SUMMARY
We demonstrate that RING finger protein MSL2 in the MOF-MSL complex is a histone ubiquitin E3 ligase. MSL2, together with MSL1, has robust histone ubiquitylation activity that mainly targets nucleosomal H2B on lysine 34 (H2B K34ub), a site within a conserved basic patch on H2B tail. H2B K34ub by MSL1/2 directly regulates H3 K4 and K79 methylation through trans-tail crosstalk both in vitro and in cells. The significance of MSL1/2 mediated histone H2B ubiquitylation is underscored by facts that MSL1/2 activity is important for transcription activation at HOXA9 and MEIS1 loci and that this activity is evolutionarily conserved in the Drosophila dosage compensation complex. Altogether, these results establish that the MOF-MSL complex possesses two distinct chromatin-modifying activities (i.e. H4 K16 acetylation and H2B K34 ubiquitylation) through MOF and MSL2 subunits. They also shed new lights on how intricate network of chromatin modifying enzymes functions coordinately in gene activation.
INTRODUCTION
Many of the changes in chromatin structure are induced by covalent modifications of histones, which are highly dynamic and frequently occur in distinct, interrelated patterns. The complex pattern of histone modifications is the result of tightly controlled activities of histone modifying enzymes as well as extensive crosstalk among them and cognate histone marks (Suganuma and Workman, 2008). One of the best examples for histone crosstalk is the ‘trans-tail regulation’ of H3 K4 and K79 methylation by H2B K120 ubiquitylation (K120ub) (Laribee et al., 2007; Osley, 2004; Weake and Workman, 2008). H2B K120ub by RAD6-RNF20/40 (also called BRE1) (Hwang et al., 2003; Robzyk et al., 2000) is clearly linked to chromatin dynamics during transcription elongation, facilitating reassembly of nucleosomes in the wake of Pol II passage (Fleming et al., 2008; Xiao et al., 2005). Importantly, it has been shown that null mutation of Rad6 or Bre1 in yeast (Laribee et al., 2007) and knockdown of RAD6 and RNF20/RNF40 in human cells (Kim et al., 2005) lead to global reduction of H3 K4 and H3 K79 methylation. This crosstalk is a direct result of H2B K120ub mediated stimulation of the H3 K4 methyltransferase SET1 and the H3 K79 methyltransferase DOT1L (Kim et al., 2009; McGinty et al., 2008). Both H3 K4 and K79 methylation function in conjunction with histone acetylation to set up optimal chromatin environment and/or to recruit factors important for active transcription.
One of the histone acetylation events that function coordinately with H3 K4 methylation is H4 K16 acetylation by MOF (also called MYST1, KAT8) (Li and Dou, 2010). MOF was originally described as an essential component of the X chromosome dosage compensation complex (DCC, also referred to as Male Specific Lethal (dMSL)) in Drosophila, causing a two-fold increase in expression of X-linked genes in male flies (Gelbart and Kuroda, 2009; Lucchesi et al., 2005). It is shown that the dMSL complex regulates gene activation at the level of transcription elongation even though the detailed mechanism remains unclear (Larschan et al., 2011). In the dMSL complex, dMOF associates with highly conserved dMSL1, 2, and 3 proteins, which are important for its acetyltransferase activity (i.e. dMSL1 and dMSL3) (Morales et al., 2004) and specific localization to male X chromosome (i.e. dMSL2) (Kelley et al., 1995). Recently, a second dMOF complex is reported in Drosophila, which contains distinct sets of proteins including a key component dMSL1v1 (also called NSL1) (Smith et al., 2005). In mammals, the MSL components for both MOF-MSL and MOF-MSL1v1 complexes are conserved (Li and Dou, 2010). Both MOF complexes are important for majority of H4 K16 acetylation in cells (Li et al., 2009) and for regulating cellular processes such as gene expression, cell cycle and DNA damage repair (Li et al., 2010). Importantly, biochemical studies documented major differences between two MOF complexes with regard to their substrate spectrum (Cai et al., 2009; Li et al., 2009). The MOF-MSL1v1 complex (MOF-MSL1v1), which was first purified with WDR5 and H3 K4 methyltransferase MLL (Dou et al., 2005), acetylates not only H4 K16 but also other lysine residues on H4 tail (Cai et al., 2009) as well as transcription factors such as p53 (Li et al., 2009). These underscore the function of MOF-MSL1v1 in transcription initiation.
In contrast to MOF-MSL1v1, the mammalian MOF-MSL complex (MOF-MSL) exclusively acetylates H4 K16 in the in vitro HAT assay (Li et al., 2009). We have previously shown that the activity of MOF-MSL is tightly regulated by MSL1 and MSL3, which are necessary and sufficient for H4 K16 acetylation (Li et al., 2009). However, this result raises an interesting question: what is the function of MSL2 in MOF-MSL? MSL2 has two evolutionarily conserved domains: a RING finger on the N-terminus and a CXC domain on the C-terminus (Fauth et al., 2010). The functions of both domains in mammalian MOF-MSL are unclear. Recently, it is reported that MSL2 promotes p53 localization to cytoplasm through an ubiquitylation-mediated mechanism (Kruse and Gu, 2009). p53 purified from cells overexpressing MSL2, but not MSL2 ΔRING, is poly-ubiquitylated at lysine residues distinct from MDM2 target sites. However, it is not clear whether p53 poly-ubiquitylation in the study is due to direct MSL2 activity or a yet uncharacterized E3 ubiquitin ligase that is regulated by MSL2. Nonetheless, it raises an interesting possibility that MSL2 may possess E3 ubiquitin ligase activity.
Using rigorous in vitro biochemical assays, along with cellular analyses, here we show that that MSL2 together with the MSL1 coiled-coil domain is a histone E3 ubiquitin ligase, mainly targeting H2B at lysine 34 (K34ub). We further show that MSL1/2 mediated H2B K34ub is important for trans-regulation of H3 K4 and K79 methylation both in vitro and in cells. It also regulates H2B K120ub by promoting chromatin association of RNF20/40. Interplay between MSL2 E3 ubiquitin ligase activity and MOF acetyltransferase activity is also explored. These findings establish that MOF-MSL possesses two distinct chromatin-modifying activities through MOF and MSL2 subunits and these two activities function together to establish the intricate network of histone marks for transcription activation.
RESULTS
MOF-MSL has E3 ubiquitin ligase activity towards nucleosomal H2B
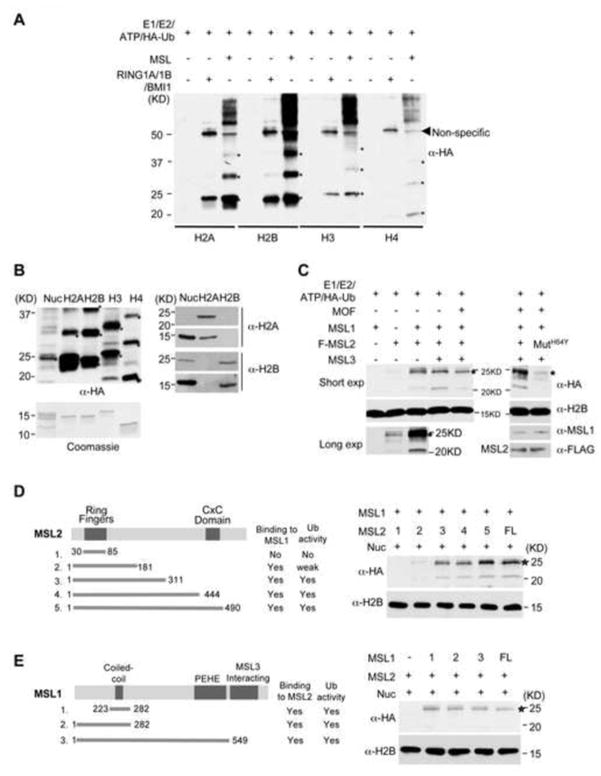
In order to test whether MSL2 has histone E3 ubiquitin ligase activity, we set up in vitro ubiquitylation assays for MOF-MSL. Recombinant E1, E2 (UBC5c), HA-ubiquitin (HA-Ub) and one of the four histones were included in each reaction. The RING1A/1B/BMI1 complex (Supplementary Figure 1A), an E3 ubiquitin ligase for H2A (Wang et al., 2004), was included as a positive control. As shown in Figure 1A, MOF-MSL showed robust E3 ubiquitin ligase activity on free histones. This activity was specific for MOF-MSL since no histone ubiquitylation was detected in its absence (Figure 1A and Supplementary Figure 1B–C). Notably, although MOF-MSL was able to ubiquitylate all four histones, it showed higher activities on free histone H2B and H2A. This was similar to the RING1A/1B/BMI1 complex, which displayed preference for H2A and H2B as well (Figure 1A and Wang et al., 2004). Interestingly, specificity of MOF-MSL improved dramatically when HeLa nucleosomes were used as the substrate. As shown in Figure 1B, MOF-MSL mainly targeted mono-ubiquitylation to nucleosomal H2B/H2A. Immunoblot using anti-H2B or anti-H2A antibody showed that MOF-MSL specifically ubiquitylated nucleosomal H2B but not H2A (Figure 1B, right panels). The increased substrate specificity for MOF-MSL on nucleosomes was similar to what was described for histone E3 ubiquitin ligases RING1A/1B/BMI1 and RNF20/40 (Kim et al., 2009; Wang et al., 2004). Of note, the activity of MOF-MSL on nucleosomes is lower than that on free histones (Figure 1B, left panel). The relatively low level of free H2B ubiquitylation (H2Bub) detected by anti-H2B antibody was due to antibody preference towards mammalian H2B. (Figure 1B, right panel). Given that MOF-MSL mainly targets mono-ubiquitylation of nucleosomal H2B, we will focus on H2B modification herein.
Figure 1. MSL2 is a histone E3 ubiquitin ligase.
(A) In vitro ubiquitylation (ub) assays for MOF-MSL and RING1A/1B/BMI1. Four recombinant Xenopus histones were used as substrates as indicated on bottom. Ubiquitylated-histones were detected by anti-HA antibody. (B) In vitro ub assays for MOF-MSL on HeLa nucleosomes and equivalent amount of Xenopus histones. The coomassie stained gel on bottom was for loading control. Ubiquitylated-histones were detected by anti-HA antibody (Left), or anti-H2A and anti-H2B antibodies as indicated (Right). Both anti-H2A and anti-H2B antibodies are raised against mammalian histones and as a result, have better signals for HeLa nucleosomes. In (A, B), positions of mono-, di-, and tri-ubiquitylated histones were marked by *. (C) In vitro ub assays for various MOF-MSL sub-complex (as indicated on top). HeLa nucleosomes were used as substrates. Ubiquitylated-histones were detected by anti-HA antibody. Immunoblots for unmodified H2B, MSL1 and Flag-MSL2 were included as the loading controls. Longer exposure for lane 1–3 was shown at bottom. (D) Left, schematic for mapping MSL2. Both RING and CXC domains were highlighted. Right, in vitro ub assay for MSL2 fragments as indicated on top. (E) Left, schematic for mapping MSL1. The coiled-coil, PEHE and MSL3 interacting domains were highlighted. Right, in vitro ub assay for MSL1 fragments as indicated on top. For (B–E), expected position for ubH2B was indicated by *, which is slight lower than the 25KD marker.
Both MSL2 and MSL1 are essential for optimal E3 ubiquitin ligase activity
The mammalian MOF-MSL has four stably associated proteins: MOF, MSL1, MSL2 and MSL3 (Smith et al., 2005). We previously demonstrated that MOF together with MSL1 and MSL3 were fully active for nucleosomal H4 K16 acetylation (Li et al., 2009). To identify the essential component(s) for histone ubiquitylation, we reconstituted and purified different MOF-MSL sub-complexes by co-infecting insect cells with different combinations of baculoviruses for each of the components (Supplementary Figure 2A). We also purified a MOF-MSL complex with an H64Y mutation (MSL2H64Y) in the second zinc cluster of the MSL2 RING finger (Supplementary Figure 2B). Although H64Y mutation is likely to at least partially unfold the RING finger, it did not affect interaction with MSL1. These complexes were tested for E3 ligase activities on HeLa nucleosomes in vitro (Figure 1C). As shown in Figure 1C, MSL2 is necessary and sufficient for H2Bub (see longer exposure). Mutation of MSL2H64Y greatly reduced its activity for H2Bub even though the MSL2H64Y-containing complex remained intact (Figure 1C and Supplementary Figure 2B). Importantly, optimal ubiquitylation activity requires MSL1, which dramatically enhanced H2Bub in vitro (Figure 1C). In fact, the activity of MSL1 and MSL2 heterodimer (referred to as MSL1/2 herein) was indistinguishable from that of the four-component MOF-MSL complex, suggesting that both MOF and MSL3 were dispensable for H2B ubiquitylation (Figure 1C).
Mapping essential domains in MSL1 and MSL2 for histone E3 ubiquitin ligase activity
We next performed detailed mapping experiments to identify essential domains in both MSL1 and MSL2 for robust H2B ubiquitylation activity. To this end, we first made five truncation mutants of MSL2: a fragment of 30-85aa that included only the RING finger and four mutants with serial deletions from C-terminus (Figure 1D). The mutant MSL2 was purified through full length His-MSL1 after co-expression (Supplementary Figure 3A) and their activities were compared to that of wild-type MSL1/2. As shown in Figure 1D, three MSL2 deletion mutants, 1-311aa, 1-444aa and 1-490aa, ubiquitylated nucleosomal H2B with the same efficiency as the full-length complex. However, the RING finger and MSL2 1-181aa had no or only weak activities (Figure 1D). This result suggests that optimal H2B ubiquitylation requires MSL2 RING finger and its C-terminal ~250 residues.
For mapping MSL1, three MSL1 polypeptides containing 223-282aa, 1-282aa, or 1-549aa were co-purified with full-length His-MSL2 and tested for in vitro H2B ubiquitylation (Figure 1E and Supplementary Figure 3B). We found that the coiled-coil domain of MSL1 was sufficient to support optimal MSL2 activity. In contrast, deleting two C-terminal domains of MSL1 (i.e. the MSL3 and PEHE/MOF interacting domains) did not affect H2B ubiquitylation (Figure 1E). This result is consistent with our observation that both MOF and MSL3 are dispensable for nucleosomal ubiquitylation activity (Figure 1C).
The MSL1/2 heterodimer ubiquitylates nucleosomal H2B on lysine 34 in vitro
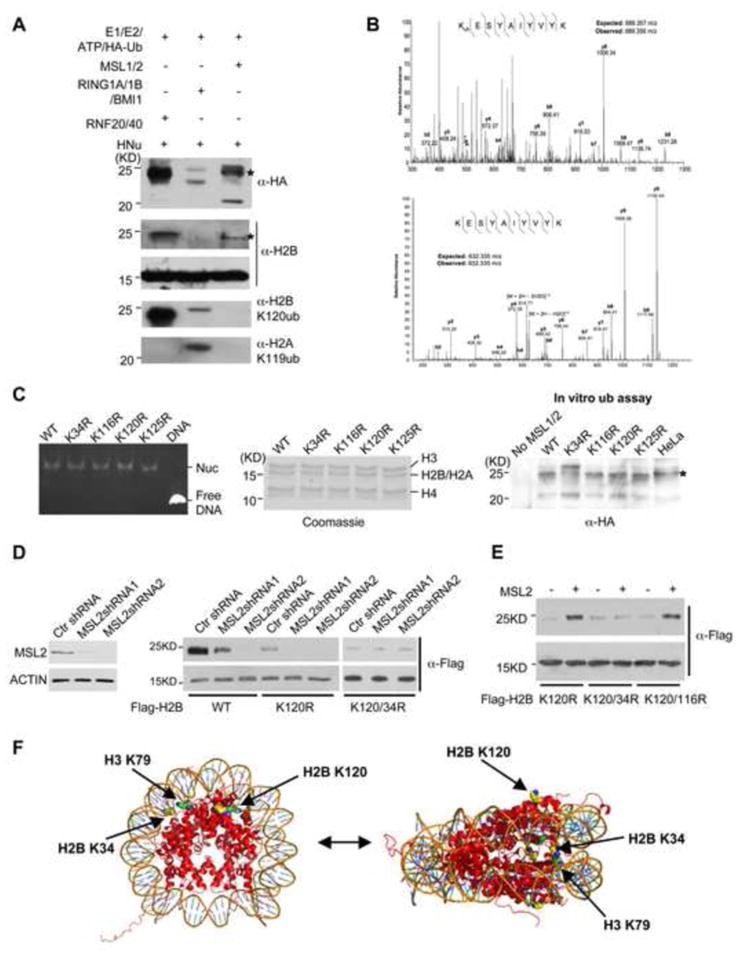
Histone H2B K120ub by RNF20/40 has been well characterized (Weake and Workman, 2008). To test whether MSL1/2 also targets H2B K120 on nucleosomes, we set up the in vitro ubiquitylation assay and used anti-H2B, anti-HA (for HA-ub) or anti-H2B K120ub antibody to probe the ubiquitylation product. RNF20/40 and RING1A/1B/BMI1 complexes were used as controls. As shown in Figure 2A, H2Bub by MSL1/2 was easily detected by immunoblot using both anti-HA and anti-H2B antibodies (Figure 2A). Surprisingly, no signal was detected for MSL1/2 by the anti-H2B K120ub antibody although the antibody readily detected H2B ubiquitylation by RNF20/40 and to a much less extent, by RING1A/1B/BMI1. As expected, anti-H2A K119ub antibody did not detect any H2Aub by MSL1/2 (Figure 2A). This result strongly suggests that MSL1/2 ubiquitylates histone H2B at a site distinct from K120.
Figure 2. MSL1/2 ubiquitylates histone H2B on K 34.
(A) In vitro ub assays for RNF20/40, RING1A/1B/BMI1 or MOF-MSL as indicated on top. HeLa nucleosomes were used as substrates. Antibodies used were indicated. (B) MS/MS spectrum of the +2 H2B 34–43 peptide ubiquitylated (top) or unmodified (bottom) on K34. For both panels, hatched lines above and below the peptide sequence denote b and y ions respectively that were successfully annotated from the spectrum. A comparison of precursor +2 ion masses from the MS spectrum is also indicated. Abbreviation: [M+2H–1H2O]+2 = molecular ion of +2 charge with loss of one water molecule; [M+2H–2H2O]+2 = molecular ion of +2 charge with loss of two water molecules. (C) Left, ethidium bromide stained gel for purified nucleosomes that contain various H2B mutations as indicated on top. Free DNA was used as the control. Middle, the coomassie stained gel for histones in assembled nucleosomes. H2A and H2B run at the same positions. Right, in vitro ub assay for MOF-MSL on various nucleosomes as indicated on top. Expected position for H2Bub was indicated by *. (D) Left, immunoblot for MSL2 after shRNA treatment. Right, ubiquitylation of Flag-H2B WT, K120R, K120R/34R in cells treated with MSL2 shRNA. (E) Ubiquitylation of Flag-H2B K120R, K120R/34R, or K120R/K116R in HeLa cells overexpressing MSL2. In (D, E), both unmodified and ubiquitylated H2B were detected by anti-Flag antibody. (F) Structure for X. laevis nucleosome core (PDB 1AOI; 2.8 Å). H2B K34, K120 and H3 K79 were indicated by arrows.
In order to identify the substrate lysine on H2B for MSL1/2, we performed mass spectrometry analyses for MSL1/2-modified recombinant H2B. The reason to use recombinant H2B as substrate despite potential low specificity is that free H2B is a much better substrate than nucleosomes, which will facilitate mass spec detection. SEQUEST searches performed on ubiquitinated H2B found four potentially modified sites in both trypsin and ArgC digested samples, which gave 63% sequence coverage. They are K34, K116, K120, and K125 respectively. No ubiquitylation site was identified by SEQUEST for H2B treated with only E1 and E2 enzymes (data not shown). Manual inspections of MS/MS spectra of modified and unmodified H2B peptides surrounding K34 were shown in Figure 2B. Among four identified H2B ubiquitylation sites, three of them, K34, K116, and K120, along with K46 and K108, have been previously reported in a large-scale protein post-translational modification study for adult mouse brains (Tweedie-Cullen et al., 2009), suggesting that they are present in vivo and are physiologically relevant.
To further investigate whether MSL1/2 targeted only a subset of these ubiquitylation sites in the context of nucleosomes, we reconstituted recombinant nucleosomes that carry H2B K34R, K116R, K120R or K125R mutation respectively (Figure 2C, top two panels). Reconstituted wild type and mutant nucleosomes were then used as substrates for in vitro ubiquitylation. Strikingly, changing H2B K34 to R abolished its ubiquitylation on nucleosomes (Figure 2C, bottom panel). A simultaneous increase of H3 and H4 ubiquitylation was observed, probably due to enhanced secondary ubiquitylation events (Figure 2C). In contrast, mutations of other lysine residues had no such effect (Figure 2C). Taken together, our results strongly support that MSL1/2 ubiquitylates nucleosomal H2B on K34 in vitro.
MSL2 ubiquitylates H2B K34 in cells
To determine whether MSL2 ubiquitylates H2B K34 in cells, we performed shRNA mediated knockdown experiment. In this experiment, we transfected cells with plasmids expressing either wild type H2B or H2B K120R after MSL2 knockdown. MSL2 expression was effectively eliminated by shRNA treatment as shown by both immunoblot (Figure 2D, left panel) and RT-PCR (Supplementary Figure 4). Several things were learned from this experiment: 1) H2B K120 was indeed the major H2Bub site in cells. Mutating H2B K120 led to dramatic reduction of total ubH2B (Figure 2D, lane 1 vs 4); 2) H2B K120R could still be ubiquitylated in cells and its ubiquitylation was significantly reduced by MSL2 knockdown (Figure 2D). In contrast, MSL2 knockdown could not further decrease ubiquitylation of the H2B K120R/34R mutant, consistent with MSL2 mainly targeting H2B K34ub; and 3) surprisingly, MSL2 knockdown led to significant reduction of overall H2Bub, which implied an effect on H2B K120ub in cells (Figure 2D). Later studies showed that this is probably due to indirect effects (see below).
In addition to knockdown experiments, we also overexpressed MSL2 with several H2B mutants: Flag-K120R, K120/34R and K120/116R respectively. H2B K120R was included in each case to highlight K120 independent ubiquitylation event. As shown in Figure 2E, although overexpression of MSL2 led to significant increase in H2Bub for both K120R and K120/116R mutants, it failed to enhance ubiquitylation of H2B K120/34R. Given the results of our in vitro assays and the cellular studies, we believe MSL1/2 is a histone E3 ubiquitin ligase that directly targets H2B K34. Importantly, only mono-ubiquitylation of H2B by MSL1/2 was detected in cells (data not shown), suggesting that MSL1/2, like RNF20/RNF40, was mainly a mono-ubiquitylase under physiological conditions.
H2B K34 on nucleosome structure
H2B K34 resides within an extremely basic, eight residue patch on H2B N-terminal tail (i.e. 27-34aa) (Supplementary Figure 5A). From the crystal structure of nucleosome core particles (Luger et al., 1997), this region of H2B passes through the minor groove channel between superhelix gyres (SHL4.5 and SHL-2.5 to -3) and is proposed to facilitate interactions with neighboring nucleosomes (Luger et al., 1997). As illustrated in Figure 2F, K34 directly faces the DNA superhelix as the H2B N-terminus exits the core particle. It is partially occluded by DNA and probably requires some conformational change in nucleosome core particle for full accessibility. Importantly, K34 is in vicinity to H3 K79 (highlighted in green, Figure 2F) and adjacent to H3 N-terminal tail. It also borders the H2A ubiquitylation site (Luger et al., 1997).
H2B K34 ubiquitylation (K34ub) directly stimulates histone H3 K4 and K79 methylation in vitro
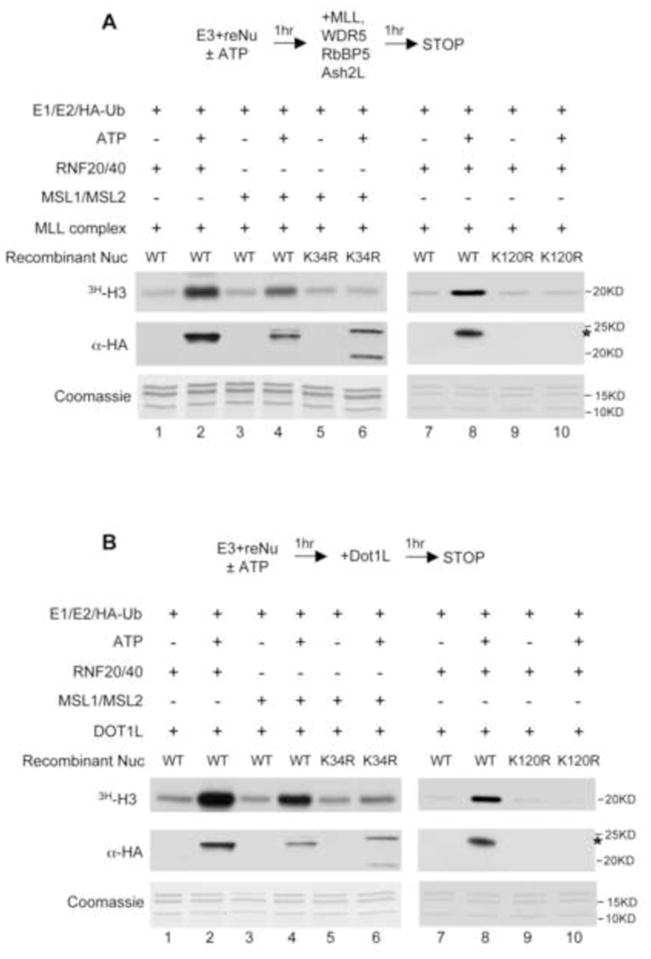
Trans-tail interactions between H2B K120ub and methylation of H3 K4 and K79 were well documented (Briggs et al., 2002; Ng et al., 2002; Sun and Allis, 2002). It was shown that H2B K120ub directly stimulates activities of MLL/SET1 family H3 K4 methyltransferases and the H3 K79 methyltransferase DOT1L, which in turn promote transcription activation (Kim et al., 2009; McGinty et al., 2008). Given H2B K34ub by MSL1/2 and its proximity to both H3 N-terminal tail and H3 K79, we asked whether H2B K34ub is also involved in trans-tail interactions similar to H2B K120ub. To this end, we used MSL1/2 or RNF20/40 treated recombinant nucleosomes as substrates for the in vitro HMT assays. In each case, ubiquitylation reaction without ATP was used as the negative control. Consistent with previous reports (Kim et al., 2009), RNF20/40-mediated ubiquitylation of wild type, but not H2B K120R nucleosomes, enhanced MLL activity (Figure 3A, lane 7–10). Interestingly, MSL1/2 was able to stimulate MLL activity in a similar fashion (Figure 3A, lane 4). The stimulation was due to H2B K34ub since it was abolished when either H2B K34 was mutated to R (lane 6) or ATP was omitted from the ubiquitylation reaction (Figure 3A, lane 3). Of note, the four-component MLL core complex (Dou et al., 2006), which was used here, was sufficient to mediate this trans-tail regulation. In addition to promote H3 K4 methylation, H2B K34ub by MSL1/2 also led to increase in DOT1L activity (Figure 3B). Mutation of H2B K34R or omission of ATP abolished MSL1/2 dependent H3 K79 methylation (Figure 3B). Immunoblots for total H2Bub showed that, consistent with Figure 2C, H2B K34R mutation resulted in enhanced ubiquitylation of H3 and H4 (Figure 3A and 3B, lane 6). However, these ubiquitylation did not affect H3 methylation in either case, suggesting certain level of specificity in ubiquitylation mediated trans-tail interactions.
Figure 3. H2Bub by MSL1/2 directly stimulates H3 K4/K79 methylation by MLL and DOT1L in vitro.
(A, B) Top, schematic for the sequential in vitro ub and HMT assays. Bottom, in vitro assays with enzymes, cofactors and substrates as indicated on top. [3H]-SAM was used as cofactor in HMT assays. Histone methylation was detected by autoradiograph and H2Bub was detected by anti-HA antibody. Coomassie stained gel for substrates was included at bottom. (A) MLL and (B) DOT1L.
H2B K34ub by MSL1/2 is important for global H3 K4me3 and H3 K79me2 in cells
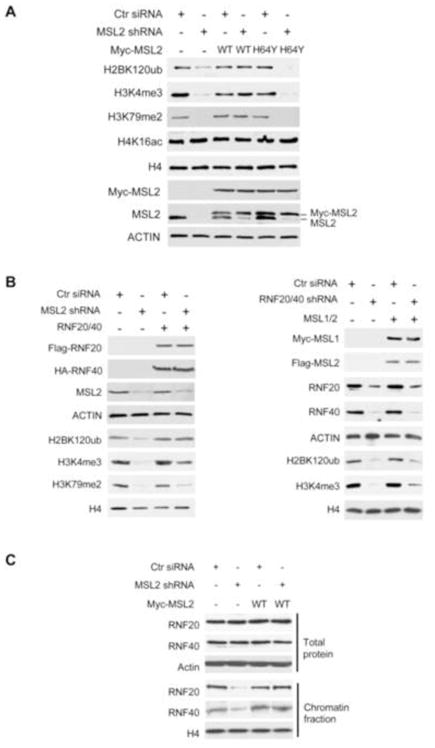
After establishing the crosstalk between H2B K34ub and H3 K4/K79 methylation in vitro, we further tested their interactions in HeLa cells. As shown in Figure 4A, MSL2 shRNA treatment resulted in significant reduction of global H3 K4me3 and H3 K79me2 compared to control siRNA treated cells, consistent with the in vitro results. Importantly, overexpression of MSL2, but not MSL2H64Y rescued reduction of H3 methylation, confirming that MSL2 activity is essential for trans-tail interaction. Reduction of H3 K4/K79 methylation was not due to indirect effect of MSL2 knockdown on expression of MLL or DOT1L genes, whose transcript levels were not affected by shRNA treatment (Supplementary Figure 4A). Consistent with previous results (Figure 2D), MSL2 knockdown led to global reduction of H2B K120ub (Figure 4A). This poses a question: are the effects on histone H3 K4 and K79 methylation in cells directly from H2B K34ub or indirectly from H2B K120ub? To address this question, we overexpressed RNF20 and RNF40 in cells treated with MSL2 shRNAs to restore H2B K120ub and then examine H3 methylation. As shown in Figure 4B, although H2B K120ub level was completely rescued by exogenous RNF20/40, H3 K4me3 and H3 K79me2 remained significantly lower than those in control cells (Figure 4B). This result argues that H2B K34ub directly stimulates activities of MLL and DOT1L in H3 methylation in cells (Figure 3A and 3B). We also performed the reciprocal experiment and knocked down RNF20/40 in cells followed by overexpression of MSL1/2. Consistently, MSL1/2 overexpression could not rescue H2B K120ub, but it partially restored H3 K4me3 (albeit at low level) (Figure 4B, lane 4 vs 2). These results suggest that MSL1/2 may function upstream of RNF20/40 in chromatin regulation.
Figure 4. H2Bub by MSL1/2 is important for H3 K4/K79 methylation in cells.
(A) HeLa cells were treated with MSL2 shRNAs followed by overexpressing Myc-tagged WT or MSL2H64Y mutant as indicated on top. (B) Left, HeLa cells were treated with MSL2 shRNAs followed by overexpressing RNF20 and RNF40. Right, HeLa cells were treated with RNF20/40 shRNAs followed by overexpressing MSL1 and MSL2. (C) Cells were fractioned into cytosol and chromatin fractions for immunoblot. (A–C) Antibodies were indicated on left.
MSL1/2 affects H2B K120ub by regulating chromatin association of RNF20/40
The observation that MSL1/2 knockdown affects H2B K120ub in cells is intriguing, especially when we failed to observe any activity by MSL1/2 for H2B K120ub in vitro (Figure 2A and 2C). To solve this discrepancy, we first looked at whether MSL1/2 affected expression and activity of RNF20/40. Although we found ~ 30% reduction of RNF20/RNF40 transcripts by MSL2 knockdown (Supplementary Figure 4A), we did not detect any changes in RNF20 and RNF40 at protein levels (Figure 4C, top panel). In addition, H2B K34ub by MSL1/2 only modestly affected RNF20/40 activity in vitro (Supplementary Figure 4B). We next examined whether MSL1/2 regulated recruitment of RNF20/40 to chromatin, a key step in regulating its activity (Laribee et al., 2007). Interestingly, MSL2 knockdown led to decrease in chromatin association for both RNF20 and RNF40, which can be completely rescued by Myc-MSL2 overexpression (Figure 4D). Although detailed mechanism for MSL1/2 dependent chromatin recruitment of RNF20/40 still needs to be worked out, our result here supports that MSL2 functions upstream of RNF20/40 and modulates H3 K4/K79 methylation both directly and indirectly.
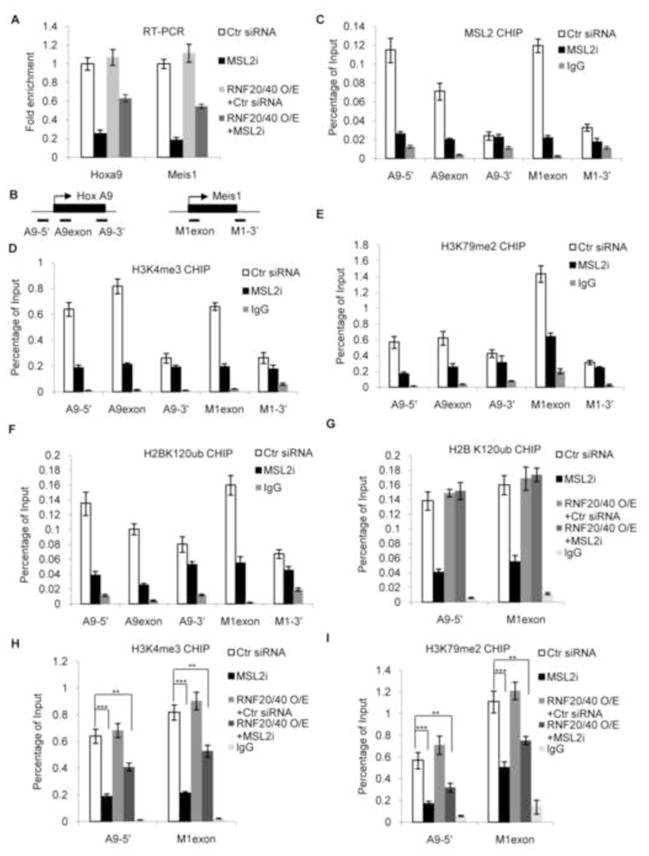
MSL1/2 regulates histone modifications at HOX gene loci
Given that MSL1/2 regulated global H2B K120ub, H3 K4me3 and H3 K79me2, we next examined its specific functions at HOX loci, whose regulation by H3 methylation and MOF was well documented (Dou et al., 2005; Milne et al., 2002). We first measured HOXA9 and MEIS1 gene expression in HeLa cells after treatment of: 1) control shRNA; 2) MSL2 shRNA; 3) RNF20/40 overexpression; and 4) MSL2 shRNA followed by RNF20/40 overexpression. Quantitative RT-PCR analysis showed that HOXA9 and MEIS1 expression was significantly down regulated (3–4 fold decrease) in MSL2 knockdown cells compared to controls (Figure 5A). This down regulation was not due to destabilization of the MOF-MSL complex (Supplementary Figure 4C). Importantly, ectopic expression of RNF20/RNF40 only partially rescued down-regulation of HOXA9 and MEIS1 (Figure 5A).
Figure 5. MSL1/2 regulates HOXA9 and MEIS1 expression through H3 methylation in cells.
HeLa cells were treated with MSL2 siRNAs with or without subsequent overexpression of RNF20/40. (A) RT-PCR for HOXA9 and MEIS1. Gene expression was normalized against GAPDH and presented as fold change against control siRNA treated sample, which is arbitrarily set at 1. (B) Primer sets for HOXA9 and MEIS1 used in ChIP assays. Black boxes, transcribed regions; Arrows, transcription start sites (Dou et al., 2005); Black lines, PCR amplified regions. (C–I) ChIP experiments using the probe sets indicated in (B). The antibody used for ChIP was indicated on top. Signals for each experiment were normalized to 5% input. Means and standard deviations (as error bars) from at least three independent experiments were presented. (H, I) Student t-test was performed for control and MSL2 knockdown cells as indicated. *, p< 0.05, **, p< 0.01 and ***, p<0.001.
To confirm that MSL2 directly binds to HOXA9 and MEIS1 loci, chromatin immunoprecipitation (ChIP) assays were performed for each gene locus (Figure 5B). MSL2 binding was detected in the transcribed regions of HOXA9 and MEIS1 with more binding at 5′ end (Figure 5C), which decreased after MSL2 shRNA treatment (Figure 5C). We next examined H3 K4me3, K79me2 and H2B K120ub by ChIP at these loci in MSL2 knockdown cells. Consistent with global changes, we found significant reduction of these marks upon loss of MSL2 (Figure 5D–5F). Importantly, although overexpression of RNF20/RNF40 fully restored H2B K120ub at 5′-region of HOXA9 and MEIS1 (Figure 5G), H3 K4/K79 methylation remained significantly lower (p<0.001) than that of control cells (Figure 5H and 5I), suggesting a direct effect of MSL2 mediated H2B K34ub on H3 K4/K79 methylation at specific gene loci. Results of RNF20/40 knockdown were consistent with previous reports (Supplementary Figure 5 and Kim et al., 2009).
Regulation of H2B K34ub and H4 K16ac in MOF-MSL
The finding of MSL1/2 ubiquitylation activity in MOF-MSL raises an interesting question: do histone ubiquitylation and acetylation activities mutually affect each other? To address this, we set up sequential in vitro HAT and Ub assays for MOF-MSL (Figure 6A and 6B). We found that prior acetylation by MOF-MSL did not affect H2B ubiquitylation, which remained the same with or without cofactor acetyl-CoA in the initial HAT assay (Figure 6A). Vice versa, prior H2B ubiquitylation by MOF-MSL did not affect H4 acetylation, which was the same with or without ATP in the initial ubiquitylation assay (Figure 6B). This result suggests that although these two histone-modifying activities reside within the same complex, they do not directly affect the enzymatic activity of each other.
Figure 6. Interplay between H4 K16ac and H2B K34ub.
(A) Top, schematic for the sequential in vitro HAT and histone ub assays. Bottom, MOF-MSL was first incubated with or without [3H]-acetyl-CoA as indicated on top. After an hour at 30°C, ATP was added to each reaction for the following histone ubiquitylation reaction. (B) MOF-MSL was first incubated with substrate, E1, E2 with or without ATP as indicated on top. After an hour at 30°C, [3H]-acetyl-CoA was added to each reaction for the following HAT reaction. In both (A) and (B), H4ac and H2Bub were detected by autoradiograph and anti-HA antibody respectively. Coomassie stained gel for nucleosomes was included. (C) In vitro HAT assays for different MOF-MSL sub-complexes as indicated on top. [3H]-acetyl-CoA was used as cofactor. H4ac was detected by autoradiograph. Immunoblot for H4 was used as the loading control. (D) Immunoblots for histone modifications using antibodies as indicated on left. Total proteins were isolated from HeLa cells after control, MSL1 or MSL3 siRNA treatment. (E) RT-PCR for cells treated with control, MSL1 or MSL3 siRNAs. Genes were indicated at bottom. Gene expression was normalized against GAPDH and presented as fold change against control siRNA treated sample, which is arbitrarily set at 1. (F–J) ChIP experiments in MSL1 or MSL3 siRNA treated cells using the indicated antibodies (on top). ChIP was quantified using the probe sets shown in Figure 5B. Signals for each experiment were normalized to 5% input. Means and standard deviations (error bars) from at least three independent experiments were presented.
Both H2B K34ub and H4 K16ac contribute to transcription regulation
We previously showed that MOF activity is tightly regulated by MSL1 and MSL3 (Li et al., 2009). MSL2, on the other hand, was dispensable for complex integrity (Supplementary Figure 2A) and for MOF activity (Figure 6C, lane 2). Since MSL1 was important for both H4 K16ac and H2B K34ub in vitro, we predicted that MSL1 knockdown would lead to global reduction of both marks in cells, which in turn led to decrease of H3 K4/K79 methylation. In contrast, MSL3 knockdown would only affect H4 K16ac. Indeed, this exactly was what we observed in cells (Figure 6D). MSL1 siRNA treatment significantly reduced global H3 K4me3, H3 K79me2 and H4 K16ac in HeLa cells (Figure 6D), which were not due to indirect effects on MLL and DOT1L expression (Figure 6E). In contrast, MSL3 knockdown only affected H4 K16ac (Figure 6D).
Different effects of MSL1 and MSL3 knockdown on histone modifications allow us to attribute respective functions of H4 K16ac and H2B K34ub in transcription regulation. To this end, we first examined HOXA9 and MEIS1 expression in cells treated with either MSL1 or MSL3 siRNAs. As shown in Figure 6F, MSL3 knockdown led to moderate decrease (30–40%) of HOXA9 and MEIS1 expression while MSL1 knockdown resulted in much more significant reduction (~50–70%), which is likely a result of simultaneous reduction of H2B K34ub and H4 K16ac. To confirm this, we performed ChIP assays for MSL2 and histone modifications at HOX and MEIS1 loci after MSL1 and MSL3 knockdown (Figure 6F–J). We found that MSL2 binding to these loci depended on MSL1 but not MSL3. Significant decrease in MSL2 binding was detected at both 5′ and 3′ transcribed regions of HOXA9 and MEIS1 after MSL1 knockdown (Figure 6F). Consistently, changes in H3 K4me3 (Figure 6H), H3 K79me2 (Figure 6I) and H2B K120ub (Figure 6J) at HOXA9 and MEIS1 were observed only after MSL1 knockdown. In contrast, comparable reduction of H4 K16ac was observed in cells treated with MSL1 or MSL3 siRNAs (Figure 6G). These results strongly argue that both H2B K34ub (and its subsequent changes in histone modifications) and H4 K16ac directly contribute to gene activation.
The histone E3 ubiquitin ligase activity of MSL1/2 is conserved in Drosophila DCC
Given the conservation of MSL1/2 to their Drosophila counterparts (Supplementary Figure 2C), we tested whether MSL1/2 ubiquitylation activity is evolutionarily conserved in fly. To this end, we co-expressed and purified a complex of dMSL1 (2-264aa) and dMSL2 (2-532aa) (Supplementary Figure 5A). In the in vitro ubiquitylation assay, Drosophila dMSL1/2 heterodimer demonstrated histone E3 ubiquitin ligase activity for nucleosomal H2B, similar to that of MSL1/2 (Figure 7A). This result suggests that Drosophila DCC, like the mammalian complex, harbors two distinct histone-modifying activities. It is interesting to note that mutations (i.e. C59A and C78A) in the second zinc cluster of dMSL2 led to male lethality in flies (Copps et al., 1998). Thus, it is important to test whether dH2B K31ub (mammalian K34 equivalent) by dMSL1/2 contributes to Drosophila dosage compensation in future.
Figure 7. Model for two mammalian MOF complexes in transcription activation.
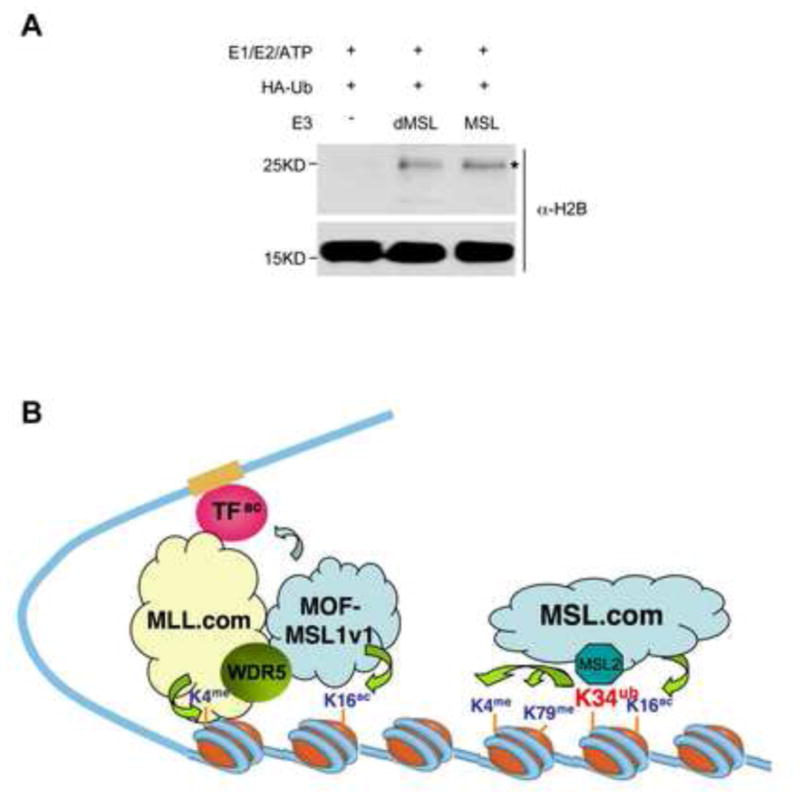
(A) In vitro ub assay for MSL1/2 and the Drosophila dMSL1/2 heterodimer as indicated on top. HeLa nucleosomes were used as the substrate. Histone ubiquitylation were detected by anti-HA antibody and expected ubH2B band is marked with *. Immunoblot for H2B was used as the loading control. (B) Model for the functions of two MOF complexes in transcription regulation. The MOF-MSL1v1 complex, which acetylates H4 and non-histone substrates, is recruited to target genes in coordination with MLL to facilitate transcription initiation. In contrast, the MOF-MSL complex functions to promote both H4 K16ac and H2B K34ub. H2B K34ub, in turn, promotes H2B K120ub, H3 K4me3 and K79me2 to facilitate transcription elongation.
DISCUSSION
Here we describe a histone E3 ligase activity for the MOF-MSL complex. We demonstrate that this activity mainly targets nucleosomal H2B K34, a site distinct from well-characterized H2B K120. We further show that both MSL2 and MSL1, but not MOF and MSL3, are indispensable for the optimal activity. Importantly, H2B K34ub by MOF-MSL directly stimulates H3 K4 and K79 methylation through trans-tail regulation. It also affects H2B K120ub by regulating RNF20/40 chromatin association. The significance of MSL1/2 mediated H2B K34ub is underscored by our revelation that MSL1/2 activity is evolutionarily conserved in Drosophila DCC. Altogether, these results shed new lights on the intricate network of chromatin modifying enzymes that often function coordinately in gene activation.
H2B K34ub by MSL1/2
Several early studies suggest that histone H2B can be ubiquitylated at sites other than K120 in vivo. For example, a proteomic study using mouse brain sample identifies five ubiquitylation sites on histone H2B, i.e. K34, K46, K108, K116 and K120 (Tweedie-Cullen et al., 2009). Ubiquitylation of endogenous yeast H2B at multiple lysine residues in addition to K123 (K120 equivalent) has also been reported (Geng and Tansey, 2008). Despite these observations, functions of these additional H2B ubiquitylation events as well as their respective Ub-conjugating enzymes and E3 ligases are largely unknown. Therefore, our study here represents the first step towards understanding the potential complexity of H2B ubiquitylation and their functions in transcription regulation beyond H2B K120ub. Comparing the activity of MSL1/2 to that of RNF20/40, several conclusions can be drawn: 1) MSL1/2 activity is lower than that of RNF20/40, contributing to low abundance of H2B K34ub in cells; 2) MSL1/2 has less strict substrate specificity, capable of weakly ubiquitylates H3 and H4 in vitro; and 3) MSL1/2 is mainly a mono-ubiquitin ligase, but it is able to add poly-ubiquitin chain to histones at lower efficiency. Given distinct substrate specificity of MSL1/2 and RNF20/40, it is likely that similar to other histone modifications (e.g. methylation, acetylation), H2B ubiquitylation is catalyzed by a multitude of E3 ubiquitin ligases with distinct substrate preference. Future studies on other RING finger-containing proteins will reveal yet uncharacterized E3 enzymes and histone targets.
H2B K34ub in trans-tail regulation
Like H2B K120ub, H2B K34ub plays important roles in regulating H3 K4/K79 methylation. It directly stimulates H3 K4 methylation by MLL and H3 K79 methylation by DOT1L in vitro (Figure 3A and 3B). This result implies that trans-tail interactions between H2Bub and H3 methylation have certain plasticity in terms of H2B ubiquitylation site requirement. This is consistent with a previous report that disulfide mimics of ubH2B (uH2Bss) at K125 and K116 sites are able to stimulate DOT1L activity in vitro (Chatterjee et al.). Given this newly demonstrated trans-tail regulation, especially that H2B K34 resides between DNA gyres, it is important to further examine the basis for H2Bub-mediated stimulation of methyltransferase activity. It would be particularly interesting to test whether H2B K34ub stimulates H3 methylation by altering intrinsic nucleosome stability and/or nucleosome breathing mode (Bohm et al., 2011). In addition to a direct effect on H3 methylation, MSL1/2 also affects H3 methylation indirectly through regulating recruitment of RNF20/40 to chromatin and thus, activity of RNF20/40 (Figure 4C). We are able to differentiate these two effects by overexpression of RNF20/40 in MSL2 knockdown cells (Figure 4 and 5). Both immunoblot and ChIP assays support that MSL1/2 regulates H3 methylation through both direct and indirect mechanisms, further strengthening the case that MSL1/2 regulates an intricate network of histone modifications for transcription activation in cells.
Both H2B K34ub and H4 K16ac contribute to MOF-MSL mediated gene activation
Since we are unable to detect any MOF-independent MSL1/2 heterodimer fraction on a size exclusion column (data not shown), it is likely that MSL1/2 always functions as an integral part of the MOF-MSL complex. Taking advantage of the fact that different histone modifying activities require different MOF-MSL components, we are able to distinguish contributions of H2B K34ub and H4K16ac to transcription activation. MSL1 knockdown, which affects both H2B K34ub and H4 K16ac, has much more profound effects on HOX gene expression than MSL3 knockdown, which only affects H4 K16ac (Figure 6). It indicates functions of MOF-MSL in transcription regulation beyond histone acetylation.
Our finding that MOF-MSL regulates H2B K120ub and H3 K79me2, two important marks for transcription elongation (Minsky et al., 2008; Roh et al., 2004), supports a role of MOF-MSL in later transcription events. Interestingly, the dMSL complex is implicated in the regulation of transcription elongation in Drosophila: 1) it is recruited to the bodies of X-linked genes; 2) recruitment of the dMSL complex to ectopic loci with X-chromosome enhancer sequences depends on active transcription (Gelbart and Kuroda, 2009); and 3) using global run-on sequencing (GRO-Seq), a recent study shows that the dMSL complex functions to facilitate progression of RNA Pol II across the bodies of active X-linked genes (Larschan et al., 2011). It would be interesting to test whether MOF-MSL in mammals regulates gene expression in the same manner and whether H2B K34ub (relative to H4 K16ac) contributes to the cascade of events associated with elongating Pol II. Future genome wide analyses of H2B K34ub and MSL1/2 in mammalian cells will be very informative to further dissect the functions of different histone modifying activities of MOF-MSL in transcription regulation.
Perspective
Several remaining issues for H2B K34ub await future studies. First, how H2B K34ub is regulated in vivo? A series of elegant yeast genetic studies and in vitro biochemical studies show that H2B K120ub is extensively regulated by factors associated with Pol II transcription. They include the PAF complex (Ng et al., 2003; Xiao et al., 2005), 19S proteasome (Ezhkova and Tansey, 2004), Pol II CTD and Kin28 kinase (Xiao et al., 2005). In addition, H2B K120ub is dynamically controlled by ubiquitin proteases such as Ubp8 and Ubp10 (Weake and Workman, 2008). Given the robust activity of MSL1/2 in vitro and low H2B K34ub level in cells, it is likely that H2B K34ub is under extensive regulation. It is important to examine whether regulation of H2B K34ub and H2B K120ub is the same or involves different mechanism despite their shared downstream effects on H3 methylation.
Second, what is the function of H2B K34ub in regulating chromatin structures? K34 resides in a highly conserved region in H2B from yeast to mammal. This region consists of a stretch of eight basic residues (Supplementary Figure 5B, blue). Like the basic patch on H4 N-terminal tail (14-20aa), this H2B region is also proposed to play important roles in formation of chromatin fibers by facilitating interactions with neighboring nucleosomes (Luger et al., 1997). This H2B basic patch overlaps with a conserved ‘HBR’ motif (30-37aa) identified in S. cerevisiae, whose deletion leads to de-repression of 8.6% of yeast genes (Parra et al., 2006). Given that both H2B K34 and H4 K16 are substrates of MOF-MSL and both reside within a basic patch, it is important to determine whether these two distinct histone modifications play any synergistic roles in regulating higher order chromatin structures.
Finally, does H2B K34ub play a role in Drosophila dosage compensation? Given the E3 ligase activity of the dMSL complex (Figure 7A), it is important to test whether dH2B K31ub marks Drosophila male X chromosome and whether inactive dMSL2 leads to defects in expression of X-linked genes in male fly. It has been reported that dMSL1/2, but not other dMOF complex components, binds to large chromosome domains defined as high affinity sites (HAS) or chromosome entry sites (CES) and initiates the spreading of H4 K16 acetylation mark along male X-chromosome (Alekseyenko et al., 2008; Straub et al., 2008). It is important to examine whether the dMSL1/2 activity plays any role in setting up these HAS/CES sites in vivo. One common feature of HAS/CES is their relatively low nucleosome density compared to other chromatin regions (Alekseyenko et al., 2008; Straub et al., 2008). In light of the E3 activity of Drosophila dMSL1/2, it is important to test whether dMSL1/2 prefers to bind nucleosome free region or this is a result of nucleosome destabilization caused by dH2B K31ub.
Experimental Procedure
Preparation of Histones, Recombinant Nucleosomes and Hela Nucleosomes
Xenopus H2A, H2B, H3, H4 and H2B mutants were expressed and purified according to Luger et al., 1997. Preparation of recombinant and HeLa nucleosomes was described previously (Dou et al., 2005).
In vitro Histone Ubiquitination, Acetylation and Methylation assays
Protocol for in vitro ubiquitylation, HAT and HMT assays was in Supplementary Info. The selection of E2 was decided empirically.
Mass Spec Analysis
Coomassie stained band corresponding to ubiquitylated H2B was subject to mass spec analyses, which was described in Supplementary Info.
Cellular assays including RT-PCR and ChIP experiments
Details for overexpression, knockdown and knockdown-rescue experiments as well as vector information are in Supplementary Info.
Supplementary Material
HIGHLIGHTS.
MSL2, a core component of the MOF-MSL complex, has E3 ubiquitin ligase activity.
MSL2 ubiquitylates histone H2B on K34, a conserved residue on H2B N-terminal tail.
H2B K34ub stimulates H3K4/K79 methylation by MLL and Dot1L respectively.
MSL2 activity is evolutionarily conserved in Drosophila dosage compensation complex.
Acknowledgments
We thank Drs. Wang for RING1/2/BMI1 viruses; Roeder and Kim for RNF20/40 and RAD6B vectors; Lucchesi and Smith for MSL1/3 antibodies; and Cadigan for Drosophila cDNA. We thank Drs Roeder and Engelke for critical reading of manuscript, and Lofgren for making Fig 2D. The work is supported by NIGMS and ACS grants to YD, NSF and NIH DP2 grants to BAG and a NSF Fellowship to BMZ.
References
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, Kuroda MI. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Toth K, Luger K, Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic acids research. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. The Journal of biological chemistry. 2009 doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nature chemical biology. 6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- Copps K, Richman R, Lyman LM, Chang KA, Rampersad-Ammons J, Kuroda MI. Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. Embo J. 1998;17:5409–5417. doi: 10.1093/emboj/17.18.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Molecular cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Molecular cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development (Cambridge, England) 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol Biol Cell. 2008;19:3616–3624. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Molecular cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995;81:867–877. doi: 10.1016/0092-8674(95)90007-1. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Molecular cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. The Journal of biological chemistry. 2009;284:3250–3263. doi: 10.1074/jbc.M805658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee RN, Fuchs SM, Strahl BD. H2B ubiquitylation in transcriptional control: a FACT-finding mission. Genes & development. 2007;21:737–743. doi: 10.1101/gad.1541507. [DOI] [PubMed] [Google Scholar]
- Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, Min J, Dou Y. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Dou Y. New perspectives for the regulation of acetyltransferase MOF. Epigenetics. 5 doi: 10.4161/epi.5.3.11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Molecular cell. 2009;36:290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annual review of genetics. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Molecular cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nature cell biology. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, Becker PB. Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. The EMBO journal. 2004;23:2258–2268. doi: 10.1038/sj.emboj.7600235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. The Journal of biological chemistry. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. The Journal of biological chemistry. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- Osley MA. H2B ubiquitylation: the end is in sight. Biochim Biophys Acta. 2004;1677:74–78. doi: 10.1016/j.bbaexp.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Parra MA, Kerr D, Fahy D, Pouchnik DJ, Wyrick JJ. Deciphering the roles of the histone H2B N-terminal domain in genome-wide transcription. Mol Cell Biol. 2006;26:3842–3852. doi: 10.1128/MCB.26.10.3842-3852.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Molecular and cellular biology. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS genetics. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tweedie-Cullen RY, Reck JM, Mansuy IM. Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. Journal of proteome research. 2009;8:4966–4982. doi: 10.1021/pr9003739. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Molecular cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Molecular and cellular biology. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.