Abstract
Emerging therapies for sensorineural hearing loss include replacing damaged auditory neurons (ANs) using stem cells. Ultimately, it is important that these replacement cells can be patient-matched to avoid immunorejection. As human induced pluripotent stem cells (hiPSCs) can be obtained directly from the patient, they offer an opportunity to generate patient-matched neurons for transplantation. Here, we used an established neural induction protocol to differentiate two hiPSC lines (iPS1 and iPS2) and one human embryonic stem cell line (hESC; H9) toward a neurosensory lineage in vitro. Immunocytochemistry and qRT-PCR were used to analyze the expression of key markers involved in AN development at defined time points of differentiation. The hiPSC- and hESC-derived neurosensory progenitors expressed the dorsal hindbrain marker (PAX7), otic placodal marker (PAX2), proneurosensory marker (SOX2), ganglion neuronal markers (NEUROD1, BRN3A, ISLET1, ßIII-tubulin, Neurofilament kDa 160), and sensory AN markers (GATA3 and VGLUT1) over the time course examined. The hiPSC- and hESC-derived neurosensory progenitors had the highest expression levels of the sensory neural markers at 35 days in vitro. Furthermore, the neurons generated from this assay were found to be electrically active. While all cell lines analyzed produced functional neurosensory-like progenitors, variabilities in the levels of marker expression were observed between hiPSC lines and within samples of the same cell line, when compared with the hESC controls. Overall, these findings indicate that this neural assay was capable of differentiating hiPSCs toward a neurosensory lineage but emphasize the need for improving the consistency in the differentiation of hiPSCs into the required lineages.
Key words: : stem cell, hiPSC, differentiation, cochlear implant, auditory neuron, deafness
Introduction
Prolonged exposure to loud noise, administration of ototoxic drugs and aging can all contribute to the degeneration of sensory hair cells and auditory neurons (ANs) in mammals. As the mammalian inner ear cells do not regenerate, the loss of these cells leads to permanent hearing loss. Presently, the cochlear implant is the only routinely available clinical treatment for deafness, but its function relies upon the presence of a healthy population of ANs. A treatment strategy to regenerate or replace damaged ANs would therefore be useful in cases of severe AN degeneration. Stem cell–based therapies for AN replacement have been extensively pursued over the last decade with promising results using human embryonic stem cells (hESCs).1–3 However, the intrinsic ethical and practical limitations associated with the use of hESCs has prompted investigation into the use of autologous stem cell types, such as nasal-4 or bone marrow-derived5–9 mesenchymal stem cells and induced pluripotent stem cells (iPSCs)10,11 for AN replacement. The use of autologous cells would avoid the need for immunosuppression treatment and eliminate the complications associated with tissue rejection by transplanting sensory neurons generated from patient cells.12
To explore the potential of human iPSCs (hiPSCs), it is vital that their capacity to differentiate into an appropriate lineage is thoroughly characterized. A reasonable approach to determine the efficiency with which they differentiate into an AN lineage is to probe for the expression of key factors involved in AN development. The inner ear originates from a non-neural ectodermal region called the otic placode.13 As development progresses, the otic placode invaginates to form an otic vesicle that gives rise to all the cell types of the inner ear, including the ANs.13 The transcription factors Pax2/8,14–16 Eya1/Six1,17–19 and Sox220–22 have been shown to upregulate expression of Neurogenin 1 (Neurog1)23 in otic progenitor cells, directing them toward a proneurosensory lineage. As development progresses, these Neurog1-positive cells delaminate from the otic vesicle to form the ANs medially and the vestibular ganglion neurons laterally.24 The delaminated neurons begin to show limited expression of Neurog1 and start to sequentially express the neurogenic differentiation factor 1 (NeuroD1),25–27 brain-specific homeobox/POU domain protein 3a (Brn3a),28–30 and transcription factor Gata331–33 during their differentiation into sensory neurons.34,35 Subsequently, the mature ANs express the characteristic pan neuronal cytoskeletal markers Neurofilament kDa 160 (NFM), ßIII-tubulin, the vesicular glutamate transporter (VGLUT1), and the tyrosine receptor kinase B (TrkB) and C (TrkC) for brain-derived neurotrophic factor (BDNF) and neurotrophin 3 (NT3).36 There have recently been several advances in specifying stem cells toward an otic lineage.37–40 For instance, Koehler and colleagues demonstrated the potential to guide murine embryonic stem cells sequentially toward a non-neural, pre-placodal and otic placode-like lineage using a complex three-dimensional culture system.40 While promising, the molecular signals and combinations of transcription factors required to prompt the differentiation of stem cell–derived otic placodal cells into ANs remains to be elucidated.
There have been a series of attempts to differentiate stem cells toward an AN-like lineage using combinations of signaling molecules [basic fibroblast growth factors (bFGFs), bone morphogenetic proteins, sonic hedgehog, and Y27632] and the neurotrophins (BDNF and NT3) involved in normal AN development.2,39,41–44 Of the studies conducted using human stem cells, it has been demonstrated that human stem cell–derived neurons express combinations of several AN developmental markers, including NEUROG 1, BRN3A, ßIII-tubulin, and NFM42; PAX2/8, FOXG1, SOX1, NESTIN, and BRN3A3; BRN3A, GATA3, and PERIPHERIN2; and PAX2, BRN3A, PERIPHERIN, and NFM.44 Importantly, we have recently reported the use of an induction protocol for generating a mixed population of neurosensory progenitors from hESCs.44 Sensory neurons derived from this protocol exhibited electrophysiological properties akin to embryonic mammalian ANs45,46 and made synapses on appropriate peripheral targets in vitro.44
The use of this protocol is supported by the recent literature indicating that placodal and neural crest–derived sensory neurons share similar molecular and phenotypic properties during development.44,46,47 Given their close association during development, it is possible that the neurons derived from this heterogeneous sphere-forming assay contain auditory-like precursors.12 This study therefore aimed to use this established protocol to determine if hiPSCs could differentiate into a neurosensory-like lineage in a similar manner to that previously reported using hESCs.44,46 We chose to characterize the potential of two hiPSC lines to differentiate into a neurosensory lineage, using hESCs as a control. To our knowledge, this is the first study reporting the potential for hiPSCs to differentiate toward an auditory neuron-like lineage.
Methods
Cell lines
The iPS (foreskin) clone 1 and clone 2, abbreviated iPS1 and iPS248 (WiCell) and the hESC line H9 (WA-09, WiCell) were used in this study. For the iPS1 cell line, cells [iPS1(A) and iPS1(B)] were obtained from two different laboratories [iPS1(A): Centre for Neural Engineering, University of Melbourne; iPS1(B): Neuroregeneration Research Unit, Centre for Eye Research Australia] but were maintained and cultured using identical experimental procedures. The passage numbers for the stem cell lines used ranged from 71 to 94 [iPS1(A)]; 55 to 92 [iPS1(B)]; 33 to 48 (iPS2); and 85 to 140 (H9). All tissue culture procedures were performed using aseptic techniques in class 2 biological safety cabinets. Stem cells were maintained at 37°C, 5% CO2 and differentiated at 37°C, 10% CO2 in humidified incubators.
Neural differentiation
The protocol used to differentiate the human stem cells toward a neurosensory lineage is depicted in Figure 1. The hESC and hiPSC cell lines were cultured as colonies on a mitotically inactivated human fibroblast feeder layer in gelatin coated organ culture dishes (BD Bioscience). The cells were cultured in Knockout Serum Replacement media (1:1 DMEM:F12 with GlutaMax, Knockout Serum Replacement, 10 mM nonessential amino acids, and 55 mM ß-mercaptoethanol; all purchased from Life Technologies) supplemented with 10 μg/mL of bFGF (Peprotech). The human stem cell colonies were routinely mechanically passaged each week.
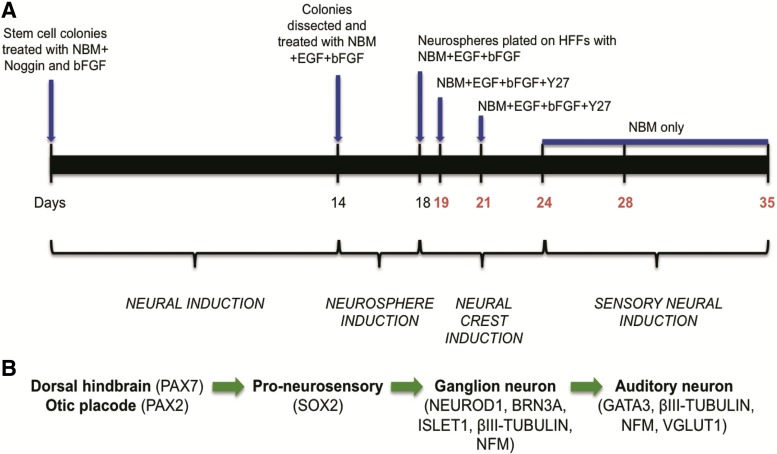
FIG. 1.
Timeline of neural differentiation and the auditory neuron developmental markers examined in vitro. (A) The hiPSCs and hESCs (control) were treated with Noggin media supplemented with bFGF for 2 weeks (neural induction). Subsequently, dissected colony pieces were treated with EGF and bFGF (neurosphere induction). The neurospheres were plated on a layer of HFF and treated with EGF and bFGF at 18 DIV, followed by treatment with Y27632 at 19 and 21 DIV (neural crest induction). From 24 DIV onward, the neurospheres were cultured in NBM only (sensory neural induction). Note: The hiPSCs and hESCs were analyzed for AN marker expression using immunochemical and qRT-PCR techniques at the time points highlighted in red. (B) The transcription factors analyzed at different stages of AN development. AN, auditory neuron; bFGF, basic fibroblast growth factor; DIV, days in vitro; EGF, epidermal growth factor; hESCs, human embryonic stem cells; HFF, human foreskin fibroblasts; hiPSCs, human induced pluripotent stem cells; NBM, neurobasal media; qRT-PCR, quantitative real time polymerase chain reaction.
To differentiate hESCs and hiPSCs toward a neural crest lineage, the stem cell colonies were transferred into Noggin media containing Neurobasal media (NBM; Neurobasal A with 1% N2, 2% B27, 2 mM l-glutamine and 0.5% penicillin/streptomycin; all purchased from Life Technologies), 500 ng/mL of Recombinant Noggin (R&D Systems) and 4 ng/mL of bFGF. The cells were cultured for 14 days without passage and Noggin media was replaced every other day. After 14 days, the Noggin media was replaced with fresh phosphate buffered saline (PBS). The colonies were mechanically cut into grids using two needles (23 gauge). The dissected pieces were washed in an organ culture dish containing NBM, epidermal growth factor (EGF; Peprotech), and bFGF (20 ng/mL each) and then transferred into a low attachment 96 well plate (1–2 pieces per well) containing NBM supplemented with EGF and bFGF (20 ng/mL each) to promote neurosphere formation. The neurospheres were maintained for a period of 4 days at 37°C with 5% CO2 in a humidified incubator.
After 18 days in vitro (DIV), the neurospheres were plated onto gelatinized organ culture dishes that contained a layer of mitotically inactivated human fibroblast feeders and NBM supplemented with EGF and bFGF (20 ng/mL each), followed by treatment with the Rho-kinase inhibitor Y27632 (25 μM; Sigma-Aldrich) at 19 and 21 DIV. From this time point onward, media was changed every other day with NBM only to promote sensory neural differentiation (24–35 DIV).
Immunocytochemistry
Neurospheres were fixed at 19–35 DIV with ice-cold 4% paraformaldehyde for 10 minutes and washed thrice in PBS (5 minutes each). The neurospheres were immunostained with the following primary antibodies: mouse α PAX7 (Developmental Hybridoma Bank; 1:20), rabbit α PAX2 (Covance; PRB-276P; 1:200), rabbit α SOX2 (Abcam; AB97959; 1:100), chicken α neurofilament-M (Millipore; AB5735; 1:1000), chicken α β-III tubulin (Millipore; AB9354; 1:1000), mouse α BRN3A (Millipore; MAB1585; 1:1000), goat α ISLET1 (Millipore; AF1837; 1:200) and rabbit α VGLUT1 (Invitrogen; 48-2400; 1:3000). The primary antibodies were prepared in primary blocking solution [0.1% Triton-X (Sigma-Aldrich) in PBS and 2% normal goat (Abacus-ALS) or donkey serum (Millipore)]. The diluted primary antibodies were added to fixed cells and stored overnight at 4°C in a humidified container. The next day, the cells were rinsed thrice in blocking solution (5 minutes each). Appropriate Alexa-Fluor labeled secondary antibodies (Life Technologies) were diluted in the secondary blocking solution [0.1% Tween (Sigma-Aldrich) in PBS and 2% goat or donkey serum] and added to the cells (200 μL per coverslip). The cells were left for 2 hours at room temperature with gentle rotation. After 2 hours, the cells were washed thrice in PBS (5 minutes each) and mounted with ProLong-Gold antifade reagent containing the nuclear stain 4′, 6-diamidino-2-phenylindole (Invitrogen). Confocal images were taken using an LSM 510 META confocal scanning laser system with a Zeiss AxioImagerZ1 microscope. Zen digital imaging software (Carl Zeiss) was used to process and analyze the images.
The immunocytochemistry results were analyzed by calculating the percentage of the total number of neurospheres that had positive immunolabeling for the markers analyzed. A minimum of eight neurospheres were analyzed for each marker and time point (19, 21, 24, 28, and 35 DIV). Statistically significant differences were evaluated using a t-test and the data presented as the mean±standard deviation from at least three independent experiments. Statistical significance was determined as follows: p<0.05 (*), p<0.01 (**), and p<0.001 (***).
Quantitative real time polymerase chain reaction
Undifferentiated and differentiating hiPSCs and hESCs were collected for quantitative real time polymerase chain reaction (qRT-PCR) at days 0 (undifferentiated), 19, 21, 24, 28, and 35. The cells were harvested by manual scraping, washed in PBS, and resuspended in RNAlater (Sigma Aldrich) for storage at −20°C. The SV Total RNA isolation kit (Promega Corporations) was used to extract total RNA from the cell samples following the manufacturer's instructions. The RNA was quantified using a NanoDrop spectrophotometer (Thermoscientific). For each sample, equal amounts of total RNA (60 ng) were reverse transcribed to single stranded cDNA using the High Capacity RNA-to-cDNA kit (Life Technologies) following the manufacturers protocol.
The qRT-PCR reaction was carried out using the TaqMan gene expression master mix (Life Technologies). The commercially available probes included PAX2 (Hs01057416_m1: FAM dye), NEUROD1 (Hs01922995_s1: FAM dye), BRN3A (POU4F1; Hs00366711_m1: FAM dye), GATA3 (Hs00231122_m1_FAM dye), and SLC17A7 (gene encoding VGLUT1; Hs00220404_m1: FAM dye), and ß-ACTIN as the housekeeper reference gene (4326315E: VIC dye, primer limited; all from Life Technologies). For each probe, a standard curve was generated from a 1:10 serial dilution of control cDNA sample. The reaction efficiencies were >97% for all the probes except for PAX2, which had a reaction efficiency of 90% (reaction efficiencies between 0.90 and 1.0 were considered acceptable, as recommended by Applied Biosystems). The experimental samples had a final cDNA concentration of 60 ng/μL and were diluted to 6 ng/μL (1:10) for the qRT-PCR experiment.
The qRT-PCR reactions were run using a RotorGene Q instrument (Qiagen) at the following thermocycler conditions: 1 cycle of 50°C for 2 minutes then 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds then 60°C for 1 minute. The reactions were run in duplex (gene of interest and housekeeping probe present in each vial). For each reaction, no template controls (water only) and reverse transcriptase negative samples (without cDNA) were included as negative controls.
The Rotor-gene 6000 software (Version 1.7, Corbitt Research) was used to analyze the data. The relative quantification of each sample was determined using the 2−ΔΔCt delta-delta analysis algorithm.49 The cycle threshold (Ct) values of the experimental samples were obtained using the thresholds from each probe's standard curves. To calculate the ΔCt value, the expression levels of the experimental samples were measured in triplicate and normalized against the endogenous control, ß-ACTIN. Next, the ΔCt values were standardized against the calibrator's ΔCt to determine the ΔΔCt value. The relative quantification was then calculated as 2−ΔΔCt. Note that for all the genes except NEUROD1, the undifferentiated cells were used as the calibrator sample. As NEUROD1 could not be detected in the undifferentiated cells, 19 DIV cells were chosen as the calibrator sample. Statistical analyses of the qRT-PCR data were performed using GraphPad Prism. The relative quantification results were analyzed using the one-way analysis of variance analysis of each gene at defined time points. A Kruskal–Wallis test was performed on the nonmatched, nonparametric data. Results are presented as the mean±standard deviation. Statistical significance was determined as follows: p<0.05 (*), p<0.01 (**), and p<0.001 (***).
In vitro electrophysiology
The stem cell–derived neurospheres and cultures of primary ANs (prepared from P4-P7 Wistar rat pups, as previously described46,50) were plated onto glass coverslips [10 mm diameter, Menzel-Glaser; precoated with polyornithine (500 μg/mL; Sigma-Aldrich) and mouse laminin (0.01 mg/mL; Invitrogen)] and grown in vitro at 37°C, 10% CO2. As previously described,44,46 coverslips were transferred to a recording chamber fitted to an AxioExaminer D1 microscope (Carl Zeiss) for electrophysiological recordings. The cells were superfused at 1–2 mL/min with a solution containing 137 mM NaCl, 5 mM KCl, 10 mM HEPES, 1 mM MgCl2, 2 mM CaCl2, 10 mM glucose (pH 7.35; 300–305 mOsmol/kg). Recordings in stem cell cultures were made from neurons with a bipolar morphology (soma diameter of ∼10 μm), between 28 and 35 DIV. Recordings were made at room temperature using borosilicate microelectrodes (2–6 MΩ; 1.0 mm O.D.; 0.58 mm I.D., Sutter) filled with an internal solution containing 115 mM K-gluconate, 10 mM HEPES, 7 mM KCl, 0.05 mM EGTA, 2 mM Na2ATP, 2 mM MgATP, and 0.5 mM Na2GTP (pH7.3; 290–295 mOsmol/kg). All chemicals were purchased from Sigma-Aldrich. Signals were recorded with a MultiClamp 700B amplifier (Molecular Devices), data acquisition system (Digidata 1440A, Molecular Devices), and AxoGraph X analysis software (AxoGraph Scientific).
Results
Both hiPSC- and hESC-derived neurosensory progenitors express auditory neural markers
The stepwise differentiation protocol and the subsequent examination of transcription factor/protein expression are shown in Figure 1. Given that the inner ear cells arise from the dorsal hindbrain region during development, we initially probed for expression of the dorsal hindbrain marker, PAX7, for gross lineage confirmation. A significant increase in the percentage of neurospheres with PAX7 immunopositive cells was observed at 24 DIV [compared with control cells at 19 DIV; iPS1(A) and iPS1(B): p<0.01, iPS2 and H9: p<0.001; Supplementary Fig. S1]. At 24 and 35 DIV, there was an increase in the expression of PAX7 relative to 19 DIV and this was consistent for all the cell lines. However, a decrease in the percentage of hESC-derived neurospheres with PAX7 positive cells was observed at 35 DIV (compared with 24 DIV), but the decrease was not statistically significant.
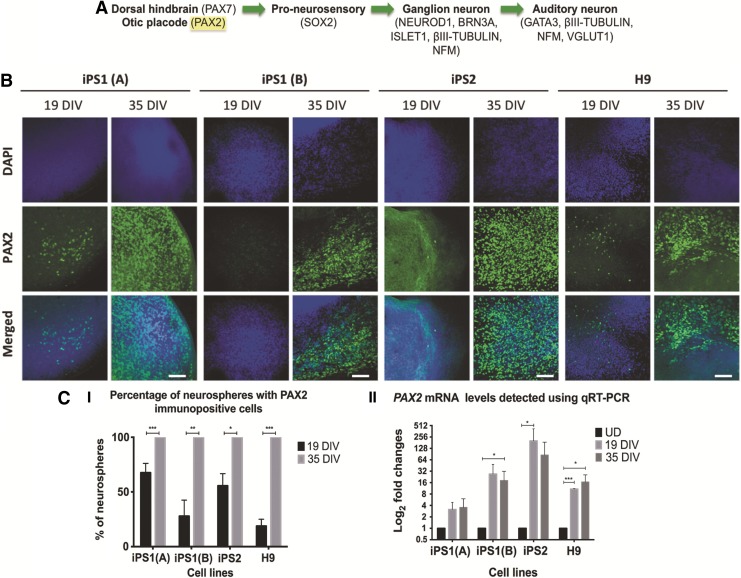
To investigate whether the hiPSC and hESC neural progenitors derived from this assay were differentiating toward an otic placodal lineage, the expression of PAX2 was also examined (Fig. 2A). Immunocytochemical results indicated that there was a significant increase in the percentages of neurospheres that had PAX2 positive cells at 35 DIV [compared with 19 DIV; iPS1(A) and H9: p<0.001, iPS1(B): p<0.01, and iPS2: p<0.05; Fig. 2B, C-I]. Notably, at 35 DIV, all neurospheres examined contained PAX2-positive cells. However, variability in the levels of PAX2 expression was noticed between the hiPSC lines (iPS1 and iPS2), between samples of the same hiPSC line [iPS1(A) and iPS1(B)], and between the hiPSCs and hESCs. To quantify the expression levels of PAX2, qRT-PCR was employed. Consistent with the immunocytochemical data, the qRT-PCR results revealed an upregulation of PAX2 in the hiPSC and hESC-derived neurosensory progenitors at 19 DIV (iPS2: p<0.05; H9: p<0.001) and 35 DIV [when compared with the undifferentiated controls; iPS1(B) and H9: p<0.05; Fig. 2C-II]. Although variabilities in the mean fold changes of PAX2 were also observed between the hiPSC lines, samples of the same hiPSC line, and between the hiPSC and hESC lines, collectively, these findings indicated that PAX2 mRNA and protein could be detected in both the hiPSC- and hESC-derived neural progenitors across different time points during differentiation.
FIG. 2.
Expression of PAX2 in the hiPSC and hESC neurospheres at 19 and 35 DIV. (A) Pax2 is expressed in otic placodal cells during the early stages of AN development. (B) Using immunostaining, an increase in the expression of PAX2 was observed at 35 DIV. Scale bar=100 μm (relative to all images). (C-I) There was a significant increase in the percentages of hiPSC and hESC neurospheres that had PAX2 immunopositive cells. Data obtained from at least three independent experiments and expressed as means±SEM of triplicates of each sample. (C-II) The qRT-PCR data show the fold changes of PAX2 in the cell lines at 19 and 35 DIV. The results were presented as fold changes relative to the endogenous control, ß-ACTIN, and the undifferentiated stem cells. Each point on the graph represents mean±SD (n=3). *p<0.05; **p<0.01; ***p<0.001. DIV, days in vitro; DAPI, 4′,6-diamidino-2-phenylindole; SD, standard deviation; SEM, standard error of the mean.
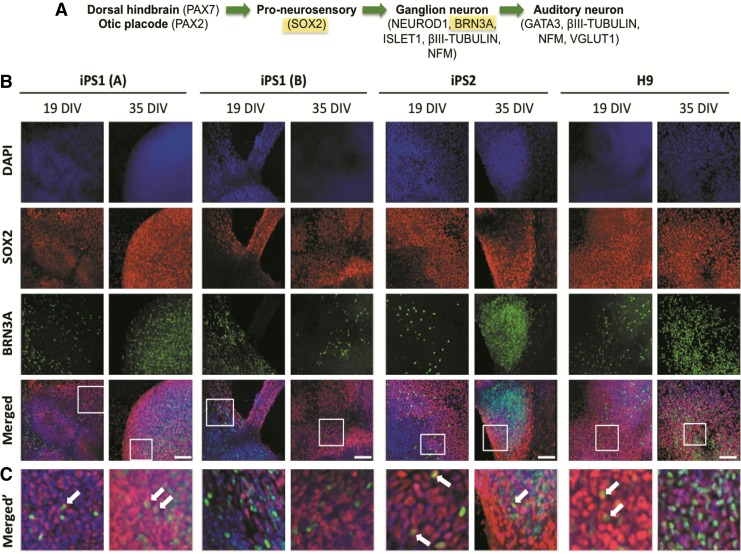
In addition to Pax2, the transcription factor Sox2 has been found to play a critical role during early AN development in promoting otic epithelial cells into a proneurosensory lineage (Fig. 3A).21,22 The coexpression of SOX2 with the sensory neural marker BRN3A was analyzed in all neurospheres at 19 and 35 DIV (Fig. 3B). We found that SOX2 was expressed in all the hiPSC and hESC neurospheres analyzed at 19 and 35 DIV. Coimmunostaining with both SOX2 and BRN3A showed subpopulations of cells that were BRN3A+/SOX2+, BRN3A−/SOX+, or BRN3A+/SOX2− (Fig. 3C). This may represent different progenitor subpopulations and/or progenitors that are at different temporal stages of differentiation.
FIG. 3.
Expression of SOX2 and BRN3A in the hiPSC and hESC neurospheres at 19 and 35 DIV. (A) SOX2 is expressed during the proneurosensory stage of AN development. (B) SOX2 immunopositive cells were observed in the hiPSC and hESC neurospheres at all the time points analyzed. (C) The box inserts of the merged images of (B) show that at 19 DIV and 35 DIV, there were some cells that were BRN3A+/SOX2+ (arrows); however, the majority were either BRN3A+/SOX2− and/or BRN3A−/SOX2+. Scale bar=100μm (all images).
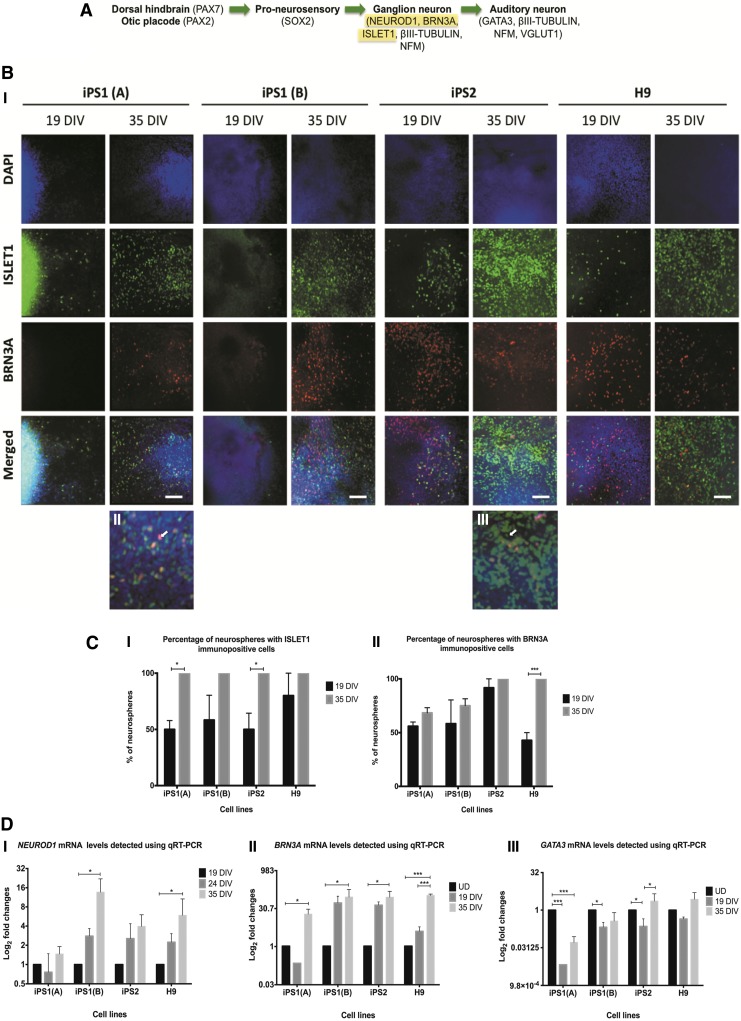
To further evaluate the potential of hiPSCs and hESCs to differentiate into an AN-like lineage, the expression of ISLET1 and BRN3A was analyzed using immunocytochemistry (Fig. 4A, B). It was found that there was an increase in the percentages of neurospheres with ISLET1 and BRN3A immunopositive cells at 35 DIV within both the hiPSC and hESC neurospheres [iPS1(A) and iPS2: p<0.05, Fig. 4C-I; H9: p<0.001, Fig. 4C-II]. The colocalization of Brn3a and Islet1 is known to be critical for sensory neural differentiation.51 Here, the presence of BRN3A+/ISLET1+immunopositive cells was noticed within the neurospheres at 35 DIV [Fig. 4B-II, B-III]. Given the differences in the relative expression levels of the sensory neural proteins between the cell lines, qRT-PCR was used to quantify the expression of NEUROD1, BRN3A, and GATA3. The qRT-PCR results revealed that NEUROD1 was upregulated in the hiPSC and hESC lines at 35 DIV [compared with the control cells at 19 DIV; iPS1(B) and H9: p<0.05; Fig. 4D-I]. Similarly, BRN3A was upregulated in the hiPSC- and hESC-derived neural progenitors at 35 DIV [compared with the undifferentiated cells; iPS1(A), iPS1(B), and iPS2: p<0.05 and H9: p<0.001; Fig. 4D-II]. Conversely, GATA3 was downregulated at 19 DIV in the hiPSC- and hESC-derived neural progenitors [compared with the undifferentiated controls; iPS1(A): p<0.001, iPS1(B) and iPS2: p<0.05; Fig. 4D-III]. However, at 35 DIV, GATA3 was significantly upregulated in the iPS2-derived neural progenitors (compared with the expression at 19 DIV; p<0.05). Consistent with the immunocytochemistry, there were differences in the fold changes of NEUROD1, BRN3A, and GATA3 between the hiPSC and hESC lines over the time course examined. Overall, the trend of AN marker expression remained similar between the hiPSC and hESC lines, but there were variabilities in the mRNA and protein expression levels of these markers between the cell lines.
FIG. 4.
Expression of NEUROD1, ISLET1, BRN3A, and GATA3 in the hiPSC and hESC neurospheres at 19 and 35 DIV. (A) NeuroD1, Islet1 and Brn3a are expressed during the ganglion neuron stage of AN development and Gata3 is expressed in ANs. (B-I) Using immunostaining, an increase in the expression of ISLET1 and BRN3A was observed at 35 DIV. Scale bar=100 μm (relative to all images). (B-II, B-III) Notably, there were several ISLET1 and BRN3A immunopositive cells at 35 DIV. (C-I, C-II) There was a significant increase in the percentages of hiPSC and hESC neurospheres that had ISLET1 and BRN3A immunopositive cells. Data obtained from at least three independent experiments and expressed as means±SEM of triplicates of each sample. (D-I) The qRT-PCR data show the fold changes of NEUROD1 in the cell lines at 24 and 35 DIV relative to the control cells at 19 DIV. (D-II) The qRT-PCR results indicate the fold changes of BRN3A in the cell lines at 19 and 35 DIV relative to the undifferentiated controls. (D-III) The fold changes of GATA3 in the cell lines at 19 and 35 DIV. The qRT-PCR results were presented as fold changes relative to the endogenous control, ß-ACTIN. Each point on the graph represents mean±SD (n=3). *p<0.05; ***p<0.001.
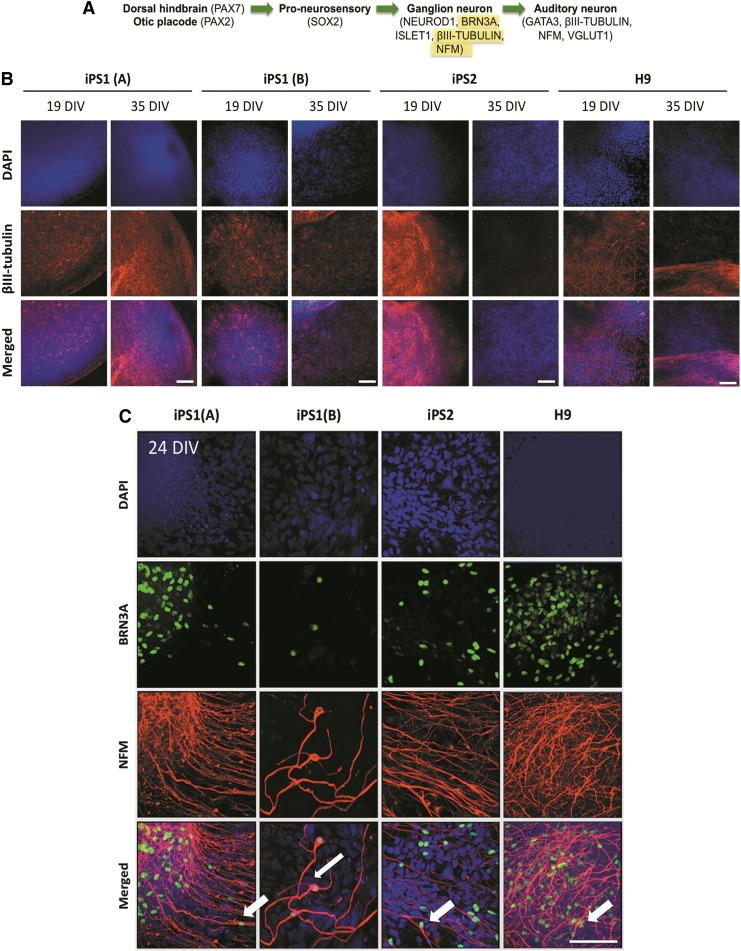
As expected, the neural specific markers ßIII-tubulin and NFM were expressed in both the hiPSC and hESC neurospheres throughout the differentiation timeline. Interestingly, a decrease in the expression levels of ßIII-tubulin was noticed at the later time points of differentiation within all cell lines (35 DIV; Fig. 5B). The decrease in ßIII-tubulin expression was most pronounced in the iPS2- and hESC-derived neurospheres. Furthermore, the presence of bipolar neurons (as determined by positive staining for BRN3A and NFM) emanating from the periphery of both the hiPSC and hESC neurospheres were observed as early as 24 DIV (Fig. 5C), supporting the differentiation to a relevant phenotype for AN replacement.
FIG. 5.
Expression of ßIII-tubulin and NFM in the hiPSC- and hESC-derived neurospheres. (A) The neural specific markers ßIII-tubulin and NFM are expressed in the ganglion neuron and at AN stages of development. (B) Both the hiPSC and hESC neurospheres showed some level of ßIII-tubulin expression over the time course examined. Note that at 35 DIV, the levels of ßIII-tubulin had declined in the hiPSC line iPS2 and hESC line H9 cell lines (arrows). Scale bars=100μm (relative to all images). (C) Bipolar BRN3A+/NFM+ neurons were observed from as early as 24 DIV in the hiPSC and hESC-derived neural progenitors (arrows). Scale bar=50 μm (relative to all images). NFM, Neurofilament kDa 160.
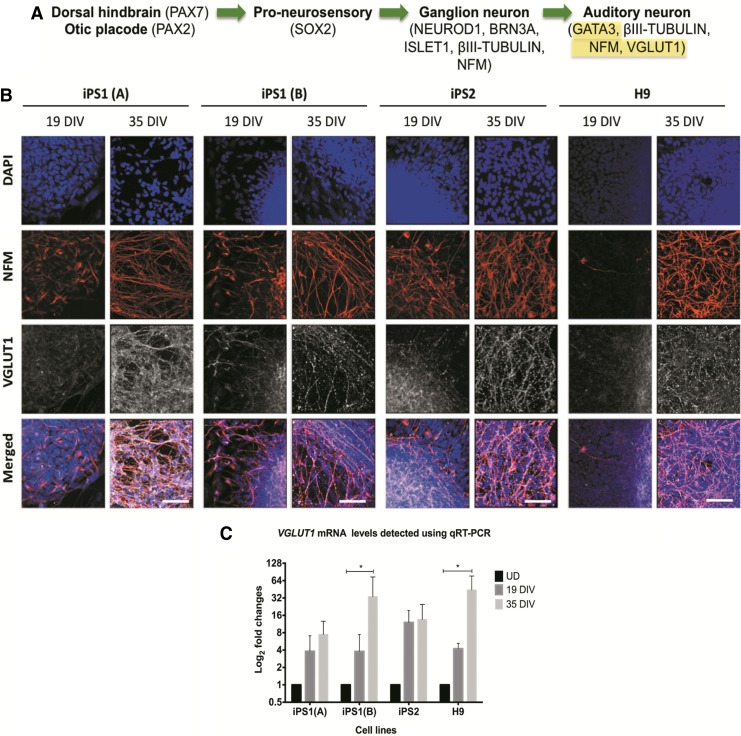
Given the glutamatergic phenotype of ANs, the temporal expression of the vesicular glutamate transporter, VGLUT1, was also analyzed in the hiPSC and hESC lines (Fig. 6A). Immunocytochemistry illustrated numerous VGLUT1-positive puncta on NFM positive neurons, in both the hiPSC and hESC neurospheres (Fig. 6B). Furthermore, by 21 DIV, all of the hESC and hiPSC neurospheres showed expression of VGLUT1 (data not shown). Accordingly, the qRT-PCR results revealed an upregulation of the gene encoding VGLUT1 (SLC17A7) at 35 DIV [when compared with the undifferentiated controls; iPS1(B) and H9: p<0.05; Fig. 6C]. However, variabilities in the fold changes of VGLUT1 were also observed between the hiPSC lines, samples of the same hiPSC line, and between the hiPSC and hESC lines. For instance, at 35 DIV, the range in fold changes of VGLUT1 expression was between 7.47 and 44 [compared with the undifferentiated controls; iPS1(A): 7.47, iPS1(B): 33.7, iPS2: 13.6, and H9: 44]. In summary, these data indicate that VGLUT1 could be detected in both the hiPSC and hESC-derived neural progenitors. However, variabilities in the absolute levels of VGLUT1 expression were found between the cell lines.
FIG. 6.
Expression of VGLUT1 in the hiPSC and hESC neurospheres at 19 and 35 DIV. (A) VGLUT1 is expressed in developing ANs. (B) Using immunostaining, an increase in the expression of VGLUT1 was observed at 35 DIV. Scale bar=50μm (relative to all images). (C) The qRT-PCR data show the fold changes of the gene encoding VGLUT1 (SLC17A7) at 19 and 35 DIV. The results are presented as fold changes relative to the endogenous control ß-ACTIN and the undifferentiated controls. Each point on the graph represents mean±SD (n=3). *p<0.05; ***p<0.001.
The hiPSC and hESC-derived neurons were electrophysiologically active
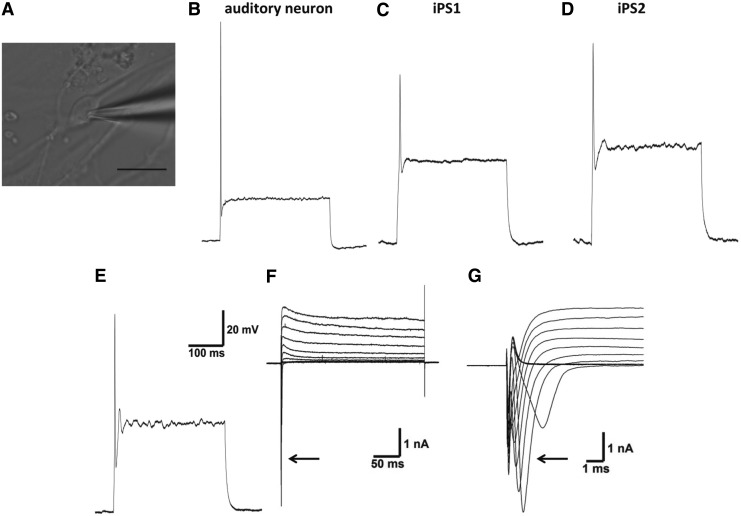
The physiological function of stem cell–derived neurons was assessed using whole cell patch clamp electrophysiological recordings (Fig. 7A). Similar to the electrical profile of early postnatal ANs, the typical pattern seen in response to membrane depolarization was a single action potential for both the hiPSC- and hESC-derived neurons (Fig. 7B, E). Voltage clamp recordings in all populations revealed a transient inward current at stimulus onset followed by a sustained outward current during membrane depolarization (Fig. 7F). The inward current was abolished in the presence of tetrodotoxin (a sodium channel blocker; Fig. 7F), while the outward component of the current could be attenuated with the addition of the potassium channel blockers tetraethylammonium and 4-aminopyridine (Fig. 7G). These findings illustrate the presence of both sodium and potassium currents (INa and IK respectively) in these populations of stem cell–derived sensory neurons.
FIG. 7.
Electrophysiological properties of hiPSC- and hESC-derived neurons. (A) Bipolar iPS2 neuron with recording microelectrode attached (Scale bar=20μm). Typical current-clamp recordings from early postnatal auditory neurons cultured in vitro (B), iPS1 (C), iPS2 (D), and H9-hESC (E) neurons. (F, G) Voltage-clamp recordings of stem cell-derived neurons reveal transient tetrodotoxin-sensitive sodium current (arrow) followed by a sustained outward potassium current evoked by membrane depolarization.
Discussion
The development of a stem cell–based therapy for deafness requires overcoming several major challenges, one being to successfully differentiate stem cells to an appropriate neurosensory lineage. It has previously been reported that exposing hESCs to human feeders and a combination of EGF, bFGF, and Y27632 prompts their differentiation into electrically active neurosensory progenitors in vitro.46 Here we aimed to determine whether hiPSCs would behave in a similar manner. It was found that the neural induction protocol used was efficient at directing hiPSCs toward a neurosensory-like lineage in vitro, confirmed by the expression of a cohort of relevant sensory neural mRNA and proteins throughout the differentiation timeline. Notably, the highest expression levels of the sensory neural markers were observed at 35 DIV, but there were differences in the absolute expression levels between the cell lines.
Human iPSCs have the potential to differentiate into a neurosensory lineage in vitro
We analyzed the expression of a cohort of AN developmental markers at different stages of differentiation using a combination of immunocytochemical and qRT-PCR techniques. To assess if the neural assay was producing dorsal, otic progenitors, expression of the dorsal hindbrain marker PAX7 and the otic placodal marker PAX2 were initially examined. The peak expression of PAX7 was observed at 35 DIV in the hiPSC neurospheres and 24 DIV in the hESC neurospheres (relative to 19 DIV), hence confirming gross lineage specification. Given that PAX7 is an early neural marker, its expression at the later time points of differentiation was surprising. However, it must be noted that this neural assay was generating a heterogeneous population of neural phenotypes at different temporal stages of differentiation; therefore, it is probable that it was producing other neural lineages (in addition to dorsal hindbrain lineages) that have later onset and persistent PAX7 expression. Furthermore, the decrease in the percentage of hESC-derived neurospheres that had PAX7 expression observed at 35 DIV is likely to reflect the specification and differentiation of the cells into more mature lineages. It has previously been reported that Pax2 has a critical role in the differentiation of ANs and their peripheral innervation in situ.16 Furthermore, Pax2 animal knockout models have reported a complete loss of ANs,14–16 clearly demonstrating the importance of this transcription factor for normal AN development. Our findings indicated that at 35 DIV, all of the neurospheres analyzed had PAX2 immunopositive cells. Consistently, qRT-PCR revealed an upregulation of PAX2 mRNA in the hiPSC and hESC lines at 35 DIV, relative to the undifferentiated cells. Overall, these results suggest that the neural populations generated from this neural assay contained correctly specified dorsal, otic progenitors.
All inner ear cells, including the cochlear hair cells, supporting cells, and sensory neurons, arise from a common neurosensory domain within the otic vesicle. The neurosensory domain of the otic vesicle can be recognized very early in development by the expression of Sox2.20 It has been reported that Sox2 is expressed in both neuronal and sensory progenitors but downregulated in differentiated hair cells and neurons.20–22 Our observations that SOX2 was highly expressed in the hiPSC and hESC-derived neurospheres throughout the differentiation timeline indicated that the neural progenitors were being specified toward a proneurosensory lineage. Furthermore, the mutual exclusivity of BRN3A and SOX2 in some cells analyzed at 19 and 35 DIV suggests that SOX2 might have been downregulated in the differentiated BRN3A positive neurons, consistent with their expression pattern during AN development. Collectively, it is evident that the sphere-forming neural differentiation assay used contained a portion of correctly specified neurosensory progenitors.
The basic helix-loop-helix transcription factors Neurog1 and NeuroD1 and the homeodomain transcription factors Brn3a and Islet1 have been found to be critical for sensory neural differentiation.51 The lack of any one of these factors has been demonstrated to impact the maturity and functionality of the sensory dorsal root ganglion neurons.51 In the cochlea, knockout of NeuroD1 or Brn3a has been shown to significantly reduce the number of ANs in mice.25–30 Furthermore, Islet1 has been indicated to have an important role in specifying sensory and neuronal lineages in the developing inner ear.52 The increase in the expression and number of cells immunopositive for ISLET1 and BRN3A protein in both the hiPSCs and hESCs at 35 DIV, indicate that the cells were differentiating into a neurosensory-like lineage. These findings were supported by the qRT-PCR data, where it was found that the sensory neural markers NEUROD1 and BRN3A were upregulated in the hiPSCs and hESCs at 35 DIV (when compared with the undifferentiated or 19 DIV controls). It was evident from these findings that a higher number of cells were differentiating into a more mature neurosensory-like lineage by 35 DIV.
The AN marker Gata3 has been found to be a critical factor involved in AN specification.31,33,53 Furthermore, in Gata3 conditional knockout mice, a significant reduction in the number of ANs in the inner ear has been reported.54,55 Using qRT-PCR, we detected GATA3 mRNA in undifferentiated hiPSCs and hESCs. The presence of GATA3 in undifferentiated human stem cells has previously been reported.56 Relative to the undifferentiated controls, GATA3 was downregulated at 19 DIV in the stem cell–derived neural progenitors, which was not unexpected given that GATA3 is also a mesodermal lineage specifier. However, at 35 DIV, an overall trend of increasing GATA3 expression was detected, particularly in the iPS2 cell line. Given that Gata3 is expressed at a more mature stage of AN development,31,33 it is plausible that the increase in expression observed at 35 DIV was due to the stem cell–derived neural progenitors achieving a more mature AN-like phenotype.
The type I afferent ANs have several characteristic traits, among them, their bipolar nature and glutamatergic phenotype. Here, we found that both the hiPSCs and hESC neurospheres contained bipolar BRN3A+/NFM+ neurons from as early as 24 DIV through to the latest time point examined (35 DIV). It has previously been indicated that mouse iPSCs transplanted into the cochlea differentiate into glutamatergic neurons (evidenced by their expression of VGLUT1), but the percentage of cells that acquired this lineage was considerably low (2.3%).10 Our data revealed an overall increase in the expression of VGLUT1 over the time course examined, with the highest expression observed at 35 DIV, hence suggesting that the cells were being specified into a glutamatergic neuronal lineage. These findings are particularly promising when considered in terms of this recent data, since bipolar and glutamatergic characteristics are key attributes for a replacement AN derived from a patient-specific source. Furthermore, given the evidence that glutamate plays a key role in the formation of peripheral synapses in the cochlea,57–59 it is possible that transplantation of a glutamatergic neuronal population would facilitate synaptogenesis in the cochlea.
Human iPSC-derived neurosensory progenitors are electrically active
In order for hiPSCs to replace ANs, they must be capable of faithfully sending electrical signals from the periphery to the brainstem. We have previously demonstrated that hESC-derived neurons obtained from this differentiation assay are capable of firing action potentials in response to depolarization and have a phasic firing pattern, similar to the profile of ANs cultured in vitro.44,46 Furthermore, the electrical profiles of the hESC-derived neurons were found to resemble those of embryonic ANs cultured in vitro.45 More specifically, the action potentials generated from hESCs were broader and exhibited higher latencies and firing thresholds, when compared to the early postnatal ANs.46 Here, we demonstrated that neurons derived from the iPS1 and iPS2 lines were also capable of firing action potentials and exhibited a phasic profile of activity. These findings were consistent with that previously reported for stem cell–derived ANs generated using the current neural assay,44,46 other differentiation paradigms,39,42 and also early post-natal ANs.50 Promisingly, these findings indicate that hiPSC lines can be differentiated into appropriate neurosensory progenitors that are similarly functionally active.
Neurosensory marker expression differed between hESC and hiPSC-derived neural progenitors
Immunocytochemical and qRT-PCR results demonstrated that all hiPSC and hESC lines analyzed produced functional neurosensory-like progenitors. However, variabilities in the levels of marker expression were observed between hiPSC lines and within samples of the same cell line, when compared with the hESC controls. Consistent with our findings, there is now accumulating evidence suggesting that hiPSCs have greater variability in their differentiation potentials when compared with their hESC counterparts.60–63 Several factors may account for these variabilities. In addition to the intrinsic differences that exist between human stem cell lines, it should be noted that hESCs and hiPSCs differ in their source (embryonic versus somatic cell) and derivation (direct isolation versus reprogramming).64 Furthermore, the two hiPSC lines used in this study were generated in the same manner, by lentiviral transfection of four transcription factors, OCT4, SOX2, NANOG, and LIN28 into skin fibroblasts.48 It has previously been reported that hiPSCs generated using the viral transfection method contain some genetic and epigenetic mutations due to genomic integration of the reprogramming transcription factors.65,66 It is plausible that these abnormalities influence their differentiation efficiency and even predispose them to differentiate into specific sublineages. Although it is difficult to conclusively determine the exact cause of the variabilities observed between the hiPSC and hESC lines, it is possible that a combination of intrinsic cell line specific variabilities and/or the presence of genomic and epigenetic instabilities in the hiPSCs may be significant contributing factors.
The variabilities observed between hiPSC lines in this study bring to light some of the challenges that may arise when using patient-specific hiPSC lines. For instance, variances may occur in the number of ANs generated from each individual and the functional integration capacity of these cells in vivo. Notably, in a recent study, it was reported that hESC-derived neural progenitors could integrate into the deaf gerbil cochlea and restore auditory function 10 weeks post transplantation.3 However, the degree of auditory brainstem response recovery between all the stem cell transplanted animals ranged from <10 dB to a complete restoration of ∼60 dB sound pressure level. In terms of translating this therapy into clinic, it would be undesirable if there were significant variances in the functional outcomes patients' achieve with the treatment. Therefore it is necessary that future studies address these variabilities prior to the clinical application of this therapy.
The presence of genomic abnormalities that may have contributed to the differentiation variabilities observed in hiPSCs has been implicated to increase their disposition to form teratomas in vivo.67 Notably, Nishimura and colleagues reported the generation of teratomas following transplantation of mouse iPSC-derived neurons into rat cochleae.11 The authors attributed the formation of teratomas in vivo to the use of iPSC lines generated using the viral transfection method. These findings highlight the necessity to explore the use of other autologous stem cell types including bone marrow- or nasal-derived mesenchymal stem cells. Whilst these stem cell types do not harbor reprogramming-induced genomic abnormalities, they have a more restricted self-renewal and differentiation capacity compared with iPSCs. Future studies using hiPSC lines generated using nonviral integration methods accompanied by regular assessment of their genomic status may overcome the risks associated with the use of hiPSCs for cell transplantation therapies.68
In conclusion, the results presented demonstrate that the described neurosensory induction protocol was capable of generating a population of appropriate bipolar hiPSC-derived neurosensory progenitors that are functionally active. These results provide the basis for future experimentation focused on assessing the survival and functional integration capacity of hiPSC-derived neurosensory progenitors in deaf animal models and represent the first step in bringing stem cells closer to clinical transplantation in the deaf cochlea. Furthermore, this neural assay could also be used as a model to interrogate the expression and interactions of other critical transcription factors involved in AN development.
Supplementary Material
Abbreviations Used
- AN
auditory neuron
- bFGF
basic fibroblast growth factor
- DIV
days in vitro
- EGF
epidermal growth factor
- hESCs
human embryonic stem cells
- hiPSCs
human induced pluripotent stem cells
- NBM
neurobasal media
- NFM
Neurofilament kDa 160
- qRT-PCR
quantitative real time polymerase chain reaction
- SD
standard deviation
Acknowledgments
We gratefully acknowledge financial support from The Garnett Passe and Rodney Williams Memorial Foundation Postgraduate Research Scholarship; NHMRC Project Grant (APP1023372); Operational Infrastructure Support from the Victorian Government; and The University of Melbourne Departments of Otolaryngology and Audiology and Speech Pathology.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Corrales CE, Pan L, Li H, et al. Engraftment and differentiation of embryonic stem cell–derived neural progenitor cells in the cochlear nerve trunk: growth of processes into the organ of corti. J Neurobiol. 2006;66:1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi F, Corrales CE, Liberman MC, et al. BMP4 induction of sensory neurons from human embryonic stem cells and reinnervation of sensory epithelium. Eur J Neurosci. 2007;26:3016–3023 [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Jongkamonwiwat N, Abbas L, et al. Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature. 2012;490:278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bas E, Van De Water TR, Lumbreras V, et al. Adult human nasal mesenchymal-like stem cells restore cochlear spiral ganglion neurons after experimental lesion. Stem Cells Dev. 2014;23:502–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo T, Johnson SA, Yoder MC, et al. Sonic hedgehog and retinoic acid synergistically promote sensory fate specification from bone marrow-derived pluripotent stem cells. Proc Natl Acad Sci U S A. 2005;102:4789–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuoka AJ, Kondo T, Miyamoto RT, et al. In vivo and in vitro characterization of bone marrow-derived stem cells in the cochlea. Laryngoscope. 2006;116:1363–1367 [DOI] [PubMed] [Google Scholar]
- 7.Matsuoka AJ, Kondo T, Miyamoto RT, et al. Enhanced survival of bone-marrow-derived pluripotent stem cells in an animal model of auditory neuropathy. Laryngoscope. 2007;117:1629–1635 [DOI] [PubMed] [Google Scholar]
- 8.Kondo T, Matsuoka AJ, Shimomura A, et al. Wnt signaling promotes neuronal differentiation from mesenchymal stem cells through activation of Tlx3. Stem Cells. 2011;29:836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duran Alonso MB, Feijoo-Redondo A, Conde de Felipe M, et al. Generation of inner ear sensory cells from bone marrow-derived human mesenchymal stem cells. Regen Med. 2012;7:769–783 [DOI] [PubMed] [Google Scholar]
- 10.Nishimura K, Nakagawa T, Ono T, et al. Transplantation of mouse induced pluripotent stem cells into the cochlea. Neuroreport. 2009;20:1250–1254 [DOI] [PubMed] [Google Scholar]
- 11.Nishimura K, Nakagawa T, Sakamoto T, et al. Fates of murine pluripotent stem cell-derived neural progenitors following transplantation into mouse cochleae. Cell Transplant. 2012; 21:763–771 [DOI] [PubMed] [Google Scholar]
- 12.Gunewardene N, Dottori M, Nayagam BA. The convergence of cochlear implantation with induced pluripotent stem cell therapy. Stem Cell Rev. 2012;8:741–754 [DOI] [PubMed] [Google Scholar]
- 13.Rubel EW. Ontogeny of structure and function in the vertebrate auditory system. In: Handbook of Sensory Physiology. Jacobsen M. (ed). Springer-Verlag: New York; pp. 135–237; 1978 [Google Scholar]
- 14.Torres M, Gomez-Pardo E, Gruss P. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development. 1996;122:3381–3391 [DOI] [PubMed] [Google Scholar]
- 15.Burton Q, Cole LK, Mulheisen M, et al. The role of Pax2 in mouse inner ear development. Dev Biol. 2004;272:161–175 [DOI] [PubMed] [Google Scholar]
- 16.Bouchard M, de Caprona D, Busslinger M, et al. Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev Biol. 2010;10:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman RA, Makmura L, Biesiada E, et al. Eya1 acts upstream of Tbx1, Neurogenin 1, NeuroD and the neurotrophins BDNF and NT-3 during inner ear development. Mech Dev. 2005;122:625–634 [DOI] [PubMed] [Google Scholar]
- 18.Ahmed M, Xu J, Xu PX. EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development. 2012;139:1965–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed M, Wong EY, Sun J, et al. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012;22:377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiernan AE, Pelling AL, Leung KK, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035 [DOI] [PubMed] [Google Scholar]
- 21.Dabdoub A, Puligilla C, Jones JM, et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A. 2008;105:18396–18401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puligilla C, Dabdoub A, Brenowitz SD, et al. Sox2 induces neuronal formation in the developing mammalian cochlea. J Neurosci. 2010;30:714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Q, Chen Z, del Barco Barrantes I, et al. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482 [DOI] [PubMed] [Google Scholar]
- 24.Lawoko-Kerali G, Rivolta MN, Lawlor P, et al. GATA3 and NeuroD distinguish auditory and vestibular neurons during development of the mammalian inner ear. Mech Dev. 2004;121:287–299 [DOI] [PubMed] [Google Scholar]
- 25.Kim WY, Fritzsch B, Serls A, et al. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jahan I, Kersigo J, Pan N, et al. Neurod1 regulates survival and formation of connections in mouse ear and brain. Cell Tissue Res. 2010;341:95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jahan I, Pan N, Kersigo J, et al. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PLoS One. 2010;5:e11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McEvilly RJ, Erkman L, Luo L, et al. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature. 1996;384:574–577 [DOI] [PubMed] [Google Scholar]
- 29.Erkman L, McEvilly RJ, Luo L, et al. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606 [DOI] [PubMed] [Google Scholar]
- 30.Huang EJ, Liu W, Fritzsch B, et al. Brn3a is a transcriptional regulator of soma size, target field innervation and axon pathfinding of inner ear sensory neurons. Development. 2001;128:2421–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karis A, Pata I, van Doorninck JH, et al. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J Comp Neurol. 2001;429:615–630 [DOI] [PubMed] [Google Scholar]
- 32.Appler JM, Goodrich LV. Connecting the ear to the brain: molecular mechanisms of auditory circuit assembly. Prog Neurobiol. 2011;93:488–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duncan JS, Fritzsch B. Continued expression of GATA3 is necessary for cochlear neurosensory development. PLoS One. 2013;8:e62046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schimmang T. Transcription factors that control inner ear development and their potential for transdifferentiation and reprogramming. Hear Res. 2013;297:84–90 [DOI] [PubMed] [Google Scholar]
- 35.Zine A, Löwenheim H, Fritzsch B. Toward translating molecular ear development to generate hair cells from stem cells. In: Adult Stem Cells. Turksen K. (ed). Springer Science: New York, pp. 111–161; 2014 [Google Scholar]
- 36.Fritzsch B, Barbacid M, Silos-Santiago I. The combined effects of trkB and trkC mutations on the innervation of the inner ear. Int J Dev Neurosci. 1998;16:493–505 [DOI] [PubMed] [Google Scholar]
- 37.Li H, Roblin G, Liu H, et al. Generation of hair cells by stepwise differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2003;100:13495–13500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshima K, Shin K, Diensthuber M, et al. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141:704–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W, Jongkamonwiwat N, Abbas L, et al. Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature. 2012;490:278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koehler KR, Mikosz AM, Molosh AI, et al. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500:217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyes JH, O'Shea KS, Wys NL, et al. Glutamatergic neuronal differentiation of mouse embryonic stem cells after transient expression of neurogenin 1 and treatment with BDNF and GDNF: in vitro and in vivo studies. J Neurosci. 2008;28:12622–12631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen W, Johnson SL, Marcotti W, et al. Human fetal auditory stem cells can be expanded in vitro and differentiate into functional auditory neurons and hair cell-like cells. Stem Cells. 2009;27:1196–204 [DOI] [PubMed] [Google Scholar]
- 43.Nayagam BA, Minter RL. A comparison of in vitro treatments for directing stem cells toward a sensory neural fate. Am J Otolaryngol. 2012;33:37–46 [DOI] [PubMed] [Google Scholar]
- 44.Nayagam BA, Edge AS, Needham K, et al. An in vitro model of developmental synaptogenesis using cocultures of human neural progenitors and cochlear explants. Stem Cells Dev. 2013;22:901–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marrs GS, Spirou GA. Embryonic assembly of auditory circuits: spiral ganglion and brainstem. J Physiol. 2012;590:2391–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Needham K, Hyakumura T, Gunewardene N, et al. Electrophysiological properties of neurosensory progenitors derived from human embryonic stem cells. Stem Cell Res. 2014;12:241–249 [DOI] [PubMed] [Google Scholar]
- 47.Huisman MA, Rivolta MN. Neural crest stem cells and their potential application in a therapy for deafness. Front Biosci (Schol Ed). 2012;4:121–32 [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 50.Needham K, Nayagam BA, Minter RL, et al. Combined application of brain-derived neurotrophic factor and neurotrophin-3 and its impact on spiral ganglion neuron firing properties and hyperpolarization-activated currents. Hearing Res. 2012;291:1–14 [DOI] [PubMed] [Google Scholar]
- 51.Dykes IM, Tempest L, Lee SI, et al. Brn3a and Islet1 act epistatically to regulate the gene expression program of sensory differentiation. J Neurosci. 2011;31:9789–9799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radde-Gallwitz K, Pan L, Gan L, et al. Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. J Comp Neurol. 2004;477:412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duncan JS, Lim KC, Engel JD, et al. Limited inner ear morphogenesis and neurosensory development are possible in the absence of GATA3. Int J Dev Biol. 2011;55:297–303 [DOI] [PubMed] [Google Scholar]
- 54.Appler JM, Lu CC, Druckenbrod NR, et al. Gata3 is a critical regulator of cochlear wiring. J Neurosci. 2013;33:3679–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo XJ, Deng M, Xie X, et al. GATA3 controls the specification of prosensory domain and neuronal survival in the mouse cochlea. Hum Mol Genet. 2013;22:3609–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levenberg S, Golub JS, Amit M, et al. Endothelial cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2002;99:4391–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seal RP, Akil O, Yi E, et al. Sensorineural deafness and seizures in mice lacking vesicular glutamate transporter 3. Neuron. 2008;57:263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruel J, Emery S, Nouvian R, et al. Impairment of SLC17A8 encoding vesicular glutamate transporter-3, VGLUT3, underlies nonsyndromic deafness DFNA25 and inner hair cell dysfunction in null mice. Am J Hum Genet. 2008;83:278–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong M, Brugeaud A, Edge AS. Regenerated synapses between postnatal hair cells and auditory neurons. J Assoc Res Otolaryngol. 2013;14:321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karumbayaram S, Novitch BG, Patterson M, et al. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells. 2009;27:806–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Q, Lu SJ, Klimanskaya I, et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712 [DOI] [PubMed] [Google Scholar]
- 62.Hu BY, Weick JP, Yu JP, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107:4335–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koyanagi-Aoi M, Ohnuki M, Takahashi K, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci U S A. 2013;110:20569–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martins-Taylor K, Xu RH. Concise review: genomic stability of human induced pluripotent stem cells. Stem Cells. 2012;30:22–27 [DOI] [PubMed] [Google Scholar]
- 65.Marchetto MC, Brennand KJ, Boyer LF, et al. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum Mol Genet. 2011;20:R109–R115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruiz S, Diep D, Gore A, et al. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:16196–16201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu X, Xu Y. Challenges to the clinical application of pluripotent stem cells: towards genomic and functional stability. Genome Med. 2012;4:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lowry WE, Quan WL. Roadblocks en route to the clinical application of induced pluripotent stem cells. J Cell Sci. 2010;123:643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.