Abstract
Since the discovery of Fibroblast Growth Factors much focus has been placed on elucidating the roles for each vertebrate FGF ligand, receptor, and regulating molecules in the context of vertebrate development, human disorders and cancer. Studies in human, mouse, Xenopus, chick, and zebrafish have gone a long way to help us understand [AS1]which FGFs are involved in which processes. However, in recent years, as more genomes are sequenced, more information is becoming available from many non-vertebrate models and a more complete picture of the FGF superfamily as a whole is emerging. In some cases less redundancy in the FGF signaling system in invertebrate models may allow for more mechanistic insights. Studies in cnidaria have highlighted how ancient FGF signaling is, and helped provide insight into the evolution of the FGF gene family. Work in C. elegans has shown that different splice forms can be used for functional specificity in invertebrate FGF signaling. Comparing FGFs from Ciona to those in vertebrates and FGFs from Tribolium to Drosophila reveals some important clues as to the process of gene loss, duplication and subfunctionalization of FGFs throughout evolution. Finally, comparing all members of the FGF ligand superfamily reveals variability in many properties, which may point to a feature of FGFs as being highly adaptable with regards to protein structure and mechanism. Further studies on FGF signaling outside of vertebrates is likely to complement work in vertebrates by contributing many insights to the FGF field as a whole and providing unexpected information that could be used for medical applications.
Introduction
Cell signaling by Fibroblast Growth Factors (FGF) is essential to the development and maintenance of animals. From their discovery in the early 1970s to today, researchers continue to detail the contributions of FGF signaling to developmental and adult metabolic processes. It has become clear that FGF signaling is not limited to a few uses, but has many functions both in developing embryos and the adult. As more genomes are sequenced and more FGF superfamily members are described, the amount of structural and functional variety within the family is becoming apparent and FGF signaling appears to be highly adaptable, helping to make possible the great variety of life forms. In this review we will first highlight the history of FGF research, the structure of the FGF signaling complex, the downstream pathways employed and major functional findings from vertebrate FGF research. Then, we will discuss findings from non-vertebrate models in the context of emerging themes in FGF superfamily research.
Historical perspective
A prelude to the discovery of FGFs was the finding in 1939 that bovine brain extracts could cause proliferation of fibroblast cell lines in vitro (Hoffman, 1940; Mohammadi et al., 2005; Trowell OA, 1939). Biochemical characterization of this mitogenic activity did not begin for another 34 years, when a factor in pituitary extracts was found to stimulate growth of 3T3 mouse fibroblast cells and was characterized as being thermolabile, sensitive to proteases and enhanced by hydrocortisone (Armelin, 1973). Gospodarowicz (1974) purified the mitogenic factor from pituitary extracts and found it was also present at higher concentrations in brain extracts. He termed this molecule Fibroblast Growth Factor and showed that with hydrocortisone FGF could stimulate DNA synthesis as effectively as crude serum (Gospodarowicz, 1974). Another important finding from these early studies was the incredible potency of FGF: the minimal effective dose was only 0.1 ng ml−1. It was also found that FGF could induce proliferation of diploid human foreskin fibroblasts and mouse fibroblast cells, showing that FGF lacks species specificity (Gospodarowicz and Moran, 1975). FGF activity was found to be due to a 15 kD molecule and was called basic FGF (bFGF) because of its high isoelectric point (pI) (Gospodarowicz, 1975; Gospodarowicz, 1978). Another molecule with FGF activity was also isolated from brain extracts and was called acidic FGF (aFGF) because of its lower pI (Maciag et al., 1979).
A number of other mitogenic proteins were subsequently found to be chemically identical to either aFGF or bFGF (Burgess and Maciag, 1989; Burgess et al., 1986; Lemmon et al., 1982; Libermann et al., 1987; Mohammadi et al., 2005). More members of the FGF family were found using several approaches and a numbering-scheme was established in which aFGF and bFGF were renamed as FGF1 and FGF2, respectively. FGF3 (INT-2) (Dickson et al., 1984), FGF4 (K-FGF/HST) (Delli Bovi and Basilico, 1987; Sakamoto et al., 1986), and FGF5 (Zhan et al., 1988) were all discovered as oncogenes. FGF6 was identified based on the similarity of its sequence to FGF4 (Marics et al., 1989). FGF7 was discovered with classical protein purification from fibroblasts and this study showed for the first time that FGFs are necessary for tissue homeostasis by enabling communication between mesenchymal and epithelial tissues (Rubin et al., 1989). FGF8 was isolated as an androgen-induced growth factor (Tanaka et al., 1992). FGF9 was found because of its ability to stimulate the growth of glia cells (Miyamoto et al., 1993).
Between 1996 and 2003, other FGFs were found through a combination of bioinformatic tools and homology-based PCR: FGF10 (Lu et al., 1999), FGF16 (Miyake et al., 1998), FGF17 (Xu et al., 1999), FGF18 (Ohbayashi et al., 1998), FGF19 (Nishimura et al., 1999), FGF20 (Kirikoshi et al., 2000), FGF22 (Nakatake et al., 2001), FGF23 (Yamashita et al., 2000), FGF24 (Draper et al., 2003). FGF11-FGF14 make up a subfamily of Fibroblast Homologous Factors (FHFs) that are not secreted and do not bind to FGF receptors (FGFR) 1–4 (Coulier et al., 1997; Smallwood et al., 1996). FHFs can also bind to heparin with high affinity like the canonical FGFs, yet despite striking structural similarity, FHFs have diverged toward interaction with a separate set of target proteins and do not share functional homology with FGFs (Olsen et al., 2003).
Today, the FGF family represents one of the largest signaling families in vertebrates, with 24 known ligands in total, although not every member is present in every vertebrate species. The first FGF receptor (FGFR) was identified in the mid-1980’s (Lee et al., 1989; Olwin and Hauschka, 1986), and since then 4 FGFRs have been found in vertebrates (Coumoul and Deng, 2003).
Structure of FGF ligands, receptors, and signaling complex
FGF ligands share a homologous core domain consisting of 120–130 amino acids ordered into 12 antiparallel β-strands (β1-β12) that are arranged into three sets of four-stranded β-sheets that fold to form a β-trefoil structure (Mohammadi et al., 2005). Additionally, they have variable length N- and C-terminal tails, which largely account for the specific biology of different FGF family members. Most FGFs have traditional signal peptides and are secreted as soluble signaling molecules. Vertebrate FGFs are also known to bind to heparan sulfate glycosaminoglycans (HSGAG) through the HSGAG binding site (HBS), located in the FGF core within the β1-β2 loop and the region between β10-β12. The elements of the HBS form a contiguous, positively charged surface.
FGF ligands bind to the FGFR family of tyrosine kinase receptors in an heparan sulfate proteoglycan(HSPG)-dependent manner. In vertebrates there are 4 FGFRs (FGFR1-FGFR4) which bind to the 24 ligands with varying degrees of promiscuity. The structure of the FGFR consists of three extracellular immunoglobulin domains (D1-D3), a transmembrane domain, and an intracellular tyrosine kinase domain. A unique feature of FGFR is the presence of an acidic, serine-rich sequence in the linker between D1 and D2, which is known as the acid box. The FGF ligands bind to the D2-D3 region of the FGFR ectodomain. The D1 and acid box are thought to play a role in receptor autoinhibition (Mohammadi et al., 2005).
A functional FGF-FGFR signaling unit consists of two 1:1:1 FGF-FGFR-HSGAG complexes that are bound together into a dimer. The ligand of each complex binds to both receptors to allow interaction with each other through a region in the D2 domain. The HSGAG incorporates into the dimer through a “basic canyon” and contributes to dimerization by binding both the ligands and the receptors (Beenken and Mohammadi, 2009). Additionally, HSGAGs stabilize FGFs against degradation, act as a storage reservoir, and can affect the radius of ligand diffusion (Häcker et al., 2005). Dimerization of FGFR allows the cytoplasmic kinase domains to become activated (Mohammadi et al., 1996).
Signaling transduction pathways utilized
Several reviews have been written detailing the research on downstream signaling pathways used by FGF signaling (Böttcher and Niehrs, 2005; Eswarakumar et al., 2005; Thisse and Thisse, 2005), and therefore we will only review briefly here this aspect of FGF signaling. FGFR-stimulation leads to tyrosine phosphorylation of Shp2 resulting in complex formation of Grb2 and its associated nucleotide exchange factor son-of-sevenless (Sos) and Grb2/Sos activate the Ras GTPase, which then activates the mitogen-activated protein kinase (MAPK) pathway. The final protein in the MAPK pathway is extracellular signal-regulated kinase (ERK) and it enters the nucleus to activate transcription factors (c-myc, AP1 and Ets-family members) that will affect FGF target genes.
The MAPK pathway is not the only pathway used by FGF signaling. Mutational analysis of tyrosine766 has shown that the phosphorylation of this tyrosine residue is essential for complex formation with and tyrosine phosphorylation of phospholipase C gamma (PLCγ) (Eswarakumar et al., 2005). PLCγ activation results in the hydrolysis of phosphatidylinositol-4,5-diphosphate (PIP2) to inositol-1,4,5-triphosphate (IP3) and the generation of two second messengers: IP3 and diacylglycerol (DAG). Recruitment to the membrane of PLCγ is mediated by binding of the Pleckstrin homology domain of PLCγ to IP3 molecules. IP3 causes a release of calcium within the cell, which stimulates GEFs that activate the Rap1 GTPase. Rap1 can assist in the maturation of intercellular junctions and mediate adhesion through the recruitment of cadherins and integrins to the plasma membrane. Signaling through the FGFR can thus result in multiple responses: cellular differentiation through Ras GTPase and cell adhesion/migration through PLCγ/Rap1 (Raaijmakers and Bos, 2009).
The PI3 Kinase/Akt pathway can be activated in three ways: (1) Gab can bind to FRS2 via Grb2, (2) the PI3 subunit p85 can bind to a phosphorylated tyrosine residue of the FGFR, and (3) activated Ras can induce membrane localization and activation of the p110 catalytic subunit of P13 kinase (Böttcher and Niehrs, 2005).
The different downstream signal transduction pathways used by FGF signaling can lead to specific cellular response in a cell-type dependent manner (Dailey et al., 2005). For instance, the ERK kinases are generally thought to be responsible for the mitogenic response of cells to FGF, while alternate MAPKs, p38 and JNK MAP kinase are usually associated with inflammatory or stress-response.
Functional information from vertebrate studies
The cumulative data [DM8]from studies on FGF signaling in vertebrate models is difficult to summarize in brief. Reviews on FGF functions in vertebrates have been written at regular intervals and include information on FGFs involved in developmental processes, adult maintenance, disorders and cancer (Beenken and Mohammadi, 2009; Ornitz and Itoh, 2001; Thisse and Thisse, 2005). Among other functions, FGFs are key regulators of development, including: mesoderm induction, gastrulation, limb development, midbrain-hindbrain patterning, and bone formation. In the mouse, FGF4 and FGF8 are required for proper migration of epiblast cells through the primitive streak. In the absence of both FGF4 and FGF8, epiblast cells move into the streak and undergo an epithelial-to-mesenchymal transition, but then most cells fail to move away from the streak (Sun et al., 1999; Thisse and Thisse, 2005). Currently it is thought FGF4 is thought to act as an attractant and FGF8 as a repellent to cells in the streak. [DM9]FGF induction of mesoderm has also been studied in Xenopus laevis, where FGF2 was first shown to have mesoderm inducing activity equivalent to the ventrovegetal signal (Slack et al., 1987). More recently, the specific roles of different spliceforms of FGF8, FGF8a and FGF8b, have been found to have different activities in the early specification of mesodermal and neural tissue in the frog. FGF8b is a potent mesoderm inducer in both explants and whole embryos while FGF8a has little effect on the development of mesoderm (Fletcher et al., 2006).
Limb Development
FGFs have been found to play key roles in the process of limb development. Formation of limb buds and their successful outgrowth is dependent upon FGF signaling and a FGF positive-feedback signaling loop between the limb mesenchyme (progress zone) and the overlying ectoderm, termed the apical ectodermal ridge (AER). FGF4, FGF8, FGF9 and FGF17 are all expressed in the AER. Combinatorial FGF mutant studies resulted in the loss of intermediate skeletal structures while the most distal and the most proximal structures remained intact, leading to a the ‘two-signal model,’ which describes limb mesenchyme initially being influenced by one signal (likely Retinoic Acid) that influence proximal cell fates and subsequently experience FGF signals from the AER establishing the distal domain. The intermediate domain would then form as a result of interactions at the domain boundary. Sonic hedgehog (Shh) is expressed in a posterior domain of the limb bid called the zone of polarizing activity (ZPA) and a positive feedback loop is established between Shh in the ZPA and FGFs in the AER. Shh is required for the induction and maintenance of Fgf4, 9, 17 and the maintenance of Fgf8, and, reciprocally, FGF signaling from the AER is required to maintain Shh expression (Duboc and Logan, 2009)
Brain Patterning
Patterning of the midbrain-hindbrain (MHB) anlage depends on an organizer activity located at the MHB junction, also known as the Isthmus. In vertebrates, FGF8 is expressed in the MHB and is a key component of its organizing activity (Crossley et al., 1996). Loss of midbrain and cerebellar tissue results in a mouse with a severe hypomorphic allele of Fgf8 (Meyers et al., 1998). FGF17 and FGF18 are also expressed in the MHB and the loss of FGF17 in mouse results in the truncation of posterior midbrain and reduced proliferation of the anterior cerebellum (Maruoka et al., 1998). FGF8 is differentially spliced to generate FGF8a and FGF8b isoforms, which are both expressed at the isthmus/MHB (Sato et al., 2001). In the chick, ectopic FGF8a causes expansion of the midbrain whereas misexpression of FGF8b transforms the midbrain into a cerebellum (Sato et al., 2001). Similarly, in the mouse, ectopic FGF8a results in expansion of the midbrain and ectopic expression of Engrailed2, whereas ectopic FGF8b leads to exencephaly and a rapid transformation of the midbrain and diencephalon into an anterior rhombomere1[AS11] fate (Liu et al., 1999). FGF8b also maintains two negative feedback loops by inducing the expression of the negative feedback FGF inhibitors Sprouty1 and Sprouty2 and repressing FGFR2 and FGFR3 (Liu et al., 2003). In Zebrafish, FGF8 is also present at the MHB and acts as a morphogen to pattern the midbrain. Acerebellar mutants, in which the FGF8 gene contains a premature stop codon, lack a functional MHB and also lacks a cerebellum (Reifers et al., 1998).
Bone Formation
FGF signaling is capable of regulating genes at all steps of osteogenesis. A point mutation in the transmembrane domain of FGFR3 was found to be the etiology of Acondroplasia, the most common genetic form of human dwarfism (Rousseau et al., 1994; Shiang et al., 1994). Missense mutations have since been found in more than 15 human bone disorders, from skeletal dysplasias to short stature. FGF2, FGF9, and FGF18 are all found in osteoblasts. Overexpression of FGF2 in mouse causes abnormal bone formation and loss-of-function of FGF2 leads to inhibition of bone formation (Coffin et al., 1995; Montero et al., 2000). FGF signaling seems to positively regulate cell proliferation and differentiation in osteogenesis. Additionally, FGFs can control apoptosis in osteoblasts when high levels of FGF signaling can reduce apoptosis in immature osteoblasts and increase the total osteoblast population.
Other developmental functions for FGFs in vertebrates have been described as well in many tissues: the nervous system, epidermis, lungs, mammary glands, somite boundaries, ear, kidney, liver, and pancreas (Coleman-Krnacik and Rosen, 1994; Delaune et al., 2005; Kobberup et al., 2010; Sawada et al., 2001; Thisse and Thisse, 2005; Wilkie, 2005).
Insights from non-vertebrate models
An Introduction to non-vertebrate FGFs and FGFRs
FGF signalling has now been described in a number of model systems outside of vertebrates including the echinoderm sea urchin Strongylcentrotus purpuratus, the urochordate ascidians Ciona intestinalis and Ciona savigny, the ecdysozoans Caenorhabditis elegans, Drosophila melanogastor, and Tribolium castaneum, and the anthozoan cnidarian, Nematostella vectensis. The relationship of these groups to vetebrates is summarized in Figure 1.
Figure 1.
Simplified tree of Metazoa phyla containing species discussed in this review.
This list will surely expand in the near future, but it is worth surveying the current described members of the FGF family outside of vertebrates (Table 1). In the sea urchin, they have identified one ligand, FGFA, and two receptors, FGFR1 and FGFR2 (Lapraz et al., 2006; McCoon et al., 1996; McCoon et al., 1998; Röttinger et al., 2008). The ligand was called FGFA because the predicted protein showed similarities to both the FGF8 and FGF9 subfamilies and phylogenetic analysis gave ambiguous results. Ciona has 6 FGF ligands and 1 receptor: Ci-FGF8/17/18, Ci-FGF11/12/13/14, Ci-3/7/10/22, Ci-FGF4/5/6, Ci-FGF9/16/20, Ci-FGFL (FGF with large molecular mass), and Ci-FGFR (Satou et al., 2002; Shi et al., 2009). In Drosophila, there are three FGF ligands: Branchless (Bnl), Thisbe (Ths), and Pyramus (Pyr). Ths and Pyr are most related to the FGF8 subfamily. Additionally, there are two FGFRs: Bnl uses the Breathless FGFR (Btl), and Ths and Pyr signal through the Heartless receptor (Htl). Tribolium has 4 FGF ligands and a single FGFR: Tc-FGF1a, Tc-FGF1b, Tc-FGF8, Tc-Branchless (Tc-Bnl), and Tc-FGFR (Beermann and Schröder, 2008). In C. elegans, there are two FGF ligands, egl-17 and LET-756, and one FGFR, egl-15. Egl-17 is most similar to the FGF8 subfamily and LET-756 to the FGF9 subfamily. In the anthoszoan cnidarian Nematostella vectensis, there are 4 ligands and 2 receptors: NvFGF8A, NvFGF8B, NvFGF1A, NvFGFa2, NvFGFRa, and NvFGFRb (Matus et al., 2007; Rentzsch et al., 2008). A probable FGFR, kringelchen, has been identified in the hydrozoan cnidarian Hydra (Sudhop et al., 2004). Two FGFRs, Dj-FGFR1 and Dj-FGFR2 have been found in the platyhelminthes planarian Dugesua japonica, rounding out representatives from all the major metazoan phyla (Ogawa et al., 2002).
Table.
| aa | kD | known functions | expressed in | references | |
|---|---|---|---|---|---|
| Ciona | |||||
| Ci-Fgf9/16/20 | 297 | 33 | mesenchyme specification, dorsal midline formation, neural induction, heart specification, trunk ventral cell migration |
vegetal hemisphere, endoderm, next to B7.5 cells |
Miyazaki et. al, 2007; Tokuoka et. al, 2004; Pasini 2006; Betrand et. al, 2003; Beh et. al, 2007; Davidson et. al, 2006; Imai et. al, 2002; Kim et. al, 2000 |
| Ci-Fgf8/17/18 | 414 | 48 | brain patterning, notochord formation, atrial placode formation |
CNS, midbrain-hindbrain boundary, trunk lateral mesenchyme |
Ikuta et. al, 2007; Imai et. al, 2002; Yasuo et. al, 2007; Imai et. al, 2009; Kourakis and Smith, 2007 |
| Ci-Fgf3/7/10/22 | 268 | 31 | notochord convergent extension | floor plate | Shi et. al, 2009 |
| Ci-Fgf4/5/6 | 229 | 27 | ? | ? | ? |
| Ci-11/12/13/14 | 182 | 21 | ? | ? | ? |
| Ci-FgfL | 658 | 75 | ? | ? | ? |
| S. purpuratus | |||||
| FGFA | 348 | 39 | migration and differentiation of PMCs | lateral ectoderm | Rottinger et. al, 2008 |
| Drosophila | |||||
| Branchless | 770 | 84 | tracheal branching, male genital disc development |
cells surrounding developing tracheal branches, male genital disc |
Sutherland et. al, 1996; Wolf et. al, 2002; Ribeiro et. al, 2004; Ahmad et. al, 2002 |
| Pyramus | 766 | 87 | mesoderm collapse & intercalation; pericardial cell specification, axonal migration |
lateral ectoderm, glial cells | Stathopoulos et. al, 2004; Klingseisen et. al, 2009; Franzdóttir et al., 2009; Kadam et. al, 2009; Gryzik et. al, 2004; McMahon et. al, 2010 |
| Thisbe | 748 | 82 | mesoderm collapse & intercalation, mesoderm differentiation, glial differentiation |
ventro-lateral ectoderm, neurons | Stathopoulos et. al, 2004; Klingseisen et. al, 2009; Franzdóttir et al., 2009; Kadam et al, 2009; Gryzik et. al, 2004; McMahon et. al, 2010 |
| Tribolium | |||||
| Tc-FGF8 | 229 | 27 | presumably brain patterning, mesoderm migration and differentiation |
early posterior pole, ectodermal stripes, foregut anlagen, CNS midbrain-hindbrain boundary |
Beerman and Schroder, 2008 |
| Tc-Branchless | 232 | 25 | likely trachael, air sac and gland formation |
between abdominal segment 10 and growth zone, growth zone, leg forming region, tracheal placodes, maxillary domain, anal ring |
Beerman and Schroder, 2008 |
| Tc-FGF1a | ? | ? | ? | ubiquitous | Beerman and Schroder, 2008 |
| Tc-FGF1b | ? | ? | ? | ubiquitous | Beerman and Schroder, 2008 |
| C. elegans | |||||
| Egl-17 | 216 | 25 | migration of sex myoblasts, essential, regulation of fluid balance, axonal migration, negative regulation of muscle membrane extension |
primary vuval cell secreted and nuclear, muscles adjacent to hypodermis |
Burdine et. al, 1997; Burdine et. al, 1998 Roubin et. al, 1999; Popovici et. al, 2006; Dixon et. al, 2006 |
| LET-756 | 425 | 50 | |||
| Nematostella | |||||
| Nv-FGF8A | ? | ? | ? | invaginating blastopore, developing pharnyx, ectodermal cells of apical tuft |
Matus et. al., 2007 |
| Nv-FGF8B | ? | ? | ? | endodermal cell at aboral end below apical tuft | Matus et. al., 2007 |
| Nv-FGF1A | ? | ? | apical ciliary organ development | aboral pole, aboral ectoderm at base of apical tuft |
Matus et. al., 2007; Rentzsch et. al, 2008 |
| Nv-FGFa2 | ? | ? | apical ciliary organ development | aboral pole | Rentzsch et. al, 2008 |
The role of FGFs in development is an ancient one
FGF signaling is an ancient cell-to-cell communication system as evidenced by its presence in the cnidaria, which split off from its sister group bilateria an estimated 600 million years ago (Rentzsch 2008). Nematostella vectensis, a sea anemone, is considered to be a representative of basal cnidarians and to have retained much of the genetic complexity contained in the cnidarian-bilaterian ancestor (Bridge et al., 1995; Bridge et al., 1992; Chourrout et al., 2006; Collins et al., 2006; Medina et al., 2001; Putnam et al., 2007; Ryan et al., 2006; Technau et al., 2005). The two FGFRs identified in Nematostella, NvFGFRa and NvFGFRb are thought to have arisen from a lineage-specific duplication, and therefore, it is thought likely that there was only 1 FGFR in the cnidarian-bilaterian ancestor (Rentzsch et al., 2008). As many as 15 putative transcripts sharing homology to FGF domains were found via bioinformatic analyses in the Nematostella genome, but so far only four have been described: NvFGF1A, NvFGFa2, NvFGF8A, NvFGF8B (Matus et al., 2007; Rentzsch et al., 2008).
In bilaterians, FGF ligands and FGF receptors are often expressed in separate germ layers or tissues and signal across epithelial-mesenchymal boundaries. Yet, in diploblastic cnidarians there is no mesoderm for FGFs to signal to/from, and so the ligands and receptors are expressed in the same domain (NvFGF1A, NvFGFa2, NvFGFRa), or in abutting ectoderm/endoderm tissues of the aboral pole (NvFGF8A, NvFGFRb).
Morpholino knockdown of NvFGF1A and NvFGFRa showed that they are required for formation of the apical organ (Rentzsch et al., 2008). Apical organs with a ciliated tuft are also present in both protostomes and deuterostomes: in the larvae of sea urchins, hemichordates, and the polychaete Platynereis, although the evolutionary relationship of cnidarian, protostomian and deuterostomian apical organs has not yet been determined. Intriguingly, FGFs or FGFRs are expressed in the region of apical organ formation in sea urchin, hemichordates and polychaetes, leading to the possibility of an ancient function in apical organ formation.
A tyrosine kinase receptor with similarity to FGFR, kringelchen, has also been identified in the hydrozoan cnidarian Hydra, where it was shown to be essential for boundary formation and tissue constriction as a prerequisite for proper bud detachment which is essential for reproduction (Sudhop et al., 2004). It has yet to be shown that this receptor can actually bind FGFs, which have not been described yet for Hydra.
Importance of tight regulation in FGF signaling
Evidence from many systems has pointed to the importance for tight regulation of FGF signaling activity, and the loss of such regulation often leads to developmental disorders and disease. A negative regulator of FGF signaling, Sprouty, was originally identified in Drosophila for its action during tracheal development (Hacohen et al., 1998). Sprouty is thought to act in a negative-feedback regulatory loop during FGF and EGF signaling (Casci et al., 1999; Kramer et al., 1999; Sivak et al., 2005). There are four mammalian Sprouty proteins and three related Spreds (Sprouty-related EVHI domain proteins). Sproutys have been found in synexpression groups with FGFs and FGFRs in other nonvertebrate systems. Nematostella Sprouty, Nv-Sprouty, is expressed in the same domain as NvFGF8A, NvFGF8B and NvFGFRa in the apical pole (Matus et al., 2007). The expression of the sea urchin sprouty largely follows that of fgfA from the late mesenchyme blastula/early gastrula to pluteus stages in bilateral regions of the ectoderm, in the PMC clusters, and at the tip of the growing arms of the larva (Röttinger et al., 2008). Two other probable FGF target genes, pea3 (Polyoma enchancer activator 3), an Ets domain transcription factor, and paired transcription factor pax2/5/8, were also expressed along with fgfA and sprouty (Röttinger et al., 2008). Sprouty proteins can have a therapeutic effect on some mouse models of disease by enhancing angiogenesis and neovascularization (formation of new blood vessels from preexisting ones) (Taniguchi et al., 2009). Many of the studies in vertebrates relied on double mouse knockouts for combinations of different Sproutys and Spreds. Studies on Sprouty proteins in nonvertebrate models may aid in the further characterization of the mechanism of regulation without the concern of redundancy.
Regulation has also been found to come from certain FGF ligands themselves when co-expressed in the same domain as the functioning ligand. In Nematostella, NvFGFa2 negatively regulates FGF signaling at the apical pole, as a morpholino against NvFGFa2 causes the expansion of the apical tuft region along with the expansion of expression of NvFGF1A and NvFGFRa (Rentzsch et al., 2008). This may be related to the function of FGFRL1 molecules (see below, Survey Approach to FGFRL1).
Multiple isoforms of FGFs and FGFRs are generated by splicing
The possible ligand-receptor combinations in vertebrates are numerous and increased by different receptor splice forms. Multiple isoforms are thought to contribute to ligand-receptor specificity and functional specificity. Several examples are also present outside of vertebrates of alternate splice forms of FGFs and FGFRs contributing to functional specificity. C. elegans has two ligands LET-756 and EGL-17 and a single receptor, EGL-15 (Birnbaum et al., 2005). EGL-15 is located on the X chromosome and encodes two isoforms, EGL-15(5A) and EGL-15(5B), which result from alternative splicing of exon 5. It has been shown genetically that the different isoforms mediate signaling through two different modules, each using a specific ligand. Egl-15(5A) interacts with egl-17 to mediate sex myoblast chemoattraction and egl-15(5B) carries out an essential function required for viability, presumably through signaling by let-756 (Goodman et al., 2003). Perhaps multiple isoforms are especially important when a single receptor is required to mediate separate functions from two different ligands.
Ciona FGF8/17/18 has two alternative forms of transcripts, that differ in their N-terminal regions (Satou et al., 2002). However, one form is missing the N-terminal region of the FGF domain and whether it is used for signaling and/or regulation is not known.
FGFs have been lost, duplicated and undergo subfunctionalization
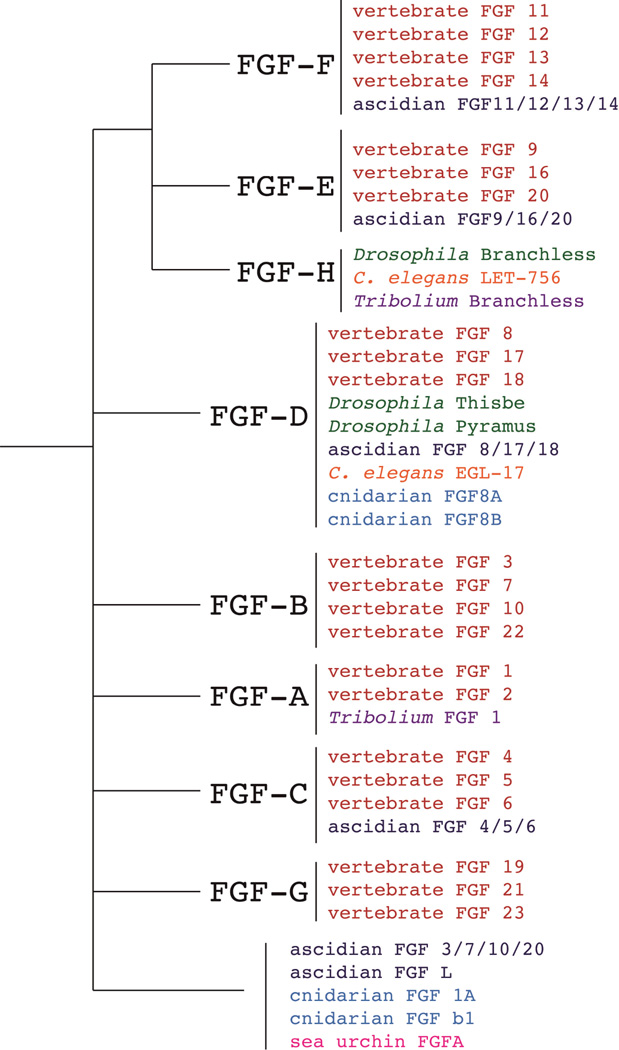
Characterizing the complement of FGF ligands in non-vertebrate taxa has provided insight into the evolution of the FGF superfamily across the Metazoa (Popovici et al., 2005). It is clear that in some lineages FGF/FGFR genes have been lost, where in other cases they have been duplicated once or multiple times. Comparisons of FGFs in Ciona to vertebrates reveals that at least two rounds of duplications of most FGF ligands and the FGFR were necessary to account for the multiple subfamily members in vertebrate genomes. It is generally thought that this is consistent with the “2R hypothesis,” which maintains that two rounds of whole genome duplication occurred at the base of vertebrate ancestry (Holland et al., 1994; Ohno, 1970).
It has been proposed that the 7 FGF subfamilies present in vertebrates (A–G) plus 1 additional subfamily lost in deuterostomes (H) represent what was once 8 proto-FGF genes in the protostome-deuterostome ancestor (Popovici et al., 2005). Ciona has six FGFs, 2 of which were confidently assigned to FGF subfamilies D and F (Ci-FGF8/17/18 and Ci-11/12/13/14) (Satou et al., 2002). Probable placement in subfamilies B, C, and E was made for an additional 3 FGFs in Ciona (Ci-FGF3/7/10/22, Ci-4/5/6, and Ci-9/16/20). The last FGF in Ciona, Ci-FGFL is characterized by its large predicted molecular mass and could not be assigned to any particular FGF subfamily with confidence. Possible assignments include grouping with other invertebrate FGFs like Branchless in subfamily H, or as a member of subfamily A, B or G (FGF1/FGF2, FGF3/7/10/22 and FGF15/FGF19/FGF21/FGF23, respectively) but its sequence has diverged beyond the similarity required for phylogenetic analysis (Popovici et al., 2005; Satou et al., 2002).
Over time duplicated genes can undergo subfunctionalization to take over different responsibilities. In some cases the combined functions of the two genes equal the function of the original gene, and sometimes the presence of a “backup” gene allows the duplicate or original gene to explore new functional space.
Ciona vs vertebrate FGFs
Many functional studies have been performed on FGFs in Ciona and comparisons to studies in vertebrates yield some important similarities (Beh et al., 2007; Bertrand et al., 2003; Davidson et al., 2006; Imai et al., 2002; Kourakis and Smith, 2007; Shi et al., 2009; Yasuo and Hudson, 2007). Ci-9/16/20 has been shown to be involved in the induction of notochord, induction of mesenchyme, and heart specification (Davidson et al., 2006; Imai et al., 2002). Ci-9/16/20 is expressed adjacent to the heart-producing B7.5 lineage and morpholino knockdown of Ci-9/16/20 results in the disruption of heart lineage markers Mesp, NoTrlc/Hand-like, Tolloid, FoxF (Davidson et al., 2006; Imai et al., 2006). FGF9 and FGF16 are also known to be involved in heart development in the mouse. Knockout mice for both FGF9 and FGF16 (but not a double mutant) have been generated and have a similar phenotype of reduced number of cardiomyoctes and smaller embryonic heart (Hotta et al., 2008; Lavine et al., 2005). FGF9 and FGF16 are thought to act synergistically to promote the proliferation of embryonic cardiomyocytes. Epicardial and endocardial FGF9/FGF16 signaling through FGFR1/FGFR2 is essential for myocardial proliferation and differentiation (Lavine et al., 2005). In this case it seems that the vertebrate paralogs FGF9 and FGF16 have retained a function in heart development (although possibly not homologous) compared to FGF9/16/20 in Ciona. FGF9 and FGF16 seem to be function redundantly at this stage of development with no subfunctionalization apparent.
Ciona FGF3/7/10/22 is expressed in the ventral midline of the neural tube and is important for convergent extension movement in the developing embryo (Shi 2009). In the Xenopus neurula FGF signaling has been implicated in axial elongation as well and possibly a similar mechanism is at play, however the details are still unclear (Sivak et al., 2005).
Ciona FGF8/17/18 is expressed in the nervous system of ascidian embryos and is thought to play a similar role to the patterning of the brain territories that FGF8/FGF17/FGF18 play in vertebrates (see midbrain-hindbrain section of introduction). Ci-FGF8/17/18 is expressed in the developing central nervous system (CNS) in a region analogous to the MHB of vertebrate embryos and has led to the hypothesis that a precursor to the organizing activity of FGF8 in the MHB in vertebrates was this region of Ci-FGF8/17/18 expression bewteen Otx and Hox genes in Ciona (Ikuta and Saiga, 2007; Imai et al., 2002). Interestingly, 3 other Ciona FGFs are also expressed in the developing CNS: Ci-9/16/20, Ci-3/7/10/22, and Ci-FGFL (Imai et al., 2002). Morpholino knockout analysis of Ci-FGF8/17/18 has revealed that this ancestor of FGF8/FGF17/FGF18 plays a central role in generating regional patterns of gene expression as morphants have altered expression of Otx, en, FoxB, Pax2/5/8, and Hox 1 (Imai et al., 2009). In vertebrates, FGF17 and FGF18 are also expressed in the mid/hindbrain in a broader domain than FGF8 that includes posterior midbrain (Maruoka et al., 1998). Loss of one copy of fgf8 in an fgf17 mutant background results in an exaggerated cerebellum phenotype (Xu et al., 2000). Ectopic FGF8 studies in the chick showed that only ectopic FGF8 leads to the expression of Engrailed-2, an early marker of mes/rhombencephalic development, Wnt1, and Fgf8 (Crossley et al., 1996). Ectopic FGF8 can also lead to expression of Engrailed-1, Pax2 and Pax5, and suppression of Otx2 expression (Liu et al., 1999; Martinez et al., 1999; Shamim et al., 1999; Sheikh and Mason, 1996). It therefore appears that FGF8, FGF17 and FGF18 have already undergone some degree of subfunctionalization in this territory and are not completely redundant.
Drosophila versus Tribolium
Recent analysis of the fully-sequenced genome of the flour beetle, Tribolium castaneum, has revealed 4 FGF ligands (Tc-FGF1a, Tc-FGF1b, Tc-FGF8, Tc-Bnl) and 1 FGF receptor (Tc-FGFR) are present (Beermann and Schröder, 2008). Tribolium and Drosophila are >300 million years diverged, yet there is some conserved microsynteny between FGF genes in the two species. The gene adjacent to pyramus (CG13197, a predicted tyrosine phosphatase) is homologous to the gene upstream of Tc-FGF8, Tc-00277.
There is only one member of the FGF8 subfamily in Tribolium, Tc-FGF8, but two in Drosophila, thisbe and pyramus. The duplication to produce thisbe and pyramus is thought to have occurred in the arthropod phylum before the radiation of insects because ths/pyr-like sequences were found in one study to be represented in both dipterans and hymenopterans (Popovici et al., 2005). However, the presence of only one FGF8 homolog in Tribolium supports a different scenario where the duplication occurred in Dipterans. Alternatively, a second FGF8-homolog may have been lost in the Tribolium genome. Genes similar to thisbe and pyramus are present in all other Drosophila genomes sequenced so far (unpublished observations), and further investigation of other insect genomes may allow us to point with greater accuracy to the time in which this gene underwent duplication.
Ours and other labs are working on piecing together the overlapping and distinct functions of pyramus and thisbe to understand how much functional redundancy remains and how far the process of subfunctionalization has gone in Drosophila. Both pyr and ths function during gastrulation, specification of mesodermal subtypes, migration of caudal visceral mesoderm, and in axonal migration and glial cell wrapping. In the axon there is a clear separation of function for pyr and ths. Glial-derived pyr modulates glial cell numbers and motility whereas neuronal-derived ths induces glial differentiation (Franzdóttir et al., 2009). Both ligands were found to influence mesoderm spreading, whereas pyr is the dominant player controlling Eve-positive cell specification in the dorsal mesoderm (Kadam et al., 2009; Klingseisen et al., 2009). It therefore seems that the subfunctionalization of pyr and ths from their insect FGF8-homolog ancestor is underway and pyr may either have some derived functions or taken over functions once performed by the single gene.
Studies in Tribolium have shown the pyr/ths homolog, Tc-FGF8, is expressed in largely the same domains as pyr/ths during embryogenesis, and so this gene is also likely involved in spreading of the mesoderm, gut development and brain regionalization (Beermann and Schröder, 2008). Tc-FGF8 is expressed in the developing brain during mid-segmentation. A stripe of Tc-FGF8 expression in each head lobe divides the brain into a larger anterior and a smaller posterior region, in a manner possibly analogous to the MHB in vertebrates. The Drosophila embryonic brain is also divided into a tripartite pattern with an anterior orthodenticle (otd) and posterior Hox domain and an intervening domain. pyr and ths, however, are not expressed in this middle region, but are expressed in one neuroblast in the anterior compartment in each hemibrain (Urbach 2007). Further functional characterization of Tribolium FGFs will undoubtedly provide even more interesting comparisons to Drosophila FGFs.
There are two members of the FGF1 subfamily in Tribolium, yet there is no member of the FGF1 (A) subfamily in Drosophila. This indicates that Drosophila has lost the FGF1 subfamily. This is corroborated by the fact that the neighboring genes (sex-lethal interactor, sin, and seven-in-absentia, sina) to FGF1a and FGF1b in Tribolium have conserved gene order in Drosophila, but FGF1 is missing in Drosophila (Beermann and Schröder, 2008). FGF1 is ubiquitously expressed and is known to play a developmental and maintenance role of neuronal tissue (Beenken and Mohammadi, 2009). Possibly other genes in Drosophila have taken over this function.
FGF variability and plasticity
FGFs are most conserved in the “core” FGF domain, however the conservation is often weak, making phylogenetic analysis difficult. Other properties of FGF ligands including secretion signals, homodimerization ability, glycosylation modifications, binding to HSPGs, and other nonconserved domains in N- and C-terminal tails, can vary from molecule to molecule. There is clearly a high level of plasticity in FGF signaling, the reason for which is unknown but likely relates to the complex networks of regulation that these molecules are involved in (Popovici et al., 2005). The 2nd and 3rd extracellular immunoglobulin (Ig) domains of the FGF receptor are involved in binding the FGF ligands. The amino acid sequence constraints imposed on Ig domains are less than for other protein domains, like kinase domains (Popovici et al., 2005). The variability in the amino acid sequence of Ig domains relates to the high degree of variability in the amino acid composition of FGF ligands (Popovici et al., 2005).
The FGF core domain is thought to be largely responsible for receptor binding. However, the N- and C-terminal tails of FGF molecules are also thought to participate in FGF ligand-receptor specificity. The N- and C-terminal tails can be of variable length. Drosophila FGF have extraordinarily long C-terminal domains compared to the average FGF family member, rendering them ~80kD in molecular weight compared to 18–30kD for the average FGF ligand. Ciona also has a FGF with a large molecular mass, called Ci-FGFL. So far Ci-FGFL has not been assigned to a particular FGF subfamily. Despite the evidence for the importance of the sequence at the N- and C- termini, the function of nonconserved domains outside the FGF domain has received little attention in most FGFs. Three notable exceptions are the study of FGF9/FGF20 and FGF23 in vertebrates and LET-756 in C.elegans.
The crystal structures of both FGF9 and FGF20 were elucidated and, unlike other FGF ligands, the N- and C-terminal regions were found to be ordered and involved in the formation of a homodimer (two FGF9 ligands or two FGF20 ligands), which obscures the receptor binding site (Kalinina et al., 2009; Plotnikov et al., 2001). The homodimerization and ratio of dimers to monomers appears to autoregulate the ligands receptor binding ability to diffuse through the ECM and bind to HSPGs (Harada et al., 2009; Kalinina et al., 2009).
FGF23 is part of a subgroup of endocrine FGFs. Full length FGF23 is 251 amino acids and is cleaved in the C-terminal tail by subtilisin-like proprotein convertases between amino acids 179 and 180. In humans, failure of this cleavage step results in secretion of additional full-length FGF23, which can cause hypophosphatemia leading to autosomal dominant hypophosphatemic rickets/osteomalacia (Benet-Pagès et al., 2004; Fukumoto, 2005).
The C-terminus of C. elegans LET-756 has been shown to contain several nuclear localization signals and the function of them appears to be shuttling LET-756 between several nuclear compartments (Popovici et al., 2006). Additionally, some nuclear localization signals are redundant, highlighting the importance of nuclear localization for LET-756, which has a viability function in C. elegans. Subnuclear localization is important for function and LET-756 may be implicated in mRNA splicing machinery and ribosome function (Popovici et al., 2006).
Recently, our lab has also undertaken the task of elucidating the function of the C-terminal domains of Ths and Pyr [AS23]in Drosophila (Tulin and Stathopoulos, in review). We found that despite their long length, these domains are not required for activity as truncated constructs removing the C-terminus are functional in an overexpression assay. Additional chimeric constructs revealed that the C-terminus of Ths, but not that of Pyr, may play a role in the rate of ligand diffusion and/or potency. We also provide evidence that Ths and Pyr are cleaved from their full-length forms into smaller FGFs in cell culture and these cleaved forms are detectable in the embryo as well. In the embryo, cleaved forms of the FGF ligands could be used to support long-range versus short-range functions as might be necessary during the sequential steps of mesoderm migration, specifically in the control of mesoderm tube collapse versus monolayer formation (McMahon et al., 2010).
Use of the survey approach in FGFRL1
The ability to survey genomes from all major metazoan phyla is a powerful tool that allows researchers to understand the degree of conservation of orthologous genes and to investigate questions about whether similar mechanisms are being used. A good example of this approach being used to study FGF signaling is seen in the study of FGFRL1 (fibroblast growth factor like 1). FGFRL1, or FGFR5, is the most recently discovered member of the FGFR family and has an ectodomain with high similarity to conventional FGFRs, but lacks the catalytic tyrosine kinase domain in the intracellular domain (Sleeman et al., 2001; Wiedemann and Trueb, 2000). FGFRL1 mutant mice die immediately after death with a hypoplastic diaphram and also display skeletal alternations, craniofacial dysplasia, heart valve defects, embryonic anemia, and defective kidney development (Baertschi et al., 2007; Catela et al., 2009; Gerber et al., 2009). Initially it was thought that FGFRL1 was limited to vertebrates, but Bertrand and colleagues have shown that there are orthologs in all metazoan phyla and suggest it may represent a conserved regulatory mechanism for attenuating FGF signaling (Bertrand et al., 2009). Some FGFRL1 orthologs have already been identified, such as FGFRL1 in sea urchin, and others remain to be further investigated, like the putative Drosophila ortholog CG31431 and the ortholog predicted in the cnidarian Nematostella. Subsequent work on FGFRL1 in cell culture and Xenopus embryos has revealed that increasing amounts of FGFR1 ectodomain is shed from primary myoblast cells when they begin differentiating into myotubes (Steinberg et al., 2010). FGFRL1 was found to bind several FGF ligands in both its membrane bound soluable state with high affinity. The affinity of FGFRL1 for FGF3 is 1 order of magitude higher than the affinity of FGF3 for its cognate receptor, FGFR2b, consistent with the model that FGFRL1 could act as a decoy receptor to sequester ligand and attenuate signaling through FGFRs. Ectopic expression of FGFRL1 in the Xenopus embryo resulted in a similar defect to that of the known phenotype of a dominant-negative form of FGFR1, XFD, and could be rescued by injection of FGFR mRNA.
The mechanism of FGF regulation by FGFRL1 type molecules appears to be widespread. The platyhelminthes planarian Dugesua japonica, has a FGFRL molecule called nou-darake, has been characterized as also having a similar phenotype as XFD in Xenopus embryos (Cebrià et al., 2002).
There are still several unknowns with respect to FGFRL1, including the identity of the protease responsible for shedding the ectodomain, the developmental processes and specific FGF receptors it acts on during normal development, and the biological importance of a polymorphism present in the human population affecting an amino acid involved in cleaving FGFR1 (Steinberg et al., 2010). It will be exciting to see if similar mechanisms of FGF regulation are present in phyla as far as Cnidaria and if work on orthologs in other models can help answer the lingering questions as to the role of FGFRL1 in regulating FGF signaling.
Conclusions and Outstanding Questions
In the context of the FGF superfamily, the mounting number of non-vertebrate FGFs is adding to our knowledge of the evolution of FGF signaling and the variety of mechanisms available to these growth factors to regulate embryonic development. Important studies from invertebrates have provided models of alternate splicing, subfunctionalization, regulation by Sprouty proteins, and structural plasticity.
FGF signaling is important for human development and human health; therefore, research will undoubtedly continue in all of the discussed areas and will likely provide targets for medical applications. Importantly, FGF signaling is an ancient metazoan cell communication mechanism predating the cnidarian – bilaterian divergence in the pre Cambrian (>600 million years ago), and is utilized by all extant taxa surveyed to date‥[DM27] This allows for a wealth of varied information that can be used in a number of ways to complement the understanding of our own biology and answer questions about how growth factor signaling has evolved and what mechanisms of signaling and regulation are possible.
Some invertebrate FGF studies have provided very specific functional information. But many studies in recently sequenced models are still based on inferences from expression patterns or simply the presence of homologous domains in the genome. Much work remains to be done to complete the details of the complex signaling and regulatory networks that are present in FGF signaling to orchestrate the grand events of embryogenesis.
Figure 2.
References
- Armelin HA. Pituitary extracts and steroid hormones in the control of 3T3 cell growth. Proc Natl Acad Sci USA. 1973;70(9):2702–2706. doi: 10.1073/pnas.70.9.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baertschi S, Zhuang L, Trueb B. Mice with a targeted disruption of the Fgfrl1 gene die at birth due to alterations in the diaphragm. FEBS J. 2007;274(23):6241–6253. doi: 10.1111/j.1742-4658.2007.06143.x. [DOI] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nature reviews Drug discovery. 2009;8(3):235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beermann A, Schröder R. Sites of Fgf signalling and perception during embryogenesis of the beetle Tribolium castaneum. Dev Genes Evol. 2008;218(3–4):153–167. doi: 10.1007/s00427-007-0192-x. [DOI] [PubMed] [Google Scholar]
- Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134(18):3297–3305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- Benet-Pagès A, Lorenz-Depiereux B, Zischka H, White KE, Econs MJ, Strom TM. FGF23 is processed by proprotein convertases but not by PHEX. Bone. 2004;35(2):455–462. doi: 10.1016/j.bone.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Somorjai I, Garcia-Fernandez J, Lamonerie T, Escriva H. FGFRL1 is a neglected putative actor of the FGF signalling pathway present in all major metazoan phyla. BMC Evol Biol. 2009;9:226. doi: 10.1186/1471-2148-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell. 2003;115(5):615–627. doi: 10.1016/s0092-8674(03)00928-0. [DOI] [PubMed] [Google Scholar]
- Birnbaum D, Popovici C, Roubin R. A pair as a minimum: the two fibroblast growth factors of the nematode Caenorhabditis elegans. Dev Dyn. 2005;232(2):247–255. doi: 10.1002/dvdy.20219. [DOI] [PubMed] [Google Scholar]
- Böttcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26(1):63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- Bridge D, Cunningham CW, DeSalle R, Buss LW. Class-level relationships in the phylum Cnidaria: molecular and morphological evidence. Mol Biol Evol. 1995;12(4):679–689. doi: 10.1093/oxfordjournals.molbev.a040246. [DOI] [PubMed] [Google Scholar]
- Bridge D, Cunningham CW, Schierwater B, DeSalle R, Buss LW. Class-level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc Natl Acad Sci USA. 1992;89(18):8750–8753. doi: 10.1073/pnas.89.18.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess WH, Maciag T. The heparin-binding (fibroblast) growth factor family of proteins. Annu Rev Biochem. 1989;58:575–606. doi: 10.1146/annurev.bi.58.070189.003043. [DOI] [PubMed] [Google Scholar]
- Burgess WH, Mehlman T, Marshak DR, Fraser BA, Maciag T. Structural evidence that endothelial cell growth factor beta is the precursor of both endothelial cell growth factor alpha and acidic fibroblast growth factor. Proc Natl Acad Sci USA. 1986;83(19):7216–7220. doi: 10.1073/pnas.83.19.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casci T, Vinós J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96(5):655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- Catela C, Bilbao-Cortes D, Slonimsky E, Kratsios P, Rosenthal N, Te Welscher P. Multiple congenital malformations of Wolf-Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. Dis Model Mech. 2009;2(5–6):283–294. doi: 10.1242/dmm.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrià F, Kobayashi C, Umesono Y, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sánchez Alvarado A, Agata K. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002;419(6907):620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- Chourrout D, Delsuc F, Chourrout P, Edvardsen RB, Rentzsch F, Renfer E, Jensen MF, Zhu B, de Jong P, Steele RE, Technau U. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature. 2006;442(7103):684–687. doi: 10.1038/nature04863. [DOI] [PubMed] [Google Scholar]
- Coffin JD, Florkiewicz RZ, Neumann J, Mort-Hopkins T, Dorn GW, Lightfoot P, German R, Howles PN, Kier A, O'Toole BA. Abnormal bone growth and selective translational regulation in basic fibroblast growth factor (FGF-2) transgenic mice. Mol Biol Cell. 1995;6(12):1861–1873. doi: 10.1091/mbc.6.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Krnacik S, Rosen JM. Differential temporal and spatial gene expression of fibroblast growth factor family members during mouse mammary gland development. Mol Endocrinol. 1994;8(2):218–229. doi: 10.1210/mend.8.2.8170478. [DOI] [PubMed] [Google Scholar]
- Collins AG, Schuchert P, Marques AC, Jankowski T, Medina M, Schierwater B. Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst Biol. 2006;55(1):97–115. doi: 10.1080/10635150500433615. [DOI] [PubMed] [Google Scholar]
- Coulier F, Pontarotti P, Roubin R, Hartung H, Goldfarb M, Birnbaum D. Of worms and men: an evolutionary perspective on the fibroblast growth factor (FGF) and FGF receptor families. J Mol Evol. 1997;44(1):43–56. doi: 10.1007/pl00006120. [DOI] [PubMed] [Google Scholar]
- Coumoul X, Deng C-X. Roles of FGF receptors in mammalian development and congenital diseases. Birth Defects Res C Embryo Today. 2003;69(4):286–304. doi: 10.1002/bdrc.10025. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martinez S, Martin GR. Midbrain development induced by FGF8 in the chick embryo. Nature. 1996;380(6569):66–68. doi: 10.1038/380066a0. [DOI] [PubMed] [Google Scholar]
- Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Reviews. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 2006;20(19):2728–2738. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132(2):299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- Delli Bovi P, Basilico C. Isolation of a rearranged human transforming gene following transfection of Kaposi sarcoma DNA. Proc Natl Acad Sci USA. 1987;84(16):5660–5664. doi: 10.1073/pnas.84.16.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C, Smith R, Brookes S, Peters G. Tumorigenesis by mouse mammary tumor virus: proviral activation of a cellular gene in the common integration region int-2. Cell. 1984;37(2):529–536. doi: 10.1016/0092-8674(84)90383-0. [DOI] [PubMed] [Google Scholar]
- Draper BW, Stock DW, Kimmel CB. Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development. 2003;130(19):4639–4654. doi: 10.1242/dev.00671. [DOI] [PubMed] [Google Scholar]
- Duboc V, Logan MP. Building limb morphology through integration of signalling modules. Curr Opin Genet Dev. 2009;19(5):497–503. doi: 10.1016/j.gde.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16(2):139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Fletcher RB, Baker JC, Harland RM. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development. 2006;133(9):1703–1714. doi: 10.1242/dev.02342. [DOI] [PubMed] [Google Scholar]
- Franzdóttir SR, Engelen D, Yuva-Aydemir Y, Schmidt I, Aho A, Klämbt C. Switch in FGF signalling initiates glial differentiation in the Drosophila eye. Nature. 2009;460(7256):758–761. doi: 10.1038/nature08167. [DOI] [PubMed] [Google Scholar]
- Fukumoto S. Post-translational modification of Fibroblast Growth Factor 23. Ther Apher Dial. 2005;9(4):319–322. doi: 10.1111/j.1744-9987.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- Gerber SD, Steinberg F, Beyeler M, Villiger PM, Trueb B. The murine Fgfrl1 receptor is essential for the development of the metanephric kidney. Dev Biol. 2009;335(1):106–119. doi: 10.1016/j.ydbio.2009.08.019. [DOI] [PubMed] [Google Scholar]
- Goodman SJ, Branda CS, Robinson MK, Burdine RD, Stern MJ. Alternative splicing affecting a novel domain in the C. elegans EGL-15 FGF receptor confers functional specificity. Development. 2003;130(16):3757–3766. doi: 10.1242/dev.00604. [DOI] [PubMed] [Google Scholar]
- Gospodarowicz D. Localisation of a fibroblast growth factor and its effect alone and with hydrocortisone on 3T3 cell growth. Nature. 1974;249(453):123–127. doi: 10.1038/249123a0. [DOI] [PubMed] [Google Scholar]
- Gospodarowicz D. Purification of a fibroblast growth factor from bovine pituitary. J Biol Chem. 1975;250(7):2515–2520. [PubMed] [Google Scholar]
- Gospodarowicz D, Moran JS. Mitogenic effect of fibroblast growth factor on early passage cultures of human and murine fibroblasts. J Cell Biol. 1975;66(2):451–457. doi: 10.1083/jcb.66.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospodarowicz DBH, Greenburg G. Purification of the fibroblast growth factor activity from bovine brain. J Biol Chem. 1978;253(10):3736–3743. [PubMed] [Google Scholar]
- Häcker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6(7):530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92(2):253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Harada M, Murakami H, Okawa A, Okimoto N, Hiraoka S, Nakahara T, Akasaka R, Shiraishi Y-I, Futatsugi N, Mizutani-Koseki Y, Kuroiwa A, Shirouzu M, Yokoyama S, Taiji M, Iseki S, Ornitz DM, Koseki H. FGF9 monomer-dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nat Genet. 2009;41(3):289–298. doi: 10.1038/ng.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R. Growth. 1940;4:361–376. [Google Scholar]
- Holland PW, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev. 1994;(Suppl):125–133. [PubMed] [Google Scholar]
- Hotta Y, Sasaki S, Konishi M, Kinoshita H, Kuwahara K, Nakao K, Itoh N. Fgf16 is required for cardiomyocyte proliferation in the mouse embryonic heart. Dev Dyn. 2008;237(10):2947–2954. doi: 10.1002/dvdy.21726. [DOI] [PubMed] [Google Scholar]
- Ikuta T, Saiga H. Dynamic change in the expression of developmental genes in the ascidian central nervous system: revisit to the tripartite model and the origin of the midbrain-hindbrain boundary region. Dev Biol. 2007;312(2):631–643. doi: 10.1016/j.ydbio.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312(5777):1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satoh N, Satou Y. Early embryonic expression of FGF4/6/9 gene and its role in the induction of mesenchyme and notochord in Ciona savignyi embryos. Development. 2002;129(7):1729–1738. doi: 10.1242/dev.129.7.1729. [DOI] [PubMed] [Google Scholar]
- Imai KS, Stolfi A, Levine M, Satou Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development. 2009;136(2):285–293. doi: 10.1242/dev.026419. [DOI] [PubMed] [Google Scholar]
- Kadam S, McMahon A, Tzou P, Stathopoulos A. FGF ligands in Drosophila have distinct activities required to support cell migration and differentiation. Development. 2009;136(5):739–747. doi: 10.1242/dev.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina J, Byron SA, Makarenkova HP, Olsen SK, Eliseenkova AV, Larochelle WJ, Dhanabal M, Blais S, Ornitz DM, Day LA, Neubert TA, Pollock PM, Mohammadi M. Homodimerization controls the fibroblast growth factor 9 subfamily’s receptor binding and heparan sulfate-dependent diffusion in the extracellular matrix. Mol Cell Biol. 2009;29(17):4663–4678. doi: 10.1128/MCB.01780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirikoshi H, Sagara N, Saitoh T, Tanaka K, Sekihara H, Shiokawa K, Katoh M. Molecular cloning and characterization of human FGF-20 on chromosome 8p21.3-p22. Biochem Biophys Res Commun. 2000;274(2):337–343. doi: 10.1006/bbrc.2000.3142. [DOI] [PubMed] [Google Scholar]
- Klingseisen A, Clark IBN, Gryzik T, Müller H-AJ. Differential and overlapping functions of two closely related Drosophila FGF8-like growth factors in mesoderm development. Development. 2009;136(14):2393–2402. doi: 10.1242/dev.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobberup S, Schmerr M, Dang M-L, Nyeng P, Jensen JN, MacDonald RJ, Jensen J. Conditional control of the differentiation competence of pancreatic endocrine and ductal cells by Fgf10. Mech Dev. 2010;127(3–4):220–234. doi: 10.1016/j.mod.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourakis MJ, Smith WC. A conserved role for FGF signaling in chordate otic/atrial placode formation. Dev Biol. 2007;312(1):245–257. doi: 10.1016/j.ydbio.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Okabe M, Hacohen N, Krasnow MA, Hiromi Y. Sprouty: a common antagonist of FGF and EGF signaling pathways in Drosophila. Development. 1999;126(11):2515–2525. doi: 10.1242/dev.126.11.2515. [DOI] [PubMed] [Google Scholar]
- Lapraz F, Röttinger E, Duboc V, Range R, Duloquin L, Walton K, Wu S-Y, Bradham C, Loza MA, Hibino T, Wilson K, Poustka A, McClay D, Angerer L, Gache C, Lepage T. RTK and TGF-beta signaling pathways genes in the sea urchin genome. Dev Biol. 2006;300(1):132–152. doi: 10.1016/j.ydbio.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8(1):85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lee PL, Johnson DE, Cousens LS, Fried VA, Williams LT. Purification and complementary DNA cloning of a receptor for basic fibroblast growth factor. Science. 1989;245(4913):4957–60. doi: 10.1126/science.2544996. [DOI] [PubMed] [Google Scholar]
- Lemmon SK, Riley MC, Thomas KA, Hoover GA, Maciag T, Bradshaw RA. Bovine fibroblast growth factor: comparison of brain and pituitary preparations. J Cell Biol. 1982;95(1):162–169. doi: 10.1083/jcb.95.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libermann TA, Friesel R, Jaye M, Lyall RM, Westermark B, Drohan W, Schmidt A, Maciag T, Schlessinger J. An angiogenic growth factor is expressed in human glioma cells. EMBO J. 1987;6(6):1627–1632. doi: 10.1002/j.1460-2075.1987.tb02410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Li JYH, Bromleigh C, Lao Z, Niswander LA, Joyner AL. FGF17b and FGF18 have different midbrain regulatory properties from FGF8b or activated FGF receptors. Development. 2003;130(25):6175–6185. doi: 10.1242/dev.00845. [DOI] [PubMed] [Google Scholar]
- Liu A, Losos K, Joyner AL. FGF8 can activate Gbx2 and transform regions of the rostral mouse brain into a hindbrain fate. Development. 1999;126(21):4827–4838. doi: 10.1242/dev.126.21.4827. [DOI] [PubMed] [Google Scholar]
- Lu W, Luo Y, Kan M, McKeehan WL. Fibroblast growth factor-10. A second candidate stromal to epithelial cell andromedin in prostate. J Biol Chem. 1999;274(18):12827–12834. doi: 10.1074/jbc.274.18.12827. [DOI] [PubMed] [Google Scholar]
- Maciag T, Cerundolo J, Ilsley S, Kelley PR, Forand R. An endothelial cell growth factor from bovine hypothalamus: identification and partial characterization. Proc Natl Acad Sci USA. 1979;76(11):5674–5678. doi: 10.1073/pnas.76.11.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marics I, Adelaide J, Raybaud F, Mattei MG, Coulier F, Planche J, de Lapeyriere O, Birnbaum D. Characterization of the HST-related FGF.6 gene, a new member of the fibroblast growth factor gene family. Oncogene. 1989;4(3):335–340. [PubMed] [Google Scholar]
- Martinez S, Crossley PH, Cobos I, Rubenstein JL, Martin GR. FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development. 1999;126(6):1189–1200. doi: 10.1242/dev.126.6.1189. [DOI] [PubMed] [Google Scholar]
- Maruoka Y, Ohbayashi N, Hoshikawa M, Itoh N, Hogan BL, Furuta Y. Comparison of the expression of three highly related genes, Fgf8, Fgf17 and Fgf18, in the mouse embryo. Mech Dev. 1998;74(1–2):175–177. doi: 10.1016/s0925-4773(98)00061-6. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ. FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol. 2007;217(2):137–148. doi: 10.1007/s00427-006-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoon PE, Angerer RC, Angerer LM. SpFGFR, a new member of the fibroblast growth factor receptor family, is developmentally regulated during early sea urchin development. J Biol Chem. 1996;271(33):20119–20125. doi: 10.1074/jbc.271.33.20119. [DOI] [PubMed] [Google Scholar]
- McCoon PE, Blackstone E, Angerer RC, Angerer LM. Sea urchin FGFR muscle-specific expression: posttranscriptional regulation in embryos and adults. Dev Biol. 1998;200(2):171–181. doi: 10.1006/dbio.1998.8943. [DOI] [PubMed] [Google Scholar]
- McMahon A, Reeves GT, Supatto W, Stathopoulos A. Mesoderm migration in Drosophila is a multi-step process requiring FGF signaling and integrin activity. Development. 2010;137(13):2167–2175. doi: 10.1242/dev.051573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M, Collins AG, Silberman JD, Sogin ML. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Natl Acad Sci USA. 2001;98(17):9707–9712. doi: 10.1073/pnas.171316998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18(2):136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Miyake A, Konishi M, Martin FH, Hernday NA, Ozaki K, Yamamoto S, Mikami T, Arakawa T, Itoh N. Structure and expression of a novel member, FGF-16, on the fibroblast growth factor family. Biochem Biophys Res Commun. 1998;243(1):148–152. doi: 10.1006/bbrc.1998.8073. [DOI] [PubMed] [Google Scholar]
- Miyamoto M, Naruo K, Seko C, Matsumoto S, Kondo T, Kurokawa T. Molecular cloning of a novel cytokine cDNA encoding the ninth member of the fibroblast growth factor family, which has a unique secretion property. Mol Cell Biol. 1993;13(7):4251–4259. doi: 10.1128/mcb.13.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol. 1996;16(3):977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16(2):107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105(8):1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatake Y, Hoshikawa M, Asaki T, Kassai Y, Itoh N. Identification of a novel fibroblast growth factor, FGF-22, preferentially expressed in the inner root sheath of the hair follicle. Biochim Biophys Acta. 2001;1517(3):460–463. doi: 10.1016/s0167-4781(00)00302-x. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Utsunomiya Y, Hoshikawa M, Ohuchi H, Itoh N. Structure and expression of a novel human FGF, FGF-19, expressed in the fetal brain. Biochim Biophys Acta. 1999;1444(1):148–151. doi: 10.1016/s0167-4781(98)00255-3. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Kobayashi C, Hayashi T, Orii H, Watanabe K, Agata K. Planarian fibroblast growth factor receptor homologs expressed in stem cells and cephalic ganglions. Dev Growth Differ. 2002;44(3):191–204. doi: 10.1046/j.1440-169x.2002.00634.x. [DOI] [PubMed] [Google Scholar]
- Ohbayashi N, Hoshikawa M, Kimura S, Yamasaki M, Fukui S, Itoh N. Structure and expression of the mRNA encoding a novel fibroblast growth factor, FGF-18. J Biol Chem. 1998;273(29):18161–18164. doi: 10.1074/jbc.273.29.18161. [DOI] [PubMed] [Google Scholar]
- Ohno S. Springer-Verlag; Berlin-Heidelberg-New York: 1970. Evolution by Gene Duplication. [Google Scholar]
- Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, Mohammadi M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem. 2003;278(36):34226–34236. doi: 10.1074/jbc.M303183200. [DOI] [PubMed] [Google Scholar]
- Olwin BB, Hauschka SD. Identification of the fibroblast growth factor receptor of Swiss 3T3 cells and mouse skeletal muscle myoblasts. Biochemistry. 1986;25(12):3487–3492. doi: 10.1021/bi00360a001. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2(3):3001–3005. doi: 10.1186/gb-2001-2-3-reviews3005. reviews3005.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov AN, Eliseenkova AV, Ibrahimi OA, Shriver Z, Sasisekharan R, Lemmon MA, Mohammadi M. Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition. J Biol Chem. 2001;276(6):4322–4329. doi: 10.1074/jbc.M006502200. [DOI] [PubMed] [Google Scholar]
- Popovici C, Fallet M, Marguet D, Birnbaum D, Roubin R. Intracellular trafficking of LET-756, a fibroblast growth factor of C. elegans, is controlled by a balance of export and nuclear signals. Exp Cell Res. 2006;312(9):1484–1495. doi: 10.1016/j.yexcr.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Popovici C, Roubin R, Coulier F, Birnbaum D. An evolutionary history of the FGF superfamily. Bioessays. 2005;27(8):849–857. doi: 10.1002/bies.20261. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317(5834):86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JH, Bos JL. Specificity in Ras and Rap signaling. J Biol Chem. 2009;284(17):10995–10999. doi: 10.1074/jbc.R800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifers F, Böhli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125(13):2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development. 2008;135(10):1761–1769. doi: 10.1242/dev.020784. [DOI] [PubMed] [Google Scholar]
- Röttinger E, Saudemont A, Duboc V, Besnardeau L, McClay D, Lepage T. FGF signals guide migration of mesenchymal cells, control skeletal morphogenesis [corrected] and regulate gastrulation during sea urchin development. Development. 2008;135(2):353–365. doi: 10.1242/dev.014282. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371(6494):252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- Rubin JS, Osada H, Finch PW, Taylor WG, Rudikoff S, Aaronson SA. Purification and characterization of a newly identified growth factor specific for epithelial cells. Proc Natl Acad Sci USA. 1989;86(3):802–806. doi: 10.1073/pnas.86.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 2006;7(7):R64. doi: 10.1186/gb-2006-7-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Mori M, Taira M, Yoshida T, Matsukawa S, Shimizu K, Sekiguchi M, Terada M, Sugimura T. Transforming gene from human stomach cancers and a noncancerous portion of stomach mucosa. Proc Natl Acad Sci USA. 1986;83(11):3997–4001. doi: 10.1073/pnas.83.11.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Araki I, Nakamura H. Inductive signal and tissue responsiveness defining the tectum and the cerebellum. Development. 2001;128(13):2461–2469. doi: 10.1242/dev.128.13.2461. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. Fgf genes in the basal chordate Ciona intestinalis. Dev Genes Evol. 2002;212(9):432–438. doi: 10.1007/s00427-002-0266-8. [DOI] [PubMed] [Google Scholar]
- Sawada A, Shinya M, Jiang YJ, Kawakami A, Kuroiwa A, Takeda H. Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development. 2001;128(23):4873–4880. doi: 10.1242/dev.128.23.4873. [DOI] [PubMed] [Google Scholar]
- Shamim H, Mahmood R, Logan C, Doherty P, Lumsden A, Mason I. Sequential roles for Fgf4, En1 and Fgf8 in specification and regionalisation of the midbrain. Development. 1999;126(5):945–959. doi: 10.1242/dev.126.5.945. [DOI] [PubMed] [Google Scholar]
- Sheikh H, Mason I. Polarising activity of FGF-8 in the avian midbrain. Int J Dev Biol Suppl. 1996;1:117S–118S. [PubMed] [Google Scholar]
- Shi W, Peyrot SM, Munro E, Levine M. FGF3 in the floor plate directs notochord convergent extension in the Ciona tadpole. Development. 2009;136(1):23–28. doi: 10.1242/dev.029157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiang R, Thompson LM, Zhu YZ, Church DM, Fielder TJ, Bocian M, Winokur ST, Wasmuth JJ. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78(2):335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- Sivak JM, Petersen LF, Amaya E. FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev Cell. 2005;8(5):689–701. doi: 10.1016/j.devcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Slack JM, Darlington BG, Heath JK, Godsave SF. Mesoderm induction in early Xenopus embryos by heparin-binding growth factors. Nature. 1987;326(6109):197–200. doi: 10.1038/326197a0. [DOI] [PubMed] [Google Scholar]
- Sleeman M, Fraser J, McDonald M, Yuan S, White D, Grandison P, Kumble K, Watson JD, Murison JG. Identification of a new fibroblast growth factor receptor, FGFR5. Gene. 2001;271(2):171–182. doi: 10.1016/s0378-1119(01)00518-2. [DOI] [PubMed] [Google Scholar]
- Smallwood PM, Munoz-Sanjuan I, Tong P, Macke JP, Hendry SH, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc Natl Acad Sci USA. 1996;93(18):9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F, Zhuang L, Beyeler M, Kälin RE, Mullis PE, Brändli AW, Trueb B. The FGFRL1 receptor is shed from cell membranes, binds fibroblast growth factors (FGFs), and antagonizes FGF signaling in Xenopus embryos. J Biol Chem. 2010;285(3):2193–2202. doi: 10.1074/jbc.M109.058248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhop S, Coulier F, Bieller A, Vogt A, Hotz T, Hassel M. Signalling by the FGFR-like tyrosine kinase, Kringelchen, is essential for bud detachment in Hydra vulgaris. Development. 2004;131(16):4001–4011. doi: 10.1242/dev.01267. [DOI] [PubMed] [Google Scholar]
- Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13(14):1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Miyamoto K, Minamino N, Takeda M, Sato B, Matsuo H, Matsumoto K. Cloning and characterization of an androgen-induced growth factor essential for the androgen-dependent growth of mouse mammary carcinoma cells. Proc Natl Acad Sci USA. 1992;89(19):8928–8932. doi: 10.1073/pnas.89.19.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Sasaki K-i, Watari K, Yasukawa H, Imaizumi T, Ayada T, Okamoto F, Ishizaki T, Kato R, Kohno R-i, Kimura H, Sato Y, Ono M, Yonemitsu Y, Yoshimura A. Suppression of Sproutys has a therapeutic effect for a mouse model of ischemia by enhancing angiogenesis. PLoS ONE. 2009;4(5):e5467. doi: 10.1371/journal.pone.0005467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U, Rudd S, Maxwell P, Gordon PMK, Saina M, Grasso LC, Hayward DC, Sensen CW, Saint R, Holstein TW, Ball EE, Miller DJ. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005;21(12):633–639. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287(2):390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Trowell OACB, Willmer EN. J Exp Biol. 1939;16:60–70. [Google Scholar]
- Wiedemann M, Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69(2):275–279. doi: 10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- Wilkie AOM. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 2005;16(2):187–203. doi: 10.1016/j.cytogfr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Xu J, Lawshe A, MacArthur CA, Ornitz DM. Genomic structure, mapping, activity and expression of fibroblast growth factor 17. Mech Dev. 1999;83(1–2):165–178. doi: 10.1016/s0925-4773(99)00034-9. [DOI] [PubMed] [Google Scholar]
- Xu J, Liu Z, Ornitz DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127(9):1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Yoshioka M, Itoh N. Identification of a novel fibroblast growth factor, FGF-23, preferentially expressed in the ventrolateral thalamic nucleus of the brain. Biochem Biophys Res Commun. 2000;277(2):494–498. doi: 10.1006/bbrc.2000.3696. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Hudson C. FGF8/17/18 functions together with FGF9/16/20 during formation of the notochord in Ciona embryos. Dev Biol. 2007;302(1):92–103. doi: 10.1016/j.ydbio.2006.08.075. [DOI] [PubMed] [Google Scholar]
- Zhan X, Bates B, Hu XG, Goldfarb M. The human FGF-5 oncogene encodes a novel protein related to fibroblast growth factors. Mol Cell Biol. 1988;8(8):3487–3495. doi: 10.1128/mcb.8.8.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]