Abstract
The mechanisms whereby different vaccines may expand distinct Ag-specific T cell clonotypes or induce disparate degrees of protection are incompletely understood. We found that several delivery modes of a model retroviral Ag, including natural infection, preferentially expanded initially rare high-avidity CD4+ T cell clonotypes, known to mediate protection. In contrast, the same Ag vectored by human adenovirus serotype 5 induced clonotypic expansion irrespective of avidity, eliciting a predominantly low-avidity response. Nonselective clonotypic expansion was caused by relatively weak adenovirus serotype 5–vectored Ag presentation and was reproduced by replication-attenuated retroviral vaccines. Mechanistically, the potency of Ag presentation determined the speed and, consequently, completion of the CD4+ T cell response. Whereas faster completion retained the initial advantage of high-avidity clonotypes, slower completion permitted uninhibited accumulation of low-avidity clonotypes. These results highlighted the importance of Ag presentation patterns in determining the clonotypic composition of vaccine-induced T cell responses and ultimately the efficacy of vaccination.
Introduction
A cardinal property of adaptive immunity is immunological memory, which protects the host against reinfection (1). This property forms the basis for vaccination, which aims at inducing an adaptive immune response that will be effective also against the natural infection (2). Indeed, vaccination-induced adaptive immunity can be highly protective against certain viral pathogens (2). However, protective immunity against some viruses (e.g., HIV-1), bacteria (e.g., Mycobacterium tuberculosis), or cancer has so far been more difficult to achieve (3, 4). T cell–mediated immunity is considered of central importance in the protection against the latter group, highlighting the need for deeper understanding of the processes underlying the induction and maintenance of a protective T cell response.
Viral vectors are an essential tool in delivering genetic material into cells for vaccination or gene therapy. However, their use in eliciting T cell immunity has revealed considerable outcome variation (5, 6). For example, a recombinant CMV strain from rhesus monkeys encoding SIV Ags was shown to induce an atypical T cell response and significant, often sterilizing immunity against SIV challenge in nonhuman primates (7). In contrast, a human adenovirus serotype 5 (Ad5) vector encoding HIV-1 Ags was found to induce largely ineffective immunity against HIV-1 infection (8, 9). Despite their importance in vaccination, the properties of viral vectors that elicit effective T cell responses are incompletely understood.
The TCR repertoire available before infection and further shaped during the response can heavily influence the protective capacity of the T cell response. A TCR repertoire comprising clonotypes with high avidity for Ag may confer distinct advantages (10). Indeed, HIV-1 control was correlated with the presence of high-avidity virus-specific CD4+ T cells (11), and comparable observations were made in other systems (12–15). The relative abundance of distinct TCR clonotypes in the virus-specific CD8+ T cell population was proposed as a significant modulator of the protective effect of HLA-B27 alleles in HIV-1 infection (16). Similarly, CD4+ T cell–mediated protection against Friend virus (FV) infection, a mouse model for retroviral infection, was found to be a property exclusively of clonotypes bearing high-avidity TCRs (17).
Selection of distinct TCR clonotypes during thymocyte development as well as during the immune response to a viral Ag is largely dictated by the intrinsic ability of each TCR to recognize Ag (18). However, several T cell–extrinsic factors were also suggested to influence the clonotypic composition of the T cell response. For example, the clonotypic composition and resulting overall avidity of the CD4+ T cell response of mice to purified pigeon cytochrome c (PCC) protein immunization was found to be modified by the coadministered adjuvant (19). Moreover, vaccination of mice with different vaccine vectors all encoding HIV-1 envelope (env) was shown to induce Ag-specific CD8+ T cells with different fine specificities and TCR usage (20).
In this study, we used a well-characterized model of the CD4+ T cell response to a retroviral Ag, in which the clonotypic composition can be monitored according to TCR avidity. Polyclonal EF4.1 TCRβ-transgenic CD4+ T cells harbor increased frequencies (on average 4%) of cells reactive with the H2-Ab–restricted env122–141 epitope within the surface unit of Friend murine leukemia virus (F-MLV) env gene (21). F-MLV is a replication-competent virus that together with the replication-defective, but pathogenic spleen focus-forming virus, form the FV, a murine retroviral complex, which causes chronic infection of the hematopoietic system (22). In EF4.1 mice, pairing of the transgenic TCRβ-chain with distinct endogenous TCRα-chains creates clonotypes with different functional avidities, and CD4+ T cells using a Vα2 chain are >30-fold more sensitive to env122–141 stimulation than are cells using other TCRα-chains (referred to as non-Vα2). Following FV infection, high-avidity Vα2 clonotypes, although a minority (∼25%) in the naive repertoire, quickly dominate the peak of the env-specific CD4+ T cell response (21, 23). We found, however, that vaccination with a replication-defective human Ad5 vector encoding F-MLV env (24) uniquely induces a predominantly low-avidity env-specific CD4+ T cell response as a result of a distinct pattern of Ag presentation driving a protracted phase of T cell expansion.
Materials and Methods
Mice
Inbred C57BL/6 (B6) and CD45.1+ congenic B6 mice were originally obtained from The Jackson Laboratory (Bar Harbor, ME). TCRβ-transgenic EF4.1 mice (21), Ifngr1−/− mice (25), Ifnar1−/− mice (26), Tnfr1−/− mice (27), Tnfrsf4−/− mice (28), Il21r−/− mice (29), Il12a−/− mice (30), and Nur77-GFP–transgenic mice (31) were all on the B6 genetic background. Mice with dendritic cell (DC)–specific deletion of MHC class II were obtained by crossing mice with a conditional H2-Ab1 allele (H2-Ab1fl) (32) with mice expressing Cre under the Cd11c promoter (33). In the latter strain, Cre-mediated recombination is observed in nearly all CD11c+ DCs, but not in CD11c− monocytes/macrophages, whereas only partial recombination is observed in CD11clow monocytes, attributed to their differentiation into DCs (33). All animal experiments were approved by the Ethical Committee of the National Institute for Medical Research and conducted according to local guidelines and U.K. Home Office regulations under the Animals Scientific Procedures Act 1986 (ASPA).
T cell purification and adoptive transfer
Single-cell suspensions were prepared from the spleens and lymph nodes of donor CD45.1+ or CD45.2+ EF4.1 mice, and CD4+ T cells were enriched using immunomagnetic positive selection (StemCell Technologies) at >96% purity. A total of 1 × 106 EF4.1 CD4+ T cells were injected in CD45.1+CD45.2+ recipients via the tail vein, resulting in engraftment of ∼8000 env-specific CD4+ T cells in the spleen. In indicated cotransfer experiments, CD4+ T cells from CD45.1+CD45.2− and CD45.1−CD45.2+ EF4.1 donor mice were mixed at equal ratios and were distinguished from each other (and from host cells) based on CD45.1 and CD45.2 expression. Where indicated, enriched EF4.1 CD4+ T cells were further purified (>98% purity) by cell sorting, performed on MoFlo cell sorters (DakoCytomation, Fort Collins, CO), according to Vα2 expression. A total of ∼1.2 × 105 Vα2 or 8.8 × 105 non–Vα2-purified EF4.1 CD4+ T cells were injected separately in recipient mice.
In vivo infection and immunization
FV stocks were propagated in vivo and prepared as 10% w/v homogenate from the spleen of 12-d-infected BALB/c mice, as previously described (23). Mice received an inoculum of ∼1000 spleen focus-forming units of FV. Stocks of B-tropic and N-tropic F-MLV (F-MLV-B and F-MLV-N, respectively) were prepared as culture supernatants of Mus dunni fibroblast cells chronically infected with the respective virus. Mice received an inoculum of ∼104 infectious units of F-MLV. Ad5.pIX-gp70 and Ad5.GFP are replication-defective Ad5-based vectors, expressing F-MLV gp70 and GFP, respectively, and stocks (24) were prepared by infection of 293A cells at a titer of 9 × 109 viral genomes/ml. Ad5.pIX-gp70 genome copies were assessed by real-time PCR using primers 5′-CTGCGCCAGAGACTGCGACGA-3′ and 5′-GACCCGGGGCAGACATAAAAT-3′, specific to the F-MLV env gene. Ad5.pIX-gp70 was administered at the indicated dilution either i.v. (in 100 μl final volume) or i.m. by injection into the leg muscle (in 40 μl final volume). To select the appropriate dose, Ad5.pIX-gp70 was titrated in vivo to the point where it induces an env-specific CD4+ T cell response of equal magnitude to that induced by FV or F-MLV-B infection (4.5 × 108 Ad5.pIX-gp70 viral genomes/mouse). For peptide immunization, mice received an i.p. injection of a total of 12.5 nmol synthetic env124–138 peptide mixed in the Sigma Adjuvant System. For a cell-based vaccine, mice were given an i.v. injection of 3 × 106 FBL-3 cells, an FV-induced tumor cell line that expresses F-MLV env, but does not produce infectious particles (34). All stocks were free of Sendai virus, murine hepatitis virus, parvoviruses 1 and 2, reovirus 3, Theiler’s murine encephalomyelitis virus, murine rotavirus, ectromelia virus, murine CMV, K virus, polyomavirus, Hantaan virus, murine norovirus, lymphocytic choriomeningitis virus, murine adenoviruses FL and K87, and lactate dehydrogenase–elevating virus (LDV). For coinfection of FV and LDV, a similarly prepared stock of FV additionally containing LDV was also used (23).
In vitro infection and transduction
The ability of different promoters to drive GFP expression was examined in B-3T3 and M. dunni fibroblast cells. For CMV promoter (CMVp)–driven GFP, the XG7 retroviral vector was used (35). For MLV long terminal repeat (LTR)–driven GFP, the env gene of the Moloney MLV genome in plasmid pNCS (36) was replaced by an in-frame promoterless bicistronic gene encoding GFP and Cre using an internal ribosome entry site. Pseudotyped retroviral particles packaging these two vectors were produced in M. dunni fibroblast cells infected with F-MLV-B as previously described (37). Assessment of GFP expression by the CMVp used in the Ad5 vector was carried out using the Ad5.GFP vector (24). Cells were transduced with the pseudotyped retroviral vectors or transfected with Ad5.GFP particles at a mean order of infection of <0.2 to minimize uptake of more than one particle per cell.
Flow cytometry
Single-cell suspensions were stained with directly conjugated Abs to surface markers, obtained from eBioscience (San Diego, CA), Caltag/Invitrogen, BD Biosciences (San Jose, CA), or BioLegend (San Diego, CA). Cell-cycle analyses were performed by intranuclear staining for the Ki-67 Ag (BD Biosciences) or Vybrant DyeCycle Violet (Invitrogen). Multicolor cytometry was performed on FACSCanto II, LSRFortessa (both from BD Biosciences), and CyAn (Dako, Fort Collins, CO) flow cytometers and analyzed with FlowJo v10 (Tree Star, Ashland, OR) or Summit v4.3 (Dako) analysis software.
Peptide–MHC class II tetramer synthesis
H2-Ab complexes with peptide-register trapped F-MLV env125–135 (corresponding to the amino acid sequence LTSLTPRCNTA) were produced and biotinylated in Drosophila melanogaster S2 cells and then combined with streptavidin-allophycocyanin (Prozyme, Hayward, CA) to make tetramers, which were used as previously described (38).
In vitro env presentation assay
DCs were examined for their ability to present to the H5 env-specific T cell hybridoma (17) during an 18-h coculture. The spleens of F-MLV–infected or Ad5.pIX-gp70–immunized mice were removed at indicated time points, treated with Liberase TL (Roche, Mannheim, Germany) for 30 min, and stained with directly conjugated Abs. DCs were purified by cell sorting as CD19−Ly6C−CD11bintCD11c+MHC class IIhi cells and used for the assay at the indicated numbers together with 105 H5 cells. Expression of CD69 on H5 cells was assessed by flow cytometry at the end of culture.
Gene expression profiling
For gene expression comparisons, Ad5.pIX-gp70–primed env-specific effector EF4.1 CD4+ T cells were purified by cell sorting as donor-type CD44hiCD4+ T cells on day 7. RNA was isolated using the QIAcube (Qiagen, Crawley, U.K.). RNA quality was assessed using the Agilent bioanalyzer (Agilent Technologies, Santa Clara, CA). Synthesis of cDNA, probe labeling, and hybridization were performed using MouseGene 1.0 ST oligonucleotide arrays (Affymetrix, Santa Clara, CA). Primary microarray data are available on the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2210. Ad5.pIX-gp70–primed effectors were compared with previously obtained data from naive EF4.1 CD4+ T cells as well as effectors primed by either FV infection or FV/LDV coinfection (23) (available under accession number E-MEXP-2950). Microarray data were analyzed with GeneSpring GX (Agilent Technologies) and Qlucore Omics 3.0 (Qlucore, Lund, Sweden). Functional gene annotations were obtained using g:Profiler (39) and Interferome v2.0 (40).
Statistical analyses
Statistical comparisons were made using SigmaPlot 12.0 (Systat Software, Erkrath, Germany). Parametric comparisons of normally distributed values that satisfied the variance criteria were made by an unpaired Student t tests. Data that did not pass the variance test were compared with a nonparametric two-tailed Mann–Whitney rank sum test.
Results
Ad5-vectored env uniquely induces low-avidity env-specific CD4+ T cells
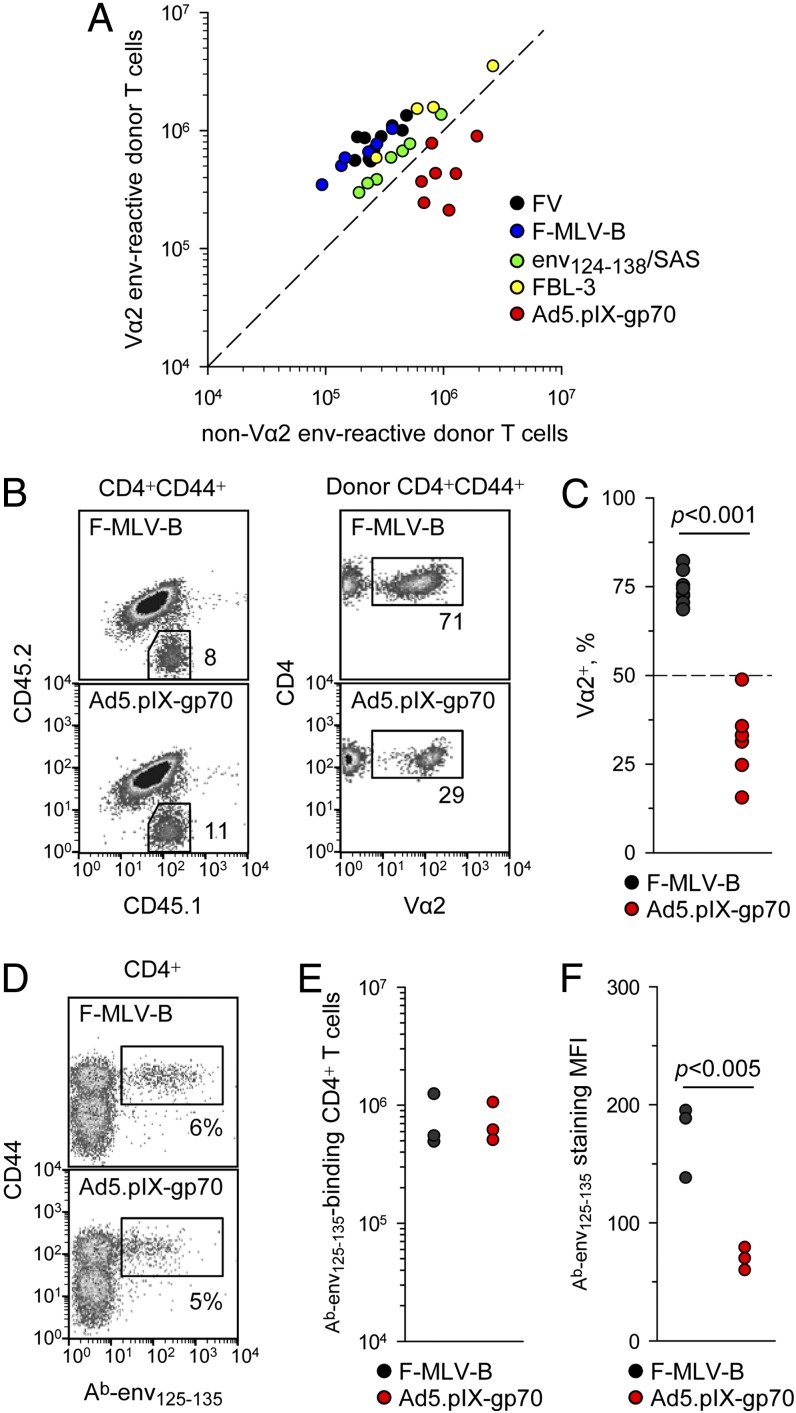
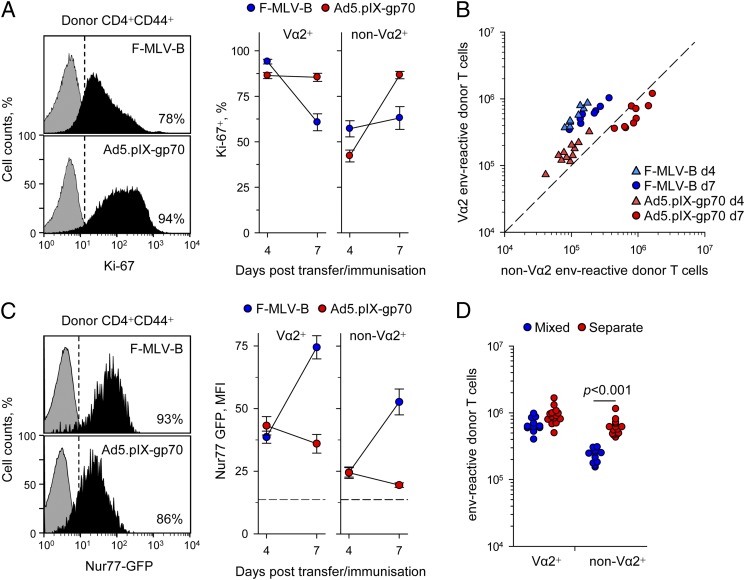
To monitor the clonotypic composition of the CD4+ T cell response to the env122–141 retroviral Ag, cohorts of allotypically marked EF4.1 TCRβ-transgenic CD4+ T cells were transferred into wild-type (WT) hosts at the time of infection or immunization and their peak response was analyzed 7 d later. Absolute numbers of high-avidity Vα2 CD4+ T cells were plotted against those of low-avidity non–Vα2 CD4+ T cells, together with the threshold at which these two populations are equal (Fig. 1A). This analysis reveals both the magnitude of the response and its composition in a single plot. At this time point, the response to FV infection was dominated (>70%) by high-avidity Vα2 CD4+ T cells (23), and this was reproduced with several different immunization methods (Fig. 1A), including F-MLV-B, env124–138 peptide in Sigma Adjuvant System, and FV-induced FBL-3 tumor cells expressing F-MLV env. In stark contrast, a replication-defective recombinant Ad5–based vaccine encoding F-MLV env (Ad5.pIX-gp70) induced predominantly non–Vα2 CD4+ T cells (Fig. 1A–C). Although both F-MLV-B and Ad5.pIX-gp70 elicited an env-specific CD4+ T cell response of comparable magnitude (Fig. 1A, 1B), the proportion of high-avidity Vα2 CD4+ T cells within this response was significantly lower for Ad5.pIX-gp70 than for F-MLV-B (Fig. 1B, 1C).
FIGURE 1.
Ad5-vectored env uniquely induces low-avidity env-specific CD4+ T cells. (A) Absolute numbers of high-avidity Vα2 or low-avidity non–Vα2 donor env-specific EF4.1 CD4+ T cells in the spleens of recipient mice 7 d after adoptive transfer and indicated immunization. The diagonal dashed line represents equal proportion of the two subsets. (B) Representative flow cytometric profiles of CD4+CD44+ T cells from F-MLV-B–infected or Ad5.pIX-gp70–immunized recipient mice (day 7), distinguishing host (CD45.1+CD45.2+) and donor (CD45.1+CD45.2−) EF4.1 cells (left) and the Vα2+ cells in the latter (right). (C) Frequency of Vα2+ cells in the same mice. (D) Representative flow cytometric staining of CD4+ T cells from F-MLV-B–infected or Ad5.pIX-gp70–immunized (day 7) nontransgenic mice with the Ab-env125–135 tetramer. (E) Absolute numbers of Ab-env125–135 tetramer-binding CD4+ T cells in the same mice. (F) Median fluorescence intensity (MFI) of Ab-env125–135 tetramer staining in the same samples. In (A), (C), (E), and (F), each symbol is an individual mouse. In (B)–(F), one of three experiments is shown.
We next confirmed that a low-avidity env-specific CD4+ T cell response to Ad5.pIX-gp70 was not only restricted to TCRβ-transgenic EF4.1 CD4+ T cells. We therefore administered F-MLV-B or Ad5.pIX-gp70 to nontransgenic WT mice and used peptide–MHC class II tetramers to identify the env-specific CD4+ T cells. Because the available Ab-env122–141 tetramer used in previous studies did not reliably identify env-specific CD4+ T cells (17, 21), we generated a new version of the reagent that included only a register-trapped core env125–135 epitope. Ab-env125–135 tetramer binding, 7 d after either F-MLV-B or Ad5.pIX-gp70 administration to WT mice, identified a comparable population of CD4+ T cells in both frequency within CD4+ T cells and absolute numbers (Fig. 1D, 1E). However, the intensity of Ab-env125–135 tetramer staining, which provides an indirect measure of TCR avidity, was significantly lower in env-specific CD4+ T cells induced by Ad5.pIX-gp70 than by F-MLV-B (Fig. 1F).
These results demonstrated that, in comparison with F-MLV-B, Ad5.pIX-gp70 induced a lower frequency of Vα2 T cells in virus-specific TCRβ-transgenic EF4.1 CD4+ T cells (Fig. 1B, 1C) and lower intensity of Ab-env125–135 tetramer binding in non–TCR-transgenic CD4+ T cells (Fig. 1D, 1F). This was despite the pronounced TCR downregulation in EF4.1 CD4+ T cells caused by F-MLV-B, but not Ad5.pIX-gp70 (Fig. 1B).
Avidity of env-specific CD4+ T cell response is determined early during priming
The cognate env peptide encoded by either Ad5.pIX-gp70 or F-MLV-B might be presented at different amounts and duration, in different proinflammatory environments, and by different APCs, even when the two viruses are administered via the same route. We therefore examined whether these parameters were responsible for differences in the resulting env-specific CD4+ T cell response.
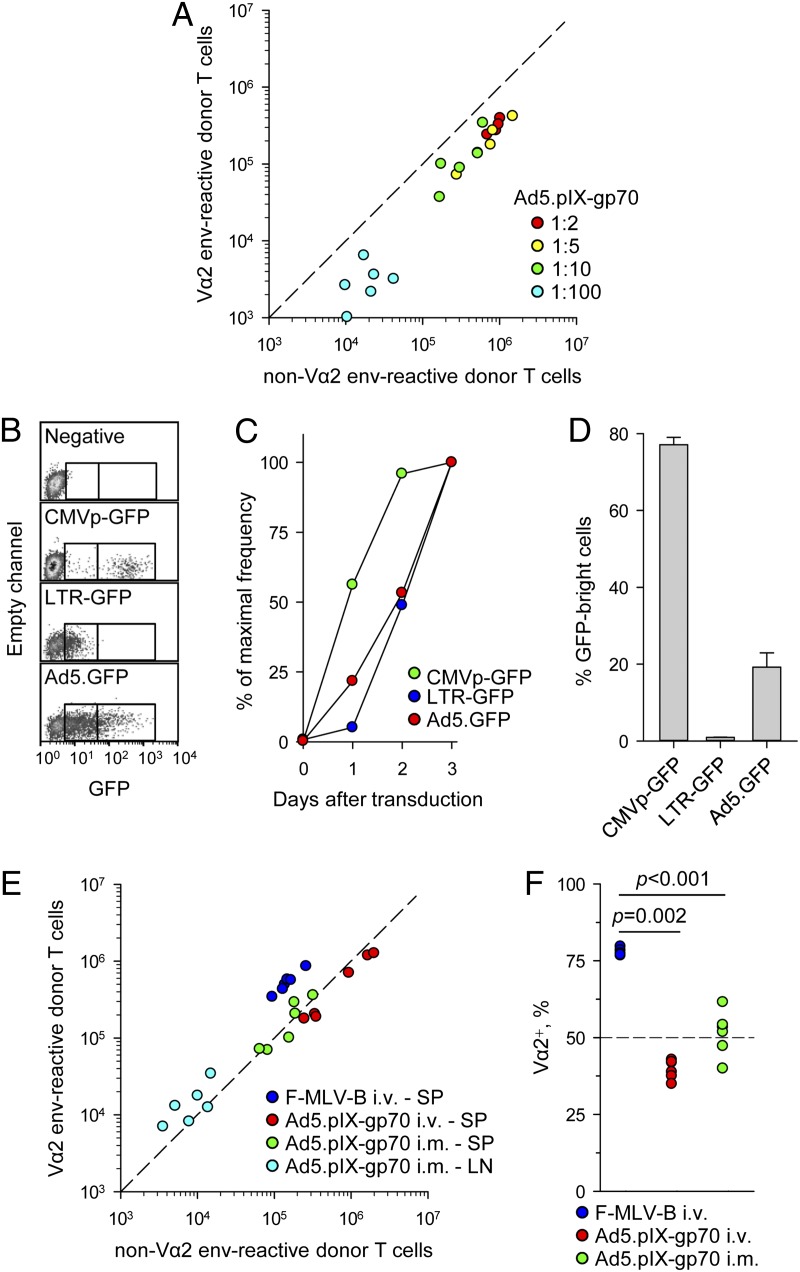
Induction of lower avidity env-specific CD4+ T cells by Ad5.pIX-gp70 was not due to excessive overall Ag production, as it was independent of the vaccine dose (Fig. 2A). However, it was theoretically possible that, on a per-cell basis, Ad5.pIX-gp70 produced excessive amounts of Ag owing to the heterologous CMVp driving env expression. Comparison of two retroviral vectors encoding GFP under either an internal CMV promoter or the native LTR confirmed the stronger in vitro activity of the former (Fig. 2B–D). An Ad5 vector encoding GFP under a CMV promoter (Ad5.GFP) produced kinetics of GFP expression that were similar to those of LTR-GFP as well as intensity that was intermediate between that of LTR-GFP and CMVp-GFP (Fig. 2B–D).
FIGURE 2.
Ad5.pIX-gp70 immunization induces low-avidity env-specific CD4+ T cells, irrespective of dose or route. (A) Absolute numbers of Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells in the spleens of recipient mice 7 d after immunization with the indicated dilutions of the standard Ad5.pIX-gp70 dose (4.5 × 108 viral genomes/mouse). (B) Representative flow cytometric profiles for B-3T3 cells transduced with retroviral vectors encoding GFP under a CMV promoter (CMVp-GFP) or the LTR (LTR-GFP) transfected with Ad5.GFP 3 d earlier or left untreated (negative). Gates mark the arbitrary populations expressing low or high amounts of GFP. (C) Kinetics of GFP expression in the same cells as in (B) plotted as the frequency of GFP+ cells during 3 d of culture expressed as a fraction of the maximum frequency on day 3. (D) Intensity of GFP expression in the same cells as in (B) on day 3 of culture, plotted as the frequency of GFPbright cells within GFP+ cells only. Results in (B)–(D) are representative of four experiments. (E) Absolute numbers of Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells in the spleens (SP) or popliteal lymph nodes (LN) of recipient mice 7 d after i.v. F-MLV-B infection or i.v. or i.m. Ad5.pIX-gp70 immunization. (F) Frequency of Vα2+ cells in total env-specific EF4.1 CD4+ T cells in spleens and lymph nodes combined from the same mice as in (E). In (A), (E), and (F), each symbol is an individual mouse.
The amount of Ag produced per cell or in total would also be expected to vary according to the cell type that is transfected following Ad5.pIX-gp70 immunization, which, in turn, depends on the route of administration. Poor representation of high-avidity Vα2 cells in splenic env-specific CD4+ T cells was also observed following i.m. Ad5.pIX-gp70 immunization (Fig. 2E, 2F). Intramuscular injection of Ad5.pIX-gp70, but not i.v. injection of either Ad5.pIX-gp70 or F-MLV-B, induced env-specific CD4+ T cells in the popliteal lymph node, which were enriched in high-avidity Vα2 cells (Fig. 2E). However, numbers of env-specific CD4+ T cells in the popliteal lymph node were an order of magnitude lower than in the spleen, and the overall frequency of Vα2 cells in total env-specific CD4+ T cells was substantially lower in mice i.m. immunized with Ad5.pIX-gp70 than those infected with F-MLV-B (Fig. 2E, 2F).
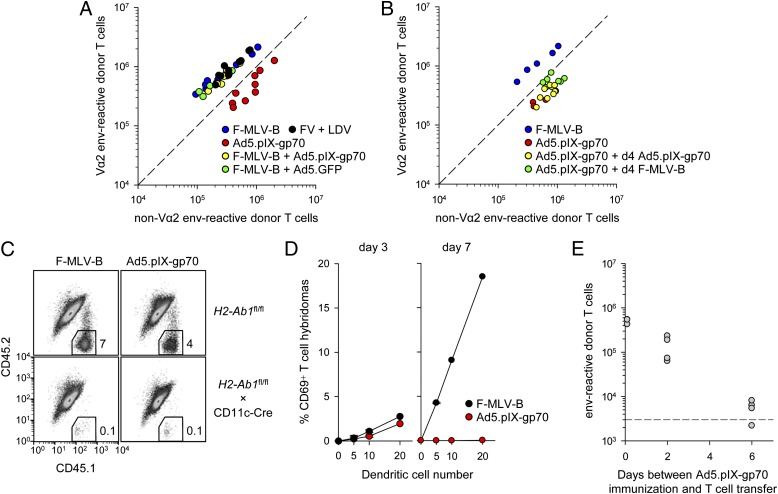
Viral infections generally induce a proinflammatory environment including the production of IFNs. However, we have previously observed that FV infection induces an exceptionally weak IFN response (41). To examine the effect of different proinflammatory environments, F-MLV-B was coinjected with either Ad5.pIX-gp70 or the control vector Ad5.GFP. In the latter combination, the Ad5.GFP vector would mimic the strong IFN induction by Ad5.pIX-gp70, but would not contribute to env presentation. Additionally, we used coinjection of FV with LDV, which has been previously shown to induce a strong IFN response (41). These experiments revealed that the ability of F-MLV-B and FV to induce a high-avidity Vα2 env-specific CD4+ T cell response was dominant and not affected by Ad5.pIX-gp70, Ad5.GFP, or LDV coinjection and the resulting proinflammatory environment (Fig. 3A).
FIGURE 3.
In vivo kinetics of Ag presentation following F-MLV-B infection or Ad5.pIX-gp70 immunization. (A) Absolute numbers of Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells in the spleens of recipient mice 7 d after immunization with the indicated combination of viruses at the time of T cell transfer. (B) Absolute numbers of Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells in the spleens of recipient mice 7 d after priming with Ad5.pIX-gp70 at the time of T cell transfer (day 0) followed by additional administration of either F-MLV-B or Ad5.pIX-gp70 on day 4. F-MLV-B infection or Ad5.pIX-gp70 immunization on day 0 was also included as control. (C) Representative flow cytometric profiles of CD4+ T cells, distinguishing host (CD45.1+CD45.2+) and donor (CD45.1+CD45.2−) cells from the spleens of either control mice (H2-Ab1fl/fl) or mice lacking DC expression of MHC class II (H2-Ab1fl/fl × CD11c-Cre) 7 d following adoptive transfer of EF4.1 T cells and F-MLV-B infection or Ad5.pIX-gp70 immunization. Data are representative of two mice per genotype per virus. (D) CD69 expression in env-specific T cell hybridomas following overnight culture with the indicated number of DCs isolated from the spleens of F-MLV-B–infected or Ad5.pIX-gp70–immunized WT mice at the indicated times after immunization. Data are pooled from two experiments. (E) Absolute numbers of total splenic env-specific EF4.1 CD4+ T cells 7 d after transfer into recipient mice that had been immunized with Ad5.pIX-gp70 0, 2, or 6 d previously. The dashed line represents recovery from nonimmunized recipients. In (A), (B), and (E), each symbol is an individual mouse.
Following F-MLV-B and Ad5.pIX-gp70 coinjection, it was not possible to establish which of the two viruses primed most CD4+ T cells. To investigate any differential effects in early priming, we analyzed the clonotypic composition of env-specific CD4+ T cells primed by Ad5.pIX-gp70 and subsequently boosted with F-MLV-B or an additional injection of Ad5.pIX-gp70 4 d later. In this experimental setting, env-specific CD4+ T cells primed by Ad5.pIX-gp70 contained predominantly non-Vα2, even when Ag presentation was continued by additional F-MLV-B or Ad5.pIX-gp70 injections (Fig. 3B). This result suggested that the clonotypic composition at the peak of the response was determined early during priming. We therefore looked for differences in early Ag presentation.
CD4+ T cell priming by both F-MLV-B and Ad5.pIX-gp70 was mediated by DCs, as it was abrogated in mice with conditional MHC class II deletion in CD11c+ DCs (Fig. 3C). Blood-borne adenoviruses are trapped in the splenic marginal zone (42), which is defective in Tnfr1−/− mice (43). Nevertheless, differential priming by the two viruses remained intact in Tnfr1−/− mice (Supplemental Fig. 1), further supporting a predominant role for DCs. Ag presentation by ex vivo splenic DCs from Ad5.pIX-gp70– or F-MLV-B–injected WT mice was comparable on day 3 after virus challenge (Fig. 3D). DC Ag presentation increased considerably by day 7 after F-MLV-B, in line with its replication kinetics, but it was entirely diminished in the case of Ad5.pIX-gp70 (Fig. 3D). Consistent with these in vitro data, cohorts of naive EF4.1 CD4+ T cells could no longer be primed when they were transferred 6 d after Ad5.pIX-gp70 immunization (Fig. 3E), which contrasted with efficient priming of EF4.1 CD4+ T cells even 35 d after FV infection (44).
Taken together, these results suggested that despite eliciting comparable peak numbers of env-specific CD4+ T cells, Ag presentation by DCs is lower in overall dose and shorter in duration following Ad5.pIX-gp70 immunization than F-MLV-B infection.
Ad5.pIX-gp70 and FV priming induce distinct proliferative phases
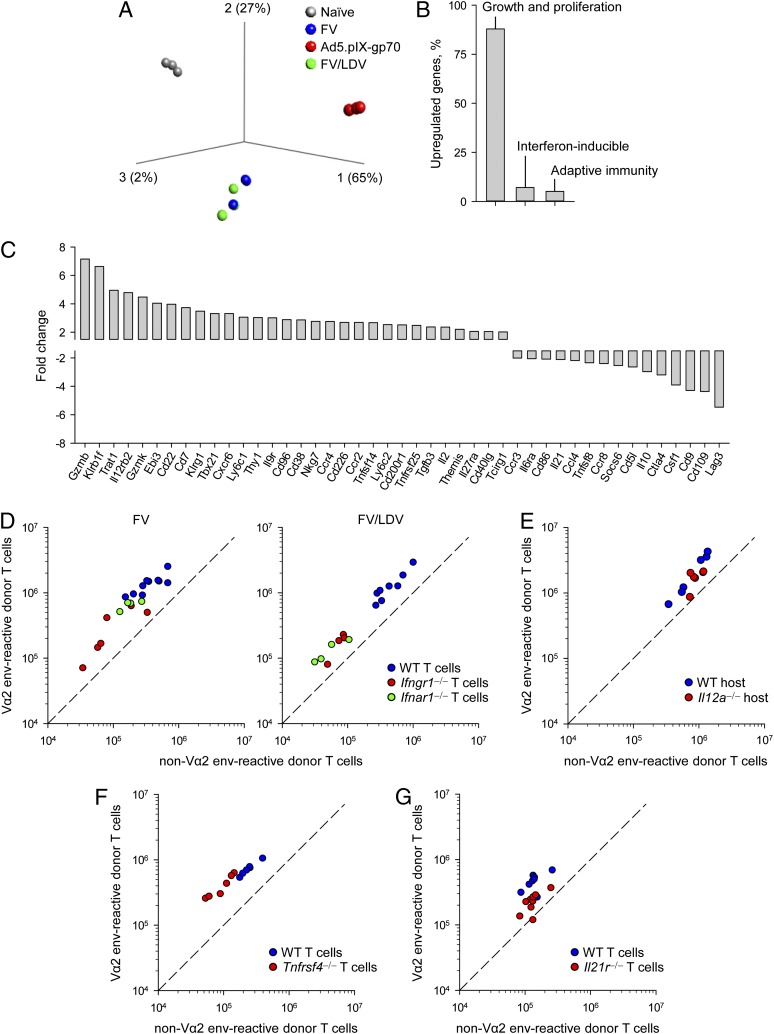
We next compared the transcriptional profiles of EF4.1 CD4+ T cells primed by FV and Ad5.pIX-gp70. To exclude transcriptional differences arising from stronger inflammatory responses we included coinfection with FV and LDV, which induces a strong IFN response and also high-avidity Vα2 env-specific CD4+ T cells. Principal component analysis (Fig. 4A) or hierarchical clustering (Supplemental Fig. 2) of the transcriptional profiles clearly separated env-specific effectors induced by Ad5.pIX-gp70, or FV (with or without LDV) and naive precursors. The vast majority (87%) of the 750 transcripts that were found in >2-fold abundance in Ad5.pIX-gp70– than in FV-induced effectors were related to cell growth, metabolism, and proliferation, with smaller proportions being IFN-inducible (7%) or related to adaptive immunity (5%) (Fig. 4B, Supplemental Tables I, II). The latter group included several molecules that have been previously implicated in the regulation of effector CD4+ T cell responses (Fig. 4C) and warranted closer inspection.
FIGURE 4.
Gene expression and reliance of EF4.1 CD4+ T cells responding to FV infection or Ad5.pIX-gp70 immunization. (A) Principal component analysis plot of triplicated samples from naive EF4.1 CD4+ T cells and day 7 effector T cells in response to FV infection, Ad5.pIX-gp70 immunization, or FV and LDV coinfection. (B) Functional profiling of the 750 genes that were found >2-fold upregulated in Ad5.pIX-gp70– than in FV-primed effector EF4.1 CD4+ T cells, expressed as a percentage of the total. (C) Fold change of the 46 adaptive immunity–related genes that were found >2-fold differentially expressed between Ad5.pIX-gp70– and FV-primed effector EF4.1 CD4+ T cells. (D) Absolute numbers of splenic Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells 7 d after cotransfer of WT and either Ifngr1−/− or Ifnar1−/− EF4.1 CD4+ T cells into recipient mice infected with FV (left) or coinfected with FV and LDV (right). (E) Absolute numbers of splenic Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells 7 d after transfer into WT or Il12a−/− recipient mice infected with FV. (F) Absolute numbers of splenic Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells 7 d after cotransfer of WT and Tnfrsf4−/− EF4.1 CD4+ T cells into recipient mice infected with FV. (G) Absolute numbers of splenic Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells 7 d after cotransfer of WT and Il21r−/− EF4.1 CD4+ T cells into recipient mice infected with FV. p = 0.003 between the frequency of Vα2+ cells in WT and Il21r−/− EF4.1 CD4+ T cells (n = 11, Mann–Whitney rank sum test). In (D)–(G), each symbol is an individual mouse.
T cell–intrinsic Ifngr1 or Ifnar1 deficiency significantly reduced the expansion of EF4.1 CD4+ T cells during FV infection and FV/LDV coinfection, according to the amount of IFNs produced (41), without affecting the dominance of Vα2 cells (Fig. 4D). Despite differential expression of Il12rb2, the gene encoding the β-chain of the heterodimeric receptor for IL-12 (Fig. 4C), EF4.1 CD4+ T cells transferred into WT or Il12a−/− hosts, unable to produce IL-12 and IL-35, exhibited similar expansion and dominance of Vα2 cells in response to FV infection (Fig. 4E). Similarly, T cell–intrinsic Tnfrsf4 deficiency reduced peak numbers of env-specific CD4+ T cells, but not the proportion of Vα2 cells (Fig. 4F). Lastly, T cell–intrinsic Il21r deficiency reduced peak numbers of env-specific CD4+ T cells and also caused a statistically significant reduction in the frequency of high-avidity Vα2 cells in response to FV infection (Fig. 4G), which was not, however, as dramatic as that seen following Ad5.pIX-gp70 immunization (Fig. 1A). These findings indicated that the clonotypic composition of the CD4+ T cell response to F-MLV env, and in particular the dominance of Vα2 cells, was relatively insensitive to factors promoting the response, such as type I or II IFN, OX40L, or IL-12/IL-35, with the possible exception of IL-21. Instead, they suggested that differences in cellular proliferation on day 7 arose from disparate kinetics of the response to Ad5.pIX-gp70 and FV.
We first confirmed that transcriptional differences in cellular proliferation-related transcripts were indeed accompanied with different proliferation rates. In line with overrepresentation of Mki67, the transcript encoding the Ag identified by the mAb Ki-67 (Supplemental Table I), on day 7 of the response, Ad5.pIX-gp70 induced higher amounts of Ki-67 protein than did F-MLV-B (Fig. 5A). Similar results were obtained when cellular proliferation was assessed by staining for DNA content (not shown). Time-course analysis of Ki-67 expression indicated that F-MLV-B induced rapid proliferation, particularly of Vα2 cells that was decelerating already by day 7, whereas Ad5.pIX-gp70 induced sustained proliferation, particularly of non–Vα2 cells, reaching maximum speed on day 7 (Fig. 5A). Differences in proliferation rates also translated in env-specific CD4+ T cell numbers. Whereas F-MLV-B induced near-peak numbers of env-specific CD4+ T cells already by day 4, with minimal further increase on day 7, numbers of env-specific CD4+ T cells were very much reduced on day 4 after Ad5.pIX-gp70 and were significantly elevated on day 7 (Fig. 5B). These results supported a model whereby high-avidity Vα2 cells exhibited an early advantage in response to both viruses (Fig. 5B). Whereas early termination of the proliferative response to F-MLV-B fixed the advantage of Vα2 cells, Ad5.pIX-gp70 permitted further T cell expansion, particularly of non–Vα2 cells, which eventually dominated the response.
FIGURE 5.
Proliferation kinetics of EF4.1 CD4+ T cells responding to FV infection or Ad5.pIX-gp70 immunization. (A) Representative flow cytometric assessment of Ki-67 expression on day 7 in donor env-specific EF4.1 CD4+ T cells (left) and frequency of Ki-67+ Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells 4 and 7 d after transfer into F-MLV-B–infected or Ad5.pIX-gp70–immunized recipients. Data are means of six to nine mice from three experiments. (B) Absolute numbers of splenic Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells 4 and 7 d after transfer into F-MLV-B–infected or Ad5.pIX-gp70–immunized recipients. (C) Representative flow cytometric assessment of Nur77-driven GFP expression on day 7 in donor env-specific EF4.1 CD4+ T cells (left) and mean fluorescence intensity (MFI) of GFP expression in Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells 4 and 7 d after transfer into F-MLV-B–infected or Ad5.pIX-gp70–immunized recipients. Data are means of eight mice from three experiments. The dashed line represents MFI of GFP signal in nonactivated cells. (D) Absolute numbers of splenic Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells 7 d after transfer into FV-infected recipients of either total EF4.1 CD4+ T cells, containing both Vα2 or non–Vα2 cells (mixed), or purified Vα2 or non–Vα2 EF4.1 CD4+ T cells separately (separate). In (B) and (D), each symbol is an individual mouse.
Reduced proliferation at the peak (day 7) of the response to FV or F-MLV-B was not due to the lack of Ag-induced TCR signaling. To measure in vivo TCR signaling strength, we used EF4.1 CD4+ T cells additionally carrying a GFP-encoding transgene driven by the promoter of the Nr4a1 gene (encoding Nur77) (31). Nearly all doubly transgenic env-specific CD4+ T cells were Nur77-GFP+ on day 7 of the response to either F-MLV-B or Ad5.pIX-gp70, indicating continued TCR signaling (Fig. 5C). Consistent with their higher functional avidities in vitro, Vα2 cells showed significantly higher intensity of GFP expression than did non-Vα2 from the same recipient, irrespective of priming virus (Fig. 5C). However, in line with viral replication and Ag presentation kinetics (Fig. 3D), GFP intensity was similarly induced by F-MLV-B or Ad5.pIX-gp70 on day 4 within either Vα2 or non–Vα2 cells and, although it remained low or decreased on day 7 after Ad5.pIX-gp70, it was significantly increased at the same time point after F-MLV-B (Fig. 5C). Thus, F-MLV-B–primed env-specific CD4+ T cells displayed reduced proliferation rates (Fig. 5A) despite receiving stronger TCR signals than did Ad5.pIX-gp70–primed cells (Fig. 5C).
The abundance of TCR signaling in env-specific CD4+ T cells following F-MLV-B infection (Fig. 5C), as well as robust high-avidity responses despite deficiencies in expansion-promoting factors (Fig. 3D–G), suggested that curtailed T cell expansion may have been due to active inhibition, particularly of non–Vα2 cells. This was tested by comparing the relative accumulation of Vα2 and non–Vα2 cells env-specific EF4.1 CD4+ T cells adoptively transferred either together or separately into FV-infected recipients (Fig. 5D). Whereas Vα2 cells transferred alone accumulated only marginally better than with non–Vα2 cells, non–Vα2 cells transferred alone accumulated at 2.7-fold higher numbers than with Vα2 cells (Fig. 5D) and were now comparable with Vα2 cells. Thus, rather than intrinsically, expansion of low-avidity non–Vα2 cells was restricted by high-avidity Vα2 cells in response to FV infection.
Induction of low-avidity env-specific CD4+ T cells by weakly priming regimens
The current observations suggested that the dominance of high-avidity Vα2 cells in response to FV involved faster initial expansion followed by inhibition of low-avidity non–Vα2 cells. In contrast, during the slower response to Ad5.pIX-gp70, there may be insufficient numbers of high-avidity Vα2 cells at early time points to effectively prevent expansion of low-avidity non–Vα2 cells. We therefore hypothesized that if a fast and strong T cell response, driven by sufficient amounts of Ag, was indeed required to promote high-avidity env-specific CD4+ T cells, then the response to FV could be directed into low avidity if Ag presentation were weaker.
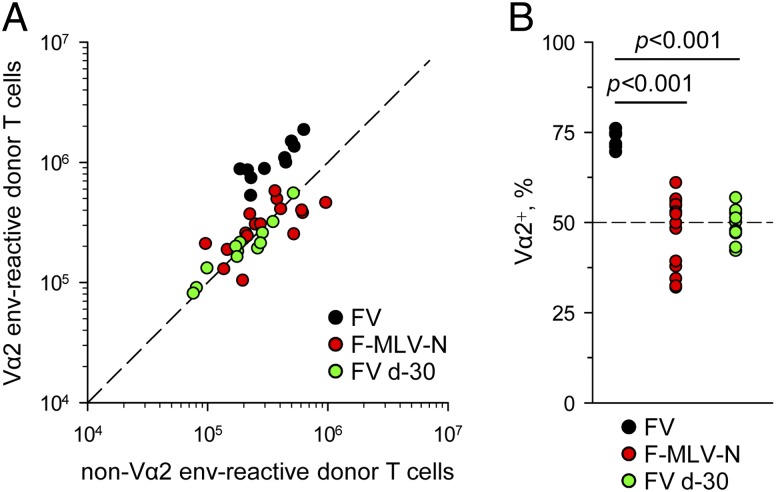
To reduce the degree of Ag presentation during FV or F-MLV-B infection we primed with F-MLV-N (45). Replication of this virus in B6 mice is attenuated by the action of Fv1b allele (46). Importantly, this prototypic retroviral restriction factor blocks F-MLV at a stage after viral entry but before integration. Therefore, F-MLV-N leads to 100 to 1000 fewer target cells infected than does F-MLV-B, but each of these infected cells produces the same amount of Ag (46). Additionally, we transferred EF4.1 CD4+ T cells into hosts that had been infected with FV 30 d prior to T cell transfer. At this time point, F-MLV-B replication is controlled to a very low and stable level, sufficient to prime a response (44). In comparison with priming during acute FV infection, priming by either acute F-MLV-N infection or during the chronic phase of FV infection led to smaller overall expansion and, notably, also to significantly lower frequencies of high-avidity Vα2 cells in the env-specific response (Fig. 6). Thus, reduced F-MLV env presentation resulted in reduced proportional representation of high-avidity env-specific CD4+ T cells.
FIGURE 6.
Effect of reduced F-MLV replication on the avidity of the env-specific CD4+ T cell response. (A) Absolute numbers of splenic Vα2 or non–Vα2 donor env-specific EF4.1 CD4+ T cells 7 d after transfer into recipient mice infected with FV or F-MLV-N at the time of T cell transfer or with FV 30 d prior to T cell transfer. (B) Frequency of Vα2+ cells in the same mice. In (A) and (B), each symbol is an individual mouse.
Discussion
Accumulating evidence suggests that the clonotypic composition of a T cell response to infection or vaccination is a critical parameter of the protective capacity of that response (11, 16, 17). It is also clear that administering the same Ag in different regimens can induce clonotypically distinct T cell responses (19, 20). By comparing a variety of immunization regimens, we have shown in the present study that the model retroviral Ag F-MLV env induced a low-avidity CD4+ T cell response when vectored by Ad5. This unique property of Ad5-vectored env held true both for TCRβ-transgenic and nontransgenic CD4+ T cells and resulted from a protracted phase of T cell proliferation of all clonotypes, irrespective of TCR avidity.
For their proliferative response, CD4+ T cells integrate signals from multiple sources and are exposed to different antigenic and inflammatory environments created by distinct types of infection or vaccination (47). These signals include costimulatory molecules and cytokines, the effect of which can also be modified by the context of T cell activation. Despite providing nonredundant signals for maximum CD4+ T cell expansion in response to FV infection, type I or II IFN, OX40L, and IL-21 were only minimally required for the dominance of high-avidity clonotypes in the residual response. This relative insensitivity to different inflammatory environments of the high-avidity CD4+ T response may represent a T cell adaptation ensuring a robust response irrespective of the infecting virus.
Despite the necessity to integrate multiple costimulatory signals, CD4+ T cells are most reliant for their expansion on Ag-derived signals (48). Continuous Ag presentation throughout the response has been shown to promote maximal CD4+ T cell responses in diverse systems (49–51). In contrast, reduced antigenic stimulation as a result of intraclonal competition has been proposed to restrain the CD4+ T cell response (52–55). The impact of TCR-emanating signals is also thought to underlie the dominance of a few clonotypes at the peak of the CD4+ T cell response in different systems (56). Similarly to the response elicited by F-MLV env (23), the peak CD4+ T cell responses to an H2-Ek–restricted PCC epitope (57) and an H2-EdAd–restricted sperm whale myoglobin epitope (58) are also dominated by initially rare high-avidity clonotypes.
The advantage of high-avidity clonotypes in all cases appears to manifest very early in the CD4+ T cell response. This is also the case following Ad5.pIX-gp70 immunization, where, at least transiently on day 4, the CD4+ T cell response is composed primarily of high-avidity clonotypes. However, our data suggest that although stronger TCR signals establish the early advantage of high-avidity clonotypes, alone they are not sufficient to maintain it. One important difference between persistent infection with a replicating virus and immunization with replication-defective vectors or purified proteins is Ag kinetics. Following F-MLV-B infection, Ag availability continued to rise to a maximum at or beyond the peak of the T cell response. This availability was also reflected in the amount of TCR signaling env-specific CD4+ T cells experienced. The use of the Nur77-driven GFP reporter transgene not only confirmed that high-avidity clonotypes received stronger TCR signals than did low-avidity ones throughout the course of the env-specific CD4+ T cell response, it also revealed a disconnect between TCR signaling and sustained T cell proliferation. The inability of strong TCR signals to maintain CD4+ T cell expansion in F-MLV-B or FV infection highlights the existence of further constrains.
Our data support a model whereby competition in CD4+ T cell expansion, when imposed early enough, preserves the initial advantage of high-avidity clonotypes. This model is also supported by the observed inhibition of low-avidity clonotypes by high-avidity counterparts in response to FV infection. Consistent with this model, in comparison with Ad5.pIX-gp70 priming, the transcriptional profile of FV-primed effector CD4+ T cells was enriched in inhibitory molecules, including CTLA-4 and LAG-3, both of which have been implicated in the control of CD4+ T cell responses through cell-extrinsic inhibitory effects on DCs (59, 60).
Alternatively, competition between high- and low-avidity clonotypes may be the result of differentiation into opposing Th subsets. Th subset commitment of effector CD4+ T cells may affect their clonal expansion, as well as their ability to persist in the memory population (61). Differentiation of CD4+ T cells into distinguishable subsets has been proposed by several studies to rely on instructive cues linked to TCR signal strength (62). Indeed, differences in Th differentiation were observed between Vα2 and non–Vα2 effector CD4+ T cells responding to FV infection (23), as well as between effector CD4+ T cells responding to FV infection or Ad5.pIX-gp70 vaccination. However, such differences were small and rather quantitative, and both Vα2 and non–Vα2 effector CD4+ T cells differentiated almost exclusively into Th1 or follicular helper T cells (23). Nevertheless, it is possible that divergent TCR signal strength-driven Th differentiation of Vα2 and non–Vα2 clonotypes is responsible for the characteristic clonotypic composition in response to different viruses. This hypothesis is currently under investigation.
Priming of the env-specific CD4+ T cell response was entirely dependent on DC-intrinsic expression of MHC class II when either FV or Ad5.pIX-gp70 was used. This finding explains the previously observed requirement for DCs in the Ab response to FV (63). The critical parameter of Ag presentation that affected the avidity of the CD4+ T cell response was not its duration, but rather the overall intensity during the first 4–6 d.
Using dispersible or depot-forming adjuvants for immunization with purified PCC, earlier pioneering work has linked the induction of higher avidity CD4+ T cell clonotypes with dispersible adjuvants containing CpG oligodeoxynucleotides or monophosphoryl lipid A (19). Our comparison of local (popliteal lymph node) and systemic (spleen) env-specific CD4+ T cell responses to i.m. Ad5.pIX-gp70 immunization demonstrated higher avidity at the local site, arguing against a requirement for Ag dispersal for high-avidity responses. Instead, common to both studies, high-avidity responses were always associated with more efficient CD4+ T cell priming. Indeed, the two adjuvants that induced the highest avidity of the CD4+ T cell response in the PCC system (CpG and, especially, monophosphoryl lipid A) also induced the biggest response (19). Assuming the magnitude of the response is proportional to the efficiency of presentation, the results from both studies suggest that the avidity of the CD4+ T cell response is promoted by fast and efficient Ag presentation. This is also supported by the following observations. First, reducing the total amount of presented F-MLV env by using attenuated vaccine F-MLV viruses or by using adoptive hosts at the chronic phase of infection, when Ag persists at very low levels, caused a concomitant decrease in the elicited CD4+ T cell response. Also, reducing the immunizing dose of Ad5.pIX-gp70 did not improve the affinity of the resulting CD4+ T cell response. Second, the extended duration of Ag presentation at later time points did not seem to contribute to a high-avidity CD4+ T cell response, as peptide immunization induced a high-avidity response, and also prolonging Ag presentation by infection with F-MLV 4 d after Ad5.pIX-gp70 priming did not restore a high-avidity CD4+ T cell response.
An important distinction to be made is between overall amounts of Ag presented by all APCs collectively and amounts produced per APC. The use of the heterologous CMV promoter driving transgene expression in Ad5 vectors (8, 9, 24) can result in higher per-cell amounts than expression driven by the native retroviral LTR. High amounts of Ag produced by a single transfected APC may compensate for the inability of replication-defective Ad5-based vectors to amplify their genome copies in the host, and may be beneficial in the induction of a protective Ab response (24). However, high Ag production per cell will have a negative impact on the avidity of the CD4+ T cell response.
High Ag density on individual APCs produced by vaccination may induce clonotypes with low avidity, which will not react with the lower amounts of Ag produced during retroviral infection or mediate effective protection against the natural infection (17). This principle was recently demonstrated with the use of vaccinia virus vectors expressing an antigenic peptide fused to either GFP or H2-Ab molecules, which create a 100-fold difference in the amount of antigenic complexes presented by individual APCs (64). In this study, high Ag density generated by fusion to H2-Ab induced CD4+ T cells with avidity and cross-reactivity patterns that were poorly induced by low Ag density (64). Therefore, attempts to maximize Ag production during vaccination should aim at generating a greater number of Ag-producing APCs, each of which, however, should not produce Ag in greater amounts than those produced per cell during natural infection.
It is now clear that distinct vectors elicit qualitatively different T cell responses, which in turn provide disparate degrees of protection against infection. Our data underpin the importance of the clonotypic composition of vaccine-induced T cell responses in determining the efficacy of vaccination.
Acknowledgments
We thank Drs. Kristin A. Hogquist and Manfred Kopf for the Nur77-GFP and Il21r−/− mice, respectively, and Dr. Oliver Wildner for the Ad5.pIX-gp70 vector. We are grateful for assistance from the Division of Biological Services, the Flow Cytometry, and Microarray Facilities at the National Institute for Medical Research.
This work was supported by Medical Research Council (United Kingdom) Grant U117581330.
The microarray data presented in this article have been submitted to the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-2210 and E-MEXP-2950.
The online version of this article contains supplemental material.
- Ad5
- adenovirus serotype 5
- B6
- C57BL/6
- CMVp
- CMV promoter
- DC
- dendritic cell
- env
- envelope
- F-MLV
- Friend murine leukemia virus
- F-MLV-B
- B-tropic F-MLV
- F-MLV-N
- N-tropic F-MLV
- FV
- Friend virus
- LDV
- lactate dehydrogenase–elevating virus
- LTR
- long terminal repeat
- MLV
- murine leukemia virus
- PCC
- pigeon cytochrome c
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Welsh R. M., Selin L. K., Szomolanyi-Tsuda E. 2004. Immunological memory to viral infections. Annu. Rev. Immunol. 22: 711–743 [DOI] [PubMed] [Google Scholar]
- 2.Zinkernagel R. M. 2003. On natural and artificial vaccinations. Annu. Rev. Immunol. 21: 515–546 [DOI] [PubMed] [Google Scholar]
- 3.McMichael A. J., Borrow P., Tomaras G. D., Goonetilleke N., Haynes B. F. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat. Rev. Immunol. 10: 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaufmann S. H., McMichael A. J. 2005. Annulling a dangerous liaison: vaccination strategies against AIDS and tuberculosis. Nat. Med. 11(4, Suppl.): S33–S44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parks C. L., Picker L. J., King C. R. 2013. Development of replication-competent viral vectors for HIV vaccine delivery. Curr. Opin. HIV AIDS 8: 402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rollier C. S., Reyes-Sandoval A., Cottingham M. G., Ewer K., Hill A. V. 2011. Viral vectors as vaccine platforms: deployment in sight. Curr. Opin. Immunol. 23: 377–382 [DOI] [PubMed] [Google Scholar]
- 7.Hansen S. G., Piatak M., Jr., Ventura A. B., Hughes C. M., Gilbride R. M., Ford J. C., Oswald K., Shoemaker R., Li Y., Lewis M. S., et al. 2013. Immune clearance of highly pathogenic SIV infection. Nature 502: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McElrath M. J., De Rosa S. C., Moodie Z., Dubey S., Kierstead L., Janes H., Defawe O. D., Carter D. K., Hural J., Akondy R., et al. Step Study Protocol Team 2008. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 372: 1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchbinder S. P., Mehrotra D. V., Duerr A., Fitzgerald D. W., Mogg R., Li D., Gilbert P. B., Lama J. R., Marmor M., Del Rio C., et al. Step Study Protocol Team 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372: 1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gras S., Burrows S. R., Turner S. J., Sewell A. K., McCluskey J., Rossjohn J. 2012. A structural voyage toward an understanding of the MHC-I-restricted immune response: lessons learned and much to be learned. Immunol. Rev. 250: 61–81 [DOI] [PubMed] [Google Scholar]
- 11.Vingert B., Perez-Patrigeon S., Jeannin P., Lambotte O., Boufassa F., Lemaître F., Kwok W. W., Theodorou I., Delfraissy J. F., Thèze J., Chakrabarti L. A., ANRS EP36 HIV Controllers Study Group 2010. HIV controller CD4+ T cells respond to minimal amounts of Gag antigen due to high TCR avidity. PLoS Pathog. 6: e1000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander-Miller M. A., Leggatt G. R., Berzofsky J. A. 1996. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc. Natl. Acad. Sci. USA 93: 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almeida J. R., Price D. A., Papagno L., Arkoub Z. A., Sauce D., Bornstein E., Asher T. E., Samri A., Schnuriger A., Theodorou I., et al. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204: 2473–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallimore A., Glithero A., Godkin A., Tissot A. C., Plückthun A., Elliott T., Hengartner H., Zinkernagel R. 1998. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J. Exp. Med. 187: 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.La Gruta N. L., Thomas P. G., Webb A. I., Dunstone M. A., Cukalac T., Doherty P. C., Purcell A. W., Rossjohn J., Turner S. J. 2008. Epitope-specific TCRβ repertoire diversity imparts no functional advantage on the CD8+ T cell response to cognate viral peptides. Proc. Natl. Acad. Sci. USA 105: 2034–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H., Ndhlovu Z. M., Liu D., Porter L. C., Fang J. W., Darko S., Brockman M. A., Miura T., Brumme Z. L., Schneidewind A., et al. 2012. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat. Immunol. 13: 691–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young G. R., Ploquin M. J., Eksmond U., Wadwa M., Stoye J. P., Kassiotis G. 2012. Negative selection by an endogenous retrovirus promotes a higher-avidity CD4+ T cell response to retroviral infection. PLoS Pathog. 8: e1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner S. J., Doherty P. C., McCluskey J., Rossjohn J. 2006. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 6: 883–894 [DOI] [PubMed] [Google Scholar]
- 19.Malherbe L., Mark L., Fazilleau N., McHeyzer-Williams L. J., McHeyzer-Williams M. G. 2008. Vaccine adjuvants alter TCR-based selection thresholds. Immunity 28: 698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda M., Wang R., Kong W. P., Kanekiyo M., Akahata W., Xu L., Matsuo K., Natarajan K., Robinson H., Asher T. E., et al. 2009. Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity. J. Immunol. 183: 2425–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antunes I., Tolaini M., Kissenpfennig A., Iwashiro M., Kuribayashi K., Malissen B., Hasenkrug K., Kassiotis G. 2008. Retrovirus-specificity of regulatory T cells is neither present nor required in preventing retrovirus-induced bone marrow immune pathology. Immunity 29: 782–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasenkrug K. J., Chesebro B. 1997. Immunity to retroviral infection: the Friend virus model. Proc. Natl. Acad. Sci. USA 94: 7811–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploquin M. J., Eksmond U., Kassiotis G. 2011. B cells and TCR avidity determine distinct functions of CD4+ T cells in retroviral infection. J. Immunol. 187: 3321–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayer W., Tenbusch M., Lietz R., Johrden L., Schimmer S., Uberla K., Dittmer U., Wildner O. 2010. Vaccination with an adenoviral vector that encodes and displays a retroviral antigen induces improved neutralizing antibody and CD4+ T-cell responses and confers enhanced protection. J. Virol. 84: 1967–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S., Hendriks W., Althage A., Hemmi S., Bluethmann H., Kamijo R., Vilcek J., Zinkernagel R. M., Aguet M. 1993. Immune response in mice that lack the interferon-γ receptor. Science 259: 1742–1745 [DOI] [PubMed] [Google Scholar]
- 26.Müller U., Steinhoff U., Reis L. F., Hemmi S., Pavlovic J., Zinkernagel R. M., Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264: 1918–1921 [DOI] [PubMed] [Google Scholar]
- 27.Rothe J., Lesslauer W., Lötscher H., Lang Y., Koebel P., Köntgen F., Althage A., Zinkernagel R., Steinmetz M., Bluethmann H. 1993. Mice lacking the tumour necrosis factor receptor 1 are resistant to TNF-mediated toxicity but highly susceptible to infection by Listeria monocytogenes. Nature 364: 798–802 [DOI] [PubMed] [Google Scholar]
- 28.Klinger M., Kim J. K., Chmura S. A., Barczak A., Erle D. J., Killeen N. 2009. Thymic OX40 expression discriminates cells undergoing strong responses to selection ligands. J. Immunol. 182: 4581–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fröhlich A., Marsland B. J., Sonderegger I., Kurrer M., Hodge M. R., Harris N. L., Kopf M. 2007. IL-21 receptor signaling is integral to the development of Th2 effector responses in vivo. Blood 109: 2023–2031 [DOI] [PubMed] [Google Scholar]
- 30.Mattner F., Magram J., Ferrante J., Launois P., Di Padova K., Behin R., Gately M. K., Louis J. A., Alber G. 1996. Genetically resistant mice lacking interleukin-12 are susceptible to infection with Leishmania major and mount a polarized Th2 cell response. Eur. J. Immunol. 26: 1553–1559 [DOI] [PubMed] [Google Scholar]
- 31.Moran A. E., Holzapfel K. L., Xing Y., Cunningham N. R., Maltzman J. S., Punt J., Hogquist K. A. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208: 1279–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashimoto K., Joshi S. K., Koni P. A. 2002. A conditional null allele of the major histocompatibility IA-β chain gene. Genesis 32: 152–153 [DOI] [PubMed] [Google Scholar]
- 33.Caton M. L., Smith-Raska M. R., Reizis B. 2007. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 204: 1653–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klarnet J. P., Kern D. E., Okuno K., Holt C., Lilly F., Greenberg P. D. 1989. FBL-reactive CD8+ cytotoxic and CD4+ helper T lymphocytes recognize distinct Friend murine leukemia virus-encoded antigens. J. Exp. Med. 169: 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bock M., Bishop K. N., Towers G., Stoye J. P. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74: 7422–7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yueh A., Goff S. P. 2003. Phosphorylated serine residues and an arginine-rich domain of the moloney murine leukemia virus p12 protein are required for early events of viral infection. J. Virol. 77: 1820–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marques R., Antunes I., Eksmond U., Stoye J., Hasenkrug K., Kassiotis G. 2008. B lymphocyte activation by coinfection prevents immune control of friend virus infection. J. Immunol. 181: 3432–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon J. J., Chu H. H., Pepper M., McSorley S. J., Jameson S. C., Kedl R. M., Jenkins M. K. 2007. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reimand J., Kull M., Peterson H., Hansen J., Vilo J. 2007. g:Profiler: a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35: W193–W200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., Hertzog P. J. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41: D1040–D1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duley A. K., Ploquin M. J., Eksmond U., Ammann C. G., Messer R. J., Myers L., Hasenkrug K. J., Kassiotis G. 2012. Negative impact of IFN-γ on early host immune responses to retroviral infection. J. Immunol. 189: 2521–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Paolo N. C., Miao E. A., Iwakura Y., Murali-Krishna K., Aderem A., Flavell R. A., Papayannopoulou T., Shayakhmetov D. M. 2009. Virus binding to a plasma membrane receptor triggers interleukin-1 α-mediated proinflammatory macrophage response in vivo. Immunity 31: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasparakis M., Kousteni S., Peschon J., Kollias G. 2000. Tumor necrosis factor and the p55TNF receptor are required for optimal development of the marginal sinus and for migration of follicular dendritic cell precursors into splenic follicles. Cell. Immunol. 201: 33–41 [DOI] [PubMed] [Google Scholar]
- 44.Pike R., Filby A., Ploquin M. J., Eksmond U., Marques R., Antunes I., Hasenkrug K., Kassiotis G. 2009. Race between retroviral spread and CD4+ T-cell response determines the outcome of acute Friend virus infection. J. Virol. 83: 11211–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dittmer U., Brooks D. M., Hasenkrug K. J. 1998. Characterization of a live-attenuated retroviral vaccine demonstrates protection via immune mechanisms. J. Virol. 72: 6554–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanz-Ramos M., Stoye J. P. 2013. Capsid-binding retrovirus restriction factors: discovery, restriction specificity and implications for the development of novel therapeutics. J. Gen. Virol. 94: 2587–2598 [DOI] [PubMed] [Google Scholar]
- 47.Jenkins M. K., Khoruts A., Ingulli E., Mueller D. L., McSorley S. J., Reinhardt R. L., Itano A., Pape K. A. 2001. In vivo activation of antigen-specific CD4 T cells. Annu. Rev. Immunol. 19: 23–45 [DOI] [PubMed] [Google Scholar]
- 48.Corse E., Gottschalk R. A., Allison J. P. 2011. Strength of TCR-peptide/MHC interactions and in vivo T cell responses. J. Immunol. 186: 5039–5045 [DOI] [PubMed] [Google Scholar]
- 49.Obst R., van Santen H. M., Mathis D., Benoist C. 2005. Antigen persistence is required throughout the expansion phase of a CD4+ T cell response. J. Exp. Med. 201: 1555–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Celli S., Lemaître F., Bousso P. 2007. Real-time manipulation of T cell-dendritic cell interactions in vivo reveals the importance of prolonged contacts for CD4+ T cell activation. Immunity 27: 625–634 [DOI] [PubMed] [Google Scholar]
- 51.Yarke C. A., Dalheimer S. L., Zhang N., Catron D. M., Jenkins M. K., Mueller D. L. 2008. Proliferating CD4+ T cells undergo immediate growth arrest upon cessation of TCR signaling in vivo. J. Immunol. 180: 156–162 [DOI] [PubMed] [Google Scholar]
- 52.Foulds K. E., Shen H. 2006. Clonal competition inhibits the proliferation and differentiation of adoptively transferred TCR transgenic CD4 T cells in response to infection. J. Immunol. 176: 3037–3043 [DOI] [PubMed] [Google Scholar]
- 53.Weaver J. M., Chaves F. A., Sant A. J. 2009. Abortive activation of CD4 T cell responses during competitive priming in vivo. Proc. Natl. Acad. Sci. USA 106: 8647–8652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitmire J. K., Benning N., Eam B., Whitton J. L. 2008. Increasing the CD4+ T cell precursor frequency leads to competition for IFN-γ thereby degrading memory cell quantity and quality. J. Immunol. 180: 6777–6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blair D. A., Lefrançois L. 2007. Increased competition for antigen during priming negatively impacts the generation of memory CD4 T cells. Proc. Natl. Acad. Sci. USA 104: 15045–15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gett A. V., Sallusto F., Lanzavecchia A., Geginat J. 2003. T cell fitness determined by signal strength. Nat. Immunol. 4: 355–360 [DOI] [PubMed] [Google Scholar]
- 57.Malherbe L., Hausl C., Teyton L., McHeyzer-Williams M. G. 2004. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity 21: 669–679 [DOI] [PubMed] [Google Scholar]
- 58.Fassò M., Anandasabapathy N., Crawford F., Kappler J., Fathman C. G., Ridgway W. M. 2000. T cell receptor (TCR)-mediated repertoire selection and loss of TCR Vβ diversity during the initiation of a Cd4+ T cell response in vivo. J. Exp. Med. 192: 1719–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker L. S., Sansom D. M. 2011. The emerging role of CTLA4 as a cell-extrinsic regulator of T cell responses. Nat. Rev. Immunol. 11: 852–863 [DOI] [PubMed] [Google Scholar]
- 60.Vignali D. A., Collison L. W., Workman C. J. 2008. How regulatory T cells work. Nat. Rev. Immunol. 8: 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pepper M., Jenkins M. K. 2011. Origins of CD4+ effector and central memory T cells. Nat. Immunol. 12: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thorborn G., Young G. R., Kassiotis G. 2014. Effective T helper cell responses against retroviruses: are all clonotypes equal? J. Leukoc. Biol. 10.1189/jlb.2RI0613-347R. [DOI] [PubMed] [Google Scholar]
- 63.Browne E. P., Littman D. R. 2009. Myd88 is required for an antibody response to retroviral infection. PLoS Pathog. 5: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanguri V., Govern C. C., Smith R., Huseby E. S. 2013. Viral antigen density and confinement time regulate the reactivity pattern of CD4 T-cell responses to vaccinia virus infection. Proc. Natl. Acad. Sci. USA 110: 288–293 [DOI] [PMC free article] [PubMed] [Google Scholar]