Abstract
Hippo-like MST1 protein kinase regulates cell growth, organ size, and carcinogenesis. Reduction or loss of MST1 expression is implicated in poor cancer prognosis. However, the mechanism leading to MST1 silencing remains elusive. Here, we report that both MYC and EZH2 function as potent suppressors of MST1 expression in human prostate cancer cells. We demonstrated that concurrent overexpression of MYC and EZH2 correlated with the reduction or loss of MST1 expression, as shown by RT-qPCR and immunoblotting. Methylation sensitive PCR and bisulfite genomic DNA sequencing showed that DNA methylation caused MST1 silencing. Pharmacologic and RNAi experiments revealed that MYC and EZH2 silenced MST1 expression by inhibiting its promoter activity, and that EZH2 was a mediator of the MYC-induced silencing of MST1. In addition, MYC contributed to MST1 silencing by partly inhibiting the expression of microRNA-26a/b, a negative regulator of EZH2. As shown by ChIP assays, EZH2-induced DNA methylation and H3K27me3 modification, which was accompanied by a reduced H3K4me3 mark and RNA polymerase II occupancy on the MST1 promoter CpG region, were the underlying cause of MST1 silencing. Moreover, potent pharmacologic inhibitors of MYC or EZH2 suppressed prostate cancer cell growth in vitro, and the knockdown of MST1 caused cells’ resistance to MYC and EZH2 inhibitor-induced growth retardation. These findings indicate that MYC, in concert with EZH2, epigenetically attenuates MST1 expression and suggest that the loss of MST1/Hippo functions is critical for the MYC or EZH2 mediation of cancer cell survival.
Keywords: epigenetics, DNA methylation, H3K27me3, MYC, EZH2 and MST1/Hippo
Introduction
MST1, encoded by the STK4 gene in chromosome 20 and related to hippo (hpo) in Drosophila, is a multifunctional protein kinase. MST1 regulates gene expression, cell growth, stem cell self-renewal, organ size, and tumorigenesis.1-5 Reduction or loss of MST1 expression is implicated the etiology of many cancers with poor prognosis,6-9 including prostate cancer (PC).10,11 PC is one of the leading causes of cancer-related deaths in men in western countries. Previously, our laboratory reported that deregulation of MST1 due to its reduced expression and posttranslational modification might play a prominent role in the disease progression.10,12 However, little is known about the mechanism underlying the reduction or loss of MST1 expression in PC cells.
MYC (c-MYC) is a transcription factor and regulates cell growth and proliferation by activating and repressing its target gene expression. Overexpression of MYC is associated with the initiation and metastatic, castration-resistant progression of PC.13-16 EZH2 (enhancer of zeste homolog 2) is a subunit of PRC2 (polycomb repressive complex 2). EZH2 is a histone methyltransferase and catalyzes the trimethylation of histone 3 at lysine 27 (H3K27me3), which causes gene silencing.17 Overexpression of EZH2 is also implicated in metastatic PC progression.18,19 DNA methyltransferases (DNMTs: DNMT1, DNMT3a, and DNMT3b) catalyze the methylation of cytosine at CpG sites in the regulatory region of genes, leading to gene inactivation. The H3K27me3 mark has been suggested to facilitate DNA methylation in cancer cells.20 DNA methylation is associated with metastatic tumor progression.21,22
MYC has been identified as a key regulator of EZH2 overexpression.23,24 MYC might promote EZH2 expression by repressing the expression of microRNAs (miR)-26a and miR-26b, which were demonstrated to be potent negative regulators of EZH2 mRNA, and by activating EZH2 promoter through the E-box, a DNA binding site for MYC.23 In addition, MYC was demonstrated to repress its target gene expression via interaction with DNMT3a and PRC2 complex.25,26 Reduction or loss of MST1 expression by DNA methylation was reported in sarcoma and glioblastoma.27,28 Despite these observations, it is unknown whether MYC and/or EZH2 biochemically and functionally intersect with MST1.
In this study, we investigated the impact of MYC and EZH2 overexpression on the MST1 gene silencing in PC cells. We demonstrated that concurrent overexpression of MYC and EZH2 attenuated MST1 expression. We found that EZH2-mediated H3K27me3 modification and DNA methylation were the underlying causes of MST1 silencing. In addition, we provide evidence that MST1 loss might play a key role in EZH2- and MYC-induced PC cell survival. Our study uncovers the new mechanism of MST1 deregulation that involves a cooperative MYC and EZH2 signaling and emphasizes the biological significance of MST1/Hippo loss in lethal disease progression.
Results
Expression of MST1, MYC, and EZH2 in prostate cancer cells
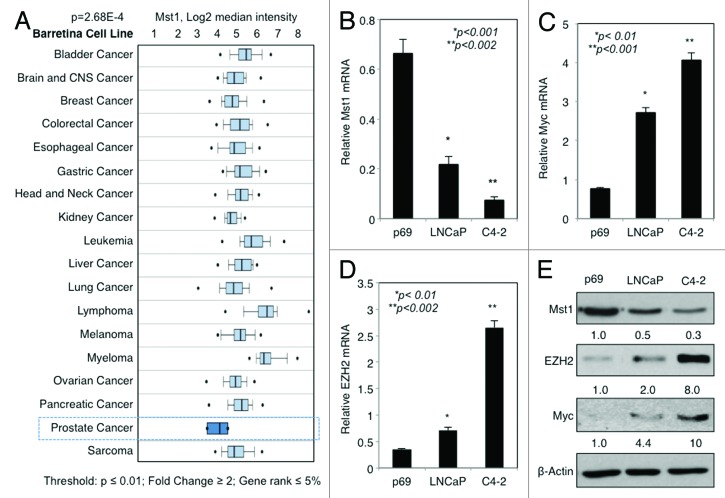
We performed Cancer Outlier Profile Analysis (COPA) of MST1 expression in multiple microarray data sets in Oncomine (www.oncomine.com). COPA revealed a reduction in MST1 expression in PC as a significant outlier (Fig. S1). In addition, analysis of the microarray data in Oncomine obtained from multiple cell lines, which were derived from major cancer types, showed that PC cells expressed the lowest levels of MST1 mRNA among others [Barretina cell line; P = 2.68E-4, fold change = -2.136, Fig. 1A, Ramaswamy multi-cancer, P = 0.003, fold change = -2.017 and Bittner multi-cancer; P = 2.51E-17, fold change = -1.551, (not shown)]. Moreover, published studies indicated that overexpression of MYC and EZH2 is frequently detected in metastatic PC.13,14,18,19 Our laboratory previously reported that MST1 protein levels declined during PC progression toward the invasive state.10 These observations let the hypothesis that overexpression of MYC and EZH2 negatively regulates MST1 expression.
Figure 1. Expression of MST1 is inversely correlated with those of MYC and EZH2 in PC cells. (A) Analysis of Oncomine microarray data for MST1 mRNA expression changes in multiple cancer cell lines. (B–E) Analysis of MST1 (B), MYC (C) and EZH2 (D) mRNA levels by RT-qPCR protein levels by western blot (WB) (E) in non-cancerous p69, cancerous LNCaP and C4–2 human prostate cells grown in serum-fed conditions. Band intensities were quantified by ImageJ software, and the data were normalized to β-Actin (loading control) and presented as fold (f) change relative to that of p69. Data are (± S.E.) from multiple experiments.
To assess the impact of MYC and EZH2 on MST1 expression, first we examined the levels of MST1, MYC, and EZH2 mRNA in p69 normal prostate epithelial cells and LNCaP PC cell models and its castration-resistant C4-2 subline using reverse transcriptase quantitative-polymerase chain reaction (RT-qPCR). The results showed that compared with noncancerous p69 cells, MST1 mRNA expression was progressively reduced in PC cell models, which was very well correlated with MYC and EZH2 overexpression (Fig. 1B–D). Similarly, as analyzed by RT-qPCR, the expression of MST1 mRNA was also inversely correlated with MYC and EZH2 overexpression in PC clinical samples compared with noncancerous counters (Fig. S2). In addition, we assessed the levels of MST1, MYC and EZH2 protein in the same cell lines by western blot. The results in Figure 1E showed that consistent with the mRNA expression, MST1 protein levels were progressively reduced, which was also inversely correlated an increased the expression of MYC and EZH2.
DNA methylation attenuates MST1 expression
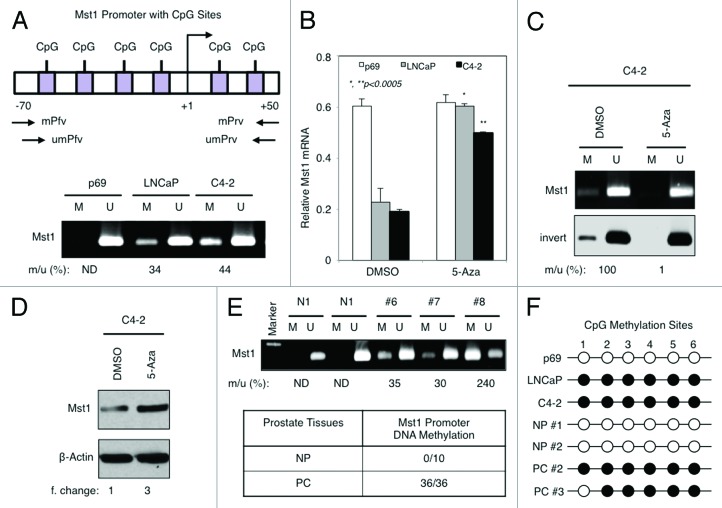
DNA methylation was suggested to reduce MST1 expression in sarcoma and glioblastoma.27,28 However, whether the similar mechanism exists in PC cells remains to be determined. To assess the methylation status of MST1 promoter in p69 normal prostate and LNCaP and C4-2 PC cells, we performed methylation sensitive PCR (MSP) using the previously published primer sets,27,28 as illustrated in Figure 2A (upper panel). The results in Figure 2A, lower panel, showed that no DNA methylation at the MST1 promoter CpG regions was observed in p69 cells, whereas PC cells displayed a variable percentage of MST1 promoter methylation: 34% in LNCaP, 44% in C4-2 cells.
Figure 2. DNA methylation at the promoter CpG sites causes MST1 silencing in PC cells. (A) Schematic representation of MST1 promoter at CpG sites spanning from -70 to +50 relative to transcriptional start site (TSS). Forward primer (Pfv) and reverse primer (Rrv) used in methylation (m) and unmethylation (um) analysis are depicted (upper panel). Bisulfite-treated genomic DNA from the indicated cells that were grown in serum-fed conditions at 80% confluence was analyzed by methylation-specific PCR (MSP). The number below the image represents the percent ratio of methylated (m) to unmethylated (u) DNA intensity quantified by ImageJ software (lower panel). (B) RT-PCR analysis of MST1 mRNA levels in p69, LNCaP, and C4–2 cells after treatment with DMSO (vehicle) or 5 µM 5-Aza deoxycytidine (5-Aza) for 72h. (C and D) MSP analysis of MST1 promoter and MST1 protein with WB in C4–2 cells after treatment with DMSO (vehicle) or 5 µM 5-Aza for 72h. The data were normalized to β-Actin protein (loading control) and presented as fold-change relative to DMSO after quantification by ImageJ. (E) A representative image of MST1 promoter methylation in human prostate clinical samples. MSP was performed with bisulfite-treated genomic DNA isolated from frozen normal prostate (n = 4) and frozen prostate tumor samples (n = 8) (upper panel). Table shows the summary of MST1 promoter methylation in cohorts of normal and PC clinical samples (lower panel). (F) Schematic representation of bisulfite-treated genomic DNA sequencing results. Bisulfite-treated DNA from p69 normal prostate epithelia, LNCaP and C4–2 PC cells, and from two normal prostate tissues and PC clinical samples were sequenced. Six CpG sites (1–6) in the amplified fragment are indicated. Black and white circles indicate methylated and unmethylated CpGs, respectively. ND, none detectable; NP, normal (noncancerous) prostate; PCa, PC. Data (± S.E.) are or multiple experiments.
To determine that DNA methylation at CpG sites is directly responsible for MST1 silencing, we assessed the levels of MST1 mRNA by RT-qPCR in p69, LNCaP, and C4–2 cells after treatment with DMSO (vehicle) or 5-Aza-2′-deoxycytidine (5-Aza), a potent pharmacologic inhibitor of DNMTs. In comparison to the DMSO, 5-Aza treatment increased MST1 mRNA 3-fold in LNCaP and 2.5-fold in C4–2 cells (Fig. 2B). This result was specific because 5-Aza did not affect MST1 mRNA expression in DNA methylation-free p69 cells (Fig. 2B). Consistently, 5-Aza effectively decreased MST1 promoter methylation in LNCaP (not shown) and C4-2 (Fig. 2C) cells. In parallel, western blot showed 3-fold increase in MST1 protein levels in 5-Aza-treated C4–2 cells relative to control DMSO (Fig. 2D).
To assess the clinical significance of the above findings, we analyzed the methylation status of MST1 promoter DNA by MSP in fresh-frozen and formalin-fixed and paraffin-embedded normal prostate tissues and PC clinical samples. None of the normal prostate specimens showed MST1 promoter DNA methylation, whereas all tumor samples analyzed displayed a variable percentage of DNA methylation (Fig. 2E; Table S3A and B). In addition, as illustrated in Figure 2F, bisulfite sequencing of genomic DNA isolated from select PC cell lines and frozen tissue samples, along with their normal counters confirming the above data showed that, indeed, MST1 promoter at CpG sites was methylated. These findings consistent with published studies,27,28 indicate that DNA methylation may play a significant role in MST1 attenuation in PC cells.
EZH2 overexpression attenuates MST1 expression
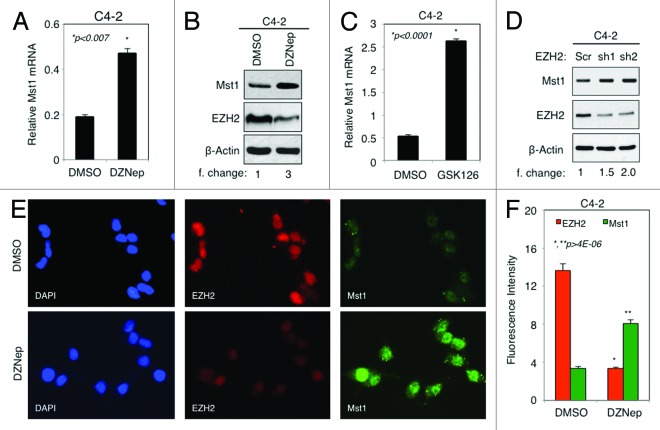
It has been suggested that EZH2 can contribute to gene silencing by mediating DNA methylation and histone modifications.20 As shown in Figure 1, EZH2 was upregulated in PC cells. To determine whether overexpression of EZH2 leads to MST1 attenuation, we assessed the levels of MST1 mRNA by RT-qPCR and protein by western blot in C4–2 cells after treatment with DMSO (vehicle) or DZNep. DZNep is a potent inhibitor of the EZH2 protein expression, by which it inhibits EZH2 functions.29 The C4–2 cell line was ideal for this experiment because it expresses much higher levels of EZH2 than the LNCaP. The results showed that treatment of cells with DZNep augmented MST1 mRNA levels 2.5-fold (Fig. 3A) and protein levels 3-fold while decreasing the expression of EZH2 protein in comparison with control DMSO (Fig. 3B).
Figure 3. Overexpression of EZH2 negatively regulates MST1 expression in PC cells. (A and B). Analysis of MST1 mRNA levels by RT-qPCR (A) and MST1, EZH2, and β-Actin protein levels by WB (B) in C4–2 cells after treatment with DMSO or 5 μM DZNep for 72h. (C) RT-qPCR analysis of MST1 mRNA levels in C4–2 cells after treatment with DMSO or 2 μM GSK126 for 72h. (D) WB analysis of MST1 protein levels in C4–2 cells after transient transfection with scramble (Src) control or two different EZH2 shRNA in lentiviral constructs for 72h. MST1 protein levels were normalized to β-Actin (loading control) and the data presented as fold (f) change relative to scramble control. (E) Co-IF staining of EZH2 and MST1 protein in C4–2 cells treated with DMSO or 5 µM DZNep for 72h in serum-fed growth conditions. Alexa Fluor 488 stained MST1 (green), Alexa Fluor 568 stained EZH2 (red), and DAPI stained cell nuclei (blue). Magnification is 20x. Micrographs are representative of multiple images from two independent experiments. (F) Graph shows signal intensity of EZH2 and MST1 signal in “E.” Data are (± S.E.) from multiple experiments.
To verify the specificity of the above finding, we used another selective and potent EZH2 inhibitor, GSK126. GSK126 attenuated the catalytic activity of EZH2 without affecting EZH2 protein levels.30 Similar to the DZNep, GSK126 caused a dose-dependent overexpression of MST1 protein (Fig. S4A). Treatment of C4–2 cells with GSK126 augmented MST1 mRNA expression about 5-fold compared with control DMSO (Fig. 3C). In addition, knockdown of EZH2 by two different EZH2 shRNAs let to an increase in MST1 protein 1.5–2.0 fold in comparison with control shRNA (Fig. 3D). These findings indicate that the effects of EZH2 on MST1 are specific.
Co-immunofluorescence (co-IF) analysis of EZH2 and MST1 protein in C4–2 cells further verified the above data that inhibition of EZH2 protein expression by DZNep caused an increased MST1 protein levels about 2-fold in comparison with control DMSO (Fig. 3E and F). Similarly, co-IF analysis of MST1 and EZH2 proteins in PC clinical samples showed that cells with normal prostate tissue architecture expressed the highest levels of MST1 protein, where EZH2 protein levels were low or undetectable (Fig. S4B, right side of the merged image). However, cells with cancerous prostate tissue architecture expressed the lowest levels of MST1 protein, where EZH2 protein levels were high (Fig. S4B, left side of the merged image). These findings suggest that EZH2 is a potent physiologic negative regulator of MST1 expression.
MYC overexpression silences MST1 expression through EZH2
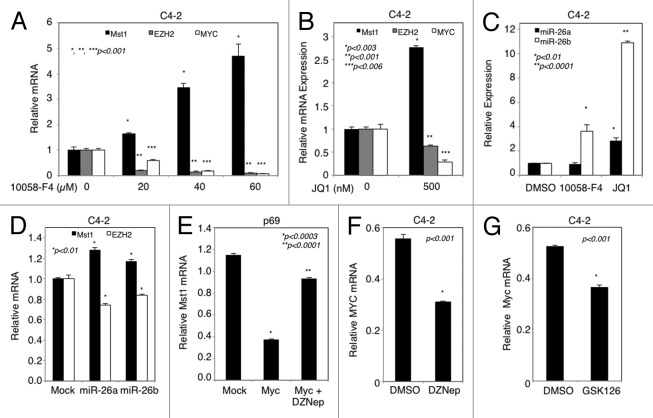
MYC was identified as a key driver of EZH2 overexpression in PC cells.23 The data in Figure 3 showed that EZH2 attenuated MST1 expression. We wanted to know whether overexpression of MYC attenuates MST1 and whether that is mediated by EZH2. To test these possibilities, we analyzed MST1, MYC and EZH2 mRNA expression by RT-qPCR (Fig. 4A) and semi-RT-qPCR (Fig. S5A) in C4-2 cells, which were treated with increasing doses of 10058-F4, a potent peptide inhibitor of MYC.31-33 The results of this experiment showed that 10058-F4 increased MST1 mRNA expression in a dose-dependent manner while decreasing the levels of MYC and EZH2 mRNA with a similar fashion (Fig. 4A; Fig. S5A).
Figure 4. Attenuation of MST1 expression by MYC through EZH2 and miR-26a/b. (A and B) Analysis of MST1, EZH2 and MYC mRNA levels by RT-qPCR in C4–2 cells after treatment with increasing doses (0, 20, 40, 60 µM) of 10058-F4 for 72h and DMSO (vehicle) or JQ1 (500 nM) for 48h in serum-fed conditions, respectively. (C) Semi-qPCR analysis of miR-26a and miR-26b mRNA levels in C4–2 cells treated either with DMSO (control), 60 µM 10058-F4 or 500 nM JQ1 for 48h in serum-fed conditions. U6 was used as an internal control. (D) Analysis of MST1 and EZH2 mRNA levels by RT-qPCR in C4–2 cells transfected with miR-26a and miR-26b mimics for 48h in serum-fed conditions. Treated cells arbitrary values were normalized to that of the untreated DMSO or Mock control. (E) Analysis of MST1 mRNA levels by RT-qPCR in p69 cells transiently transfected with mock (vector) or MYC expression construct and then treated with DMSO (vehicle) or 5 µM DZNep. (F and G) QPCR analysis of MYC mRNA levels in C4–2 cells treated with DMSO or 5 µM DZNep (G) and with DMSO or 2 µM GSK126 for 72h. Data are (± S.E.) from multiple experiments.
To verify the negative effect of MYC on MST1 mRNA expression, we used another pharmacologic inhibitor of MYC, JQ1, which is a selective and potent small-molecule bromodomain inhibitor. JQ1 was demonstrated to downregulate MYC transcriptional functions.34,35 We showed that treatment of C4–2 cells with JQ1 (500 nM) attenuated MYC and EZH2 expression that was accompanied by an increased MST1 mRNA expression, as assed by RT-qPCR (Fig. 4B). MYC was demonstrated to promote EZH2 overexpression by suppressing the expression of miR-26a and miR-26b in several prostate cancer cell lines23 and other cancer cells.36,37 To demonstrate the role of miR-26a/b in MYC-regulated EZH2 overexpression in C4-2 cells, we assessed the levels of miR-26a and miR-26b mRNA levels in 10058-F4 and JQ1 treated C4-2 cells by semi-qPCR. Although 10058-F4 upregulated only miR-26b expression, JQ1 increased both miR-26a and miR-26b mRNA levels (Fig. 4C; Fig. S5B). To further verify the link between MYC, EZH2 and MST1, we transiently overexpressed miR-26a or miR-26b mimetic in C4-2 cells and showed that overexpression of both miR-26a and miR-26b significantly reduced EZH2 mRNA expression that was correlated with the upregulation of MST1 mRNA expression (Fig. 4D). These findings establish that MYC regulates EZH2 overexpression through miR-26a/b to attenuate MST1.
To further verify the specificity of the above findings, we transiently transfected p69 normal prostate cells with control vector (mock) or MYC expression construct, followed by treatment with DMSO or DZNep. The p69 cell line was used in this experiment because it expresses high levels of MST1 and low levels of both MYC and EZH2 protein in comparison to LNCaP cells. Ectopic expression of MYC in p69 cells resulted in the overexpression of EZH2 mRNA (Fig. S5C and D, respectively) that was accompanied by the downregulation of MST1 mRNA expression (Fig. 4E). Consistently, administration of DZNep reversed MYC-induced MST1 attenuation (Fig. 4E), indicating that MYC signals through EZH2 to silence MST1.
A recent study suggests that EZH2 is able to promote MYC expression by attenuating miR-494 expression, which was shown to inhibit MYC expression in aggressive B-cell lymphomas.26 Our data in Figure 1 showed that MYC and EZH2 were concurrently upregulated in PC cells and that the upregulation of MYC promoted EZH2 expression (see Fig. 4A; Fig. S5C and D). Thus, we wanted to assess whether EZH2 promotes MYC overexpression in C4–2 cells after treatment with DMSO, DZNep or GSK126 potent inhibitor of EZH2. The results showed that inhibition of EZH2 by pharmacologic inhibitors significantly decreased MYC mRNA expression in comparison with DMSO control, as assessed by RT-qPCR (Fig. 4F and G).
EZH2-mediated DNA methylation and histone modification suppress MST1 promoter activation
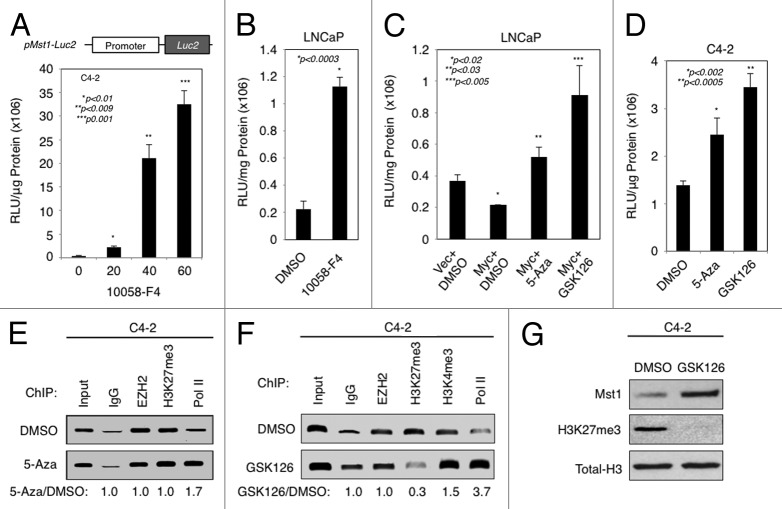
To determine whether MYC and EZH2 suppress MST1 promoter activity to attenuate its expression, we generated a MST1 promoter luciferase reporter construct and assessed its activity in C4-2 cells after treatment with increasing doses of 10058-F4. Figure 5A demonstrates that addition of 10058-F4 increased MST1 promoter activity in a dose-dependent fashion. Similarly, the 10058-F4 caused an increase in MST1 promoter activation in LNCaP cells compared with control DMSO (Fig. 5B). Effects of MYC on MST1 promoter were specific because the ectopic expression of MYC in LNCaP cells suppressed the activation of MST1 promoter, and addition of 5-Aza or GSK126 abolished that (Fig. 5C). Likewise, treatment of C4-2 cells with DMSO, 5-Aza, or GSK126 resulted in the activation of MST1 promoter 2 to 2.5-fold, respectively, compared with control DMSO (Fig. 5D). Correspondingly, inhibition of EZH2 attenuated MST1 promoter DNA methylation 50% with GSK126 (not shown) and 70% with DZNep compared with control DMSO (Fig. S5E).
Figure 5. EZH2-mediated chromatin remodeling and DNA methylation attenuates MST1 promoter activation and expression. (A) MST1 promoter luciferase reporter activity in C4–2 cells after transient transfection with pMST1-Luc2 reporter or mock vector, followed by treatment with increasing doses (0, 20, 40, 60 µM) of 10058-F4 for 48h. Relative luciferase units (RLUs) were normalized to total protein and then normalized to the RLU of vector control. (B) Activity of MST1 promoter luciferase reporter in LNCaP cells transiently transfected with pMST1-Luc2 reporter or mock vector construct and then treated with DMSO (vehicle) or 60 µM 10058-F4 in serum-fed conditions. Luciferase assay was performed at 48h post treatment. (C) MST1 promoter luciferase reporter activity in LNCaP cells transiently co-transfected with pMST1-Luc2 reporter with mock (vector) or MYC expression construct and then treated with DMSO (vehicle), 5 µM 5-Aza, or 2 µM GSK126. Luciferase assay was performed at 48h post-transfection. (D) MST1 promoter luciferase reporter activity in C4–2 cells that were transiently transfected with pMST1-Luc2 reporter or mock vector construct and then treated with DMSO (vehicle), 5 µM 5-Aza, or 2 µM GSK126. Luciferase assay was performed at 48h post treatment. (E and F) Chromatin immunoprecipitation (ChIP) analysis of the EZH2, H3K27me3, H3K4me3, RNAP II and IgG (control) enrichment on MST1 promoter. C4-2 cells were treated with DMSO (vehicle), 5 µM 5-Aza or 2 µM GSK126 in serum-fed conditions at 48h. Bound DNA was analyzed by PCR. Band intensities quantified by ImageJ are presented as fold change relative to DMSO. (G) Levels of total MST1, H3K27me3 or H3 protein in C4-2 cells that were treated with DMSO or 2 µM GSK126. Western blot was performed using antibodies to corresponding proteins at 48h post-treatment. Data (± S.E.) are representative of three independent experiments.
Trimethylation of histone 3 at Lys27 (H3K27me3) has been suggested to facilitate DNA methylation, thereby leading to gene silencing.38 We employed chromatin immunoprecipitation (ChIP) assays to determine the status of a gene silencing H3K27me3 and a gene activating H3K4me3 marks as well as the presence of EZH2 and RNA Polymerase II (RNAP II) on MST1 promoter at the CpG region. In comparison to control DMSO, treatment of cells with 5-Aza augmented the RNAP II occupancy (lane 5) without significantly affecting the enrichment of EZH2 (lane 3) and H3K27me3 (lane 4) on the MST1 promoter CpG region (Fig. 5E). However, Figure 5F demonstrates that treatment of C4–2 cells with GSK126 decreased the H3K27me3 enrichment (lane 4) without affecting the EZH2 occupancy (lane 3), and a decrease in H3K27me3 enrichment was associated with the enrichment of H3K4me3 mark (lane 5) and the RNAP II occupancy (lane 6). Accordingly, western blot showed that the GSK126 caused a decrease in H3K27me3 protein signal that was accompanied by an increase in MST1 protein expression. This observation was specific because the treatment did not affect the expression of total H3 protein levels (Fig. 5G). Altogether, these findings indicate that MYC-induced EZH2 signaling mediates H3K27me3 modification and DNA methylation, thereby lead to MST1 silencing.
MST1 silencing attenuates MYC or EZH2 inhibitor-induced cell growth retardation
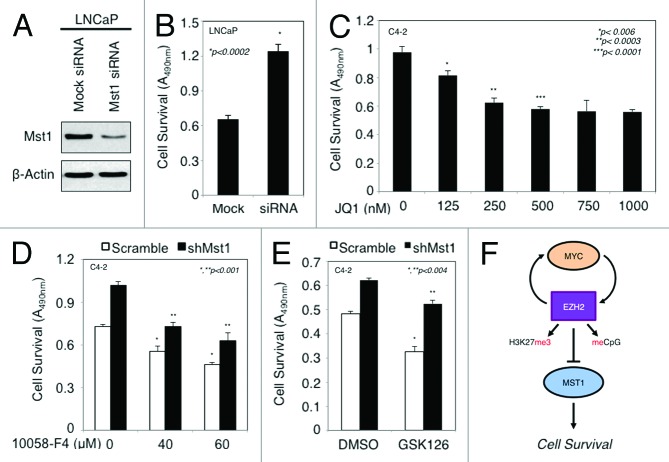
First, we examined the significance of MST1 loss in the contribution of androgen-independent PC cell growth in vitro, given that castration-resistant C4–2 cells expresses about 50% less MST1 than the parental LNCaP cells. As shown in Figure 6A and B, the knockdown of MST1 by small interfering RNA (siRNA) significantly increased an androgen-independent LNCaP cell growth compared with mock siRNA control in vitro (P < 0.0002). These findings, in line with our previous observations,10,11 suggest that the loss of MST1 expression is biologically relevant and may be associated with lethal disease progression.
Figure 6. Suppression of MST1 expression causes androgen independence and resistance to MYC or EZH2 inhibitor-induced growth retardation. (A and B) Effects of MST1 knockdown on an androgen-independent LNCaP cell growth in vitro. LNCaP cells were transiently transfected with the pool of mock siRNA or MST1 siRNA. WB (A) and MTS assays (B) were performed at 48 h post-transfection in serum-starved growth conditions. (C) Analysis of cell growth by MTS assay in C4–2 cells treated with doses (0, 125, 250, 500, 750 or 1000 nM) of JQ1 in serum-fed conditions for 48h. (D and E) Analysis of C4–2 cell growth by MTS. C4–2 cells were transiently transfected with scramble (Src) shRNA and the MST1 shRNA, followed by treatment with doses (0, 40, or 60 µM) of 10058-F4 in serum-fed conditions for 48h (C) or with DMSO (vehicle) or 2 µM GSK126 for 72h (D). Data (± S.E.) are of multiple repeats. (F) A proposed model summarizing the findings.
We previously reported that the gain of MST1 function sensitized C4-2 cells to a known experimental cancer drug.11 To determine whether MYC or EZH2 functionally intersect with MST1 in PC cells, we performed a cell viability assay under varying conditions. First, we showed that administration of JQ1, which caused upregulation of MST1 expression (see Fig. 5B), suppressed C4–2 cell viability in vitro in a dose-dependent manner (Fig. 6C). Second, we showed that administration of 10058-F4 suppressed C4-2 cell viability in vitro in a dose-dependent manner, and knockdown of MST1 by shRNA reversed the 10058-F4-induced growth retardation relative to control shRNA (Fig. 6D). Third, GSK126 treatment significantly reduced C4-2 cell viability in vitro, and likewise, silencing of MST1 reversed the GSK126 cytotoxic effects (P < 0.01) (Fig. 6E). These findings indicate that both MYC and EZH2 functionally intersect with growth regulatory activity of MST1.
Discussion
This study, to our knowledge, is the first to demonstrate that MYC and EZH2 cooperate to attenuate MST1 expression in PC cells through histone modification and DNA methylation. The model in Figure 6F summarizes the findings. We provided convincing evidence to support this model. We demonstrated that overexpression of both EZH2 and MYC was correlated with the reduction of MST1 expression. We showed that DNA methylation caused the reduction of MST1 expression. We also showed that overexpression of MYC and EZH2 suppressed MST1 promoter activity, thereby attenuated its expression. Our data revealed that MYC and EZH2 reciprocally regulate each other’s expression and that EZH2 functioned as a key mediator of MST1 attenuation by MYC. We also demonstrated that MYC promoted EZH2 expression by partly inhibiting miR-26a/b, leading MST1 silencing. Using multiple approaches, we demonstrated that DNA methylation and H3K27me3 modification that was mediated by EZH2 contributed to MST1 silencing. In addition, our data showed that knockdown of MST1 by RNAi caused resistance to androgen deprivation and EZH2 or MYC inhibitor-induced cell growth retardation. Thus, this study discloses a new mechanism of MST1 regulation that involves a cooperative MYC and EZH2 signaling. Our data suggest that cross talk between MST1, MYC, and EZH2 may have important implications in PC cell survival and perhaps lethal disease progression.
Our study showed that MYC overexpression increased EZH2 expression and that EZH2 induction could also induced MYC overexpression in PC cells. Our observations, which are consistent with the published studies,23,24,26 indicate that there exist a positive feedback mechanism between MYC and EZH2 in regulating each other’s expression. EZH2 has recently been demonstrated to function as a prominent transcriptional co-activator for androgen receptor (AR) and a mediator of castration-resistant PC cell growth in vitro and tumor xenografts in mice.19 It is well established that AR is a potent oncogene for PC initiation and metastatic castration-resistant progression. In addition, a study let by Gao et al.39 showed that overexpression of MYC reversed the anti-growth effects of AR suppression.39 We identified MST1 as a potent negative regulator of AR signaling and suppressor of PC cell growth.10,11 Besides, MST1 has recently been shown to be an important mediator of the AR transcriptional repression by the SAFB1 transcriptional co-repressor.40 Here, we demonstrated that castration-resistant C4-2 PC cells and its metastatic C4-2B (not shown) subline expressed about 50% less MST1 than the parental LNCaP, and that silencing of MST1 significantly increased androgen-independent LNCaP cell growth in vitro. Our data showed that MYC attenuated MST1 expression via signaling EZH2. These observations suggest that deregulation of MST1/Hippo may play a prominent role in the emergence of castration-resistant lethal disease progression.
MYC is identified as an important regulator of stem cell biology and it was demonstrated to control the equilibrium between self-renewal and differentiation of embryonic stem cells (ESCs) and hematopoietic stem cells. Also, MYC was shown to induce and maintain PC stem cells (PCSCs).41,42 EZH2 is known to prevent the differentiation of ESCs,43 though its role in PCSCs is yet to be determined. MST1 was demonstrated to induce ESC differentiation.44 PCSCs are shown to be resistant to castration and conventional chemotherapy.42 Although the underlying mechanism of how MYC modulates PCSC expansion remains elusive, it is possible that the loss of MST1 expression by MYC has a significant role in PCSC propagations and functions. Currently, we are actively investigating these possibilities, but it is not the scope of this study.
It appears that MYC regulates the target gene expression by multiple mechanisms that include the recruitment of DNMT3A to the target gene promoter,25 binding to transcription factors, which can transform an activator complex into an inhibitor complex,45 interacting with HDAC3 and SUZ12, a member of PRC2 complex, and regulating microRNAs.26 As illustrated in Figure 6F, our data showed that MYC and EZH2 positively regulates each other’s expression, and the upregulation of EZH2 promotes H3K27me3 mark and DNA methylation at the MST1 promoter CpG sites that causes loss or reduction of MST1 expression, leading to an enhanced cell survival. Nevertheless, our data suggest that MYC could inhibit MST1 expression independently from or in parallel with EZH2, given the fact that inhibition of EZH2 partially attenuated the MST1 promoter DNA methylation (Fig. S5E), and yet, inhibition of MYC abolished MST1 promoter DNA methylation (not shown). Therefore, a future investigation aimed at detailing the direct transcriptional regulation of MST1 by MYC is warranted.
In summary, the current study describes a new mechanism of deregulation of MST1 functions in PC cells in addition to our previously described mechanisms that is mediated by PI3K/AKT and mTOR signaling.10,12 The mechanism that we have described here involves MYC and EZH2 signaling that epigenetically silence MST1 expression. The results of this study suggest that the MYC-EZH2-MST1 signaling axis may have important implications for PC cell survival and is a promising cancer drug target for therapeutic interventions.
Materials and Methods
Cell Cultures, Plasmids, and Chemicals
Human normal p69 prostate epithelial cells and LNCaP and its subline C4-2 and C4-2B PC cell models were grown in RPMI 1640 cell culture medium supplemented with 10% FBS and 1% Pens/Strep.10,11 EZH2 and scramble shRNA lentiviral plasmids were kindly provided by Dr. Jeong-Ho Kim (The George Washington University Medical Center). MST1 and scramble shRNA in pGFP-V-RS retroviral plasmids were obtained from Origene. To construct the MST1 promoter luciferase reporter plasmid, the MST1 promoter region (-951 to +2 base pair (bp) relative to transcriptional start site (+1) was amplified by PCR from a human BAC clone carrying 20q13.12 chromosome loci (RP11–169A6, BACPAC Resources) using MST1 promoter specific cloning primers listed in Table S1. The PCR product was cloned into the KpnI and HindIII sites in the pGL4.20 [luc2/Puro] vector (Promega). The final product was designated as pMst1-Luc2. 5-Aza-2’-deoxycytidine (5-Aza) (A3656, Sigma-Aldrich), 3-Deazaneplanocin A (DZNep) (13828, Cayman, Ann Arbor, MI), dimethylsulfoxide (DMSO) and 10058-F4 (F3680, Sigma-Aldrich, St. Louis, MO) and GSK126 (1346574–57–9, ChemieTek) were obtained.
RNA and Protein Analysis
Total RNAs were isolated using RNeasy mini kit (Qiagen, Inc.) and microRNAs were isolated using mirVANA miRNA isolation kit (Life Technologies), respectively. Reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) was performed with Fast SYBR Green Master Mix (Invitrogen) using Applied Biosystems 7500 RT-PCR system (Life Technologies/Applied Biosystems) to analyze RNA expression. All primers used in expression analysis were listed in Table S1. Protein Analysis with western blotting was performed as described previously.10,12 Antibodies used in this study were listed in Table S2.
DNA methylation assay
The EZ DNA Methylation-Direct kit (Zymo Research) was used to analyze the methylation status of MST1 promoter in PC cell lines and frozen or formalin-fixed and paraffin-embedded (FFPE) human prostate tumor samples using methylation and unmethylation MST1 promoter specific primers as described.27
Cell transfection, cell viability, and luciferase reporter assays
Plasmids were introduced into cells using Lipofectamine 2000 transfection reagent (Invitrogen) as described.11 Small interfering RNA (siRNA) for MST1 or scrambled control siRNA, miRIDIAN Mimic hsa-mir-26a, miRIDIAN Mimic hsa-mir-26b and miRIDIAN microRNA Mimic Negative Control #1 obtained from Thermo Scientific (Dharmacon RNAi Technologies) was transfected using DharmaFECT-2 transfection reagent (Thermo Scientific) at a final concentration of 50nM. Cell viability assay was conducted using a CellTiter 96 AQueous system (Promega) as described.12 MST1 promoter luciferase reporter assay was performed using the Luciferase Assay System (Promega) and a BMG Labtech microplate reader as described.12
Immunofluorescence and Microscopy
For co-staining EZH2 and MST1, C4-2 cells were treated with DMSO (vehicle) or 5 µM DZNep for 72h. Cells and formalin-fixed and paraffin-embedded prostate tissues were incubated with antibodies listed in Table S2. Images were taken by fluorescence microscopy (Nikon Eclipse Ti model; USA) at 20× magnification. All human clinical samples were analyzed according to a protocol approved by Internal Review Board at Cedars-Sinai Medical Center.
Chromatin immunoprecipitation (ChIP)
Briefly, C4-2 cells grown in 10% FBS were treated with DMSO, 5 µM 5-Aza, or 2 µM GSK126 for 48h. The crosslinked DNA was precipitated by anti-EZH2, -H3K27me3, -H3K4me3 (07-473, Millipore), or –RNAP II (05-623, Millipore) antibody. Bound and eluded DNA was analyzed by semi-qPCR using primer pair specific to MST1 promoter surrounding CpG sites from -70 to +50 relative to TSS listed in Table S1.
Statistical analysis
Values are expressed as mean ± SD. An unpaired t test was conducted to analyze for differences between control and test. Statistical significance was set at P ≤ 0.05.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Drs Peter Jones, Susan Kasper, and Leland Chung for critically evaluating the data and carefully reading the manuscript, Gina Chia-Yi Chu for technical assistance, and Gary Mawyer for editing the manuscript. This study was partly supported by grants from the National Center for Advancing Translational Sciences (UL1TR000124), the New York Academy of Medicine, The Jesse and Donna Garber Foundation, and Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center.
Glossary
Abbreviations:
- MST1
mammalian serine/threonine kinase 1
- H3K27me3
trimethylation of histone 3 at lysine 27
- H3K4me3
trimethylation of histone 3 at lysine 4
- DNMTs
DNA methyltransferases
- EZH2
enhancer of zeste homolog 2
- PRC2
polycomb repressive complex 2
- ChIP
chromatin immunoprecipitation
- MSP
methylation sensitive polymerase chain reaction
- 5-Aza
5′-Aza-2’-deoxycytidine
- DZNep
3-deazaneplanocin A
- RLU
relative luminescence unit
References
- 1.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–91. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 2.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107:1437–42. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A. 2010;107:1431–6. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–38. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, Dawson D, Willis JE, Markowitz SD, Camargo FD, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A. 2011;108:E1312–20. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiao M, Wang Y, Xu X, Lu J, Dong Y, Tao W, Stein J, Stein GS, Iglehart JD, Shi Q, et al. Mst1 is an interacting protein that mediates PHLPPs’ induced apoptosis. Mol Cell. 2010;38:512–23. doi: 10.1016/j.molcel.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 7.Steinmann K, Sandner A, Schagdarsurengin U, Dammann RH. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep. 2009;22:1519–26. doi: 10.3892/or_00000596. [DOI] [PubMed] [Google Scholar]
- 8.Minoo P, Zlobec I, Baker K, Tornillo L, Terracciano L, Jass JR, Lugli A. Prognostic significance of mammalian sterile20-like kinase 1 in colorectal cancer. Mod Pathol. 2007;20:331–8. doi: 10.1038/modpathol.3800740. [DOI] [PubMed] [Google Scholar]
- 9.Lin X, Cai F, Li X, Kong X, Xu C, Zuo X, Yang Q. Prognostic significance of mammalian sterile 20-like kinase 1 in breast cancer. Tumour Biol. 2013;34:3239–43. doi: 10.1007/s13277-013-0895-8. [DOI] [PubMed] [Google Scholar]
- 10.Cinar B, Fang PK, Lutchman M, Di Vizio D, Adam RM, Pavlova N, Rubin MA, Yelick PC, Freeman MR. The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. EMBO J. 2007;26:4523–34. doi: 10.1038/sj.emboj.7601872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cinar B, Collak FK, Lopez D, Akgul S, Mukhopadhyay NK, Kilicarslan M, Gioeli DG, Freeman MR. MST1 is a multifunctional caspase-independent inhibitor of androgenic signaling. Cancer Res. 2011;71:4303–13. doi: 10.1158/0008-5472.CAN-10-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collak FK, Yagiz K, Luthringer DJ, Erkaya B, Cinar B. Threonine-120 phosphorylation regulated by phosphoinositide-3-kinase/Akt and mammalian target of rapamycin pathway signaling limits the antitumor activity of mammalian sterile 20-like kinase 1. J Biol Chem. 2012;287:23698–709. doi: 10.1074/jbc.M112.358713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nupponen NN, Kakkola L, Koivisto P, Visakorpi T. Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol. 1998;153:141–8. doi: 10.1016/S0002-9440(10)65554-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visakorpi T, Kallioniemi AH, Syvänen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–7. [PubMed] [Google Scholar]
- 15.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and Prostate Cancer. Genes Cancer. 2010;1:617–28. doi: 10.1177/1947601910379132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38. doi: 10.1016/S1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 17.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 18.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 19.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 21.Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog. 2002;33:163–71. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 22.De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–67. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669–83. doi: 10.18632/oncotarget.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaur M, Cole MD. MYC acts via the PTEN tumor suppressor to elicit autoregulation and genome-wide gene repression by activation of the Ezh2 methyltransferase. Cancer Res. 2013;73:695–705. doi: 10.1158/0008-5472.CAN-12-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenner C, Deplus R, Didelot C, Loriot A, Viré E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–46. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Zhao X, Fiskus W, Lin J, Lwin T, Rao R, Zhang Y, Chan JC, Fu K, Marquez VE, et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell. 2012;22:506–23. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Seidel C, Schagdarsurengin U, Blümke K, Würl P, Pfeifer GP, Hauptmann S, Taubert H, Dammann R. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46:865–71. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- 28.Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 31.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 32.Huang MJ, Cheng YC, Liu CR, Lin S, Liu HE. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp Hematol. 2006;34:1480–9. doi: 10.1016/j.exphem.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 33.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–9. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 34.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Möller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–12. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 37.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, Chen YC, Peng Y, Yao KT, Kung HF, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–33. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 38.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 39.Gao L, Schwartzman J, Gibbs A, Lisac R, Kleinschmidt R, Wilmot B, Bottomly D, Coleman I, Nelson P, McWeeney S, et al. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS One. 2013;8:e63563. doi: 10.1371/journal.pone.0063563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhopadhyay NK, Kim J, You S, Morello M, Hager MH, Huang WC, et al. Scaffold attachment factor B1 regulates the androgen receptor in concert with the growth inhibitory kinase MST1 and the methyltransferase EZH2. Oncogene. 2013 doi: 10.1038/onc.2013.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Civenni G, Malek A, Albino D, Garcia-Escudero R, Napoli S, Di Marco S, Pinton S, Sarti M, Carbone GM, Catapano CV. RNAi-mediated silencing of Myc transcription inhibits stem-like cell maintenance and tumorigenicity in prostate cancer. Cancer Res. 2013;73:6816–27. doi: 10.1158/0008-5472.CAN-13-0615. [DOI] [PubMed] [Google Scholar]
- 43.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–18. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gartel AL, Shchors K. Mechanisms of c-myc-mediated transcriptional repression of growth arrest genes. Exp Cell Res. 2003;283:17–21. doi: 10.1016/S0014-4827(02)00020-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.