ABSTRACT
The majority of bacteria detected in the nostril microbiota of most healthy adults belong to three genera: Propionibacterium, Corynebacterium, and Staphylococcus. Among these staphylococci is the medically important bacterium Staphylococcus aureus. Almost nothing is known about interspecies interactions among bacteria in the nostrils. We observed that crude extracts of cell-free conditioned medium from Propionibacterium spp. induce S. aureus aggregation in culture. Bioassay-guided fractionation implicated coproporphyrin III (CIII), the most abundant extracellular porphyrin produced by human-associated Propionibacterium spp., as a cause of S. aureus aggregation. This aggregation response depended on the CIII dose and occurred during early stationary-phase growth, and a low pH (~4 to 6) was necessary but was not sufficient for its induction. Additionally, CIII induced plasma-independent S. aureus biofilm development on an abiotic surface in multiple S. aureus strains. In strain UAMS-1, CIII stimulation of biofilm depended on sarA, a key biofilm regulator. This study is one of the first demonstrations of a small-molecule-mediated interaction among medically relevant members of the nostril microbiota and the first description of a role for CIII in bacterial interspecies interactions. Our results indicate that CIII may be an important mediator of S. aureus aggregation and/or biofilm formation in the nostril or other sites inhabited by Propionibacterium spp. and S. aureus.
IMPORTANCE
Very little is known about interspecies interactions among the bacteria that inhabit the adult nostril, including Staphylococcus aureus, a potential pathogen that colonizes about a quarter of adults. We demonstrated that coproporphyrin III (CIII), a diffusible small molecule excreted by nostril- and skin-associated Propionibacterium spp., induces S. aureus aggregation in a manner dependent on dose, growth phase, and pH. CIII also induces S. aureus to form a plasma-independent surface-attached biofilm. This report is the first description of a role for CIII in bacterial interspecies interactions at any human body site and a novel demonstration that nostril microbiota physiology is influenced by small-molecule-mediated interactions.
INTRODUCTION
The pathobiont (1) Staphylococcus aureus resides primarily in human nostrils and colonizes at least a quarter of the U.S. population (2, 3). (A pathobiont is a member of the microbiota with the capacity to cause significant disease; this distinguishes it from benign or beneficial members lacking virulence potential.) The nostrils (also known as anterior nares) are the openings into the nasal vestibules, which are covered by skin that harbors sweat and pilosebaceous glands and exhibit a characteristically acidic pH (4) (Fig. 1). Henceforth, we refer to the nostrils and nasal vestibules simply as “nostrils.” In contrast to the nostrils, the epithelium of the more posterior nasal cavity, beginning at the limen nasi, produces mucus and has a pH close to neutral (5). Within the body, the nostrils are a unique environment with distinct exposures, environmental characteristics, and microbiota (6–11).
FIG 1 .
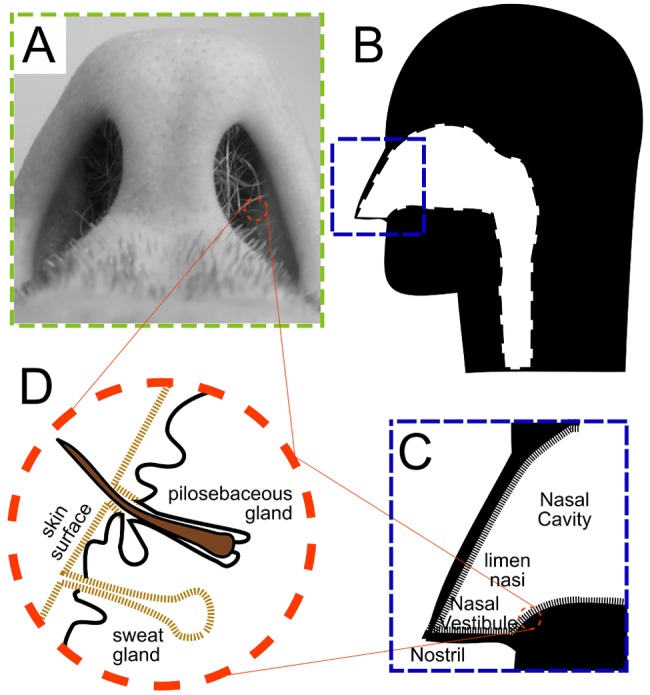
The nostrils (anterior nares) open onto the nasal vestibules, which are lined by skin, complete with sweat and pilosebaceous glands. (A) Inferior view of the nostrils and nasal vestibules; (B) sagittal illustration of the nasal cavity; (C) the limen nasi forms a boundary between the nasal vestibule and posterior aspects of the nasal cavity, which have a mucosal epithelial lining; (D) the nasal vestibule epithelium (skin) contains sweat and pilosebaceous glands and does not secrete mucus.
Although generally harmless, S. aureus nostril colonization is associated with an increased risk of S. aureus infection (12–15). An estimated 80% of S. aureus bloodstream isolates match the patient’s nostril strain (16, 17). The true global burden of S. aureus infections, which range in severity from mild skin infection to life-threatening invasive disease (e.g., bacteremia, endocarditis, and pneumonia), is unknown. However, from 2005 to 2011, methicillin-resistant S. aureus (MRSA) alone is estimated to have caused over 80,000 cases of invasive disease and over 10,000 deaths yearly in the United States (18). To date, S. aureus has eluded repeated efforts at vaccine development (19, 20). Thus, a deeper understanding of S. aureus and its membership in human nostril microbiota is an important public health issue.
Within the nostril ecosystem, S. aureus is part of a mixed-species microbial community, with three genera accounting for the majority of bacteria detected in most adults: Staphylococcus, Corynebacterium, and Propionibacterium (6–11). Few studies have examined nostril-associated Corynebacterium and Propionibacterium. In particular, there is a surprising lack of data about the interactions that can occur among members of these three genera and how these impact the growth and behavior of S. aureus.
We hypothesized that Propionibacterium spp. produce small molecules that mediate interspecies interactions with other nostril-associated bacteria. Due to its medical importance, we selected S. aureus as a candidate interactor. We identified coproporphyrin III (CIII) as an abundant S. aureus aggregation-inducing molecule in Propionibacterium conditioned medium (CM) and characterized this response. Previous studies establish that CIII is the most abundant porphyrin detected in sebaceous material from human skin, supporting its presence on nostril surfaces (21, 22). Porphyrins include a range of biologically important molecules (e.g., chlorophyll, vitamin B12, hemoglobin), and the porphyrin biosynthetic pathway is highly conserved (23–25) (see Fig. S1 in the supplemental material, adapted from reference 26). Nevertheless, the biological function(s) of some porphyrins, e.g., coproporphyrins, are poorly understood. This work represents the application of an experimental hypothesis generated from microbiome enumeration data to novel observations of the physiology and molecular biology of a clinically relevant bacterium, S. aureus.
RESULTS
Propionibacterium spp. induce S. aureus coaggregation via extracellular CIII.
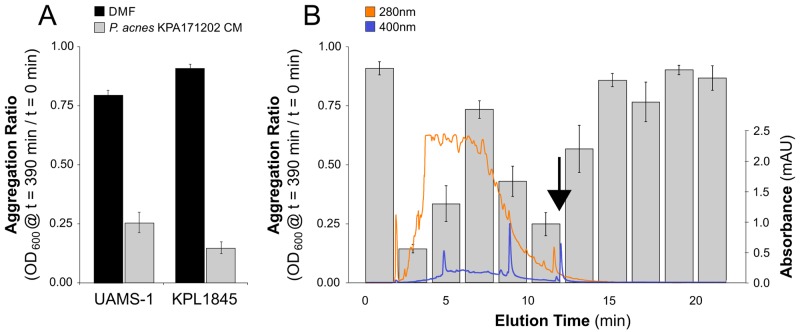
Members of the Actinobacteria, which includes Propionibacterium, are well-known producers of bioactive small molecules (27, 28). We hypothesized that cutaneous Propionibacterium spp. produce extracellular small molecules affecting S. aureus. The significant difference in the rate of growth to stationary phase (1 day for S. aureus compared to over 5 days for Propionibacterium spp.) limited cocultivation in liquid. Therefore, to test this hypothesis, a crude extract of conditioned medium (CM) from the genome-sequenced strain Propionibacterium acnes KPA171202 was concentrated, lyophilized, and resolubilized in dimethylformamide (DMF). This CM extract was added to cultures of two different S. aureus strains: a commonly studied clinical isolate, UAMS-1, and a primary nostril isolate, KPL1845 (Table 1). The addition of P. acnes KPA171202 CM extract resulted in aggregation of both strains, yielding a significantly lower aggregation ratio (after 390 min) than that of a DMF-only control (Fig. 2A; two-tailed, paired t test, P < 0.001). (Aggregation ratio is the optical density at 600 nm [OD600] of a standing liquid culture at a given time posttreatment addition divided by the original OD600 of that culture immediately before treatment addition [t = 0]). These data indicate that P. acnes KPA171202 secretes into the environment an active principle that induces S. aureus aggregation.
TABLE 1 .
Bacteria used in this study, isolation source, and genotypes
| Genus and species | Strain/isolate name | Source (human) | Genotype | Reference |
|---|---|---|---|---|
| Corynebacterium accolens | DSM44278 | Cervix | NAa | [52] |
| C. accolens | KPL1855 | Nostril | NA | This study |
| Escherichia coli | MG1655 | Feces | NA | [53] |
| Propionibacterium acnes | KPA171202 (DSM16379) | Skin | NA | [45] |
| P. acnes | KPL1849 | Nostril | NA | This study |
| Propionibacterium avidum | DSM4901 | Feces | NA | [54] |
| Propionibacterium granulosum | DSM20700 | Unknown | NA | [55] |
| P. granulosum | KPL1844 | Nostril | NA | This study |
| Staphylococcus aureus | UAMS-1 | Osteomyelitis | NA | [56] |
| S. aureus | UAMS-929 | NA | sarA::tet | [57] |
| S. aureus | SC-01 (UAMS-732) | Human | NA | [58, 59] |
| S. aureus | Newman | Unknown | NA | [60] |
| S. aureus | HG003 | Sepsis | RN1 derivative; rsbU tcaR repaired | [61] |
| S. aureus | KPL1845 | Nostril | NA | This study |
| S. aureus | JE2 | Skin/soft tissue infection | USA300 LAC without p01 and p03 | [62] |
| Staphylococcus epidermidis | DSM20044 | Nostril | NA | [63] |
| S. epidermidis | KPL1815 | Nostril | NA | This study |
NA, not applicable.
FIG 2 .
Extracts of P. acnes KPA171202 conditioned medium (CM) induce S. aureus aggregation. (A) Addition of a crude extract of P. acnes KPA171202 CM to stationary-phase cultures of S. aureus grown in SSD0Fe induced aggregation; (B) HPLC fractions (denoted by elution time) of P. acnes KPA171202 CM were added to S. aureus KPL1845 cultures. The 400-nm (blue line) and 280-nm (orange line) absorbance during HPLC elution are shown. The black arrow indicates the 400-nm-absorbing HPLC peak included in the 10- to 12-min fraction. Histogram bars represent the mean S. aureus aggregation ratio for each elution fraction or sample. All data are from five independent experiments; error bars are standard errors of the means (SEM).
To identify the small molecule(s) that induced S. aureus aggregation, we fractionated the crude P. acnes KPA171202 CM extract by preparative high-performance liquid chromatography (HPLC). Testing each 2-min fraction for induction of S. aureus KPL1845 aggregation revealed several active fractions (Fig. 2B). The 10- to 12-min fraction was notable for its strong aggregation-inducing activity and its markedly lower chemical complexity than that of earlier-eluting active fractions. Examination of the different absorption spectra within this 10- to 12-min fraction revealed a relatively abundant peak with an absorption maximum at 400 nm that eluted at the end of the 10- to 12-min fraction, tailing into the less-active 12- to 14-min fraction (Fig. 2B, arrow).
Since porphyrins are well known for having an absorption maximum at 400 nm, we next investigated the potential involvement of extracellular porphyrins as a mediator of the Propionibacterium-produced S. aureus aggregation-inducing activity. To this end, we performed an additional analysis using a primary nostril isolate of Propionibacterium (KPL1844) putatively identified to the species level as Propionibacterium granulosum and isolated from the same nostril as S. aureus KPL1845. In an effort to enhance the amount of extracellular porphyrins, we added 5-aminolevulinic acid (5-ALA) to the medium; 5-ALA is the first committed precursor in porphyrin biosynthesis (reviewed in reference 26; see also Fig. S1 in the supplemental material) and is known to increase extracellular porphyrin production by Propionibacterium spp. (29). The crude CM extract of Propionibacterium sp. KPL1844 grown in the presence of 5-ALA also induced aggregation of both S. aureus KPL1845 and UAMS-1 (see Fig. S2A in the supplemental material). When this extract was fractionated and assayed, as described above, the 10- to 12-min fraction again exhibited strong aggregation-inducing activity and harbored a peak with an absorption maximum at 400 nm (see Fig. S2B). Comparison of the absorption spectra from this fractionation with that from P. acnes KPA171202 revealed that the 400-nm-absorbing peak was one of the most abundant common peaks in the 10- to 12-min fractions. All these data led us to hypothesize that a Propionibacterium-produced extracellular porphyrin might mediate S. aureus aggregation activity.
To determine if the compound with an absorption maximum at 400 nm in the 10- to 12-min fractions was a porphyrin, we developed an analytical HPLC method for separation of four commonly occurring porphyrins (see Fig. S3A in the supplemental material). This method was optimized to enable the separation of the regioisomers CIII and CI, as a previous study reported that CIII and CI are the most abundant porphyrins present in 403-nm-absorbing P. acnes and P. granulosum CM extracts (29). The 10- to 12-min P. acnes KPA171202 fraction was analyzed with this HPLC method and found to contain a doublet of peaks with absorption maxima at 400 nm with the same retention time as that of CI and CIII (see Fig. S3B). These peaks were consistent with CI/CIII by mass spectrometry analysis (observed [M + H]+ = 655.2; monoisotopic molecular weight for CI/CIII = 654.27; see Fig. S3C). To determine which regioisomer was most abundantly produced by representatives of common nostril bacteria, we used the analytical HPLC method to separate and quantify extracellular CI and CIII from crude extracts of cell-free CM (CFCM) of representative strains of nostril bacteria. Production of extracellular CIII per milligram of dry cell weight exceeded CI levels at least 3-fold for all analyzed strains (Fig. 3, gray [CIII] and black [CI] bars). Propionibacterium spp. (P. acnes and P. granulosum) were the most prominent coproporphyrin producers, yielding amounts that were at least 2-fold greater than those produced by Corynebacterium accolens (Fig. 3). Although no CI production was detected for the Staphylococcus spp. (S. aureus and Staphylococcus epidermidis), they did produce low levels of CIII (ranging from 0.7 to 9.4 pmol/mg dry cells, Fig. 3). High-level production of extracellular CIII was specific to Propionibacterium spp.
FIG 3 .
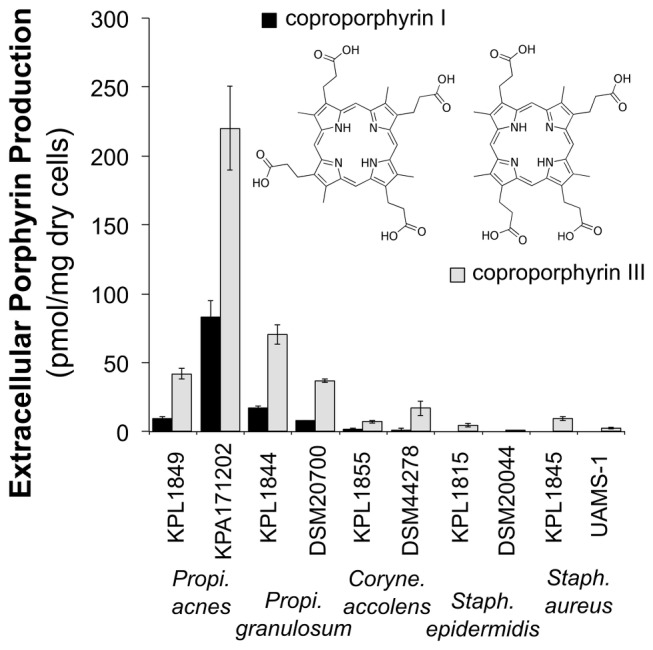
Production of extracellular coproporphyrin I and III by primary nostril isolates (KPL) and standard strains (inset, chemical structures). Extracellular coproporphyrin production estimated from HPLC fractionations. Identification to the species level of the primary nostril isolates (KPL1815, KPL1844, KPL1845, KPL1849, and KPL1855) is described in Text S1, Table S1, and Fig. S7 in the supplemental material. Histogram bars represent the means from three biological replicates; error bars are SEM.
Purified exogenous CIII induces S. aureus aggregation in a dose-dependent manner.
We proceeded to focus on determining the effect of CIII on S. aureus for two reasons. First, the above-described data indicated that CIII is the most abundant coproporphyrin produced by cutaneous Propionibacterium spp. Second, CIII is the most abundant porphyrin detected in material from pilosebaceous glands on human skin (21, 22), which indicates that CIII is likely the most abundant porphyrin on skin, including in the nostrils.
Addition of authentic CIII at 50 µM induced aggregation of both S. aureus KPL1845 and UAMS-1 (Fig. 4A, gray symbols) compared to that of a solvent-only control (Fig. 4A, black symbols). A solution of 50 µM CIII in DMF is colored pink; 4 h after adding CIII to our S. aureus aggregation assay, the majority of the pink coloration was associated with aggregated cells at the bottom of the cuvette, leaving the medium clear. From this observation, we hypothesized that S. aureus coaggregates with CIII. Microscopic examination of S. aureus UAMS-1 cells showed groups of two and four cells in DMF-treated culture (Fig. 4B), in contrast to larger multicellular aggregates in the presence of CIII (Fig. 4C). S. aureus-CIII coaggregation was dose dependent in terms of both the rate and total amount of aggregation (see Fig. S4 in the supplemental material). In addition to UAMS-1 and KPL1845, we tested other S. aureus strains (e.g., Newman); however, the aggregation assay was unable to detect increased aggregation in the presence of CIII for these strains due to a high level of self-aggregation in the presence of solvent alone.
FIG 4 .
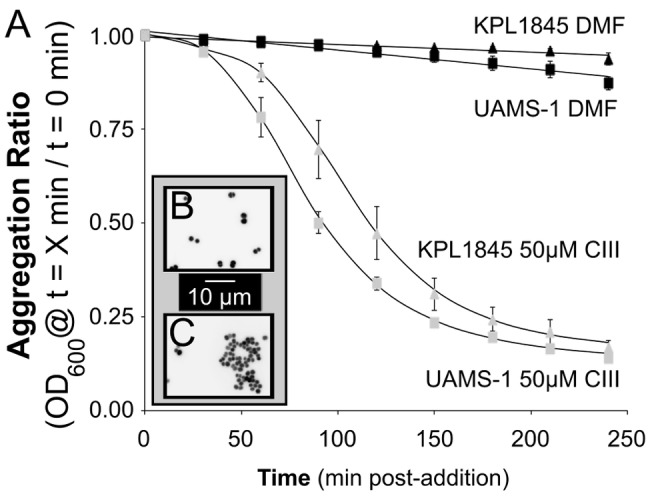
Coproporphyrin III induces S. aureus aggregation in culture. (A) DMF (black symbols) does not induce aggregation; 50 µM CIII (gray symbols) induces aggregation. Data points are the means from five biological replicates of aggregation assays done after 19 h of growth. Error bars are SEM. (Inset) Epifluorescence microscopy of Syto-9-stained UAMS-1 cultures 4 h after addition illustrates lack of aggregation with only DMF (B) and aggregation into multicellular clusters with 50 µM CIII (C).
High concentrations of some porphyrins (e.g., heme) are toxic to S. aureus (30). Examination of S. aureus cells using live-dead staining showed no statistical difference in the percentage of dead cells between solvent-only (DMF) and CIII-treated cultures (Fig. S5; two-tailed, paired t test, P > 0.3). These live-dead staining data indicate that exogenous CIII at the concentrations utilized here did not have a significantly deleterious effect on S. aureus survival.
CIII-mediated S. aureus coaggregation occurs in early stationary phase.
We combined a 24-h growth curve with the aggregation assay to better understand the relationship between growth phase of S. aureus UAMS-1 in the semidefined, low-iron medium SSD0Fe and aggregation potential in response to exogenous CIII. In SSD0Fe, S. aureus UAMS-1 exhibited diauxic growth, which was modeled using a published equation (31) (Fig. 5, open squares). S. aureus did not detectably coaggregate with CIII until early stationary phase (Fig. 5, gray squares). Onset of aggregation correlated with the concomitant drop in the culture pH to a range of 6 to 6.5 (Fig. 5, top).
FIG 5 .
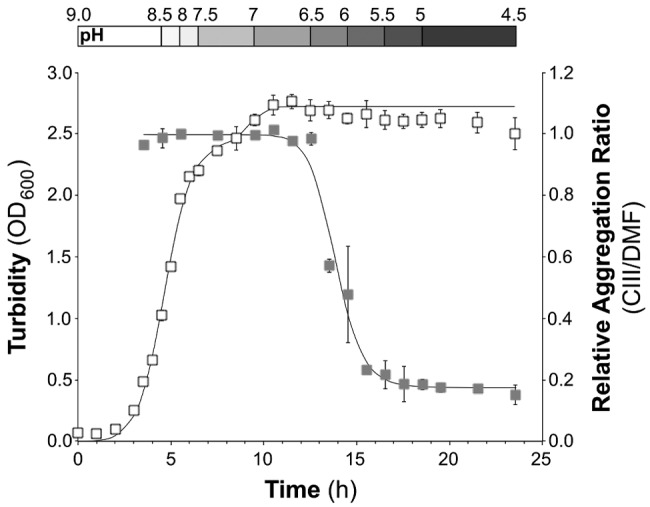
S. aureus-CIII coaggregation is growth phase dependent. S. aureus UAMS-1 exhibited diauxic growth in SSD0Fe over 24 h (white squares). At each time point, cells were harvested to assess the response to CIII addition; “relative aggregation ratio” (gray squares) is the aggregation ratio with 50 µM CIII over the aggregation ratio with DMF alone 4 h postaddition. The measured pH range of the culture at each time point is depicted above the graph. All values are the means from three biological replicates; error bars are SEM.
CIII aggregation in S. aureus CFCM requires both acidic pH and an additional S. aureus product(s).
Because some porphyrins self-aggregate at acidic pH (32), the observed correlation between the drop in culture pH and CIII-S. aureus coaggregation led us to question the role of pH in this response. To study CIII self-aggregation in SSD0Fe, we tested the impact of pH on CIII aggregation kinetics without S. aureus addition. SSD0Fe was either adjusted to the pH detected in S. aureus cultures at a point in stationary phase corresponding to the maximal aggregation response (i.e., pH of ~4.5 to 5.0 per Fig. 5) or left at its original pH (~8 to 8.5). CIII did not self-aggregate in fresh SSD0Fe at a pH of 5 at concentrations and time scales used in our assays (Fig. 6 and Fig. S6 in the supplemental material, open squares).
FIG 6 .
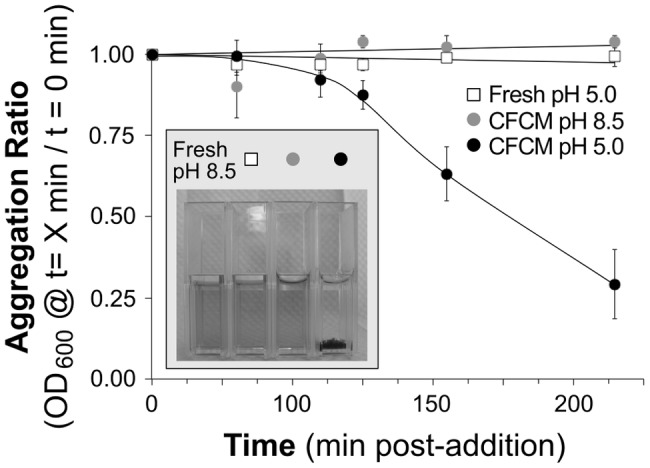
S. aureus UAMS-1 cell-free conditioned medium induces CIII aggregation at a low pH. CM from a 19-h UAMS-1 culture was filter sterilized; then the pH was either left unadjusted (black circles) or adjusted to 8.5 with NaOH (gray circles). Fresh SSD0Fe was adjusted to pH 5.0 with HCl (white squares). CIII (FC of 100 µM) was added to each condition and aggregation was measured. Graph points represent the mean aggregation ratio from three experiments, each using medium prepared on a different date; error bars are SEM (inset). A photo taken 72 h post-CIII addition shows CIII visibly aggregated at the bottom of the cuvette with CFCM pH 5.0 but remained in solution in the other three conditions.
Next, we explored whether growth medium conditioned by S. aureus would induce CIII aggregation in the absence of cells. The addition of CIII to stationary-phase S. aureus CFCM (pH 4.5 to 5) resulted in visible CIII aggregation within 3 h (Fig. 6 and Fig. S6 in the supplemental material, black circles). Raising the pH of this stationary-phase CFCM from its acidic baseline (4.5 to 5.0) up to 8.0 to 8.5 abrogated CIII aggregation (Fig. 6 and Fig. S6 in the supplemental material, gray circles). In fact, CIII did not aggregate within 7 days in high-pH S. aureus CFCM, low-pH SSD0Fe, or normal-pH SSD0Fe. From all of these data, we concluded that although a low pH was necessary for CIII aggregation, it was not sufficient in this assay. Similarly, although S. aureus conditioning of the medium was necessary to induce CIII aggregation, it too was not sufficient in this assay. Rather, the induction of aggregation required both acidic pH and an additional S. aureus product(s).
Purified exogenous CIII also induces S. aureus surface-attached biofilm formation.
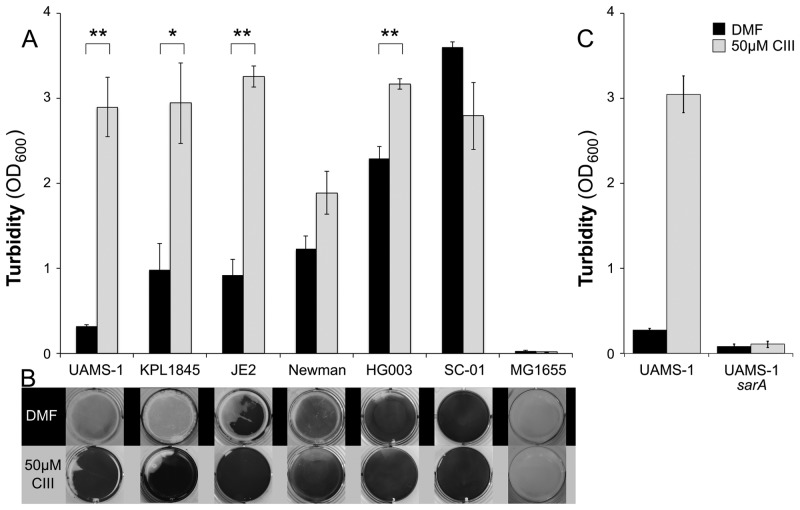
S. aureus is known for its ability to both self-aggregate and form surface-attached biofilms (33, 34). Aggregation does not guarantee biofilm formation on a surface, as this also requires surface attachment. When assayed, exogenous CIII (Fig. 7A, gray) induced S. aureus biofilm formation on an abiotic surface, compared to DMF alone (Fig. 7A, black). UAMS-1 is known to require plasma precoating of plastic surfaces to form robust biofilm (35). So it was striking that CIII-induced UAMS-1 surfaced-adhered biofilm formation did not require precoating of the plastic surface with plasma. We intentionally did not precoat wells for this assay, as plasma proteins are not expected to be present on intact nostril and skin surfaces. Using this surface-attached biofilm assay, we observed an effect of CIII addition on a number of S. aureus strains that could not be effectively studied by the aggregation assay, due to robust baseline self-aggregation (Fig. 7A and B). Although the majority of strains tested exhibited a clear increase in biofilm formation in response to CIII, a few did not (SC-01 and Newman), demonstrating strain-to-strain variation in the S. aureus response to CIII. Escherichia coli MG1655, a human-associated bacterium that is not indigenous to the nostril, also grew in and acidified SSD0Fe but did not form biofilm either in the absence or presence of CIII (Fig. 7), providing support that low pH, which is a characteristic of human skin surfaces, is not alone sufficient to cause coaggregation of CIII with a different bacterial species.
FIG 7 .
CIII induces S. aureus biofilm formation in culture; in strain UAMS-1, this induction requires sarA. (A) For four S. aureus isolates, cell density (biofilm formation) was significantly greater in the presence of 50 µM CIII (gray bars; two-tailed, paired t test with Bonferroni correction: **, P < 0.05; *, P < 0.06) compared to DMF alone (black bars). E. coli MG1655 did not form biofilm under these assay conditions. Data are the means from five biological replicates; error bars represent SEM. (B) Representative photos taken of individual wells after 20 min of 1% crystal violet staining. (C) S. aureus UAMS-1 formed a biofilm in response to culture with SSD0Fe and 50 µM CIII (gray bars) but was unable to form a biofilm in identical conditions when the sarA gene was interrupted. Data represent the means from three biological replicates; error bars are SEM.
CIII-mediated biofilm formation requires S. aureus components needed for the general biofilm response.
The sarA gene encodes a key transcriptional regulator of biofilm formation in some S. aureus strains, including UAMS-1 (35–37). As expected, a UAMS-1 sarA-deficient mutant was defective in surface-associated biofilm formation compared to the wild type. The addition of CIII did not alter the sarA mutant biofilm deficit (Fig. 7C). This demonstrates that CIII-induced biofilm formation requires sarA-regulated cellular factors that are part of the standard biofilm response.
DISCUSSION
This work breaks new ground by demonstrating that a small molecule produced by human-associated Propionibacterium spp. can impact the behavior of the medically important pathobiont S. aureus. Specifically, we demonstrated that, in culture, Propionibacterium-produced coproporphyrin III (CIII) induces S. aureus coaggregation and biofilm formation. To our knowledge, this is the first demonstration of a Propionibacterium-produced small molecule altering the behavior of S. aureus (or any other Staphylococcus spp.) and one of only a few reports of S. aureus-Propionibacterium interaction (38, 39).
The relevance of our findings to the in vivo ecology of Propionibacterium and S. aureus is supported by reports that CIII is the predominant porphyrin present in sebaceous material collected from inflamed pilosebaceous follicles, i.e., acne lesions, in which Propionibacterium spp. are abundant (21, 22). Additionally, the CIII response observed in culture occurs in a pH range (4.5 to 6.5) that is physiologically relevant in the habitats where S. aureus is naturally most abundant; the pH at various skin sites is acidic, generally ranging from 4.5 to 6, with the nostril surface pH around 5.5 (4). Stationary-phase S. aureus cells responded to CIII-induced coaggregation. This is consistent with the requirement in strain UAMS-1 for a SarA-regulated factor(s) and raises the question of whether early stationary phase in batch culture is similar to the physiological state of bacteria on skin/nostril surfaces. Surprisingly, CIII-mediated induction of Staphylococcus biofilm formation in culture was independent of precoating plastic with plasma proteins, an indication that this response could occur on surfaces free of plasma, such as skin. This finding also indicates that in the presence of Propionibacterium spp., S. aureus biofilm formation does not require binding of plasma proteins by surface-expressed proteins.
In addition to its relevance to S. aureus research, we have addressed a long-standing question about Propionibacterium physiology. More than three decades after reports that human-associated Propionibacterium spp. produce extracellular coproporphyrins (29, 40), this work establishes a role for one of these molecules, coproporphyrin III, as a mediator of interspecies interactions among members of the nostril and skin microbiota. Although it has long been known that Propionibacterium spp. produce and excrete porphyrins, the biological roles of extracellular porphyrins and the mechanisms by which these act remain poorly understood. Schaller and colleagues implicated Propionibacterium-produced CIII in stimulation of interleukin 8 (IL-8) production by keratinocytes (21). Possibly, CIII secretion could be involved in divalent metal ion scavenging, similar to its proposed CuII acquisition function in the soil bacterium Paracoccus denitrificans (41). Alternatively, extracellular porphyrins could play a role in electron shuttling, analogous to the secretion of flavins or the system cell surface decaheme cytochrome nanowires used by Shewanella spp. (42–44). A genomic analysis of P. acnes porphyrin biosynthesis reveals that the coproporphyrinogen III oxidase hemN (PPA0911 in P. acnes KPA171202) is not clustered with the rest of the heme biosynthetic operon (hemABCDEHLY, PPA0301 to PPA0310) (45). This gene organization suggests that the porphyrin production phenotype might be adaptive and could provide P. acnes with opportunities for differential regulation of its porphyrin biosynthesis pathways (see Fig. S1 in the supplemental material).
In addition, suitable hypotheses are necessary to address the evolutionary significance of CIII-mediated S. aureus aggregation and biofilm formation. One possibility is that increased coaggregation with CIII might benefit S. aureus, as biofilm development has been demonstrated to provide biofilm-associated bacteria with increased survival in hostile environments. For example, bacterial biofilms show greater resistance to antibiotic exposure, extracellular enzymes (e.g., those produced by neighboring bacteria), and the adaptive immune response (33, 46–48). Additionally, the relationship between S. aureus and Propionibacterium might not be simply antagonistic. For example, rather than just inducing single-species S. aureus microcolonies, CIII might promote mixed biofilm formation with Propionibacterium spp. and/or other nostril/skin community members.
Future research is needed to elucidate the mechanism by which CIII coaggregates with S. aureus cells. We have demonstrated that this interaction requires S. aureus factors that are regulated, at least in UAMS-1, by the transcription factor SarA. In UAMS-1, SarA is critical for biofilm formation, suggesting that the CIII interactor(s) also plays a general role in UAMS-1 biofilm formation. The CIII interaction with S. aureus requires both S. aureus-produced factors and an acidic pH in the normal range of nostril and skin pH. This acidic pH requirement could indicate that CIII, which should be negatively charged under these conditions, must be in a particular state to undergo a specific interaction with an S. aureus factor(s) or that CIII might interact nonspecifically with positively charged cell surface-expressed proteins on S. aureus (but not on E. coli). Future experiments will be directed at determining the molecular mechanisms of the CIII-S. aureus interaction.
Approaching each human body site as an ecosystem implies the existence of a network of interspecies interactions, many of which could be mediated by small molecules. We speculate that S. aureus responds with altered behavior/physiology to a number of bacterium-produced small molecules that are excreted by its microbial neighbors at its primary sites of colonization: nostrils, skin, and oropharynx. Understanding the molecular mechanisms of this network of interactions, and the ramifications for the local community of removing or adding species, is likely to be essential to developing an ecologically sound approach for managing the composition of nostril microbiota to promote long-term health, e.g., a “parks management” approach (49).
MATERIALS AND METHODS
Bacterial strains, cultures, and media.
Bacteria were streaked on agar medium from frozen stocks 1 to 7 days before each assay’s start depending on the species (Table 1). Staphylococcus spp. were grown aerobically using tryptic soy broth (TSB); Corynebacterium spp. were grown aerobically using brain heart infusion (BHI) with 1% Tween 80 (BHIT); Propionibacterium spp. were grown anaerobically using BHI. Complex media were BD brand (Becton, Dickinson, and Co.). Low-iron medium (SSD0Fe) used to grow S. aureus for aggregation and biofilm assays was prepared using published recipes (50, 51) as detailed in Text S1 in the supplemental material. Routine culture of antibiotic-requiring strains was with 10 µg/ml erythromycin.
Ethics statement.
Primary nostril isolates were collected as part of an ongoing protocol to study the bacterial composition of adult nostril microbiota that was initially approved by the Harvard Medical School Committee on Human Studies (8) and is currently approved by the Forsyth Institute Institutional Review Board (IRB). All participants were adults, 21 years or older, who, after receiving both a verbal and a written explanation of the study, provided verbal informed consent prior to nostril swabbing.
Preparation of Propionibacterium sp. conditioned medium crude extract and HPLC fractionation.
The conditioned medium (CM) of 2 liters of BHI (pH 6.1) Propionibacterium sp. cultures (37°C for 90 h, anaerobic) was extracted with Diaion HP-20 beads (Sigma-Aldrich) by being stirred in a light-protected environment. The beads were washed with distilled H2O (dH2O) and extracted 3 times with MeOH, and combined extracts were dried down. The resulting dry crude extract was dissolved in 10 ml MeOH, and a 1-ml aliquot was filtered and separated by preparative HPLC (1200 series; Agilent Technologies) on a semipreparative C18 column (Phenomenex; Luna 5-µM C18, 250 by 10 mm) at a flow rate of 7 ml/min, using a linear 20-min gradient of 10% to 100% acetonitrile in water (both solvents containing 0.1% trifluoroacetic acid [TFA]). For further details regarding the extraction and separation, see Text S1 in the supplemental material.
Analytical HPLC porphyrin quantitation from cell-free conditioned medium.
For each strain, 5-ml cultures were grown at 37°C to stationary phase under the following conditions: S. aureus and S. epidermidis, TSB, 18 h, aerobic; C. accolens, BHIT, 30 h, aerobic; P. granulosum and P. acnes, BHI, 50 h, anaerobic. CM and cell pellets were separated by centrifugation (3,250 × g, 15 min, 4°C). Cell pellets were washed with dH2O and then lyophilized to determine the dry cell mass for standardization of the production results. Intracellular porphyrin levels were determined by combining the three lyophilized cell pellets of each strain and homogenizing them in 5 ml ethyl acetate and acetic acid (3:1, vol/vol). Each CM was filtered (0.22-µm pore size) and extracted with an equal volume of ethyl acetate and acetic acid (3:1, vol/vol). The organic phase from the above-described cell pellet or CM extractions was collected, all solvent was evaporated, and the dried residue was resuspended in 250 µl acetone and MeOH (1:1, vol/vol).
Fifty microliters of each suspension was injected into analytical HPLC equipment (1200 series; Agilent Technologies) on a C18 column (Phenomenex; Luna 5-µM C18, 100 by 4.6 mm) with detection at λ = 400 nm. Mobile phase A was 0.1% TFA in dH2O; mobile phase B was 0.1% TFA in acetonitrile. Separation was obtained at a flow rate of 1 ml/min with a gradient program that started at 10% B, changing to 25% B in 3 min, followed by an increase to 45% B over 40 min. After this, the solvent composition was increased to 100% B over an additional 7 min followed by a 5-min wash at 100% B. Porphyrin levels of CM or cell pellets (below detection limit) were quantified based on a standard curve that was generated using a dilution series of known porphyrin stock solutions, as described in detail in Text S1 in the supplemental material.
S. aureus aggregation assay.
Frozen stocks were streaked on TSB agar and grown aerobically at 37°C for 24 to 48 h. Colonies were picked and resuspended in 1 ml SSD0Fe to an optical density at 600 nm (OD600) of ≈1. For each experimental replicate/condition/culture, 50 µl from this resuspension was mixed with 950 µl of fresh SSD0Fe and placed into a sterile 1.7-ml microcentrifuge tube. Culture tubes were incubated at 37°C for 19 h with shaking at 200 rpm. After incubation, 10 µl of either a concentrated coproporphyrin III dihydrochloride solution (CIII; C654-3; Frontier Scientific) solubilized in dimethylformamide (DMF; Sigma-Aldrich) or DMF alone was mixed into each tube. The DMF final concentration (FC) was always 1% (vol/vol). Absorbance readings were made at OD600 over a defined period of time with an LED-based spectrophotometer (Amersham Biosciences Ultrospec 10). Each aggregation datum at time x was calculated as a ratio: OD600 at time x/OD600 at time 0 min (compound addition). All aggregation assay data presented are averages from at least three biological replicates. Aggregation trends were either fit to the four-parameter nonlinear regression model y = d + [(a − d)/(1 + (x/c)b)] by minimizing the mean squared error (MSE) between the real and model values using the “Solver” tool in Microsoft Excel (Frontline Systems Inc.) or fit using a linear model.
S. aureus UAMS-1 growth, aggregation, and pH measurements.
S. aureus UAMS-1 colonies were grown as described above. Colonies were picked and resuspended in 2 ml of SSD0Fe to an OD600 of ≈1. This was mixed with 38 ml of SSD0Fe and 1-ml aliquots dispensed into 40 sterile, 1.7-ml microcentrifuge tubes prior to incubation at 37°C for up to 23.5 h with shaking at 200 rpm. At intervals during growth, one or more tubes were removed and the OD600 was measured; if the OD600 was ≥1, the culture was diluted in fresh SSD0Fe to permit measurement in the linear range of the spectrophotometer. The growth curve of S. aureus UAMS-1 in SSD0Fe was fit with the diauxic model y = aαbα + aβbβ, where aα = (1 − e−x/c)/(1 − e−x/c + e−x/d) and aβ = (1 − e−x/e)/(1 − e−x/e + e−x/f); additionally, bα + bβ = 1 (31). As described above, the “Solver” tool was used to minimize the MSE between observed and predicted values.
At each time point where OD600 was measured, pH was also measured using strips of both pH 2 to 9 (BDH, VWR) and pH 4 to 7 (Merck KGaA). Aggregation assays were started periodically throughout the growth curve. Similar to that described above, 1% (vol/vol) DMF or CIII (FC of 50 µM) in DMF was mixed into the culture and the OD600 was measured. For each time point on the growth curve, the relative aggregation ratio was calculated as the aggregation ratio of the CIII-exposed culture after 4 h divided by the aggregation ratio of the DMF-exposed culture after 4 h. The relative aggregation curve for UAMS-1 was fit using a four-parameter nonlinear regression model as described above.
S. aureus biofilm assay.
Each well of a polystyrene, flat-bottom, tissue culture-treated, 12-well plate (BD Biosciences) was filled with 1.9 ml fresh SSD0Fe, 0.1 ml of bacterial culture at an OD600 of ≈1 prepared as described above, and 10 µl of either a 10 mM CIII solution in DMF (FC of 50 µM) or DMF alone. Each plate was incubated standing at 37°C for 24 h; then, the plate was agitated by hand for 30 s to resuspend unattached, sedimented cells. Medium and planktonic cells were removed, and each well was washed with 0.5 ml of fresh SSD0Fe (pH adjusted to 4 to 5 with 1 N HCl) and agitated by hand for 30 s. The liquid was again removed, and each well was again washed and agitated and the liquid was removed as described before. After two washes, 1 ml of fresh SSD0Fe (pH of ≈8) was added to each well. Wells were scraped with a sterile plastic spatula or pipette tip to remove all surface-adhered cells. The cell mix from each well was transferred to a plastic cuvette, and the OD600 was measured.
SUPPLEMENTAL MATERIAL
Supplemental Methods. Download
Portions of the porphyrin biosynthetic pathway are highly conserved among bacteria. Black arrows represent biosynthetic step(s) in porphyrin biosynthesis. Heme, in many bacteria, as well as cobalamin or chlorophyll, in some bacteria, are the major products of this pathway, in contrast to coproporphyrin III and I, which are considered side products (red arrows). Download
Extracts of Propionibacterium granulosum KPL1844 (putative identification to the species level) conditioned medium (CM) induce S. aureus aggregation. (A) Addition of a crude extract of P. granulosum KPL1844 CM to stationary-phase cultures of S. aureus grown in SSD0Fe induced aggregation. (B) HPLC fractions (denoted by elution time) of P. granulosum KPL1844 CM added to cultures of S. aureus UAMS-1 (gray bars). The 400-nm (blue line) and 280-nm (orange line) absorbances during HPLC elution are shown. The black arrow indicates the 400-nm-absorbing HPLC peak included in the 10- to 12-min fraction. Histogram bars indicate the mean S. aureus aggregation ratio for each elution fraction or sample. Data are from five independent experiments; error bars are SEM. Download
Identification of naturally occurring porphyrins in conditioned medium by HPLC and mass spectrometry analysis. (A) Separation of porphyrin standards (at concentrations given in Text S1 in the supplemental material) by analytical HPLC with detection at 400 nm (uroporphyrin III [UIII], coproporphyrin I and III [CI and CIII], protoporphyrin IX [PIX]). (B) Separation of compounds in the 10- to 12-min elution from P. acnes KPA171202 by analytical HPLC with detection at 400 nm. (C) Mass spectrometry identifies the 400-nm-absorbing compounds in the 10- to 12-min fraction as CI/CIII ([M + H]+ = 655.2 and [M + 2H]2+ = 328.2; monoisotopic molecular weight for CI/CIII = 654.27). Download
S. aureus aggregation in response to CIII is dose dependent. S. aureus UAMS-1 (a) and KPL1845 (b) were grown for 19 h in SSD0Fe prior to addition of the indicated CIII concentrations (0 to 100 µM). Data represent the means from five biological replicates; error bars represent SEM. Download
Coproporphyrin III coaggregation with S. aureus does not significantly alter cell viability. Following an aggregation assay, S. aureus cells were then stained with Syto-9 and propidium iodide and counted using epifluorescence microscopy. Data represent the average percentage of dead cells (propidium iodide stained) from five biological replicates. Error bars are SEM. There was no significant difference between solvent-only controls and CIII-treated samples for either S. aureus UAMS-1 or KPL1845 (two-tailed, paired t test, P > 0.3). Download
S. aureus KPL1845 cell-free conditioned medium induces coproporphyrin III aggregation at a low pH. S. aureus KPL1845 was grown in SSD0Fe for 19 h prior to filter sterilization of spent medium. After filtration, the pH was either left unadjusted (black circles) or adjusted to 8.5 with NaOH (gray circles). Fresh SSD0Fe medium was adjusted to a pH of 5.0 with HCl (white squares). CIII (FC of 100 µM) was added to each condition and aggregation was measured. Graph points represent mean aggregation ratios from three experiments each using medium prepared on a different date; error bars are SEM. (Inset) A photo taken 72 h post-CIII addition shows CIII visibly aggregated at the bottom of the cuvette with CFCM pH 5.0 but remained in solution in the other three conditions. Download
Phylogenetic data support the assignment of KPL1855 as putative Corynebacterium accolens. Slightly truncated 16S (~1,300-nucleotide [nt]) and rpoB (~3,150-nt) sequences from previously identified Corynebacterium spp., KPL1855, and the outgroup Rhodococcus equi were concatenated and subjected to phylogenetic reconstruction using Bayesian (Ba), maximum likelihood (ML), and maximum parsimony (MP) methods. The tree represents the 50% majority-rule consensus tree from the Bayesian analysis, with nodes depicted as having greater than 0.6 posterior probability and greater than 60% support by either ML or MP bootstrapping. The blue highlight represents a well-conserved C. accolens clade determined by these reconstruction methods. The bar represents 0.1 substitutions per site. Download
Species-typing for primary nostril KPL Propionibacterium isolates.
ACKNOWLEDGMENTS
This work was supported in part by NIH through the Forsyth Postdoctoral Training in Oral Health Research T32 DE007327 (M.S.W.), DE020751 (K.P.L.), AI101018 (K.P.L., M.A.F.), and OD007290 (M.A.F.) and by a Boston Children’s Hospital Career Development Fellowship (K.P.L.), a Fundación Ramón Areces Fellowship (I.F.E.), a Medical Research Program grant from the WM Keck Foundation (M.A.F.), and a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (M.A.F.). Genomic data used to identify KPL strains to the species level were generated in part with U.S. federal funds from the NIH Human Microbiome Project (HMP), the Common Fund, NIH, DHHS, as part of the HMP.
We thank Mark S. Smeltzer, Roberto Kolter, and Michael Gilmore for published strains, Ashlee Earl for advice on genomic DNA preparation, and Evan E. Pagano, Beck Jacobson, and Kelsey L. Goguen for assistance with genus identification of KPL strains and preparation of genomic DNA. Rebecca J. Case, Lindsey Bomar, Matthew M. Ramsey, Carla Cugini, and Amanda Wollenberg contributed constructive criticism to the manuscript, to its benefit. Thanks to Jorge Frias Lopez, Mary Ellen Davey, and their labs for input.
Footnotes
Citation Wollenberg MS, Claesen J, Escapa IF, Aldridge KL, Fischbach MA, Lemon KP. 2014. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. mBio 5(4):e01286-14. doi:10.1128/mBio.01286-14.
REFERENCES
- 1. Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625. 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- 2. Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, Fosheim G, McDougal LK, Chaitram J, Jensen B, Fridkin SK, Killgore G, Tenover FC. 2006. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J. Infect. Dis. 193:172–179. 10.1086/499632 [DOI] [PubMed] [Google Scholar]
- 3. Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. 2008. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J. Infect. Dis. 197:1226–1234. 10.1086/533494 [DOI] [PubMed] [Google Scholar]
- 4. McShane D, Davies JC, Davies MG, Bush A, Geddes DM, Alton EW. 2003. Airway surface pH in subjects with cystic fibrosis. Eur. Respir. J. 21:37–42. 10.1183/09031936.03.00027603 [DOI] [PubMed] [Google Scholar]
- 5. Washington N, Steele RJ, Jackson SJ, Bush D, Mason J, Gill DA, Pitt K, Rawlins DA. 2000. Determination of baseline human nasal pH and the effect of intranasally administered buffers. Int. J. Pharm. 198:139–146. 10.1016/S0378-5173(99)00442-1 [DOI] [PubMed] [Google Scholar]
- 6. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. 2010. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 1(3):e00129-00110. 10.1128/mBio.00129-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. 2010. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 5:e10598. 10.1371/journal.pone.0010598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, Becker K, Pieper DH. 2010. A poke into the diversity and associations within human anterior nare microbial communities. ISME J. 4:839–851. 10.1038/ismej.2010.15 [DOI] [PubMed] [Google Scholar]
- 11. HMP Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5:751–762. 10.1016/S1473-3099(05)70295-4 [DOI] [PubMed] [Google Scholar]
- 13. Kluytmans JA, Mouton JW, Ijzerman EP, Vandenbroucke-Grauls CM, Maat AW, Wagenvoort JH, Verbrugh HA. 1995. Nasal carriage of Staphylococcus aureus as a major risk factor for wound infections after cardiac surgery. J. Infect. Dis. 171:216–219. 10.1093/infdis/171.1.216 [DOI] [PubMed] [Google Scholar]
- 14. Luzar MA, Coles GA, Faller B, Slingeneyer A, Dah GD, Briat C, Wone C, Knefati Y, Kessler M, Peluso F. 1990. Staphylococcus aureus nasal carriage and infection in patients on continuous ambulatory peritoneal dialysis. N. Engl. J. Med. 322:505–509. 10.1056/NEJM199002223220804 [DOI] [PubMed] [Google Scholar]
- 15. Nouwen J, Schouten J, Schneebergen P, Snijders S, Maaskant J, Koolen M, van Belkum A, Verbrugh HA. 2006. Staphylococcus aureus carriage patterns and the risk of infections associated with continuous peritoneal dialysis. J. Clin. Microbiol. 44:2233–2236. 10.1128/JCM.02083-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study group. N. Engl. J. Med. 344:11–16. 10.1056/NEJM200101043440102 [DOI] [PubMed] [Google Scholar]
- 17. Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus noncarriers. Lancet 364:703–705. 10.1016/S0140-6736(04)16897-9 [DOI] [PubMed] [Google Scholar]
- 18. CDC 2005-2011. Active bacterial core surveillance Report, emerging infections program network, methicillin-resistant Staphylococcus aureus. CDC, Atlanta, GA. http://www.cdc.gov/abcs/reports-findings/surv-reports.html [Google Scholar]
- 19. Proctor RA. 2012. Challenges for a universal Staphylococcus aureus vaccine. Clin. Infect. Dis. 54:1179–1186. 10.1093/cid/cis033 [DOI] [PubMed] [Google Scholar]
- 20. Jansen KU, Girgenti DQ, Scully IL, Anderson AS. 2013. Vaccine review: “Staphyloccocus aureus vaccines: problems and prospects.” Vaccine 31:2723–2730. 10.1016/j.vaccine.2013.04.002 [DOI] [PubMed] [Google Scholar]
- 21. Schaller M, Loewenstein M, Borelli C, Jacob K, Vogeser M, Burgdorf WH, Plewig G. 2005. Induction of a chemoattractive proinflammatory cytokine response after stimulation of keratinocytes with Propionibacterium acnes and coproporphyrin III. Br. J. Dermatol. 153:66–71. 10.1111/j.1365-2133.2005.06530.x [DOI] [PubMed] [Google Scholar]
- 22. Borelli C, Merk K, Schaller M, Jacob K, Vogeser M, Weindl G, Berger U, Plewig G. 2006. In vivo porphyrin production by P. acnes in untreated acne patients and its modulation by acne treatment. Acta Derm. Venereol. 86:316–319. 10.2340/00015555-0088 [DOI] [PubMed] [Google Scholar]
- 23. Heinemann IU, Jahn M, Jahn D. 2008. The biochemistry of heme biosynthesis. Arch. Biochem. Biophys. 474:238–251. 10.1016/j.abb.2008.02.015 [DOI] [PubMed] [Google Scholar]
- 24. Cavallaro G, Decaria L, Rosato A. 2008. Genome-based analysis of heme biosynthesis and uptake in prokaryotic systems. J. Proteome Res. 7:4946–4954. 10.1021/pr8004309 [DOI] [PubMed] [Google Scholar]
- 25. Panek H, O’Brian MR. 2002. A whole genome view of prokaryotic haem biosynthesis. Microbiology 148:2273–2282. [DOI] [PubMed]
- 26. Leeper FJ. 1989. The biosynthesis of porphyrins, chlorophylls, and vitamin B12. Nat. Prod. Rep. 6:171–203. 10.1039/np9890600171 [DOI] [PubMed] [Google Scholar]
- 27. Clardy J, Fischbach MA, Walsh CT. 2006. New antibiotics from bacterial natural products. Nat. Biotechnol. 24:1541–1550. 10.1038/nbt1266 [DOI] [PubMed] [Google Scholar]
- 28. Baltz RH. 2008. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 8:557–563. 10.1016/j.coph.2008.04.008 [DOI] [PubMed] [Google Scholar]
- 29. Lee WL, Shalita AR, Poh-Fitzpatrick MB. 1978. Comparative studies of porphyrin production in Propionibacterium acnes and Propionibacterium granulosum. J. Bacteriol. 133:811–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109–119. 10.1016/j.chom.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liquori AM, Monroy A, Parisi E, Tripiciano A. 1981. A theoretical equation for diauxic growth and its application to the kinetics of the early development of the sea urchin embryo. Differentiation 20:174–175. 10.1111/j.1432-0436.1981.tb01173.x [DOI] [PubMed] [Google Scholar]
- 32. Scolaro LM, Castriciano M, Romeo A, Patane S, Cefalo E, Allegrini M. 2002. Aggregation behavior of protoporphyrin IX in aqueous solutions: clear evidence of vesicle formation. J. Phys. Chem. B 106:2453–2459. 10.1021/jp013155h [DOI] [Google Scholar]
- 33. Otto M. 2008. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322:207–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cassat JE, Smeltzer MS, Lee CY. 2014. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus. Methods Mol. Biol. 1085:195-211. 10.1007/978-1-62703-664-1_12 [DOI] [PubMed] [Google Scholar]
- 35. Beenken KE, Blevins JS, Smeltzer MS. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206–4211. 10.1128/IAI.71.7.4206-4211.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penadés JR, Lasa I. 2003. SarA and not SigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075–1087. 10.1046/j.1365-2958.2003.03493.x [DOI] [PubMed] [Google Scholar]
- 37. Cheung AL, Nishina KA, Trotonda MP, Tamber S. 2008. The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40:355–361. 10.1016/j.biocel.2007.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lo CW, Lai YK, Liu YT, Gallo RL, Huang CM. 2011. Staphylococcus aureus hijacks a skin commensal to intensify its virulence: immunization targeting beta-hemolysin and CAMP factor. J. Invest. Dermatol. 131:401–409. 10.1038/jid.2010.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, Gallo RL, Huang CM. 2013. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One 8:e55380. 10.1371/journal.pone.0055380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cornelius CE, III, Ludwig GD. 1967. Red fluorescence of comedones: production of porphyrins by Corynebacterium acnes. J. Invest. Dermatol. 49:368–370. 10.1038/jid.1967.54 [DOI] [PubMed] [Google Scholar]
- 41. Anttila J, Heinonen P, Nenonen T, Pino A, Iwaï H, Kauppi E, Soliymani R, Baumann M, Saksi J, Suni N, Haltia T. 2011. Is coproporphyrin III a copper-acquisition compound in Paracoccus denitrificans? Biochim. Biophys. Acta 1807:311–318. 10.1016/j.bbabio.2010.12.014 [DOI] [PubMed] [Google Scholar]
- 42. Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U. S. A. 105:3968–3973. 10.1073/pnas.0710525105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. von Canstein H, Ogawa J, Shimizu S, Lloyd JR. 2008. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74:615–623. 10.1128/AEM.01387-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clarke TA, Edwards MJ, Gates AJ, Hall A, White GF, Bradley J, Reardon CL, Shi L, Beliaev AS, Marshall MJ, Wang Z, Watmough NJ, Fredrickson JK, Zachara JM, Butt JN, Richardson DJ. 2011. Structure of a bacterial cell surface decaheme electron conduit. Proc. Natl. Acad. Sci. U. S. A. 108:9384–9389. 10.1073/pnas.1017200108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brüggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A, Hujer S, Dürre P, Gottschalk G. 2004. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305:671–673. 10.1126/science.1100330 [DOI] [PubMed] [Google Scholar]
- 46. López D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398. 10.1101/cshperspect.a000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Anderson GG, O’Toole GA. 2008. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 322:85–105. 10.1007/978-3-540-75418-3_5 [DOI] [PubMed] [Google Scholar]
- 48. Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. 2011. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 186:6585–6596. 10.4049/jimmunol.1002794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. 10.1126/science.1224203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lindsay JA, Riley TV. 1994. Staphylococcal iron requirements, siderophore production, and iron-regulated protein expression. Infect. Immun. 62:2309–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- 52. Neubauer M, Šourek J, Rýc M, Boháček J, Mára M, Mňuková J. 1991. Corynebacterium accolens sp. nov., a Gram-positive rod exhibiting satellitism, from clinical material. Syst. Appl. Microbiol. 14:46–51. 10.1016/S0723-2020(11)80360-7 [DOI] [Google Scholar]
- 53. Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. 10.1126/science.277.5331.1453 [DOI] [PubMed] [Google Scholar]
- 54. Moore WEC, Holdeman LV. 1970. In Cato EP, Cummins CS, Holdeman LV, Johnson JL, Moore WEC, Smibert RM, Smith LD. (ed), Outline of clinical methods in anaerobic bacteriology. Virginia Polytechnic Institute, Anaerobe Laboratory, Blacksburg, VA [Google Scholar]
- 55. Johnson JL, Cummins CS. 1972. Cell wall composition and deoxyribonucleic acid similarities among the anaerobic coryneforms, classical propionibacteria, and strains of Arachnia propionica. J. Bacteriol. 109:1047–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Blevins JS, Beenken KE, Elasri MO, Hurlburt BK, Smeltzer MS. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470–480. 10.1128/IAI.70.2.470-480.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goetz MB, Mulligan ME, Kwok R, O’Brien H, Caballes C, Garcia JP. 1992. Management and epidemiologic analyses of an outbreak due to methicillin-resistant Staphylococcus aureus. Am. J. Med. 92:607–614. 10.1016/0002-9343(92)90778-A [DOI] [PubMed] [Google Scholar]
- 59. Tenover FC, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hébert GA, Hill B, Hollis R. 1994. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32:407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miller KD, Hetrick DL, Bielefeldt DJ. 1977. Production and properties of Staphylococcus aureus (strain Newman D2C) with uniform clumping factor activity. Thromb. Res. 10:203–211. 10.1016/0049-3848(77)90002-0 [DOI] [PubMed] [Google Scholar]
- 61. Herbert S, Ziebandt AK, Ohlsen K, Schäfer T, Hecker M, Albrecht D, Novick R, Götz F. 2010. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 78:2877–2889. 10.1128/IAI.00088-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4(1):e00537-00512. 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hugh R, Ellis MA. 1968. The neotype strain for Staphylococcus epidermidis. Evans, Winslow 1916. Int. J. Syst. Bacteriol. 18:231–239 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods. Download
Portions of the porphyrin biosynthetic pathway are highly conserved among bacteria. Black arrows represent biosynthetic step(s) in porphyrin biosynthesis. Heme, in many bacteria, as well as cobalamin or chlorophyll, in some bacteria, are the major products of this pathway, in contrast to coproporphyrin III and I, which are considered side products (red arrows). Download
Extracts of Propionibacterium granulosum KPL1844 (putative identification to the species level) conditioned medium (CM) induce S. aureus aggregation. (A) Addition of a crude extract of P. granulosum KPL1844 CM to stationary-phase cultures of S. aureus grown in SSD0Fe induced aggregation. (B) HPLC fractions (denoted by elution time) of P. granulosum KPL1844 CM added to cultures of S. aureus UAMS-1 (gray bars). The 400-nm (blue line) and 280-nm (orange line) absorbances during HPLC elution are shown. The black arrow indicates the 400-nm-absorbing HPLC peak included in the 10- to 12-min fraction. Histogram bars indicate the mean S. aureus aggregation ratio for each elution fraction or sample. Data are from five independent experiments; error bars are SEM. Download
Identification of naturally occurring porphyrins in conditioned medium by HPLC and mass spectrometry analysis. (A) Separation of porphyrin standards (at concentrations given in Text S1 in the supplemental material) by analytical HPLC with detection at 400 nm (uroporphyrin III [UIII], coproporphyrin I and III [CI and CIII], protoporphyrin IX [PIX]). (B) Separation of compounds in the 10- to 12-min elution from P. acnes KPA171202 by analytical HPLC with detection at 400 nm. (C) Mass spectrometry identifies the 400-nm-absorbing compounds in the 10- to 12-min fraction as CI/CIII ([M + H]+ = 655.2 and [M + 2H]2+ = 328.2; monoisotopic molecular weight for CI/CIII = 654.27). Download
S. aureus aggregation in response to CIII is dose dependent. S. aureus UAMS-1 (a) and KPL1845 (b) were grown for 19 h in SSD0Fe prior to addition of the indicated CIII concentrations (0 to 100 µM). Data represent the means from five biological replicates; error bars represent SEM. Download
Coproporphyrin III coaggregation with S. aureus does not significantly alter cell viability. Following an aggregation assay, S. aureus cells were then stained with Syto-9 and propidium iodide and counted using epifluorescence microscopy. Data represent the average percentage of dead cells (propidium iodide stained) from five biological replicates. Error bars are SEM. There was no significant difference between solvent-only controls and CIII-treated samples for either S. aureus UAMS-1 or KPL1845 (two-tailed, paired t test, P > 0.3). Download
S. aureus KPL1845 cell-free conditioned medium induces coproporphyrin III aggregation at a low pH. S. aureus KPL1845 was grown in SSD0Fe for 19 h prior to filter sterilization of spent medium. After filtration, the pH was either left unadjusted (black circles) or adjusted to 8.5 with NaOH (gray circles). Fresh SSD0Fe medium was adjusted to a pH of 5.0 with HCl (white squares). CIII (FC of 100 µM) was added to each condition and aggregation was measured. Graph points represent mean aggregation ratios from three experiments each using medium prepared on a different date; error bars are SEM. (Inset) A photo taken 72 h post-CIII addition shows CIII visibly aggregated at the bottom of the cuvette with CFCM pH 5.0 but remained in solution in the other three conditions. Download
Phylogenetic data support the assignment of KPL1855 as putative Corynebacterium accolens. Slightly truncated 16S (~1,300-nucleotide [nt]) and rpoB (~3,150-nt) sequences from previously identified Corynebacterium spp., KPL1855, and the outgroup Rhodococcus equi were concatenated and subjected to phylogenetic reconstruction using Bayesian (Ba), maximum likelihood (ML), and maximum parsimony (MP) methods. The tree represents the 50% majority-rule consensus tree from the Bayesian analysis, with nodes depicted as having greater than 0.6 posterior probability and greater than 60% support by either ML or MP bootstrapping. The blue highlight represents a well-conserved C. accolens clade determined by these reconstruction methods. The bar represents 0.1 substitutions per site. Download
Species-typing for primary nostril KPL Propionibacterium isolates.