio is reflected by the size of the boxes.
Under short-term AN, the abundance of all quantified PSII subunits (PsbA, PsbB, PsbC, PsbD, PsbE -P1|2|3, and PsbR) were decreased in pgrl1 compared with the wild type. P. patens possesses a high number of isoforms for the PSI core subunits (Busch et al., 2013). As observed for PSII core subunits, the reaction center proteins of PSI, PsaA and PsaB, were down-regulated in anaerobic pgrl1 compared with the anaerobic wild type (Fig. 3). Isoforms of the subunits of PSI PsaD1|2|3|4, PsaE1|2, PsaF1|3|4, PsaG1|2, PsaH1|2, and PsaK1|2|3 were reported as a protein group and were down-regulated in the pgrl1 mutant compared with the wild type under AN. An exception was PsaG1. Structurally, PsaG is involved in Lhca1 binding to the PSI complex in pea (Pisum sativum; Amunts et al., 2007).
Isoforms of LHCII major and minor antenna complex proteins behaved differentially, as they were found to be slightly down- or up-regulated, also pointing to remodeling of the antenna (Supplemental Fig. S4). By contrast, most isoforms of LHCI polypeptides were slightly down-regulated in pgrl1 compared with the wild type under AN (Supplemental Fig. S4). Only the P. patens Lhca2.2 was more abundant in the pgrl1 mutant compared with the wild type.
CEF, where electrons are shunted from ferredoxin back to PSI, has been considered to occur via two pathways: (1) the PGRL1/PGR5-dependent pathway or (2) the NDH-dependent pathway (see above). Interestingly, subunits of the NDH complex, NdhH, NdhI, NdhK, NdhM, NdhS, and NdhU exhibited significant up-regulation in pgrl1 compared with the wild type under AN (Fig. 3). Moreover, the subunits NdhA and NdhJ, as well as PnsL5, a subunit of the NDH subcomplex lumenal location in P. patens (Armbruster et al., 2013), appeared to be unchanged in expression in the pgrl1 compared with the wild type under AN. The cyt b6f complex proteins (Photosynthetic Electron Transfer A [PETA], PETB, and PETD) displayed no variance in abundance within the observed error in the pgrl1 relative to the wild type, while the Rieske subunit, the plastocyanin isoforms 1 and 2, and ferredoxin-NADPH oxidoreductase were significantly accumulated to higher levels in pgrl1.
In contrast to other previously analyzed organisms, which contain one gene copy of CAS, the genome of P. patens encodes three CAS gene copies (XP_001782854.1 and XP_001779309.1 published by Weinl et al. [2008], and XP_001771640.1). All three CAS proteins were identified in this experiment, providing the first evidence that the three PpCAS genes are expressed in P. patens. The abundance of the CAS isoforms was slightly increased in pgrl1 compared with the wild type under AN (Fig. 3).
PSBS or LHCSR proteins are involved in the generation of qE (Li et al., 2000; Peers et al., 2009) in higher plants or green algae, respectively. P. patens uses both types of regulatory proteins to operate NPQ (Alboresi et al., 2010). PSBS remained constant in the pgrl1 mutant when compared with the wild type under AN (Fig. 3). Equally, LHCSR1 abundance did not significantly differ between genotypes. On the other hand, LHCSR2 exhibited down-regulation in pgrl1 compared with the wild type. In P. patens, zeaxanthin has a major influence on LHCSR-dependent NPQ (Pinnola et al., 2013). The accumulation of the key enzyme of the xanthophyll cycle, namely violaxanthin de-epoxidase (VDE), did not change in expression within the observed error in the P. patens pgrl1 when compared with the wild type. Among the subunits of chloroplastic ATPase complex, the CF0a, CF0b, and CF0c subunits exhibited lower relative abundance in pgrl1 (Supplemental Fig. S4). CF1α and CF1β were slightly down-regulated in pgrl1 compared with the wild type, respectively.
The Anaerobic Growth Phenotype Is Confirmed by a C. reinhardtii pgrl1 Knockout Mutant
The quantitative proteomic analyses of wild-type and pgrl1 responses to short-term anaerobic conditions revealed significant differences between the proteomes of pgrl1 and the wild type. It is likely that those changes are due to different acclimation responses and/or already caused by stress-dependent protein degradation, giving a possible explanation for the severe growth phenotype of pgrl1 in long-term acclimation. The proteomic data revealed that, in the P. patens pgrl1 mutant, PSI and PSII are decreased, whereas components of the NDH-dependent CEF machinery are up-regulated or remain unchanged under anoxia. At the same time, PSBS and LHCSR1 are not changed in abundance, whereas LHCSR2 expression level is decreased. Yet, such protein expression changes do not allow long-term acclimation to anoxic growth conditions, suggesting a crucial role of PGRL1 in this process.
Taken together, these data indicate that CEF and NPQ are functionally linked in P. patens. This strict relationship has been already proposed in C. reinhardtii. In Arabidopsis, regulation of NPQ by ATPases was reported (Avenson et al., 2005); yet, a tight link between CEF and NPQ could not be ruled out. Moreover, our data also indicate that PGRL1-dependent CEF is essential for low-oxygen stress responses, in agreement with previous suggestion for the alga. It appears therefore that a parallel can be traced between the photosynthetic acclimation responses of these two organisms. To test this hypothesis, we reinvestigated the physiological connection between NPQ and CEF by constructing a pgrl1 npq4 double mutant (Supplemental Fig. S5) by crossing the pgrl1 mutant (Tolleter et al., 2011) with the npq4 mutant that lacks the LHCSR3 genes and therefore shows no qE component of the NPQ (Peers et al., 2009).
To minimize genetic background effects and allow a more reliable comparison, the original pgrl1 (Tolleter et al., 2011) and npq4 (Peers et al., 2009) mutants were first backcrossed several times to the reference wild-type strain CC124 (or CC125) and then crossed together. The insertion of the aminoglycoside phosphotransferase AphVIII gene disruption cassette and the lack of the LHCSR3.1 gene were confirmed by PCR for both parental and backcrossed pgrl1 and npq4, respectively. To check for changes in protein amounts, strains were first grown under low light (LL; 20 µE m–2 s–1) in Tris-acetate phosphate (TAP) medium until they reached the exponential phase (2–3 × 106 cells mL–1) and then were shifted for 24 h to HL (200 µE m–2 s–1) in a minimal high salt medium (HSM) devoid of acetate. These conditions are known to trigger strong expression of LHCSR3 proteins (Peers et al., 2009). As expected, single mutants lacked either PGRL1 or LHCSR3, and the double mutant showed absence of both proteins. Interestingly, the npq4 mutant displayed increased expression of the PGRL1 protein relative to wild-type levels, whereas LHCSR3 expression in pgrl1 was comparable (Supplemental Fig. S5). NPQ data clearly confirmed that the C. reinhardtii pgrl1 mutant had impaired qE induction, as we observed for the P. patens pgrl1 placed either under anaerobic or HLC conditions. This reduction in NPQ accompanied by comparable LHCSR3 expression levels has already been reported for C. reinhardtii (Tolleter et al., 2011), and it was ascribed to a reduction in PGRL1-dependent CEF, which is required for the generation of a thylakoid transmembrane proton gradient involved in the establishment of qE. However, in contrast to published data (Tolleter et al., 2011), NPQ in pgrl1 cells cultivated for 24 h under HL was first induced but then severely diminished (Supplemental Fig. S5). Interestingly, the pgrl1 npq4 double mutant displayed a massive reduction in NPQ, which was barely detectable after a 24-h HL HSM treatment (Supplemental Figs. S5 and S6). At LL intensities, the amount of oxidizable P700 was almost equal in the wild type, pgrl1, npq4, and pgrl1 npq4. However, the quantity of oxidizable PSI primary donor (P700) was significantly diminished in the pgrl1 and pgrl1 npq4 mutant compared with the wild type and npq4 after 24-h HL, as already reported for the C. reinhardtii pgr5 mutant (Johnson et al., 2014; Supplemental Fig. S6). Notably, the amounts of oxidizable P700 in the double pgrl1 npq4 mutant were more reduced after 24-h HL compared with the level of oxidizable P700 in pgrl1. Thus, the quantity of oxidizable P700 was more susceptible to HL in double pgrl1 npq4 than in the single pgrl1 mutant. The fact that the amounts of oxidizable P700 were significantly diminished in the pgrl1 strain after 24-h HL likely explains the drop in the NPQ trace, as photosynthetic electron transfer and thereby acidification of the lumen (Supplemental Fig. S6) are hampered by restricted availability of electron acceptors at the reducing site of PSI, while photoinhibition of PSI may also contribute.
The impact on NPQ observed in the double mutant demonstrates that in C. reinhardtii, both PGRL1 and LHCSR3 are required and complementary for proper induction of qE under HL stress. Mutants that constitutively accumulate zeaxanthin still require low luminal pH for the induction of the qE (Niyogi, 1999), suggesting that low pH has additional roles in addition to activation of the xanthophyll cycle (Müller et al., 2001). Moreover, the severe diminishment in the amounts of oxidizable special chlorophyll a dimer in the reaction center of PSI (P700) in the double mutant indicates that LHCSR3 is functionally involved in protection of PSI under HL stress.
To test whether a lack of the two proteins impacts photosynthetic performance under anoxia, functional measurements were performed and revealed no significant differences when measuring the contribution of LEF and CEF together in the absence of DCMU. On the other hand, electrochromic carotenoid band shift relaxation in presence of DCMU revealed significantly lower CEF activity for all mutant strains tested, with pgrl1 and pgrl1 npq4 showing higher statistically significant differences than npq4 compared with wild-type levels (Supplemental Fig. S7). Interestingly, pgrl1 and the double mutant showed a comparable reduction in CEF rates, suggesting that the enhanced sensibility observed when the npq4 mutation is added to the pgrl1 background does not depend on CEF.
To substantiate the physiological implications of the observed reductions in NPQ levels, we performed an in-depth screening where the wild type and the two single and the double mutants were subjected to different combinations of light (20, 50, and 200 µE m–2 s–1), carbon sources (TAP versus HSM), and oxygen levels (oxic versus anoxic; Fig. 4).
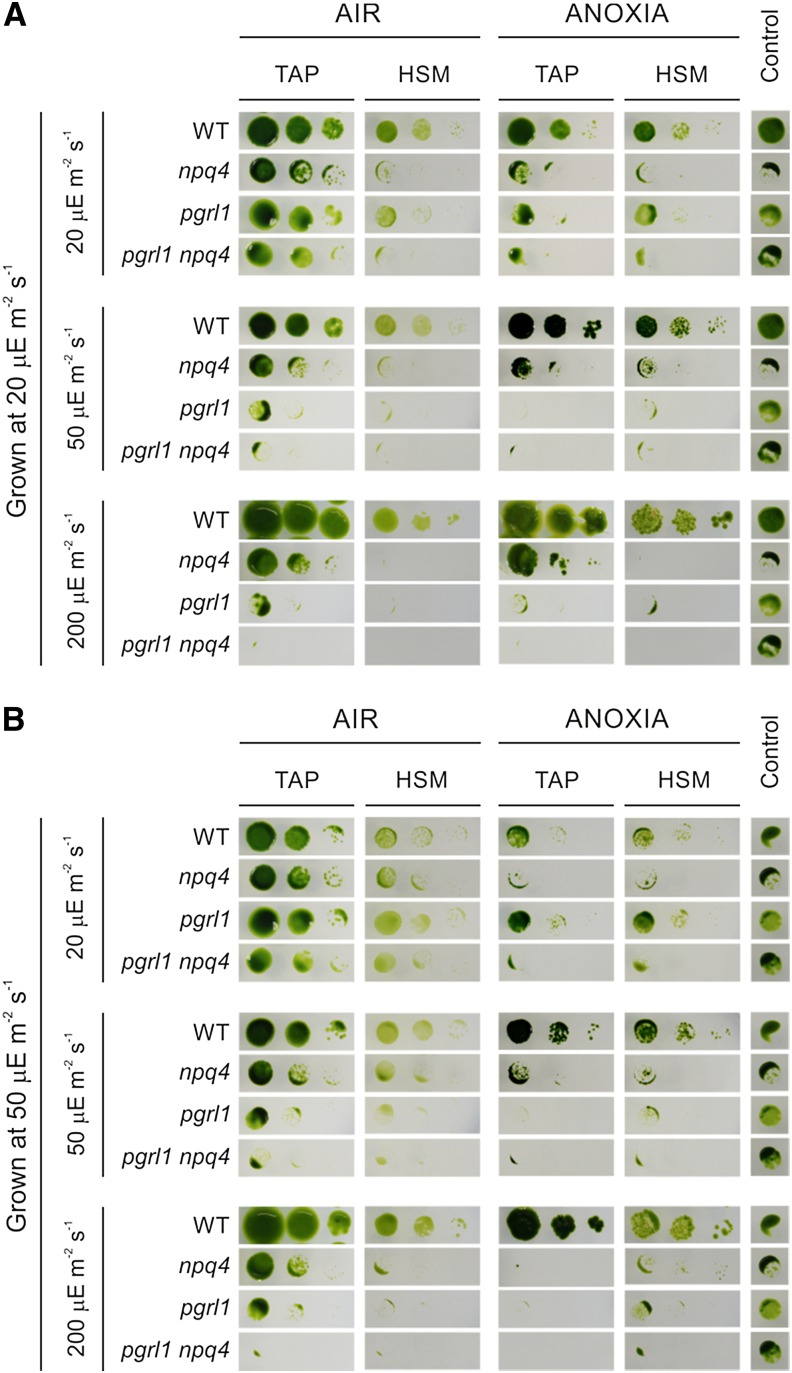
Figure 4.
Anaerobic growth phenotype of pgrl1, npq4, and pgrl1 npq4 mutants in the presence of different carbon and light sources. C. reinhardtii strains were grown in liquid TAP medium under low (A) or normal (B) light conditions until they reached the exponential phase (1.5–3 × 106 cells mL–1). Cells were then counted three times for accuracy, and their concentration was equalized to 1 × 106 cells mL–1). Five microliters of undiluted (1 × 106 cells mL–1) and 1:10 (0.1 × 106 cells mL–1) and 1:100 (0.01 × 106 cells mL–1) dilutions (left to right) were spotted on the surface of TAP or HSM agar plates, allowed to dry, and subsequently incubated under aerobic (AIR) or anoxic (ANOXIA) conditions in the presence of different light intensities (20, 50, or 200 µE m–2 s–1) for 10 to 14 d. As an internal control, undiluted (1 × 106 cells mL–1) cultures of each strain were also allowed to grow heterotrophically in the dark for 3 weeks. Repetition of the experiment with three independent biological replicates yielded essentially identical results. WT, Wild type.
Strains were maintained on solid TAP medium and then transferred to liquid TAP (approximately 50 mL) and grown for 2 to 3 d at 25°C and 20 µE m–2 s–1 (Fig. 4A) or 50 µE m–2 s–1 (Fig. 4B) photosynthetically active radiation constant irradiance. When cultures reached the exponential phase (1.5–3 × 106 cells mL–1), they were equalized to a cell concentration of 1 × 106 cells mL–1. Serial dilutions (1:10 and 1:100) were also included so that every 5-µL spot corresponded to an initial number of 5,000 (undiluted), 500 (1:10), or 50 (1:100) cells, thus allowing the visual monitoring of growth performances over a 2-week period. As a control, undiluted (1 × 106 cells mL–1) cultures were spotted on TAP plates and allowed to grow for 4 weeks in the dark (control; see “Materials and Methods” for a detailed description of the experiment).
Under permissive conditions (i.e. LL TAP growth in air [AIR]), strain performances were overall comparable (Fig. 4). Increasing the light intensities without altering the carbon source (TAP) or the oxygen (AIR) parameters still resulted in evident pgrl1 growth defects, which were exacerbated in the pgrl1 npq4 double mutant. In line with the NPQ data reported in Supplemental Figure S5, a shift from LL TAP to HL HSM under aerobic conditions resulted in growth inhibition for npq4, pgrl1, and pgrl1 npq4. The sensitivity of the npq4 strain when shifted from LL to HL conditions has already been reported (Peers et al., 2009), and in our experiment, it appears to be somehow influenced by the light intensity at which the starting culture was grown, as shown by the higher tolerance of the mutant grown at 50 µE m–2 s−–1 (Fig. 4B).
As in the P. patens pgrl1 (Fig. 1), growth performances of the C. reinhardtii pgrl1 mutant were strongly impaired under anoxia at light intensities equal or higher than 50 μE m–2 s–1, regardless of the carbon source used (Fig. 4). The npq4 strain also displayed a reduction in growth but appears to be more tolerant than pgrl1 at the same light intensities. Notably, lack of both PGRL1 and LHCSR3 proteins results in acute sensitivity to anaerobic conditions, as shown by a strong decrease in the pgrl1 npq4 anoxic growth already at 20 μE m–2 s–1 (Fig. 4).
Before undertaking physiological and molecular analyses, we performed independent HL aerobic and anaerobic growth tests with liquid media (Supplemental Fig. S8). These experiments confirmed that pgrl1 npq4 was most susceptible to the conditions applied.
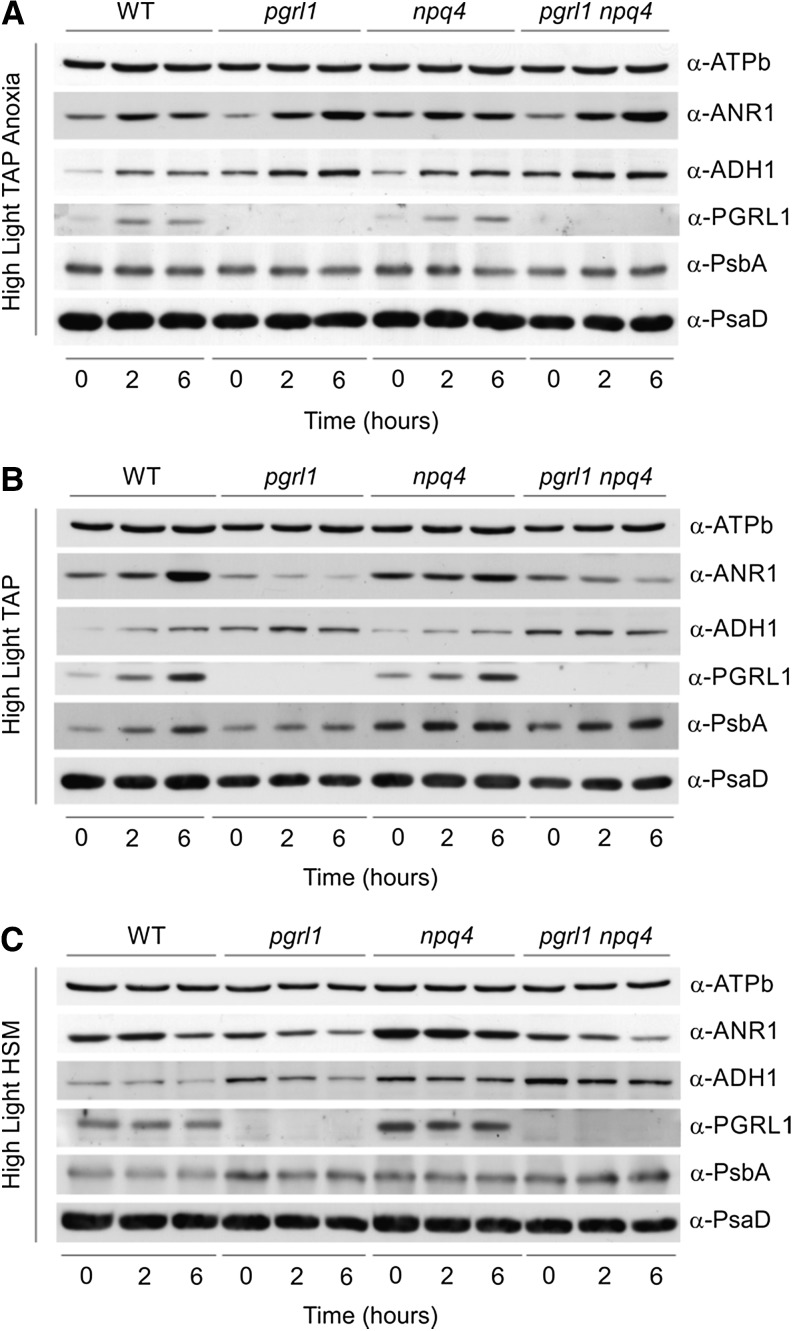
To tackle molecular mechanisms responsible for the different fitness observed in the mutant strains, we assessed short-term responses at the protein level in three different conditions: HL TAP anoxia, HL TAP, and HL HSM (Fig. 5).
Figure 5.
Protein expression in pgrl1, npq4, and pgrl1 npq4 mutant strains under HL TAP anoxia, HL TAP, and HL HSM. C. reinhardtii strains were grown in liquid TAP medium under LL conditions until they reached the exponential phase (1.5–3 × 106 cells mL–1). Cells were then pelleted, resuspended either in TAP (A and B) or HSM (C) at a concentration of 1 × 106 cells mL–1, and exposed to anoxic (A) or aerobic (B and C) conditions in the presence of 200 µE m–2 s–1 light. Cell samples were collected 0, 2, and 6 h from the beginning of the treatment. Total proteins were extracted, and 35 µg was loaded on each lane of a 13% polyacrylamide SDS-PAGE and blotted on nitrocellulose membranes. Levels of expression for ANR1, ADH1, PGRL1, PsbA, and PsaD were assessed by incubation with specific antibodies. α-ATPase subunit b antibodies were used as a loading control. WT, Wild type.
The CEF supercomplex formed under anoxia in C. reinhardtii involves, among others, PGRL1, the Ca2+ sensor CAS, and the newly characterized protein ANR1, whose expression is required for anaerobic growth in the green alga. Interestingly, ANR1 appears to be an algal-specific protein, with orthologs found only in Volvox carteri and Coccomyxa subellipsoidea, both of which possess iron hydrogenase enzymes.
Upon anaerobic conditions, ANR1 expression increased in all strains, with pgrl1 and pgrl1 npq4 showing higher expression levels after 6 h (Fig. 5A). The higher ANR1 expression levels observed in the strains devoid of PGRL1 could be interpreted as a compensatory mechanism aimed at the rescuing of the impaired CEF. Similarly, increased ANR1 and PGRL1 expression levels relative to the wild type are observed already after 4 h of anoxia in npq4, whose CEF rates were shown to be slightly reduced (Fig. 5A; Supplemental Fig. S7). As for ANR1, a minor increase in CAS levels was detected in thylakoids of anoxia-acclimated pgrl1 cells (Supplemental Fig. S9), confirming the P. patens pgrl1 proteomic data (Fig. 3).
Expression of the bifunctional alcohol/acetaldehyde dehydrogenase (ADH1) has been shown to be involved in the control of acclimation responses to anoxia and iron starvation (Magneschi et al., 2012; Höhner et al., 2013). Both the single and double mutant showed increased ADH1 expression throughout the anaerobic treatment relative to wild-type levels. Higher ADH1 expression levels in the double mutant are likely triggered to alleviate the even harsher reducing poise conditions.
DISCUSSION
In this work, we took advantage of homologous recombination in P. patens to generate a pgrl1 knockout mutant and assess the physiological relevance of the protein in the acclimation to environmental stresses in the moss. Moreover, we generated a pgrl1 npq4 double mutant in C. reinhardtii. Together with single PGRL1 and LHCSR3 knockout strains, the double mutant was used for phenotypic and physiological comparative analyses. Our data demonstrate that PGRL1 is crucial for both C. reinhardtii and P. patens acclimation to HL and anaerobic conditions, suggesting that the functional importance of the protein has been conserved during evolution of the green lineage (Karpowicz et al., 2011). Furthermore, the data demonstrate a link between CEF and NPQ required for successful acclimation to HL and anoxic stress. Given the complementarity of CEF and NPQ in C. reinhardtii, we suggest that PGRL1-driven CEF contributed to the evolution of PSBS-dependent qE in the land plants.
PGRL1 Is Required for Survival under Anoxic Conditions in C. reinhardtii and P. patens
As already observed for Arabidopsis (DalCorso et al., 2008) and C. reinhardtii (Petroutsos et al., 2009; Tolleter et al., 2011; Supplemental Fig. S7), the physiological data as well as the electron transfer measurements of CEF obtained for the P. patens pgrl1 knockout mutant (Figs. 1 and 2) clearly indicate that PGRL1 is also required for efficient NPQ and CEF under HLC stress and anoxia in the moss. A novel aspect, however, is the inability of the PGRL1-deficient P. patens and C. reinhardtii lines to survive under anaerobic growth conditions (Figs. 1 and 4). Acclimation to low-oxygen stress is a common strategy during life of the microalga C. reinhardtii and the moss P. patens, as oxygen availability might be frequently restricted by environmental conditions. C. reinhardtii is usually found in eutrophic shallow ponds that are rich in biomass and therefore subjected to periods of anoxia (Peers and Niyogi, 2008). Moreover, P. patens is, due to its small size, frequently subjected to flooding, which will also impair the availability to oxygen. An anoxic phenotype has recently been reported for C. reinhardtii anr1 artificial microRNA knockdown strains (Terashima et al., 2012). As PGRL1, ANR1 is required for efficient CEF under an anaerobic growth regime, and both proteins are part of the same CEF supercomplex (Terashima et al., 2012). Finazzi et al. (2002) showed that CEF was activated under dark anaerobic settings in C. reinhardtii. Activation of CEF is a strategy to overcome a limitation at the PSI acceptor side (Munekage et al., 2002), which proceeds, as low oxygen availability will create a reducing environment in the chloroplast. Activation of CEF will force cycling of electrons around the PSI and cyt b6f complexes and will thereby decrease the flow of electrons into the NADPH pool, acidify the thylakoid lumen, and produce ATP. The luminal acidification, on the other hand, will slow down the oxidation of plastoquinol at the luminal site (Qo) of the cyt b6f complex, which is considered to be the rate-limiting step of linear photosynthetic electron transfer (Stiehl and Witt, 1969). This is consistent with the fact that ETR (Fig. 1) is enhanced when CEF is diminished, as already pointed out by Tolleter et al. (2011). In C. reinhardtii, activation of [FeFe]-hydrogenase isoform1 (HYDA1) is believed to function as an electron valve and control the chloroplast redox poise under oxygen deficiency (Grossman et al., 2011; Hemschemeier and Happe, 2011). As outlined by Mazor et al. (2012), protons could have served as a sink for excess electrons produced by photosynthesis, when oxygenic photosynthesis suffered a shortage of oxidized electron acceptors in early-earth reductive atmosphere. Thus, hydrogenase might have evolved to sustain ATP production by LEF under an anoxic atmosphere, implicating that oxygen produced by PSII is readily consumed by respiration. In this respect, CEF and hydrogenase both drive ATP production. Notably, in contrast to the PGRL1-deficient mutants, the lack of HYDAs in a hydEF1 mutant C. reinhardtii strain (Dubini et al., 2009) did not impact growth under anoxic conditions (Terashima et al., 2012). This mutant utilizes reverse tricarboxylic acid cycle to dispose reducing equivalents via production of succinate under dark anoxia (Dubini et al., 2009). On the contrary, diminishment of CEF by deletion of pgrl1 is deleterious and can apparently not be metabolically compensated, indicating a central role of CEF in cellular bioenergy management under anoxia. Importantly, the quantitative proteomic data obtained from the P. patens wild type and pgrl1 mutant showed an up-regulation of subunits of the chloroplast NDH complex (Fig. 3) involved in NAD(P)H-dependent reduction of the plastoquinone pool. Although there is no proof for chloroplast NDH activity in P. patens, Munekage et al. (2004) demonstrated that the PGR5/PGRL1- and NDH-dependent CEF pathways are complementary and required for efficient photosynthesis in Arabidopsis. Thus, up-regulation of NDH complex subunits in absence of PGRL1 could be interpreted as a compensatory effect underpinning the importance of CEF in P. patens under anoxia. Notably, the amounts of two plastocyanin isoforms increased in the P. patens pgrl1 mutant, while cyt b6f complex subunits remained stable. As described above, electron transfer at the Qo site of the cyt b6f complex has been described as the limiting step of photosynthetic electron transfer (Stiehl and Witt, 1969). Plastocyanin has also been considered a key component that adjusts to control the electron transport rate (Schöttler et al., 2004). Thus, its increase in the absence of pgrl1 points to a necessity to adjust possible limitations of the electron transfer yield, likely to compensate for depletion in ATP caused by diminished CEF in absence of pgrl1. Induction of CEF is observed in mosses under anoxic conditions accompanied by an extensive light-dependent quenching of chlorophyll fluorescence (Heber et al., 2000). Analogous to what was described in algae, CEF and NPQ go hand in hand in mosses. Importantly, a hallmark of the described pgrl1 phenotypes in Arabidopsis and C. reinhardtii and P. patens (DalCorso et al., 2008; Tolleter et al., 2011) is a decreased NPQ, likely due to diminished acidification of the thylakoid lumen. NPQ is diminished, although NDH subunits and plastocyanin are up-regulated in the P. patens pgrl1 mutant, suggesting that in the absence of PGRL1, lumen acidification is diminished, as observed for C. reinhardtii and Arabidopsis (see above). LHCSR proteins are required for efficient qE in C. reinhardtii and P. patens, and mechanistically, their quenching is linked to an acidified thylakoid lumen (Niyogi et al., 1997; Petroutsos et al., 2011; Pinnola et al., 2013). As evidenced by the proteomic data (Fig. 3), VDE was not down-regulated in pgrl1. In addition to LHCSRs and PSBS (Alboresi et al., 2010), VDE is required for qE in the moss, as formation of qE is strictly zeaxanthin dependent (Pinnola et al., 2013). Hence, stable expression of VDE will contribute to formation of qE in pgrl1 and is therefore not responsible for diminished qE in pgrl1 (Figs. 1F and 2). Surprisingly, a reduction in protein amount of LHCSR2 was observed in the P. patens pgrl1 (Fig. 3). Interestingly, this was paralleled by decreased amounts in PSI and PSII proteins, pointing to an overall impact of the PGRL1 knockout on the photosynthetic machinery in P. patens. The different regulation of LHCSR1 and LHCSR2 might indicate functional diversification as suggested for C. reinhardtii LHCSR1 and LHCSR3 (Peers et al., 2009). As seen in P. patens pgrl1, deletion of PGRL1 in C. reinhardtii caused a clear decrease in PSI amounts (Fig. 5; Supplemental Figs. S6 and S9) and diminished the quantity of oxidizable P700. Thus, in addition to the impact on CEF, also seen in the decreased amounts of oxidizable P700, and NPQ, the deletion of pgrl1 changes composition of thylakoid membrane proteins. Furthermore, with an increase in NDH complexes in P. patens, it induces alternative electron transfer pathways similar to what was described for C. reinhardtii, where respiratory protein complexes and respiratory activity (Petroutsos et al., 2009; Tolleter et al., 2011) were induced in the absence of PGRL1. However, none of the described compensatory adaptations in C. reinhardtii (Petroutsos et al., 2009; Tolleter et al., 2011) or P. patens can rescue the described anoxic and HL phenotypes, implying that PGRL1 is required for robustness and buffering of stress.
PGRL1 and LHCSR3 Are Required and Complementary for Proper Induction of qE under HL Stress and Photoprotection as Well as for Survival under Low Oxygen
It is evident that in C. reinhardtii, the lack of LHCSR3 in a pgrl1 background strongly affects HL and anaerobic responses, suggesting that this peculiar component of qE offers an additional layer of protection in the alga. Thus, our data generated for the pgrl1 npq4 C. reinhardtii double mutant clearly shows that PGRL1 and LHCSR3 are required and complementary for proper induction of qE under HL stress. Moreover, the data demonstrate that PGRL1 and LHCSR3 are required and complementary for photoprotection and for survival under low oxygen. Here, it is of note that the quantity of oxidizable P700 is even more diminished in the pgrl1 npq4 double mutant compared with pgrl1 (Supplemental Fig. S6), suggesting that LHCSR3 may have an additional role in protecting PSI. Nonetheless, it is apparent that the loss of PGRL1 has more severe consequences compared with the loss of LHCSR3 (Fig. 4), therefore suggesting that sufficient acidification of the thylakoid lumen, as seen for the P. patens pgrl1 (Figs. 1 and 2), is an essential prerequisite for efficient qE, as previously shown for C. reinhardtii (Petroutsos et al., 2011). The fact that some CEF activity remains in the pgrl1 P. patens and C. reinhardtii mutants strongly suggests that a minimum of CEF activity is required for qE formation and for successful HL acclimation. Thus, CEF and NPQ are interrelated and required for survival of photosynthetic organism in an ever-changing light environment in the presence but also in the absence of oxygen.
PGRL1 Is a Capacitor That Contributes to Phenotypic Evolution of Terrestrial Plants
In evolutionary terms, both PGRL1 and LHCSR proteins are required for robustness under stress. Generally, robustness provides the capacity to accumulate cryptic genetic variation, which can develop into a source providing phenotypic variation and therefore increase the ability to evolve (Waddington, 1957). The release of cryptic genetic variation and its contribution to phenotypic evolution via impairment of robustness is also known as capacitance. A capacitor is a biological switch capable of revealing previously cryptic heritable variation (Rutherford and Lindquist, 1998). The concept of capacitance was strengthened by experiments where hidden genetic variation was released when cellular buffering mechanisms were challenged by stress (for review, see Badyaev, 2005). Accordingly, PGRL1 and LHCSR3 are likely to be capacitors. Remarkably, LHCSR genes were lost from genomes of vascular plants (Koziol et al., 2007), while PGRL1 is conserved during evolution of the green lineage (Karpowicz et al., 2011) and could therefore be considered a capacitor for terrestrial plant evolution. During the evolution of terrestrial plants, land plants evolved a novel PSBS-dependent qE mechanism before losing the ancestral LHCSR-specific mechanism found in algae. In this respect, it is highly interesting that PSBS expression remained stable, while LHCSR2 was down-regulated in the pgrl1 knockout in P. patens. As CEF and NPQ are interrelated, evolution of terrestrial plants may have been driven by their capacity to functionally compensate mutations affecting each of the two mechanisms. In this scenario, evolution of PSBS-dependent qE would have evolved together with PGRL1-driven CEF, as PSBS is kept to balance an impact in PGRL1-driven CEF in the land-colonizing pioneer moss P. patens.
MATERIAL AND METHODS
Plant Material and Culture Conditions
Physcomitrella patens derived from a wild-type specimen collected in Gransden Wood in Huntingdonshire, United Kingdom (Ashton and Cove, 1977) was grown axenically either in liquid or solid KNOP medium (Reski and Abel, 1985) consisting of 250 mg L–1 of KH2PO4, 250 mg L–1 of KCl, 250 mg L–1 of MgSO4 × 7 H2O, 1,000 mg L–1 of Ca(NO3)2 × 4 H2O, and 12.5 mg L–1 of FeSO4 × 7 H2O, pH 5.8; solid medium was supplemented with 0.8% (w/v) agar and, if needed, for protonema, overlaid with a cellophane disc. For metabolic labeling, Ca(14NO3)2 × 4 H2O in solid and liquid media was substituted with Ca(15NO3)2 × 4 H2O (Cambridge Isotope Laboratories). Control plants were cultured in a growth chamber at 25°C ± 1°C under a 16-h/8-h light/dark photoperiod with a light intensity of 60 µE m–2 s–1. For HLC stress tests, gametophores or 7-d-old protonema were shifted from control light conditions to 450 μE m–2 s–1 at 4°C in liquid or on solid KNOP medium. Short-term AN was induced by bubbling the liquid cultures with argon for 7 h in the presence of 60 µE m–2 s–1 light, whereas long-term AN was performed by incubating plants on petri dishes inside an anaerobic bag (GENbox anaer, bioMérieux SA) for 1 month at 60 µE m–2 s–1 light.
Chlamydomonas reinhardtii pgrl1 (Tolleter et al., 2011) and npq4 (Peers et al., 2009) mutants were backcrossed four times with the wild-type strain CC124 (nit2–, mt–) and then mated together to generate the pgrl1 npq4 double mutant. For the screening of crossed progeny, we assessed insertion of the AphVIII resistance marker within the PGRL1 coding sequence with primers AphVIII_Fw1 (5′-TCGGGCCGGAGTGTTCC-3′) and PGRL1_Rev (5′-TTACGCAGCGGCCTTAGCC-3′), producing an amplification product only in pgrl1 strains. For npq4, lack of the genomic LHCSR3.1 gene was verified by primers gLHCSR3.1_Fw (5′-GCACAGCAAAGAACAGCATC-3′) and gLHCSR3.1_Rev (5′-CCTTCAGGTTGTGGTGTCAA-3′), which amplifies part of the LHCSR3.1 gene in the wild type but not in npq4 strains. Backcrossed lines verified by PCR were subsequently analyzed by immunoblot for the absence of the respective protein. Wild-type, pgrl1, npq4, and pgrl1 npq4 strains were maintained on TAP medium, pH 7.0, solidified with 1.2% (w/v) agar at 25°C and 50 µE m–2 s–1 photosynthetically active, constant irradiance. For spot test experiments, cells maintained on solid TAP medium were transferred to liquid TAP medium (approximately 50 mL) and grown for 2 d at 25°C in presence of 20 or 50 µE m–2 s–1 photosynthetically active, constant irradiance. After 2 d, cells typically reaching the exponential phase (1.5–3 × 106 cells mL–1) were counted three times, pelleted at 4,000g for 5 min, and resuspended in TAP at a final concentration of 1 × 106 cells mL–1. Five microliters of these cultures and their 1:10 and 1:100 dilutions were spotted on TAP or HSM plates, allowed to dry, and placed under aerobic or anoxic conditions in the presence of different light intensities (20, 50, or 200 µE m–2 s–1) for 10 to 14 d. Anaerobic bags (GENbag anaer, bioMériuex, 45 534) were used to create an anaerobic environment. As an internal control, undiluted (1 × 106 cells mL–1) cultures of each strain were also allowed to grow aerobically in the dark for 4 weeks on TAP plates. Repetition of the experiment with three independent biological replicates yielded essentially identical results.
For a tolerance test in liquid media, after 2-d growth in liquid TAP at 25°C in presence of 20 µE m–2 s–1 photosynthetically active, constant irradiance, cells were counted, and their concentration was equalized to 0.1 × 106 cells mL–1 (TAP) or 1 × 106 cells mL–1 (HSM). Cultures were allowed to grow at 25°C under aerobic (120-rpm shaking) or anoxic (N2 bubbling) conditions for 2 to 3 d in the presence of 200 µE m–2 s–1 constant irradiance.
Chlorophyll Fluorescence Measurements
Fluorescence was measured at room temperature using a Maxi-Imaging PAM Chlorophyll Fluorometer (Heinz Walz GmbH). Plants were dark adapted for 40 min (20 min for C. reinhardtii strains) at room temperature before each measurement. The effective photochemical quantum yield of PSII was measured as PSII yield [Y(II) = (Fm′ – F)/Fm′] and total NPQ was calculated as (Fm – Fm′)/Fm′. ETR was calculated as I × 0.5 × Y(II), where I is the intensity (in µmols photons m–2 s–1) of the incident light. The variable fluorescence Fv was calculated as Fv = Fm – Fo, and Fv/Fm was used to evaluate the maximum PSII fluorescence in the dark-adapted state. The proportion of PSII centers that are closed, 1 − qP, was calculated as 1 – [Fm – Fm′/(Fm′ - Fo′)], where Fm and Fm′ represent the maximum PSII fluorescence in the dark-adapted state and in any light-adapted state, respectively, and Fo and Fo′ represent the minimum PSII fluorescence in the dark-adapted state and after light exposure, respectively.
Absorption Change Measurements
The relaxation kinetics of the carotenoid electrochromic band shift were measured at 520 nm (Witt, 1979; Joliot and Joliot, 2002) using a pump-and-probe LED-based JTS 10 spectrophotometer (BioLogic). The signal at 520 nm was corrected by subtracting the signal at 546 nm to deconvolute the signal band shift from other contributions due to redox changes of cyt f (Terashima et al., 2012). The measurement was performed in absence and in presence of the PSII inhibitor DCMU with final concentration of 10 μm. Under such conditions, the sustained steady-state electron transfer occurring in the light is attributed to CEF around PSI. Postillumination recovery kinetics at 520 nm were recorded and used to determine CEF rates. Results were expressed as electrons per second per PSI upon normalization to the PSI amount. The amount of PSI was determined from the amplitude of the electrochromic shift signal upon excitation with a saturating single-turnover flash (5-ns laser pulse, provided by an Nd:YAG pumping a dye cell filled with 4-(dicyanomethylene)-2-methyl-6-(4-dimethylaminostyryl)-4H-pyran). The measurements were performed in a cuvette, where liquid protonema was mixed with a buffer (20 mm HEPES-KOH, pH 7.2, 10 mm KCl) containing 20% (w/v) Ficoll to avoid sedimentation during measurements. P700 measurements were conducted using a JTS 10 spectrophotometer (BioLogic) as described by Johnson et al. (2014).
Pppgrl1-Neomycin Phosphotransferase II Disruption Construct
For the P. patens disruption construct, the marker cassette containing P35S-neomycin phosphotransferase II (nptII)-CamVTer was excised from pMBL6 vector (GenBank DQ228131.1) using HindIII. The genomic PpPGRL1 sequence (locus XP_001772047.1) was PCR amplified using gene-specific primers containing specific attachment B sites (forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACGAATAC GGAGAGGGAGTCG-3′; reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTGCACAGTTGTCTGTTCCTCA-3′) required for the cloning with the Gateway Cloning BP Reaction Kit (Invitrogen) and cloned into the destination vector pDONR201. The 1,967-bp fragment of the selection marker cassette P35S-nptII-CamVTer was inserted into a unique HindIII restriction site in the PpPGRL1gene, generating the pBK1 (Pppgrl1-nptII) replacement vector. To increase the efficiency of homologous recombination, pBK1 was digested with ClaI, which cuts once in the vector backbone, prior to P. patens transformation.
Generation and Molecular Characterization of pgrl1 Knockout Mutants in P. patens
Moss transformation was performed using particle bombardment as described in Cho et al. (1999 with minor modifications. Briefly, 4- to 6-d-old protonema were transformed by a helium-driven PDS-1000/He Particle Gun (Bio-Rad) with 7.6 MPa rupture discs (Bio-Rad). For every round of six bombardments, 3 mg of gold particle (Bio-Rad) 0.6 µm in diameter was coated with 5 µg of DNA of the pBK1 disruption construct. Particles were first vortexed with 25 µL of CaCl2 (2.5 m) and 10 µL of spermidine (0.1 m) for 3 min. Afterwards, they were pelleted and washed once with 140 µL of 70% (v/v) ethanol and then with 140 µL of 100% (v/v) ethanol. Washed coated particles were subsequently centrifuged and resuspended in 24 µL of 100% (v/v) ethanol. For each shot, 8 µL of the particle suspension was used. Resistant plant selection started 3 d after transformation by transferring plants on culture media supplemented with the selection marker G418 (50 μg mL–1; Sigma-Aldrich) for 2 weeks. To select for stable transformants, plants were then first placed for 2 weeks onto a medium without antibiotic and subsequently exposed to a second 2-week selection treatment. To confirm integration, DNA was isolated from resistant plants as described in Schween et al. (2002. Several independent PCR reactions were performed on purified genomic DNA using primers: P1 (5′-TCTCATTGCCCTAGGTTTGG-3′) and P2 (5′-TTCAGCCAAAGGGCTCTCTA-3′) to detect the disruption of the wild-type PpPGRL1 gene, P3 (5′-TAAAAAATCAAGTGATGTTATCCA-3′) and P4 (5′-GGCAATGGAATCCGAGGAGGT-3′) to confirm the integration of the transgene at the 5′ end, and P5 (5′-ATTGAACAAGATGGATTGCACGC-3′) and P6 (5′-AGGAACTGAGAGTACATATGGTGA-3′) to approve the integration at the 3′ end. All PCR fragments were sequenced. Plants showing the expected PCR fragments were selected for further analysis.
Southern-Blot Analysis
Genomic DNA was isolated using a modified cetyl-trimethyl-ammonium bromide method. Ten micrograms of DNA was digested for 10 h with EcoRV, which does not cut within the selection marker cassette. After electrophoresis on a 0.7% (w/v) agarose gel, DNA was transferred to a positively charged nylon membrane (Roche) by high-salt capillary transfer. Labeling, hybridization, and detection of fragments carrying nptII sequences were performed as described in the Roche DIG application manual. The digoxigenin-labeled nptII hybridization probe was generated by PCR labeling (primer pair: nptII-coding sequence [CDS]-forward, 5′-ATTGAACAAGATGGATTGCACGC-3′ and nptII-CDS-reverse, 5′-TCAGAAGAACTCGTCAAGAAGGCG-3′) using the random-primed labeling mix from Roche and Phusion polymerase provided by Finnzymes-Thermo Scientific.
RNA Analysis/Reverse Transcriptase PCR
Total RNA from moss tissue was isolated using the TRIzol kit (Sigma) according to the manufacture’s instructions. Purified RNA was treated with DNase I using the RNase-Free DNase Kit from Qiagen. RNA integrity was checked on 1% (w/v) agarose gel containing ethidium bromide. Complementary DNA synthesis was performed using the iScript cDNA Synthesis Kit (Bio-Rad). Reverse transcriptase PCR was performed to show the absence of the PpPGRL1 transcript in the knockout mutant. Each reaction mix contained 0.5 µm of each primer and complementary DNA corresponding to 4 ng of RNA in the reverse transcriptase reaction. ACTIN2, the endogenous reference gene from P. patens, was used as control (Lunde et al., 2006). Primers annealing to the coding sequence of PpPGRL1 (CDS PGRL1-forward, 5′-CCATCCAACAACGTCAAA TG-3′ and CDS PGRL1-reverse, 5′-TTCAGCCAAAGGGCTCTCTA-3′) were used. The identity of the products was verified by sequencing. The reaction conditions for the Rotor-Gene version 4.6 (Corbett Research) were 95°C for 7 min, followed by cycles of 95°C for 10 s, 55°C for 30 s, and 72°C for 30 s, for up to a total of 40 cycles. The 2(−Delta Delta cycle threshold) method was used to calculate expression levels from fluorescence data (Livak and Schmittgen, 2001). Results are presented as fold change in mRNA abundance, normalized to the endogenous reference gene (described above) relative to the sample from cells grown under control growth conditions.
Protein Isolation, SDS-PAGE, and Immunoblot Analysis
Three hundred to 500 mg of frozen P. patens protonema or gametophores was harvested and frozen in liquid nitrogen. Samples were homogenized with a Retsch MM300 mill. Grinded tissue was resuspended in 50 mm Tris buffer (pH 8.0) containing 10 mm EDTA, 2% (w/v) SDS, 1 mm phenylmethylsulfonyl fluoride, and 1 mm benzamidine-HCl. Protein concentrations were determined by the BCA Assay (Thermo Fisher Scientific) as described by the manufacturer. For PAGE of proteins, pelleted cells were resuspended in 2% SDS and 1 mm β-mercaptoethanol and then heated for 10 min at 65°C. Solubilized proteins were separated by SDS-PAGE according to Laemmli (1970) and stained with Coomassie Brilliant Blue. C. reinhardtii western blot shown in Figure 5 was obtained with 35 µg of total protein extract, whereas blots reported in Supplemental Figure S9 were obtained with 8 µg [80 µg for CAS and plastoquinone-reducing type II NAD(P)H dehydrogenase] of protein from isolated thylakoid membranes. Antibodies against C. reinhardtii. ATPase subunit b (1:5,000), ADH1 (1:5,000), and PsbA (1:10,000) were purchased by Agrisera. Antibodies against ANR1 were purchased from Eurogentec and used at 1:1,000 dilution. Antibodies against CAS, PGRL1 (62), LHCSR3 (Naumann et al., 2007), and PsaD (Naumann et al., 2005) were used at 1:1,000 dilution. Antibodies against Nuclear Division Arrest2 (1:10,000) were a kind gift of Dr. Pierre Cardol (University of Liège).
Analysis of Proteins via Liquid Chromatography-Tandem Mass Spectrometry
Protein analyses by liquid chromatography-tandem mass spectrometry were performed to relatively quantify the proteome of the different strains and conditions using samples from two independent protonema harvests (set1 and set2) and labeling with 15N. The wild-type P. patens protonema was metabolically labeled with Ca(15NO3)2, and pgrl1 protonema was labeled with Ca(14NO3)2. Differentially labeled tissues were shifted to AN by argon bubbling for 7 h, harvested, mixed based on equal protein content (40 µg from each strain), and fractionated by SDS-PAGE according to Laemmli (1970). After SDS-PAGE fractionation and Coomassie Brilliant Blue staining, single SDS-PAGE bands were excised, tryptic in-gel digested, and analyzed as in Terashima et al., 2010 and Höhner et al., 2013. mzML files associated with this manuscript may be downloaded from http://www.peptideatlas.org/PASS/PASS00384.
Protein Identification from Mass Spectrometric Data
Proteomics data evaluation and quantitation were performed individually for each biological replicate (set1 and set2) using Proteomatic (Specht et al., 2011) and pymzML (Bald et al., 2012). Protein identification was conducted using the database search engines OMSSA (version 2.1.9; Geer et al., 2004) and X! Tandem (version 2012.10.01.1; Craig and Beavis, 2004). Searches were conducted against the Phytozome P. patens proteome version v1.6 (Rensing et al., 2008) combined with the EMBL proteomes of the chloroplast and mitochondria (RefSeq_20140218 and RefSeq_20140218, respectively). The parent mass error was set to 20 parts per million, and the fragment mass error was set to 0.5 D. Oxidation of Met was used a variable modification, and a maximum number of two missed cleavages was allowed. Linear precursor charge dependency was set for OMSSA, and noise suppression was enabled for X! Tandem. Statistical assessment of the quality of peptide spectral matches was conducted using a target/decoy approach (68) and qvality (version 2.02; Käll et al., 2009). Final posterior error probability for peptide spectral match was less than 0.01.
Quantitative Proteome Data Analysis
Quantification was conducted as described in Barth et al. (2014) using pyQms and piqDB. Similar to accurate mass time tags used in Superhirn (Mueller et al., 2007), piqDB is used to define accurate isotopic elution windows, which are used to quantify stringently. A quality assessment of the retention time alignment procedure can be visualized by comparing the size of all peptide elution windows. Supplemental Figure S10 shows such visualization as a density plot of all observed retention time windows (n = 7,535) before (red) and after (blue) alignment, which highlights the high quality of the alignment. Please refer to Barth et al. (2014 for more information about piqDB and other procedures. Heat maps were created using pyGCluster (Jaeger et al., 2013). All protein ratios were log2 transformed.
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession number(s) listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The P. patens pgrl1 knockout mutant harbors a single copy of the disruption construct in the genome, resulting in the lack of PpPGRL1 transcripts and protein expression.
Supplemental Figure S2. Phenotypic comparison of the P. patens wild type and the pgrl1 during the developmental stages of the haploid gametophore grown under optimal conditions.
Supplemental Figure S3. Phenotype and time course of NPQ and Y(II) of P. patens wild-type and pgrl1 gametophores grown for 1 month under optimal conditions (as control for long-term AN experiment).
Supplemental Figure S4. Relative abundance of LHCII, LHCI, and ATPase subunits in the P. patens pgrl1 compared with the wild type after short-tern anoxia.
Supplemental Figure S5. Generation of the C. reinhardtii pgrl1 npq4 double mutant.
Supplemental Figure S6. Acceptor-side inhibition of PSI in pgrl1 npq4 and pgrl1 cells under HL conditions accounts for impairment of NPQ.
Supplemental Figure S7. CEF activity is impaired in the pgrl1, npq4, and pgrl1 npq4 strains.
Supplemental Figure S8. Susceptibility of pgrl1, npq4, and pgrl1 npq4 mutants to HL conditions and AN in liquid media.
Supplemental Figure S9. Protein expression analyses of C. reinhardtii wild-type and pgrl1 thylakoids under HL TAP anoxia, HL TAP, and HL HSM conditions.
Supplemental Figure S10. Retention time quality assessment shown as density plots for all observed retention time windows, i.e. (n = 7,535) before and after alignment.
Supplemental Table S1. Full detail and the identified and quantified proteins in set1 and set2.
Supplementary Material
Acknowledgments
We thank Friederike König from the University of Bremen for the gift of antibodies.
Glossary
- NPQ
nonphotochemical quenching
- CEF
cyclic electron flow
- cyt
cytochrome
- Fv/Fm
maximum photochemical efficiency of PSII in the dark-adapted state
- AN
anaerobiosis
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- HLC
high-light cold
- LL
low light
- HL
high light
- TAP
Tris-acetate phosphate
- Fm′
maximum PSII fluorescence in the light-adapted state
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. HI 739/8.1 and HI 739/9.1 to M.H.), the Alexander von Humboldt Stiftung/Foundation, the Marie Curie Initial Training Network AccliPhot (FP7–PEPOPLE–2012–ITN, grant no. 316427 to G.F.), the Grenoble Alliance for Integrated Structural Cell Biology (grant no. ANR–10–LABEX–04 Labex GRAL to G.F.), the French Agence Nationale de la Recherche (grant no. ANR–12–BIME–0005 DiaDomOil to D.P.), and the VILLUM Center of Excellence Plant Plasticity.
The online version of this article contains Web-only data.
References
- Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T. (2010) Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc Natl Acad Sci USA 107: 11128–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alric J. (2010) Cyclic electron flow around photosystem I in unicellular green algae. Photosynth Res 106: 47–56 [DOI] [PubMed] [Google Scholar]
- Amunts A, Drory O, Nelson N. (2007) The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature 447: 58–63 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Rühle T, Kreller R, Strotbek C, Zühlke J, Tadini L, Blunder T, Hertle AP, Qi Y, Rengstl B, et al. (2013) The PHOTOSYNTHESIS AFFECTED MUTANT68-LIKE protein evolved from a PSII assembly factor to mediate assembly of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell 25: 3926–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1959) Conversion of light into chemical energy in photosynthesis. Nature 184: 10–21 [DOI] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ. (1977) The isolation and preliminary characterization of auxotrophic and analogue resistant mutants in the moss Physcomitrella patens. Mol Gen Genet 154: 87–95 [Google Scholar]
- Avenson TJ, Cruz JA, Kanazawa A, Kramer DM. (2005) Regulating the proton budget of higher plant photosynthesis. Proc Natl Acad Sci USA 102: 9709–9713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badyaev AV. (2005) Stress-induced variation in evolution: from behavioural plasticity to genetic assimilation. Proc Biol Sci 272: 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bald T, Barth J, Niehues A, Specht M, Hippler M, Fufezan C. (2012) pymzML: Python module for high-throughput bioinformatics on mass spectrometry data. Bioinformatics 28: 1052–1053 [DOI] [PubMed] [Google Scholar]
- Barth J, Bergner SV, Jaeger D, Niehues A, Schulze S, Scholz M, Fufezan C. (2014) The interplay of light and oxygen in the reactive oxygen stress response of Chlamydomonas reinhardtii dissected by quantitative mass spectrometry. Mol Cell Proteomics 13: 969–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham JA, Benson AA, Calvin M. (1950) The path of carbon in photosynthesis. J Biol Chem 185: 781–787 [PubMed] [Google Scholar]
- Busch A, Petersen J, Webber-Birungi MT, Powikrowska M, Lassen LM, Naumann-Busch B, Nielsen AZ, Ye J, Boekema EJ, Jensen ON, et al. (2013) Composition and structure of photosystem I in the moss Physcomitrella patens. J Exp Bot 64: 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraretto L, Formentin E, Teardo E, Checchetto V, Tomizioli M, Morosinotto T, Giacometti GM, Finazzi G, Szabó I. (2013) A thylakoid-located two-pore K+ channel controls photosynthetic light utilization in plants. Science 342: 114–118 [DOI] [PubMed] [Google Scholar]
- Cho SH, Chung YS, Cho SK, Rim YW, Shin JS. (1999) Particle bombardment mediated transformation and GFP expression in the moss Physcomitrella patens. Mol Cells 9: 14–19 [PubMed] [Google Scholar]
- Cornic G, Bukhov NG, Wiese C, Bligny R, Heber U. (2000) Flexible coupling between light-dependent electron and vectorial proton transport in illuminated leaves of C3 plants. Role of photosystem I-dependent proton pumping. Planta 210: 468–477 [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato R, Leister D. (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Dubini A, Mus F, Seibert M, Grossman AR, Posewitz MC. (2009) Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J Biol Chem 284: 7201–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi G, Rappaport F, Furia A, Fleischmann M, Rochaix JD, Zito F, Forti G. (2002) Involvement of state transitions in the switch between linear and cyclic electron flow in Chlamydomonas reinhardtii. EMBO Rep 3: 280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. (2004) Open mass spectrometry search algorithm. J Proteome Res 3: 958–964 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Catalanotti C, Yang W, Dubini A, Magneschi L, Subramanian V, Posewitz MC, Seibert M. (2011) Multiple facets of anoxic metabolism and hydrogen production in the unicellular green alga Chlamydomonas reinhardtii. New Phytol 190: 279–288 [DOI] [PubMed] [Google Scholar]
- Heber U, Bilger W, Bligny R, Lange OL. (2000) Phototolerance of lichens, mosses and higher plants in an alpine environment: analysis of photoreactions. Planta 211: 770–780 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A, Happe T. (2011) Alternative photosynthetic electron transport pathways during anaerobiosis in the green alga Chlamydomonas reinhardtii. Biochim Biophys Acta 1807: 919–926 [DOI] [PubMed] [Google Scholar]
- Hertle AP, Blunder T, Wunder T, Pesaresi P, Pribil M, Armbruster U, Leister D. (2013) PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol Cell 49: 511–523 [DOI] [PubMed] [Google Scholar]
- Höhner R, Barth J, Magneschi L, Jaeger D, Niehues A, Bald T, Grossman A, Fufezan C, Hippler M. (2013) The metabolic status drives acclimation of iron deficiency responses in Chlamydomonas reinhardtii as revealed by proteomics based hierarchical clustering and reverse genetics. Mol Cell Proteomics 12: 2774–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AG, Rosso D, Savitch LV, Stachula P, Rosembert M, Oquist G, Hurry V, Hüner NP. (2012) Implications of alternative electron sinks in increased resistance of PSII and PSI photochemistry to high light stress in cold-acclimated Arabidopsis thaliana. Photosynth Res 30: 191–206 [DOI] [PubMed] [Google Scholar]
- Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J. (2010) Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Jaeger D, Barth J, Niehues A, Fufezan C. (2013) pyGCluster, a novel hierarchical clustering approach. Bioinformatics 30: 896–898 [DOI] [PubMed] [Google Scholar]
- Johnson X, Steinbeck J, Dent RM, Takahashi H, Richaud P, Ozawa S, Houille-Vernes L, Petroutsos D, Rappaport F, Grossman AR, et al. (2014) Proton gradient regulation 5-mediated cyclic electron flow under ATP- or redox-limited conditions: a study of ΔATpase pgr5 and ΔrbcL pgr5 mutants in the green alga Chlamydomonas reinhardtii. Plant Physiol 165: 438–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Joliot A. (2002) Cyclic electron transfer in plant leaf. Proc Natl Acad Sci USA 99: 10209–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot PA, Finazzi G. (2010) Proton equilibration in the chloroplast modulates multiphasic kinetics of nonphotochemical quenching of fluorescence in plants. Proc Natl Acad Sci USA 107: 12728–12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Storey JD, Noble WS. (2009) QVALITY: non-parametric estimation of q-values and posterior error probabilities. Bioinformatics 25: 964–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz SJ, Prochnik SE, Grossman AR, Merchant SS. (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J Biol Chem 286: 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol AG, Borza T, Ishida K, Keeling P, Lee RW, Durnford DG. (2007) Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol 143: 1802–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Leister D, Shikanai T. (2013) Complexities and protein complexes in the antimycin A-sensitive pathway of cyclic electron flow in plants. Front Plant Sci 4: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lunde C, Baumann U, Shirley NJ, Drew DP, Fincher GB. (2006) Gene structure and expression pattern analysis of three monodehydroascorbate reductase (Mdhar) genes in Physcomitrella patens: implications for the evolution of the MDHAR family in plants. Plant Mol Biol 60: 259–275 [DOI] [PubMed] [Google Scholar]
- Magneschi L, Catalanotti C, Subramanian V, Dubini A, Yang W, Mus F, Posewitz MC, Seibert M, Perata P, Grossman AR. (2012) A mutant in the ADH1 gene of Chlamydomonas reinhardtii elicits metabolic restructuring during anaerobiosis. Plant Physiol 158: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor Y, Toporik H, Nelson N. (2012) Temperature-sensitive PSII and promiscuous PSI as a possible solution for sustainable photosynthetic hydrogen production. Biochim Biophys Acta 1817: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Mitchell P. (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191: 144–148 [DOI] [PubMed] [Google Scholar]
- Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Müller M. (2007) SuperHirn: a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7: 3470–3480 [DOI] [PubMed] [Google Scholar]
- Müller P, Li XP, Niyogi KK. (2001) Non-photochemical quenching: a response to excess light energy. Plant Physiol 125: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T. (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T. (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Naumann B, Stauber EJ, Busch A, Sommer F, Hippler M. (2005) N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J Biol Chem 280: 20431–20441 [DOI] [PubMed] [Google Scholar]
- Niyogi KK. (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50: 333–359 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Bjorkman O, Grossman AR. (1997) Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G, Niyogi KK. (2008) Pond scum genomics: the genomes of Chlamydomonas and Ostreococcus. Plant Cell 20: 502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Peltier G, Tolleter D, Billon E, Cournac L. (2010) Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth Res 106: 19–31 [DOI] [PubMed] [Google Scholar]
- Petroutsos D, Busch A, Janssen I, Trompelt K, Bergner SV, Weinl S, Holtkamp M, Karst U, Kudla J, Hippler M. (2011) The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 23: 2950–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D, Terauchi AM, Busch A, Hirschmann I, Merchant SS, Finazzi G, Hippler M. (2009) PGRL1 participates in iron-induced remodeling of the photosynthetic apparatus and in energy metabolism in Chlamydomonas reinhardtii. J Biol Chem 284: 32770–32781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnola A, Dall’Osto L, Gerotto C, Morosinotto T, Bassi R, Alboresi A. (2013) Zeaxanthin binds to light-harvesting complex stress-related protein to enhance nonphotochemical quenching in Physcomitrella patens. Plant Cell 25: 3519–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Reski R, Abel WO. (1985) Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta 165: 354–358 [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. (1998) Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342 [DOI] [PubMed] [Google Scholar]
- Savitch LV, Ivanov AG, Gudynaite-Savitch L, Huner NP, Simmonds J. (2011) Cold stress effects on PSI photochemistry in Zea mays: differential increase of FQR-dependent cyclic electron flow and functional implications. Plant Cell Physiol 52: 1042–1054 [DOI] [PubMed] [Google Scholar]
- Schöttler MA, Kirchhoff H, Weis E. (2004) The role of plastocyanin in the adjustment of the photosynthetic electron transport to the carbon metabolism in tobacco. Plant Physiol 136: 4265–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schween G, Fleig S, Reski R. (2002) High-throughput-PCR screen of 15,000 transgenic Physcomitrella plants. Plant Mol Biol Rep 40: 34–47 [Google Scholar]
- Shikanai T. (2014) Central role of cyclic electron transport around photosystem I in the regulation of photosynthesis. Curr Opin Biotechnol 26: 25–30 [DOI] [PubMed] [Google Scholar]
- Specht M, Kuhlgert S, Fufezan C, Hippler M. (2011) Proteomics to go: Proteomatic enables the user-friendly creation of versatile MS/MS data evaluation workflows. Bioinformatics 27: 1183–1184 [DOI] [PubMed] [Google Scholar]
- Stiehl HH, Witt HT. (1969) Quantitative treatment of the function of plastoquinone in phostosynthesis. Z Naturforsch B 24: 1588–1598 [DOI] [PubMed] [Google Scholar]
- Terashima M, Petroutsos D, Hüdig M, Tolstygina I, Trompelt K, Gäbelein P, Fufezan C, Kudla J, Weinl S, Finazzi G, et al. (2012) Calcium-dependent regulation of cyclic photosynthetic electron transfer by a CAS, ANR1, and PGRL1 complex. Proc Natl Acad Sci USA 109: 17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima M, Specht M, Naumann B, Hippler M. (2010) Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol Cell Proteomics 9: 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Ghysels B, Alric J, Petroutsos D, Tolstygina I, Krawietz D, Happe T, Auroy P, Adriano JM, Beyly A, et al. (2011) Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH (1957) The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. Allen and Unwin, London [Google Scholar]
- Weinl S, Held K, Schlücking K, Steinhorst L, Kuhlgert S, Hippler M, Kudla J. (2008) A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytol 179: 675–686 [DOI] [PubMed] [Google Scholar]
- Whatley FR, Tagawa K, Arnon DI. (1963) Separation of the light and dark reactions in electron transfer during photosynthesis. Proc Natl Acad Sci USA 49: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt HT. (1979) Energy conversion in the functional membrane of photosynthesis. Analysis by light pulse and electric pulse methods. The central role of the electric field. Biochim Biophys Acta 505: 355–427 [DOI] [PubMed] [Google Scholar]
- Wraight CA, Crofts AR. (1970) Energy-dependent quenching of chlorophyll α fluorescence in isolated chloroplasts. Eur J Biochem 17: 319–327 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.