Abstract
ORCTL3 is a member of a group of genes, the so-called anticancer genes, that cause tumour-specific cell death. We show that this activity is triggered in isogenic renal cells upon their transformation independently of the cells’ proliferation status. For its cell death effect ORCTL3 targets the enzyme stearoyl-CoA desaturase-1 (SCD1) in fatty acid metabolism. This is caused by transmembrane domains 3 and 4, which are more efficacious in vitro than a low molecular weight drug against SCD1, and critically depend on their expression level. SCD1 is found upregulated upon renal cell transformation indicating that its activity, while not impacting proliferation, represents a critical bottleneck for tumourigenesis. An adenovirus expressing ORCTL3 leads to growth inhibition of renal tumours in vivo and to substantial destruction of patients’ kidney tumour cells ex vivo. Our results indicate fatty acid metabolism as a target for tumour-specific apoptosis in renal tumours and suggest ORCTL3 as a means to accomplish this.
Keywords: Anticancer gene, cell death, ORCTL3, stearoyl-CoA desaturase
INTRODUCTION
Recent studies have led to the emergence of a new class of genes with a specific anti-cancer activity. Upon ectopic expression these factors cause the specific destruction of tumour cells by various forms of cell death such as apoptosis, autophagy, or mitotic catastrophe, while normal cells are spared. Among those genes are apoptin, mda-7/IL-24, par-4, and also TRAIL.1-5 Some of the factors are in clinical trials and show promising results.6 Their modes of action are currently under intense investigation fuelled by the expectation that they reveal novel therapeutic interference options. We have recently presented ORCTL3 as one of the members of this growing, functionally defined gene family that we collectively named “anticancer genes”.6 ORCTL3 was isolated in a systematic screen for such genes and its transfection into numerous tumourigenic cells induced apoptosis, while normal and primary cells remained healthy.7 How this is accomplished remained unknown.
Recently, the metabolism of tumour cells has intensely been studied for differences to normal cells with the expectation that this will lead to novel targets and treatment options.8,9 Several studies indicated that fatty acid metabolism is changed in malignant cells, which is mainly interpreted as a consequence of the increased requirement of lipids for their excessive proliferation.10 Indeed, most conventional anticancer compounds target actively proliferating cells and their efficiency, as well as their side effects, are correlated with enhanced proliferation. However, many tumour cells, in particular cancer stem cells, do not feature changes in their proliferation rate. Hence, the usefulness of targeting fatty acid metabolism for tumour treatment is currently unknown.
Renal cancer is the fourteenth most common cancer worldwide, with an estimated 273,500 new cases diagnosed in 2008. So far the therapy of renal tumours relies mainly on surgery and there is hardly any systemic drug treatment that can be used against advanced renal tumours.11 The majority of tumors eventually become refractory even to novel targeted therapies.12 The survival rate is only around 50% within the first five years after diagnosis. Hence, there is an urgent need to discover novel treatment options.
Here we show that ORCTL3 is activated for apoptosis induction when renal cells become transformed, independently of the proliferation status of the cells. For its apoptosis effect ORCTL3 targets stearoyl-CoA desaturase, an enzyme that introduces a double bond in the fatty acid stearic acid. We have found that ORCTL3 exerts its tumour-specific effect on renal cancer cells in vitro, in vivo and ex vivo.
RESULTS
ORCTL3 is activated for apoptosis induction by transformation
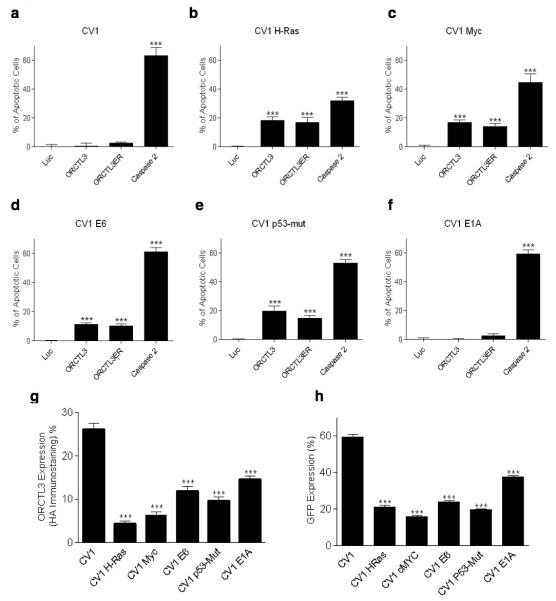
We chose an isogenic system of transformed versus normal renal cells in order to implement the most stringent test for ORCTL3’s tumour specificity. To focus on a proven transformation system we turned to the primate renal CV-1 cells, which have extensively been used in the past and become transformed by individual tumourigenic mutations.13 The sequencing of kidney cancer genomes has revealed considerable genetic inter- and intra-tumour heterogeneity of renal tumours.14,15 Because of this we asked whether ORCTL3 can target changes emerging from renal cell transformation per se rather than changes based on specific mutations found in subpopulations of renal tumours. We transfected these cells with myc, an activated allele of H-ras, and also with a mutant allele of p53, which establish cell transformation by various signalling pathways and have previously been connected with at least some human kidney cancers.16-18 We also transfected the viral oncogenes E6 and E1A that have long been used for a partial transformation of cells.19 In order not to introduce a bias by picking individual colonies, we used pools of transfected cells with a range of expression levels of the transfected genes to more accurately recapitulate the genetic heterogeneity in tumours.20 These mutations caused morphological changes, ranging from profound, especially for H-ras and myc, which lost their contact inhibition, to more subtle changes such as for E1A, which mostly resembled their wild type (WT) CV-1 counterparts (Supplementary Figure S1a,b). Nevertheless, all tumourigenic mutations led to the immortalisation of the cells, while the parental CV-1 cells cease proliferation after about 10 passages. Transfection of WT CV-1 cells with a number of known pro-apoptotic genes such as RIP1, caspase-8, ANT1, and caspase-2 caused efficient cell death confirming the integrity of apoptosis signalling pathways in these cells (Supplementary Figure S1c, S2). The transformed as well as the WT CV-1 cells were then transfected with an expression construct for ORCTL3 and caspase-2 as a positive control. In parallel a fusion construct of ORCTL3 with an ER retention signal (ORCTL3-ER) was introduced that was found to generate higher apoptosis levels.7 In the WT CV-1 cells we detected no appreciable apoptosis with both ORCTL3 constructs (Figure 1a), while caspase-2 was an efficient apoptosis inducer indicating, as before (Supplementary Figure S1c, S2), the intact apoptosis sensitivity of these cells. In contrast, when ORCTL3 was transfected into the transformed CV-1 cells we observed significant apoptosis induction with all cells harbouring tumourigenic mutations, except the E1A transfected cells, which correlated with their minor transformed phenotype (Figure 1b-f and Supplementary Figure S1a). No further increase of apoptosis induction was observed with the ORCTL3-ER construct. In some of the transformed cells such as those transfected by H-ras the general apoptosis inducer caspase-2 was less efficient, in agreement with reports that attribute an apoptosis-inhibiting activity to this oncogene.21 The lack of apoptosis induction in the WT CV-1 cells was not due to a lower expression level of ORCTL3 in those cells. Rather, we detected a considerably higher transfection efficiency and consequently a higher expression of ORCTL3 in WT CV-1 cells than in their transformed counterparts (Figure 1g,h).
Figure 1. ORCTL3 induces apoptosis in transformed but not in normal CV-1 kidney cells.
(a-f) ORCTL3, ORCTL3ER, Luc, and Caspase 2 were transfected into the H-ras-, myc-, E6-, and P53-transformed CV-1 cells as well as into E1A- and empty vector-transfected CV-1 cells. Apoptosis was monitored after 24 hours. Data represent the means of independent experiments ± SD, (n=3). (g) ORCTL3 is more efficiently expressed in WT CV-1 compared to transformed CV-1 cells. WT and transformed CV-1 cells transfected with HA-tagged ORCTL3 were harvested 24 hours after transfection and stained with an Alexa-488 conjugated antibody against HA. Data represent the means of independent experiments ± SD, (n=3). (h) Cells transfected with a plasmid expressing GFP were harvested 24 hours after transfection and assayed by flow cytometry to quantify the GFP signals. Each bar represents the mean ± SD, (n=3).
We compared ORCTL3 with the other anticancer genes apoptin, par-4, and mda-7/IL-24. Like ORCTL3, they remained ineffective for apoptosis induction in parental CV-1 cells (Supplementary Figure S3a-f). Par-4 became an apoptosis inducer in all CV-1 cell variants with tumourigenic mutations, as was the case for apoptin with the exception of p53-mut transformed cells and for IL-24/mad-7 with the exception of E6 and myc transformed cells. Just like ORCTL3, all three anticancer genes remained inactive in E1A transformed cells. A similar trend with higher absolute apoptosis values was observed when high amounts of the recombinant TRAIL ligand were exogenously added to the cells (Supplementary Figure S3g).
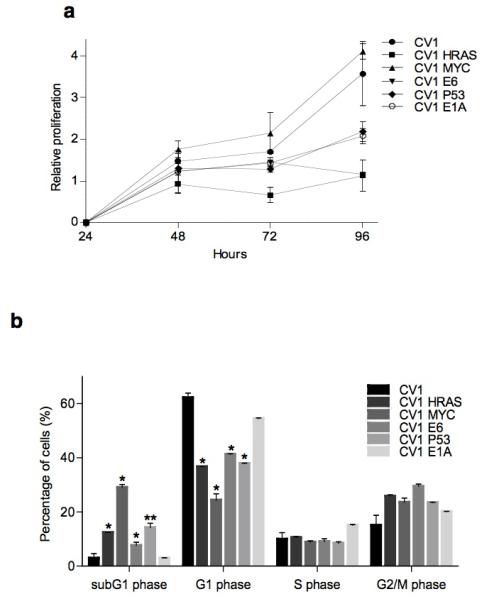
As a first step to determine the cell characteristics that underlie ORCTL3’s specificity we assessed the proliferation status of WT and transformed CV-1 cells and found that in an MTT assay only myc-transformed cells seemed to replicate slightly faster than WT CV-1 cells. All other tumourigenic mutations caused a slower proliferation (Figure 2a). Cell cycle analysis revealed that cells with tumourigenic mutations, with the exception of E1A, had a reduced population of cells in the G1 phase and an increase in subG1 cells indicating that some of them underwent spontaneous apoptosis (Figure 2b).
Figure 2. Analysis of cell proliferation and cell cycle in WT CV-1 and transformed CV-1 cells.
(a) Cell proliferation evaluated at different time points by the MTT assay. Results represent the means ± standard deviations (n=6). (b) The percentage of normal and transformed CV-1 cells in subG1, G1, S and G2/M phase of the cell cycle was determined by analysis with the cell cycle platform of the Flowjo software. The means ± standard deviations are shown (n=3).
ORCTL3 causes ARCosome activation and an incomplete ER stress response
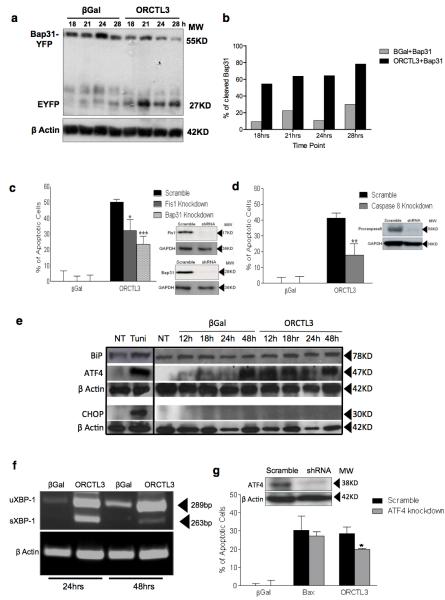
We recently characterised the ARCosome, a caspase-8-activation complex that physically connects the ER with mitochondria and consists of mitochondrial Fis1 and Bap31 at the ER.22 Upon recruitment of caspase-8 to the ARCosome Bap31 is cleaved and activates downstream signals for apoptosis. ORCTL3 induces its apoptosis signal from the ER7 and the physical proximity of the ARCosome to ORCTL3 led us to test the involvement of this apoptosis sensor. Co-transfection of ORCTL3 led to the cleavage of a fusion protein of Bap31-EYFP after 18 hours (50%), a time point at which no overt apoptosis could be observed (Figure 3a,b and Supplementary Figure S4a) suggesting that it is an early event of the apoptosis signalling by ORCTL3. In agreement with this, when we individually inactivated the three components of the ARCosome, we observed a reduction of apoptosis induced by ORCTL3 (Figure 3c,d). Moreover, ORCTL3 could interact with Bap31 in a co-immunoprecipitation experiment (Supplementary Figure S4b). However, transfection of the ARCosome activator Fis1 elicited cell death in WT CV-1 cells indicating that this complex is not exerting the tumour-specific effect. We previously observed an accumulation of the ER stress sensor ATF4 but not of BiP/Grp78 as a signature of ORCTL3-transfected tumour cells.7 We extended these results and also did not detect the upregulation of the proapototic transcription factor CHOP, a ATF4 target gene in the canonical ER stress response (Figure 3e), while ATF3 was found to accumulate upon ORCTL3 expression (Supplementary Figure S4c). The same ER stress response was observed when we assessed Bap31, ATF4, ATF3, and BiP in H-ras transformed CV1 cells. Normal CV1 cells, on the other hand, showed a much reduced Bap31 cleavage and no increase of the other ER stress markers (Supplementary Figure S4d). Nevertheless, in Hela cells the mRNA of XBP-1 was spliced upon ORCTL3 expression, a signal more proximal to the chaperone BiP (Figure 3f). The knock-down of ATF4 led to a weak but significant reduction of ORCTL3 apoptosis (Figure 3g). These results indicate that some, but not all, consequences of canonical ER stress are instigated by ORCTL3 expression.
Figure 3. ORCTL3 induces apoptosis via the ARCosome and elicits an incomplete ER stress.
(a) Expression constructs for Bap31-EYFP and ORCTL3 or β-gal were co-transfected into HeLa cells in a 1:3 ratio, cellular extracts were probed for cleavage of the Bap31-GFP fusion protein with an anti-GFP antibody at the indicated time points. (b) the percentage of cleaved product to the total signal of the Bap31-EYFP fusion constructs was quantified after scanning in the Western blots. Each bar represents the mean ± SD (n=2). (c,d) HeLa cells harbouring shRNA constructs against the indicated genes (inserts) were transfected with an expression construct for ORCTL3 and apoptosis was scored after 24 hours. Raw data were normalised to transfection efficiency estimated by GFP for each cell type. Data represents the means ± SD (n=6). (e) Protein levels of the ER stress factors Grp78/Bip chaperone protein, ATF4/CREB2, and CHOP were determined in Hela cells at different times post-transfection of either ORTCL3 or β-galactosidase using 5 ng/μl tunicamycin for eight hours as a positive control and ß-actin and GAPDH as loading controls. (f) After transfection of an expression plasmid for ORCTL3 the conversion of XBP-1 from its unspliced form (uXBP-1) into its spliced form (sXBP-1) was monitored by RT-PCR with β-actin as a control. (g) HeLa cells were stably transfected with an shRNA construct against ATF4 (insert), followed by introducing an expression construct for ORCTL3, Bax or β-gal. After 24 hours apoptosis was quantified evaluated by flow cytometry analysis. Data represent means ± SD (n=3).
ORCTL3 targets stearoyl-CoA desaturase for tumour-specific apoptosis induction
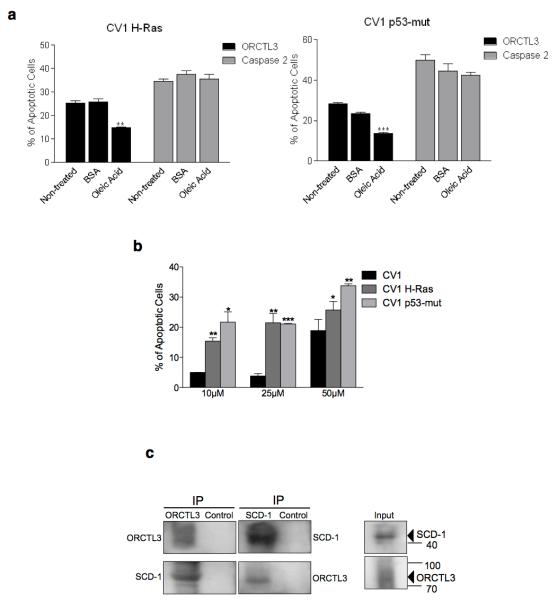
Downregulation of stearoyl-CoA desaturase (SCD) has recently been revealed to cause apoptosis in lung and colon tumour cells lines,23 which is mediated by a cellular response that, like ORCTL3, leads to an incomplete ER stress response with ATF4 upregulation but not BiP.24 Because of this and since both the SCD and ORCTL3 proteins are localised to the ER membrane, we tested whether this enzyme is a target of ORCTL3 for apoptosis induction. We added BSA-complexed oleic acid, the enzymatic product of SCD, to the cells after they were transfected with ORCTL3 or caspase-2 as a control. ORCTL3-induced apoptosis in p53- and H-ras-transformed CV-1 cells was significantly reduced, while cell death caused by the upregulation of caspase-2, Bax, and caspase-8 remained unaffected (Figure 4a and Supplementary Figure S5a,b). The transformed Hela and 293T cell lines also showed a reduced apoptosis by ORCTL3 upon addition of oleic acid (Supplementary Figure S5c). We observed an increase of p53- and H-ras-transformed cells undergoing apoptosis over their normal WT CV-1 counterparts when an inhibitor of SCD1 (CAY10566) was applied (Figure 4b). The application of this SCD1 inhibitor also caused Bap31 cleavage in the H-ras transformed CV1 cells but not in the normal CV1 cells (Supplementary Figure 5d).
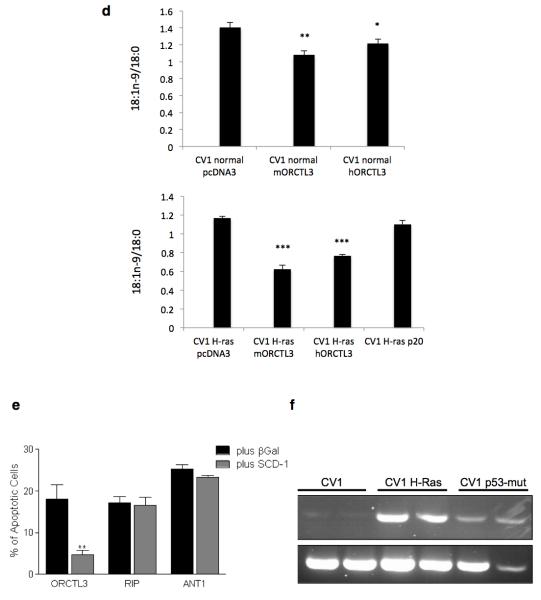
Figure 4. ORCTL3 targets stearoyl-CoA desaturase.
(a) H-ras and p53mut transformed CV-1 cells were transiently cotransfected with GFP and luciferase, ORCTL3, or caspase 2 in a 1:3 ratio and cultivated with either 100 μM BSA-conjugated oleic acid or BSA for 24 hours. Luciferase-control background was subtracted. Data are means ± SD (n=4). (b) The SCD1 inhibitor CAY10566 or equal volumes of DMSO were applied to WT as well as H-ras and p53mut transformed CV-1 cells and apoptosis was quantified after 48 hours by fluorescein diacetate (FDA) staining. (c) ORCTL3-HA was transfected into 293T cells (right, input). Cell lysates were immunoprecipitated with an α-HA antibody (left panels) that revealed multiple, probably glycosylated, forms of ORCTL3 or with an antibody against the endogenous SCD1 (middle panels). The immunoprecipitates were analysed using appropriate antibodies. (d) The desaturation index (18:1n-9/18:0) upon transfection of the indicated expression constructs into normal (top panel) and H-ras transfected (bottom panel) CV1 cells. (e) 293T cells were transfected with ORCTL3, RIP1 and ANT1 with either β-Gal or a plasmid expressing SCD1 in a 3:1 ratio. 48 hours post-transfection, apoptosis was assessed by staining the cells with DiOC6/PI and analysing by flow cytometry. Data represent the means ± SD (n=3). Raw data were normalised to transfection efficiency estimated by GFP. (f) Endogenous SCD1 mRNA level was semi-quantitatively determined in dublicates by RT-PCR in WT-, H-ras-, and P53-mut-transformed CV-1 cells.
Furthermore, co-immunoprecipitation revealed an interaction between ORCTL3 and SCD as we could detect endogenous SCD1 by western blot when we immunoprecipitated overexpressed ORCTL3. Conversely, when the endogenous SCD1 was immunoprecipitated, we detected the overexpressed ORCTL3 as a discrete band, rather than a smear as detected in the input probably representing differentially glycosylated forms of the ORCTL3 transporter (Figure 4c). Moreover, the expression of both mouse and human ORCTL3 reduced the desaturation index (18:1n-9/18:0 ratio) in normal and transformed CV1 cells, with the mouse gene being slightly more effective (Figure 4d). The reduction of the desaturation index in H-ras transformed CV1 cells was not a consequence of apoptosis induction as the pro-apopotic p20 Bap31 fragment did not lead to a significant change in the desaturation index. We also measured the fatty acid composition in normal and H-rad transformed CV-1 cells (Supplementary Figure S5e). Co-transfection of SCD1 reduced ORCTL3’s apoptosis induction but not cell death by RIP, ANT1, p20 and par-4 expression (Figure 4e and Supplementary Figure S5f,g). We also observed the co-localisation of transfected mouse and human ORCTL3 with endogenous SCD1 (Supplementary figure S6). These results indicated that SCD activity is required for the survival of transformed, but not WT CV-1 cells. In agreement with this, we found that the transformed CV-1 cells harbour an upregulated SCD1 compared with normal cells (Figure 4f). We also tested the expression level of SCD5 and found a pronounced upregulation of this enzyme in H-ras transformed CV1 cells (Supplementary Figure S7a). Moreover, we detected an interaction between mouse and ORCTL3 with SCD5 (Supplementary Figure 7b).
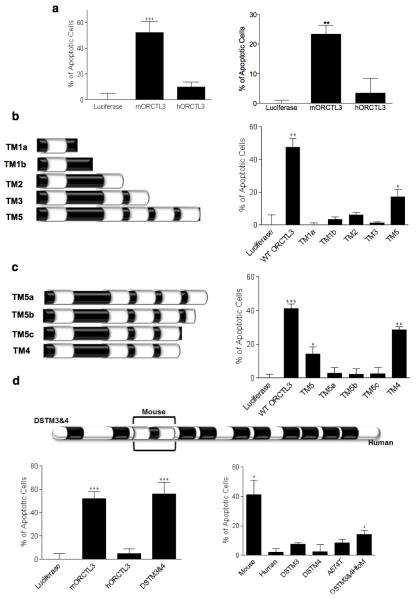
ORCTL3’s apoptosis induction is specific for the mouse gene and critically depends on transmembrane domains 3 and 4
The mouse ORCTL3 gene and its human homologue are 73% identical at the protein level (84% similarity). Surprisingly, when we transfected human ORCTL3, we did not detect appreciable apoptosis induction in transformed HeLa and 293T cells (Figure 5a). Even titration experiments revealed that the degree of cell death attainable with human ORCTL3 remained significantly below the mouse ORCTL3 (Supplementary Figure S8a). We investigated the accumulation of the protein in transfected cells and found that mouse ORCTL3 was considerably more stable (Supplementary Figure S8b). Moreover, the human ORCTL3 interacted slightly weaker than mouse ORCTL3 with SCD1 (Supplementary figure S8c). To determine whether it is the accumulation and interaction of the proteins or whether the mouse ORCTL3 also comprises unique pro-apoptotic sequences, we mapped the domain(s) responsible for this differential apoptosis effect starting with the smallest ORCTL3 deletion construct that still induces apoptosis comprising the 5 N-terminal transmembrane domains (TM5).7 While only the construct TM5 caused apoptosis, construct TM2 was nevertheless expressed to an appreciable level as assessed by FACS analysis to monitor sub-populations with various expression levels (Figure 5b and Supplementary Figure S8d). These data suggested that the TM domains 3, 4, and 5 are required for apoptosis induction. Since the construct TM3 was unstable, we focused our further fine-mapping on the TM domains 4 and 5. We found that when the TM5 was partially or fully deleted (TM5b, TM5c) or when the loop between the 5th and the 6th TM was deleted (TM5a), the protein was rendered unstable and inactive for apoptosis induction. A construct harbouring the first four TM domains (TM4) was even more efficient than the larger construct TM5 for apoptosis induction (Figure 5c). Apoptosis correlated with the expression levels of the constructs (Supplementary Figure S8e). The minimal construct TM4 could still be inhibited by SCD1 co-transfection (Supplementary Figure 8f). Given that the first four TM domains were sufficient for apoptosis (TM4) and the first two TM domains were stable but not active for apoptosis, we concentrated in a new round of mapping experiments on the third and the forth TM domain. It is known that during protein synthesis pairs of TM domains are co-inserted into the ER membrane.25 However, neither the two TM domains on their own (TM34) nor when fused to the N-terminal signal sequence of ORCTL3 (NTM34), or when attached to two arginine residues, which when flanking α-helical transmembrane domains ensure their correct orientation in the membrane (NPosTM34), could elicit apoptosis (Supplementary Figure S8g). All of these constructs were found not to be stable (Supplementary Figure S8h).
Figure 5. Analysis of ORCTL3’s apoptosis domain.
(a) Expression constructs for human and mouse ORCTL3 were transfected into HeLa (left) and 293T (right) cells and apoptosis quantified after 24 hours by DiOC6/PI and flow cytometry analysis. The means±SD are shown (n=3). (b,c) Schematic representation of the respective ORCTL3 deletion mutants (left). Upon their transfection the extent of apoptosis was quantified by DiOC6/PI and FACS analysis, compared to Luciferase as a background control, and normalised with GFP transfection efficiency (right). The histogram shows the means ± SD,(n=3). (d) Schematic representation of the swap mutant of the mouse TM 3 and 4 domains into the human ORCTL3 (“DSTM3&4”) (top). The indicated constructs were transfected into HeLa cells and the extent of apoptosis was quantified after 24 hours by DiOC6/PI staining and FACS analysis, compared to Luciferase as a background control, and normalised with GFP transfection efficiency. The histograms show the means ± SD (n=3) (bottom).
We speculated that the two TM domains from mouse ORCTL3 could enable human ORCTL3 to initiate apoptosis when put into the human sequence context. Figure 5d (top and left bottom panel) reveals that the chimera (DSTM3&4) caused apoptosis as efficiently as the mouse ORCTL3. The stability of the construct was substantially increased over the human ORCTL3 (Supplementary Figure S8i). A sequence comparison indicated that both TM domains comprise a number of amino acids that differ between mouse and human and that the loop between the two TMs contains one exchange (Supplementary Figure S8j). We swapped the TMs individually and converted the loop domain of the human ORCTL3 into the mouse sequence (Supplementary Figure S8k). None of these constructs were capable of apoptosis induction or accumulated to an appreciable level (Figure 5d right bottom panel). When we inserted the two human TM domains into the mouse ORCTL3 gene (DSTM3&4HtoM), despite its expression level being comparable with the mouse protein (Supplementary Figure S8l), only a very modest apoptosis was observed (Figure 5d, right bottom panel). We then asked whether apoptosis is a consequence of an incompatibility of the mouse ORCTL3 protein in a human cellular environment but the mouse ORCTL3 gene was also active for cell death in mouse N2a cells, in which it also accumulated to a higher level than its human equivalent (Supplementary Figure S9).
An ORCTL3-expressing adenovirus causes tumour growth inhibition in vivo and retains its specificity for malignant cells explanted from patients
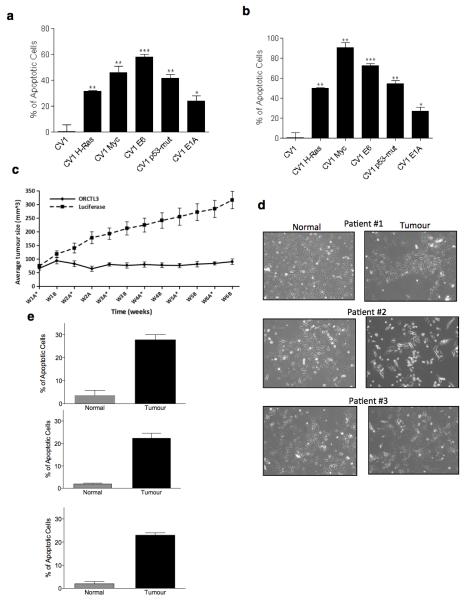
As the above mapping results highlighted the requirement of sufficient accumulation of ORCTL3 for apoptosis induction, we used an adenovirus vector, which can express genes to high levels. Based on our observation that ORCTL3, when retained at the ER, is more efficient for apoptosis,7 as also detected with the TM4 deletion mutant (Supplementary Figure S10a), we inserted an ORCTL3-ER construct into the adenovirus genome. We first adjusted the infection conditions so that we achieved equal expression in WT CV-1 cells and the E1A-transfected variants. The same titres were then used to express ORCTL3 in the other transformed cells. Figure 6a shows that ORCTL3 still did not cause cell death in the non-transformed CV-1 cells but all transformed cells showed considerable cell death after 24 hours, including the E1A-transformed CV-1 cells that had not responded to the transfection of the ORCTL3 expression plasmid (Figure 1f). When the extent of apoptosis was normalised to the expression levels (Supplementary Figure S10b), it became evident that ORCTL3 could induce cells death in 91% of myc-transformed CV-1 cells, in 72% of E6-transformed cells, 50% of H-ras-, 55% of p53-transformed cells, 20% of E1A-expressing cells and none in the WT CV-1 cells (Figure 6b). This efficiency and specificity was supported by the apoptosis effects when the cells were infected twice, which likewise did not indicate any toxicity in the WT CV-1 cells (Supplementary Figure S10c). To evaluate the effects of ORCTL3 in an in vivo xenograft model we made use of the human clear cell renal carcinoma cells Caki-2. In vitro experiments showed that these cells succumb to cell death upon ORCTL3 expression by the adenovirus, particularly when infected repeatedly (Supplementary Figure S10d,e). 2×106 of these cells were injected into the flanks of nude mice and after the tumours reached approximately 50mm3 the ORCTL3-expressing adenovirus or a luciferase control virus were injected once every 6 days and the size of the tumours was determined. Figure 6c and Supplementary S10f,g show that in all tumours ORCTL3 inhibited growth compared to the increase in tumour size seen with the luciferase control over a period of six weeks after which the animals were sacrificed.
Figure 6. Effects of an ORCTL3-expressing adenovirus on transformed cells in vitro, in vivo and on primary human renal tumour cells ex vivo.
(a) WT CV-1 and oncogene-transfected CV-1 cells were infected with an ORCTL3ER-expressing adenovirus at suitable multiplicities of infection (MOI) for equal expression level of the protein using GFP and Luciferase-expressing adenoviruses as infection and negative control, respectively. 24 hours post-infection, apoptosis was quantified by fluorescein diacetate and comparing the ratios of unstained cells to total cell number between different cell types. Toxicity induced by the viral infection itself (luciferase) was subtracted. Data are means ± SD (n=3). (b) Cell death induced by the ORCTL3-expressing adenovirus shown in (a) was normalized to the expression levels of ORCTL3 determined by immunostaining against HA as shown in Fig S8B. (c) Caki-2 renal carcinoma cells were engrafted bilaterally into the flanks of nu/nu BALB/c mice. ORCTL3 or luciferase expressing adenovirus were weekly (*) injected intratumouraly. The size of the tumours was determined by caliper measurements twice a week. Error bars indicate the standard error of the means. (d) Morphology of representative normal and tumour cells explanted from three ccRCC patients. (e) The cells in (d) were infected with an MOI of 5000 of ORCTL3- and luciferase-expressing adenovirus and apoptosis was quantified by DiOC6/PI and flow cytometry analysis. Data are the means of representative experiments, patient #1:n=3; patients #2 and #3:n=2. The background level generated by luciferase infection was subtracted from each reading.
The ORCTL3-expressing adenovirus was then used on explanted primary renal tumour and normal cells obtained from the same patient. A total of six tissue specimens (3 from normal cortex and 3 from advanced kidney cancers of Fuhrman grade 3) were maintained in culture. Clear cell renal cell carcinoma (ccRCC) was diagnosed by a specialised pathologist in all cases upon inspection of the tissues taken from the same tumorous lesions used for the primary cell cultures. Similar to what has been reported26 normal kidney epithelial cells showed higher proliferation rates than tumour cells and displayed a homogeneous population of cells with cobblestone appearance, whilst ccRCC cells displayed flattened polygonal morphology and many lipid vesicles in the cytoplasm (Figure 6d). The required concentration of the virus was optimised by infecting normal and tumour cells and examining the expression of ORCTL3 or GFP (Supplementary Figure S10h). Figure 6e shows that in primary tumour cells from all patients ORCTL3 caused a specific apoptosis-inducing signal (27, 22, and 23% cell death) but exerted negligible toxicity in normal renal cells.
DISCUSSION
Anticancer genes have attracted considerable interest based on the expectation that the specificity of their proteins would aid in defining their mode of action and reveal additional targets in cancer treatment. For some anticancer genes, such as apoptin and par-4, several aspects of their downstream signalling have been elucidated.27,28 For ORCTL3 we identified here an effector - the fatty acid metabolism enzyme steaoryl-CoA desaturase (SCD) - as a target responsible for ORCTL3’s tumour-specific pro-apoptotic effect in renal tumours. This connection was discovered based on ORCTL3’s activation of an incomplete ER stress response, which comprises the activation of ATF4 but not BiP (Figure 3e), and which was also found when SCD was downregulated to initiate apoptosis.24 This cell death was found to cause the demise of all tested tumour cells in one24 but not in another study23 suggesting that the cellular transformation scenario in which this enzyme can be targeted had not yet been established.
We have found kidney cancers as a tumour model that responds to the inhibition of this metabolic enzyme. This was indicated when individual tumourigenic mutations were introduced into renal cells in an otherwise isogenic cell system (Figure 1, 6a,b) and when kidney tumours were studied ex vivo (Figure 6e). Previous work from our group had already shown that ORCTL3 is non-toxic to primary renal cells.7 So far the therapy of renal tumours relies mainly on surgery. As they often do not respond to chemotherapy and radiotherapy, there is hardly any systemic treatment against disseminated renal tumours.11 Consequently, renal tumours constitute a model in which conventional drugs are failing and only recently have targeted therapy treatments against VEGF and mTOR been approved for ccRCC. Nevertheless, the vast majority of tumors eventually also become refractory.12
Studies on changes in the metabolism of cancer cells currently define a dynamic field. The commonly held view is that the increase in the proliferation makes it imperative for malignant cells to alter the metabolism of ATP production, macromolecule synthesis, redox status, and fatty acids. Because of their increased proliferation, tumour cells are thought to become dependent on (addicted to) desaturated fatty acids through their increased requirement of phospholipids that constitute membranes.10 Our results represent a new paradigm as the cells with tumourigenic mutations did not feature an increased proliferation rate (Figure 2) but nevertheless became sensitive to the inhibition of the metabolic enzyme and succumbed to ORCTL3. Most established antineoplastic agents act against rapidly-proliferating cells, which, however, is one of the reasons why these compounds also exert considerable side effects in, for example, blood cells. Our data indicate that ORCTL3 does not merely target cells with a higher proliferation rate (Figure 2).
SCD catalyzes the introduction of a double bond between carbons 9 and 10 of saturated fatty acids and in particular palmitic (16:0) and stearic (18:0) acid to yield the monounsaturated fatty acids palmitoleic (16:1) and oleic (18:1) acid, respectively. The presence of double bonds in FA has been shown to increase the fluidity of the membrane, a feature that has been implicated in facilitating tumour and metastasis formation.29,30 However, how specific these effects are for tumour cells remained unknown. By comparing isogenic normal with transformed renal cells we have shown here for the first time that inhibition of SCD is synthetic lethal with transformation by various tumorigenic mutations (Figure 1, 4b, 6a,b). Interestingly, the transformation of these cells is also accompanied by the upregulation of its transcript (Figure 4f) confirming this gene as a treatment target, even though this was not accompanied by an increase of the desaturation index (Figure 4d).
The role of SCD in obesity has been the focus of many studies most of which concluded that targeted inhibition of SCD1 is effective in preventing diet-induced obesity 31 thus establishing SCD1 as a therapeutic target for this indication. This finding in combination with our data presented here suggest an interesting connection between obesity and cancer through SCD1. Various studies concluded that obesity was associated with an increased risk of developing cancer or metastasis.32-34 The abnormally elevated levels of SCD1 linked to the metabolic alterations found in obesity, as well as diabetes, and the upregulated levels of SCD1 in cancer cells, suggests SCD1 to be a molecular link between cancer and obesity.35 Its inhibition is positively correlated to preventing obesity31 and in this study we propose that its inhibition by ORCTL3 overexpression also results in specific toxicity to renal cancer cells.
We found ORCTL3 in a complex with Bap31 (Supplementary Figure S4b), a component of the previously identified ARCosome22 and its apoptosis induction depends on the components of this protein complex (Figure 3c,d). Various results presented here indicate that the tumour-specific effect of ORCTL3 is accomplished through SCD inhibition and that the activation of the ARCosome is a downstream effect: The pro-apoptotic cleavage product of Bap31 cannot be inhibited for apoptosis induction by SCD1 co-transfection (Supplementary Figure S5g) and does not impact on the desaturation index (Figure 4d). Also, caspase-8, which is activated in the ARCosome22, is not inhibitable by the addition of oleic acid and is not tumour-specific (Supplementary Figure S5b).
High affinity compounds against SCD have been already developed. In agreement with the assumed higher specificity of anticancer genes and their encoded proteins, in vitro ORCTL3 elicited higher apoptosis and lower non-specific activity (background in normal tissue) than the high affinity SCD1 inhibitor CAY10566 (Figure 4b and Figure 6b). For a therapeutic application of ORCTL3 it is essential to efficiently introduce the gene into target cells. This is evident by the increased apoptosis exerted by the adenovirus expressing ORCTL3 (Figure 1 and 6) and the difference between the human and the mouse ORCTL3, which seems to be caused by the increased stability in combination with a higher affinity to SCD1 of the mouse protein in cells (Figure 5, Supplementary Figure S8c). The stronger apoptosis activity of mouse ORCTL3 was reflected by a slightly increased reduction of the desaturation index (Figure 4d) suggesting that a critical enzyme inhibition has to be reached for cell death. The ORCTL3-expressing adenovirus was more efficient in vitro (Figure S8e) than in vivo (Figure 6c) in reducing the tumour cells. However, the hemorrhagic tissue observed in the tumours could have obscured the in vivo effects, as indicated by the more pronounced size difference when the tumours were removed from the animals (Figure S10g). Gene therapy of tumours is still evolving but has recently made progress 36 and this work establishes proof of concept and biological validation of an ORCTL3-based therapeutic application.
Previously we found that ORCTL3 is downregulated in a number of tumours suggesting that it is a tumour suppressor gene, which when re-expressed in tumour cells, re-constitutes a pro-apoptotic pathway that is initiated by oncogenes. However, since the human ORCTL3 is less efficient for apoptosis induction, it is more likely that the overexpression of mouse ORCTL3 creates a neomorphic mutation in the tumour cells. The finding that the transfer of the 3rd and 4th TM domain from human to mouse retained its stability but did not lead to apoptosis (Figure 5d) indicates that the two mouse TM domains have another function than just providing sufficient protein stability. This is a surprising finding as interactions with other proteins, if any, should be more conserved in the same species. Our data indicate that for high protein stability at least two mouse TM domains, either the 1/2 or the 3/4 pair in the correct context, are required. Accumulation within cells, however, is insufficient for efficient apoptosis induction as exemplified by the chimeric constructs with the human 3/4 TM domains (Figure 5d). Hence, for efficient apoptosis the mouse TM domains 3/4 are necessary.
We have in this work focused on renal cancer, but our previous data indicate that ORCTL3 is active against a wide variety of cancers.7 Future studies have to address the range of tumours that can be targeted with ORCTL3.
MATERIALS AND METHODS
DNA constructs
The mouse WT ORCTL3 and its ER fusion construct have been described.7 Deletion mutants and domain swapping were generated using suitable primers and recombinant PCR with Phusion® High-Fidelity DNA Polymerase (Finnzymes). PCR products were subcloned into the mammalian expression vector pcDNA3 (Invitrogen) and fused to an HA-tag at the C-terminal ends either by conventional restriction digestion and subsequent ligation using T4 DNA ligase (Promega) or by PCR Cloning using In-Fusion™ Advantage Kit (Clontech Laboratories, Inc.). All subcloning products were sequenced to verify the correct sequence. Plasmid DNA was isolated using Invitrogen’s maxi-prep kit according to the manufacturer’s instructions. Human ORCTL3 and SCD1 cDNAs were obtained from OriGene Technologies, Inc.
Cell culture
CV-1 cells (ATCC no: CCL-7), which are untransformed normal kidney fibroblasts, HeLa cells (ATCC no: CCL-2), HEK293T cells,7 and N2a cells (M.Sastre, Imperial College) were cultured in DMEM (Sigma-Aldrich) supplemented with 10% fetal calf serum (Sigma-Aldrich). CV-1 cells were cotransfected by a plasmid containing the G418 resistance gene and an empty control vector or the gene of interest in a 1:9 ratio using Xfect transfection reagent (Clontech) and selected using 2.5mg/ml G418 (Gibco). HeLa cells were transfected with JetPEI (Peqlab) or Effectene (Qiagen) and HEK293T cells were transfected with Xfect (Clontech) according to the manufacturer’s instructions. All cell lines were validated by short tandem repeat (STR) DNA profiles prior to the start of the project.
MTT assay
CV-1 cells were cultured in a 96-well plate at a density of 5×103 cells/well containing 100μl complete phenol red-free DMEM medium (Sigma-Aldrich). 20 μl of 5 mg/ml MTT solution (Sigma-Aldrich) was added to each well and the manufacturer’s protocol followed.
Cell cycle analysis
Cell cycle analysis was performed by propidium iodide (PI) staining and flow cytometry. Cells were cultured for 72 hours in 24-well plates in complete DMEM. Cells were harvested, washed, and stained with PI lysis buffer (10μg/ml PI (Sigma-Aldrich), 0.05% sodium citrate (Sigma-Aldrich), 0.05% Triton (Sigma-Aldrich) in PBS). The fluorescent signal was detected with FACS and cell cycle distribution was analysed using Flowjo 7.6.2 software.
In vivo experiment
Caki-2 cells were obtained from ATCC and cultured in McCoy’s 5A medium with 10% FCS. At 24hours and 4hours before inoculation, the cells in culture were fed with fresh medium. At the time of inoculation cells were harvested, resuspended in media-free of serum and antibiotics, and kept under sterile conditions in sterile bijous. To form bilateral tumours 2×106 cells were inoculated subcutaneously in a volume of 100 l into the right and the left flank of each female nude mice. When tumour size reached 50-80mm3 ORCTL3- or luciferase-expressing adenovirus (50 l of 5×109 viral particles) was injected intratumouraly once every 6 days to investigate the therapeutic potential of ORCTL3.
Collection and isolation of primary renal cells
Human kidney tissue samples were obtained from patients undergoing radical nephrectomy for renal cell carcinoma at Charing Cross Hospital (London). Tissue was collected from areas macroscopically identified as normal or tumourous immediately after extracting the specimen by an expert pathologist (tissue bank licence number 12275). Isolation procedure was performed within an hour of tissue removal according to an established protocol 26.
Adenoviral infection
Three different adenoviruses (constructed and produced by Welgen, Inc) were used expressing ORCTL3-ER cDNA, luciferase as a negative control, and GFP as an infection efficiency control. Cells were plated at low density (20×103 cells/well of 24 well plate) and cultured overnight. On the day of infection, cells from one representative well were counted. Viruses at an original concentration of 1×1012 virions/ml were diluted in 2% FCS DMEM to obtain the required multiplicity of infection (MOI). 200μl of virus-containing medium was added to the cells, which were incubated for 7 hours. After which the cells were washed and kept in 10% DMEM until analysed. Primary renal carcinoma cells from patient #3 lacking the coxsackievirus and adenovirus receptor (CAR) were infected with the help of AdBooster (Vector Biolabs). The appropriate amount of the adenovirus was incubated with the recommended amount of the reagent in 2% FCS medium for 30 minutes at room temperature on a rotating wheel. The same procedure of the viral infection above was followed.
Immunofluorescence
Cells transfected with HA-tagged plasmids were fixed with 2% paraformaldehyde (Sigma-Aldrich) 24 hrs post transfection. Fixed cells were stained with Alexa flour 488-labellebd monoclonal antibody against HA (Covance Inc.) as described 22.
Apoptosis quantification
Apoptosis in HeLa, HEK293T, CaKi2 and primary cells was quantified using 3,3′-dihexylocarbocyanine iodide (DiOC6)/propidium iodide (PI) double staining and flow cytometry as described.37 Apoptosis in normal CV1 and transformed CV1 was determined by phenotype quantification as described.7 For the FDA assay harvested cells were resuspended in 200μl of 2μg/ml FDA (Sigma-Aldrich) in PBS, incubated for 15 min at 37° C, and analysed on flow cytometer with excitation at 488nm using FL1 channel.
Cell lysate preparation
Cell were lysed in 100μl RIPA buffer (1% Nonidet P-40, 50 mM Tris pH 8 (Sigma-Aldrich), 150 mM sodium chloride (Sigma-Aldrich), 0.1% SDS (Biorad), 0.5% deoxycholate (Sigma-Aldrich)) containing 1x protease inhibitor (Thermo Scientific) and incubated on ice for 30 minutes. Cell lysates were centrifuged at 13,300 rpm for 30 minutes at 4°C.
Western blot analysis
Using BCA protein assay (Sigma Aldrich), equal amounts of proteins were denatured with 1X protein loading buffer (ThermoFisher Scientific) at 100°C for 10 minutes. Samples were electrophoresed in an SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Millipore) by semidry electroblotting (Bio-Rad) as described.22 Membranes were probed with primary antibodies for HRas (1:200; sc-520, Santa Cruz Biotechnology), c-Myc (1:200; sc-789, Santa Cruz Biotechnology), p53 (1:500; sc-126, Santa Cruz Biotechnology), COX2 (1:1000; clone CX229, Caymann Chemicals), E6 (sc-460, Santa Cruz Biotechnology), E1A (1:1000, clone M73, Millipore), ATF4 (1:250, sc-200, Santa Cruz Biotechnology), CHOP (1:1000, sc-7351, Santa Cruz Biotechnology), SCD1 (1:300, sc30435, goat polyclonal, Santa Cruz), β-actin (1:1000, A1978, Sigma-Aldrich), BiP (1:250, sc-1050, Santa Cruz Biotechnology), HA (1:1000, H6908 Sigma), SCD-5 (AP53809PU-N, 2BScientific Ltd), ATF-3 (sc-188(C-19), Santa Cruz), and against GAPDH (1:1500, sc-32233, Santa Cruz). The antibody against Bap31 was kindly provided by Dr Gordon Shore. Antibody binding was detected by enhanced chemiluminescence (Thermo Sientific) and visualised on film.
Co-immunoprecipitation
After harvesting the cells and washing them once with PBS, they were lysed in 500μl Tris/Triton buffer (20 mM Tris-HCl pH 7.4, 150mM NaCl, 0.5 mM EDTA, 0.5 mM EGTA, 1% triton X-100) together with protease inhibitors on ice for 30 mins. Cell lysates were then processed as described22 with the relevant antibody -HA (H3663, mouse monoclonal, Sigma) (1:100) or -SCD1 (sc30435, goat polyclonal, Santa Cruz) (1:100). Isotype-specific antibodies were used in the controls.
Fatty acid analysis
Total fatty acid composition of frozen CV1 cells was determined by gas chromatography following total lipid extraction and transesterification according to Lepage and Roy (1986) with modifications38. Briefly, 1 ml of methanol/toluene (4:1) was added to frozen CV1 pellets followed by vortexing and sonification. Acetyl chloride (100 μl) was added to cell lipid extract suspension then incubated for 2 hours at 60°C. Extraction suspensions were cooled to room temperature and 2.5 ml of 6% K2CO3 solution was added to each sample. The organic phase was then transferred to GC vials, evaporated to dryness under a stream of nitrogen then dissolved in 300 μl of n-hexanes. Samples were then analysed by gas chromatography (Agilent Technologies, UK; GC-7890A) equipped with a BPX70 capillary column (SGE Analytical Science, UK; 25 m × 0.22 mm (ID) and 25 μm film thickness). Fatty acid composition was expressed as a percentage of the total of all fatty acids (weight % or wt%).
Statistical analysis
Statistical analysis was performed using the unpaired student’s t-test. Data were obtained from n>3 experiments for every figure unless otherwise specified and were regarded as statistically significant if P < 0.05 (*), 0.01 (**), and 0.001 (***) based on student’s t-test.
Supplementary Material
ACKNOWLEDGEMENTS
G. A. was supported by the Ministry of Higher Education in Saudi Arabia. E.P. was supported by Cancer Research UK, W.C by the Development and Promotion of Science and Technology Talents Project (DPST), Royal Thai Government, Thailand and M-S. H. by a stipend from AstraZeneca Ldt. We thank Joel Abraham for in vivo work and the Imperial’s tissue bank, Dr David Hrouda, Ms Taneisha McFarlane, and Prof Gerry Thomas, for help. Tissue samples were provided by the Imperial College Healthcare NHS Trust Tissue Bank. Other investigators may have received samples from these same tissues. The research was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. We thank Mr Robert Gray for lipid extraction and primary GC analysis.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Danen-Van Oorschot AA, Fischer DF, Grimbergen JM, Klein B, Zhuang S, Falkenburg JH, et al. Apoptin induces apoptosis in human transformed and malignant cells but not in normal cells. Proc Natl Acad Sci U S A. 1997;94:5843–5847. doi: 10.1073/pnas.94.11.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su ZZ, Madireddi MT, Lin JJ, Young CS, Kitada S, Reed JC, et al. The cancer growth suppressor gene mda-7 selectively induces apoptosis in human breast cancer cells and inhibits tumor growth in nude mice. Proc Natl Acad Sci U S A. 1998;95:14400–14405. doi: 10.1073/pnas.95.24.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle. 2011;10:3153–3158. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nalca A, Qiu SG, El-Guendy N, Krishnan S, Rangnekar VM. Oncogenic Ras sensitizes cells to apoptosis by Par-4. J Biol Chem. 1999;274:29976–29983. doi: 10.1074/jbc.274.42.29976. [DOI] [PubMed] [Google Scholar]
- 5.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 6.Grimm S, Noteborn M. Anticancer genes: inducers of tumour-specific cell death signalling. Trends Mol Med. 2010;16:88–96. doi: 10.1016/j.molmed.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Irshad S, Mahul-Mellier AL, Kassouf N, Lemarie A, Grimm S. Isolation of ORCTL3 in a novel genetic screen for tumor-specific apoptosis inducers. Cell Death Differ. 2009;16:890–898. doi: 10.1038/cdd.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30:671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 11.Linehan WM, Yang JC, Rini B. Cancer of the kidney. In: DeVita VTea., editor. Cancer: Principles and practice of oncology. Lippincott Williams & Wilkins; New Haven, CT: 2011. pp. 1161–1182. [Google Scholar]
- 12.Singer EA, Gupta GN, Srinivasan R. Update on targeted therapies for clear cell renal cell carcinoma. Current opinion in oncology. 2011;23:283–289. doi: 10.1097/CCO.0b013e32834479c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen FC, Girardi AJ, Gilden RV, Koprowski H. INFECTION OF HUMAN AND SIMIAN TISSUE CULTURES WITH ROUS SARCOMA VIRUS. Proc Natl Acad Sci U S A. 1964;52:53–59. doi: 10.1073/pnas.52.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita J, Kraus MH, Onoue H, Srivastava SK, Ebi Y, Kitamura Y, et al. Activated H-ras oncogenes in human kidney tumors. Cancer Res. 1988;48:5251–5255. [PubMed] [Google Scholar]
- 17.Gordan JD, Lal P, Dondeti VR, Letrero R, Parekh KN, Oquendo CE, et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14:435–446. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noon AP, Vlatkovic N, Polanski R, Maguire M, Shawki H, Parsons K, et al. p53 and MDM2 in renal cell carcinoma: biomarkers for disease progression and future therapeutic targets? Cancer. 2010;116:780–790. doi: 10.1002/cncr.24841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nevels M, Dobner T. Determination of the transforming activities of adenovirus oncogenes. Methods in molecular medicine. 2007;131:187–195. doi: 10.1007/978-1-59745-277-9_13. [DOI] [PubMed] [Google Scholar]
- 20.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. Embo J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwasawa R, Mahul-Mellier AL, Datler C, Pazarentzos E, Grimm S. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2011;30:556–568. doi: 10.1038/emboj.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan-Lappe SE, Tucker LA, Huang X, Zhang Q, Sarthy AV, Zakula D, et al. Identification of Ras-related nuclear protein, targeting protein for xenopus kinesin-like protein 2, and stearoyl-CoA desaturase 1 as promising cancer targets from an RNAi-based screen. Cancer research. 2007;67:4390–4398. doi: 10.1158/0008-5472.CAN-06-4132. [DOI] [PubMed] [Google Scholar]
- 24.Minville-Walz M, Pierre AS, Pichon L, Bellenger S, Fevre C, Bellenger J, et al. Inhibition of stearoyl-CoA desaturase 1 expression induces CHOP-dependent cell death in human cancer cells. PLoS ONE. 2010;5:e14363. doi: 10.1371/journal.pone.0014363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wessels HP, Spiess M. Insertion of a multispanning membrane protein occurs sequentially and requires only one signal sequence. Cell. 1988;55:61–70. doi: 10.1016/0092-8674(88)90009-8. [DOI] [PubMed] [Google Scholar]
- 26.Valente MJ, Henrique R, Costa VL, Jeronimo C, Carvalho F, Bastos ML, et al. A rapid and simple procedure for the establishment of human normal and cancer renal primary cell cultures from surgical specimens. PLoS ONE. 2011;6:e19337. doi: 10.1371/journal.pone.0019337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burikhanov R, Zhao Y, Goswami A, Qiu S, Schwarze SR, Rangnekar VM. The tumor suppressor Par-4 activates an extrinsic pathway for apoptosis. Cell. 2009;138:377–388. doi: 10.1016/j.cell.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohn JL, Zhang YH, Aalbers RI, Otto N, Den Hertog J, Henriquez NV, et al. A tumor-specific kinase activity regulates the viral death protein Apoptin. J Biol Chem. 2002;277:50820–50827. doi: 10.1074/jbc.M208557200. [DOI] [PubMed] [Google Scholar]
- 29.Zeisig R, Koklic T, Wiesner B, Fichtner I, Sentjurc M. Increase in fluidity in the membrane of MT3 breast cancer cells correlates with enhanced cell adhesion in vitro and increased lung metastasis in NOD/SCID mice. Arch Biochem Biophys. 2007;459:98–106. doi: 10.1016/j.abb.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 30.Funaki NO, Tanaka J, Kohmoto M, Sugiyama T, Ohshio G, Nonaka A, et al. Membrane fluidity correlates with liver cancer cell proliferation and infiltration potential. Oncol Rep. 2001;8:527–532. doi: 10.3892/or.8.3.527. [DOI] [PubMed] [Google Scholar]
- 31.Brown JM, Rudel LL. Stearoyl-coenzyme A desaturase 1 inhibition and the metabolic syndrome: considerations for future drug discovery. Current opinion in lipidology. 2010;21:192–197. doi: 10.1097/MOL.0b013e32833854ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 33.Ceschi M, Gutzwiller F, Moch H, Eichholzer M, Probst-Hensch NM. Epidemiology and pathophysiology of obesity as cause of cancer. Swiss medical weekly. 2007;137:50–56. doi: 10.4414/smw.2007.11435. [DOI] [PubMed] [Google Scholar]
- 34.Gong Z, Agalliu I, Lin DW, Stanford JL, Kristal AR. Obesity is associated with increased risks of prostate cancer metastasis and death after initial cancer diagnosis in middle-aged men. Cancer. 2007;109:1192–1202. doi: 10.1002/cncr.22534. [DOI] [PubMed] [Google Scholar]
- 35.Igal RA. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31:1509–1515. doi: 10.1093/carcin/bgq131. [DOI] [PubMed] [Google Scholar]
- 36.Muthana M, Rodrigues S, Chen YY, Welford A, Hughes R, Tazzyman S, et al. Macrophage Delivery of an Oncolytic Virus Abolishes Tumor Regrowth and Metastasis After Chemotherapy or Irradiation. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-3056. [DOI] [PubMed] [Google Scholar]
- 37.Mahul-Mellier AL, Pazarentzos E, Datler C, Iwasawa R, Abuali G, Lin B, et al. De-ubiquitinating protease USP2a targets RIP1 and TRAF2 to mediate cell death by TNF. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lepage G, Roy CC. Direct transesterification of all classes of lipids in a one-step reaction. J Lipid Res. 1986;27:114–120. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.