Abstract
The U1A/U2B″/SNF family of small nuclear ribonucleoproteins uses a phylogenetically conserved RNA recognition motif (RRM1) to bind RNA stemloops in U1 and/or U2 small nuclear RNA (snRNA). RRMs are characterized by their α/β sandwich topology, and these RRMs use their β-sheet as the RNA binding surface. Unique to this RRM family is the tyrosine-glutamine-phenylalanine (YQF) triad of solvent-exposed residues that are displayed on the β-sheet surface; the aromatic residues form a platform for RNA nucleobases to stack. U1A, U2B″, and SNF have very different patterns of RNA binding affinity and specificity, however, so here we ask how YQF in Drosophila SNF RRM1 contributes to RNA binding, as well as to domain stability and dynamics. Thermodynamic double-mutant cycles using tyrosine and phenylalanine substitutions probe the communication between those two residues in the free and bound states of the RRM. NMR experiments follow corresponding changes in the glutamine side-chain amide in both U1A and SNF, providing a physical picture of the RRM1 β-sheet surface. NMR relaxation and dispersion experiments compare fast (picosecond to nanosecond) and intermediate (microsecond-to-millisecond) dynamics of U1A and SNF RRM1. We conclude that there is a network of amino acid interactions involving Tyr-Gln-Phe in both SNF and U1A RRM1, but whereas mutations of the Tyr-Gln-Phe triad result in small local responses in U1A, they produce extensive microsecond-to-millisecond global motions throughout SNF that alter the conformational states of the RRM.
Introduction
The U1A/U2B″/SNF family of metazoan proteins is found in the U1 and/or U2 snRNPs (1–4). In vertebrates, the U1A protein is bound to Stemloop II (SLII) of U1 small nuclear RNA (snRNA) in the U1 small nuclear ribonucleo protein (snRNP), and the U2B″ protein is bound to SLIV of U2 snRNA in the U2 snRNP. In other metazoans, there is a single protein, exemplified by Drosophila SNF, that binds to both SLII and SLIV. Proteins in the U1A/U2B″/SNF family have two RNA recognition motifs (RRMs), but only the N-terminal RRM1 is necessary for RNA recognition (5–8).
An RRM structure is a member of the larger class of ferredoxin folds, with a distinctive β1αβ2β3αβ4 secondary structure and a global α/β tertiary fold (Fig. 1). Within this fold class, RRMs are identified by their RNP sequences found in β1 (RNP2) and β3 (RNP1) (9). An unusual feature of the conserved RNPs is the presence of two or three aromatic amino acids displayed on the surface of the β-sheet, which are used for stacking with RNA nucleobases. U1A/U2B″/SNF RRMs display tyrosine in RNP2 and glutamine and phenylalanine in RNP1, which is a YQF triad unique to this family of RRMs. Fig. 1 illustrates the position of Y13-Q54-F56 in crystal structures of U1A RRM1 alone (Fig. 1 B) and bound to SLII (Fig. 1 C) (10). The solution NMR structure of RRM1 alone (11) locates its Q54 side chain with respect to Y13 by nuclear Overhauser effects (NOEs) from tyrosine aromatic protons to glutamine NH2 protons (Fig. S1 in the Supporting Material). The average distance between the Tyr13 ring and Gln54 NH2, calculated from the 43 NMR structures, is 2.9 ± 0.4 Å (Protein Data Bank (PDB) code 1fht (11)), consistent with a direct hydrogen bond between the hydroxyl oxygen (Y13) and amide side-chain proton (Q54). In the original cocrystal (10), there is a different hydrogen-bonding pattern: the Tyr13 hydroxyl group makes a hydrogen bond to the side-chain carbonyl oxygen of Gln54, whereas the Gln54 side-chain amide makes hydrogen bonds to the main-chain carbonyls of Lys50 and Arg52. Lys50 and Arg52 lie in the region of flexible loop 3 (between β2 and β3), which becomes ordered upon RNA binding. SNF and U1A RRM1s have the YQF triad and identical RNP sequences (β1 and β3), but their flanking amino acids in loop 3 and in β2 and β4 vary (Fig. 1 D). The network including the YQF triad will differ in the two proteins.
Figure 1.
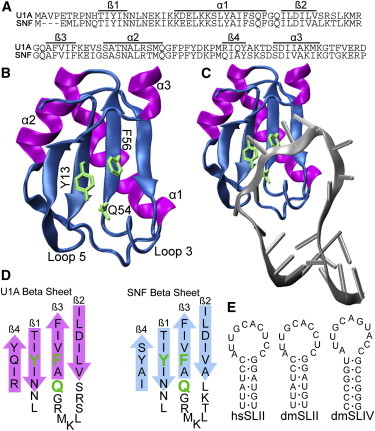
Structural characterization of U1A and SNF proteins. (A) Alignment of U1A and SNF RRM1. (B and C) U1A RRM 1 (PDB code 1urn) with the YQF triad indicated in green (B), and bound to SLII RNA (C) (10). (D) Organization of β-sheets with the YQF triad indicated in green. (E) RNA stemloops used in RNA binding experiments. hs, Homo sapiens; dm, Drosophila melanogaster.
The first cocrystal of U1A RRM1 bound to SLII (1990 (10)) showed the single-stranded loop of the RNA draped over the β-sheet of the RRM (Fig. 1 C), and the cocrystal of U2B″ RRM1 bound to SLIV (and to U2A′ protein) showed the same geometry (12). Notably, the U1A and U2B″ RRM structures in these cocrystals are nearly identical. In both complexes, the RNA loop nucleobases are stacked with tyrosine and phenylanine. Although this stacking interaction does not alone confer any sequence specificity to recognition of RNA by RRMs, the RNA is held in a particular orientation on the surface of the β-sheet that facilitates specific RNA-amino acid interactions, so the position of the aromatic amino acids in RNP1 and RNP2 is important for RNA binding. There is no structure of an SNF RRM1-RNA complex, but NMR solution structures of free SNF RRM1 (13) show it to be nearly identical to U1A RRM1.
SLII and SLIV RNAs are phylogenetically conserved in metazoans (14) and have similar loop sequences (Fig. 1 E). These 10 or 11 nucleotide loops are unstructured in solution (15), facilitating the RRM loop 3 insertion and nucleobase stacking with tyrosine/phenylalanine. The most significant difference between the RNAs is the loop-closing basepair: SLII has a conserved CG pair, whereas Drosophila SLIV has a wobble UG pair (human SLIV has a noncanonical UU pair). The RNA loops share the sequence AUUGCA on the 5′ side, which is specifically recognized by both U1A (16) and SNF (17).
Although their RNP sequences are identical, Drosophila SNF and human U1A have distinct RNA binding properties. In vitro experiments showed that human U1A RRM1 binds only SLII (for wild-type SLII, KD = 4.9 ± 0.1 × 10−11 M in 150 mM NaCl, 1 mM MgCl2, and 10 mM sodium cacodylate, pH 7.0) (18). Full-length SNF binds to SLII with higher affinity than to SLIV (KD (SLII) = 7.0 ± 6 × 10−10 in 250 mM KCl and 1 mM MgCl2, and KD (SLIV) = 4.5 ± 0.4 × 10−9 in 100 mM KCl, 1 mM MgCl2, and 10 mM sodium cacodylate, pH 8) (17). Certainly some of the binding properties of the two RRMs are attributable to sequence differences between U1A and SNF. In particular, the sequence of β2-loop 3 is known to contribute to RNA affinity/specificity (19–21). Here, we focus on their common YQF triad and its contribution to RRM properties.
To characterize the properties of SNF and U1A, we use NMR relaxation and dispersion methods, thermodynamic coupling, and RNA binding to study the interactions and dynamics of the RRMs with YQF perturbations. Our goal is to look for RRM properties that could explain the different molecular binding mechanisms of SNF and U1A and, more generally, to understand how RRMs use conserved sequences to bind different RNAs.
Methods
Protein purification
Purification of SNF and its mutants has been previously described (17). All mutations were introduced via QuickChange Mutagenesis (Agilent, Santa Clara, CA). Briefly, protein constructs were isolated from Escherichia coli BL-21(DE3) cells (Invitrogen, Carlsbad, CA) that had been transformed with a plasmid carrying the protein of interest under control of the TAC promoter. Cells were grown in Luria broth medium at 37°C and induced at OD600 = 0.6–0.8 with 1 mM isopropyl β-D-1-thiogalactopyranoside, then grown for an additional 4 h at 30°C. Proteins for NMR experiments were grown in M9 minimal media supplemented with either 15NH4Cl or 13C-glucose. Cells were pelleted and stored at −70°C until lysis in 50 mM sodium acetate, pH 5.3, 100 mM NaCl, 2 mM EDTA, and 8.5% sucrose supplemented with protease inhibitor cocktail (Sigma, St. Louis, MO), phenylmethanesulfonyl fluoride, and DNase II. Cell suspension was passed through a French press four times and then centrifuged. Lysate was passed over an SP-XL sepharose fast protein liquid chromatography column (GE Healthcare, Fairfield, CT) preequilibrated in 20 mM sodium cacodylate, pH 7, washed with 0 M and 100 mM NaCl, and eluted over a 100–400 mM NaCl gradient. Fractions containing protein were collected, concentrated, and buffer-exchanged into 10 mM sodium cacodylate, pH 7, 50 mM KCl, and 5 mM EDTA.
Protein circular dichroism/denaturation
A Jasco J715 spectropolarimeter was used to record circular dichroism spectra. All spectra were recorded at room temperature in buffer containing 50 mM KCl and 10 mM cacodylate, pH 7, for samples containing 20 μM protein. Circular dichroism spectra of each purified RRM (in 0 M urea) are consistent with a canonical RRM secondary structure.
For chemical denaturation, mean residue ellipticity at 221 nm (SNF) or 223 nm (U1A) was monitored as a function of denaturant. Unfolding curves were fit in KaleidaGraph to a two-state folding model using the linear extrapolation method of Santoro and Bolen (22). The unfolding free energy was averaged from two independent experiments and uncertainty was propagated from the fits. U1A RRM1 constructs were denatured in guanidine HCl, but SNF RRM1 constructs were much less stable, so urea was used. Even using urea, the lower (native) baseline of SNF RRM1 mutants was difficult to obtain, so these domains were stabilized by the addition of 0.5 M trimethylamine N-oxide (TMAO) in denaturation solutions. To calculate the folding free energy for the mutants in the absence of TMAO, the difference in wild-type SNF RRM1 (ΔGno TMAO (−3.5 kcal/mol) – ΔGTMAO (−4.2 kcal/mol) = 0.7 kcal/mol) was added to all calculated values. There is an assumption that the effect of TMAO on RRM stability is the same for wild-type and mutants.
RNA binding experiments
Most RNA binding was assessed by fluorescence and conducted using an SLM 8000 fluorometer. Temperature was maintained at room temperature using a circulating water bath. Binding buffer included 20 μg/mL bovine serum albumin in 10 mM potassium phosphate (pH 8.0) and indicated concentration of KCl and MgCl2. Fluorescein was attached at the 5′ end of the chemically synthesized RNA (IDT, San Jose, CA), to make 6-FAM-dSLII and 6-FAM-dSLIV (d indicates the Drosophila sequence). (FAM does not alter binding affinity; data not shown). RNAs were used at concentrations of 0.2 nM in all experiments. Acid-washed cuvettes were blocked with buffer with bovine serum albumin for 1 h. Emission and excitation wavelengths were set to 520 nm and 490 nm, respectively, with no polarizers. Fluorescence intensity was recorded as a function of total protein concentration and binding curves were fit to a standard Langmuir isotherm using KaleidaGraph (Synergy Software, Reading, PA). Binding of U1A Q54F to SLII was measured at pH 7 using nitrocellulose filter binding, as described previously (17) and binding curves were fit to a standard Langmuir isotherm using KaleidaGraph.
NMR spectroscopy
NMR data were acquired on 500 and 700 MHz Varian Inova spectrometers with z-axis pulsed-field-gradient triple-resonance Nalorac and Varian probes, respectively. High-field data were collected at the National Magnetic Resonance Facility (NMRFAM, Madison, WI) on a 900 MHz Varian spectrometer equipped with a z-axis pulsed-field-gradient triple-resonance cold probe. Wild-type and mutant protein samples included 20 mM sodium cacodylate (pH 6.5), 50 mM KCl, 2 mM EDTA, and 10% 2H2O at 22°C. Temperature was calibrated against a standard methanol NMR sample (23). 2,2-dimethyl-2-silapentane-5-sulfonate was added for chemical-shift reference. Assignments for U1A proteins were published previously (24,25). SNF chemical-shift assignments (13) were further refined with SNF mutant chemical shifts under our experimental conditions using 3D 15N TOCSY heteronuclear single-quantum coherence (HSQC) and15N NOESY-HSQC experiments (26). SNF asparagine/glutamine sidechain primary amide NH2 resonance assignments were refined with 3D 15N TOCSY-HSQC and 15N NOESY-HSQC where possible and confirmed with SNF mutants Q7H and Q33R. The U1A RRM1 mutant Q54F was assigned using established 3D triple-resonance backbone NMR experiments (HNCACB and CBCA(CO)NH) (27). NMR spectra were processed in NMRPipe (28) and analyzed in NMRViewJ (29).
The effective rotational correlation time, τc, was used as a check for intermolecular association in one-dimensional [15N, 1H]-TRACT experiments (30). Wild-type and mutant SNF proteins gave similar τc values: wild-type SNF, 6.3 ns; SNF(F53Y), 6.3 ns; SNF(Y10F), 6.8 ns; and SNF(Y10F/F53Y), 7.1 ns. The new U1A(Q54F) mutant gave τc = 5.8 ns, similar to that of U1A(Q54E) (5.9 ns) (25). TRACT experiments with U1A RRM1 gave τc = 6.3 ns.
The weighted change in backbone amide chemical shifts between wild-type and mutant proteins was calculated as:
where ΔδHN and ΔδN are the measured differences between wild-type and mutant chemical shift for 1HN and 15N, respectively, in parts per million, and n = 0.154 is a normalization factor for the15N chemical-shift range (31).
Heteronuclear 15N-{1H} NOE data were acquired as a pair of spectra, with and without proton saturation (32). Spectra with proton saturation included 3 s of relaxation delay and 3 s of saturation, whereas spectra without saturation included a 6 s relaxation delay between scans. Two sets of NOE spectra were collected and the peak intensity ratios (I/I0) were used to give the steady-state NOE. Errors in the NOE were determined by propagation of the baseplane noise.
Relaxation dispersion profiles were acquired with a relaxation-compensated Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence (33) at 700 1H MHz and 22°C for all proteins and mutants, and in addition at 900 MHz for wild-type U1A and SNF. Sixteen spectra with CPMG νCPMG values from 50 to 1000 Hz, Trelax = 40 ms, and one reference spectrum (no CPMG period) were collected. The relaxation delay was 2.5 s and R2,app values were determined as (34):
where Trelax is the total time for the CPMG refocusing period (40 ms) and I(νCPMG) and I0 are the peak intensities with and without CPMG (νCPMG) refocusing periods, respectively. Relaxation dispersion profiles were fit to a two-state model with GUARDD in MATLAB (35). Values of ΔR2, (the enhancement of the intrinsic R2 due to conformational exchange) were calculated from endpoints of the CPMG experiment:
ΔR2 values were calculated for all backbone amides, but full CPMG relaxation dispersion analyses were calculated only for selected residues. Errors were determined by the propagation of baseplane noise in ΔR2 experiments, and by repeat spectra in the GUARDD analysis of relaxation dispersion data.
Results
Thermodynamic coupling
We used thermodynamic double-mutant cycles to investigate pairwise coupling free energies between the conserved amino acids in the YQF triad on the surface of SNF RRM1 β-sheet in the free and bound states of the protein. If two sites are thermodynamically coupled, perturbations at one or both sites can result in a change in the interaction free energy (the pairwise coupling free energy (ΔGc)) (36–38). Briefly, the first perturbation is described as site j and the second perturbation as site i. A pairwise coupling cycle (see Fig. 2 B), is used to graphically describe the relationship between the wild-type protein and all three mutants (SNF and its Y10F (j), F53Y (i), and Y10F/F53Y (ij) constructs; mutations in U1A (Y13F (j), F56Y (i), and Y13F/F56Y (ij)) are taken from our previous study (25)). We emphasize that these are perturbations designed to disrupt single putative interactions (e.g., hydrogen bonding), rather than mutations that change the character of the site.
Figure 2.
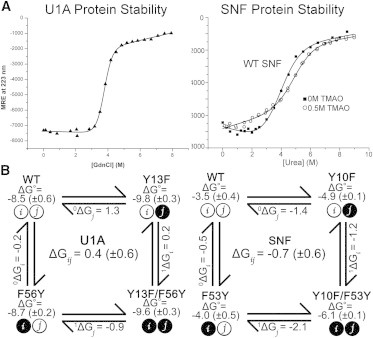
RRM unfolding and thermodynamic cycles. (A) Equilibrium unfolding of U1A and SNF RRM1. U1A was denatured in guanidine (25) and SNF in urea in the presence or absence of 0.5 M TMAO in 10 mM sodium cacodylate (pH 7) and 50 mM KCl, 22°C. (B) Thermodynamic pairwise coupling cycle of U1A (left) and SNF (right). ΔG° values for SNF are corrected for TMAO.
The coupling free energy, ΔGc = ΔGij, quantifies the change in local communication upon perturbation of the individual sites (Y10F and F53Y). Using the nomenclature of DiCera (36), ΔGij = 1ΔGi − 0ΔGi = 1ΔGj − 0ΔGj, where the superscript indicates the perturbation alone (0) or in the background of the second perturbation (1). Nonzero values of the pairwise coupling free energy indicate linkage between the two sites. A negative coupling free energy (ΔGc < 0) indicates that the perturbations have resulted in a loss of communication, whereas positive coupling free energy (ΔGc > 0) suggests that the perturbations have resulted in increased interaction free energy. By examining the pairwise coupling energy of the free protein using the folding free energy as a thermodynamic probe, we measure linkage between the two sites that could be indicative of preorganization of the binding surface. In the RNA-bound states of the protein, we use RNA binding free energies as a thermodynamic probe. The difference in the calculated pairwise coupling free energies in the bound and free states shows how interactions between the sites have changed upon complex formation.
RRM stability
The standard free energies of protein folding (ΔG°folding) for U1A, SNF, and YQF mutants provide the data for construction of a thermodynamic cycle. Protein stability of U1A and SNF RRM1 was measured using chemical denaturation (Fig. 2). Wild-type SNF has ΔG°folding = −3.5 (± 0.3) kcal/mol, which represents a very significant decrease in protein stability compared to wild-type U1A (ΔG°folding = −8.5 (± 0.6) kcal/mol) (18). Due to the relatively weak folding free energy of SNF and its mutants, 0.5 M TMAO was included in all the denaturation solutions. The effect was to raise the stability slightly (ΔΔG°TMAO = 0.7 kcal/mol), which was sufficient to allow more accurate fitting of the native baseline of the denaturation curves (Fig. 2 A, right). For comparison of SNF and U1A folding free energies, and for calculation of thermodynamic coupling, +0.7 kcal/mol was added to the measured values of all SNF folding free energies.
The thermodynamic cycle for free U1A RRM1 is shown in Fig. 2 B, left, with the folding free energies indicated in Table 1. Calculating the coupling free energy between sites Y13 and F56 shows weak positive coupling (1ΔGi − 0ΔGi = 0.2 − −0.2 kcal/mol = ΔGc = +0.4 (± 0.6) kcal/mol). SNF RRM1 exhibits weak negative Y10/F53 pairwise coupling (Fig. 2 B): (1ΔGi − 0ΔGi = −1.2 − −0.5 kcal/mol = ΔGc = −0.7 (± 0.6) kcal/mol). The flanking amino acids clearly influence the pairwise coupling of the conserved YQF triad, with the result that the surface organization of SNF differs fundamentally from that of U1A.
Table 1.
SNF and U1A folding free energy
| U1A RRM1a | ΔG°fold (kcal/mol) | ΔΔG°fold (kcal/mol) |
|---|---|---|
| WT | −8.5 (± 0.6) | — |
| Y13F | −9.8 (± 0.3) | −1.3 (± 0.3) |
| F56Y | −8.7 (± 0.2) | −0.2 (± 0.3) |
| Y13F/F56Y | −9.6 (± 0.3) | −1.1 (± 0.3) |
| ΔGc = 0.4 (± 0.6) | ||
| SNF RRM1 | ΔG°fold (kcal/mol) corrected for TMAO | ΔΔG°fold (kcal/mol) |
| WT | −3.5 (± 0.4) | — |
| Y10F | −4.9 (± 0.1) | −1.4 (± 0.4) |
| F53Y | −4.0 (± 0.5) | −0.5 (± 0.6) |
| Y10F/F53Y | −6.1 (± 0.1) | −2.6 (± 0.6) |
| ΔGc = −0.7 (± 0.6) |
ΔΔG°fold = ΔG° mutant − ΔG° WT. aData for U1A were reported previously (25). All chemical denaturation was conducted at 22°C at the buffer conditions indicated.
b150 mM NaCl, 10 mM sodium cacodylate, pH 7.0, 1 mM MgCl2; melted with GdnHCl.
¶ 50 mM KCl, 10 mM potassium phosphate, pH 7.0, 0.5 M TMAO; melted with urea.
The glutamine residue in the YQF triad is unusual among RRMs, and in previous experiments, we replaced it with glutamate or asparagine in U1A RRM1 (25). Here, we replaced it in U1A RRM1 with phenylalanine, which is more commonly found in this position in RRMs in other families (9). The resulting U1A YFF RRM was thermodynamically more stable than the wild-type by ∼1.7 kcal/mol (ΔG°folding = −10.3 ± 0.4 kcal/mol). This protein was used in experiments to represent the properties of a more canonical RRM triad in the context of this family. It also clarifies the unique contributions of the glutamine in the YQF triad. We were unable to express an SNF construct with the analogous mutation, which we assume is due to folding problems in E. coli.
RNA binding
The YQF triad is at the center of the RNA binding surface of these RRMs, and although there are no sequence-specific interactions between these amino acids and the RNA, the RNA nucleobases must stack with tyrosine and phenylalanine in a defined orientation. Our previous experiments with U1A YQF mutants showed that substitution of these residues can have a profound effect on RNA binding affinity, so here we repeated those experiments with SNF to compare their contribution to binding affinity.
The standard Gibbs free energies for RNA binding were determined for each RRM mutant using fluorescence of Drosophila FAM-dmSLII or FAM-dmSLIV (Table 2) in titration assays. Y10F and F53Y swaps on the binding surface of SNF RRM1 resulted in RNA binding affinities so low that it was difficult to accurately quantify them under the salt conditions used for the wild-type protein. For these technical reasons, we used the full-length SNF protein for all protein-RNA interaction assays. The added affinity of the full-length protein appears to be due to salt-dependent electrostatic interactions (17), which will not interfere with our analysis of Y and F perturbations on the domain surface. The U1A Q54F protein bound SLII so weakly that it had to be measured in very low salt, precluding its use in coupling studies. In 50 mM KCl and 10 mM sodium cacodylate, pH 7, it bound to SLII with KD = 8.5 ± 1.4 × 10−8 M.
Table 2.
SNF and U1A binding to SLII and SLIV
| U1A RRM1a | KD hsSLII (M)b | ΔG°bind (kcal/mol) hsSLII | ΔΔG°bind (kcal/mol) |
|---|---|---|---|
| WT | 4.9 ± 0.1 × 10−11 | −13.9 (± 0.1) | — |
| Y13F | 4.9 ± 0.1 × 10−9 | −11.2 (± 0.1) | +2.7 (± 0.2) |
| F56Y | 5.2 ± 0.1 × 10−10 | −12.5 (± 0.1) | +1.4 (± 0.2) |
| Y13F/F56Y | 1.3 ± 0.1 × 10−6 | −7.9 (± 0.1) | +6.0 (± 0.2) |
| ΔGc = +1.9 (± 0.3) | |||
| SNF FL | KD dmSLII (M) | ΔG°bind (kcal/mol) dmSLII | ΔΔG°bind (kcal/mol) |
| WT | 7.0 ± 6 × 10−10 | −12.5 (± 0.5) | — |
| Y10F | 7.3 ± 0.5 × 10−8 | −9.7 (± 0.04) | +2.8 (± 0.5) |
| F53Y | 3.3 ± 0.2 × 10−8 | −10.2 (± 0.04) | +2.3 (± 0.5) |
| Y10F/F53Y | 8.3 ± 0.7 × 10−7 | −8.3 (± 0.1) | +4.2 (± 0.5) |
| ΔGc = −0.9 (± 0.5) | |||
| SNF FL | KD dmSLIV (M)§ | ΔG°bind (kcal/mol) dmSLIV | ΔΔG°bind (kcal/mol) |
| WT | 4.5 ± 0.4 × 10−9 | −11.4 (± 0.1) | — |
| Y10F | 4.5 ± 0.2 × 10−8 | −10.0 (± 0.03) | +1.4 (± 0.1) |
| F53Y | 3.6 ± 0.3 × 10−8 | −10.1 (± 0.1) | +1.3 (± 0.1) |
| Y10F/F53Y | 4.0 ± 0.2 × 10−7 | −8.7 (± 0.03) | +2.7 (± 0.1) |
| ΔGc = 0.0 (± 0.1) |
ΔΔG°bind = ΔG° mutant − ΔG° WT.
aData for U1A were reported previously (25). All binding was conducted at 22°C in the buffer conditions indicated: † 150 mM NaCl, 10 mM sodium cacodylate pH 7.0, 1 mM MgCl2. ¶ 250 mM KCl, 10 mM potassium phosphate pH 8.0, 1mM MgCl2. § 100 mM KCl, 10 mM potassium phosphate pH 8.0, 1mM MgCl2.
To quantify the contribution of the YQF triad to RNA binding, we compare the binding free energies of the single and double mutants. The RNA binding free energy change is quantified as ΔΔG°binding = ΔG°binding(mutant) − ΔG°binding(wild-type) for each protein:RNA complex. If the two sites Y and F are not coupled in the bound state (i.e., they are independent), then the sum of ΔΔG°binding for each single protein mutant will be equal to the difference for the double mutant. If this is not the case, then the sites are not independent and there is coupling between them.
When SLII RNA is bound by U1A Y13F and F56Y mutants, ΔΔG°binding(Y13F) + ΔΔG°binding(F56Y) = +4.1 kcal/mol, whereas for the double mutant U1A(Y13F/F56Y), ΔΔG°binding = +6.0 kcal/mol. The effect of the double mutation is not additive, and we interpret this difference as evidence of coupling of the two sites when RNA is bound. The Y/F pairwise coupling free energy for SLII binding to U1A RRM1 is the difference between the two values, ΔGc° = +1.9 (± 0.3) kcal/mol. The pairwise coupling between these residues has increased almost fivefold in the complex from their coupling in the free protein. Note that coupling can be direct or indirect, and the difference seen here could contain a contribution from the RNA.
When SNF constructs bind to SLII, ΔΔG°binding(Y10F) + ΔΔG°binding(F53Y) = +5.1 (± 0.5) kcal/mol, which is measurably greater than the free-energy change for binding by the double mutant SNF(Y10F/F53Y), ΔΔG° = +4.2 (± 0.5) kcal/mol. Again the effect of the mutations is nonadditive, indicating that the sites are not acting independently. Calculation of the pairwise coupling free energy between the sites when SLII RNA is bound yields ΔGc° (SLII) = −0.9 (± 0.5) kcal/mol. However, when SNF binds to SLIV the effects of the two mutations are additive: ΔΔG°binding(Y10F) + ΔΔG°binding(F53Y) = +2.7 (± 0.1) is equal to SNF(Y10F/F53Y) ΔΔG° = +2.7 (±0.1) kcal/mol. The coupling free energy (ΔGc° (SLIV) = 0 (± 0.08)) indicates that the sites are acting independently. We conclude that 1), the contribution of SNF YQF to RNA binding is determined by the RNA sequence; and 2), that the RNA can alter the coupling of Y/F on the protein surface.
Backbone structural perturbations
A trivial explanation for the loss of RNA binding affinity upon mutation of tyrosine and phenylalanine is that the RRM structure has been perturbed. Here, we use NMR to compare the structures of wild-type SNF to SNF RRM1 Y10F, F53Y, and Y10F/F53Y. The 1H/15N backbone amide resonances of SNF are all broad with weak intensity, indicating that the RRM is sampling conformational space on the intermediate exchange regime of the chemical-shift timescale. The Y/F mutations in SNF do not alter this spectral property. The 1H-15N HSQC spectra for wild-type N-terminal RRM1 domains of U1A and SNF are compared in Fig. S2. Spectra of the mutants were readily assigned by reference to the wild-type proteins and so indicate that their tertiary structures are not significantly perturbed.
The mutations are a simple swap of the tyrosine and phenylalanine side chains on the surface of the β-sheet, but the swap will have changed the environment of flanking amino acids. To assess the extent of changes, we first compared chemical shifts of the backbone amides. (We assigned backbone amide chemical shifts for all U1A and SNF RRMs in our NMR buffer (20 mM sodium cacodylate, pH 6.5, 50 mM KCl, 2 mM EDTA, and 10% 2H2O at 22°C)). Amide backbone chemical-shift changes of U1A RRM1 Y13F, F56Y, and Y13F/F56Y are localized to two dominant regions of the protein: in loop 3/β3 and to a lesser extent at the C terminus of β4 and α3 (Figs. 3 and S3). The magnitude of the changes was greatest with proteins that contained the Y13F substitution and was minimal for the F56Y protein. The new U1A Q54F mutation resulted in the greatest magnitude and extent of chemical-shift changes, encompassing most of the β-sheet surface of U1A RRM1, including residues in β1, loop 1, loop 5, loop 3, and C-terminal sites (Fig. S4).
Figure 3.
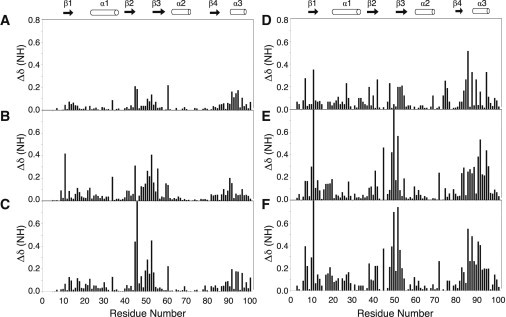
Backbone amide weighted chemical-shift differences plotted by residue for wild-type and mutant U1A and SNF RRM1 proteins. (A) U1A(F56Y). (B) U1A(Y13F). (C) U1A(F56Y/Y13F). (D) SNF(F53Y). (E) SNF(Y10F). (F) SNF(Y10F/F53Y). Secondary structural elements are shown above the plots for reference. Protein conditions were 300 μM, 22°C, in 50 mM KCl, 2 mM EDTA, and 20 mM sodium cacodylate, pH 6.5, 10% 2H2O. These data are mapped onto RRM structures in Fig. S3.
The Y10F/F53Y substitutions in SNF resulted in more widespread chemical-shift changes, encompassing β1, loop 1, loop 3, the β4/α3 junction, and the entire α3, especially prominent for Y10F mutants (Fig. 3). Backbone amide chemical-shift changes in SNF mutants are greater in magnitude compared to those in the corresponding mutants of U1A. We can interpret these data in terms of structural changes to the RRM, particularly in the case of the Y10F/F53Y mutant, where α3 resonances are shifted, suggesting that the entire helix has moved. The extent of the chemical-shift changes in SNF suggest that there is a broad network of residue interactions across the surface of the RRM (notably excluding α1 and α2) that was not observed in U1A.
We think it is significant that substantial chemical-shift perturbations were observed in loop 3 upon substitution of Y13F. Since loop 3 is responsible for RNA recognition, changes in the environment of its amino acids could have consequences for its interactions with nucleotides. We conclude that there is physical communication between loop 3 and the YQF triad in U1A and in SNF that is (somehow) responsible for weaker RNA binding.
Side-chain amides probe surface conformations
Interchanging the tyrosine and phenylalanine residues in the YQF triad led to weaker RNA binding of both U1A and SNF but did not report on the resultant properties of glutamine 54. To probe the consequences of Y/F swaps for Q54/51 in U1A/SNF, we monitored the primary amide chemical shifts (NH2) of asparagine and glutamine side chains (Fig. 4). U1A RRM1 has 10 Asn/Gln residues and SNF has 11. These residues are conserved, with the exception of residue 7 in the N-terminal tail, which is glutamine in SNF and histidine in U1A; SNF has Gln80, and U1A has Gln85. Four of the N/Q residues are on the RNA binding surface of U1A and SNF proteins, including Q54/51 (U1A/SNF) in the YQF triad.
Figure 4.
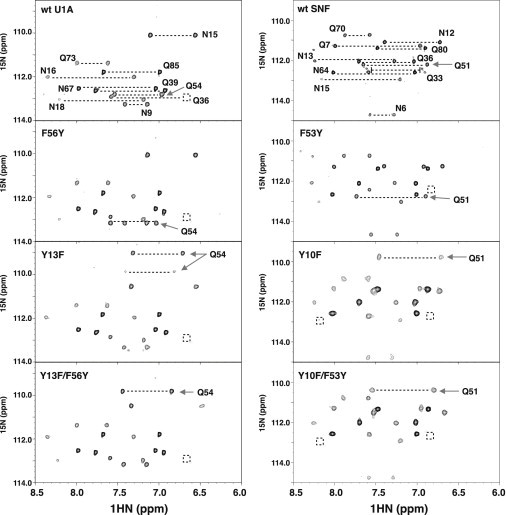
{1H}-15N HSQC spectra of Gln/Asn side-chain NH2 in wild-type and mutant U1A (left column) and SNF (right column) RRM1 proteins. Alternate side-chain conformations of wild-type SNF(Q33) are noted. Boxed regions indicate missing resonances from the NH2 pair at this contour level. Arrows show the NH2 side-chain peaks of U1A(Gln54) or SNF(Gln51). Protein conditions were 300 μM, 22°C, in 50 mM KCl, 2 mM EDTA, 20 mM sodium cacodylate, pH 6.5, 10% 2H2O.
The side-chain amide spectra of all U1A RRMs are well dispersed, and the two nonequivalent protons are observed for most amides. The Q54F mutant spectrum was nearly identical to that of the WT (Fig. S4). The greatest NH2 chemical-shift changes occur primarily at two positions, Q54 and N15, on the RRM surfaces of other U1A mutants (Fig. 4). Asn15 in β1 senses environmental effects of the mutation as evidenced by small changes in its 15N side-chain shifts. In the U1A(F56Y) mutant, the Gln54 NH2 pair moves ∼0.5 ppm (15N). Mutations with Y13F in the single and double mutants shift the Gln54 NH2 pair by ∼3.0 and 3.5 ppm (15N), respectively. More significantly, in the Y13F mutant, there are two pairs of Gln54 side-chain amide resonances, indicative of slow exchange between two conformations.
SNF side-chain amides are more crowded (Fig. 4). One contributor is Q33, located at the C-terminal end of α1, which has three discernible pairs of NH2 peaks in the spectrum of wild-type SNF. In SNF(F53Y), the Gln51 NH2 pair moves only slightly (by ∼0.5 ppm in 15N). Large perturbations to the Gln51 NH2 chemical shift arise from mutants containing the Y10F substitution: in the double mutant, Gln51 NH2 moves almost 2 ppm, and in Y10F, it moves ∼2.5 ppm (15N). Also present are changes in the NH2 pair corresponding to the neighboring Asn12 side chain, which moves ∼0.5 ppm in the Y10F mutants.
Responses of the Q54/51 and N15/13 side chains to the U1A and SNF mutants are strikingly similar. We propose that this result provides evidence of a specific interaction between two members of the unique YQF triad: specifically, a hydrogen bond between Q54/51-NH2 and Y13/10-OH. Its loss in the Y-to-F swap leads Q54/51 to find another hydrogen-bond acceptor nearby (perhaps N15/N13). The subsequent rearrangement of the RNA binding surface correlates with weaker RNA binding, which we suspect is due to both a structural and dynamic change in the RRM.
Fast (nanosecond to picosecond) timescale backbone dynamics
Since the Y/F swaps led to chemical-shift changes in the RRM backbone, we wanted to know if there was also a change in its global or local dynamics. To probe the rapid backbone amide dynamics of the RRMs, we used 15N relaxation experiments (T1, T2, 1H/15N NOE).
In general, we found that the fast-timescale dynamics of U1A and its mutants were unchanged throughout the body of the RRMs (Fig. S5). The {1H}-15N heteronuclear NOE (hetNOE) data report fast-timescale (nanosecond to picosecond) backbone dynamics and were used as a comparative method to look for changes in the proteins. We find that loop 3 shows enhanced backbone motions in all U1A proteins, {1H}-15N NOE ∼ 0.65, as do the C-terminal α-helix and N- and C-terminal residues ({1H}-15N NOE < 0.65).
We have collected T1 and T2 data for wild-type SNF, but because the resonances for SNF and the Y/F mutants are consistently broad and weak, we did not pursue them for the mutants. To avoid nonspecific intermolecular interactions, the protein concentration was 300 μM, which exacerbated the signal/noise problem. Although the SNF hetNOE data show more error (due to the weaker peak intensity), the wild-type and mutant data look very similar. The motions in the loops appear to be enhanced, although clearly the N-terminal residues and the C-terminal α3 have more rapid motions than does the body of the RRM.
Intermediate (microsecond to millisecond) timescale motions/conformational exchange
The NMR spectral characteristics of U1A and SNF RRM1 provide an immediate indication that the conformational-exchange properties of the two domains are different. A comparison of linewidths of the backbone amide resonance of U1A and SNF RRM1 {1H}-15N HSQC shows that U1A RRM1 resonances are sharp and intense, whereas those of SNF are broad and weak. This property of SNF RRM1 is also apparent in the full-length SNF protein (shown in a stacked plot of a portion of the full-length SNF HSQC (Fig. S6)), where resonances in RRM1 are less intense and broadened compared to those of RRM2. We used NMR CPMG experiments to compare the micro- to millisecond dynamics of U1A and SNF RRM1 and then compared the wild-type RRM dynamics to the Y/F mutants, looking for correlations with RNA binding affinity.
Both wild-type RRMs exhibit conformational exchange in the microsecond-to-millisecond regime in selected regions of the domain, but the exchange is more extensive for SNF. These data are compared in Fig. S7 in the plots of ΔR2 (ΔR2 was calculated from the difference in the endpoints of a CPMG relaxation dispersion experiment). We use ΔR2 to provide an estimate of motions on the microsecond-to-millisecond timescale, since it represents the line broadening from conformational exchange added to the intrinsic R2. These data are mapped onto the structures in Fig. 5.
Figure 5.
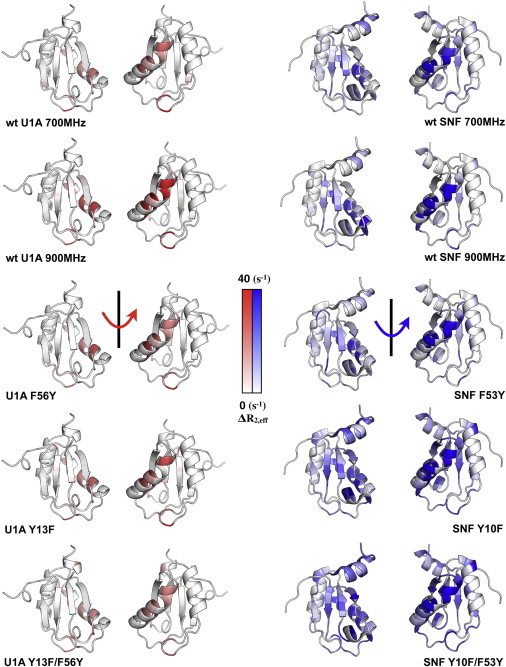
Intermediate-exchange ΔR2 for wild-type and mutant U1A (left column) and SNF (right column) RRM1 proteins. Data are plotted on the structures of U1A (PDB code 1urn) and SNF (PDB code 2k3k). In this U1A structure, the third α-helix was truncated, whereas in the SNF NMR structure, it is shown in one of its possible orientations (13).
In our experimental conditions, measured conformational exchange in wild-type U1A is localized to α1 with ΔR2 = 5–10 s−1 at 700 MHz (10–20 s−1 at 900 MHz) and to loop 5 (ΔR2 = 20 s−1). The same pattern is observed in the U1A(Q54F) (Fig. S5) and U1A(F56Y) RRMs. U1A(Y13F) increases the exchange rates slightly and extends them to loop 3, an effect that persists in U1A(Y13F/F56Y).
In general, exchange measured by ΔR2 in SNF on this intermediate timescale increases relative to that of U1A, with involvement of more of the RRM loops and body, a result consistent with its backbone peak intensities and linewidths. In detail, conformational exchange on this timescale in SNF encompasses α1, loop 3, and loop 5, with ΔR2 = 10–25 s−1. SNF(F53Y) dynamics are similar to those in the wild-type RRM, with an increase in exchange rates in α1 and loop 5. As with U1A(Y13F), SNF(Y10F) enhances the dynamics in α1 and loop 3 but now includes β1 (Fig. 5); this pattern is shared by SNF(Y10F/F53Y), but exchange rates are more rapid.
To quantify the conformational exchange occurring in these regions, backbone CPMG relaxation dispersion experiments were performed for wild-type U1A and SNF proteins (33,39). Relaxation dispersion data were fit to a two-state exchange model to compare the motional exchange rates, kex. Although this model is certainly inadequate to describe the conformational ensemble of SNF, the apparent kex values are used to compare the dynamical processes. Increased ΔR2 in regions of α1 and loop 5 were found in both U1A and SNF, so CPMG relaxation dispersion data in these regions were analyzed. Fig. 6 shows fits to α1 residues A29 and Q33 (SNF) and Y31 and Q36 (U1A), where kex ∼ 3000 s−1 for U1A and 1400 s−1 for SNF. In the side-chain amide spectrum of SNF (Fig. 4), Q33 had three pairs of resonances; here, we see that its backbone amide is also in exchange, although on a faster timescale. In U1A, residues from loop 5 could be fit to give kex ∼ 3000 s−1, but those residues in SNF loop 5 showed more complicated decay curves that could not be fit with this model.
Figure 6.
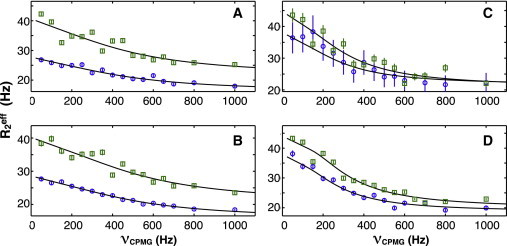
CPMG relaxation dispersion curves for U1A Y31 (A) and Q36 (B) and SNF A29 (C) and Q33 (D) at 900 (open squares) and 700 MHz (open circles), upper and lower curves, respectively. Lines are fits by GUARDD to a two-state model which gave estimated kex of 2900 s−1 (A), 3005 s−1 (B), 1190 s−1 (C), and 1550 s−1 (D). Errors were estimated from intensity analysis of duplicate spectra. To see this figure in color, go online.
The CPMG data clearly show that the two RRMs undergo different intrinsic motions in these experimental conditions. U1A α1 and loop 5 and SNF α1, loop 5, and loop 3 exhibit local intermediate dynamics. Whereas these local motions in U1A are relatively insensitive to Y/F swaps, the same mutations in SNF produce significant responses that extend beyond the local regions. Our conclusion is that there is a network of interactions connecting both sides of the RRM (β-sheet and α-helices). More experiments are necessary to map the network in the free proteins and to see how it changes in their bound states.
Discussion
The U1A/U2B″/SNF family of RRMs is not characteristic of most RRMs, since this family recognizes a complicated RNA sequence displayed in a preferred (hairpin) structural context with nanomolar affinity. To accomplish this selectivity, the RNA binding surface of the RRM had to specialize not only to select its preferred target but to reject others. Our results clearly identify the phylogenetically conserved YQF triad on the β-sheet surface as a critical element for establishment of a productive RNA binding surface. By comparison, common protein RRMs with their analogous triads include hnRNP A1 (RRM1 FFF and RRM2 FFF), PABP (RRM1 YYY, RRM2 FYF, RRM3 YFF, and RRM4 YFF), ASF/SF2 (YFF), nucleolin (RRM1 FFY, RRM2 LIY, RRM3 VYF, and RRM4 FFF), and SC35 (KFF) (9); these proteins have loosely defined RNA specificity (they are promiscuous) and often rely on multiple RRMs for affinity and selectivity. What we learn from the U1A/U2B″/SNF family is that the RNA binding surface of an RRM can become highly specialized. The RRM scaffold can be loosely organized (an ensemble) (SNF) or tightly packed (U1A), but it must maintain a network of connected residues that together regulate geometry and dynamics of the RNA binding surface. The YQF triad of the U1A/U2B″/SNF family appears to be the nexus of the network (40,41) that defines the structure/function/dynamics of these RRMs.
We compared Drosophila SNF RRM1 with human U1A RRM1 from the perspective of the protein stability, structure, and dynamics, as well as RNA binding, to develop a model of their functions. These two proteins represent the two phylogenetic contexts within this family: not only do metazoan organisms with a single SNF protein far outnumber organisms with selective U1A and U2B″ proteins, but the human proteins are examples of late-evolving RRMs found in jawed vertebrates (14). Therefore, Drosophila SNF is a prototype of the more common RRMs that bind two RNA targets, whereas human U1A has become specialized. SNF is more similar to the ancestral protein of this family (14), and its RNA binding surface represents the more ancient version of the protein. A comparison of their properties shows how RRMs in this family utilize both their common and unique elements to bind RNAs.
Protein dynamics, YQF, and RNA binding
U1A and SNF RRM1 both bind U1 SLII and have identical RNP sequences. However, their protein properties are quite different: the folding free energy of U1A RRM1 is −5 kcal/mol more favorable than that of SNF RRM1, and whereas U1A is best described as having a single dominant conformation (or at least in fast exchange with other conformations on the NMR chemical-shift timescale), SNF RRM1 is best described as a conformational ensemble. Where the Y/F swaps within the YQF triad introduce moderate local changes in the dynamics of U1A, the same swaps in SNF have widespread effects throughout the entire RRM. Of particular note is the increase in microsecond-to-millisecond motions that result in SNF upon Y10F substitution, effectively increasing the conformational heterogeneity (the ensemble) of SNF structures in the free protein.
Is there a functional connection between protein dynamics and the YQF triad? We suggest that there is, and that it differs between U1A and SNF. We interpret the chemical-shift changes in the Gln54/51 side-chain amide in Y-to-F swaps to indicate that it has lost its original hydrogen-bonding partner (Tyr13/10). Breaking this connection in U1A leads to locally enhanced mobility of loop 3, the β4/α3 junction, and α3. The implication is that these sites are physically connected (by direct or water-mediated hydrogen bonds, nonpolar packing, or electrostatics). These are the three parts of the protein that make sequence-specific contact with SLII, and we posit that their preorganization is necessary for RNA binding. In contrast, the Y-Q interaction in SNF acts as a staple to restrain the microsecond-to-millisecond dynamics of nearly the entire RNA binding surface. Loss of the Y-Q interaction increases the rate of conformational exchange between the members of the ensemble of SNF structures and likely also increases its ensemble complexity.
In the conformational capture model of RNA-protein interactions, the RNA is typically considered to be sampling conformations (its structure is best described as a conformational ensemble) (42,43). The protein must select the RNA structure that accommodates binding from among the ensemble; this model seems appropriate for the binding of U1A to SLII (44). However, when the protein is also sampling conformations, as is SNF, then binding requires mutual conformational capture. We suggest that this model applies to SNF-SLII and SNF-SLIV, and that when SNF dynamics are enhanced in the Y10F constructs, the process becomes less efficient and leads to weaker RNA binding. We predict that the conformational ensembles in the mutant SNF proteins will result in a more unfavorable entropy of binding, which can be experimentally tested by isothermal titration calorimetry measurements.
Kinetics experiments using surface plasmon resonance have been used to compare the on and off rates of U1A binding to SLII (45). Among the U1A mutants used in those experiments were Tyr13Phe and Phe56Tyr (in 150 mM NaCl at 22°C). Results showed that either substitution slowed the on rate by twofold compared to wild-type U1A, but that the off rates were significantly different. We predict that SNF Y10F mutants would have much slower on rates, if there is a change in the distribution of binding-competent SNF structures in the conformational ensemble of the Y10F mutants.
The Y10F swap in SNF also results in more extensive dynamics (ΔR2) on the intermediate timescale, involving both the β strands and α1. These data lead to our conclusion that the two sides of the RRM are more efficiently coupled in SNF than in U1A. The SNF Y10F/F53Y RRM shows the greatest response of α1 dynamics, and indeed of the entire domain; there is an increase in the ΔR2 rates overall. U1A is far more resistant to such changes and effectively maintains the wild-type pattern and rates. The network that couples the RNA binding surface, which we suspect is mediated by the Tyr-Gln interaction, seems to also couple to the back side of SNF RRM.
We suggest that the network propagates through the protein backbone and side chains to preorganize the RNA binding surface. This is not to say that the surface is fixed and static; loop 3 has significant mobility on the picosecond-to-nanosecond timescale, as does α1. We propose that the YQF triad is at the center of this network, holding it all together and restricting independent motions. Disruptions of the network lead to weaker RNA binding for both U1A and SNF. However, based on the observed negative thermodynamic coupling between tyrosine and phenylalanine in free U1A RRM1 and their positive thermodynamic coupling in free SNF RRM1, the YQF triad does not have the same function in U1A and SNF. In the complex, YQF appears to differentially modulate the RNA binding surface of SNF depending on which RNA is bound, as these sites appear to be entirely uncoupled in the SNF-SLIV complex. In other words, the RNA binding surface of SNF is controlled by RNA and tuned for specific recognition.
Several computational studies of U1A RRM1 in the free (46) and bound (41,47,48) states have identified residues that are part of networks with collective motions. The YQF sequences feature prominently in both states of the RRM, and in the complex bound to SLII, the networks included nucleobases. Analysis of short (10 ns) molecular dynamics trajectories of RRM1-SLII led Kormos et al. (41) to conclude that there were dynamical contacts throughout the RRM that contributed to RNA binding. The distribution of the networks across the RNA binding surface included loop 3 and the C-terminal region, in agreement with our previous thermodynamic pairwise coupling data that linked those sites (18,25). Other sites of dynamical contact were found in β1, β4, and loop 1 (41), which span the RNA binding surface. The networks that we now describe for both U1A and (particularly) SNF are more extensive than previously reported, as they encompass both sides of the RRM. These slower motions would not be captured in basic MD experiments but might be detected using other computational methods. Although the NMR data show that changes in backbone dynamics are correlated with site-specific mutations of the RRM, only the simulations can determine whether the correlations are causative and whether the motions are indeed collective. We expect that we will see dramatic differences in the networks in SNF and U1A both free and bound to RNA.
Conclusion
U1A and SNF have taken separate evolutionary paths to arrive at the same end: they both bind U1 snRNA SLII with nanomolar affinity. However, although U1A binds only SLII, SNF also binds SLIV. Here, we show that these RRMs are fundamentally different in their intrinsic backbone dynamics and folding free energies, yet they maintain extensive networks of intraresidue communications centered around their unique conserved tyrosine-glutamine-phenylalanine triad on the surface of the β-sheet. As such, they provide an intriguing example of the variations that can be accommodated in this small domain, and they may lead to new RNA binding mechanisms.
Acknowledgments
We thank Dr. Changguo Tang for assistance and maintenance of the NMR spectrometers. We thank Dr. Patrick Loria for the relaxation-compensated CPMG pulse sequence.
This study made use of the National Magnetic Resonance Facility at Madison (NMRFAM), which is supported by National Institutes of Health grants P41RR02301 (Biomedical Research Training Program/National Center for Research Resources) and P41GM10399 (National Institute of General Medical Sciences). We thank Dr. Marco Tonelli for help at NMRFAM. This work was supported by the National Institutes of Health (1R01 GM096444 to K.B.H.).
Supporting Material
References
- 1.Will C.L., Lührmann R. snRNP structure and function. In: Krainer A.R., editor. Eukaryotic mRNA Processing. IRL Press; Oxford, United Kingdom: 1997. pp. 130–173. [Google Scholar]
- 2.Scherly D., Boelens W., Mattaj I.W. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B” and their cognate RNAs. Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- 3.Harper D.S., Fresco L.D., Keene J.D. RNA binding specificity of a Drosophila snRNP protein that shares sequence homology with mammalian U1-A and U2-B″ proteins. Nucleic Acids Res. 1992;20:3645–3650. doi: 10.1093/nar/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polycarpou-Schwarz M., Gunderson S.I., Mattaj I.W. Drosophila SNF/D25 combines the functions of the two snRNP proteins U1A and U2B′ that are encoded separately in human, potato, and yeast. RNA. 1996;2:11–23. [PMC free article] [PubMed] [Google Scholar]
- 5.Scherly D., Boelens W., Mattaj I.W. Identification of the RNA binding segment of human U1 A protein and definition of its binding site on U1 snRNA. EMBO J. 1989;8:4163–4170. doi: 10.1002/j.1460-2075.1989.tb08601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boelens W., Scherly D., van Venrooij W.J. Analysis of in vitro binding of U1-A protein mutants to U1 snRNA. Nucleic Acids Res. 1991;19:4611–4618. doi: 10.1093/nar/19.17.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz-Freyermuth C., Query C.C., Keene J.D. Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem-loop II of U1 RNA. Proc. Natl. Acad. Sci. USA. 1990;87:6393–6397. doi: 10.1073/pnas.87.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boelens W., Scherly D., van Venrooij W.J. A weak interaction between the U2A′ protein and U2 snRNA helps to stabilize their complex with the U2B″ protein. Nucleic Acids Res. 1991;19:455–460. doi: 10.1093/nar/19.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birney E., Kumar S., Krainer A.R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oubridge C., Ito N., Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 11.Avis J.M., Allain F.H., Neuhaus D. Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol. 1996;257:398–411. doi: 10.1006/jmbi.1996.0171. [DOI] [PubMed] [Google Scholar]
- 12.Price S.R., Evans P.R., Nagai K. Crystal structure of the spliceosomal U2B″-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 13.Hu J., Cui G., Xia B. Structure and novel functional mechanism of Drosophila SNF in sex-lethal splicing. PLoS ONE. 2009;4:e6890. doi: 10.1371/journal.pone.0006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams S.G., Harms M.J., Hall K.B. Resurrection of an Urbilaterian U1A/U2B″/SNF protein. J. Mol. Biol. 2013;425:3846–3862. doi: 10.1016/j.jmb.2013.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rau M., Stump W.T., Hall K.B. Intrinsic flexibility of snRNA hairpin loops facilitates protein binding. RNA. 2012;18:1984–1995. doi: 10.1261/rna.035006.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall K.B. Interaction of RNA hairpins with the human U1A N-terminal RNA binding domain. Biochemistry. 1994;33:10076–10088. doi: 10.1021/bi00199a035. [DOI] [PubMed] [Google Scholar]
- 17.Williams S.G., Hall K.B. Coevolution of Drosophila snf protein and its snRNA targets. Biochemistry. 2010;49:4571–4582. doi: 10.1021/bi100374d. [DOI] [PubMed] [Google Scholar]
- 18.Kranz J.K., Hall K.B. RNA binding mediates the local cooperativity between the β-sheet and the C-terminal tail of the human U1A RBD1 protein. J. Mol. Biol. 1998;275:465–481. doi: 10.1006/jmbi.1997.1441. [DOI] [PubMed] [Google Scholar]
- 19.Bentley R.C., Keene J.D. Recognition of U1 and U2 small nuclear RNAs can be altered by a 5-amino-acid segment in the U2 small nuclear ribonucleoprotein particle (snRNP) B″ protein and through interactions with U2 snRNP-A′ protein. Mol. Cell. Biol. 1991;11:1829–1839. doi: 10.1128/mcb.11.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsamba P.S., Bayramyan M., Laird-Offringa I.A. Complex role of the β2-β3 loop in the interaction of U1A with U1 hairpin II RNA. J. Biol. Chem. 2002;277:33267–33274. doi: 10.1074/jbc.M200304200. [DOI] [PubMed] [Google Scholar]
- 21.Rimmele M.E., Belasco J.G. Target discrimination by RNA-binding proteins: role of the ancillary protein U2A′ and a critical leucine residue in differentiating the RNA-binding specificity of spliceosomal proteins U1A and U2B″. RNA. 1998;4:1386–1396. doi: 10.1017/s1355838298981171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolen D.W., Santoro M.M. Unfolding free energy changes determined by the linear extrapolation method. 2. Incorporation of ΔG° N-U values in a thermodynamic cycle. Biochemistry. 1988;27:8069–8074. doi: 10.1021/bi00421a015. [DOI] [PubMed] [Google Scholar]
- 23.Cavanagh J., Fairbrother W.J., Rance M. 2nd ed. Elsevier Academic Press; San Diego: 2007. Protein NMR Spectroscopy: Principles and Practice. [Google Scholar]
- 24.Showalter S.A., Hall K.B. Altering the RNA-binding mode of the U1A RBD1 protein. J. Mol. Biol. 2004;335:465–480. doi: 10.1016/j.jmb.2003.10.055. [DOI] [PubMed] [Google Scholar]
- 25.Kranz J.K., Hall K.B. RNA recognition by the human U1A protein is mediated by a network of local cooperative interactions that create the optimal binding surface. J. Mol. Biol. 1999;285:215–231. doi: 10.1006/jmbi.1998.2296. [DOI] [PubMed] [Google Scholar]
- 26.Zhang O., Kay L.E., Forman-Kay J.D. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 27.Muhandiram D.R., Kay L.E. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B. 1994;103:203–216. [Google Scholar]
- 28.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Johnson B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 30.Lee D., Hilty C., Wüthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J. Magn. Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 31.Mulder F.A., Schipper D., Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 32.Farrow N.A., Muhandiram R., Kay L.E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 33.Loria J.P., Rance M., Palmer A.G. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 1999;121:2331–2332. [Google Scholar]
- 34.Mulder F.A., Skrynnikov N.R., Kay L.E. Measurement of slow (μs-ms) time scale dynamics in protein side chains by 15N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 35.Kleckner I.R., Foster M.P. GUARDD: user-friendly MATLAB software for rigorous analysis of CPMG RD NMR data. J. Biomol. NMR. 2012;52:11–22. doi: 10.1007/s10858-011-9589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Cera E. Cambridge University Press; Cambridge, United Kingdom: 1995. Thermodynamic Theory of Site-Specific Binding Processes in Biological Macromolecules. [Google Scholar]
- 37.Di Cera E., Dang Q.D., Ayala Y.M. Molecular mechanisms of thrombin function. Cell. Mol. Life Sci. 1997;53:701–730. doi: 10.1007/s000180050091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.di Cera E. Site-specific analysis of mutational effects in proteins. Adv. Protein Chem. 1998;51:59–119. doi: 10.1016/s0065-3233(08)60651-8. [DOI] [PubMed] [Google Scholar]
- 39.Palmer A.G., 3rd, Kroenke C.D., Loria J.P. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 40.Anunciado D., Agumeh M., Baranger A.M. Characterization of the dynamics of an essential helix in the U1A protein by time-resolved fluorescence measurements. J. Phys. Chem. B. 2008;112:6122–6130. doi: 10.1021/jp076896c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kormos B.L., Baranger A.M., Beveridge D.L. A study of collective atomic fluctuations and cooperativity in the U1A-RNA complex based on molecular dynamics simulations. J. Struct. Biol. 2007;157:500–513. doi: 10.1016/j.jsb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haller A., Rieder U., Micura R. Conformational capture of the SAM-II riboswitch. Nat. Chem. Biol. 2011;7:393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 43.Leulliot N., Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 44.Pitici F., Beveridge D.L., Baranger A.M. Molecular dynamics simulation studies of induced fit and conformational capture in U1A-RNA binding: do molecular substates code for specificity? Biopolymers. 2002;65:424–435. doi: 10.1002/bip.10251. [DOI] [PubMed] [Google Scholar]
- 45.Law M.J., Chambers E.J., Laird-Offringa I.A. Kinetic analysis of the role of the tyrosine 13, phenylalanine 56 and glutamine 54 network in the U1A/U1 hairpin II interaction. Nucleic Acids Res. 2005;33:2917–2928. doi: 10.1093/nar/gki602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Showalter S.A., Hall K.B. A functional role for correlated motion in the N-terminal RNA-binding domain of human U1A protein. J. Mol. Biol. 2002;322:533–542. doi: 10.1016/s0022-2836(02)00804-5. [DOI] [PubMed] [Google Scholar]
- 47.Showalter S.A., Hall K.B. Correlated motions in the U1 snRNA stem/loop 2:U1A RBD1 complex. Biophys. J. 2005;89:2046–2058. doi: 10.1529/biophysj.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kormos B.L., Baranger A.M., Beveridge D.L. Do collective atomic fluctuations account for cooperative effects? Molecular dynamics studies of the U1A-RNA complex. J. Am. Chem. Soc. 2006;128:8992–8993. doi: 10.1021/ja0606071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


