Abstract
Replication and segregation of genetic information is an activity central to the well-being of all living cells. Concerted mechanisms have evolved that ensure that each cellular chromosome is replicated once and only once per cell cycle and then faithfully segregated into daughter cells. Despite remarkable taxonomic diversity, these mechanisms are largely conserved across eubacteria, although species specific distinctions can often be noted. Here, we provide an overview of the current state of knowledge about maintenance of the chromosome structure in Pseudomonas aeruginosa. We focus on global chromosome organization and its dynamics during DNA replication and cell division. Special emphasis is made on contrasting these activities in P. aeruginosa and other bacteria. Among unique P. aeruginosa features are the presence of two distinct autonomously replicating sequences and multiple condensins, which suggests existence of novel regulatory mechanisms.
Keywords: condensins, MksBEF, SMC, chromosome structure, Pseudomonas aeruginosa, PA4685
In the 1970’s Booker and Loutit proposed that Pseudomonas aeruginosa strain PAO1 has two chromosomes, based on conjugational linkage studies (Booker & Loutit, 1974). This later proved incorrect. There is, of course, only one chromosome in P. aeruginosa, whereas the manifestation of the two linkage groups was caused by the now well-known phenomenon of clustering of housekeeping genes near the origin of replication and accessory genes near the terminus (Nichols, et al., 2011). However, although our understanding of bacterial genome organization greatly improved since then, we still know far less about chromosome maintenance in P. aeruginosa than in its more celebrated relatives Escherichia coli, Caulobacter crescentus and Bacillus subtilis. This short review attempts to take stock of our present knowledge about replication and organization of P. aeruginosa chromosome with the focus on global chromosome dynamics.
The chromosome of P. aeruginosa (6.3 Mb for strain PAO1) is about one third longer than that of a typical laboratory strain of E. coli (4.6 Mb) or B. subtilis (4.2 Mb). The evolutionary origins of the extra sequence are yet to be determined. What is clear, however, is that the greater length of the P. aeruginosa chromosome results from genetic complexity rather than gene duplication, which allows this bacterium to colonize diverse niches (Stover, et al., 2000, Silby, et al., 2011). Encoded within this additional DNA is a variety of biosynthetic enzymes, transport systems, transcription factors and signal-response regulators, which allow this bacterium not only to strive in diverse environments but also display a remarkable plasticity of gene expression and ability to differentiate into metastable populations (Stover, et al., 2000, Lee, et al., 2006). Additional layers of complexity can be also found in global chromosome organization and segregation; however, their contribution to the cell physiology is only beginning to emerge.
DNA polymerases
Bioinformatics analysis identifies five DNA polymerases in PAO1. Four of them are homologous to the E. coli polymerases I through IV (Table I), whereas no homolog can be found to the Y-family UmuDC translesion polymerase. UmuDC, also known as Pol V, is induced as a part of SOS response and, following RecA-dependent activation, supports DNA synthesis across damaged DNA (Sutton & Walker, 2001, McHenry, 2011). Although this type of replication is highly mutagenic, it also allows cells survive heavy damage to DNA.
Table I.
DNA polymerases in E. coli and P. aeruginosa
| Polymerase | E. coli | P. aeruginosa |
|---|---|---|
|
| ||
| Pol I | PolA | PA5493 |
| Pol II | PolB | PA1886 |
| Pol IIIα | DnaE | PA3640 |
| Pol IV | DinB | PA0923 |
| Pol V | UmuC | none |
| DnaE2- | none | PA0669 |
The fifth P. aeruginosa polymerase, DnaE2 (PA0669), belongs to the Pol IIIα family and is broadly spread among several subdivisions of eubacteria (Timinskas, et al., 2014). DnaE2 is non-essential, at least in planktonic cells, and contributes to error-prone DNA repair (Sanders, et al., 2006). The other two genes in the dnaE2 operon share high similarity to a Y-family DNA polymerase (PA0670) and an SOS-induced inhibitor of cell division SulA (PA0671). These data indicate that DnaE2 might play the same role in P. aeruginosa as UmuDC in E. coli and is likely responsible for SOS-induced DNA repair.
Compared to its E. coli counterpart, the replicative DNA polymerase Pol III lacks the theta subunit of its core polymerase. In contrast to the rest of the subunits, the psi subunit of the clamp loader (PA4679) displays little homology to its E. coli counterpart and is misannotated in the primary databases as is the start codon of the gene (Jarvis, et al., 2005). Based on protein expression and copurification studies, the correct 5′ end of the gene was assigned to the in-frame UUG codon located 135 bp upstream from the database prediction. Both the theta and psi subunits are non-essential in E. coli (McHenry, 2011), indicating that their primary function is in coordination of DNA synthesis with other cellular activities rather than in DNA synthesis itself. Accordingly, the rate of DNA replication is not impaired by their absence. Indeed, PAO1 transfers the entire chromosome during conjugation in 75 min (O’Hoy & Krishnapillai, 1987). For comparison, E. coli K-12 transfers its chromosome in 100 min. At least in E. coli, a similar rate of DNA synthesis is observed during chromosome replication. Given that DNA replication must keep up with DNA transfer, these data imply that, at least during conjugation, DNA synthesis occurs twice as fast in PAO1 that in E. coli!
The origin of replication
Several key chromosomal loci have been mapped in P. aeruginosa genome by now. These include the origin of replication, OriC (Yee & Smith, 1990, Jiang, et al., 2006), ParS sites, which are required for correct chromosome partitioning (Bartosik, et al., 2004, Livny, et al., 2007), and the dif sites, where XerCD recombinase resolves chromosome dimers. No ter system, which ensures termination of chromosome replication, has been described so far. Likewise, no proteins with significant homology to Tus or RTP, which facilitate termination of replication in E. coli and B. subtilis, respectively, can be found in PAO1 genome (Lewis, et al., 1990, Bastia, et al., 2008). The lack of a close ortholog might not be surprising given the low sequence and even structure similarity between Tus and RTP (Bussiere, et al., 1995, Kamada, et al., 1996) and suggests that pseudomonads employ their own unique system, if any, to limit chromosome over-replication.
Bacterial origins of replication are several hundred base pairs long and located mostly in intergenic regions. Initiation of DNA replication is triggered by the binding of DnaA to its targets followed by loading of the helicase DnaB onto the nearby AT-rich repeats also known as the DNA unwinding element, DUE (Kornberg & Baker, 1992, Mott & Berger, 2007). The consensus sequence for the DnaA box is TTATNCACA with only one or two mismatches in it found in diverse bacteria (Mott & Berger, 2007, Zakrzewska-Czerwinska, et al., 2007). A typical origin contains between two and five closely spaced DnaA boxes and two or three tandemly arranged AT-rich repeats. An exception to this rule is found in alfa-proteobacteria where as few as two DnaA boxes and with significant deviation from the consensus sequence can suffice for initiation of chromosome replication (Ioannidis, et al., 2007, Shaheen, et al., 2009). An inspection of numerous bacterial genomes revealed that the density of DnaA boxes serves as a good predictor of the location of the replication origin (Mackiewicz, et al., 2004). Notably, the DnaA boxes do not have to be continuously located since they are brought together during origin activation via DNA bridging activity of DnaA. The split organization of OriC was reported for B. subtilis, where elements of the origin can be found both up- and downstream of dnaA (Moriya, et al., 1992, Smits, et al., 2011).
Several clusters of DnaA boxes are often found elsewhere on the chromosome. Chromatin immunoprecipitation studies revealed that these clusters indeed serve as a high affinity binding site for DnaA and contribute to correct timing of DNA replication in B. subtilis (Smits, et al., 2011, Okumura, et al., 2012).
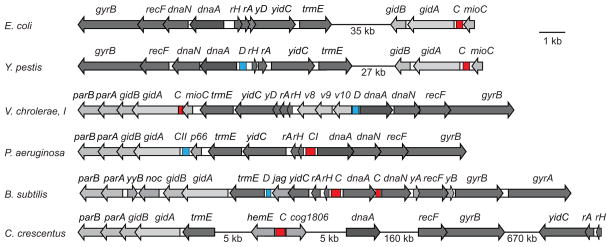
Aside from the DnaA boxes, the origins of replication are poorly conserved and show detectable homology only among closely related species. Their genomic context displays greater stability. Inspection of multiple genomes reveals existence of two characteristic cassettes that harbor oriC regions (Fig. 1). The first of them, found in E. coli and closely related γ-proteobacteria, carries oriC between mioC and gidA (glucose inhibited cell division) genes in the mioC-oriC-gidA-gidB cassette (von Meyenburg, et al., 1982, Ogawa & Okazaki, 1991). gidA and gidB are involved in posttranslational modification of, respectively, tRNA and 16S RNA (Okamoto, et al., 2007, Moukadiri, et al., 2009). The mechanism that leads to inhibition of cell division upon disruption of these genes remains unknown. This lays grounds for an intriguing possibility that these proteins are involved in cell cycle control of DNA replication (although other mechanisms cannot be ruled out). Curiously, some bacteria carry parA and parB genes within this cassette, whose involvement in chromosome replication and segregation is far better established. Notably, this arrangement of the genes is conserved even in bacteria that initiate chromosome replication from other loci (Fig. 1).
Fig. 1.
Comparison of genomic context for various bacterial origins of replication. The origin of replication (C, red) and a DnaA box clusters with DUE (D; blue) are often found embedded within the dnaA or gidA cassettes. rH, rpmH; rA, rnpA; yD, yidD; v8, VC0008; v9, VC0009; v10, VC0010; p66, PA5566; yyB, yyaB; yA, yaaA; yB, yaaB.
The second cassette contains oriC between divergently expressed rpmH and dnaA, which encode ribosomal protein L34 and replication initiator DnaA. The intergenic space between rpmH and dnaA can be often found even in species that initiate replication elsewhere (Fig. 1). In many bacteria, including P. aeruginosa, the two gene clusters are located together and face in opposite directions (Briggs, et al., 2012). Curiously, a genetic screen for autonomously replicating sequences, ARS, identified both these cassettes as a potential origin of replication (Yee & Smith, 1990). The two elements, named oriCI and oriCII, contain five DnaA boxes and two (oriCII) or three (oriCI) AT-rich 13-mer repeats. Although only one of the origins, oriCI, is essential in planktonic bacteria, any of them can support propagation of an otherwise origin-less plasmid (Jiang, et al., 2006). In contrast, the E. coli rpmH-dnaA intergenic region carries only one consensus DnaA box, which is consistent with previous failures to find alternative autonomously replicating sequences in E. coli K-12.
Pseudomonads are not the only bacteria to harbor more than one DnaA box cluster complete with a plausible DUE. A similar arrangement can be found in Vibrio cholerae and Yersinia pestis, which are presumed to initiate replication from the cluster upstream from gidA (Fig. 1). This comparison reveals that migration of OriC from the dnaA to gidA cassette is a process distinct from the large chromosome rearrangement that split the two cassettes apart in enterobacteria. In firmicutes, the origin proximal DnaA box cluster is also found in the vicinity of trmE, but not downstream from it as in γ-Proteobacteria but upstream, next to the firmicute specific gene jag (Fig. 1).
Notably, the two cassettes do not exhaust potential locations for oriC. In C. crescentus and other α-proteobacteria, for example, the origin of replication is found between divergent hemE (encoding an uroporphyrine decarboxylase; CC_3763) and cog1806 (a putative PEP synthetase regulatory protein; CC_0001) genes (Shaheen, et al., 2009). It is unclear, however, whether or not this distinction points to independent evolutionary origins of the C. crescentus Cori. Indeed, the hemE-oriC-cog1806 fragment is located only 5 kb upstream from the trmE-gidAB-parAB cassette and dnaA, and such migration could potentially occur in a single recombination event.
Initiation of DNA replication
The reason why PAO1 does not use oriCII is unclear and suggests the existence of additional control elements. Several such systems have been described in various bacteria. The first one involves dam DNA methylation coupled to the activity of SeqA (Slater, et al., 1995). The E. coli oriC contains multiple GATC sites, which are methylated at N6 position of adenines in both DNA strands by dam methylase prior to replication. Curiously, the origin-less intergenic rpmH-dnaA region in enterobacteria also contains multiple dam methylation sites. DNA replication converts the fully methylated GATC into their hemimethylated versions (Zyskind & Smith, 1986), which, in turn, recruit SeqA and become sequestered from DnaA and dam methylase (Slater, et al., 1995, Brendler & Austin, 1999). This system provides a time delay needed for replication to advance before DNA is fully methylated again and the next round of replication is initiated (von Freiesleben, et al., 2000). As a result, it blocks premature initiation of DNA replication and is essential for coordination of chromosome replication and segregation (Riber & Lobner-Olesen, 2005). Inactivation of this pathway results in excessive initiation of DNA replication, increased DNA damage and asynchronous chromosome replication (Boye, et al., 1996). Notably, this system is unique to enterobacteria and a subset of γ-proteobacteria and is not found in pseudomonads (Brezellec, et al., 2006).
The second system was described in C. crescentus and involves cell-type specific control of DNA replication. C. crescentus undergoes asymmetric cell division producing a surface-attached stalked cell and a mobile swarmer cell (Domian, et al., 1997, McAdams & Shapiro, 2003). The swarmer cells do not replicate their chromosomes until they differentiate into a stalked cell. This is accomplished with the help of CtrA (cell cycle transcription regulator A) protein, which is expressed in swarmer but not stalked cells (Domian, et al., 1997). The C. crescentus origin of replication, Cori, contains five CtrA binding sites, which are spread throughout Cori (Siam & Marczynski, 2000). Binding of CtrA to Cori apparently blocks DnaA binding and initiations of replication. A similar system has been recently identified in B. subtilis, where a master regulator of sporulation Spo0A was shown to inhibit DNA replication (Castilla-Llorente, et al., 2006). These examples indicate that control of DNA replication at the level of initiation might be widespread among bacteria undergoing differentiation.
Yet another potential link to cell physiology is implied by the presence of binding sites for histone-like proteins within oriC. The E. coli oriC contains one each binding site for FIS (factor for inversion stimulation) and IHF (integration host factor) (Filutowicz & Roll, 1990, Gille, et al., 1991) and, similarly, an IHF binding site is found in the C. crescentus Cori (Siam, et al., 2003). Besides their initially recognized role in site-specific recombination, these proteins also serve as nucleoid organizing proteins and were also implicated in regulation of gene expression (Browning, et al., 2010, Dillon & Dorman, 2010, Rimsky & Travers, 2011). The ability of IHF and FIS to modulate DNA reactions stems from DNA bending that accompanies their binding to DNA (Pan, et al., 1994, Rice, et al., 1996). Owing to such bending, distant DNA sites could be brought together in a proper orientation that would favor- or preclude- a macromolecular nucleoprotein assembly needed for a given reaction. FIS and IHF indeed contribute to DNA replication in E. coli since their inactivation, while not affecting cell viability, disrupts the synchrony of the origin firing (Ryan, et al., 2004). This mechanism gives the cell the means to link DNA replication to its growth stage. Indeed, the abundance of the nucleoid proteins varies depending on growth phase or environmental conditions and could conceivably be used to adjust replication rate to fit the environment (Browning, et al., 2010, Dillon & Dorman, 2010, Rimsky & Travers, 2011). It should be noted here that the control of replication initiation rather elongation is preferred from the cell fitness point of view since it helps the cell to direct its resources into production of complete genomes and thereby maximize its survival rate.
None of the replication control systems described here has been identified so far in P. aeruginosa. It seems very likely, however, that they exist. Indeed, a recent FROS (fluorescent repressor operator system) microscopy study revealed a highly coordinated progression of replication forks in PAO1 (Vallet-Gely & Boccard, 2013), which implies synchronous firing of all replication forks across the cell and, by extension, the existence of mechanisms that preclude premature initiation. Likewise, the intricately controlled propensity of P. aeruginosa to differentiate into various planktonic and adherent forms as well as its ability to withstand hostile environment suggests high efficiency of the bacterium in marshalling resources to increase its fitness and persistence.
Local chromatin structure
The global structure of bacterial chromosome is established in concerted action of numerous DNA binding and remodeling activities (Fig. 2). The major nucleoid associated proteins (NAPs) were identified and thoroughly characterized in E. coli (reviewed in (Browning, et al., 2010, Dillon & Dorman, 2010, Rimsky & Travers, 2011)). Most of them, including HU (heat unstable nucleoid protein), IHF, FIS, Dps (DNA binding protein from starved cells) and Hfq (host factor for Qβ replicase), have close homologs in P. aeruginosa (Stover, et al., 2000). There is no H-NS or StpA in PAO1, although the H-NS-like MvaT and MvaU appear to function as their homologs (Vallet-Gely, et al., 2005). NAPs are typically recognized through their tight DNA association and the resultant copurification with the chromosome during cell fractionation (Murphy & Zimmerman, 1997, Ohniwa, et al., 2011). Contrary to the initial views, NAPs primarily function as global transcription regulators, whereas their contribution to DNA packing owes mostly to their abundance and the ability to bend or bridge DNA. Accordingly, inactivation of NAPs seldom has noticeable effects on cell physiology unless their primary function is affected. For example, a recent report linked synthetic lethality of MvaU and MvaT to activation of Pf4 prophage in the mutant cells (Castang & Dove, 2012).
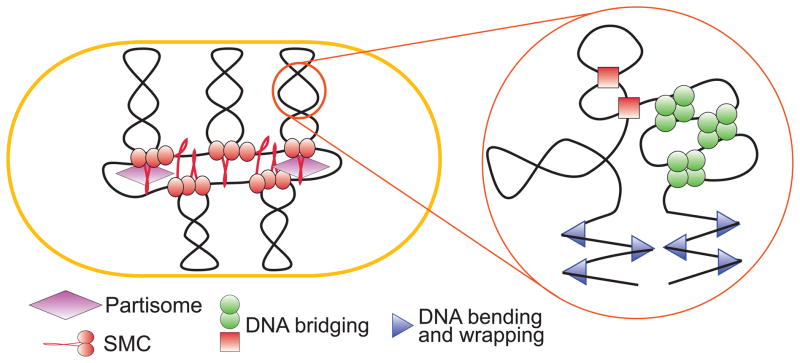
Fig. 2.
A working model of bacterial chromosome. Chromosome structure is stabilized by various NAPs that bend and bridge DNA, supercoiling, which energizes and compacts DNA, and condensins, which stabilize giant loops and tether them to extrachromosomal elements.
DNA supercoiling is another major factor that affects compactness and activity of bacterial chromosome (Cozzarelli & Wang, 1990). Owing to its double helical nature, DNA is constantly unwound and rewound by numerous information processing enzymes such as DNA or RNA polymerases. To avoid the ensuing potentially staggering entanglement problems, the cell carries a battery of special enzymes, DNA topoisomerases, which remove the generated topological links (Corbett & Berger, 2004, Wang, 2009). P. aeruginosa carries the same full complement of DNA topoisomerases as originally identified in E. coli and is expected to exhibit the same regulatory mechanisms. While highly efficient in general, topoisomerases fall behind in highly transcribed regions, especially in the context of divergent promoters, which gives rise to local waves of supercoiling (Wu, et al., 1988, Rovinskiy, et al., 2012).
The net activity of topoisomerases maintains cellular DNA underwound by about 5% throughout the chromosome. About half of the resulting DNA supercoiling is constrained by the bound nucleoid associated proteins, whereas the rest is absorbed by DNA twisting and writhing (Bliska & Cozzarelli, 1987). The effect of supercoiling on DNA activity is two-fold. First, the altered shape of supercoiled DNA dramatically changes statistics of intersegment collisions, which, in turn, markedly affects activity of many DNA processing enzymes (Vologodskii & Cozzarelli, 1996). Similarly, DNA supercoiling provides a powerful driving force for DNA decatenation and thereby contributes to chromosome segregation (Rybenkov, et al., 1997, Alexandrov, et al., 1999, Jun & Mulder, 2006). Second, DNA supercoiling favors recruitment of proteins that untwist DNA upon binding (Vologodskii & Cozzarelli, 1994). Being a global property, DNA supercoiling affects activity of the entire chromosome. Expression of about 10% of E. coli genes changes in response to variations in DNA supercoiling (Peter, et al., 2004). Inside the cell, diffusion of DNA supercoiling is limited by the bound proteins to within ~10 kb stochastically formed topological domains (Postow, et al., 2004). As a result, DNA supercoiling is non-uniformly distributed throughout the DNA and can significantly deviate from the average around actively transcribed genes or the progressing replication fork (Rovinskiy, et al., 2012).
Whereas the local chromatin structure is largely opportunistic and dedicated to support of regulated gene expression within cellular confines, the global folding of the chromosome ensures spatial coordination of chromosome replication with other cellular activities, most notably, cell division. Such coordination is needed to ensure that exactly two copies of genome are produced during each round of replication and then passed one each to the daughter cells. Precise mechanism how this is achieved is yet to be understood. However, some themes are beginning to emerge. Two of such widely spread systems, condensins and ParABS, are discussed below.
Chromosome dynamics during segregation
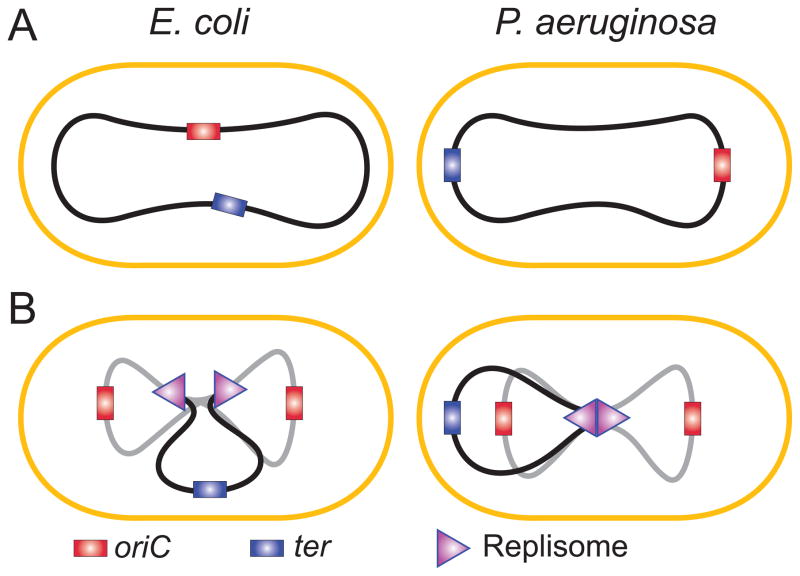
Examination of several bacteria using FROS microscopy revealed ordered arrangement of the chromosome within the cell. This was observed for E. coli (Bates & Kleckner, 2005, Nielsen, et al., 2006, Espeli, et al., 2008), B. subtilis (Teleman, et al., 1998), C. crescentus (Viollier, et al., 2004) and, recently, for P. aeruginosa (Vallet-Gely & Boccard, 2013). In all cases, genomic coordinate of the DNA was found to correlate with its subcellular location. Curiously, two typical arrangements emerged. In non-growing E. coli, the origin of replication is found in the middle of the cell, whereas the two chromosome arms are aligned along the two halves of the cell. In B. subtilis, C. crescentus and P. aeruginosa, the alignment is longitudinal, with oriC and ter located at the opposite poles and the arms linearly stretching along the cell length (Fig. 3A). Thus, global chromosome dynamics in bacteria is decided not by their phylogenetic proximity but should be traceable to the presence or absence of a particular genetic marker.
Fig. 3.
Subcellular organization of the chromosome in E. coli and P. aeruginosa prior to (A) or during (B) replication. Newly replicated DNA is shown in gray.
This distinction becomes less pronounced once replication begins. In all tested bacteria, the two daughter origins move into the opposite halves of the cell soon after their formation. In E. coli and P. aeruginosa, they settle close to ¼ and ¾ positions (0.2 and 0.8 in P. aeruginosa), i.e. the places that will become the middle of the daughter cells (Fig. 3B), In C. crescentus, one copy of the origin remains at the cell pole throughout the cell cycle whereas the other, once formed, migrates to the opposite pole (Jensen & Shapiro, 1999). There is no clarity yet whether this motion is powered by some sort of a mitotic apparatus or simply caused by topological repulsion of two growing unlinked polymer chains. The replicated clockwise and counterclockwise arms of the chromosome follow the origins to be orderly placed in the daughter cells. The ter region has to pass through the middle of the cell, where the FtsK DNA translocase is located. The activity of FtsK is required in order to align the dif sites on dimeric chromosomes and allow XerCD catalyzed resolution of the dimer (Aussel, et al., 2002).
Despite many similarities, chromosome segregation did not proceed identically in all species. One discrepancy was related to location of the replisomes. The E. coli replisomes move around the cell presumably tracking the DNA (Reyes-Lamothe, et al., 2008). In P. aeruginosa, replisomes stay in the middle of the cell for most of the replication cycle (Vallet-Gely & Boccard, 2013). In this respect, P. aeruginosa behave closer to B. subtilis than E. coli. Also unlike in E. coli, segregation of the replicated regions occurred progressively, without any discontinuity. Thus, the existence of sister chromatid cohesion at the snap regions that was observed in E. coli (Joshi, et al., 2011) appears to be a species dependent phenomenon.
Global chromosome architecture
In many bacterial genomes, two systems, condensins and ParABS, are routinely found as the key factors responsible for global folding of the chromosome. The emerging data indicate that the two systems cooperate with each other as well as DNA replication to yield a functional chromosome. Mutations in condensins or ParABS lead to massive chromosome disorganization and are lethal in some species.
Condensins are multisubunit cytoplasmic proteins that link the global and local chromatin dynamics in organisms ranging from bacteria to humans (Cobbe & Heck, 2004, Graumann & Knust, 2009, Gruber, 2011). They contain at their core a dimer of the characteristically V-shaped SMC (structural maintenance of the chromosome) proteins. SMC proteins consist of the ABC type ATPase globular domain connected via a long coiled-coil to the hinge domain (Melby, et al., 1998, Matoba, et al., 2005). Exact architecture of the complex is unclear since both V- and I-shaped molecules can be found in solution (Matoba, et al., 2005). The globular domain undergoes ATP-sandwiched dimerization and is responsible for ATP-modulated interaction with DNA whereas the function of the hinge and the coiled-coil is less clear (Woo, et al., 2009). The accessory subunits interact in a dynamic, ATP-controlled manner with the globular domain of the SMC (Hirano & Hirano, 2004, Lammens, et al., 2004, Petrushenko, et al., 2006, Woo, et al., 2009).
The primary activity of SMCs is ATP-controlled DNA bridging which allows them to act as macromolecular clamps that bring distant DNA segments together (Strick, et al., 2004, Cui, et al., 2008, Petrushenko, et al., 2010), In principle, this activity could give rise to the chromosome scaffold that organizes the DNA into a set of giant loops (Cui, et al., 2008). In this sense, the protein could be viewed as an intermediate between local and global chromatin folding. The actual mechanism of the protein is even more complex. Condensins are not uniformly distributed across the DNA but form distinct foci at the conspicuous ¼ and ¾ positions (Ohsumi, et al., 2001, She, et al., 2007, Minnen, et al., 2011, Badrinarayanan, et al., 2012, Kleine Borgmann, et al., 2013), which points to their possible association with the replication or chromosome positioning machinery. Mutational analysis indicated that the focal localization of condensins is essential for their activity and that the accessory subunits play a central role in it (Shin, et al., 2009, She, et al., 2013). In contrast, the scaffolding activity of SMCs could be accomplished by the SMC core alone (Cui, et al., 2008). Given that both DNA and non-SMC binding to the SMC core is controlled by ATP, a variety of models can be devised that envision coordination between DNA binding by SMCs and their subcellular localization.
Recent data suggested an alternative or, perhaps, supplemental explanation why recruitment of condensins to the quarter foci is essential for chromosome organization. The E. coli condensin, MukBEF, was found to associate with the ParC subunit of topo IV, and this association was confirmed as essential and stimulatory for the topo IV activity (Hayama & Marians, 2010, Li, et al., 2010). Perhaps, recruitment of topo IV to replication fork facilitates decatenation of the daughter chromatids and, thereby, promotes both progression of the DNA replication and chromosome segregation.
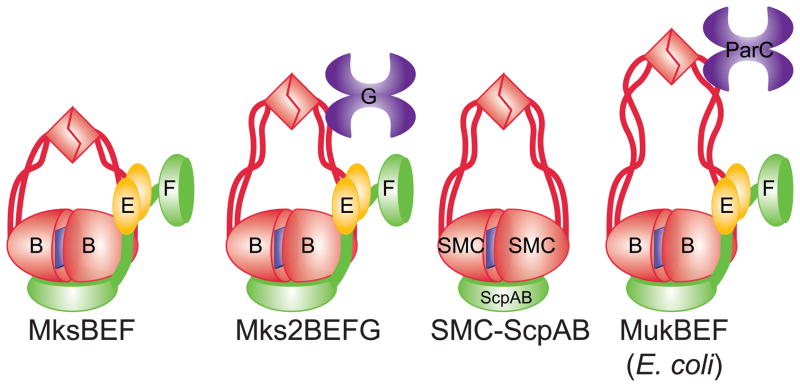
Intriguingly, P. aeruginosa carries several condensins, which sets it apart from the archetypal laboratory strains of E. coli and B. subtilis (Petrushenko, et al., 2011). Until recently, two families of condensins were known in bacteria. Several orders of γ-proteobacteria, including Enterobacteriales, Vibrionales and Pasteurelleles, encode MukBEF, whereas almost all other eubacteria carry the SMC-ScpAB complex (Hiraga, et al., 2000, Cobbe & Heck, 2004). Despite significant sequence divergence, the two proteins are apparently similar in function and structure. In addition, a third family of condensins, MksBEF (MukBEF-like SMCs), was recently discovered (Petrushenko, et al., 2011). MksBEF has the same operon organization as MukBEF and sometimes can be traced to the E. coli MukBEF based on homology. However, MksBs have shorter coiled coil than MukB and display low sequence conservation (Fig. 3). Several families of MksBEFs were identified with barely detectable homology to each other, mostly among outliers, suggesting that the proteins evolved independently.
P. aeruginosa strain PAO1 encodes two condensins, SMC-ScpAB and MksBEF, and the third condensin MksBEFG2 is found in the more virulent strain UCBPP-PA14 (Table II; Fig. 4). Of note, the correct start codon of mksB2, GUG, is found 168 bp upstream from its predicted position (unpublished data). Compared to PAO1, UCBPP-PA14 carries about 200 kb of extra genome, which appears to be remnants of a prophage that are now split into several pathogenicity islands (Lee, et al., 2006). Finding MksBEF2 in one of these islands points to potential evolutionary origins of condensins and suggests that the proteins could have been involved in packing of large extrachromosomal genomes. The P. aeruginosa condensins perform distinct, partially overlapping functions, although their precise role is under investigation (Petrushenko, et al., 2011). At least in planktonic bacteria, faithful chromosome partitioning requires the SMC-ScpAB complex, whereas MksBEF is expendable. Curiously, a widely known deletion in PA4684 and PA4685 that now spread throughout many subclones of PAO1 (Dotsch, et al., 2009) is located in the mksBEF operon and encompasses mksE and mksF.
Table II.
Condensins in P. aeruginosa strains PAO1 and UCBPP_PA14
| Condensin | PAO1 | UCBPP_PA14 |
|---|---|---|
|
| ||
| SMC/ScpA/B | PA1527/PA3197/PA3198 | PA14_44680/PA14_22840/PA14_22860 |
| MksF/E/B | PA4684/PA4685/PA4686 | PA14_61960/PA_61980/PA14_61990 |
| MksF2/E2/B2/G2 | none | PA14_03250/PA14_03260/PA14_03270/PA14_03285 |
Fig. 4.
Comparison of the E. coli and P. aeruginosa condensins. The P. aeruginosa condensins differ in the length of their coiled coil region, which are all shorter than in MukB. One of the condensins, Mks2BEFG, encodes a Toprim protein MksG and is postulated to form a similar complex with MksB2 as ParC with MukB.
Mitotic apparatus
Long being controversial, the bacterial mitotic apparatus has finally materialized in the body of the ParABS system (reviewed in (Szardenings, et al., 2011, Mierzejewska & Jagura-Burdzy, 2012)). This system consists of three elements. ParA protein (known as Spo0J in B. subtilis) is a cytoskeletal ATPase highly prone to oligomerization in vitro and in vivo (Fogel & Waldor, 2006, Ringgaard, et al., 2009, Ptacin, et al., 2010). ParB (Soj in B. subtilis) is a sequence specific DNA binding protein that serves as an adaptor for ParA. ParS is a cis acting DNA stretch that recruits ParB. This system is found in genomes of many bacteria, often as a part of the oriC cassette, as well as in low copy plasmids both in Gram-negative and Gram-positive bacteria (Livny, et al., 2007). Chromosome (or plasmid) segregation is accomplished with the help of the pulling forces within ParB filament that stretches between the sister ParS sites or, perhaps, connects ParS to anchor proteins on cell poles. The system is completely portable and can be used to stabilize low copy number plasmids in foreign bacteria.
The consensus ParS sequence, TGTTCCACGTGGAACA, is highly conserved in diverse bacteria (Livny, et al., 2007). In P. aeruginosa, two of such have been found, both within several kb from oriC. When up to two substitutions to the consensus are allowed, 10 putative sites can be found in the PAO1 chromosome (Bartosik, et al., 2004). Four of them are located close to the origin, two each at about 500 kb from oriC both counter- and clockwise from it, and two are located in the ter region. The functional significance of the perfect matches was verified when the predicted ParABS cassette was found to stabilize plasmids in E. coli (Bartosik, et al., 2004).
ParA and ParB are non-essential in PAO1, although their inactivation leads to dramatic chromosome disorganization and increased frequencies of the chromosome partition defects (Bartosik, et al., 2009, Vallet-Gely & Boccard, 2013). It is tempting to speculate that these defects develop due to the loss of the mitotic forces that push sister chromosomes apart. This conclusion, however, needs further verification, since the activity of chromosomally encoded ParABS is integrated into other genome duplication functions. In B. subtilits, for example correct loading of the SMC-ScpAB condensin onto the chromosome requires functional ParABS with correctly positioned ParS sites (Gruber & Errington, 2009). Similarly, ParABS is involved in correct timing of DnaA-mediated initiation of DNA replication (Murray & Errington, 2008).
Concluding remarks
Most of the key systems involved in replication and segregation of P. aeruginosa chromosome have been mapped and at least initially characterized. This makes this bacterium an attractive model system for further studies of chromosome dynamics. A word of caution here is a rather high frequency of misannotated start codons in public databases.
The replication origin region consists of the contiguously located dnaA and gidA cassettes complete with the DUE-containing DnaA box clusters. Both these elements can serve as an OriC in one bacterium or another and support propagation of origin-less plasmids in pseudomonads. Pseudomonads are the only documented bacterial system with two ARS on the same chromosome. The function of OriCII, if any, is unknown as are the structural determinants that render it dormant.
Multiple chromosome maintenance systems are yet unidentified. Prominently missing is the knowledge on systems that ensure synchronous initiation of chromosome replication or its control in response to developmental needs.
Spatial chromosome dynamics bears greater resemblance to B. subtilis than E. coli. The chromosome layout is longitudinal, not transeversal; the DNA polymerase stays long at midcell during replication. However, no evidence for polar attachment of the origin has emerged so far.
P. aeruginosa encodes multiple condensins, from both the SMC and MukBEF superfamilies’. These condensins apparently play distinct roles. Intriguingly, inactivation of condensins has only mild impact on P. aeruginosa viability, which points to existence on redundant mechanisms in global chromosome packing.
Most of research has been focused on planktonic bacteria. Virtually nothing is known about how cell differentiation affects chromosome maintenance. The existence of potentially redundant systems raises questions about their possible role in differentiation.
Acknowledgments
This work was supported by Grants 1049755 from the National Science Foundation and AI094124 from the National Institutes of Health.
Footnotes
The authors have no conflict of interests to declare.
References
- Alexandrov AI, Cozzarelli NR, Holmes VF, et al. Mechanisms of separation of the complementary strands of DNA during replication. Genetica. 1999;106:131–140. doi: 10.1023/a:1003749416449. [DOI] [PubMed] [Google Scholar]
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science. 2012;338:528–531. doi: 10.1126/science.1227126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosik AA, Mierzejewska J, Thomas CM, Jagura-Burdzy G. ParB deficiency in Pseudomonas aeruginosa destabilizes the partner protein ParA and affects a variety of physiological parameters. Microbiology. 2009;155:1080–1092. doi: 10.1099/mic.0.024661-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosik AA, Lasocki K, Mierzejewska J, Thomas CM, Jagura-Burdzy G. ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. Journal of bacteriology. 2004;186:6983–6998. doi: 10.1128/JB.186.20.6983-6998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia D, Zzaman S, Krings G, Saxena M, Peng X, Greenberg MM. Replication termination mechanism as revealed by Tus-mediated polar arrest of a sliding helicase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12831–12836. doi: 10.1073/pnas.0805898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska JB, Cozzarelli NR. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987;194:205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Booker RJ, Loutit JS. The order of replication of chromosomal markers in Pseudomonas aeruginosa strain 1. I Marker frequency analysis by transduction. Genetical research. 1974;23:145–153. doi: 10.1017/s0016672300014762. [DOI] [PubMed] [Google Scholar]
- Boye E, Stokke T, Kleckner N, Skarstad K. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:12206–12211. doi: 10.1073/pnas.93.22.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendler T, Austin S. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. The EMBO journal. 1999;18:2304–2310. doi: 10.1093/emboj/18.8.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezellec P, Hoebeke M, Hiet MS, Pasek S, Ferat JL. DomainSieve: a protein domain-based screen that led to the identification of dam-associated genes with potential link to DNA maintenance. Bioinformatics. 2006;22:1935–1941. doi: 10.1093/bioinformatics/btl336. [DOI] [PubMed] [Google Scholar]
- Briggs GS, Smits WK, Soultanas P. Chromosomal replication initiation machinery of low-G+C-content Firmicutes. Journal of bacteriology. 2012;194:5162–5170. doi: 10.1128/JB.00865-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Grainger DC, Busby SJ. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Bussiere DE, Bastia D, White SW. Crystal structure of the replication terminator protein from B. subtilis at 2.6 A. Cell. 1995;80:651–660. doi: 10.1016/0092-8674(95)90519-7. [DOI] [PubMed] [Google Scholar]
- Castang S, Dove SL. Basis for the essentiality of H-NS family members in Pseudomonas aeruginosa. Journal of bacteriology. 2012;194:5101–5109. doi: 10.1128/JB.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla-Llorente V, Munoz-Espin D, Villar L, Salas M, Meijer WJ. Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication. The EMBO journal. 2006;25:3890–3899. doi: 10.1038/sj.emboj.7601266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbe N, Heck MM. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol Biol Evol. 2004;21:332–347. doi: 10.1093/molbev/msh023. [DOI] [PubMed] [Google Scholar]
- Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- Cozzarelli NR, Wang JC. DNA topology and its biological effects. Cold Spring Habor Laboratory Press; Cold Spring Harbor, New York: 1990. [Google Scholar]
- Cui Y, Petrushenko ZM, Rybenkov VV. MukB acts as a macromolecular clamp in DNA condensation. Nat Struct Mol Biol. 2008;15:411–418. doi: 10.1038/nsmb.1410. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Dotsch A, Pommerenke C, Bredenbruch F, Geffers R, Haussler S. Evaluation of a microarray-hybridization based method applicable for discovery of single nucleotide polymorphisms (SNPs) in the Pseudomonas aeruginosa genome. BMC Genomics. 2009;10:29. doi: 10.1186/1471-2164-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Mercier R, Boccard F. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Molecular microbiology. 2008;68:1418–1427. doi: 10.1111/j.1365-2958.2008.06239.x. [DOI] [PubMed] [Google Scholar]
- Filutowicz M, Roll J. The requirement of IHF protein for extrachromosomal replication of the Escherichia coli oriC in a mutant deficient in DNA polymerase I activity. The New biologist. 1990;2:818–827. [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes & development. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H, Egan JB, Roth A, Messer W. The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic acids research. 1991;19:4167–4172. doi: 10.1093/nar/19.15.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann PL, Knust T. Dynamics of the bacterial SMC complex and SMC-like proteins involved in DNA repair. Chromosome Res. 2009;17:265–275. doi: 10.1007/s10577-008-9014-x. [DOI] [PubMed] [Google Scholar]
- Gruber S. MukBEF on the march: taking over chromosome organization in bacteria? Mol Microbiol. 2011;81:855–859. doi: 10.1111/j.1365-2958.2011.07764.x. [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Hayama R, Marians KJ. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:18826–18831. doi: 10.1073/pnas.1008140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Ichinose C, Onogi T, Niki H, Yamazoe M. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells. 2000;5:327–341. doi: 10.1046/j.1365-2443.2000.00334.x. [DOI] [PubMed] [Google Scholar]
- Hirano M, Hirano T. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. Embo J. 2004;23:2664–2673. doi: 10.1038/sj.emboj.7600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis P, Dunning Hotopp JC, Sapountzis P, et al. New criteria for selecting the origin of DNA replication in Wolbachia and closely related bacteria. BMC Genomics. 2007;8:182. doi: 10.1186/1471-2164-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis TC, Beaudry AA, Bullard JM, Ochsner U, Dallmann HG, McHenry CS. Discovery and characterization of the cryptic psi subunit of the pseudomonad DNA replicase. The Journal of biological chemistry. 2005;280:40465–40473. doi: 10.1074/jbc.M508310200. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci U S A. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Yao S, Helinski D, Toukdarian A. Functional analysis of two putative chromosomal replication origins from Pseudomonas aeruginosa. Plasmid. 2006;55:194–200. doi: 10.1016/j.plasmid.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Joshi MC, Bourniquel A, Fisher J, Ho BT, Magnan D, Kleckner N, Bates D. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2765–2770. doi: 10.1073/pnas.1019593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K. Structure of a replication-terminator protein complexed with DNA. Nature. 1996;383:598–603. doi: 10.1038/383598a0. [DOI] [PubMed] [Google Scholar]
- Kleine Borgmann LA, Ries J, Ewers H, Ulbrich MH, Graumann PL. The bacterial SMC complex displays two distinct modes of interaction with the chromosome. Cell reports. 2013;3:1483–1492. doi: 10.1016/j.celrep.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA replication. W. H. Freeman and Co; New York: 1992. [Google Scholar]
- Lammens A, Schele A, Hopfner KP. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr Biol. 2004;14:1778–1782. doi: 10.1016/j.cub.2004.09.044. [DOI] [PubMed] [Google Scholar]
- Lee DG, Urbach JM, Wu G, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ, Ralston GB, Christopherson RI, Wake RG. Identification of the replication terminator protein binding sites in the terminus region of the Bacillus subtilis chromosome and stoichiometry of the binding. Journal of molecular biology. 1990;214:73–84. doi: 10.1016/0022-2836(90)90147-E. [DOI] [PubMed] [Google Scholar]
- Li Y, Stewart NK, Berger AJ, et al. Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc Natl Acad Sci U S A. 2010;107:18832–18837. doi: 10.1073/pnas.1008678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. Journal of bacteriology. 2007;189:8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz P, Zakrzewska-Czerwinska J, Zawilak A, Dudek MR, Cebrat S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic acids research. 2004;32:3781–3791. doi: 10.1093/nar/gkh699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba K, Yamazoe M, Mayanagi K, Morikawa K, Hiraga S. Comparison of MukB homodimer versus MukBEF complex molecular architectures by electron microscopy reveals a higher-order multimerization. Biochem Biophys Res Commun. 2005;333:694–702. doi: 10.1016/j.bbrc.2005.05.163. [DOI] [PubMed] [Google Scholar]
- McAdams HH, Shapiro L. A bacterial cell-cycle regulatory network operating in time and space. Science. 2003;301:1874–1877. doi: 10.1126/science.1087694. [DOI] [PubMed] [Google Scholar]
- McHenry CS. DNA replicases from a bacterial perspective. Annual review of biochemistry. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzejewska J, Jagura-Burdzy G. Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid. 2012;67:1–14. doi: 10.1016/j.plasmid.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol. 2011;81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- Moriya S, Atlung T, Hansen FG, Yoshikawa H, Ogasawara N. Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Molecular microbiology. 1992;6:309–315. doi: 10.1111/j.1365-2958.1992.tb01473.x. [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nature reviews Microbiology. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- Moukadiri I, Prado S, Piera J, Velazquez-Campoy A, Bjork GR, Armengod ME. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic acids research. 2009;37:7177–7193. doi: 10.1093/nar/gkp762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LD, Zimmerman SB. Isolation and characterization of spermidine nucleoids from Escherichia coli. J Struct Biol. 1997;119:321–335. doi: 10.1006/jsbi.1997.3883. [DOI] [PubMed] [Google Scholar]
- Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Molecular microbiology. 2006;62:331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- O’Hoy K, Krishnapillai V. Recalibration of the Pseudomonas aeruginosa strain PAO chromosome map in time units using high-frequency-of-recombination donors. Genetics. 1987;115:611–618. doi: 10.1093/genetics/115.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Okazaki T. Concurrent transcription from the gid and mioC promoters activates replication of an Escherichia coli minichromosome. Molecular & general genetics : MGG. 1991;230:193–200. doi: 10.1007/BF00290668. [DOI] [PubMed] [Google Scholar]
- Ohniwa RL, Ushijima Y, Saito S, Morikawa K. Proteomic analyses of nucleoid-associated proteins in Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus. PLoS One. 2011;6:e19172. doi: 10.1371/journal.pone.0019172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi K, Yamazoe M, Hiraga S. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE- MukB complex in Escherichia coli cells. Mol Microbiol. 2001;40:835–845. doi: 10.1046/j.1365-2958.2001.02447.x. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Tamaru A, Nakajima C, et al. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Molecular microbiology. 2007;63:1096–1106. doi: 10.1111/j.1365-2958.2006.05585.x. [DOI] [PubMed] [Google Scholar]
- Okumura H, Yoshimura M, Ueki M, Oshima T, Ogasawara N, Ishikawa S. Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic acids research. 2012;40:220–234. doi: 10.1093/nar/gkr716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan CQ, Feng JA, Finkel SE, Landgraf R, Sigman D, Johnson RC. Structure of the Escherichia coli Fis-DNA complex probed by protein conjugated with 1,10-phenanthroline copper(I) complex. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1721–1725. doi: 10.1073/pnas.91.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrushenko ZM, Lai CH, Rybenkov VV. Antagonistic interactions of kleisins and DNA with bacterial condensin MukB. J Biol Chem. 2006;281:34208–34217. doi: 10.1074/jbc.M606723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrushenko ZM, She W, Rybenkov VV. A new family of bacterial condensins. Mol Microbiol. 2011;81:881–896. doi: 10.1111/j.1365-2958.2011.07763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrushenko ZM, Cui Y, She W, Rybenkov VV. Mechanics of DNA bridging by bacterial condensin MukBEF in vitro and in singulo. Embo J. 2010;29:1126–1135. doi: 10.1038/emboj.2009.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nature cell biology. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riber L, Lobner-Olesen A. Coordinated replication and sequestration of oriC and dnaA are required for maintaining controlled once-per-cell-cycle initiation in Escherichia coli. Journal of bacteriology. 2005;187:5605–5613. doi: 10.1128/JB.187.16.5605-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- Rimsky S, Travers A. Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr Opin Microbiol. 2011;14:136–141. doi: 10.1016/j.mib.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovinskiy N, Agbleke AA, Chesnokova O, Pang Z, Higgins NP. Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS genetics. 2012;8:e1002845. doi: 10.1371/journal.pgen.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan VT, Grimwade JE, Camara JE, Crooke E, Leonard AC. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Molecular microbiology. 2004;51:1347–1359. doi: 10.1046/j.1365-2958.2003.03906.x. [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Vologodskii AV, Cozzarelli NR. The effect of ionic conditions on the conformations of supercoiled DNA. II Equilibrium catenation. J Mol Biol. 1997;267:312–323. doi: 10.1006/jmbi.1996.0877. [DOI] [PubMed] [Google Scholar]
- Sanders LH, Rockel A, Lu H, Wozniak DJ, Sutton MD. Role of Pseudomonas aeruginosa dinB-encoded DNA polymerase IV in mutagenesis. Journal of bacteriology. 2006;188:8573–8585. doi: 10.1128/JB.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen SM, Ouimet MC, Marczynski GT. Comparative analysis of Caulobacter chromosome replication origins. Microbiology. 2009;155:1215–1225. doi: 10.1099/mic.0.025528-0. [DOI] [PubMed] [Google Scholar]
- She W, Wang Q, Mordukhova EA, Rybenkov VV. MukEF is required for stable association of MukB with the chromosome. J Bacteriol. 2007;189:7062–7068. doi: 10.1128/JB.00770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She W, Mordukhova E, Zhao H, Petrushenko ZM, Rybenkov VV. Mutational analysis of MukE reveals its role in focal subcellular localization of MukBEF. Mol Microbiol. 2013;87:539–552. doi: 10.1111/mmi.12112. [DOI] [PubMed] [Google Scholar]
- Shin HC, Lim JH, Woo JS, Oh BH. Focal localization of MukBEF condensin on the chromosome requires the flexible linker region of MukF. FEBS J. 2009;276:5101–5110. doi: 10.1111/j.1742-4658.2009.07206.x. [DOI] [PubMed] [Google Scholar]
- Siam R, Marczynski GT. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. The EMBO journal. 2000;19:1138–1147. doi: 10.1093/emboj/19.5.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siam R, Brassinga AK, Marczynski GT. A dual binding site for integration host factor and the response regulator CtrA inside the Caulobacter crescentus replication origin. Journal of bacteriology. 2003;185:5563–5572. doi: 10.1128/JB.185.18.5563-5572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. Pseudomonas genomes: diverse and adaptable. FEMS microbiology reviews. 2011;35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- Smits WK, Merrikh H, Bonilla CY, Grossman AD. Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. Journal of bacteriology. 2011;193:640–648. doi: 10.1128/JB.01253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Strick TR, Kawaguchi T, Hirano T. Real-time detection of single-molecule DNA compaction by condensin I. Curr Biol. 2004;14:874–880. doi: 10.1016/j.cub.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Sutton MD, Walker GC. Managing DNA polymerases: coordinating DNA replication, DNA repair, and DNA recombination. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8342–8349. doi: 10.1073/pnas.111036998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szardenings F, Guymer D, Gerdes K. ParA ATPases can move and position DNA and subcellular structures. Current opinion in microbiology. 2011;14:712–718. doi: 10.1016/j.mib.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Graumann PL, Lin DCH, Grossman AD, Losick R. Chromosome arrangment within a bacterium. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- Timinskas K, Balvociute M, Timinskas A, Venclovas C. Comprehensive analysis of DNA polymerase III alpha subunits and their homologs in bacterial genomes. Nucleic acids research. 2014;42:1393–1413. doi: 10.1093/nar/gkt900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I, Boccard F. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS genetics. 2013;9:e1003492. doi: 10.1371/journal.pgen.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11082–11087. doi: 10.1073/pnas.0502663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. From The Cover: Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. PNAS. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii AV, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- Vologodskii AV, Cozzarelli NR. The effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U, Krekling MA, Hansen FG, Lobner-Olesen A. The eclipse period of Escherichia coli. The EMBO journal. 2000;19:6240–6248. doi: 10.1093/emboj/19.22.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Meyenburg K, Jorgensen BB, Nielsen J, Hansen FG. Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Molecular & general genetics : MGG. 1982;188:240–248. doi: 10.1007/BF00332682. [DOI] [PubMed] [Google Scholar]
- Wang JC. A journey in the world of DNA rings and beyond. Annual review of biochemistry. 2009;78:31–54. doi: 10.1146/annurev.biochem.78.030107.090101. [DOI] [PubMed] [Google Scholar]
- Woo JS, Lim JH, Shin HC, et al. Structural Studies of a Bacterial Condensin Complex Reveal ATP-Dependent Disruption of Intersubunit Interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- Wu H-Y, Shyy S, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yee TW, Smith DW. Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1278–1282. doi: 10.1073/pnas.87.4.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Czerwinska J, Jakimowicz D, Zawilak-Pawlik A, Messer W. Regulation of the initiation of chromosomal replication in bacteria. FEMS microbiology reviews. 2007;31:378–387. doi: 10.1111/j.1574-6976.2007.00070.x. [DOI] [PubMed] [Google Scholar]
- Zyskind JW, Smith DW. The bacterial origin of replication, oriC. Cell. 1986;46:489–490. doi: 10.1016/0092-8674(86)90873-1. [DOI] [PubMed] [Google Scholar]