Abstract
A previous NMR structure of the duplex revealed an unusually stable RNA internal loop with three consecutive sheared GA pairs. Here, we report NMR studies of two duplexes, (replacing a UG with a UA closing pair) and (replacing the middle GA with an AA pair). An unusually stable loop with three consecutive sheared GA pairs forms in the duplex . The structure contrasts with that reported for this loop in the crystal structure of the large ribosomal subunit of Deinococcus radiodurans [Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A. (2001) Cell 107, 679–688]. The middle AA pair in the duplex rapidly exchanges orientations resulting in alternative base stacking and pseudo-symmetry with exclusively sheared pairs. The internal loop is 2.1 kcal/mol less stable than the internal loop at 37 °C. Structural, energetic, and dynamic consequences upon functional group substitutions within related 3 × 3 and 3 × 6 internal loops are also reported.
Non-canonical pairs within the internal loops of RNA are important elements for folding and function. Understanding the sequence dependent folding free energy and dynamics of internal loops can facilitate prediction of structure (1, 2), dynamics, and functional significance from sequence.
AA and GA can form isosteric sheared-type (trans Hoogsteen/sugar edge A–A or A–G) non-canonical pairs (Figure 1a) (3–9). Typically, the AA pair is thermodynamically destabilizing but the GA pair is stabilizing (7, 10–13). Depending on sequence context, GA often forms a sheared pair, but AA is more flexible (Figure 1). Two A’s can potentially switch base pairing orientation in a sheared AA pair (i.e. trans Hoogsteen/sugar edge A1-A2 or A2-A1) without loss of base–base hydrogen bonding. In a sheared GA pair, the equivalent interchange of bases would result in the loss of the two hydrogen bonds between G and A in a sheared GA pair.
Figure 1.
Schematic representation of (a) different sheared pairs, and (b) a GA and various AA pairs mentioned in this paper. The hydrogen bonds between base and backbone are not shown. Note that two conformations with one base-base hydrogen bond are possible for an AA pair because the amino group of either A can form the hydrogen bond. Only one such conformation is possible for the PA and IA pairs because neither P nor I have amino groups.
The duplex, (P1 = purine riboside), contains an unusually stable and relatively abundant internal loop, (9). The NMR structure of this duplex reveals three consecutive sheared GA pairs (trans Hoogsteen/sugar edge A–G) with separate stacks of three G’s (G4/G5/G14 in the major groove) and three A’s (A6/A15/A16 in the minor groove), which are closed by wobble UG (cis Watson-Crick/Watson-Crick U–G) and Watson-Crick CG pairs (9). (Throughout the paper, each top strand is written from 5′ to 3′ in going from left to right. Numbering starts at the left most (5') nucleotide of the top strand and ends at the left most (3') nucleotide of the bottom strand.)
Helix 68 of the crystal structure of the large ribosomal subunit of Deinococcus radiodurans contains a loop that has only one sheared GA pair (shown in bold) (14). There is less hydrogen bonding and the base stacking pattern is equivalent to A6/G5/A16 in the minor groove instead of the A6/A15/A16 found in the NMR structure for the equivalent loop with a UG rather than UA closing pair.
Here, we report NMR and thermodynamic studies of (A17 duplex) and (A5 duplex) to determine the effects of replacing a UG closing pair with UA or a middle GA pair with AA, respectively, relative to (3GA duplex) (Figure 2). NMR restrained molecular dynamics reveals a conformation of three consecutive sheared GA pairs for the loop in . A5 and A15 in rapidly exchange positions forming alternative sheared AA pairs (i.e. exchanging between trans Hoogsteen/sugar edge A15-A5 and trans Hoogsteen/sugar edge A5-A15) flanked by sheared GA pairs. The exchanging AA pair results in alternative base stacking of A6/A15/A16 or A6/A5/A16 in the minor groove. The flexibility of alternative orientations of a middle adenine base edge in the minor groove, i.e. from A15 (N3-C2-N1) (as observed in and ) to A5 (N1-C2-N3), might provide switching between different binding partners for dynamic functions.
Figure 2.
Secondary structure, numbering, and abbreviations for the duplexes studied previously (9, 13) and here. Lower case “a” represents deoxyadenosine. The value to the right of each duplex is the free energy increment in kcal/mol for formation of the internal loop at 37 °C, at pH7 in 1 M NaCl.
Functional group substitutions (atomic mutations) have been extensively used for studying elements of molecular recognition in RNA (10, 15–24). Here, the structural, energetic, and dynamic consequences of functional group substitutions are explored by studying duplexes of the form , where R and Q are various purine nucleotides (Figures 1a and 2). Single predominant conformations form in the a5, P5, I5, and I15 duplexes. Functional group substitutions also facilitate interpretation of NMR data. The thermodynamic effects of functional group substitutions within 3 × 6 internal loops are also reported.
MATERIALS AND METHODS
Oligonucleotide Synthesis and Purification
Oligonucleotides were synthesized using the phosphoramidite method (25, 26) and purified as described previously (9, 12). CPG supports and phosphoramidites were acquired from Proligo, Glen Research, or ChemGenes. The mass of all oligonucleotides was verified by ESI MS with a Hewlett Packard 1100 LC/MS Chemstation. Purities were checked by reverse phase HPLC or analytical TLC on a Baker Si500F silica gel plate (250 µm thick) and all were greater than 95% pure.
UV Melting Experiments and Thermodynamics
Concentrations of single-stranded oligonucleotides were calculated from the absorbance at 280 nm at 80 °C and extinction coefficients predicted from those of dinucleotide monophosphates and nucleosides (27, 28) with the RNAcalc program (http://www.meltwin.com) (29). The extinction coefficients were estimated by replacing purine riboside, 2,6-diaminopurine riboside, deoxyadenosine, and 2'-O-methyl adenosine with adenosine; and replacing inosine and deoxyguanosine with guanosine. Although extinction coefficients differ upon functional group substitutions, individual nucleotides contribute only a small portion of the oligomer extinction and thus do not significantly affect thermodynamic measurements. UV melting buffer conditions were 1.0 M NaCl, 20 mM sodium cacodylate, and 0.5 mM disodium EDTA at pH 7 or 80 mM NaCl, 10 mM sodium phosphate, 0.5 mM disodium EDTA, pH 7. Curves of absorbance at 280 nm versus temperature were acquired using a heating rate of 1 °C/min with a Beckman Coulter DU640C spectrophotometer having a Peltier temperature controller.
Melting curves were fit to a two-state model with the MeltWin program (http://www.meltwin.com), assuming linear sloping baselines and temperature-independent ΔH° and ΔS° (29–31). Additionally, the temperature at which half the strands are in duplex, TM, at total strand concentration, CT, was used to calculate thermodynamic parameters for non-self-complementary duplexes according to (32):
| (1) |
Here R is the gas constant, 1.987 cal/mol·K. All of the ΔH° values from TM−1 versus ln(CT/4) plots and from the average of the fits of melting curves to two-state transitions agree within 15%, suggesting that the two-state model is a reasonable approximation for these transitions. The equation ΔG°37 = ΔH° − (310.15)ΔS° was used to calculate the free energy change at 37 °C (310.15 K).
NMR Sample Preparation
With minor modification, sample preparation was similar to that previously reported (7, 9). The sample buffer conditions were 80 mM NaCl, 10 mM sodium phosphate, 0.5 mM disodium EDTA, pH 5.1 for H2O, pD 7.3 for D2O for and pH 5.4 for H2O, pD 6.8 for D2O for . Exchangeable proton spectra at pH 6.0 for were very similar to those at pH 5.1. Moreover, chemical shifts and critical loop NOEs involving non-exchangeable protons were essentially the same in water at pH 5.1 and pD 7.3. Total volumes were 300 µL with 90:10 (v:v) H2O:D2O for exchangeable proton spectra and 99.996% D2O (Cambridge Isotope Laboratories) for non-exchangeable spectra. The total duplex concentrations were ~2 mM. The total duplex concentrations of other sequences were 0.5–1.2 mM.
NMR Spectroscopy
Unless otherwise noted, all exchangeable and non-exchangeable proton spectra were acquired on a Varian Inova 500 MHz (1H) spectrometer (33). One-dimensional imino proton spectra were acquired with an S pulse sequence (33) with a sweep width of 12 kHz and temperatures ranging from 0 to 55 °C. SNOESY spectra were recorded with a 150 ms mixing time at 5 and 30 °C. NOESY spectra of samples in D2O were acquired at 30 °C with 100, 200, and 400 ms mixing times. TOCSY spectra were acquired at 30°C with 8, 20, and 40 ms mixing times. Natural abundance 1H–13C HMQC specta for and were acquired with a 5000 Hz spectral width for proton and 15000 Hz spectral width for carbon. The 1H–31P HETCOR and natural abundance 1H–13C HSQC spectra were acquired on a Varian Inova 600 MHz (1H) spectrometer. The 1D 1H-decoupled 31P spectra (referenced to external standard of 85% H3PO4 at 0 ppm) were acquired on a Bruker Avance 500 MHz (1H) spectrometer at 30 °C. Proton spectra were referenced to H2O or HDO at a known temperature dependent chemical shift relative to 3-(trimethylsilyl) tetradeutero sodium propionate (TSP). The Felix (2000) software package (Molecular Simulations Inc.) was used to process 2D spectra.
Restraint Generation
Very similar restraints were generated for (Supporting Information Table S1) as for (9). For and , 15 hydrogen bond restraints limiting proton and hydrogen-bond acceptor distances within 1.8 to 2.5 Å were applied for the five Watson-Crick GC pairs, but no hydrogen bond restraints were used within the loop and UG or UA pair. Dihedral angles of residues in the Watson-Crick stems and UG or UA pair were loosely restrained: α (0±120°), β (180±30°), γ (60±30°), δ (85±30°), ε (−140±40°), ζ (0±120°) (ζ was mistakenly given as ξ in ref (9).), and χ (−170±40°). For loop residues, glycosidic bond dihedral angles, χ’s, were loosely restrained (−120±90°) because there was no indication of a syn glycosidic conformation. For the structural modeling of , the δ dihedral angle for G5 was restrained to be C2′-endo with δ (160±30°), and for A6, G14, U10, and P20, the δ dihedral angles were restrained to cover both C2′-endo and C3′-endo conformations with δ (122.5±67.5°).
Two sets of distance and dihedral angle restraints (set I: A6/A15/A16 and set II: A6/A5/A16) were run for because NOEs were inconsistent with a single structure. The previous NMR structure of (9) facilitated segregation of restraints for structural modeling. For inter-proton distance restraints that differ for A6/A15/A16 and A6/A5/A16 structural modeling, lower and upper bounds were loosened; all other restraints are the same for sets I and II (Supporting Information Table S2).
Two sets of δ dihedral angle restraints were generated for loop residues (G4, A5, A6, G14, A15, and A16) in . For set I (A6/A15/A16), the δ dihedral angle for A5 was restrained to be C2′-endo with δ (160±30°), all other loop residues and the two 3′-dangling residues, U10 and P20, were restrained to cover both C2′-endo and C3′-endo conformations with δ (122.5±67.5°). For set II (A6/A5/A16), all the loop residues were restrained to cover both C2′-endo and C3′-endo conformations, with the other dihedral angle restraints the same as those of set I (A6/A15/A16) and .
In summary, a total of 222 distance restraints (110 intra-nucleotide, 112 inter-nucleotide) including hydrogen bond restraints, and 98 dihedral angle restraints were used for the structural modeling of (Supporting Information Table S1). For the structural modeling of , a total of 250 distance restraints (128 intra-nucleotide, 122 inter-nucleotide) including hydrogen bond restraints, and 98 dihedral angle restraints were used for the structural modeling of set I (A6/A15/A16), and a total of 249 distance restraints (128 intra-nucleotide, 121 inter-nucleotide) including hydrogen bond restraints, and 98 dihedral angle restraints were used for the structural modeling of set II (A6/A5/A16) (Supporting Information Table S2).
Structural Modeling
NMR restrained molecular dynamics and energy minimization was done with the Discover 98 package on a Silicon Graphics computer. An A-form like RNA starting structure was generated with the Biopolymer module of Insight II (2000). The AMBER 95 force field (34) was used with addition of flat-bottom restraint pseudo-potentials, with force constants of 25 kcal/(mol•Å2) for NOE distance restraints and 50 kcal/(mol•rad2) for torsion angle restraints and with a maximum force of 1000 kcal/mol. Group-based summation with an 18 Å cutoff was used for calculating van der Waals interactions. The cell-multipole method (35), with distance dependent dielectric constant (ε = 2r), was used for calculating electrostatic interactions. The progression of the structure simulation was the same as previously reported (7, 9, 17). Several figures were generated with the PyMOL program (36).
RESULTS
Functional Group Substitutions and Thermodynamics of Molecular Recognition
Measured thermodynamic parameters for several duplexes and internal loops with and without functional group substitutions are listed in Tables 1 and 2, respectively. Most were measured at 1 M NaCl to allow comparison to existing databases, but four were also measured in the 80 mM NaCl buffer used for most NMR experiments. The lower salt makes duplex formation less favorable on average by 3.41±0.02 kcal/mol at 37 °C, which is consistent with a sequence independent salt effect. Measured thermodynamic parameters for formation of the internal loops (Table 2) are calculated according to the following equation which relies on the nearest neighbor model for predicting duplex stability (37):
| (2a) |
For example,
| (2b) |
Here, is the measured value of the duplex containing the internal loop (Table 1), is the measured value of the duplex without the loop (9), and is the free energy increment for the nearest neighbor base stack interaction interrupted by the internal loop (1, 31). ΔH°loop and ΔS°loop are calculated similarly. All the thermodynamic parameters used in this calculation are derived from TM−1 versus ln(CT/4) plots (eq 1). When eq 2a was applied to 2 × 2 nucleotide internal loops of non-canonical pairs flanked by different stems, the values for ΔG°37, loop for a given loop sequence differed by an average of 0.40 kcal/mol (2). The model should be even better for sequences with identical stems.
Table 1.
Measured Thermodynamic Parameters for Duplex Formation in 1 M NaCl, pH7 and in 80 mM NaCl if Listed in Parentheses.
| TM−1 vs ln(CT/4) plots (eq 1) | Average of melt curve fits | |||||||
|---|---|---|---|---|---|---|---|---|
| Sequences | −ΔH° (kcal/mol) |
−ΔS° (eu) |
−ΔG°37 (kcal/mol) |
Tma (°C) |
−ΔH° (kcal/mol) |
−ΔS° (eu) |
−ΔG°37 (kcal/mol) |
Tma (°C) |
| GGUGGAGGCU PCCGAMGCCG |
108.4±3.5 | 302.9±10.4 | 14.47±0.24 | 61.5 | 105.1±1.2 | 293.0±3.6 | 14.24±0.12 | 61.6 |
| GGUGGAGGCU PCCGDAGCCG |
104.2±3.1 | 290.9±9.4 | 13.96±0.22 | 60.8 | 105.4±3.0 | 294.4±9.0 | 14.04±0.23 | 60.8 |
| GGUGgAGGCU PCCGAAGCCG |
103.9±3.1 | 290.8±9.3 | 13.68±0.20 | 59.9 | 100.1±3.0 | 279.3±8.9 | 13.44±0.21 | 60.0 |
| GGUGGAGGCUb PCCGAAGCCG |
94.3±8.2 (82.2±3.4) |
261.2±24.5 (233.5±10.7) |
13.26±0.57 (9.79±0.10) |
60.8 (49.8) |
94.5±2.4 (87.8±4.5) |
261.9±7.2 (251.1±14.1) |
13.27±0.25 9.94±0.15 |
60.8 (49.5) |
| GGUGGAGGCUb PCCAAAGCCG |
92.7±2.2 (84.6±4.3) |
258.0±6.6 (243.2±13.6) |
12.64±0.13 (9.15±0.13) |
58.9 (46.9) |
93.2±3.9 (81.9±9.3) |
259.7±12.0 (234.9±29.2) |
12.67±0.23 (9.09±0.30) |
58.9 (47.0) |
| GGCGGAGGCUb PCCGAAGCCG |
81.2±7.0 (56.1±1.9) |
223.4±21.2 (152.6±6.1) |
11.92±0.47 (8.74±0.05) |
59.1 (49.7) |
77.8±5.8 (60.7±8.3) |
213.1±17.7 (167.2±26.3) |
11.76±0.36 (8.88±0.16) |
59.3 (49.5) |
| GGUGDAGGCU PCCGAAGCCG |
93.6±1.9 | 262.9±5.9 | 12.09±0.11 | 56.6 | 89.4±4.4 | 250.1±13.6 | 11.88±0.22 | 56.7 |
| GGUGIAGGCU PCCGAAGCCG |
90.7±2.5 | 254.3±7.7 | 11.86±0.14 | 56.4 | 89.6±5.0 | 250.8±15.3 | 11.84±0.27 | 56.5 |
| GGUGAAGGCU PCCGAIGCCG |
90.6±3.4 | 255.0±10.4 | 11.52±0.18 | 55.1 | 92.0±3.4 | 259.0±10.2 | 11.61±0.20 | 55.1 |
| GGUGAAGGCU PCCGAMGCCG |
92.6±3.8 | 261.7±11.7 | 11.47±0.19 | 54.5 | 90.3±4.6 | 254.4±14.1 | 11.38±0.23 | 54.6 |
| GGUGAAGGCU PCCGDAGCCG |
89.2±1.8 | 250.8±5.6 | 11.42±0.09 | 55.0 | 88.5±4.6 | 248.6±14.1 | 11.41±0.23 | 55.1 |
| GGUGPAGGCU PCCGAAGCCG |
88.9±2.1 | 250.5±6.4 | 11.17±0.10 | 54.1 | 84.3±2.4 | 236.6±7.5 | 10.97±0.10 | 54.2 |
| GGUGAAGGCUb PCCGAAGCCG |
84.2±6.1 (75.7±3.3) |
235.7±18.6 (219.4±10.4) |
11.12±0.32 (7.64±0.05) |
54.9 (41.6) |
86.5±5.4 (74.9±3.6) |
242.8±16.9 216.8±11.4 |
11.23±0.25 (7.64±0.11) |
54.8 (41.7) |
| GGUGaAGGCU PCCGAMGCCG |
88.7±3.4 | 250.9±10.6 | 10.93±0.16 | 53.2 | 85.5±2.4 | 240.9±7.3 | 10.79±0.14 | 53.3 |
| GGUGaAGGCU PCCGAAGCCG |
86.7±1.5 | 244.2±4.7 | 10.92±0.07 | 53.5 | 82.9±1.5 | 232.7±4.8 | 10.75±0.07 | 53.6 |
| GGUGGA GGCUc PCCGAAGAAACCG |
90.8±1.9 | 259.1±6.0 | 10.47±0.07 | 51.1 | 84.5±4.2 | 239.4±13.0 | 10.25±0.19 | 51.3 |
| GGUGgA GGCU PCCGAAGAAACCG |
89.4±2.9 | 256.1±8.9 | 9.97±0.09 | 49.4 | 81.3±5.2 | 230.8±16.2 | 9.74±0.20 | 49.7 |
| GGUGIAGGCU PCCGAIGCCG |
80.7±1.4 | 228.9±4.4 | 9.70±0.05 | 49.7 | 78.5±1.9 | 222.2±5.9 | 9.61±0.08 | 49.7 |
| GGUGIA GGCU PCCGAAGAAACCG |
80.3±4.1 | 231.7±13.0 | 8.47±0.09 | 44.7 | 71.8±2.5 | 204.8±7.8 | 8.31±0.14 | 44.9 |
| GGUGDA GGCU PCCGAAGAAACCG |
70.0±3.4 | 199.3±11.0 | 8.17±0.06 | 44.4 | 65.2±2.9 | 184.0±9.3 | 8.12±0.13 | 44.8 |
| GGUGAA GGCU PCCGAAGAAACCG |
73.6±4.3 | 211.1±13.7 | 8.07±0.08 | 43.6 | 67.8±7.1 | 192.8±22.5 | 8.01±0.12 | 43.9 |
| GGUGaA GGCU PCCGAAGAAACCG |
72.7±4.8 | 209.0±15.3 | 7.91±0.09 | 43.0 | 66.1±3.3 | 187.7±10.4 | 7.84±0.14 | 43.3 |
| GGUGPA GGCU PCCGAAGAAACCG |
66.1±3.9 | 188.7±12.3 | 7.57±0.07 | 42.0 | 59.2±2.2 | 166.6±7.3 | 7.56±0.14 | 42.5 |
| GGUGGCUb PCCGCCG |
75.6±3.9 | 205.0±11.7 | 12.05±0.28 | 61.4 | 80.8±2.3 | 220.5±7.0 | 12.39±0.17 | 61.3 |
Table 2.
Measured Thermodynamic Parameters for Internal Loop Formation in 1 M
| NaCl, pH7a | ||||
|---|---|---|---|---|
| Sequence | ΔG°37, loop (kcal/mol) |
ΔH°loop (kcal/mol) |
ΔS°loop (eu) |
ΔΔG°37, loop (kcal/mol) |
| GGUGGAGGCU PCCGAMGCCG |
−3.83±0.59 | −38.4±9.9 | −111.4±30.3 | −1.21b |
| GGUGGAGGCU PCCGDAGCCG |
−3.32±0.58 | −42.0±9.7 | −99.4±30.0 | −0.70b |
| GGUGgAGGCU PCCGAAGCCGc |
−3.04±0.57 | −33.9±9.7 | −99.3±29.9 | −0.42b |
| GGUGGAGGCUc PCCGAAGCCG |
−2.62±0.78 [−2.39] |
−24.3±12.4 | −69.7±37.5 | 0.00b |
| GGUGGAGGCUc PCCAAAGCCG |
−2.27±0.59 [−1.44] |
−23.9±9.7 | −69.5±29.5 | − |
| GGUGDAGGCU PCCGAAGCCG |
−1.45±0.54 | −23.6±9.4 | −71.4±29.0 | 1.17b, −0.97d |
| GGUGIAGGCU PCCGAAGCCG |
−1.22±0.55 | −20.7±9.6 | −62.8±29.4 | 1.40b, −0.74d |
| GGUGAAGGCU PCCGAIGCCG |
−0.88±0.56 | −20.6±9.8 | −63.5±30.3 | 1.74b, −0.40d |
| GGUGAAGGCU PCCGAMGCCG |
−0.83±0.57 | −22.6±10.0 | −70.2±30.8 | −0.35d |
| GGUGAAGGCU PCCGDAGCCG |
−0.78±0.54 | −27.0±9.4 | −59.3±29.0 | −0.30d |
| GGUGPAGGCU PCCGAAGCCG |
−0.53±0.54 | −18.9±9.4 | −59.0±29.1 | 2.09b, −0.05d |
| GGUGAAGGCUc PCCGAAGCCG |
−0.48±0.57 [−0.03] |
−14.2±11.1 | −44.2±34.0 | 0.00d |
| GGUGaAGGCU PCCGAMGCCG |
−0.29±0.56 | −26.5±9.8 | −59.4±30.4 | 0.19d |
| GGUGaAGGCU PCCGAAGCCG |
−0.28±0.54 | −16.7±9.3 | −52.7±28.8 | 0.20d |
| GGUGGA GGCUe PCCGAAGAAACCG |
0.17±0.54 [0.20] | −20.8±9.4 | −67.6±29.0 | 0.00f |
| GGUGgA GGCU PCCGAAGAAACCG |
0.67±0.54 | −19.4±9.6 | −64.6±29.8 | 0.50f |
| GGUGIAGGCU PCCGAIGCCG |
0.94±0.54 | −10.7±9.3 | −37.4±28.8 | − |
| GGUGIA GGCU PCCGAAGAAACCG |
2.17±0.54 | −10.3±10.1 | −40.2±31.2 | 2.00f, −0.40g |
| GGUGDA GGCU PCCGAAGAAACCG |
2.47±0.54 | 0.0±9.8 | −7.8±30.5 | 2.30f, −0.10g |
| GGUGAA GGCU PCCGAAGAAACCG |
2.57±0.54 [2.56] | −3.6±10.2 | −19.6±31.6 | 0.00g |
| GGUGaA GGCU PCCGAAGAAACCG |
2.73±0.54 | −2.7±10.4 | −17.5±32.3 | 2.56f, 0.16g |
| GGUGPA GGCU PCCGAAGAAACCG |
3.07±0.54 | 3.9±10.0 | 2.8±31.0 | 2.90f, 0.50g |
Experimental error for ΔG°37, ΔH°, and ΔS° for the canonical stems are estimated as 4%, 12%, and 13.5%, respectively, according to ref (31). There is less error in comparisons between these sequences because the stems are either identical or different by only one or two base pairs. Values in square brackets are predicted according to ref (13);
Compared with from ref (9);
From ref (9);
Compared with from ref (9);
From ref (13);
Compared with from ref (13);
Compared with .
Functional Group Substitutions, NMR Assignments, and Structural Features
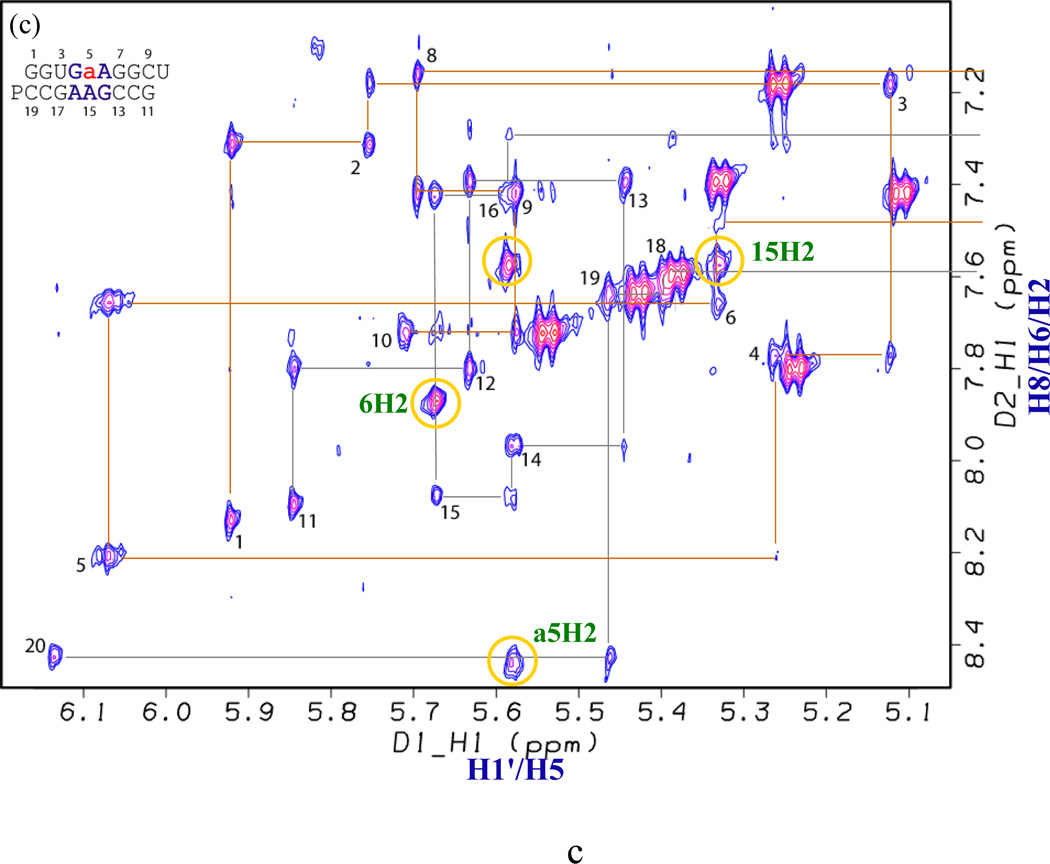
The base – (H1′/H5) “NOESY walk” regions of the 400 ms NOESY spectra at 30 °C are shown in Figure 3. NMR resonances were assigned essentially as described previously (7, 9, 38, 39). Comparison of spectra with those of the duplex (9) facilitates NMR assignments of and (see Supporting Information Tables S1, S2, and S3 for assignments and restraints used for structural modeling). U3A17 in forms a Watson-Crick pair as indicated by a strong NOE between U3H3 and A17H2. The imino proton of U3H3 is relatively broad and shifted upfield (12.53 ppm) (13) relative to the usual range of 13 to 15 ppm for a Watson-Crick UA pair. A similar upfield shift (11.75 ppm) was observed in the 2 × 2 loop (40). Three consecutive sheared GA pairs form in , similar to (Figures 4 and 5) (9). Several medium to strong NOEs, which are similar to NOEs observed for , define the loop structure, e.g. G14H2'–G5H1, A6H1'-A15H2, A15H1'-A6H2, A16H1'-A15H2, G7H1'–A6H2, and A17H1'–A16H2 (compare to G17H1'–A16H2 in ) (Figure 3a and Table 3) (9). In the loop of , G4 and G5 have C3'-endo and C2'-endo sugar puckers, respectively, and G14 is populated in both conformations as evidenced by TOCSY (Supporting Information Figure S1a) and NOESY (Figure 3a) spectra. The same sugar puckers are found in (9).
Figure 3.
(H8/H6/H2)-(H1′/H5) region of the 400 ms mixing time NOESY spectra of duplexes (Figure 2) (a) A17, (b) A5, (c) a5, (d) P5, (e) I5, and (f) I15 at 30 °C in 80 mM NaCl, 10 mM sodium phosphate, 0.5 mM disodium EDTA, pD 7 except I5 duplex at pD 6. For A5 sequence in panel (b), yellow and green circles connected by gray lines identify related cross peaks of major and minor conformations, respectively. Yellow and green circles in other spectra identify cross peaks related to those in circles of the same color for A5 duplex.
Figure 4.
Major groove stereo view of , with A6/A15/A16 stack in minor groove shown in green, and G4/G5/G14 stack in major groove shown in red. Hydrogen and non-bridging oxygen atoms are deleted for clarity.
Figure 5.
Comparison of minor groove view of the crystal and NMR structures. Residues stacking in minor groove are shown in sticks and labeled in bold. Hydrogen and non-bridging oxygen atoms are deleted for clarity. Top panel: NMR and crystal structures of the internal loop . Three G’s and three A’s in the loop are shown in red and green, respectively. Bottom panel: NMR structures of the duplex with an alternating middle sheared AA pair.
Table 3.
Distance Restraints Involving A5 and A15 Residues for the Structural Modeling of a and comparison with that of (9).
| Distance restraints that differ for the structural modeling of A6/A15/A16 and A6/A5/A16 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A6/A15/A16 | A6/A5/A16 | ||||||||||
| Distance (Å) |
Distance (Å) |
||||||||||
| Atom1 | Atom2 | Lower | Upper | NMR | Modelb | Atom1 | Atom2 | Lower | Upper | NMR | Modelb |
| G4H3' | A5H8 | 1.80 | 5.00 | 3.35 | 3.50/4.73 | G14H3' | A15H8 | 1.80 | 5.00 | 2.79 | 4.47/2.80 |
| G4H8 | A5H8 | 3.40 | 6.00 | 4.86 | 5.41/6.68 | G14H8 | A15H8 | ×c | × | × | 6.68/5.51 |
| A5H1' | A6H8 | 1.80 | 4.12 | 3.17 | 3.11/4.98 | A15H1' | A16H8 | 1.80 | 4.34 | 3.34 | 5.20/2.75 |
| G14H1' | A5H2 | 1.80 | 5.00 | 4.62/10.32 | G4H1' | A15H2 | 1.80 | 5.00 | 10.22/4.23 | ||
| G14H2' | A5H2 | 2.00 | 5.00 | 2.21/9.41 | G4H2' | A15H2 | 1.80 | 5.00 | 9.98/2.27 | ||
| A5H2 | A15H8 | 2.00 | 5.00 | 2.30/10.41 | A5H8 | A15H2 | 1.80 | 5.00 | 10.47/2.35 | ||
| A5H4' | A16H2 | 2.83 | 5.26 | 4.05 | 4.02/5.38 | A15H4' | A6H2 | 3.00 | 6.00 | 5.52/4.44 | |
| A6H1' | A15H2 | 1.80 | 4.86 | 3.74 | 2.55/7.66 | A16H1' | A5H2 | 1.80 | 5.00 | 7.11/2.88 | |
| A15H1' | A6H2 | 1.80 | 4.14 | 3.19 | 2.84/6.60 | A5H1' | A16H2 | 1.80 | 5.00 | 6.08/2.74 | |
| G14H1' | A15H8 | 2.00 | 5.50 | 4.23 | 4.69/5.99 | G4H1' | A5H8 | 1.80 | 5.52 | 4.24 | 5.73/4.82 |
| A16H1' | A15H2 | 1.80 | 4.30 | 3.31 | 2.80/7.39 | A6H1' | A5H2 | 1.80 | 5.00 | 7.75/2.80 | |
| A15H8 | A16H8 | 3.08 | 5.73 | 4.41 | 4.08/6.16 | A5H8 | A6H8 | 3.27 | 6.00 | 4.67 | 6.58/4.06 |
| A15H2' | A16H8 | 1.80 | 3.99 | 3.07 | 3.32/4.64 | A5H2' | A6H8 | 1.80 | 4.66 | 3.58 | 5.07/3.13 |
| A15H3' | A16H8 | 1.80 | 5.00 | 2.48/4.80 | A5H3' | A6H8 | 1.80 | 4.91 | 3.77 | 4.86/2.47 | |
| Distance restraints that are the same for A6/A15/A16 and A6/A5/A16 | |||||||||||
| Distance (Å) | Distance (Å) | ||||||||||
| Atom1 | Atom2 | Lower | Upper | NMR | Modelb | Atom1 | Atom2 | Lower | Upper | NMR | Modelb |
| G4H2' | A5H8 | 1.80 | 5.00 | 4.86 | 3.08/3.10 | G14H2' | A15H8 | 1.80 | 4.76 | 3.66 | 2.83/3.44 |
| A5H1' | A5H2' | 1.80 | 3.94 | 3.03 | 2.85/2.45 | A15H1' | A15H2' | 1.80 | 3.65 | 2.80 | 2.61/2.71 |
| A5H1' | A5H3' | 1.80 | 4.81 | 3.70 | 3.70/3.56 | A15H1' | A15H3' | 1.80 | 5.00 | 4.19 | 3.66/3.69 |
| A5H1' | A5H4' | 1.80 | 4.41 | 3.39 | 3.36/3.16 | A15H1' | A15H4' | 1.80 | 4.42 | 3.40 | 3.32/3.27 |
| A5H1' | A5H8 | 1.80 | 4.97 | 3.82 | 3.72/3.41 | A15H1' | A15H8 | 1.80 | 5.04 | 3.88 | 3.48/3.68 |
| A5H2' | A5H8 | 1.80 | 3.19d | 2.45 | 2.10/3.97 | A15H2' | A15H8 | 1.80 | 5.00 | 2.79 | 4.11/2.03 |
| A5H3' | A5H8 | 1.80 | 5.00 | 3.35 | 4.09/3.42 | A15H3' | A15H8 | 1.80 | 4.27 | 3.28 | 3.42/3.89 |
| Distance restraints involving G5 and A15 residues for the structure modeling of
(9)e | |||||||||||
| Distance (Å) |
Distance (Å) |
||||||||||
| Atom1 | Atom2 | Lower | Upper | NMR | Model | Atom1 | Atom2 | Lower | Upper | NMR | Model |
| G4H3' | G5H8 | 2.41 | 4.47 | 3.44 | 3.87 | G14H3' | A15H8 | × | × | × | 4.50 |
| G4H8 | G5H8 | 2.36 | 6.00 | 3.37 | 5.69 | G14H8 | A15H8 | × | × | × | 6.35 |
| A5H1' | A6H8 | 2.30 | 4.27 | 3.29 | 3.55 | A15H1' | A16H8 | 3.28 | 6.00 | 4.69 | 4.81 |
| G14H1' | A5H2 | -f | - | - | - | G4H1' | A15H2 | × | × | × | 8.77 |
| G14H2' | G5H1 | 2.50 | 5.84 | 4.17 | 4.05 | G4H2' | A15H2 | × | × | × | 8.74 |
| A5H2 | A15H8 | - | - | - | - | G5H8 | A15H2 | × | × | × | 10.38 |
| G5H4' | A16H2 | × | × | × | 5.21 | A15H4' | A6H2 | × | × | × | 5.49 |
| A6H1' | A15H2 | 2.15 | 3.99 | 3.07 | 2.45 | A16H1' | A5H2 | - | - | - | - |
| A15H1' | A6H2 | 2.13 | 3.95 | 3.04 | 2.92 | G5H1' | A16H2 | × | × | × | 6.99 |
| G14H1' | A15H8 | 3.27 | 6.00 | 4.67 | 4.44 | G4H1' | A5H8 | × | × | × | 6.35 |
| A16H1' | A15H2 | 2.06 | 3.83 | 2.94 | 2.53 | A6H1' | A5H2 | - | - | - | - |
| A15H8 | A16H8 | × | × | × | 4.82 | G5H8 | A6H8 | × | × | × | 7.16 |
| A15H2' | A16H8 | 1.94 | 3.60 | 2.77 | 2.20 | G5H2' | A6H8 | × | × | × | 5.47 |
| A15H3' | A16H8 | 2.39 | 4.44 | 3.42 | 2.87 | G5H3' | A6H8 | × | × | × | 5.14 |
| G4H2' | G5H8 | 2.73 | 5.07 | 3.90 | 3.66 | G14H2' | A15H8 | 2.02 | 3.74 | 2.88 | 2.50 |
| G5H1' | G5H2' | 2.23 | 4.14 | 3.19 | 2.99 | A15H1' | A15H2' | 2.30 | 4.26 | 3.28 | 2.73 |
| G5H1' | G5H8 | 3.29 | 6.00 | 4.70 | 3.91 | A15H1' | A15H8 | 3.15 | 5.85 | 4.50 | 3.74 |
| G5H2' | G5H8 | 1.43 | 2.66 | 2.05 | 2.25 | A15H2' | A16H1' | 3.06 | 5.68 | 4.37 | 3.55 |
| G5H3' | G5H8 | 2.46 | 4.57 | 3.52 | 4.05 | A15H3' | A15H8 | 2.09 | 3.88 | 2.99 | 3.14 |
| G5H1 | G4H1 | 2.34 | 5.46 | 3.90 | 3.52 | A15H2 | A6H8 | 3.84 | 6.00 | 5.48 | 4.70 |
| G5H1 | G14H1 | 2.50 | 7.00 | 5.41 | A15H2 | A16H2 | 3.49 | 6.00 | 4.99 | 4.54 | |
All other distance restraints for both structural modelings are identical and are provided in Supporting Information.
Distances measured for the averaged structure of A6/A15/A16 followed by A6/A5/A16;
Cross-peaks not observed and labeled with ×;
Loosened upper bound to 4.50 Å for the structure modeling of A6/A5/A16;
The distances in the columns of models are measured from a representative structure of (9);
not applied and labeled with -.
Similarity of sugar edges (6) of adenosine, purine riboside, and inosine (Figure 1) facilitates H2 assignments of middle purine-purine pairs. Also, the NOEs of G14H1'/H2'-A5H2 in (Figure 3b) are similar to those of G14H1'/H2'–G5H1 as present for (9) and (Table S1). Both sets of NOEs, G14H1'/H2'–I5H2 and G14H1'/H2'–I5H1 (data not shown), are observed in the I5 duplex, (Figure 3e). The chemical shift of I5H1, 12.1 ppm (Supporting Information Figure S2) is in agreement with the formation of a sheared IA pair (41). Downfield chemical shifts for inosine imino protons beyond 14 ppm were observed in face-to-face IA pairs (4, 42).
While comparison of NOESY spectra (Figure 3, Table 3, and Supporting Information Table S2) indicates very similar base pairing and stacking geometries with three sheared-type purine-purine pairs for (a5 duplex), (P5 duplex), and (I5 duplex), the NMR spectra provide evidence for two populations of structures for the middle A5A15 pair in (A5 duplex) (Figure 6). For example, in addition to NOEs of R5H2-G14H1'/H2' (R is any purine), A6H2-A15H1', A15H2-A6H1', and A15H2 -A16H1' (shown in yellow circles) for the a5, P5, and I5 duplexes, one extra set of NOEs A15H2- G4H1'/H2', A16H2-A5H1', A5H2-A16H1', and A5H2- A6H1' (shown in green circles) are present for (Figure 3). Moreover, on the basis of TOCSY (Supporting Information Figure S1) and NOESY (Figure 3) spectra, sugar puckers of a5, P5, and I5 are C2'-endo as indicated by strong H1'–H2' couplings, which corresponds to the C2'-endo conformation of the G5 sugar pucker in (9). Sugar puckers for A15 are C3'-endo in each of these duplexes with the possible exception of the a5 duplex. For , however, A5 and A15 are populated in both C2'-endo and C3'-endo conformations with A5 having higher C2'-endo population than A15. Evidently, the middle A5A15 pair is more populated in trans Hoogsteen/sugar edge A15-A5 than in trans Hoogsteen/sugar edge A5-A15 and related conformations. This is in agreement with relatively stronger NOEs (Figure 3b) observed for A5H2-G14H1'/H2', A6H2- A15H1', A15H2- A6H1', and A15H2- A16H1' (shown in yellow circles) than A15H2- G4H1'/H2', A16H2- A5H1', A5H2- A16H1', and A5H2-A6H1' (shown in green circles), respectively, in . Presence of a single set of chemical shifts for indicates that the middle A5A15 pair is alternating rapidly (fast exchange on the NMR time scale), with the sugar edge of either A5 or A15 on the base pairing edge of the other, forming trans Hoogsteen/sugar edge A15-A5 (shown in yellow) or A5-A15 (shown in green) and related pairs (Figure 5).
Figure 6.
Major groove stereo views of two alternating structures of with A6/A15/A16 or A6/A5/A16 stacks in minor groove. Two G’s and four A’s in the loop are shown in red and green, respectively. Hydrogen and non-bridging oxygen atoms are deleted for clarity. The hydrogen bonding shown for the AA pair in the minor conformation is similar to that shown in Figure 1a, but a variety of hydrogen bonding patterns are seen in the ensemble of structures generated with the restraints from NMR data.
NMR spectra acquired at 1 M NaCl indicate that the structural and dynamical properties of the A5 loop are similar at 80 mM and 1 M NaCl (Figure 3b and S3, respectively). Chemical shifts show only modest salt dependence and the same pattern of NOEs for the major and minor conformations are observed, including the A5H2-A15H8 (major) and A15H2-A5H8 (minor) cross peaks (data not shown).
Relatively downfield chemical shifts of H2 protons on the base pairing edge (Figure 1) are observed for P5H2, 9.30 ppm (compared to P20H2, 8.16 ppm (9)), a5H2, 8.44 ppm (compared to A15H2, 7.57 ppm in the a5 duplex), and I5H2, 8.69 ppm, in , , and , respectively (Figure 3 and Table 4). Such downfield chemical shifts of H2 on the base pairing edge of sheared purine-purine pairs are expected due to ring current de-shielding effects (Figure 1a), as observed previously in 2 × 2 loops: A4H2, 8.19 ppm in (compared to 7.88 ppm for A5H2, which is not on a base pairing edge) and P4H2, 8.97 ppm in (18).
Table 4.
Chemical shifts (ppm) and full widths (Hz) at half height of H2 peaks of the central loop residues, 5H2 and 15H2, in A5, a5, I5, P5, and I15 duplexes at 30 °C in 80 mM NaCl. G2H8 from the stem is included for reference. Error limits are listed in parentheses.
| Linewidths (Hz) | Chemical shifts (ppm) | |||||
|---|---|---|---|---|---|---|
| Duplex | 5H2 | 15H2 | G2H8 | 5H2 | 15H2 | G2H8 |
| A5 | 15.0 (1.5)a | 13.1 (1) | 4.1 (1) | 8.28 | 7.83 | 7.32 |
| a5 | 12.0 (2) | 6.2 (1.5) | 3.8 (1) | 8.44 | 7.57 | 7.31 |
| P5 | 10.0 (1) | 4.3 (1) | 3.6 (0.5) | 9.30 | 7.69 | 7.33 |
| I5 | 7.1 (1) | 4.1 (1) | 4.4 (0.5) | 8.69 | 7.55 | 7.35 |
| I15 | 4.4 (1) | 12.0 (1.5) | 4.8 (0.5) | 7.51 | 8.55 | 7.29 |
If the chemical shift of the A5H2 resonance in the A5 duplex differs by 0.87 ppm between major and minor conformations and the fraction of A5 duplexes in the major conformation ranges between 0.6 and 0.9, then a rough calculation (67, 68) assuming an inherent linewidth of 4 Hz suggests the rate of exchange between the two conformations of the A5 duplex is between 20,000 and 65,000 s−1.
On the edge that is not base paired, relatively upfield chemical shifts of H2 protons are observed: A15H2 chemical shifts are 7.69, 7.57, and 7.55 ppm in , , and , respectively (Figure 3 and Table 4). These can be compared to the relatively further upfield chemical shifts of 7.13 and 7.16 ppm for A15H2 protons in and , respectively, which might reflect stronger base pairing and better stacking of the motif with three consecutive sheared GA pairs, resulting in larger ring current shielding effects from A6 and A16. Intermediate chemical shifts of A5H2 (8.28 ppm) and A15H2, (7.83 ppm) are observed in , which is consistent with a rapidly alternating sheared A5A15 pair.
The I15 duplex provides further evidence for two structures of the A5 duplex. Several features in the NMR spectra suggest that the loop conformation in the I15 duplex is most similar to the less populated conformation observed in the A5 duplex. NOEs observed in the I15 duplex that were weak in A5 duplex, and not observed at all in a5, P5, or I5 duplexes include 15H2-G4H1', A16H2–5H1', 5H2-A16H1', 5H2-A6H1', and 15H2–5H8 (Figure 3). NOEs not observed in I15 duplex, but observed in and all other duplexes include 5H2-G14H1', A6H2–15H1', 15H2-A6H1', and 15H2-A16H1'. The relatively downfield chemical shift of I15H2 (8.55 ppm) and the relatively upfield shift of A5H2 (7.51 ppm) are consistent with those protons being at the A5-I15 base-pairing edge and out in the minor groove, respectively, as discussed for the other duplexes. A strong scalar coupling, I15H1'-H2', and a large downfield 31P shift of A16 (2.75 ppm) indicate a C2'-endo ribose conformation at I15. In contrast, A5H1'-H2' scalar coupling is much smaller and the 31P shift of A6 is 1.10 ppm. These contrast with a5, P5, and I5 duplexes which show strong 5H1'-H2' coupling and weak 15H1'-H2' coupling. Moreover, 31P shifts of I5 duplex are 0.86 and 2.58 ppm for A16 and A6, respectively. Additionally, the G4H1'-H2' and G14H1'-H2' couplings are moderate and zero, respectively, in the I15 duplex while they are zero and moderate, respectively, in a5, P5, and I5 duplexes. The moderate couplings probably indicate dynamic interconversion of sugar puckers. The 31P chemical shifts for the A5 duplex are 1.41 and 2.20 ppm for A16 and A6, which suggest that the A5 duplex is more populated in a conformation similar to a5, P5, and I5 duplexes.
There is a large chemical shift difference between the H2 resonances for the central purine-purine paris, and the linewidths of these resonances are consistent with two rapidly interconverting structures for the A5 duplex (Table 4). The linewidths for A5H2 and A15H2 are both about 14 Hz for the A5 duplex. In contrast, the H2 resonance not at the base pairing edge in a5, P5, I5, and I15 duplexes has an average linewidth of 4.75±1 Hz, which is approximately the same as the average of the stem resonance G2H8 (4.1±0.5 Hz). This is consistent with a model in which duplexes having a single conformation have a relatively narrow linewidth, while the A5 duplex resonances are broadened due to switching between two conformations. The base pairing edge H2 resonance in a5, P5, I5, and I15 duplexes has average linewidth of 10.2±2.3 Hz. This may indicate that this residue is slightly less stable than the pairing partner on the other strand, and/or that the chemical shift of the proton in this position is more sensitive to slight structural fluctuations.
Structural statistics for and
A total of 24 of 40 modeled structures (PDB: 2DD1) were selected for analysis of . The average root-mean-square deviation of all selected structures to the average structure for all atoms is 0.80 ± 0.15 Å. No distance or dihedral angle restraint violations were greater than 0.2 Å or 2°, respectively. The average of the final energies at 300 K from the force field is − 428.0 ± 4.9 kcal/mol.
A total of 27 of 40 modeled structures (PDB: 2DD2) were selected for analysis of the A6/A15/A16 structure of . The average root-mean-square deviation of all selected structures to the average structure for all atoms is 0.69 ± 0.21 Å. Two distance and one dihedral angle restraint violations were greater than 0.2 Å and 2°, respectively. The average of the final energies at 300 K from the force field is −430.6 ± 5.7 kcal/mol.
A total of 18 of 40 modeled structures (PDB: 2DD3) were selected for analysis of the A6/A5/A16 structure of . The average root-mean-square deviation of all selected structures to the average structure for all atoms is 0.97 ± 0.25 Å. One distance and no dihedral angle restraint violations were greater than 0.2 Å and 2°, respectively. This minor conformation is less convergent than the other structures due to loosened restraints in the loop region. The average of the final energies at 300 K from the force field is −423.4 ± 6.3 kcal/mol.
Other known AA geometries are not consistent with the NMR data
As illustrated in Figure 1b, several non-sheared AA pairs have been observed in crystal and NMR structures (6, 43–49). These potential structures can be compared to the NMR spectra for .
A potential A-zipper motif, as seen in a DNA internal loop and NMR structure of an RNA tetraloop receptor (50, 51), which would place A5 and A15 in stacking on each other, is ruled out because no NOE of A5H2 – A15H1' or A15H2–A5H1' is observed in .
Both AH2 protons are exposed in the minor groove for a trans Watson-Crick/Hoogsteen A–A pair (Figure 1b) (6, 43). This conformation is ruled out for the middle AA pair in because it would not give the observed cross-strand G4H1'/H2'–A15H2 and G14H1'/H2'–A5H2 cross-peaks (Figure 3b, Table 3, and Supporting Information Table S2).
The cis Watson-Crick/Watson-Crick A–A conformation (6, 45), with both AH2 protons exposed in the minor groove, is ruled out for the middle AA in because it would not give all the observed G14H1'/H2'-5H2, A6H1'-A15H2, A15H1'-A6H2, A16H1'-A15H2 (shown in yellow circles); and G4H1'/H2'-A15H2, A16H1'-A5H2, A5H1'-A16H2, and A6H1'-A5H2 (shown in green circles) cross-peaks (Figure 3b).
A trans Hoogsteen/Hoogsteen A–A pair (Figure 1b) (6, 46–49) is ruled out for the middle AA in because there is no indication of a syn glycosidic conformation as evidenced by A5H1'-A5H8 and A15H1'-A15H8 cross-peaks and because the 14H1'/H2'– 5H2 and 4H1'/H2'–15H2 cross-peaks seen in are not expected for a trans Hoogsteen/Hoogsteen A–A with two AH2 protons exposed in the minor and major groove, respectively.
DISCUSSION
Understanding relationships between sequence, energetics, structure, dynamics, and function can facilitate rapid extraction of the information encoded in the constantly expanding databases of RNA sequences. The internal loop is a common RNA motif where such relationships are not fully understood (9, 12, 52–54). Detailed understanding of interactions such as hydrogen bonding and base stacking in internal loops will allow prediction of the contributions of internal loops to RNA folding and function.
Three Consecutive Sheared GA Pairs in
The previous NMR structure of reveals three consecutive sheared GA pairs in the unusually stable internal loop (9). Formation of three consecutive sheared GA pairs in (−2.27 kcal/mol) (Figure 4 and 5), similar to (−2.62 kcal/mol), is consistent with the thermodynamic stabilities (Table 2) and the occurrences of both loops in helix 41a of small subunit rRNA (52). (Throughout the paper, the values in parenthesis after the duplex are the measured free energy at 37 °C for loop formation in 1 M NaCl unless otherwise noted.)
In contrast to the NMR structures, helix 68 of the crystal structure of D. radiodurans large subunit rRNA contains a loop that has only one sheared GA pair (shown in bold) (14). The major difference is that the corresponding G5 and A15 bases are shifted, opposite to a sheared GA pair, to the minor and major groove, respectively. This results in the loss of hydrogen bonding, and in a corresponding base stacking pattern equivalent to A6/G5/A16 in the minor groove, instead of the A6/A15/A16 stacking pattern found in the NMR structure (Figure 5). Several critical cross-strand NOEs define the stacking pattern of A6/A15/A16 in the NMR structure with three consecutive sheared GA pairs, e.g. A15H2–A6H1', A15H1'–A6H2, and A15H2–A16H1' (Figure 3a, Supporting Information Table S1). The distances between the protons in each pair exceed 5 Å in the crystal structure (PDB: 1NKW) when hydrogens are added (Supporting Information Table S1). Interestingly, the A6/G5/A16 stacking pattern in the crystal structure (Figure 5, top panel) is similar to the A6/A5/A16 stacking pattern determined for the minor NMR structure of (Figure 5, bottom panel), although fewer hydrogen bonds are formed in the crystal structure (14).
There are several differences between the environments of the loop in the crystal and in NMR buffer. The crystals were grown from ribosomal subunits in 10 mM MgCl2, 60 mM NH4Cl, 5 mM KCl, and 10 mM HEPES, pH 7.8 (14). The NMR buffer has 80 mM NaCl and 10 mM sodium phosphate, pD 6.8. It would be surprising, however, if Mg2+ shifted the local structure. The thermodynamics of the 3GA duplex (Figure 2) were essentially the same in 1 M NaCl and in 10 mM MgCl2, 150 mM KCl (9). It is quite possible, however, that other interactions in the ribosomal subunit crystal are strong enough to break hydrogen bonds and rearrange stacking. There is no tertiary interaction with or protein binding to the loop in the crystal, but the loop, , which is directly 5' to the UA closing pair of , has tertiary interactions with helix 75 by consecutive A-minor interactions. Similar A-minor tertiary interactions are observed in the crystal structures of the large ribosomal subunits of Haloarcula marismortui and E. coli between helix 75 and helix 68 (55, 56). While long range effects may affect local structure, it may also be difficult to determine such fine details in a large crystal refined to 3.1 Å.
Tandem sheared GA pairs closed by UA Watson-Crick pairs have been reported in an NMR structure of (40). The sugars of G’s in are in C2'-endo conformation. In the loop, , of the A17 duplex, G4 and G5 have C3'-endo and C2'-endo sugar puckers, respectively, and G14 is populated in both conformations as evidenced by the TOCSY spectrum (Supporting Information Figure S1a). Evidently, a C2'-endo sugar pucker for G is not required to form a sheared GA pair in a motif.
Two Alternating Structures for
The sheared AA pair in is rapidly exchanging between alternative conformations (Figure 5, bottom panel and Figure 6). This exchange is consistent with an intrinsically flexible AA pair with fewer hydrogen bonds than a GA pair. The pseudo-symmetry of the dynamic AA pair allows an estimate of the lower limit for the exchange rate. The a5 duplex has only one conformation and it is the same as the major conformation of the A5 duplex. The chemical shifts for a5H2 and A15H2 in this conformation are 8.44 and 7.57 ppm, respectively. With the assumption that the minor conformation of the A5 duplex would result in an A5H2 chemical shift of 7.57 ppm, the lower limit for the exchange rate is estimated as 0.87 (500) = 435 s−1. A calculation based on linewidths (57, 58) of H2 resonances suggests an even faster exchange rate of between 20,000 and 65,000 s−1 (Table 4) for estimates of the major conformation ranging from 90% to 60 % of the population. Fast exchange has also been detected between syn and anti G’s in a single GG pair in an RNA duplex (17). A recent theoretical study has provided insight into possible mechanisms for such rapid exchange in the absence of duplex dissociation (59).
In contrast to , duplexes a5, P5, I5, and I15 all have a single predominant conformation. As indicated by NOE patterns (Figure 3), there is little structural change for a5, P5, and I5 duplexes relative to (9) and to the major conformation of the A5 duplex. NMR data for the I15 duplex suggest that its structure differs from that of a5, P5, and I5 duplexes, but resembles the minor structure of the A5 duplex.
The duplex with a deoxyadenosine (a5), (−0.28 kcal/mol), has a single structure with a trans Hoogsteen/sugar edge A15-a5 pair (Figure 1). This is consistent with the fact that G5 is predominantly in C2'-endo sugar pucker conformation with a trans Hoogsteen/sugar edge A15-G5 pair in (−2.62 kcal/mol) (9). Presumably, the a5 duplex has a single structure because the deoxy sugar favors C2'-endo sugar pucker which thus favors a single conformation similar to that of (9). This is also consistent with the observation that the deoxy g5 substitution in (−3.04 kcal/mol) enhances loop stability by 0.42 kcal/mol despite loss of hydrogen bonds G5 (2'-hydroxyl) – G14 (imino/amino). The opposite change in thermodynamic stability is observed for (0.67 kcal/mol) compared with (0.17 kcal/mol). Perhaps the greater flexibility of a 3 × 6 loop negates the necessity for a C2'-endo sugar.
Previous NMR studies showed no orientation exchange for the tandem sheared AA pairs of (7), presumably because switching the orientation would result in making the backbone too narrow for the adjacent Watson-Crick pair (60, 61). Evidently, switching the side where backbone narrowing occurs is not a problem when an AA pair is flanked by sheared GA pairs.
It has been pointed out that might be a potential groove binding and/or intercalation site (43). The alternating sheared AA pair in could potentially serve as a switch between different binding partners for dynamic functions because the smooth N1-C2-N3 edge of either A5 or A15 is presented differently in the minor groove in alternative orientations (Figure 5, bottom panel).
Energetics of molecular recognition
The consistency of structures for the 3GA, I5, P5, and a5 duplexes (Figures 2 and 3) provides models for studying the interactions determining the energetics of a 3 × 3 loop with three sheared pairs. The P5 duplex (−0.53 kcal/mol) is thermodynamically similar to the a5 duplex (−0.28 kcal/mol) and to (−0.48 kcal/mol) (Table 2). This is in agreement with the formation of a sheared PA pair (trans Hoogsteen/sugar edge A15-P5) without loss of hydrogen bonds compared with a sheared AA pair (Figure 1).
The I5 duplex (−1.22 kcal/mol) is 1.40 kcal/mol less stable than (−2.62 kcal/mol). Similar destabilizations of 1.74 and 2.00 kcal/mol are observed with the G to I substitutions in (−0.88 kcal/mol) and (2.17 kcal/mol), respectively (Table 2). This is presumably primarily due to the loss of hydrogen bonds of G5 amino to A15N7 and to the A15 non-bridging oxygen in the IA pair compared to GA (Figure 1). The free energy of about 1.5 kcal/mol attributed to two hydrogen bonds is a lower limit because subtle rearrangement of three-dimensional structure is expected upon inosine substitution and this can strengthen the remaining hydrogen bonds (62).
Interestingly, (−1.22 kcal/mol) is more stable than (−0.53 kcal/mol), (−0.48 kcal/mol), and (−0.28 kcal/mol) (Table 2 and Figure 2), even though the number of base–base hydrogen bonds is expected to be the same (Figure 1). A water-mediated hydrogen bond between the G imino proton and the non-bridging oxygen of A was predicted (15) and observed in a crystal structure of a GNRA tetraloop (63). A similar water-mediated hydrogen bond might exist between I5H1 and an A15 non-bridging oxygen in the I5 duplex, . This water-mediated hydrogen bond might explain the extra stability of the I5 duplex relative to P5, A5, and a5 duplexes (Figure 2).
The D5 (2,6-diamino purine) duplex, (−1.45 kcal/mol), is about 1.2 kcal/mol less stable than (−2.62 kcal/mol) (Table 2 and Figure 2), even though no base–base hydrogen bonds are lost (Figure 1). The destabilizing effect upon substituting D5 for G5 may also be due to loss of the proposed water-mediated hydrogen bond between G5H1 and an A15 non-bridging oxygen (15, 63). It is also possible that the 2-amino group of G is a better hydrogen bond donor than that of D (2,6-diamino purine) because of relatively larger positive partial charges on the G amino hydrogens (64). Larger destabilization of 2.3 kcal/mol is observed for (2.47 kcal/mol) compared with (0.17 kcal/mol). Perhaps the greater flexibility of the size asymmetric loop allows binding of a water molecule in a GA pair to be more favorable. In contrast, D16 substitution for A16 to give (−3.32 kcal/mol) and (−0.78 kcal/mol) stabilizes the loop by −0.70 kcal/mol and −0.30 kcal/mol, respectively, even though the 6-amino group on D (2,6-diamino purine) has essentially the same partial charges as those of A (64). Possibly the extra amino group on D allows better stacking and/or extra hydrogen bonding to the backbone.
A 2'-O-methyl substitution favors the C3'-endo sugar conformation (65–67). The 2'-O-methyl A15 substitution in (−3 83 kcal/mol) and (−0.83 kcal/mol) stabilizes the duplexes by −1.21 and −0.35 kcal/mol relative to (− 2.62 kcal/mol) and (−0.48 kcal/mol), respectively (Table 2). Part of the reason for the smaller effect in the loop may be that the 2'-O-methyl substitution limits the loop to a single conformation. If the two conformations of the natural loop have equal concentrations, then the dynamics would favor loop formation by ΔG°37 = −TΔS = −310(1.987)ln2 = −0.4 kcal/mol.
The motif within other internal loops
Adjacent sheared GA and AA pairs are also found in the loops and in helix 89 of the large ribosomal subunits of H. marismortui (45) and E. coli (56), respectively. In this case, the bold A pairs with its Hoogsteen edge and the CC pair is in a cis Watson-Crick bifurcated conformation (45) according to Leontis-Westhof nomenclature (6). The same loop sequence occurs in helix 41a of E. coli 16S rRNA with similar three dimensional conformation (56).
Other motifs have non-sheared pairs. Only face-to-face (cis Watson-Crick/Watson-Crick) purine-purine pairs (Figure 1) form in the loop in helix 23 of the crystal structure of T. thermophilus 16S rRNA (44), which is consistent with previous NMR studies that imino GA (face-to-face, cis Watson-Crick/Watson-Crick A–G) is favored in the motif (68). The crystal structure of a symmetric 4×4 loop shows little base overlap for the nearest neighbors with sheared GA (trans Hoogsteen/sugar edge A–G) pairs and trans Watson-Crick/Hoogsteen A–A pairs (43). (In the trans Watson-Crick/Hoogsteen A–A pairs, the A paired with Hoogsteen edge is shown in bold (Figure 1).) Evidently, is an intrinsically flexible structure within size-symmetric internal loops.
The free energy increment for at 37 °C in 1 M NaCl is 0.96 kcal/mol as calculated from the measurement of the duplex (13). In contrast, internal loops with consecutive GA pairs are very stable, e.g. (−4.27 kcal/mol) (13), which is consistent with extensive stacking and hydrogen bonding as observed in the crystal structure of the loop with four sheared GA pairs (69). Previous thermodynamic studies showed that the destabilizing 2 × 2 loop (1.2 kcal/mol) with two sheared AA pairs has similar base pairing and stacking geometries but fewer hydrogen bonds than (−0.7 kcal/mol) with two sheared GA pairs (2, 5, 7, 8). Evidently, the thermodynamic and structural effects of replacing a GA pair with an AA pair are context dependent.
A recently proposed “reverse kink-turn” motif involves a size asymmetric 2 × 5 internal loop, , with a sheared GA followed by a symmetric AA pair (trans Hoogsteen/Hoogsteen A–A) (Figure 1b) (49). Such a conformation is also observed in some Loop E motifs, (47, 48). The glycosidic bond of the A (in bold) 3' to the G of the sheared GA pair is in a syn conformation. Thus it might facilitate the packing between two stems via major grooves, with the smooth N1-C2-N3 edge of the bold A flipped to the major groove. Different detailed structures of an AA pair adjacent to a sheared GA pair in nearest neighbors with the bold A paired on its Hoogsteen side are also observed in kink-turn motifs (70) within internal loops, such as kt-11 (trans Watson-Crick/Hoogsteen A–A), and multibranch loops such as kt 94/99 (trans Hoogsteen/Sugar edge A–A), kt 4/5 (trans Hoogsteen/Hoogsteen A-A, with the A in a syn glycosidic conformation). These kink-turns facilitate local and long-range tertiary interactions (70). In these cases, the A 3' to the G of a sheared GA pair prefers to base pair with its Hoogsteen edge. Evidently, the nearest neighbor is intrinsically flexible compared with the motif of consecutive GA pairs (13) in both size symmetric and asymmetric internal loops.
Supplementary Material
ACKNOWLEDGEMENT
We thank B. Tolbert for help with the PyMOL program and critical reading of the manuscript; Prof. R. Kierzek and Dr. E. Kierzek for discussions on synthesis of oligonucleotides with modified nucleotides.
Footnotes
This work was supported by NIH grant GM22939 (D. H. T.)
Protein Data Bank entry: 2DD1 (A17 duplex); 2DD2 and 2DD3 (A5 duplex)
Abbreviations: a, deoxyadenosine; CT, total concentration of oligonucleotide strands; D, 2,6-diaminopurine riboside; g, deoxyguanosine; I, inosine; M, 2'-O-methyl adenosine; P, purine riboside; R, any purine nucleotide; TM, melting temperature in kelvin; Tm, melting temperature in degrees Celsius.
SUPPORTING INFORMATION AVAILABLE
Tables listing chemical shift assignments, tables of NMR distance restraints, and figures of TOCSY, one-dimensional proton spectra (9–14.5 ppm), and 2D NOESY spectrum at 1 M NaCl are available. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 2.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Zon G, Wilson WD. NMR and molecular modeling evidence for a GA mismatch base pair in a purine rich DNA duplex. Proc. Natl. Acad. Sci. U. S. A. 1991;88:26–30. doi: 10.1073/pnas.88.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SantaLucia J, Jr, Turner DH. Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics. Biochemistry. 1993;32:12612–12623. doi: 10.1021/bi00210a009. [DOI] [PubMed] [Google Scholar]
- 6.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Znosko BM, Burkard ME, Schroeder SJ, Krugh TR, Turner DH. Sheared Aanti·Aanti base pairs in a destabilizing 2 × 2 internal loop: The NMR structure of 5'(rGGCAAGCCU)2. Biochemistry. 2002;41:14969–14977. doi: 10.1021/bi020326f. [DOI] [PubMed] [Google Scholar]
- 8.Jang SB, Baeyens K, Jeong MS, SantaLucia J, Jr, Turner DH, Holbrook SR. Structures of two RNA octamers containing tandem G·A base pairs. Acta Crystallogr. D. 2004;60:829–835. doi: 10.1107/S0907444904003804. [DOI] [PubMed] [Google Scholar]
- 9.Chen G, Znosko BM, Kennedy SD, Krugh TR, Turner DH. Solution structure of an RNA internal loop with three consecutive sheared GA pairs. Biochemistry. 2005;44:2845–2856. doi: 10.1021/bi048079y. [DOI] [PubMed] [Google Scholar]
- 10.SantaLucia J, Kierzek R, Turner DH. Functional group substitutions as probes of hydrogen bonding between GA mismatches in RNA internal loops. J. Am. Chem. Soc. 1991;113:4313–4322. [Google Scholar]
- 11.Peritz AE, Kierzek R, Sugimoto N, Turner DH. Thermodynamic study of internal loops in oligoribonucleotides: Symmetrical loops are more stable than asymmetric loops. Biochemistry. 1991;30:6428–6436. doi: 10.1021/bi00240a013. [DOI] [PubMed] [Google Scholar]
- 12.Chen G, Znosko BM, Jiao XQ, Turner DH. Factors affecting thermodynamic stabilities of RNA 3 × 3 internal loops. Biochemistry. 2004;43:12865–12876. doi: 10.1021/bi049168d. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Turner DH. Consecutive GA pairs stabilize medium size RNA internal loops. Biochemistry. 2006 doi: 10.1021/bi052060t. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic Eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 15.SantaLucia J, Kierzek R, Turner DH. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992;256:217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- 16.Disney MD, Turner DH. Molecular recognition by the Candida albicans group I intron: Tertiary interactions with an imino G·A pair facilitate binding of the 5' exon and lower the KM for guanosine. Biochemistry. 2002;41:8113–8119. doi: 10.1021/bi020102x. [DOI] [PubMed] [Google Scholar]
- 17.Burkard ME, Turner DH. NMR structures of r(GCAGGCGUGC)2 and determinants of stability for single guanosine-guanosine base pairs. Biochemistry. 2000;39:11748–11762. doi: 10.1021/bi000720i. [DOI] [PubMed] [Google Scholar]
- 18.Znosko BM, Burkard ME, Krugh TR, Turner DH. Molecular recognition in purine-rich internal loops: Thermodynamic, structural, and dynamic consequences of purine for adenine substitutions in 5 ‘(rGGCAAGCCU)2. Biochemistry. 2002;41:14978–14987. doi: 10.1021/bi0203278. [DOI] [PubMed] [Google Scholar]
- 19.Schroeder SJ, Fountain MA, Kennedy SD, Lukavsky PJ, Puglisi JD, Krugh TR, Turner DH. Thermodynamic stability and structural features of the J4/5 loop in a Pneumocystis carinii group I intron. Biochemistry. 2003;42:14184–14196. doi: 10.1021/bi0301587. [DOI] [PubMed] [Google Scholar]
- 20.Broda M, Kierzek E, Gdaniec Z, Kulinski T, Kierzek R. Thermodynamic stability of RNA structures formed by CNG trinucleotide repeats. Implication for prediction of RNA structure. Biochemistry. 2005;44:10873–10882. doi: 10.1021/bi0502339. [DOI] [PubMed] [Google Scholar]
- 21.Moody EM, Feerrar JC, Bevilacqua PC. Evidence that folding of an RNA tetraloop hairpin is less cooperative than its DNA counterpart. Biochemistry. 2004;43:7992–7998. doi: 10.1021/bi049350e. [DOI] [PubMed] [Google Scholar]
- 22.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 23.Szewczak LBW, DeGregorio SJ, Strobel SA, Steitz JA. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 2002;9:1095–1107. doi: 10.1016/s1074-5521(02)00239-9. [DOI] [PubMed] [Google Scholar]
- 24.Blount KE, Uhlenbeck OC. The structure-function dilemma of the hammer head ribozyme. Annu. Rev. Biophys. Biomolec. Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 25.Usman N, Ogilvie KK, Jiang MY, Cedergren RJ. Automated chemical synthesis of long oligoribonucleotides using 2'-O-silylated ribonucleoside 3'-O-phosphoramidites on a controlled-pore glass support: Synthesis of a 43-nucleotide sequence similar to the 3'-half molecule of an Escherichia coli formylmethionine tRNA. J. Am. Chem. Soc. 1987;109:7845–7854. [Google Scholar]
- 26.Wincott F, Direnzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borer PN. In: Handbook of Biochemistry and Molecular Biology: Nucleic Acids. 3rd Ed. Fasman GD, editor. Cleveland, OH: CRC Press; 1975. pp. 589–595. [Google Scholar]
- 28.Richards EG. In: Handbook of Biochemistry and Molecular Biology: Nucleic Acids. 3rd Ed. Fasman GD, editor. Cleveland, OH: CRC Press; 1975. pp. 596–603. [Google Scholar]
- 29.McDowell JA, Turner DH. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 30.Petersheim M, Turner DH. Base-stacking and base-pairing contributions to helix stability: Thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 31.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 32.Borer PN, Dengler B, Tinoco I, Jr, Uhlenbeck OC. Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 1974;86:843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- 33.Lukavsky PJ, Puglisi JD. RNAPack: An integrated NMR approach to RNA structure determination. Methods. 2001;25:316–332. doi: 10.1006/meth.2001.1244. [DOI] [PubMed] [Google Scholar]
- 34.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 35.Ding HQ, Karasawa N, Goddard WA. Atomic level simulations on a million particles: The cell multipole method for coulomb and London nonbond interactions. J. Chem. Phys. 1992;97:4309–4315. [Google Scholar]
- 36.DeLano WL. The PyMOL User's Manual. CA, USA: DeLano Scientific San Carlos; 2002. [Google Scholar]
- 37.Gralla J, Crothers DM. Free energy of imperfect nucleic acid helices .3. small internal loops resulting from mismatches. J. Mol. Biol. 1973;78:301–319. doi: 10.1016/0022-2836(73)90118-6. [DOI] [PubMed] [Google Scholar]
- 38.Varani G, Tinoco I. RNA structure and NMR spectroscopy. Q. Rev. Biophys. 1991;24:479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- 39.Varani G, Aboulela F, Allain FHT. NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spectrosc. 1996;29:51–127. [Google Scholar]
- 40.Heus HA, Wijmenga SS, Hoppe H, Hilbers CW. The detailed structure of tandem G·A mismatched base-pair motifs in RNA duplexes is context dependent. J. Mol. Biol. 1997;271:147–158. doi: 10.1006/jmbi.1997.1158. [DOI] [PubMed] [Google Scholar]
- 41.Uesugi S, Oda Y, Ikehara M, Kawase Y, Ohtsuka E. Identification of I–A mismatch base pairing structure in DNA. J. Biol. Chem. 1987;262:6965–6968. [PubMed] [Google Scholar]
- 42.SantaLucia J., Jr . Ph.D. Thesis: The Role of Hydrogen Bonding in the Thermodynamics and Structure of Mismatches in RNA Oligonucleotides. Rochester, NY: University of Rochester; 1991. [Google Scholar]
- 43.Baeyens KJ, DeBondt HL, Pardi A, Holbrook SR. A curved RNA helix incorporating an internal loop with GA and AA non-Watson-Crick base pairing. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12851–12855. doi: 10.1073/pnas.93.23.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 45.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 46.Correll CC, Munishkin A, Chan YL, Ren Z, Wool IG, Steitz TA. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 48.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 49.Strobel SA, Adams PL, Stahley MR, Wang JM. RNA kink turns to the left and to the right. RNA. 2004;10:1852–1854. doi: 10.1261/rna.7141504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou SH, Zhu L, Reid BR. Sheared purine·purine pairing in biology. J. Mol. Biol. 1997;267:1055–1067. doi: 10.1006/jmbi.1997.0914. [DOI] [PubMed] [Google Scholar]
- 51.Butcher SE, Dieckmann T, Feigon J. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997;16:7490–7499. doi: 10.1093/emboj/16.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. The Comparative RNA Web (CRW) Site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mathews DH, Schroeder SJ, Turner DH, Zuker M. In: The RNA World. 3rd edition. Gesteland RF, Cech TR, Atkins JF, editors. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2005. pp. 631–657. [Google Scholar]
- 54.Schroeder SJ, Burkard ME, Turner DH. The energetics of small internal loops in RNA. Biopolymers. 1999;52:157–167. doi: 10.1002/1097-0282(1999)52:4<157::AID-BIP1001>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 55.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 angstrom resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 57.Gutowsky HS, Holm CH. Rate processes and nuclear magnetic resonance spectra 2. Hindered internal rotation of amides. J. Chem. Phys. 1956;25:1228–1234. [Google Scholar]
- 58.Legault P, Pardi A. Unusual dynamics and pKa shift at the active site of a lead-dependent ribozyme. J. Am. Chem. Soc. 1997;119:6621–6628. [Google Scholar]
- 59.Mathews DH, Case DA. J. Mol. Biol. 2006. Nudged elastic band calculation of minimal energy paths for the conformational change of a GG non-canonical pair. In press. [DOI] [PubMed] [Google Scholar]
- 60.Gautheret DF, Konings D, Gutell RR. A major family of motifs involving G·A mismatches in ribosomal RNA. J. Mol. Biol. 1994;242:1–8. doi: 10.1006/jmbi.1994.1552. [DOI] [PubMed] [Google Scholar]
- 61.Wu M, SantaLucia J, Jr, Turner DH. Solution structure of (rGGCAGGCC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry. 1997;36:4449–4460. doi: 10.1021/bi9625915. [DOI] [PubMed] [Google Scholar]
- 62.Jucker FM, Heus HA, Yip PF, Moors EHM, Pardi A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 63.Batey RT, Doudna JA. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry. 2002;41:11703–11710. doi: 10.1021/bi026163c. [DOI] [PubMed] [Google Scholar]
- 64.Ornstein RL, Fresco JR. Correlation of crystallographically determined and computationally predicted hydrogen bonded pairing configurations of nucleic acid bases. Proc. Natl. Acad. Sci. U. S. A. 1983;80:5171–5175. doi: 10.1073/pnas.80.17.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adamiak DA, Milecki J, Popenda M, Adamiak RW, Dauter Z, Rypniewski WR. Crystal structure of 2'-O-Me(CGCGCG)(2), an RNA duplex at 1.30 angstrom resolution. Hydration pattern of 2'-O-methylated RNA. Nucleic Acids Res. 1997;25:4599–4607. doi: 10.1093/nar/25.22.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Popenda M, Biala E, Milecki J, Adamiak RW. Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)(2) and 2'-O-Me(CGCGCG)(2) under low salt conditions. Nucleic Acids Res. 1997;25:4589–4598. doi: 10.1093/nar/25.22.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kierzek E, Ciesielska A, Pasternak K, Mathews DH, Turner DH, Kierzek R. The influence of locked nucleic acid residues on the thermodynamic properties of 2'-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 2005;33:5082–5093. doi: 10.1093/nar/gki789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu M, Turner DH. Solution structure of (rGCGGACGC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry. 1996;35:9677–9689. doi: 10.1021/bi960133q. [DOI] [PubMed] [Google Scholar]
- 69.Jovine L, Hainzl T, Oubridge C, Scott WG, Li J, Sixma TK, Wonacott A, Skarzynski T, Nagai K. Crystal structure of the Ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA. Structure Folding Des. 2000;8:527–540. doi: 10.1016/s0969-2126(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 70.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.