Summary
To mount an immune response, T lymphocytes must successfully search for foreign material bound to the surface of antigen-presenting cells. How T cells optimize their chances of encountering and responding to these antigens is unknown. T cell motility in tissues resembles a random or Levy walk and is regulated in part by external factors including chemokines and lymph node topology, but motility parameters such as speed and propensity to turn may also be cell-intrinsic. Here we found that the unconventional Myosin 1g (Myo1g) motor generates membrane tension, enforces cell-intrinsic meandering search and enhances T-DC interactions during lymph node surveillance. Increased turning and meandering motility, as opposed to ballistic motility, is enhanced by Myo1g. Myo1g acts as a “turning motor” and generates a form of cellular “flânerie”. Modeling and antigen challenges show that these intrinsically-programmed elements of motility search are critical for the detection of rare cognate antigen presenting cells.
Introduction
Search is a universal requirement in many biological systems: from a predator strategy to locate prey, to the meandering search that T cells undertake to identify foreign peptides presented by major histocompatibility complex (MHC) molecules on antigen-presenting cells (APC). This latter search has been described as having features of a Brownian random walk (Miller et al., 2003; Preston et al., 2006) or a Levy walk (Harris et al., 2012). The efficiency of this random-like motility pattern observed for T cell in lymph nodes (LN) has been heavily modeled (Beauchemin et al., 2007; Beltman et al., 2009; Textor et al., 2011); however, perturbing cellular motility patterns in vivo has not previously been possible.
To optimize initial detection of antigens, a T cell must balance migration speed with the need to dwell in a given location for long enough to detect bona-fide signaling complexes and become activated. Furthermore, it must meander sufficiently to fully explore a region before departing to scan neighboring areas. The apparently random motility of T cells in tissues may arise through the combinations of three main mechanisms. First, the curved underlying stromal network of LN may guide motility in convoluted patterns matching these structures (Bajenoff et al., 2006; Katakai et al., 2004). Second, the LN is seeded with micropatterns of highly localized and variable chemokine gradients (Bromley et al., 2008). Finally, it has been suggested that cell-intrinsic mechanisms would control T cell interstitial migration and contribute to tissue surveillance (Mrass et al., 2010), but direct evidence for this is lacking.
The intrinsic rate of T cell motility is determined by the rate of actin polymerization (Serrador et al., 1999; Vicente-Manzanares et al., 2002) coupled with the bundling actions of molecules such as crosslinked myosin IIA (Jacobelli et al., 2009). Motility under some (Overstreet et al., 2013) but not all (Friedl et al., 1998; Jacobelli et al., 2010; Woolf et al., 2007) 3D environments requires the coordinated activity of integrins, presumably to transmit sufficient force to pull nuclei through restrictive spaces or move against flow. Actin polymerization rate can be inhibited by tension of the cell membrane (Oster and Perelson, 1987). Cell intrinsic control of directional persistence, the tendency not to turn, is less clear. While chemokines again may function as guidance cues, T cells in 3D environments show an intrinsic propensity to “weave” (Jacobelli et al., 2010), a feature shared with many other amoeboid cells including neutrophils (Inoue and Meyer, 2008) and Dictyostelium (Andrew and Insall, 2007; Fukui, 2002). How this plays out in random search strategies such as those undertaken by T cells is yet to be examined.
Class I myosins are the largest group of unconventional myosins (Coluccio, 2008; Kim and Flavell, 2008). They are monomeric motor proteins that interact with actin filaments within cells and, through lipid-binding C-terminal domains, associate with cellular membranes (Greenberg and Ostap, 2013; McConnell and Tyska, 2010). These associations generate membrane tension in at least one isoform, Myosin1a (Nambiar et al., 2009). Additionally, it has been suggested that these motors may act to sense forces on the membrane and actively oppose them (Laakso et al., 2008). How this family contributes to cellular motility remains largely undiscovered.
In this study, we identified Myosin 1g (Myo1g) as a prominent Class I myosin motor highly expressed in murine T cells. We found that Myo1g transiently accumulates in discrete areas at the plasma membrane of migrating cells or when membranes are deformed. Although T cells genetically deleted for Myo1g had global reduction in membrane tension, their homeostatic tissue distribution and responsiveness to TCR engagement was indistinguishable from wild-type cells. However, deficient cells moved faster and straighter. This combination of phenotypes allowed for side-by-side empirical and in silico modeling of the critical features of T cell search. While Myo1g−/− cells covered territory more quickly due to increased velocities and straighter paths, this ultimately proved to be a deficit, specifically for detection of rare antigens. This highlights that random walk motility is tuned, in part from within, and generates optimal combinations of speed and local dwell time.
Results
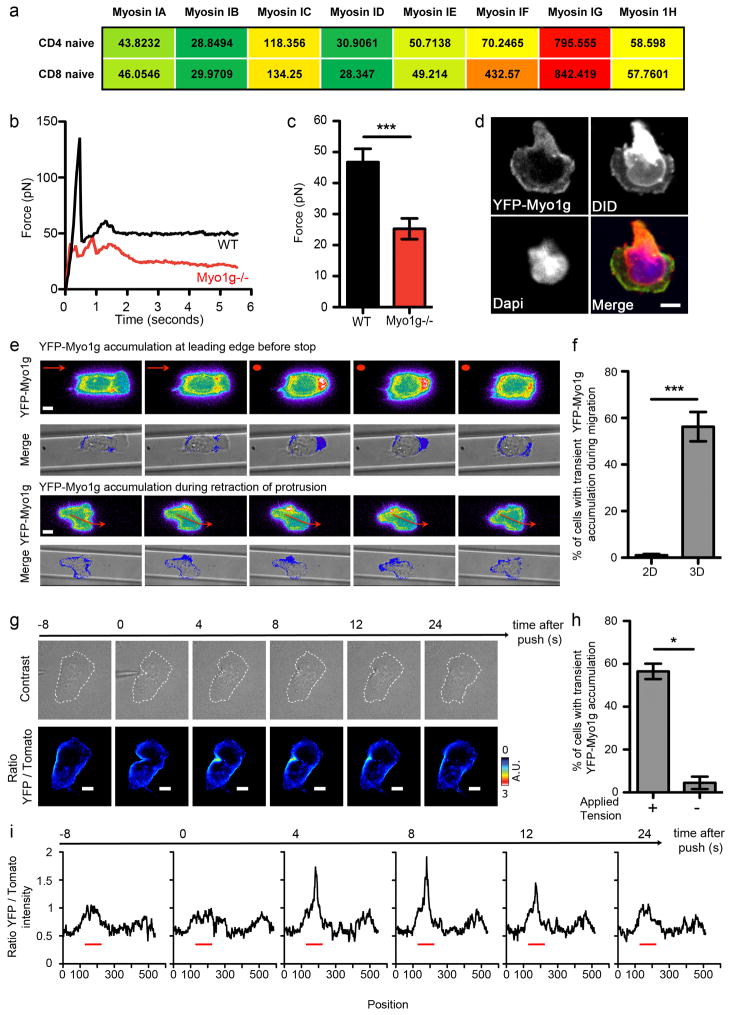
Myo1g is a hematopoietic specific Myosin (Olety et al., 2010; Patino-Lopez et al., 2010) and the most heavily transcribed Class I Myosin in naïve CD4 and CD8 T cells (Fig. 1a). To understand the function of this Class I Myosin in T cells, we generated mice lacking this motor protein by targeted mutagenesis (Fig. S1a–b). Offspring were viable and healthy and western blot confirmed the loss of Myo1g protein (Fig. S1c). The cellularity and composition of thymus, spleens and LNs from KO animals were grossly normal (Fig. S1d and data not shown).
Figure 1. Myo1g regulates membrane tension and transiently accumulates during T cell migration in 3D environment or after membrane tension perturbation.
a- Heat map showing Class I Myosins gene expression in naïve CD4 and CD8 T cells from LNs. Data are microarray data from the Immgen resource. b- Individual tether force records of WT (black) and Myo1g−/− (red) T cells. c- Average steady state tether force measurements on WT (black) and Myo1g−/− (red) T cells (n=5). ***p<0.001. (d–f) YFP-Myo1g expressing OTI cell blasts were harvested at day 4 to 5 after activation. d- Snapshot showing representative YFP-Myo1g localization in cells migrating on ICAM-1 coated coverslips (“2D”). Upper left: YFP; Upper right: membrane labeling DID; Lower left: DAPI; Lower right: Merge. Scale bar = 3μm. e- Images represent individual time points chosen from live imaging time-courses of YFP-Myo1g expressing T cells migrating in ICAM-1-coated microchannels (“3D”). YFP-Myo1g fluorescence is shown in pseudo-color (YFP-Myo1g), or overlayed with brightfield images (Merge). Scale bar = 3μm. Arrows indicate direction of migration, circles indicate a stopped cell. f- Quantification of transient YFP-Myo1g accumulation in cell migrating in 2D versus 3D environment. Cell was scored positive if it displayed any localized increased in fluorescence intensity during the imaging period. Data correspond to 2 independent experiments. n>50, ***p<0.001. (g–i) YFP-Myo1g expressing T cell blasts mTomato mice were allowed to migrate on ICAM-1-coated coverslip. Mechanical forces were applied by “pushing” the cell with a needle tip. Cells were imaged at 2–4″ intervals, using a 63X objective. g- Snapshots showing representative YFP-Myo1g accumulation at the site where mechanical stress was applied. Upper panel: DIC; Lower panel: ratio between YFP and Tomato fluorescence intensity is shown in pseudo-color. A.U.: Arbitrary Unit. Scale bar = 3μm. h- Quantification of transient YFP-Myo1g local accumulation as in (f) before and after application of mechanical forces. Data correspond to 4 independent experiments. *p<0.05, n=32. i- Graphs show the ratio between YFP and membrane (mTomato) fluorescence intensities along the cell surface at different times following application of membrane pressure. Site of pressure is depicted by a red line.
See also Figure S1.
Myosin I isoforms have a prominent role as force-sensitive motors, are located on cell membranes and can regulate membrane tension (Gillespie and Cyr, 2004; Laakso et al., 2008; Nambiar et al., 2009). We performed tether force assays which demonstrated that Myo1g deficiency resulted in a dramatic reduction in the initial force required to pull membrane away from the cortex as well as a reduction in the force required to continue elongating a tether (Fig. 1b–c). Consistent with a direct effect of Myo1g on membrane tension, a YFP-Myo1g fusion co-localized with the membrane marker DiD when T cells were plated onto integrin-coated glass substrates (2D) (Fig. 1d and Fig. S1e), suggesting that Myo1g was anchored to the plasma membrane. However, when T cells were plated in complex 3-dimensional microchannels (Jacobelli et al., 2010) (3D), which mimic the spatial constraints experienced by T cells in vivo, we observed additional transient local accumulations (Fig. 1e–f). Enrichment of YFP-Myo1g at the leading edge typically preceded motility arrest and accumulation at side edges coincided with turns; in these cases, the portion of membrane in which Myo1g had accumulated would ultimately retract and another portion of the leading edge dominated the direction of motility (Fig. 1e).
The difference between Myo1g localization during 2D and 3D migration suggested that Myo1g accumulated in 3D in response to additional and/or asymmetric pressure generated through contacts within the 3D environment. Indeed, when we applied local pressure to cells migrating in 2D environment with a needle tip, YFP-Myo1g was recruited to the site of pressure, often accompanied by accumulation of a membrane-targeted tdTomato (Fig. 1g and Movie S1). The ratio between YFP-Myo1g and membrane fluorescence intensities was increased at the site of pressure application (Fig. 1h–i), suggesting that Myo1g accumulation was not solely due to membrane accumulation. Consistent with this, transient accumulation during 3D migration required a functional motor domain of Myo1g (Fig. S3b).
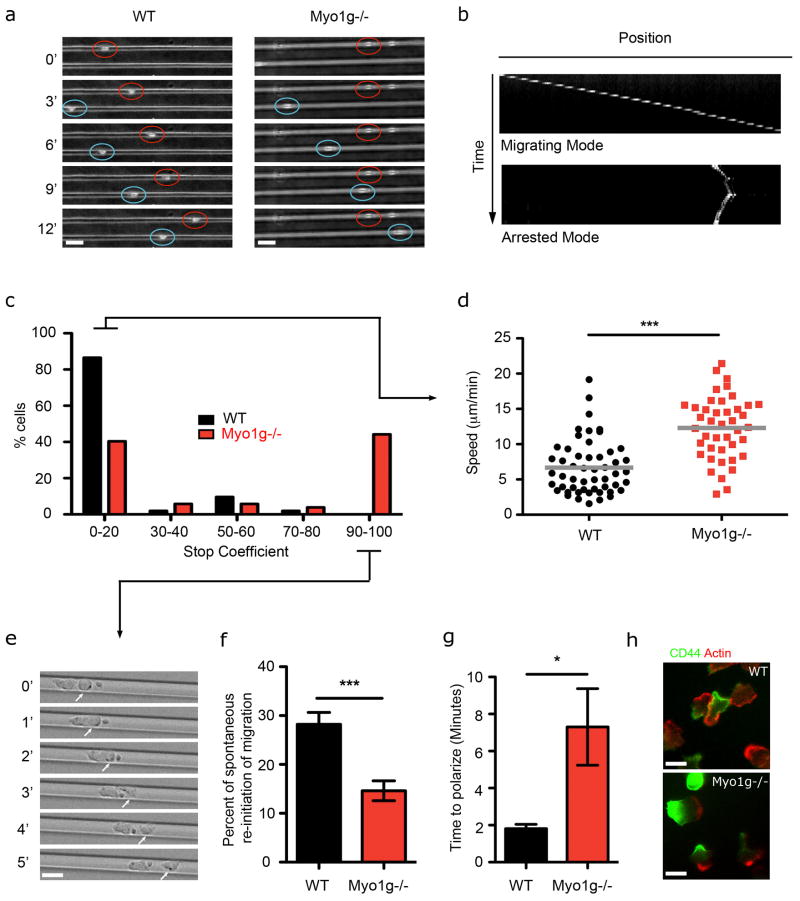
To test whether Myo1g controls T cell migration in 3D environment, we used microchannels. These channels give control over confinement and adhesiveness (Jacobelli et al., 2010) while eliminating other external factors, allowing us to investigate a cell intrinsic mode of T cell migration. We first analyzed T cell behavior in channels with relatively narrow widths that we have previously shown to optimize T cell speed (Jacobelli et al., 2010) (Fig. 2a, Movie S2). When T cells migrated through an 8μm-wide microchannel, two major behaviors could be observed, as cells alternated between migrating and arrested phases (Fig. 2b). Typically, WT T cells would arrest for only short periods; 86% of WT T cells would either migrate continuously, or stop for less than 20% of the time they were imaged and then resume migration (Fig. 2a–c). While 10.5% of WT T cells were arrested for at least half of the time they were imaged, 53% of Myo1g−/− T cells did so (Fig. 2a–c). Interestingly, this stopping behavior was not apparent during 2D migration (not shown), reminiscent of the absence of Myo1g transient and local accumulation in 2D (Fig. 1d and 1f), and suggesting that migration in 2D and 3D environment are differentially regulated. However, when motile, T cells from Myo1g−/− mice moved nearly twice as fast as T cells from WT mice (Fig. 2d).
Figure 2. Myo1g controls T cell speed and re-initiation of migration.
(a–f) T cells isolated from WT or Myo1g−/− mice were introduced into ICAM-1 coated microchannels. a- Brightfield images represent individual time points chosen from live imaging time-courses. Red and blue circles outline T cell positions; numbers indicate time in minutes. Scale bar = 10μm. b- Kymograph-basis for analysis of T cell migration versus arrest. Kymographs are selected examples. c- Graph shows the percentage of WT (black) and Myo1g−/− (red) cells stopping. The stop coefficient corresponds to percentage of time a cell was arrested over total time this cell was imaged. Data represent one representative experiment of 2 independent experiments (n=50). d- Graph shows mean velocities for WT (black) and Myo1g−/− (red) T cells under 3D confinement. Data correspond to 3 independent experiments. ***p<0.001. e- Snapshots showing a cell re-initiating polarization and migration when pushed by another migrating cell. Numbers represent time in minutes. Scale bar = 10μm. f- Graph shows the percentage of arrested WT (black) or Myo1g−/− (red) cells that re-initiated migration without any external stimuli, compared to cells that were pushed by another cell (n=62). Cells that did not move at all during the imaging period were excluded. Data correspond to 3 independent experiments. *p<0.05. (g–h) WT or Myo1g−/− naïve T cells migrating on ICAM-1 coated coverslips were treated with blebbistatin for 10 min and the inhibitor was washed away. g- Graph shows the time necessary for the cells to re-initiate polarization (n=120). Data correspond to 3 independent experiments. *p<0.05. h- Cells were fixed 20 min after washing the inhibitor and stained for CD44 and Actin. Pictures are representative of 2 independent experiments. Scale bar = 10μm.
See also Figure S2.
We noticed in microchannels containing more than one T cell that arrested cells were able to re-initiate migration when they encountered and were pushed by migrating cells (Movie S3, Fig. 2e and data not shown). In the absence of external force, we found that spontaneous re-initiation of migration was defective in Myo1g−/− T cells (Fig. 2f). Consistent with this concept, we found that Myo1g deletion resulted in an increased percentage of cells with rounded morphology when in microchannels (Fig S2a). To test whether Myo1g facilitated re-initiation of polarization and symmetry breaking, we challenged cells to re-polarize following wash-out of Blebbistatin, a drug which acutely causes cells to round up (Movie S4). In this context, Myo1g−/− T cells required approximately three times as long to re-initiate polarization compared to their control counterparts (Fig. 2g). Similar delay was observed when other chemicals were used to inhibit polarization, such as Nocodazole (data not shown). Myo1g−/− T cells were ultimately able to polarize, as confirmed by localization of CD44 at the uropod and enrichment of phalloidin stain at the leading edge (Fig. 2h). Finally, because Myo1g−/− T cells migrate faster, we also analyzed the function of Myo1g on the opposite transition, i.e whether cells deficient in Myo1g would have a defect in transitioning from a polarized to a rounded morphology. We quantified in 2D (a condition in which we did not see any increased stopping due to Myo1g deficiency) whether Myo1g−/− cells would have the tendency to stay polarized longer, which should be reflected by the percentage of cells polarized at any time point and did not find any difference in this measure (Fig. S2b). Together, these data demonstrate that while these cells do not have a global defect in polarization, they are delayed in generating a leading edge from a rounded morphology. Such data is consistent with a requirement for asymmetric application of membrane tension in amoeboid motility (Houk et al., 2012).
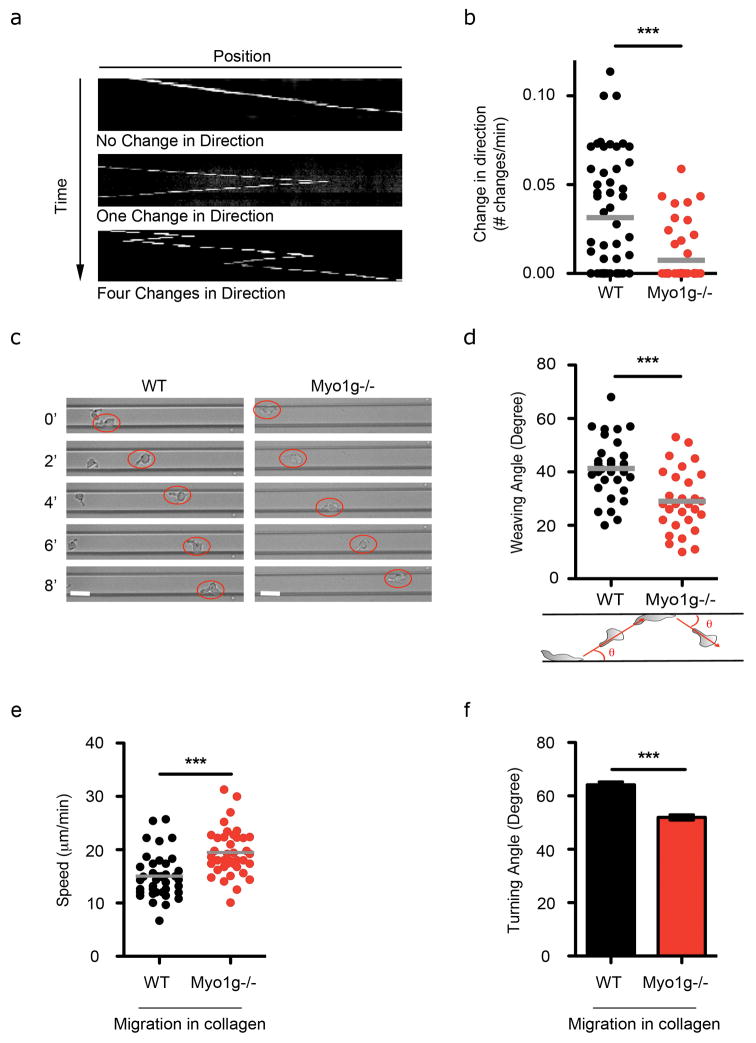
Because we often observed Myo1g transiently accumulating along cell margins as a cell veers away from that space, we speculated that Myo1g might encourage T cell turning. We therefore quantified the frequency at which a given cell would change direction while migrating in thin microchannels. We found that Myo1g−/− cells underwent approximately 3 times fewer complete changes in direction (“u-turns”) (Fig. 3a–b). In larger microchannels, activated T cells weave between opposing walls rather than crawling forward along a single wall (Jacobelli et al., 2010) (Fig. 3c, Movie S5). The weaving angle as T cells turned away from each sidewall was significantly shallower for Myo1g−/− cells as compared to controls (41.3°+/−2 for WT and 29°+/−2 for Myo1g−/− and Fig. 3d). We confirmed these results by analyzing T cell migration in 3D collagen matrices. Again, Myo1g deletion induced increased speed (Fig. 3e), decreased turning angle (Fig. 3f) but also increased rounded morphology (Fig. S2c), herein demonstrating that T cells retain intrinsic migration features even when within a fiber network. Finally, we confirmed that decreased weaving angle of Myo1g−/− T cells as they turn was not solely due to increased speed (Fig. S2d). Taken together, this analysis reveals a role for Myo1g in regulating intrinsic speed as well as the propensity to turn.
Figure 3. Myo1g regulates autonomous T cell weaving during migration in 3D.
(a–d) T cells isolated from WT or Myo1g−/− mice were introduced into ICAM-1 coated microchannels. a- Kymograph-basis for analysis of change in direction in 8μm-wide microchannels. Kymographs are selected examples. b- Change in direction (number of time of cell changed direction in 1 minute) of WT (black) and Myo1g−/− (red) T cells migrating in 8μm-wide microchannels. Data are from 3 independent experiments. ***p<0.001. c- Brightfield images of live imaging time-courses of cells migrating in 20μm-wide microchannels. Red circles outline T cell positions during weaving; numbers indicate time in minutes. Scale bar = 10μm. d- Mean weaving angle (angle a given cell bounces from one wall to the opposite wall) of WT (black) and Myo1g−/− (red) T cells migrating in 20um microchannels. Data are from 3 independent experiments. ***p<0.001. (e–f) T cells isolated from WT or Myo1g−/− mice were embedded and allowed to migrate in collagen lattices. Graphs show mean speed (e) and turning angle (f) of migrating WT (black) and Myo1g−/− (red) T cells. Data correspond to 2 independent experiments. ***p<0.001.
See also Figure S3.
We then investigated whether Myo1g- regulation of cell migration could be attributed to membrane tension. In a first set of experiments, we artificially increased membrane tensions in cells migrating in 2D environments by adding a hypotonic buffer which causes swelling and increased tension (Keren, 2011). This treatment reduced and therefore rescued cell speed of Myo1g−/− cells (Fig. S3a). We noted that WT cells were also sensitive to hypotonic treatment, suggesting that additional tensioning systems may exist in T cells. In a second set of experiments, we tested whether a Myo1g mutant that was properly localized at the plasma membrane, but did not transiently accumulate during migration in 3D (Fig. S3b–c), was able to rescue migration of Myo1g−/− cells. This motor-deficient mutant (IQ-Tail) could not link actin to the plasma membrane. Whereas full-length (FL) Myo1g restored T cell speed (Fig. S3d) and T cell weaving angles (Fig. S3e) of Myo1g−/− T cells migrating in microchannels to wild-type values, the mutant IQ-Tail did not. Altogether, we conclude that T cells have an intrinsic mode of migration regulated by Myo1g in a motor dependent manner, most likely through global and discrete regulation of membrane tension.
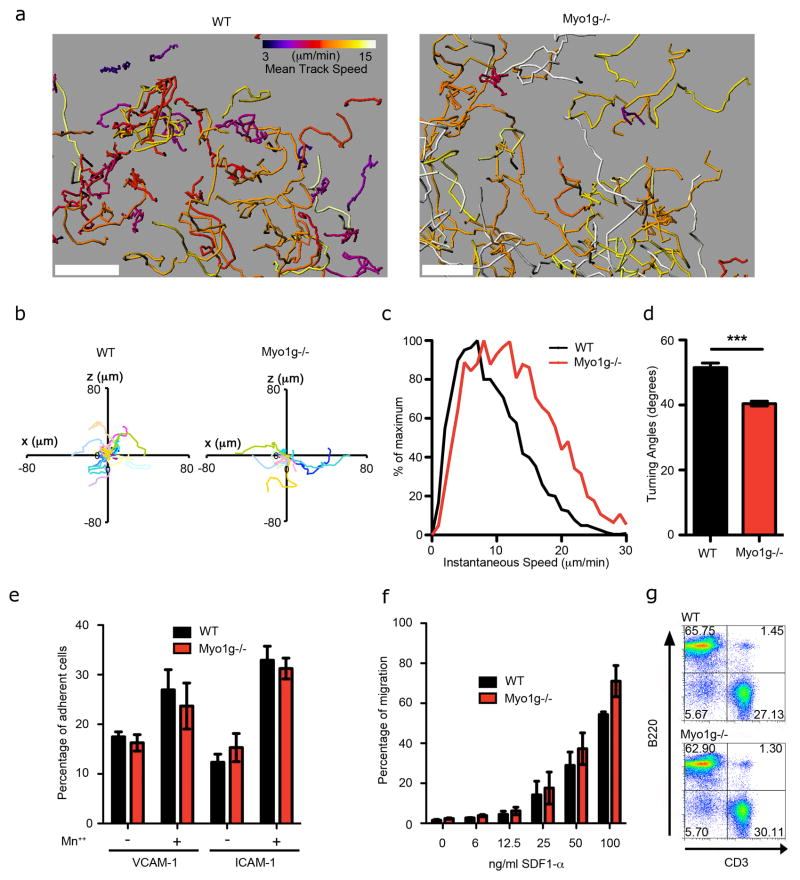
Meandering by naïve T cells in secondary lymph organs is not mechanistically well-understood (Germain et al., 2012; Mempel et al., 2004; Miller et al., 2003). To address whether Myo1g regulates random walk motility, WT and Myo1g−/− T cells were tracked in the T cell zone of LNs where they were found in the same intra-nodal space (Fig. 4a, Movie S6 and data not shown). As we observed in vitro (Fig. 3), motility tracks demonstrated a strong tendency for Myo1g−/− cells to move in straighter paths through the LN (Fig. 4a–b). Quantification of their speed showed that they also moved approximately 50% faster on average (Fig. 4a and c). Further track analysis showed that the average turning angle between 30-second frames was reduced from 52 degrees for WT T cells to 40 degrees for the Myo1g−/− T cells (Fig. 4d). We never observed enhanced arrest in Myo1g−/− cells in LNs, presumably due to the multitude of mechanical pushing forces generated in this environment, or due to local chemokine gradients.
Figure 4. Myo1g generates meandering interstitial migration.
(a–d) Migratory behavior of fluorescently labeled WT and Myo1g−/− OTI cells inside WT LNs. a- Image of a region of the T cell zone showing tracks for WT (left) and Myo1g−/− (right) T cells. Tracks are color-coded according to the mean track speed. Scale-bar is 50μm. b- Representative tracks in the xy plane of control T cells (left plot) and Myo1g−/− T cells (right plot) over a 15 min period. Starting coordinates are set to the origin. Units are in micrometers. Each colored line represents a single T cell track. c- Instantaneous velocity profiles of WT (black) and Myo1g−/− (red) OTI cells. Histograms show the relative frequency distribution of the two populations. d- The median turning angle was calculated for each cell and plotted for each group (WT; black and Myo1g−/−; red). Data are from 3 independent experiments. ***p<0.001. e- Percentage (%) of WT (black) and Myo1g−/− (red) T cells that adhered to integrin-coated plates. f- WT (black) and Myo1g−/− (red) OTI T cells were labeled with CFSE and CMTMR, respectively, mixed at a ratio 1:1 and subjected to a Boyden chamber assay. Percentage of cells that migrated towards the indicated dose of SDF1α was quantified by flow cytometry. The data is representative from 3 independent experiments. g- Flow cytometry analysis of T-cell (CD3+) and B Cell populations (B220+) in LNs.
See also Figure S4.
Although our analysis of in vitro migration in simplified 3D environments demonstrated that Myo1g regulated an autonomous mode of cell migration (Fig. 2–3), we verified that differences in migration patterns in vivo were not influenced by a defect in responding to external factors such as integrin ligands or chemokines. Myo1g−/− T cells expressed normal levels of integrins LFA-1 and chemokine receptor CCR7 expression (data not shown), they were as competent as their WT counterparts to adhere to integrin ligands (Fig. 4e), and to respond to chemokines such as SDF1α (Fig. 4f and Fig. S4a–b). Normal overall response to tissue was also supported by the observation that B and T cell populations appear at normal frequencies (Fig. 4g) and numbers (data not shown) in LNs of Myo1g−/− animals, and that homing to LNs was not impaired by Myo1g deficiency (data not shown).
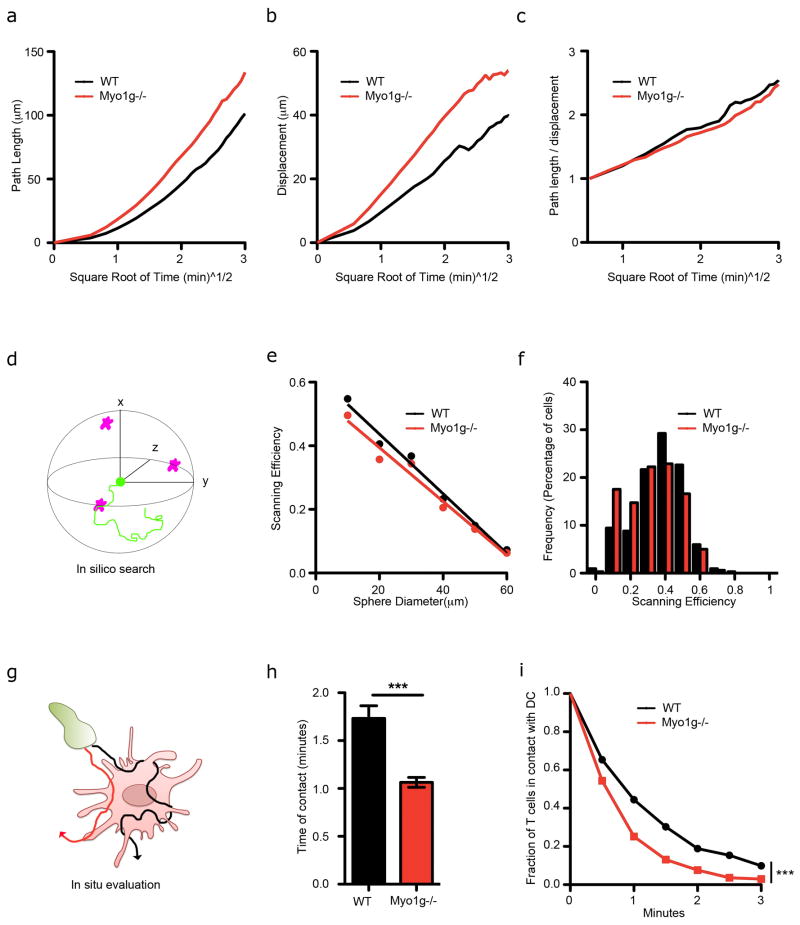
Based on these data, Myo1g deficient T cells were more “ballistic”, i.e they could cover territory faster as assessed by path length (Fig. 5a) but simultaneously expanded the radius of their search (i.e. underwent displacement) at a faster rate (Fig. 5b). Since the ratio of these, loosely representing the degree of saturation of volumes actually surveyed versus potentially surveyed, was similar between WT and KO cells (Fig. 5c), we hypothesized that their efficiency for running into relatively fixed targets might not be improved. We thus initially evaluated LN surveillance (Search) by modeling and quantifying the efficiency with which WT and Myo1g−/− T cell tracks, taken from live-imaging data in LNs, would intersect randomly placed in silico “APC targets” inside a sphere, placing the starting point of T cell tracks at the centroid (Fig. 5d). Previous modeling showed that increased T cell speed (without altering turning patterns) increases LN surveillance (Beltman et al., 2007), but over hundreds of simulations, we found only subtle differences in the frequency with which 20–30 minute tracks from WT and Myo1g−/− intersected in silico targets (Fig. 5e–f). Similar effects were seen when path segments were assembled into artificial 12-hour tracks (Fig. S4c–f) and these results concur with data on other biological search strategies in which ballistic motility is not necessarily more efficient (Reynolds and Bartumeus, 2009).
Figure 5. Defective LN surveillance of Myo1g−/− T cells.
(a–c) Migratory behavior of fluorescently labeled WT and Myo1g−/− OTI cells inside WT LNs. Representative experiment showing path length (a), mean displacement (b) and the ratio between path length and mean displacement (c) of WT (black) and Myo1g−/− (red) OTI cells over square root of time. (d–f) Lymph Node Search. d- T cell tracks start at the origin of a sphere where targets are placed randomly. Track is scored positive if it crosses a target. e- Scan efficiency (probability a track touches a target) relative to the diameter of the sphere. f- Frequency of scan efficiency for a sphere diameter of 20 um. (g–i) DC Evaluation. g- The time a T cell (green) was in contact with a single DC (pink) was deduced from 2P data (see Experimental procedures). Red and black lines illustrate trajectories of T cells. h- Average time WT or Myo1g−/− T cell contact a DC. ***p<0.001. n=50 i- Fraction of cells still in contact with a DC over time. ***p<0.001. n=70.
See also Figure S4.
Reasoning that fewer turns and faster migration might regulate the dwell time for surveying targets (illustrated in Fig. 5g), we also performed live-imaging of WT and Myo1g−/− T cells in excised LNs from mice that had been transferred with labeled activated DCs (without antigen), and quantified the time spent in contact for each T-DC encounter (which we refer to as Evaluation). T cells deficient for Myo1g spent less time in contact with DCs (Fig. 5h). Off-rate analysis showed that only 25% of Myo1g−/− T cells were in contact with a DC for at least 1 minute, compared to 45% of control cells (Fig. 5i and Fig. S4g). We conclude that migration patterns generated by Myo1g amplifies the time a T cell scans a DC while otherwise giving rise to similar efficiencies of “search”.
What is the significance of Myo1g and this motility pattern for T cells? T cells specific for a particular antigen are rare in the naive repertoire (Blattman et al., 2002). Additionally, cellular re-polarization and recognition of antigens on the surface of APC lags about ~1 min after initial contact (Krummel et al., 2000; Wulfing et al., 1997). Given that Myo1g deficiency reduced the time a T cell would evaluate a potential APC and that there was a 55% reduction in the percentage of T cells that were in contact with a DC for more than 1 minute (Fig 5), we sought to take advantage of Myo1g−/− T cells to determine whether an autonomous mode of T cell migration optimized the efficiency of recognizing and responding to DCs.
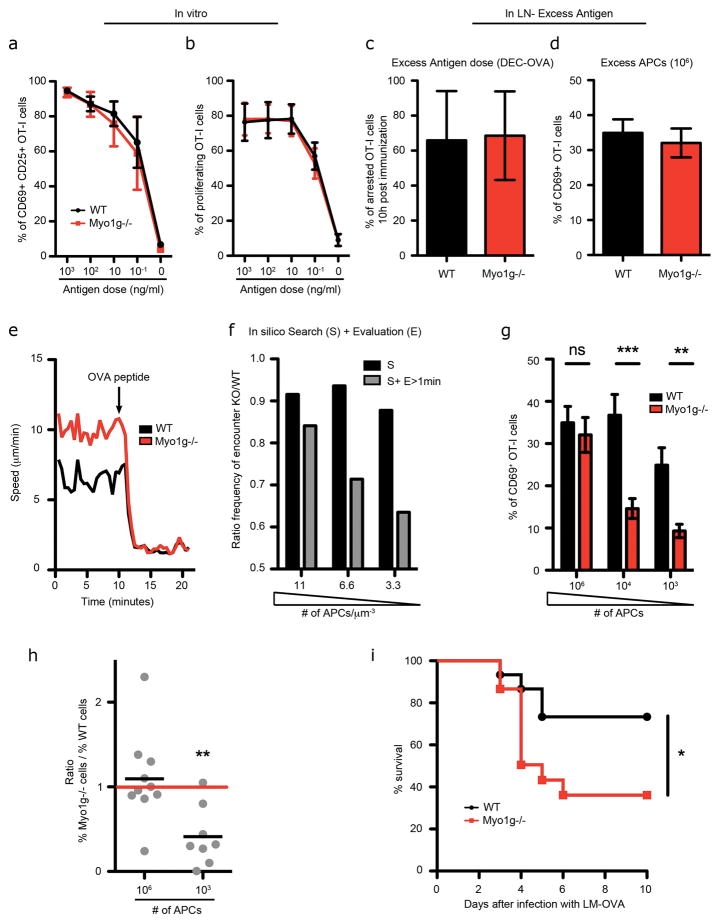
We first established that T cell receptor (TCR) triggering by cognate antigen was not defective in Myo1g−/− T cells by the following criterions: 1.) When WT and Myo1g−/− OTI T cells, specific for ovalbumin peptides, were challenged with antibodies against CD3 and CD28, they activated equivalently, as measured by up-regulation of the activation marker CD69 or by proliferation (Fig. S5a–b). 2.) Activation (Fig. 6a) and proliferation (Fig. 6b) of these same cells in response to APCs pulsed with their cognate ovalbumin peptide was indistinguishable, even using limiting concentrations of antigen. 3.) Proximal calcium signaling was equivalent (data not shown). 4.) Motility arrest and CD69 up-regulation was equivalent between WT and Myo1g−/− OT-I cells when antigens were presented on a large population of APCs in vivo (Fig. 6c–d). 5.) Symmetry breaking necessary for synapse formation during antigen recognition was not impaired in vitro, as accessed by TCR microcluster dynamics (Fig. S5c–d and Movie S7). 6.) The rate of symmetry breaking during antigen recognition was also not impaired in vivo, as WT and Myo1g−/− T cells slow their speed, stop and rounded up at the same rate following Ag recognition (Fig. 6e and Movie S8). Thus, Myo1g deletion did not alter antigen recognition per se.
Figure 6. Cell-intrinsic migration regulated by Myo1g is important for immune activation against rare antigens.
Percentage of WT (black) and Myo1g−/− (red) OTI cells positive for CD25 and CD69 24 hours (a) or percentage of proliferating cells 72 hours (b) after activation in vitro with BMDCs pulsed with different doses of OVA peptide. Data are from 3 independent experiments. (c–d) Mice bearing WT or Myo1g−/− OTI cells were immunized with a high dose of Antigen. c- Mean Arrest Coefficient (percent of time a cell has a speed ≤2um/min) of WT (black) and Myo1g−/− (red) OTI cells arrested in LN 8 hours after immunization with DEC-OVA. Data are from 2 independent experiments. d- Percentage of OTI cells positive for CD69 24 hours post-immunization with 106 OVA pulsed APCs. e- Fluorescently labeled WT and Myo1g−/− OTI cells were transferred in WT recipients and LNs were explanted after 16 hours. Cells were imaged by 2-Photon microscopy for 10 minutes. OVA peptide was then added to the media perfusing the LN and cells were imaged for another 10–15 minutes. Graph shows a representative example (out of 3 independent experiments) of WT (black) and Myo1g−/− (red) cell speed over time (n>50 for each cell type). OVA peptide addition is depicted by an arrow. f- LN Search was assessed as in Figure 5d–f. Graph shows the ratio of the frequency of encounter of KO tracks over WT tracks relative to target density. The efficiency of the search was calculated including or not an Evaluation > 1min. (g–h) Mice bearing WT or Myo1g−/− OTI cells were immunized with the indicated number of OVA pulsed BMDCs (APCs). Percentage of OTI cells positive for CD69 24 hours post-immunization (g) or ratio between the percentage of Myo1g−/− cells and percentage of WT cells 6 days post-immunization (h). Data are from 4 independent experiments. **p<0.01 and ***p<0.001. i- Mice (n = 10) were transferred with 5 × 103 WT (black) or Myo1g−/− (red) OT-I cells and were challenged with a sub-lethal dose of LM-OVA (0.25× the half-maximal lethal dose (LD50)). Graph shows mouse survival over time.
See also Figure S5.
We then performed in silico efficiency modeling based on tracks from Figure 5 including a factor that takes into account an Evaluation or requisite mean detection time (Campos et al., 2013) for a T cell to find and respond to displayed pMHC on the APC. As expected, Myo1g tracks were comparatively less efficient when the model required T cells to undertake an evaluation period that was equal or greater (data not shown) to the observed ~1 minute lag time for T cells to signal in response to APC contact (Fig. 6f) (Krummel et al., 2000; Wulfing et al., 1997). In predator-prey searches, larger numbers of prey can compensate for poor detection. In this case, for minimal interaction times equal to or greater than the measured lag time (1 minute), the decreased antigen detection efficiency of Myo1g−/− T cells is predicted to be rescued by increasing target density.
Based on these predictions, we determined whether Myo1g was required, empirically, for T cells to find and respond to rare APCs. WT or Myo1g−/− OT-I cells were transferred into mice, followed by immunization with graded numbers of APCs. One day after immunization, when transferred DCs had reached the LN (Fig. S5e), equivalent fractions of WT and Myo1g−/− OTI cells were activated when immunizing with a high number of DCs (Fig. 6g). In contrast, the fraction of Myo1g−/− OTI cells that were successfully triggered when a low number of APCs was transferred was decreased by 62% compared to WT OTI cells (Fig. 6g). This effect perpetuated out into the generation of clonal expansion; Myo1g−/− OTI T cells had expanded to the same extent as WT counterpart when a large number of APCs was introduced (Fig. 6h – 106 APCs), but APCs were rarer, Myo1g−/− OTI T cells showed a 59% reduction in numbers compared to controls (Fig. 6h – 103 APCs). Finally, we examined whether search behavior regulated by Myo1g and optimized for discovering rare APCs was actually required for an adaptive immune response to infection. Mice bearing a physiological number of WT or Myo1g−/− OTI cells were infected with a dose of Listeria Monocytogenes expressing OVA, below the LD50. Whereas 75% of mice bearing control OTI survived after challenge, only 35% of mice transferred with OTI deficient in Myo1g did so (Fig. 6i). We thus concluded that Myo1g and the motility patterns it imparts, though dispensable for antigen recognition per se, are nevertheless important for efficient surveillance.
Discussion
In this study, we provide direct genetic evidence that turning in T cells is in part “hardwired” and that a membrane-bound Myosin 1 isoform supplies much of the basis for this behavior. Myo1g−/− T cells, which showed altered migration patterns in vitro and in vivo, are less efficient in scanning and evaluating APC. Motility pattern, therefore, is a critical determinant of immune sensitivity to foreign antigens.
Myo1g is localized at the plasma membrane, and displays transient accumulation during migration in 3D. While significantly more biophysical data will be required to determine whether Myo1g is recruited by other molecules to membranes or is ubiquitous and autonomously force-sensing, our data clearly show that Myo1g is the source of a significant amount of the tension on the T cell membrane. Differential dynamic localization of Myo1g in T cells migrating in 2D or 3D also suggests that Myo1g is particularly important in 3D. This follows on other emerging data that 2D and 3D motility have significantly different requirements and modes (Friedl et al., 2012). We hypothesize that global membrane tension, regulated, at least in part, by Myo1g, attenuates T cell speed, whereas localized changes in membrane tension by Myo1g is involved in T cell turning. Consistent with this, Myo1g−/− T cells migrating in a uniform soft media (2D) have increased speed compared to WT cells, but equivalent path straightness (data not shown).
Myo1g function is likely complemented by other proteins such as those modulating the site of actin polymerization (Dang et al., 2013). The identity, role and relationship to Myo1g of such other molecules will need to be elucidated. Modulation of tension at different sites and times also leads to apparent opposite phenotypes. For instance, deletion of Myo IIA in Dictyostelium induces a loss of cortical tension (as opposed to membrane tension), which is correlated with decreased cell speed (Jay et al., 1995) and in T cells, loss of Myosin II leads to over-adherence and slow motility in vivo (Jacobelli et al., 2010). Establishing exact localization during migration and binding partners of those regulators is probably, here again, the key to our understanding of how tension within different regions orchestrates cell migration.
TCR triggering by T cells lacking Myo1g is unaffected in a multitude of assays, except when T cells are exposed to very low numbers of APCs. Infections with virus may generate far fewer than 100 APCs per LN (Usherwood et al., 1999) and early protective response to viruses limit both dissemination and ensuing destruction. Levels of cognate APC in many immunizations and infection models are in the higher range of our assay and thus are likely over-saturating by the measures we have used.
Search is a feature of many biological systems and it is well understood that ballistic modes of predatory searching, as by some sea birds, produces a search that is significantly incomplete (Miramontes et al., 2012). The phenotype of Myo1g−/− T cells and the modeling of their “Search” versus “Evaluation” dynamics provide an enlarged framework for additional understanding of immune search and surveillance. This work would suggest that future modeling will profit from including a parameter equivalent to the pre-exponential (or frequency) factor from chemical kinetics (Zhdanov et al., 1988). That factor ‘k’ partially encompasses the likelihood that two or more chemical reactants bump into each other in a favorable orientation to facilitate the generation of products. In the case of T cell search, our study suggests that an optimum exists between high T cell speed (to increase the chance a T cell comes across a target), and the slower process of meandering (which puts the length of T-DC contacts in a range consistent with generation of signaling).
Cell motility in vivo is undoubtedly influenced by features of the LN itself, like the (fibroblastic reticular cell) FRC network and local chemotaxis. Our results do not necessarily negate the potential role of FRC in guiding motility. Since the FRC network of LNs is highly convoluted (Bajenoff et al., 2006; Katakai et al., 2004), straighter paths by Myo1g−/− cells compared to WT cells in the same region might still represent cells that are loosely attached and “guided” by these fibers but which take more straight choices at each FRC junction. In comparison to our findings, computational analysis suggests that the guidance afforded by the FRC network has only a minor effect on the probability to find rare APC (Graw and Regoes, 2012).
Since Myo1g is not required for chemotaxis, the straighter paths taken by knockout cells may imply that chemokines are not a prominent part of directional guidance cues for early antigen searches in the T cell zone. That situation likely changes when T cells get some degree of initial triggering and up-regulate chemokine receptors such as CCR5 (Castellino et al., 2006; Hugues et al., 2007) or CXCR3 (Hu et al., 2011); cells that up-regulate these in response to successful detection of an initial pMHC encounter may thereby be subsequently guided rather than relying on unguided search. Thus, the search that may matter in the case of rare-antigen detection relates to the ability of a T cell to find and successfully respond to its very first pMHC-bearing APC and to subsequently reach a threshold of engagement(s) for chemokine receptor expression. Beyond that, the chemokines generated by other successful clones may help a T cell to find additional APCs or cells already identified by clones responding in parallel.
This work exemplifies an unappreciated role for an unconventional class I myosin in the cell biology of motility. In mice and humans, class I myosins can be sub-divided in short-tailed forms (Myo1a, b, c, d, g, and h) and long-tailed (amoeboid) forms (Myo1e and f). Of these, the long form Myo1f, and the short forms Myo1c and Myo1g are enriched in lymphocytes (Patino-Lopez et al., 2010). Long-form Myo1f has been implicated in neutrophil adhesion (Kim et al., 2006), but in its case this is likely due to its association with vesicular membranes leading to changes in integrin function. The loss of Myo1g did not affect integrin adhesion in T cells, suggesting that the specificity of the tail domain likely determines the relevant function for the particular myosin. Myo1c is strongly expressed in B cells, in which it participates in cytoskeleton rearrangements and antigen presentation (Maravillas-Montero et al., 2011).
To conclude, we provide evidence that Myo1g is a master regulator of membrane tension in T cells and is required for optimal meandering and successful LN surveillance.
Experimental Procedures
Local application of pressure
Activated T cells generated from WT or mTomato mice were transduced with YFP-Myo1g. ICAM-1-coated chambers were obtained by coating 8-well chambers (Lab-Tek) with 5μg/ml ICAM-1-Fc (R&D systems) in PBS for 1 h at 37C. 4–8 days after activation, cells were allowed to migrate on ICAM-1-coated coverslips for at least 2h. Mechanical stress was locally applied by using a glass probe with an inside diameter of 0.5mm. Most cells demonstrated continual viability after pushing and any cell that appeared damaged was not scored in this analysis. T cells were imaged for 2 min at intervals of 2–4″ with a modified microscope (Axiovert 200M; Carl Zeiss, Inc.) with Plan-Neofluar 63x objective (Carl Zeiss, Inc.).
Microchannel fabrication and imaging
Microchannel fluidic devices were fabricated and cells were loaded in microchannels as already described (see supplemental Methods) (Faure-Andre et al., 2008; Jacobelli et al., 2010). T cells crawling in the microchannels were imaged for 60 min at intervals of 1 min. Metamorph software (Molecular Devices) was used for calculation of cell speed and directionality. For analysis of the localization of YFP-Myo1g constructs in migrating T cells, cells were imaged for 5 min at intervals of 5″ intervals. Images were acquired with an inverted Zeiss with Yokogawa CSU-10 Spinning Disk. The imaging and control software used was MetaMorph (MDS Analytical Technologies). A minimum of 30 cells per microchannel width per treatment condition were analyzed.
Re-initiation of polarization and polarization quantification
Coverslips were coated with 5ug/ml ICAM-1 in PBS for 1h at 37C. T cell blasts generated from WT or Myo1g−/− mice were allowed to migrate on ICAM-1-coated coverslips. Cells were treated with Blebbistatin (racemic mix, 100μM of the racemate, Calbiochem) for 10 min and the inhibitor was washed away. T cells were imaged for 20 min at intervals of 10″ with a modified microscope (Axiovert 200M; Carl Zeiss, Inc.) with Plan-Neofluar 20x objective (Carl Zeiss, Inc.). For quantification of cell polarization, cells migrating on ICAM-1 coverslip (or alternately 20 min after washing Blebbistatin away) were fixed in PBS 1% PFA 15 min at RT and permeabilized with 0.05% Saponin for 5 min at RT. Cells were stained with CD44-FITC and Phalloidin-Alexa555 in PBS 2% BSA for 20 min at RT.
2-Photon imaging of explanted lymph nodes
WT and Myo1g−/− OTI cells were labeled with 2μM CFSE and 20μM CMTMR, respectively, ad-mixed and 3×106 total cells were transferred to WT recipient. Switching dyes did not affect results (data not shown).
For DC scanning experiments, 1×106 LPS-activated BMDCs from Act-CFP mice were injected subcutaneously in the footpad or the flank at the same time of T cell transfer.
For analysis of cell during the arrest phase, mice were immunized subcutaneously in the footpad or the flank with 2ug anti-DEC205 conjugates that were produced in house (Bonifaz et al., 2002; Gerard et al., 2013) together with 10ug anti-CD40 (Clone 1C10, eBiosciences).
Popliteal, Inguinal and Axillary LNs were taken out 16 to 24 hours after T cell transfer, or 8 hours after DEC-OVA immunization, and immobilized on coverslips with the hilum facing away from the objective. Time-lapse imaging was performed with a custom resonant-scanning instrument containing a four-photomultiplier tube (Hamamatsu) operating at video rate, as described (Friedman et al., 2010). Each xy plane spanned 288μm × 240μm at a resolution of 0.60μm per pixel. Images of up to 35 xy planes with 3-μm z-spacing were acquired every 30″ for 30 min.
Imaris (Bitplane) and Matlab software (Mathworks) were used to quantify T cell migration behavior. To characterize contact parameters between T cells and DCs, tracks and surface of T cells and DCs were generated and the dwell time of interaction between surfaces was analyzed as previously described (Gerard et al., 2013): T cell–DC cell interaction was defined as the close association of a given OT-I cell surface with a DC surface for at least 1 min. A threshold of 4 μm between cell edges was used, which account for low fluorescence frequently encountered at cell edges, which fit manual quantification (data not shown).
Track simulation
To overcome deficiencies in cell-based track analyses (Beltman et al., 2009; Textor et al., 2011), long duration tracks were simulated from shorter duration T cell tracks. Trajectories generated by tracking T cells that transited in LNs in the absence of antigen were pooled into control and knockout cell groups. The instantaneous displacements (the x, y and z displacements measured for each 30 second sampling interval) were calculated from all tracks in the two groups. Simulated tracks for WT and Myo1g−/− cells were then generated by randomly sampling the instantaneous displacements and cumulatively summing the selected displacements. 128 control and knockout tracks lasting 12 hours were simulated.
Track encounter efficiency or Search efficiency
To estimate the efficiency with which T cell tracks encountered a stationary dendritic cell, hypothetical target dendritic cells were placed in space at an average distance d from the T cell tracks’ common origin. The distance d, in the range of 5–60 μm (experimental data), or 50–600 μm (simulated tracks), was supplied as an input parameter to the simulation. Dendritic cell positions were generated by sampling from a random uniform distribution with a mean of 2d, which placed the target dendritic cells, on average, d μm from the T cell tracks’ origin. The distance for each target was combined with two random angles on the interval [0, 2π) to generate a spherical coordinate for each dendritic cell. A capture distance, defined as the sum of the dendritic cell radius and the T cell radius was used to determine if the T cell encountered a hypothetical dendritic cell. The dendritic cell radius (10 μm) was specified as an input parameter to the simulation, while the T cell radius was a randomly chosen value from the list of T cell radii measured as part of the in vivo tracking analysis. A T cell was considered to encounter a dendritic cell if at any point in the track, the distance between the T cell and dendritic cell positions was less than the capture distance. The simulation was repeated for 50 trials, with new dendritic cell positions generated for each trial.
Dwell duration or Evaluation
To estimate the amount of time that T cells spent near dendritic cells in the absence of antigen, the track encounter efficiency simulation was modified to calculate the number of successive simulation frames that a T cell remained within the capture distance of a dendritic cell for each encounter. The distribution of the dwell durations was calculated, with a single frame encounter assumed to represent a 30 second dwell time, a two frame encounter assumed to represent a 60 second dwell time, etc.
Statistical analysis
Data were expressed as mean +/− SEM, unless specified. Comparisons between groups were analyzed with the t-test, one-way or two-way Anova test, using GraphPad Prism software.
Supplementary Material
Highlights.
Myo1g regulates membrane tension and features of T cell migration.
T cells migrate in an intrinsic manner even when guided by fiber/stromal network.
Faster and straighter ballistic T cell migration is an inefficient search strategy.
Search mode matters for finding rare antigens and mount an adaptive response.
Acknowledgments
We thank the Biological Imaging Development Center personnel for technical assistance with imaging. We thank G.A. Bizarri, F. Bartumeus, G. Altan-Bonnet, D. Barber D. Erle and S. Dumont for helpful comments and suggestions and the ImmGen consortium for array data. We thank M.B. Headley for critical reading of the manuscript. This work was supported by grants from NIH (DK075555), American Heart Association to MJT, The NIH Intramural Research Program to SS and NIH (AI052116) to MFK.
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
Author Contribution
A.G. designed and performed the experiments unless specified. G.P-L. generated and characterized the thymic and peripheral populations in Myo1g−/− mice, performed adhesion assay, and together with A.G. made initial observation of increased migration speed of Myo1g−/− cells in vitro. F.J.T assisted with cSMAC assays. Y.L. and K.B. participated in Myo1g−/− mice characterization and P.B. wrote MATLAB scripts for extrapolating T cell tracks and quantifying search and evaluation efficiency. S.S. conceived and participated in design and interpretation of some experiments and M.F.K. conceived and participated in design and interpretation of all experiments. R.N. and M.J.T. performed analyses of membrane tension; A.G and M.F.K. wrote, revised and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nature cell biology. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- Bajenoff M, Egen JG, Koo LY, Laugier JP, Brau F, Glaichenhaus N, Germain RN. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25:989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin C, Dixit NM, Perelson AS. Characterizing T cell movement within lymph nodes in the absence of antigen. J Immunol. 2007;178:5505–5512. doi: 10.4049/jimmunol.178.9.5505. [DOI] [PubMed] [Google Scholar]
- Beltman JB, Henrickson SE, von Andrian UH, de Boer RJ, Maree AF. Towards estimating the true duration of dendritic cell interactions with T cells. Journal of immunological methods. 2009;347:54–69. doi: 10.1016/j.jim.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltman JB, Maree AF, Lynch JN, Miller MJ, de Boer RJ. Lymph node topology dictates T cell migration behavior. The Journal of experimental medicine. 2007;204:771–780. doi: 10.1084/jem.20061278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattman JN, Antia R, Sourdive DJ, Wang X, Kaech SM, Murali-Krishna K, Altman JD, Ahmed R. Estimating the precursor frequency of naive antigen-specific CD8 T cells. The Journal of experimental medicine. 2002;195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaz L, Bonnyay D, Mahnke K, Rivera M, Nussenzweig MC, Steinman RM. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. The Journal of experimental medicine. 2002;196:1627–1638. doi: 10.1084/jem.20021598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley SK, Mempel TR, Luster AD. Orchestrating the orchestrators: chemokines in control of T cell traffic. Nature immunology. 2008;9:970–980. doi: 10.1038/ni.f.213. [DOI] [PubMed] [Google Scholar]
- Campos D, Bartumeus F, Mendez V. Search times with arbitrary detection constraints. Phys Rev E. 2013;88 doi: 10.1103/PhysRevE.88.022101. [DOI] [PubMed] [Google Scholar]
- Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440:890–895. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- Coluccio LM. Myosin I. In: Coluccio LM, editor. Myosins: A superfamily of molecular motors. The Netherlands: Springer; 2008. [Google Scholar]
- Dang I, Gorelik R, Sousa-Blin C, Derivery E, Guerin C, Linkner J, Nemethova M, Dumortier JG, Giger FA, Chipysheva TA, et al. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature. 2013;503:281–284. doi: 10.1038/nature12611. [DOI] [PubMed] [Google Scholar]
- Faure-Andre G, Vargas P, Yuseff MI, Heuze M, Diaz J, Lankar D, Steri V, Manry J, Hugues S, Vascotto F, et al. Regulation of dendritic cell migration by CD74, the MHC class II-associated invariant chain. Science. 2008;322:1705–1710. doi: 10.1126/science.1159894. [DOI] [PubMed] [Google Scholar]
- Friedl P, Entschladen F, Conrad C, Niggemann B, Zanker KS. CD4+ T lymphocytes migrating in three-dimensional collagen lattices lack focal adhesions and utilize beta1 integrin-independent strategies for polarization, interaction with collagen fibers and locomotion. European journal of immunology. 1998;28:2331–2343. doi: 10.1002/(SICI)1521-4141(199808)28:08<2331::AID-IMMU2331>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Friedl P, Sahai E, Weiss S, Yamada KM. New dimensions in cell migration. Nature reviews Molecular cell biology. 2012;13:743–747. doi: 10.1038/nrm3459. [DOI] [PubMed] [Google Scholar]
- Friedman RS, Beemiller P, Sorensen CM, Jacobelli J, Krummel MF. Real-time analysis of T cell receptors in naive cells in vitro and in vivo reveals flexibility in synapse and signaling dynamics. The Journal of experimental medicine. 2010;207:2733–2749. doi: 10.1084/jem.20091201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y. Mechanistics of amoeboid locomotion: signal to forces. Cell biology international. 2002;26:933–944. doi: 10.1006/cbir.2002.0959. [DOI] [PubMed] [Google Scholar]
- Gerard A, Khan O, Beemiller P, Oswald E, Hu J, Matloubian M, Krummel MF. Secondary T cell-T cell synaptic interactions drive the differentiation of protective CD8+ T cells. Nature immunology. 2013;14:356–363. doi: 10.1038/ni.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain RN, Robey EA, Cahalan MD. A decade of imaging cellular motility and interaction dynamics in the immune system. Science. 2012;336:1676–1681. doi: 10.1126/science.1221063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PG, Cyr JL. Myosin-1c, the hair cell’s adaptation motor. Annual review of physiology. 2004;66:521–545. doi: 10.1146/annurev.physiol.66.032102.112842. [DOI] [PubMed] [Google Scholar]
- Graw F, Regoes RR. Influence of the fibroblastic reticular network on cell-cell interactions in lymphoid organs. PLoS computational biology. 2012;8:e1002436. doi: 10.1371/journal.pcbi.1002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MJ, Ostap EM. Regulation and control of myosin-I by the motor and light chain-binding domains. Trends in cell biology. 2013;23:81–89. doi: 10.1016/j.tcb.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TH, Banigan EJ, Christian DA, Konradt C, Tait Wojno ED, Norose K, Wilson EH, John B, Weninger W, Luster AD, et al. Generalized Levy walks and the role of chemokines in migration of effector CD8+ T cells. Nature. 2012;486:545–548. doi: 10.1038/nature11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148:175–188. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, Kagari T, Clingan JM, Matloubian M. Expression of chemokine receptor CXCR3 on T cells affects the balance between effector and memory CD8 T-cell generation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E118–127. doi: 10.1073/pnas.1101881108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugues S, Scholer A, Boissonnas A, Nussbaum A, Combadiere C, Amigorena S, Fetler L. Dynamic imaging of chemokine-dependent CD8+ T cell help for CD8+ T cell responses. Nature immunology. 2007;8:921–930. doi: 10.1038/ni1495. [DOI] [PubMed] [Google Scholar]
- Inoue T, Meyer T. Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PloS one. 2008;3:e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobelli J, Bennett FC, Pandurangi P, Tooley AJ, Krummel MF. Myosin-IIA and ICAM–1 regulate the interchange between two distinct modes of T cell migration. J Immunol. 2009;182:2041–2050. doi: 10.4049/jimmunol.0803267. [DOI] [PubMed] [Google Scholar]
- Jacobelli J, Friedman RS, Conti MA, Lennon-Dumenil AM, Piel M, Sorensen CM, Adelstein RS, Krummel MF. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA-regulated adhesions. Nature immunology. 2010;11:953–961. doi: 10.1038/ni.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay PY, Pham PA, Wong SA, Elson EL. A mechanical function of myosin II in cell motility. Journal of cell science. 1995;108(Pt 1):387–393. doi: 10.1242/jcs.108.1.387. [DOI] [PubMed] [Google Scholar]
- Katakai T, Hara T, Lee JH, Gonda H, Sugai M, Shimizu A. A novel reticular stromal structure in lymph node cortex: an immuno-platform for interactions among dendritic cells, T cells and B cells. International immunology. 2004;16:1133–1142. doi: 10.1093/intimm/dxh113. [DOI] [PubMed] [Google Scholar]
- Keren K. Cell motility: the integrating role of the plasma membrane. European biophysics journal: EBJ. 2011;40:1013–1027. doi: 10.1007/s00249-011-0741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SV, Flavell RA. Myosin I: from yeast to human. Cellular and molecular life sciences: CMLS. 2008;65:2128–2137. doi: 10.1007/s00018-008-7435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SV, Mehal WZ, Dong X, Heinrich V, Pypaert M, Mellman I, Dembo M, Mooseker MS, Wu D, Flavell RA. Modulation of cell adhesion and motility in the immune system by Myo1f. Science. 2006;314:136–139. doi: 10.1126/science.1131920. [DOI] [PubMed] [Google Scholar]
- Krummel MF, Sjaastad MD, Wulfing C, Davis MM. Differential clustering of CD4 and CD3zeta during T cell recognition. Science. 2000;289:1349–1352. doi: 10.1126/science.289.5483.1349. [DOI] [PubMed] [Google Scholar]
- Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I can act as a molecular force sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maravillas-Montero JL, Gillespie PG, Patino-Lopez G, Shaw S, Santos-Argumedo L. Myosin 1c participates in B cell cytoskeleton rearrangements, is recruited to the immunologic synapse, and contributes to antigen presentation. J Immunol. 2011;187:3053–3063. doi: 10.4049/jimmunol.1004018. [DOI] [PubMed] [Google Scholar]
- McConnell RE, Tyska MJ. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends in cell biology. 2010;20:418–426. doi: 10.1016/j.tcb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- Miller MJ, Wei SH, Cahalan MD, Parker I. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2604–2609. doi: 10.1073/pnas.2628040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramontes O, Boyer D, Bartumeus F. The effects of spatially heterogeneous prey distributions on detection patterns in foraging seabirds. PloS one. 2012;7:e34317. doi: 10.1371/journal.pone.0034317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrass P, Petravic J, Davenport MP, Weninger W. Cell-autonomous and environmental contributions to the interstitial migration of T cells. Seminars in immunopathology. 2010;32:257–274. doi: 10.1007/s00281-010-0212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar R, McConnell RE, Tyska MJ. Control of cell membrane tension by myosin-I. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11972–11977. doi: 10.1073/pnas.0901641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olety B, Walte M, Honnert U, Schillers H, Bahler M. Myosin 1G (Myo1G) is a haematopoietic specific myosin that localises to the plasma membrane and regulates cell elasticity. FEBS letters. 2010;584:493–499. doi: 10.1016/j.febslet.2009.11.096. [DOI] [PubMed] [Google Scholar]
- Oster GF, Perelson AS. The physics of cell motility. Journal of cell science Supplement. 1987;8:35–54. doi: 10.1242/jcs.1987.supplement_8.3. [DOI] [PubMed] [Google Scholar]
- Overstreet MG, Gaylo A, Angermann BR, Hughson A, Hyun YM, Lambert K, Acharya M, Billroth-Maclurg AC, Rosenberg AF, Topham DJ, et al. Inflammation-induced interstitial migration of effector CD4(+) T cells is dependent on integrin alphaV. Nature immunology. 2013;14:949–958. doi: 10.1038/ni.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino-Lopez G, Aravind L, Dong X, Kruhlak MJ, Ostap EM, Shaw S. Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent pleckstrin homology (PH) domain (Myo1PH) The Journal of biological chemistry. 2010;285:8675–8686. doi: 10.1074/jbc.M109.086959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope C, Kim SK, Marzo A, Masopust D, Williams K, Jiang J, Shen H, Lefrancois L. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- Preston SP, Waters SL, Jensen OE, Heaton PR, Pritchard DI. T-cell motility in the early stages of the immune response modeled as a random walk amongst targets. Physical review E, Statistical, nonlinear, and soft matter physics. 2006;74:011910. doi: 10.1103/PhysRevE.74.011910. [DOI] [PubMed] [Google Scholar]
- Reynolds AM, Bartumeus F. Optimising the success of random destructive searches: Levy walks can outperform ballistic motions. J Theor Biol. 2009;260:98–103. doi: 10.1016/j.jtbi.2009.05.033. [DOI] [PubMed] [Google Scholar]
- Serrador JM, Nieto M, Sanchez-Madrid F. Cytoskeletal rearrangement during migration and activation of T lymphocytes. Trends in cell biology. 1999;9:228–233. doi: 10.1016/s0962-8924(99)01553-6. [DOI] [PubMed] [Google Scholar]
- Textor J, Peixoto A, Henrickson SE, Sinn M, von Andrian UH, Westermann J. Defining the quantitative limits of intravital two-photon lymphocyte tracking. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12401–12406. doi: 10.1073/pnas.1102288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usherwood EJ, Hogg TL, Woodland DL. Enumeration of antigen-presenting cells in mice infected with Sendai virus. J Immunol. 1999;162:3350–3355. [PubMed] [Google Scholar]
- Vicente-Manzanares M, Sancho D, Yanez-Mo M, Sanchez-Madrid F. The leukocyte cytoskeleton in cell migration and immune interactions. International review of cytology. 2002;216:233–289. doi: 10.1016/s0074-7696(02)16007-4. [DOI] [PubMed] [Google Scholar]
- Woolf E, Grigorova I, Sagiv A, Grabovsky V, Feigelson SW, Shulman Z, Hartmann T, Sixt M, Cyster JG, Alon R. Lymph node chemokines promote sustained T lymphocyte motility without triggering stable integrin adhesiveness in the absence of shear forces. Nature immunology. 2007;8:1076–1085. doi: 10.1038/ni1499. [DOI] [PubMed] [Google Scholar]
- Wulfing C, Rabinowitz JD, Beeson C, Sjaastad MD, McConnell HM, Davis MM. Kinetics and extent of T cell activation as measured with the calcium signal. The Journal of experimental medicine. 1997;185:1815–1825. doi: 10.1084/jem.185.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanov VP, Pavlicek J, Knor Z. Preexponential Factors for Elementary Surface Processes. Catal Rev. 1988;30:501–517. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.