Abstract
A hormetic response is characterized by an opposite effect in small and large doses of chemical exposure, often resulting in seemingly beneficial effects at low doses. Here, we examined the potential mechanisms underlying the hormetic response of Daphnia magna to the energetic trinitrotoluene (TNT). Daphnia magna were exposed to TNT for 21 days and a significant increase in adult length and number of neonates was identified at low concentrations (0.002 – 0.22 mg/L TNT) while toxic effects were identified at high concentrations (0.97 mg/L TNT and above). Microarray analysis of D. magna exposed to 0.004, 0.12, and 1.85 mg/L TNT identified effects on lipid metabolism as a potential mechanism underlying hormetic effects. Lipidomic analysis of exposed D. magna supported the hypothesis that TNT exposure affected lipid and fatty acid metabolism, showing that hormetic effects could be related to changes in polyunsaturated fatty acids known to be involved in Daphnia growth and reproduction. Our results show that Daphnia exposed to low levels of TNT presented hormetic growth and reproduction enhancement while higher TNT concentrations had an opposite effect. Our results also show how a systems approach can help elucidate potential mechanisms of action and adverse outcomes.
INTRODUCTION
Hormesis is a dose response phenomenon characterized by a stimulation of an organismal response (e.g., growth, reproduction) at low doses of a chemical, usually with a maximum stimulation of 30–60% over controls, and inhibition of the response (toxicity) at a higher dose 1. This phenomenon has been observed across a wide range of contaminants and classes of organisms 2. Hormesis is thought to be overcompensation to an alteration in homeostasis. That is, an organism more than compensates for an initial disruption and/or damage caused by a chemical insult, leading to a net stimulatory (i.e., hormetic) response. This phenomenon challenges the paradigm that a chemical is not active in biologically meaningful ways below its toxicity threshold 1. Because the vast majority of toxicological experimental designs are focused on treatment concentrations well above the no effect level, low-dose stimulatory responses are often not explored or reported.
Hormetic responses to energetic compounds have been previously reported, but to our knowledge, the specific mechanism(s) of action of such responses have not been explored. In terrestrial systems, hormetic responses to energetic compounds have been observed in invertebrates and plants. Energetic compounds are compounds with a high amount of stored chemical energy that can be released such as explosives, pyrotechnics, propellants or fuels. These compounds, including TNT, have been reported to contaminate land, water and retired ammunition plants. Their released into the environment can happen during several stages of production, storage, transport, usage, and final dispersal or disposal. Energetic compounds such as TNT have been detected in surface water of numerous military installations, at concentrations as high as 3.38 mg/L for TNT 3. These compounds undergo extensive transformation in aquatic systems. Therefore, aquatic organisms may be exposed not only to energetic compounds released to the environment but also to their transformation products 3. Stimulation of juvenile production in the potworm (Enchytraeus crypticus) has been observed upon exposure to TNT 4 and 1,3,5-trinitrobenzene (TNB) 5. In aquatic systems, hormetic responses to energetic compounds have been observed in algae, daphnids, and fish 6. Bailey et al. 7 reported a significant increase in D. magna reproduction in sublethal exposures to 2,4,6-trinitrotoluene (TNT). Bentley et al. 8 saw an increase in egg production in fathead minnows exposed to hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX).
Observations of apparent beneficial effects can confound hazard and risk assessments of a chemical. A mechanistic understanding of low concentration effects of energetic compounds will enable more accurate predictions of the effects and environmental risks of these chemicals in the environment. The water flea Daphnia magna is presumably the invertebrate species most used in ecotoxicology and together with its close relative D. pulex is used as a model for environmental genomics research 9. The purpose of the present study was to investigate the mechanism underlying hormetic responses in D. magna exposed to the energetic compound TNT. To do so, D. magna were exposed to different TNT concentrations for 21 days and effects on length and reproduction were measured. We also measured effects on gene expression to generate a hypothesis to elucidate potential hormetic mechanisms. We then measured lipid levels in order to test the hypothesis that lipid metabolism was involved in the hormetic response, as suggested by gene expression analysis.
EXPERIMENTAL
The 2,4,6-trinitrotoluene (TNT) was obtained from an internal U.S. Army Engineer Research and Development Center (ERDC) stock. TNT was added to moderately hard reconstituted water made according to U.S. EPA 10 methods using methanol (≤ 0.5 ml methanol/L water) as a solvent carrier. All D. magna endpoint comparisons were made to a methanol solvent control.
Range finding D. magna 21-day bioassays
Two range finding 21-d D. magna chronic toxicity tests (range finder A and range finder B) designed to identify the hormetic range of D. magna responses to TNT were carried out in basic accordance with U.S. EPA 11 guidance. Daphnia magna younger than 24 h old and obtained from in-house cultures at ERDC (Vicksburg, MS) were used to initiate the bioassay. Test containers were 50 ml beakers with a test solution volume of 40 ml. These were static renewal tests, with renewals being performed three times a week. One D. magna was addd per test chamber and 10 test chambers per treatment level. Daphnia magna were fed 0.48 ml per day of a 1:1 mixture of the green alga P. subcapitata and YCT, a mixture of yeast, ground cereal leaves, and trout chow fish food. Measured endpoints were D. magna survival, growth by length and dry weight, measured after drying for 24 h in a 60°C oven, and reproduction as young per surviving organism. Length was measured by capturing a digital image of D. magna at test breakdown and using the Image-Pro Plus (Version 7.0, Media Cybernetics, Inc., Rockville, MD, USA) image analysis software. Length was measured along the exterior dorsal curvature from the posterior edge of the eye spot to the base of the anal spine. Statistical significance of D. magna response variables was determined using a one-way analysis of variance (ANOVA, p < 0.05) and a Dunnett’s post-hoc test using SigmaStat statistical software (version 3.5, Systat Software, Inc., Chicago, IL, USA). Nominal treatment levels for range finder A were 0.06, 0.1, 0.3, 0.5, 1, and 2 mg/L and 0.004, 0.008, 0.02, 0.03, 0.06, 0.13 mg/L for range finder B.
Daphnia magna exposures for microarray and lipid analysis
In order to provide enough tissue mass for microarray and lipid analysis, additional exposures were performed in 1 L beakers with a solution volume of 800 mL. Twenty <24 h old D. magna were added to each beaker, with six replicate beakers per treatment. The organisms in these 1 L beaker mass exposures were fed 9.6 mL of the P. subcapitata / YCT mixture daily. Thus, the ratios of one organism per 40 mL of exposure water and 0.48 ml of food per 40 mL of exposure water were maintained in the mass exposures. In the microarray mass exposure, nominal treatment levels were 0.004, 0.02, 0.12, 0.44, and 1.85 mg/L. These exposure levels were chosen to represent concentrations exhibiting hormetic and toxic responses in the range finding exposures. Based on the results of the testing in the 50 ml beakers, organisms from three treatments were selected for analysis of gene expression: 0.004, 0.12, and 1.85 mg/L. These treatments were selected to represent a hormetic concentration (0.004 mg/L), a toxic concentration (1.85 mg/L), and an intermediate concentration (0.12 mg/L). In the lipid analysis mass exposure, the same three treatment levels were tested. Daphnia magna for lipid or microarray analysis were removed from the exposure media, flash frozen in liquid nitrogen, and stored at −80°C until lipids or total RNA were extracted.
Lipid analysis
Five biological replicates per treatment with 2–5 daphnids per replicate were used for lipid analysis. Sample homogenization was performed using a handheld homogenizer in a chloroform/methanol (2:1 v/v) mixture followed by lipid extraction using the method developed by Bligh and Dyer 12. After extraction, samples were dried with nitrogen and sent to the Kansas Lipidomics Research Center (KLRC, Kansas State University, Manhattan, KS) for lipid analysis. There, an automated electrospray ionization-tandem mass spectrometry approach was used, and data acquisition and analysis were carried out as described previously 13 with modifications. The lipid samples were dissolved in 1 ml chloroform. An aliquot of 100 to 450 μl of extract in chloroform was used. Polar lipid profiles were obtained using precursor and neutral loss scans. Phosphatidylcholine (PC), lysophosphatidylcholine (LPC), sphingomyelin (SM), phosphatidylserine (PS), phosphatidylethanolamine (PE) and lysophosphatidylethanolamine (PE) were analyzed as singly-charged positive [M+H]+ ions, while monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), phosphatidylglycerol (PG), phosphatidylinositide (PI), phosphatidic acid (PA), and PS were analyzed as singly-charged [M + NH4]+ ions, and lysophosphatidylglycerol (LPG) as negative [M-H] − ions. Precise amounts of internal standards, obtained and quantified as previously described 14 were added. The sample and internal standard mixture were combined with solvents, such that the ratio of chloroform/methanol/300 mM ammonium acetate in water was 300/665/35, and the final volume was 1.4 ml. Unfractionated lipid extracts were introduced by continuous infusion into the ESI source on a triple quadrupole MS/MS (API 4000, Applied Biosystems, Foster City, CA). The background of each spectrum was subtracted, the data were smoothed, and peak areas integrated using a custom script and Applied Biosystems Analyst software, and the data were isotopically deconvoluted. Peaks corresponding to the target lipids in these spectra were identified and molar amounts calculated in comparison to the two internal standards on the same lipid class, except for PI, which was quantified in relation to a single internal standard. Ether-linked (alk(en)yl,acyl) lipids were quantified in comparison to the diacyl compounds with the same head groups without correction for response factors for these compounds as compared to their diacyl analogs. The free fatty acids (FFA) were detected using negative MS1 scan mode for 80 cycles. The declustering potential of −100 and entrance potential was −14, −4.5 kV were applied to the electrospray capillary, the curtain gas was set at 20 (arbitrary units), and the two ion source gases were set at 45 (arbitrary units). The FFA standard was 1 nmol 15:0 fatty acid. To correct for chemical or instrumental noise in the samples, the molar amount of each lipid metabolite detected in the “internal standards only” spectra was subtracted from the molar amount of each metabolite calculated in each set of sample spectra. Finally, the data were corrected for the fraction of the sample analyzed and normalized to the mg protein and number of Daphnias to produce data in the units mg lipid/daphnia. Statistical significance was analyzed using Sigma Stat version 3.1. To identify significantly changed lipids, one-way ANOVA was applied to lipid values (mg lipid/daphnia) followed by pairwise comparison using Fisher LSD (p<0.05).
RNA extraction
Six replicates consisting of 2–5 daphnids per treatment were used for RNA extraction. Total RNA was isolated from samples using RNeasy kits following the manufacturer’s protocol (Qiagen, Valencia, CA, USA) and was DNAase treated. RNA quality was assessed with an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and all samples had a RNA Integrity Number, RIN ≥ 7. The RNA quantity was determined using a Nanodrop® ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). Total RNA was stored at −80°C until analyzed using oligonucleotide microarrays.
Microarray Analysis
Custom Daphnia magna 15,000-probe microarrays (GPL13761) were purchased from Agilent (Palo Alto, CA) with array hybridizations performed using the Agilent single color protocol. A total of 24 arrays were used, with 6 biological replicates per each dose (solvent control, 0.004 mg/L, 0.12 mg/L, and 1.85 mg/L). One array for the 0.12mg/L treatment did not work properly and was removed from the analysis. The cRNA synthesis, labeling, amplification and hybridizations were performed following the manufacturer’s kits and protocols (One Color Microarray-based Gene Expression Analysis Quick Amp Labeling version 5.7; Agilent, Palo Alto, CA). After hybridizing for 17 h, microarrays were washed and then scanned with a Surescan High-Resolution DNA Microarray scanner G2505 C (Agilent, CA, USA). Data were extracted from microarray images using Feature Extraction software v10.7 (Agilent, CA, USA). Raw microarray data from this study have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo; GSE43960).
Microarray Data Normalization and Analysis
Raw microarray data were imported into GeneSpring version GX11 (Agilent, Santa Clara, CA, USA). Raw data were log2 transformed and normalized using quantile normalization, followed by median scaling across all samples. To identify differentially expressed genes (DEGs), one-way Analysis of Variance (ANOVA) test was performed followed by pair-wise Tukey’s HSD (Honestly Significant Difference) test (p<0.05). Functional analysis of DEGs was performed using Ingenuity Pathway Analysis (IPA, Redwood City, CA, USA). Hierarchical clustering was performed with GeneSpring. Venn diagrams were performed with Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Benchmark Concentration Analysis
The DEGs obtained from ANOVA were used for a transcriptional benchmark concentration (BMC) analysis using the BMDExpress software 15 as described by Thomas et al. 16. This approach was used to identify the dose-response changes in the functional categories allowing the identification of processes that had different dose-response characteristics 16, particularly those that followed a pattern consistent with the hormetic response. The genes were fit to four different concentration-response models – linear, 2° polynomial, 3° polynomial, and power models. Each model was run with the benchmark response factor (amount required to shift the mean transcriptional response of the control distribution such as the treated distribution contains 11% in a single tail) set to 1.349 multiplied by the standard deviation in the control sample. For model selection, a nested likelihood ratio test was performed on linear, 2nd polynomial, and 3rd polynomial models. Following the developers’ recommendations16, the 3rd polynomial model was selected based on the significantly improved p<0.05. Probe sets with BMC value greater than the highest concentration (1.85mg) were removed from the further analysis. The final probe sets obtained from the BMD analysis tool that showed a concentration-response behavior and had a BMC value lower than 1.85 were used for functional enrichment analysis using IPA (Redwood City, CA), focusing on pathways and toxicological functions, two different classifications used by IPA. The BMC values for the genes involved in the enriched pathways and GO terms were averaged to obtain a single BMC value for the each biological term. Functional enrichment analysis was also performed on the gene lists obtained from post hoc analysis of each dose. The genes in the enriched terms were then searched against the selected model for the BMC values, which were then averaged to obtain a single BMC value for each term in each dose.
Aqueous TNT Chemistry
Aqueous TNT samples were separated and quantified by high performance liquid chromatography (HPLC) following a modified version of the U.S. Environmental Protection Agency SW-486 Method 8330 (U.S. EPA on-line). The quantified compounds included TNT, TNB, the TNB transformation products (dinitroanilines, DNAs), by concurrent detection of 2,4-DNA and 3,4-DNA isomers, and the TNT transformation products (aminodinitrotoluenes, 2-ADNT and4-ADNT), and diaminonitrotoluenes (DANTs, concurrent detection of 2,4-DANT and 2,6-DANT isomers). Analyses were conducted with an Agilent 1100 Series HPLC (Palo Alto, CA) equipped with a Supelco (Bellefonte, PA, USA) RP-Amide C-16 column and a photodiode array detector. Sample injection volume was 100 μl with a flow rate of 1ml per minute and column temperature of 45 °C using an isocratic mobile phase consisting of 55% methanol and 45% water. Absorbance was measured at 230 and 254 nM. Peak identification was based on retention time with spectral analysis confirmation. The nominal and measured TNT concentrations for the three different exposures can be found in SI Table S1.
RESULTS AND DISCUSSION
Hormetic response
We detected a hormetic response in the number of neonates per survivor and length (Fig 1a and 1b). The number of neonates increased significantly in the 0.06, 0.12 and 0.22 mg/L TNT treatments with the greatest stimulation of reproduction (an increase of 39.7%) being observed at the 0.12 mg/L treatment (Fig 1a). Reproduction was significantly decreased by 89.5% at the 0.97 mg/L treatment, and reproduction did not occur at the 1.85 mg/L TNT treatment (Fig 1a). Daphnia length increased significantly (up to 12.5%) at 0.002, 0.004, 0.02, 0.04, 0.06, 0.12, 0.22, and 0.97 mg/L TNT and significantly decreased by 16.7% at 1.85 mg/L TNT (Fig 1b and 1c).
Figure 1.
a) Number of neonates per survivor (as % of control treatment) in Daphnia exposed to TNT from range-finding exposure A; b) Daphnia length (as % of control treatment) in Daphnia exposed to TNT from range-finding exposure A; c) Daphnia length (as % of control treatment) in Daphnia exposed to TNT from range-finding exposure B.
Deciphering the Hormetic Mechanism through Transcriptomics
Transcriptional analysis was performed at 0, 0.004, 0.12, and 1.85 mg/L TNT to capture changes involved in the hormetic and toxic responses due to TNT exposure. Four samples of the 24 total samples were determined to be poor quality and were removed from further analysis. A complete list of DEGs can be found in SI Tables S2–S5. A total of 271, 512, and 1,297 genes were differentially expressed in the 0.004, 0.12 and 1.85 mg/L TNT treatments respectively. Clustering of all samples using hierarchical analysis (SI Figure S1) showed very unique and different gene expression patterns for both the control and 1.85 mg/L treatments, while the 0.004 and the 0.12 mg/L treatments were more similar to each other, consistent with the majority of hormetic effects observed.
A total of 72 DEGs were common among the three TNT treatments, roughly a third of all DEGs for the lowest dose, suggesting a conserved mechanism of TNT toxicity among all doses, regardless of the final outcome (SI Figure S2). Due to the limited functional annotation of the D. magna gene set, only three of those could be mapped to known genes: methylmalonyl-CoA mutase (mut), adenosylmethionine decarboxylase (samd), and mesencephalic astrocyte-derived neutrophic factor (manf).
The enzyme mut transcript, which was significantly up-regulated at all three TNT concentrations, catalyzes the isomerization of methylmalonyl-CoA to succinyl CoA and is involved in key metabolic pathways. The substrate of mut is mostly derived from propionyl-CoA, a product of the catabolism and digestion of several aminoacids, cholesterol or odd-chain fatty acids. The enzyme’s product, succinyl-CoA, is a key molecule of the tricarboxylic acid (TCA) cycle, which generates energy through the catabolism of sugars, fats, and proteins. Effects of TNT and its degradation products on glycolisis, gluconeogenesis and the TCA cycle have been previously shown in other species such as quail and rat 17,18.
Adenosylmethionine decarboxylase (samd), down-regulated in all three TNT concentrations, plays an essential regulatory role in the polyamine biosynthetic pathway by generating the n-propylamine residue required for the synthesis of spermidine and spermine from putrescein 19. Spermidine is a polyamine involved in cellular metabolism that inhibits the neuronal nitric oxide synthase (nos) 20. Up-regulation of nos has been suggested as an adaptation mechanism to the increase in oxidative stress during TNT exposure 21. Down-regulation of samd could accordingly be part of an effort to decrease nos inhibition.
The mesencephalic astrocyte-derived neutrophic factor (manf) is an evolutionarily conserved neurotrophic factors that support dopaminergic neurons. This family is evolutionary well conserved among multicellular organisms 22,23. Recent studies in Drosophila suggest the involvement of manf in metabolism, membrane transport and transporters, oxidative phosphorylation, mitochondrial function, and protein ubiquitination, which is consistent with some of the effects detected in Daphnia by functional analysis 24.
The results from the functional analysis for all treatments can be found in SI Tables S6–S8. Interestingly, fatty acid metabolism was affected at the lowest and highest doses, while lipid metabolism was significantly enriched at all doses (p≤0.05). Aryl hydrocarbon receptor (AhR) signaling appeared to be important in all three treatments, although with p>0.05. The peroxisome proliferator-activated receptor α (PPARα)/RXRα activation as well as the NRF2-mediated oxidative stress appeared to be important at the two highest doses (p>0.05). While most analysis techniques calculate p-values to identify the significance of the involvement of a given pathway in an experimental condition, recent approaches suggest that pathways with insignificant p-values can be biologically meaningful 25. We therefore consider these pathways might be of interest even though their p-value cannot be considered statistically significant. Furthermore, these three pathways, as well as lipid metabolism, have been shown to be involved in the toxicity of TNT and derivatives in other species such as quail and rat 17,18. These results indicate that fatty acid and lipid metabolism, as well as AhR receptor and PPARα might be important in the hormesis and/or toxicity as a result of TNT exposure.
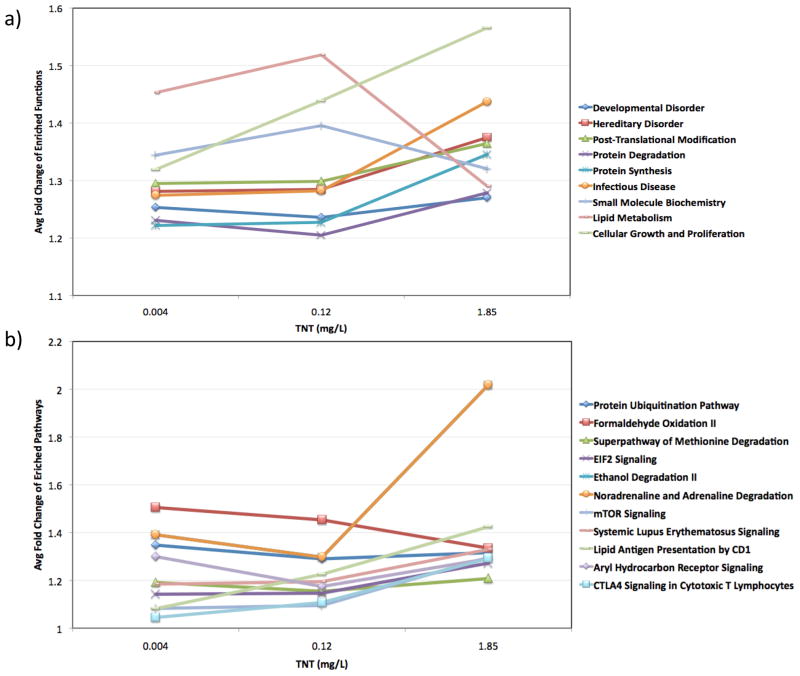
Transcriptional BMC analysis was then used to elucidate the mechanisms involved in the hormetic response as well as those involved in the switch from hormetic to toxic response. This type of analysis has been suggested as being extremely useful to find genes/pathways that follow a dose-dependent response and to detect apical points of departure from normal 16. The results from this analysis (Figure 2) suggested that lipid metabolism was involved in the first hormetic response, as this category clearly followed a pattern similar to a hormetic response. Other pathways such as noradrenaline and adrenaline degradation, cellular growth and proliferation, AhR signaling, mTOR signaling, protein degradation, or EIF2 signaling increased at the higher dose, suggesting that they might be involved in the toxic response.
Figure 2.
Results from the transcriptional Benchmark Concentration analysis showing significant toxicological functions (a) and pathways (b) used for functional enrichment after TNT exposure.
Linkage of Predicted Hormetic Mechanism to Physiological Outputs
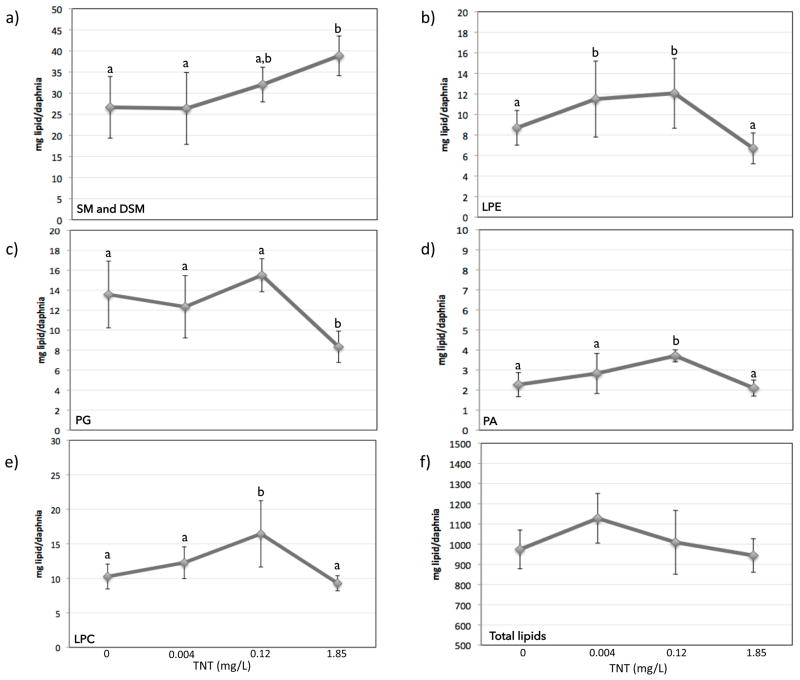
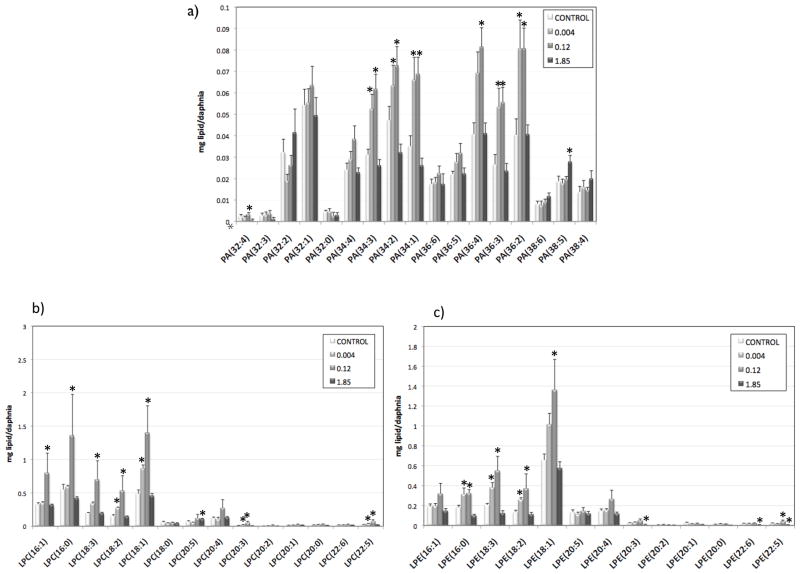
In order to test the hypothesis that lipid metabolism was involved in hormesis due to TNT exposure, we performed lipidomic analysis in all treatments. The amount of total lipids (mg lipid/daphnia) followed a curve similar to the hormetic response (Figure 3f), although the differences were not statistically significant. We then compared the different species of lipids, five of which were significantly changed by TNT exposure (Figure 3): sphingomyelin and dihydrosphingomyelin (SM and DSM, Fig 3a), lysophosphatidyl-ethanolamines (LPE, Fig 3b), phosphatidylglycerols (PGs, Fig 3c), phosphatidic acid (PA, Fig 3d), and lysophosphatidylcholines (LPC, Fig 3e). Three of these species (PA, LPC, and LPE) followed a curve similar to the hormetic response, increasing at the lower doses and decreasing at the highest dose. We then looked at the individual components of these three families (Figure 4). In the three cases, shorter molecules with fewer carbons were more abundant than longer molecules. This was particularly true at the lower doses, implicating impacts on degradation of longer lipid molecules in the hormetic response, consistent with transcriptomic observations of the importance of lipid metabolism in the TNT hormetic response. This degradation could also be related to an increased energy demand of the organisms due to increased length and reproduction.
Figure 3.
Concentration of different lipids (mg lipid/daphnia) at different TNT concentration. Letters indicate statistical significance after pairwise comparison (p<0.05). The lipids shown are: a) Total sphingomyelin (SM) and dihydrosphingomyelin (DSM), b) Total lysophosphatidylethanolamine (LPE), c) Total phosphatidylglycerol (PG), d) Total phosphatidic acid (PA), e) Total lysophosphatidylcholine (LPC), f) Total lipids
Figure 4.
Concentration of different lipid isomers (mg lipid/daphnia) at different TNT concentration (significant changes indicated by *, p<0.05). A) phosphatidic acid isomers; b) lysophosphatidylethanolamine isomers; c) lysophosphatidylcholine isomers.
Phosphatidic acid is present at very low concentrations in animal cells and it is fundamental in the biosynthesis and metabolism of triacylglycerols and phospholipids and as a signaling molecule 26. Levels of the PA species PA(34:1), (34:2), (34:3), (36:2), (36:3) and (36:4), where (x:y) equals (total carbons:total double bonds), were increased at the lower doses, while PA(38:5) was increased at the highest dose. Although the exact role of PA as a signaling molecule is still largely unknown, it has been reported to be involved in the mechanical regulation of skeletal muscle growth through the mammalian target of rapamycin (mTOR) signaling 27,28. Target of rapamycin is an evolutionary conserved sensor of essential nutrients needed for the synthesis of biological molecules 29 and it has been shown to require PA for the stability of mTOR complexes and their activity30. It has been suggested that TOR is dependent on PA to sense the presence of sufficient phospholipids for cell growth 31. The TOR pathway is evolutionarily conserved and has been shown to be present in many species including yeast, Drosophila and Daphnia 32,33. Furthermore, the conservation of the PA-binding domain in TOR, from yeast to mammals, suggests that PA requirement by TOR was a very early evolutionary event 31. In human cells, PA is bound to the nuclear receptor steroidogenic factor 1 (sf1), stimulates sf1-dependent transcription of cyp17 and induces expression of cyp17 and several other steroidogenic genes 34. Although there is evidence that crustaceans synthesize steroid hormones from cholesterol 35, the molecular details of steroidogenesis in crustaceans remains mostly conjectural 36. Nevertheless, orthologs of enzymes that metabolize insect steroids have been identified in Daphnia 36–38. While the above-mentioned effects of PA on muscle growth and steroidogenesis have been observed in mammals, some of these functions could be similar or evolutionarily conserved to the functions of PA in Daphnia, suggesting there could be a link between changes on PA levels and the observed effects on growth and reproduction in Daphnia.
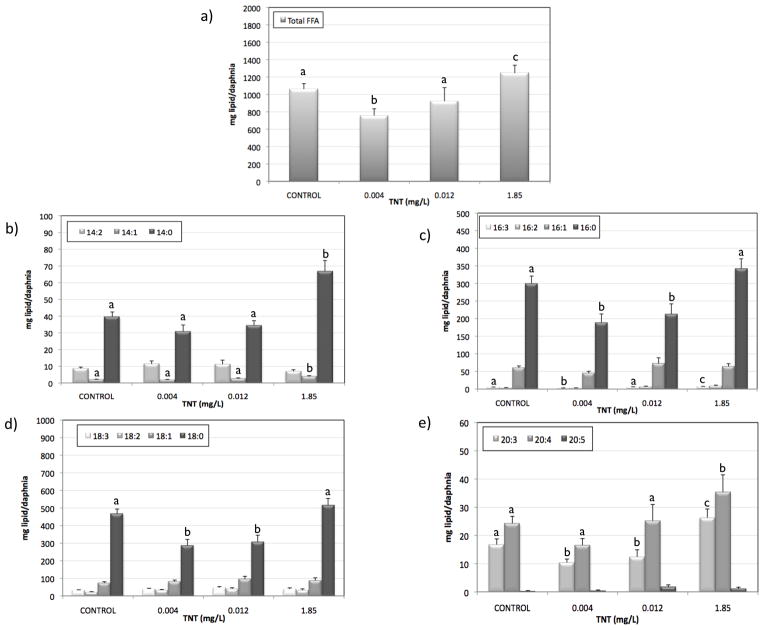
Hydrolysis of PEs and PCs produces LPEs and LPCs respectively through removal of one of the fatty acid groups. Levels of LPEs were significantly increased at 0.004 mg/L TNT, and levels of both LPCs and LPEs were significantly increased at 0.12 mg/L TNT. This is consistent with results observed in rat liver 18, where levels of LPC and LPE were increased after TNT exposure. These results suggest that low TNT levels stimulate an increase in hydrolysis of PC and PE to LPCs, LPEs, and polyunsaturated chains that may be metabolized to eicosanoids. While no significant change in total PC and PE levels was detected, several individual PC and PE isomers such as PE(38:5), PE(38:4), PE(42:10), PC(38:5), PC(38:6), and PC(42:10) followed a trend consistent with increased hydrolysis of PC and PE (SI Figure S3). The most common fatty acid combinations that would form these PC and PE isomers would most likely correspond to the fatty acid combinations 16:0/22:4, 16:0/22:6, 18:2/20:3, 18:1/20:4, 18:0/20:5, 18:0/20:4, 20:4/22:6 (Welti, personal communication). Comparison of these combinations to the increased LPEs and LPCs species (Fig 4) indicated that the polyunsaturated chains resulting from the hydrolysis of PCs and PEs would likely correspond to palmitic acid (16:0), stearic acid (18:0), dihomo-gamma-linoleic acid (DGLA, 20:3), arachidonic acid (ARA; 20:4), and eicospentaenoic acid (EPA, 20:5). Free fatty acid analysis showed a decrease in total FFA at the lowest TNT concentration and an increase at the highest concentration (Fig. 5a). Figure 5 shows the levels of different FFA that showed significant changes by TNT treatment: 14 carbon atoms FFA (Fig 5b), 16 carbon atoms FFA (Fig 5c), 18 carbon atoms FFA (Fig 5d), and 20 carbon atoms FFA (Fig 5e). Palmitic acid, stearic acid, and DGLA levels decreased at the lower TNT concentrations, while FFA 14:1, 14:0, DGLA and ARA increased at the highest concentration. It is worth noting that food composition can have major effects on the biochemical composition of Daphnia. Particularly, the polyunsaturated fatty acid composition present in Daphnia generally reflect that of their food sources 39.
Figure 5.
concentration of different free fatty acids (mg lipid/daphnia) at different TNT concentrations. Letters indicate statistical significance after pairwise comparison (p<0.05). Letters are not added to FFA without any significant changes. The free fatty acids are: a) total free fatty acids; b) 14:0, 14:1, 14:2 free fatty acids; c) 16:0, 16:1, 16:2, 16:3 free fatty acids; d) 18:0, 18:1, 18:2, 3:3 free fatty acids, e) 20:3, 20:4, and 20:5 free fatty acids.
Eicosanoids are cell signaling molecules derived from C20 polyunsaturated fatty acids (DGLA, ARA, and EPA) and have an important role in the regulation of essential functions including reproduction 40,41. There is evidence suggesting that ARA and EPA support growth and reproduction in Daphnia because they are precursors of eicosanoids 42,43. Several invertebrate studies have investigated the potential roles of eicosanoids demonstrating that both prostanoids and lipoxygenase products are important in oogenesis (especially vitellogenesis) and embryogenesis 44–46. For example, prostaglandin E2 initiates egg-laying behavior in several insect species, where it seems to regulate muscle contractions in the ovarian musculature 47. Eicosanoids have also been identified as important mediators in arthropod immune system 48 and ion transport 41. There is evidence suggesting that the eicosanoid machinery is present in Daphnia 40,49 and that eicosanoids play vital roles in processes key to daphnid reproduction and survival 40,50,51. The fact that DGLA levels are decreased at the hormetic concentrations and increased at the highest concentration suggests that the daphnids could be using DGLA to synthesize eicosanoids at the hormetic concentrations but were not able to do it at the highest concentration. That would be consistent with the increased growth and reproduction at the hormetic concentrations.
Our results show how a systems approach using toxicogenomics can help elucidate the physiological outcomes driven by molecular changes. Exposure of D. magna to low TNT doses resulted in a hormetic response leading to increased growth and reproduction, while higher TNT doses had toxic effects. Functional analyses suggested the involvement of lipid and fatty acid metabolism, AhR signaling, or mTOR signaling in TNT’s effects. The BMC analysis suggested that lipid metabolism could be involved in the hormetic response observed at low TNT concentrations. Lipidomic analysis supported this hypothesis by showing the increase of several LPC and LPE isomers at the lower TNT doses, which imply the release of several polyunsaturated fatty acids. Free fatty acid analysis showed the decrease of several FFA at the hormetic doses, suggesting there could be an increase in energetic demands and/or an increase in eicosanoid synthesis, which could be linked to Daphnia growth and reproduction.
Supplementary Material
Acknowledgments
Lipid analysis was performed at the Kansas Lipidomics Research Center (KLRC) supported by NSF grants MCB 0455318 and 0920663 and DBI 0521587, and NSF EPSCoR grant EPS-0236913 with matching support from the State of Kansas through Kansas Technology Enterprise Corporation and Kansas State University. The KLRC is also supported by K-INBRE (NIH Grant P20 RR16475). The authors would like to thank Drs Mary Roth and Ruth Welti for advise on lipidomics data interpretation. This work was funded by the US Army Environmental Quality Research Program (including BAA 11-4838). Permission for publishing this information has been granted by the Chief of Engineers.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Hierarchical clustering of all the differentially expressed genes per sample (SI-Figure S1), Venn diagram of the differentially expressed genes by the three TNT doses (SI-Figure S2), Concentration of different lipids (mg lipid/daphnia) at different TNT doses (SI-Figure S3), TNT concentrations in the three different exposures (SI-Table S1), differentially expressed genes for all treatments (SI-Table S2), differentially expressed genes for TNT 0.004 mg/L (SI-Table S3), differentially expressed genes for TNT 0.12 mg/L (SI-Table S4), differentially expressed genes for TNT 1.85 mg/L (SI-Table S5), functional analysis for TNT 0.004 mg/L (SI-Table S6), functional analysis for TNT 0.12 mg/L (SI-Table S7), functional analysis for TNT 1.85 mg/L (SI-Table S8). This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Calabrese EJ, Baldwin LA, Holland CD. Hormesis: a highly generalizable and reproducible phenomenon with important implications for risk assessment. Risk Anal. 1999;19:261–281. doi: 10.1023/a:1006977728215. [DOI] [PubMed] [Google Scholar]
- 2.Calabrese1 EJ, Baldwin LA. Hormesis: A Generalizable and Unifying Hypothesis. Critical Reviews in Toxicology. 2001;31:353–424. doi: 10.1080/20014091111730. [DOI] [PubMed] [Google Scholar]
- 3.Marion Nipper RSCAGRL. Ecotoxicology of explosives. 2009. pp. 1–39. [Google Scholar]
- 4.Kuperman RG, Checkai RT, Simini M, Phillips CT, Kolakowski JE, Kurnas CW. Weathering and aging of 2,4,6-trinitrotoluene in soil increases toxicity to potworm Enchytraeus crypticus. Environ Toxicol Chem. 2005;24:2509. doi: 10.1897/04-513r.1. [DOI] [PubMed] [Google Scholar]
- 5.Kuperman RG, Checkai RT, Simini M, Phillips CT, Kolakowski JE, Kurnas CW. Toxicities of dinitrotoluenes and trinitrobenzene freshly amended or weathered and aged in a sandy loam soil to enchytraeus crypticus. Environ Toxicol Chem. 2006;25:1368. doi: 10.1897/05-475r1.1. [DOI] [PubMed] [Google Scholar]
- 6.Steevens JA, Duke BM, Lotufo GR, Bridges TS. Toxicity of the explosives 2,4,6-trinitrotoluene, hexahydro-1,3,5-trinitro-1,3,5-triazine, and octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine in sediments to Chironomus tentans and Hyalella azteca: Low-dose hormesis and high-dose mortality. Environ Toxicol Chem. 2002;21:1475–1482. [PubMed] [Google Scholar]
- 7.Bailey HC, Spanggord RJ, Javitz HS, Liu DHW. Toxicity of TNT Wastewaters to Aquatic Organisms. Chronic Toxicity of LAP Wastewater and 2,4,6-Trinitrotoluene. 1985;3 [Google Scholar]
- 8.Bentley RE, Dean JW, Ells SJ, Hollister TA, LeBlanc GA, Sauter S, Sleight BH. U S Army Medical Res Develop Command Frederick, M. D, editor. Laboratory Evaluation of the Toxicity of Cyclotrimethylene Trinitramine (RDX) to Aquatic Organisms. 1977. [Google Scholar]
- 9.Piña BB, Barata CC. A genomic and ecotoxicological perspective of DNA array studies in aquatic environmental risk assessment. Aquat Toxicol. 2011;105:40–49. doi: 10.1016/j.aquatox.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Technology, U. E. O. O. O. S. A. Methods for Measuring the Acute Toxicity of Effluents and Receiving Waters to Freshwater and Marine Organisms. 2002. pp. 1–39. [Google Scholar]
- 11.United States Environmental Protection Agency. Ecological Effects Test Guidelines. 1996. pp. 1–12. [Google Scholar]
- 12.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 13.Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci US A. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welti R. Profiling Membrane Lipids in Plant Stress Responses. Journal of Biological Chemistry. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 15.Yang L, Allen BC, Thomas RS. BMDExpress: a software tool for the benchmark dose analyses of genomic data. BMC Genomics. 2007;8:387. doi: 10.1186/1471-2164-8-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas RS, Allen BC, Nong A, Yang L, Bermudez E, Clewell HJ, Andersen ME. A Method to Integrate Benchmark Dose Estimates with Genomic Data to Assess the Functional Effects of Chemical Exposure. Toxicol Sci. 2007;98:240–248. doi: 10.1093/toxsci/kfm092. [DOI] [PubMed] [Google Scholar]
- 17.Rawat A, Gust KA, Deng Y, Garcia-Reyero N, Quinn MJ, Johnson MS, Indest KJ, Elasri MO, Perkins EJ. From raw materials to validated system: the construction of a genomic library and microarray to interpret systemic perturbations in Northern bobwhite. Physiol Genomics. 2010;42:219–235. doi: 10.1152/physiolgenomics.00022.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng Y, Meyer SA, Guan X, Escalon BL, Ai J, Wilbanks MS, Welti R, Garcia-Reyero N, Perkins EJ. Analysis of common and specific mechanisms of liver function affected by nitrotoluene compounds. PLoS ONE. 2011;6:e14662. doi: 10.1371/journal.pone.0014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Poelje PD, Snell EE. Pyruvoyl-dependent enzymes. Annual review of biochemistry. 1990 doi: 10.1146/annurev.bi.59.070190.000333. [DOI] [PubMed] [Google Scholar]
- 20.Hu J, Mahmoud MI, el-Fakahany EE. Polyamines inhibit nitric oxide synthase in rat cerebellum. Neurosci Lett. 1994;175:41–45. doi: 10.1016/0304-3940(94)91073-1. [DOI] [PubMed] [Google Scholar]
- 21.Kumagai Y, Kikushima M, Nakai Y, Shimojo N, Kunimoto M. Neuronal nitric oxide synthase (NNOS) catalyzes one-electron reduction of 2,4,6-trinitrotoluene, resulting in decreased nitric oxide production and increased nNOS gene expression: implication for oxidative stress. Free Radical Biology and Medicine. 2004;37:350–357. doi: 10.1016/j.freeradbiomed.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 22.Palgi M, Lindström R, Peränen J, Piepponen TP, Saarma M, Heino TI. Evidence that DmMANF is an invertebrate neurotrophic factor supporting dopaminergic neurons. Proc Natl Acad Sci US A. 2009;106:2429–2434. doi: 10.1073/pnas.0810996106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindholm P, Saarma M. Novel CDNF/MANF family of neurotrophic factors. Devel Neurobio. 2010:NA–NA. doi: 10.1002/dneu.20760. [DOI] [PubMed] [Google Scholar]
- 24.Palgi M, Greco D, Lindström R, Auvinen P, Heino TI. Gene expression analysis of Drosophilaa Manf mutants reveals perturbations in membrane traffic and major metabolic changes. BMC Genomics. 2012;13:134. doi: 10.1186/1471-2164-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donato M, Draghici S, Tomoiaga A, Westfall P, Xu Z, Romero R. A method for analysis and correction of cross-talk effects in pathway analysis. Neural Networks (IJCNN), The 2012 International Joint Conference on Computational Intelligence; 2012. pp. 1–7. [Google Scholar]
- 26.Leray C. Introduction to Lipidomics. CRC Press; 2012. [Google Scholar]
- 27.Hornberger TA, Sukhija KB, Chien S. Regulation of mTOR by mechanically induced signaling events in skeletal muscle. Cell Cycle. 2006;5:1391–1396. doi: 10.4161/cc.5.13.2921. [DOI] [PubMed] [Google Scholar]
- 28.Hornberger TA, Chu WK, Mak YW, Hsiung JW, Huang SA, Chien S. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc Natl Acad Sci US A. 2006;103:4741–4746. doi: 10.1073/pnas.0600678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polak P, Hall MN. mTOR and the control of whole body metabolism. Current Opinion in Cell Biology. 2009;21:209–218. doi: 10.1016/j.ceb.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 30.Foster DA. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007 doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- 31.Foster DA. Phosphatidic acid and lipid-sensing by mTOR. Trends in Endocrinology & Metabolism. 2013;24:272–278. doi: 10.1016/j.tem.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dam TJP, Zwartkruis FJT, Bos JL, Snel B. Evolution of the TOR Pathway. J Mol Evol. 2011;73:209–220. doi: 10.1007/s00239-011-9469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boucher P, Ditlecadet D, Dubé C, Dufresne F. Unusual duplication of the insulin-like receptor in the crustacean Daphnia pulex. BMC Evol Biol. 2010;10:305. doi: 10.1186/1471-2148-10-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D, Urs AN, Allegood J, Leon A, Merrill AH, Sewer MB. Cyclic AMP-Stimulated Interaction between Steroidogenic Factor 1 and Diacylglycerol Kinase Facilitates Induction of CYP17. Molecular and Cellular Biology. 2007;27:6669–6685. doi: 10.1128/MCB.00355-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lachaise F, Le Roux A, Hubert M, Lafont R. The molting gland of crustaceans: localization, activity, and endocrine control (a review) Journal of Crustacean Biology. 1993 [Google Scholar]
- 36.Rewitz KF, Gilbert LI. Daphnia Halloween genes that encode cytochrome P450s mediating the synthesis of the arthropod molting hormone: Evolutionary implications. BMC Evol Biol. 2008;8:60. doi: 10.1186/1471-2148-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markov GV, Tavares R, Dauphin-Villemant C, Demeneix BA, Baker ME, Laudet V. Independent elaboration of steroid hormone signaling pathways in metazoans. Proc Natl Acad Sci US A. 2009;106:11913–11918. doi: 10.1073/pnas.0812138106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldwin WS, Marko PB, Nelson DR. The cytochrome P450 (CYP) gene superfamily in Daphnia pulex. BMC Genomics. 2009;10:169. doi: 10.1186/1471-2164-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brett MT, Müller-Navarra DC, Ballantyne AP, Ravet JL, Goldman CR. Daphnia fatty acid composition reflects that of their diet. Limnol Oceangr. 2006;51:2428–2437. [Google Scholar]
- 40.Heckmann LH, Sibly RM, Timmermans MJ, Callaghan A. Outlining eicosanoid biosynthesis in the crustacean Daphnia. Front Zool. 2008;5:11. doi: 10.1186/1742-9994-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanley DW. Eicosanoids in invertebrate signal transduction systems. Princeton University Press; Princeton, NJ: 2000. [Google Scholar]
- 42.Martin-Creuzburg D, Wacker A, Basen T. Interactions between limiting nutrients: Consequences for somatic and population growth of Daphnia magna. Limnol Oceangr. 2010;55:2597–2607. [Google Scholar]
- 43.Schlotz N, Sørensen JG, Martin-Creuzburg D. The potential of dietary polyunsaturated fatty acids to modulate eicosanoid synthesis and reproduction in Daphnia magna: a gene expression approach. Comparative Biochemistry and Physiology - Part A: Molecular & Integrative Physiology. 2012;162:449–454. doi: 10.1016/j.cbpa.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Silver RB, Oblak JB, Jeun GS, Sung JJ, Dutta TC. Leukotriene B4, an arachidonic acid metabolite, regulates intracellular free calcium release in eggs and mitotic cells of the sand dollar (Echinaracnius parma) Biol Bull. 1994;187:242–244. doi: 10.1086/BBLv187n2p242. [DOI] [PubMed] [Google Scholar]
- 45.Sagi A, Silkovsy J, Fleisher-Berkovich S, Danon A, Chayoth R. Prostaglandin E2 in Previtellogeic Ovaries of the Prawn Macrobrachium rosenbergii: Synthesis and Effect on the Level of cAMP. Gen Comp Endocrinol. 1995;100:308–313. doi: 10.1006/gcen.1995.1161. [DOI] [PubMed] [Google Scholar]
- 46.Medeiros MN, Mendonça LH, Hunter AL, Paiva-Silva GO, Mello FG, Henze IP, Masuda H, Maya-Monteiro CM, Machado EA. The role of lipoxygenase products on the endocytosis of yolk proteins in insects: participation of cAMP. Arch Insect Biochem Physiol. 2004;55:178–187. doi: 10.1002/arch.10129. [DOI] [PubMed] [Google Scholar]
- 47.Loher W, Ganjian I, Kubo I, Stanley-Samuelson D, Tobe SS. Prostaglandins: Their role in egg-laying of the cricket Teleogryllus commodus. Proc Natl Acad Sci US A. 1981;78:7835–7838. doi: 10.1073/pnas.78.12.7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller JS, Nguyen T, Stanley-Samuelson DW. Eicosanoids mediate insect nodulation responses to bacterial infections. Proc Natl Acad Sci US A. 1994;91:12418–12422. doi: 10.1073/pnas.91.26.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, Bauer DJ, Caceres CE, Carmel L, Casola C, Choi JH, Detter JC, Dong Q, Dusheyko S, Eads BD, Frohlich T, Geiler-Samerotte KA, Gerlach D, Hatcher P, Jogdeo S, Krijgsveld J, Kriventseva EV, Kultz D, Laforsch C, Lindquist E, Lopez J, Manak JR, Muller J, Pangilinan J, Patwardhan RP, Pitluck S, Pritham EJ, Rechtsteiner A, Rho M, Rogozin IB, Sakarya O, Salamov A, Schaack S, Shapiro H, Shiga Y, Skalitzky C, Smith Z, Souvorov A, Sung W, Tang Z, Tsuchiya D, Tu H, Vos H, Wang M, Wolf YI, Yamagata H, Yamada T, Ye Y, Shaw JR, Andrews J, Crease TJ, Tang H, Lucas SM, Robertson HM, Bork P, Koonin EV, Zdobnov EM, Grigoriev IV, Lynch M, Boore JL. The Ecoresponsive Genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayashi Y, Heckmann LH, Callaghan A, Sibly RM. Reproduction recovery of the crustacean Daphnia magna after chronic exposure to ibuprofen. Ecotoxicology. 2008;17:246–251. doi: 10.1007/s10646-008-0191-3. [DOI] [PubMed] [Google Scholar]
- 51.Heckmann LH, Sibly RM, Connon R, Hooper HL, Hutchinson TH, Maund SJ, Hill CJ, Bouetard A, Callaghan A. Systems biology meets stress ecology: linking molecular and organismal stress responses in Daphnia magna. Genome Biol. 2008;9:R40. doi: 10.1186/gb-2008-9-2-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.