Abstract
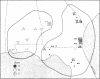
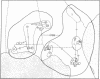
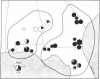
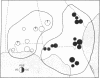
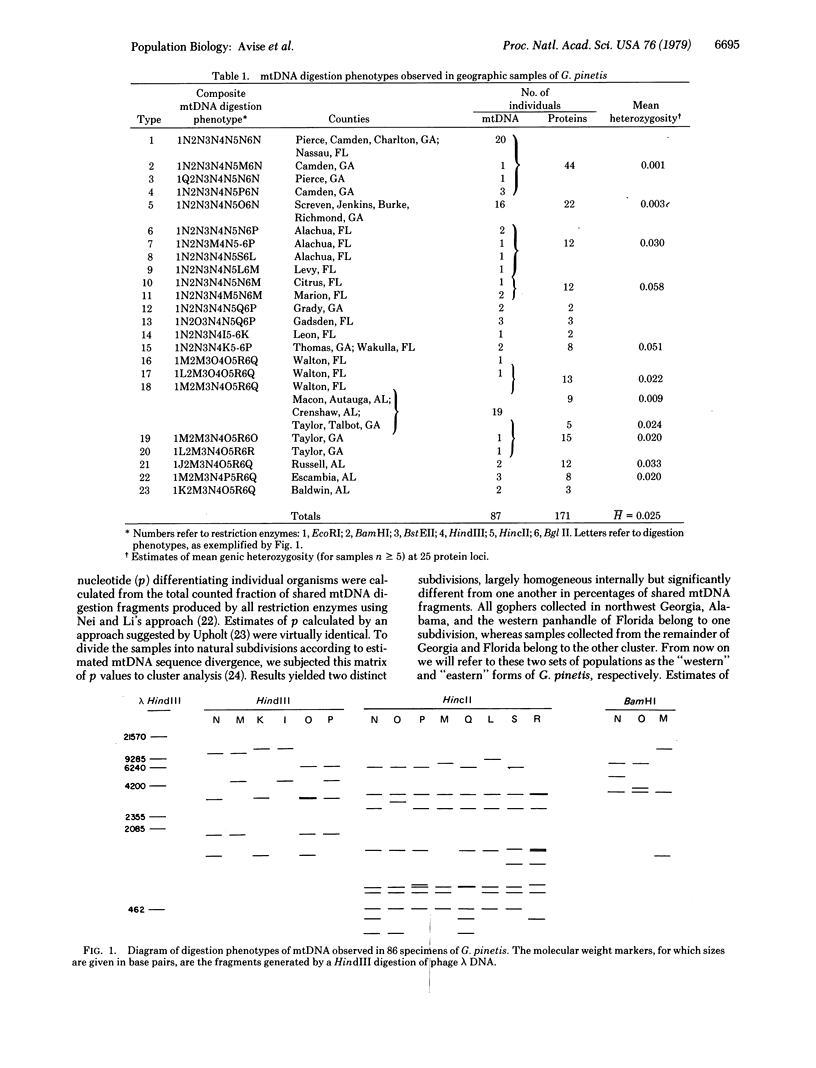
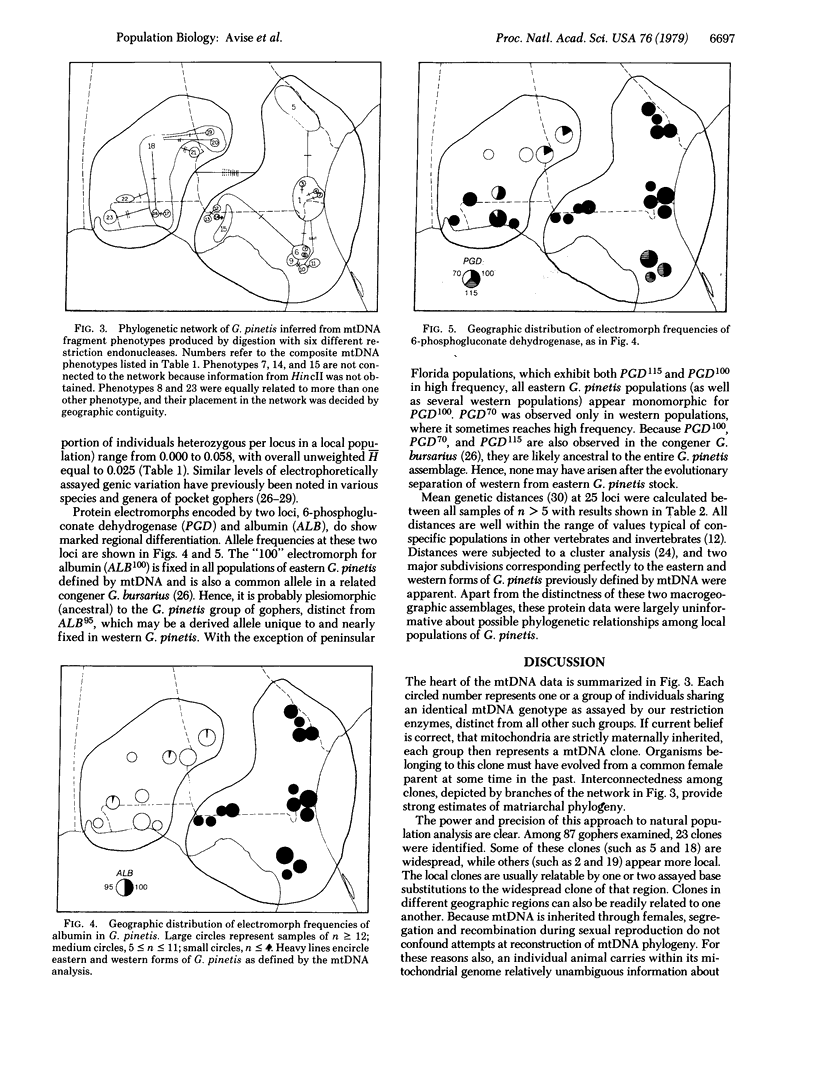
Restriction endonuclease assay of mitochondria DNA (mtDNA) and standard starch-gel electrophoresis of proteins encoded by nuclear genes have been used to analyze phylogenetic relatedness among a large number of pocket gophers (Geomys pinetis) collected throughout the range of the species. The restriction analysis clearly distinguishes two populations within the species, an eastern and a western form, which differ by at least 3% in mtDNA sequence. Qualitative comparisons of the restriction phenotypes can also be used to identify mtDNA "clones" within each form. The mtDNA clones interconnect in a phylogenetic network which represents an estimate of matriarchal phylogeny for G. pinetis. Although the protein electrophoretic data also differentiate the eastern and western forms, the data are of limited usefulness in establishing relationships among more local subpopulations. The comparison between these two data sets suggests that restriction analysis of mtDNA is probably unequalled by other techniques currently available for determining phylogenetic relationships among conspecific organisms.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Lansman R. A., Shade R. O. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics. 1979 May;92(1):279–295. doi: 10.1093/genetics/92.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala F. J., Powell J. R., Tracey M. L., Mourão C. A., Pérez-Salas S. Enzyme variability in the Drosophila willistoni group. IV. Genic variation in natural populations of Drosophila willistoni. Genetics. 1972 Jan;70(1):113–139. doi: 10.1093/genetics/70.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W. DNA restriction and modification mechanisms in bacteria. Annu Rev Microbiol. 1971;25:153–176. doi: 10.1146/annurev.mi.25.100171.001101. [DOI] [PubMed] [Google Scholar]
- Boyer H. W. Restriction and modification of DNA: enzymes and substrates. Introductory remarks. Fed Proc. 1974 May;33(5):1125–1127. [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Wright J. W. Mitochondrial DNA analyses and the origin and relative age of parthenogenetic lizards (genus Cnemidophorus). Science. 1979 Mar 23;203(4386):1247–1249. doi: 10.1126/science.424751. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Lansman R. A., Clayton D. A., Cohen S. N. Studies of mouse mitochondrial DNA in Escherichia coli: structure and function of the eucaryotic-procaryotic chimeric plasmids. Cell. 1975 Oct;6(2):231–244. doi: 10.1016/0092-8674(75)90014-8. [DOI] [PubMed] [Google Scholar]
- Coyne J. A. Lack of genic similarity between two sibling species of drosophila as revealed by varied techniques. Genetics. 1976 Nov;84(3):593–607. doi: 10.1093/genetics/84.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Blackler A. W. Maternal and cytoplasmic inheritance of mitochondrial DNA in Xenopus. Dev Biol. 1972 Oct;29(2):152–161. doi: 10.1016/0012-1606(72)90052-8. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G. B. Hidden alleles at the alpha-glycerophosphate dehydrogenase locus in Colias butterflies. Genetics. 1976 May;83(1):149–167. doi: 10.1093/genetics/83.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. L., Ohta T. Polyallelic mutational equilibria. Genetics. 1975 Apr;79(4):681–691. doi: 10.1093/genetics/79.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon A. M., de Vos W. M., Bakker H. The heterogeneity of rat-liver mitochondrial DNA. Biochim Biophys Acta. 1978 Jun 22;519(1):269–273. doi: 10.1016/0005-2787(78)90079-5. [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. S., Lewontin R. C., Felton A. A. Genetic heterogeneity within electrophoretic "alleles" of xanthine dehydrogenase in Drosophila pseudoobscura. Genetics. 1976 Nov;84(3):609–629. doi: 10.1093/genetics/84.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. S., Lewontin R. C., Felton A. A. Genetic heterogeneity within electrophoretic "alleles" of xanthine dehydrogenase in Drosophila pseudoobscura. Genetics. 1976 Nov;84(3):609–629. doi: 10.1093/genetics/84.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upholt W. B., Dawid I. B. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977 Jul;11(3):571–583. doi: 10.1016/0092-8674(77)90075-7. [DOI] [PubMed] [Google Scholar]
- Upholt W. B. Estimation of DNA sequence divergence from comparison of restriction endonuclease digests. Nucleic Acids Res. 1977;4(5):1257–1265. doi: 10.1093/nar/4.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]