Abstract
Inducible systems for gene expression emerge as a new class of artificial vectors offering temporal and spatial exogenous control of gene expression. However, most inducible systems are less efficient in vivo and lack the target-organ specificity. In the present study, we have developed and optimized an oligonucleotide-based inducible system for the in vivo control of transgenes in the liver. We generated a set of simple, inducible plasmid-vectors based on the addition of four units of liver-specific miR-122 target sites to the 3′untranslated region of the gene of interest. Once the vector was delivered into hepatocytes this modification induced a dramatic reduction of gene expression that could be restored by the infusion of an antagomir for miR-122. The efficiency of the system was tested in vivo, and displayed low background and strong increase in gene expression upon induction. Moreover, gene expression was repeatedly induced even several months after the first induction showing no toxic effect in vivo. By combining tissue-specific control elements with antagomir treatment we generated, optimized and validated a robust inducible system that could be used successfully for in vivo experimental models requiring tight and cyclic control of gene expression.
Keywords: cell type specific, liver expression, long-term gene expression, reporter system
Introduction
Inducible systems for gene expression are a new class of artificial vectors that offer the advantage of temporal and spatial exogenous control of gene expression. The ability to exogenously control gene expression, in vivo, is highly important in biomedical research.1,2,3,4 There are at least three areas where gene inducible systems are crucial. Firstly, gene regulation technologies contribute in the efforts to understand the role of specific gene products in fundamental biological processes in both normal development and disease states.1 Secondly, such technologies are important in bioprocess engineering where they offer the advantage of fine-tuning expression of protein or RNA pharmaceuticals, including conditional control for best production performance.5 Thirdly, gene inducible systems have the potential to become in the future, a new form of treatment.6,7,8
There are several inducible systems for gene expression largely in use today, such as the Cre-lox system.9 This system allows very tight gene expression control, and is used when only a single transition from on-off or off-on state is desired. Once the inducer, the Cre-recombinase is present in the cell, it will alter irreversibly the DNA sequence that contains the lox P sites, and thus, permanently changes gene expression.
Another highly used inducible system is based on the tetracycline-responsive element.2,10 Compared to Cre-lox, the tetracycline-inducible system can be designed in such a way that it can be reversibly turned on or off when desired. However, some of the disadvantages of the tet-system are its large size and the gene expression leakiness in vivo. Thus, even when gene expression is supposed to be in the off state, there is still a considerable level of protein production. Hence, although the existing systems can already be used in several applications, a versatile, inducible system that has tissue or organ specificity, low or no leakiness, and can be used repeatedly, is required.
MicroRNAs (miRs) are small intracellular molecules, on average 22 nucleotides long, that play an important role in endogenous gene regulation.11-13 They act mainly as posttranscriptional regulators by silencing genes.11,12,14 It has been shown that every tissue has a miR signature.15,16,17 Depending on the intracellular miR abundance and the level of complementarity between the miRs and their target sites, the gene silencing can vary up to a few hundred-fold. Intelligent vectors have been designed so that endogenous miRs suppress transgene expression in hematopoietic lineages thereby reducing immunosuppression and enabling stable gene transfer.18,19,20
Such systems, containing one ore more complementary sites to endogenous miRs in their 3′-UTRs18,21 were successfully employed for gene silencing given that the levels of endogenous miRs are high. For instance, miR-122 is abundant in hepatocytes (between 50 and 70,000 units/cell) and in malignant transformed hepatoma cell lines, such as Huh7 and controls hundreds of genes via translational suppression.
In this work, we have designed and tested a new inducible genetic system based on the presence of miR-122 target sites in the 3′-untranslated region (3′-UTR) of different vectors. These vectors are silenced in the liver by endogenous miR-122 and activated once the endogenous miRs are blocked by the use of an antagomir.21,22,23
Results
AmiR122 selectively upregulates transcripts with miR-122 target sites
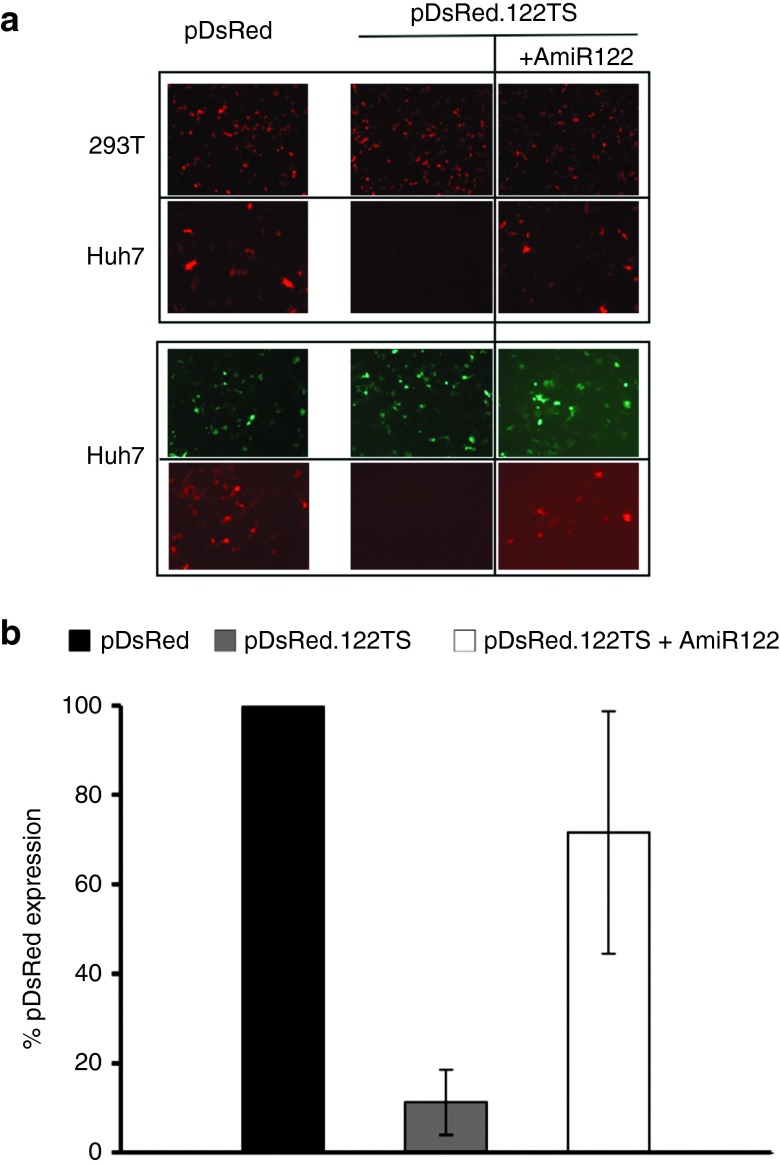
We first designed a DsRed expressing vector containing four complementary target sites for miR-122 (Table 1). Next, we tested the specific regulatory effect of this miR by comparing the DsRed expression in cells expressing, or lacking, miR-122. Downregulation was only present in Huh7 cells when using the plasmid containing miR-122 target sites (Figure 1a,b). Moreover, when adding the AmiR122, we could restore the expression of the gene containing the miR-122 target sites. The expression was unimpaired by the presence of a control antogomir, AmiR142 (Supplementary Figure S1a), and in the control 293T cells lacking miR-122 (Figure 1a).
Table 1. Plasmid characteristics and antagomir sequences.
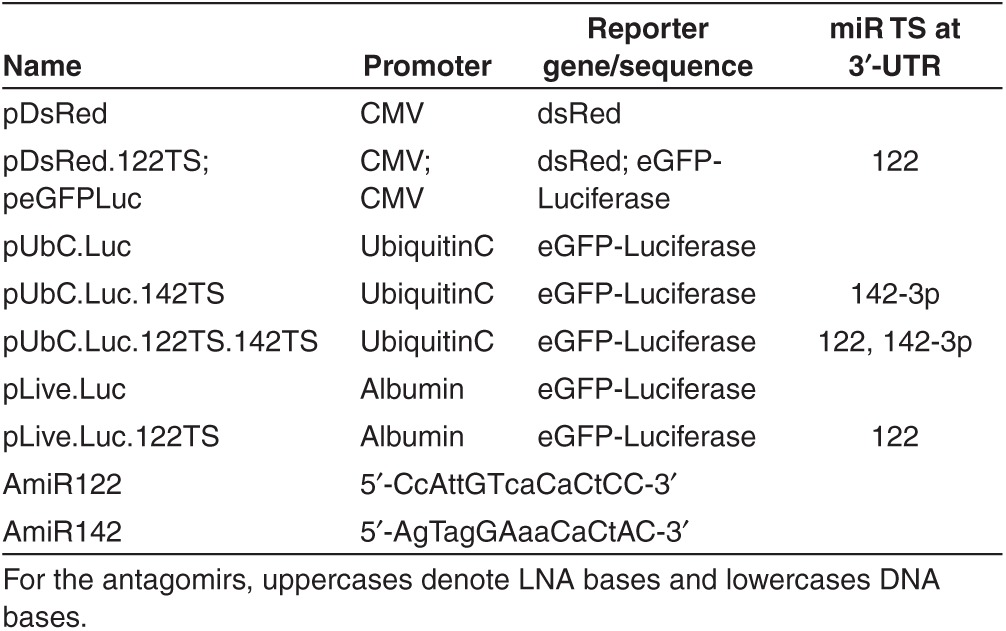
Figure 1.
Effects of miR-122 and AmiR122 on plasmid expression in transfected cells. (a) Representative microscopic images. Upper panel: 293T cells (first row) and Huh7 cells (second row) were transfected with plasmids with, or without, miR-122 target sites (pDsRed.122TS and pDsRed, respectively). In 293T cells the expression profile is unaffected by the presence of miR target sequences because endogenous miR-122 is lacking, while in Huh7 cells the protein expression encoded by plasmids containing miR122 TS (pDsRed.122TS) is highly downregulated (middle). The protein expression of pDsRed.122TS can be restored to levels similar to pDsRed by using AmiR122 (right). Lower panel: Cotransfection of peGFPLuc with either pDsRed (left) or pDsRed.122TS (right) plasmids in Huh7 cells. First row shows the peGFPLuc expression while the second row shows the pDs.Red and pDs.Red.122TS expression. (b) Quantification of the miR-122 and AmiR122 effect on pDsRed plasmids expression in Huh7 cells. The DsRed expressing cells were counted from three different photomicrographs. Error bars represent standard deviation of triplicates.
AmiR122 regulates gene expression dose-dependently in vivo
The next step was to generate a vector that could easily be used in vivo. Viral promoters, such as the CMV promoter show a high initial expression but short-lasting gene expression when used in vivo. However, endogenous promoters, such as, ubiquitin C promoter induce a high and long-lasting level of gene-expression in the liver.24 Consequently, we cloned the ubiquitin C promoter into the peGFPLuc vector25 by replacing the CMV promoter. It has been reported that adding four target sites of miR-142-3p at the 3′-UTR region of an exogenous gene in a lentiviral vector contributed to the in vivo-maintained expression of an exogenous gene18 by impairing its expression in hematopoietic cells, thereby preventing an immune response. Thus, apart from the four target sites for miR-122, we added four target sites of miR-142-3p at the 3'UTR region of the new vector, creating the pUbC.Luc.122TS.142TS and pUbC.Luc.142TS plasmids (Table 1). When testing this vector in the myeloid U937 cell line which expresses elevated levels of miR-142, there was more than an 80% decrease in gene expression from the vector containing the miR-142-3p target sites (Supplementary Figure S1b).
Making use of the highly efficient delivery method of hydrodynamic injections, we coinjected the vector encoding the eGFPLuc fusion construct carrying the miR-122 and 142-3p target sites (pUbC.Luc.122TS.142TS) with the increasing doses of AmiR122. The pLacZ plasmid (expressing β-galactosidase) was also coinjected to normalize the measured luciferase activity for differences in plasmid delivery.
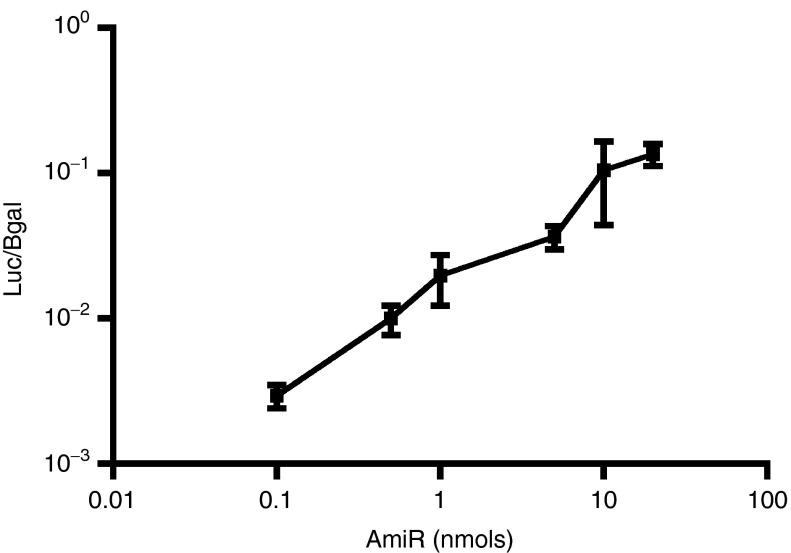
We used different doses of AmiR122 ranging from 0.1 to 20 nmol/injection (Figure 2). Increasing the dose of AmiR blocked endogenous miR-122 and, thus, induced expression from the pUbC.Luc.122TS.142TS plasmid. Interestingly, the use of only 0.1 nmol of AmiR122 induced a 10-fold increase in luciferase expression. For the following studies, 20 nmol of AmiR, the dose that gave the maximum effect, was used.
Figure 2.
Dose-dependent induction of pUbC.Luc.122TS expression by co-injection with AmiR122. NMRI mice were injected hydrodynamically with increasing doses of AmiR122 together with a mix of two plasmids, 5 µg/animal of the luciferase expressing plasmid containing miR122-target sites (pUbC.Luc.122TS) and 1 µg/animal of the control plasmid expressing β-galactosidase (Bgal), lacking miR target sites. Each value represents the average of ratios between luciferase and Bgal measured on liver extract, 1 day after treatment, from three different animals (±SD).
AmiR122 enables repeated induction of gene expression
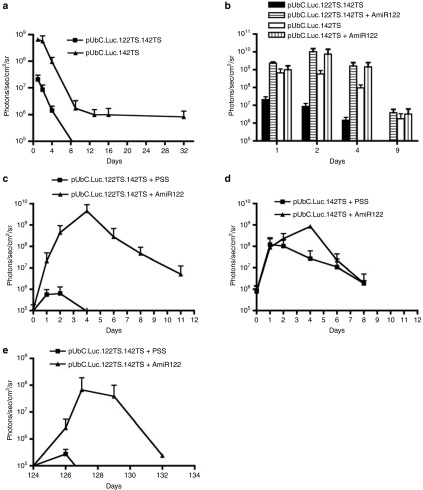
Hydrodynamic-based delivery is the gold standard method for introducing plasmids into the mouse liver.26,27 The expression profile of the transgenes usually follows a two-phase curve: an initial strong downregulation is seen within the first ~10 days followed by a slow and constant decrease afterwards. In order to determine the degree of downregulation that can be mediated by endogenous miR-122, we compared two different vectors with or without miR-122 target sites, using hydrodynamic injection into NMRI mice. The difference in expression was in the range of 40-fold during the first 8 days (Figure 3a). After this interval, the signal from the group injected with the vector containing miR-122 target sites was too weak to be quantified, being close to the background values of the IVIS 100 instrument.
Figure 3.
In vivo luciferase expression profiles from pUbC.Luc plasmids containing or lacking target sites for miR-122, with, or without, injection of AmiR122. NMRI mice were hydrodynamically injected with 5 µg of plasmids with complementary target sites for both miR-122 and miR-142-3p (pUbC.Luc.122TS.142TS) or for mir-142-3p only (pUbC.Luc.142TS), and followed over time. Each value represents the average of the treated group (+SD). Plotted values are above the background signal of the IVIS machine of 1 × 105 photons/sec/cm2/sr. (a) Luciferase expression in mice injected with plasmids containing target sites for miR-122 and/or miR-142-3p and followed over time (n ≥ 4). (b) Luciferase expression in mice co-injected with AmiR122 and followed over several days (n ≥ 3). (c) Effect of AmiR122 administration on plasmid containing miR-122 target sites. A number of eight animals were preinjected with pUbC.Luc.122TS.142TS. After 1 month, four animals/group were hydrodynamically injected with either 20 nmol of AmiR122 in physiological salt solution (PSS) or with PSS only (n = 4). (d) Effect of AmiR122 administration on plasmid without miR-122 target sites. A number of eight animals were preinjected with pUbC.Luc.142TS. After 1 month, four animals/group were hydrodynamically injected with either 20 nmol of AmiR122 in PSS or with PSS only (n ≥ 3). (e) The effect of a second AmiR administration 124 days from the date when the plasmid with miR-122 target sites was hydrodynamically injected, and 90 days after the first dose of AmiR122. A second dose of 20 nmol of antagomir in PSS or PSS only was hydrodynamically delivered (n ≥ 3).
Furthermore, in order to test the rescue of luciferase expression mediated by miR-122, we hydrodynamically coinjected the plasmids with AmiR122. The coadministration restored the luciferase expression from the miR-controlled plasmid (pUbC.Luc.122TS.142TS) to the same levels as what was expressed from the plasmid lacking the miR-122 target sites (pUbC.Luc.142TS) (Figure 3b). The AmiR122-induced upregulation was maximal 1–4 days after delivery, and was present even after 9 days. Interestingly, coadministration of AmiR122 and the control plasmid without the miR122-target sites induced a lower and slightly delayed (detected at days 2–4) increase in luciferase expression (Figure 3b).
An important feature of any inducible systems is the ability to promote gene expression at a later time point. The animals hydrodynamically injected with plasmids were followed up to 32 days (Figure 3a). At this point, each group of mice was divided into two subgroups with four animals per group. One subgroup was hydrodynamically injected with a dose of 20 nmol of AmiR122 (in physiological salt solution) while the control group was hydrodynamically injected with only physiological salt solution. Before the injections, the mice with the plasmid containing miR-122 target sites had no, or very low, expression. After injecting the AmiR122, the luciferase expression dramatically increased until day 4, reaching a level of >104 times higher than seen in control mice treated with physiological salt solution only (Figure 3c; Supplementary Figure S2). The effect slowly decreased over the following days. The saline-treated animals showed a small, 10 times boost in luciferase expression that was detectable only for 4 days.
As a control for possible nonspecific effects due to either the hydrodynamic injection itself, or to indirect effects of the AmiR122 administration, we infused two groups of animals pretreated with the control-plasmid lacking the miR-122 target sites, 32 days after initial plasmid administration. We could identify a clear but transient effect of the hydrodynamic injection itself, namely a boost of gene expression from the preinjected plasmids (Figure 3d), similar to what has been reported before.28 In addition, there was a clear, 1.5 orders of magnitude, increase in Luc expression at day 4, in the group injected with AmiR122, but apart from that, both groups showed similar expression profiles.
Once an inducible vector is present in a tissue, it would be very useful if the expression could be induced at different time points and more than once. Consequently, after inducing the expression 32 days after plasmid inoculation, a second test of inducibility was initiated 3 months later, 124 days after the plasmid injection (Figure 3e). Even if the inducibility was approximately one order of magnitude lower than after 32 days, likely secondary to the general decrease in gene expression and to the hepatocytes turn over,29 an almost 103-fold boost in gene expression was observed from the AmiR122-treated animals as compared to the control group.
Liver-specific promoter, enhancer and untranslated regions suppress “unspecific” hydrodynamics-induced gene expression
Apart from the promoter used, several other regulatory elements are important when designing highly and stable expressing plasmids. Using several regulatory endogenous elements, Wolff's laboratory designed a new generation of plasmids, named pLive that have the ability to prolong the transgene expression, at high levels for more than 1 year.30,31 We transferred the eGFPLuc fusion gene from the UbC-plasmids into the pLive vector, obtaining a plasmid designated pLive.Luc (Table 1). Into this plasmid, we also added 4 miR-122 target sites to the 3′UTR region, obtaining pLive.Luc.122TS vector. Both these plasmids were tested in mice to determine the level of downregulation induced by miR-122. We have not generated any pLive plasmid containg miR-142-3p target sites, since there was no significant difference for the in vivo long-term expression among plasmids that contained, or lacked, miR-142-3p target sites (Supplementary Figure S3). The downregulation of pLive.Luc.122TS was at similar levels (40–100 times) as seen when using the UbC-based plasmids (Figure 4a).
Figure 4.
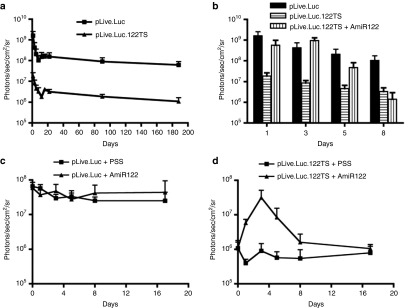
In vivo luciferase expression profiles from pLive.Luc plasmids containing or not target sites for miR-122, with or without injection of AmiR122. Balb/c mice were hydrodynamically injected with 5 µg of plasmids and followed over time. Each value represents the average of the treated group (+SD). Plotted values are above the background signal of the IVIS machine of 1 × 105 photons/sec/cm2/sr. (a) Luciferase expression in mice injected with plasmids that lack (pLive.Luc) or contain target sites for miR-122 (pLive.Luc.122TS) (n ≥ 4). (b) Luciferase expression in mice co-injected with pLive.Luc and AmiR122 in comparison with expression from mice injected with pLive.Luc.122TS or pLive.Luc only. Expression was monitored for 8 days (n = 4). (c) Effect of AmiR122 administration on the expression from plasmids without miR-122 target sites. A number of eight animals were preinjected with pLive.Luc plasmid. After 6 months, four animals/group were hydrodynamically injected with either 20 nmol of AmiR122 in physiological salt solution (PSS) or with PSS only. (d) Effect of AmiR122 administration on the expression from plasmids with miR-122 target sites. A number of eight animals were preinjected with pLive.Luc.122TS plasmid. After 6 months, four animals/group were hydrodynamic injected with either 20 nmols of AmiR122 in PSS or with PSS only.
Next, we tested the capacity of the AmiR122 to induce the expression from pLive.Luc.122TS plasmid. Animals were either coinjected with plasmid and AmiR122, or injected with plasmid alone. As expected, the AmiR122 induced an increased luciferase expression, with levels resembling those found in control animals injected with pLive.Luc, lacking the miR-122 target sites (Figure 4b). Similar to what was found for the UbC plasmid, the effect of AmiR122 lasted for more than 4 days.
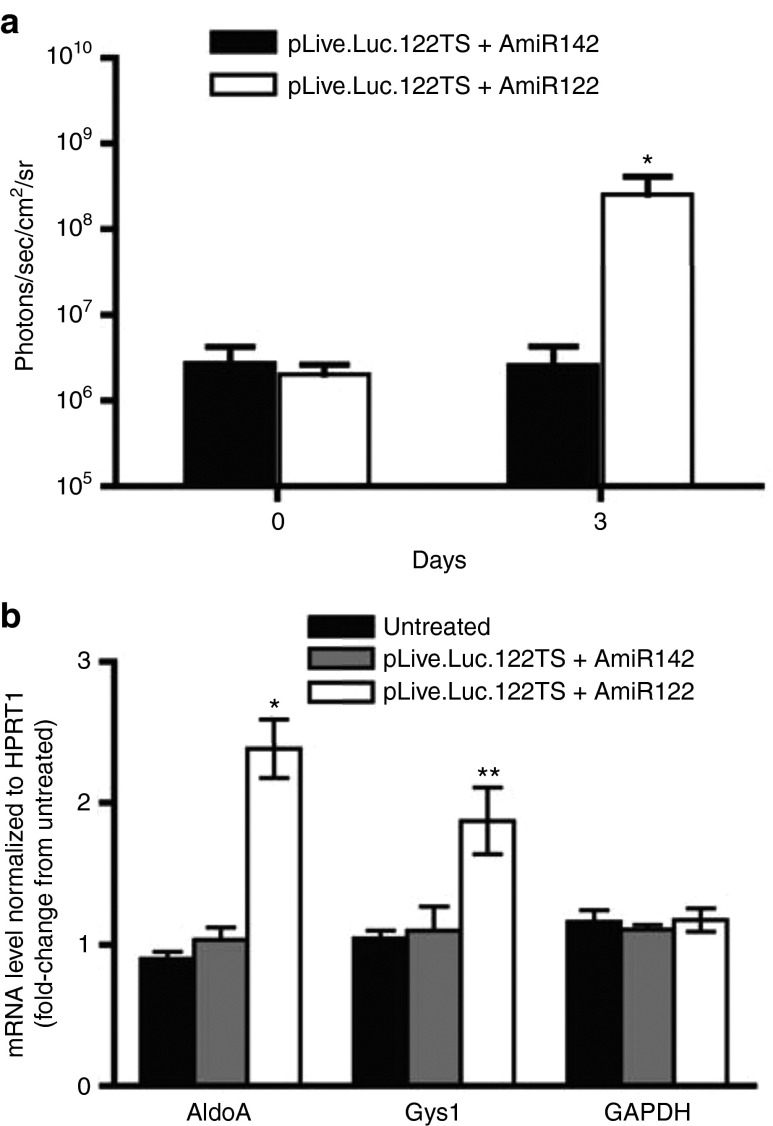
In order to assay if the pLive.Luc.122TS could be induced at a later time point, we then treated the two groups of animals 6 months after plasmid administration using the same strategy as for the pUbC.Luc-based vectors (Figure 3c,d). Importantly, compared to the pUbC.Luc treatment, when using the pLive.Luc vector there was no unspecific inducible effect neither from AmiR122 nor upon saline injection (Figure 4c). When the same treatment was applied on animals preinjected with the pLive.Luc.122TS vector, there was a boost in expression only from the group receiving the antagomir (Figure 4d). The expression from this group was maximal at day 3, reaching the same level as seen in animals receiving the control plasmid, pLive.Luc. Furthermore, to verify the specific effect of AmiR122 in inducing the pLive.Luc.122TS expression, the unrelated AmiR142 was also tested. As seen in (Figure 5a), it was only the AmiR122 that induced expression, thus demonstrating the specificity of the antagomir treatment. This effect was specific due to a decrease in miR-122 as demonstrated by a significant upregulation of miR-122 target proteins such as AldoA and Gys1 (Figure 5b).
Figure 5.
Expression of pLive.Luc.122TS and miR-122 target genes in the liver after AmiR122 or AmiR142 infusion. (a) Luciferase expression in Balb/c mice co-injected with pLive.Luc.122TS and AmiR122 or with pLive.Luc.122TS and AmiR142. Each value represents the average of the treated group (+SD, n = 4). A number of eight animals were preinjected with pLive.Luc.122TS plasmid. After 1 month, four animals/group were hydrodynamic injected with 20 nmol of either AmiR122 or AmiR142 in physiological salt solution. Luciferase expression was monitored for 3 days. (b) The expression of AldoA, Gys1 and GAPDH was assayed on liver extracts at day 3 after antagomir infusions using qRT-PCR. Each value represents the average of the treated group (± SD, n = 3). *P < 0.5, **P < 0.001 AmiR122 versus AmiR142.
To identify possible acute liver toxicity of the antagomir treatment, we measured basic toxicity indicators, such as creatine phosphokinase, aspartate aminotransferase (AST) and alanine aminotransferase (ALT) 3 days after oligonucleotide administration (Supplementary Figure S4). No indication of acute toxicities either in the AmiR122- or AmiR142-treated groups was observed.
Discussion
We describe a new, inducible system for the control of gene expression in hepatocytes both in vitro and in vivo. One of the most appealing features of this system is its simplicity: it is based on the addition of 4 units of miR-122 target sites, an ~120 bp sequence, to the 3′UTR region of the gene of interest. Compared to the most known inducible vectors,2,9,10,32,33,34 this design is by far the simplest. Furthermore, one major advantage over the tetracycline-based inducible vectors is the absence of transactivator.
The efficiency of the system was tested in vivo and displayed up to between 104- and 105-fold increase in gene expression, upon induction. Using the first generation of vectors, this level of inducibility was partially due to the unspecific effect of the hydrodynamic injection per se, similar to what has been reported before.28 This phenomenon was not only due to the blocking of miR-122 acting on the specific target sites in the reporter-mRNA, instead, the lower and somewhat delayed effect seen on the expression from plasmids lacking the specific target sites indicates that this outcome was in part also dependent on other, indirect, effects. Thus, blocking of miR-122 seems also to affect other molecules involved in expression of the ubiquitin-C driven reporter construct. Importantly, using the second generation vectors based on the pLive design,30,31 we could avoid these unwanted effects. Compared to the described inducible vectors, this very high range of inducibility in vivo is only achieved by the Cre-lox system. However, our system in contrast to the Cre-lox system is reversible. Another advantage of the miR-based inducible system is that gene expression can be induced repeatedly, even several months after the first induction. In addition, when testing the toxicity of the AmiR122, our new antagomir-based system shows no apparent toxic effect in vivo.
The inducer of the described inducible system, AmiR122, is a short 15-mer LNA-DNA oligonucleotide, and is already in clinical phase II by Santaris Pharma A/S, under the name Miravirsen (SPC3649) for the treatment of chronic hepatitis C virus infection.21 Even though in this study we have only used hydrodynamic infusion as a delivery method, the AmiR122 can be delivered using intravenous administration (as in the mentioned human trial).
This inducible system was optimized for controlling transgenes expression in the liver. There are several possible applications having the liver as the main target organ. Some examples include hepatitis viral infections and liver tumors that are extremely difficult to treat. Using vectors that are under the control of miR-122 to express therapeutic proteins, one can combine gene therapy with oligonucleotide-based therapy to fine-tune protein expression. In case of hepatathis C virus infection, the inclusion of other transgenes that can protect the hepatocytes or eliminate the virus particles could in future potentially be regarded as an alternative to the current inefficient treatments. Another potential therapeutic strategy could be against cancer. MiR122 is considerably down-regulated in particular types of hepatocarcinomas.35 Using vectors similar to the one described here, but expressing cell-toxic transgenes under the control of a hepatocyte-specific promoter, one could induce apoptosis selectively in the hepatic tumoral cells and not in normal hepatocytes. In this scenario, it might be necessary to fine-tune the system using antagomirs if the tumor cells resisted the toxicity, owing to the remaining low levels of miR122. However, here the balance needs to be carefully monitored to avoid as much as possible toxicity in normal hepatocytes. Another reason why the duration of the antagomir activity should be carefully adjusted is because there is an increase evidence showing modified signaling pathways such as the interferon pathway that are potentially involved in several diseases.36,37,38,39
The specificity of our miR-based inducible system was tested in vitro and also in vivo. The presence of the miR target sites restricts the downregulation to those cells that have high levels of the corresponding endogenous miRs. In order to limit the expression only to selected cell populations, tissue-specific promoters can be used. Thus, to reduce the background expression, it is important to use a tissue-specific promoter that is active only in those cells for which the miR target sites in the vector are designed. In our case, in order to have a selective, inducible system, the preferred vector contains a liver-specific promoter and in combination with miR-122 target sites in the 3′UTR.
In this work, we have used the hydrodynamic technique to deliver plasmids with, or without, the corresponding inducer, AmiR122. Unspecific effects of the hydrodynamic technique consisting of the induction of endogenous and exogenous gene expression, were shown before28 and we have also observed this phenomenon in our studies. This side effect can be avoided if the inducer can be non-hydrodynamically injected21,23 or, as we demonstrate, by the use of tissue-specific control elements, which are insensitive to hydrodynamic changes. To our knowledge, this has not been reported before. Combining the same tissue-specific elements and the minicircle (MC) technology may further improve the outcome, since MC vectors have shown robust long-term expression in hepatocytes.40 The use of chemical-based carriers, such as cell-penetrating peptides for oligonucleotide delivery41 or encapsulated liposomes42 may also obviate the need for hydrodynamic delivery of the AmiR. Furthermore, the technique is not limited to plasmids, since it should also be possible to use viral vectors with hepatic tropism for the delivery.43,44
By combining tissue-specific control elements with antagomir treatment, it was possible to generate a versatile, inducible system, which allows in vivo repeated and tight induction of gene expression in the liver, even after long-time periods.
Materials and methods
Plasmids and oligonucleotides. The plasmid pDsRed, encodes the dsRed fluorescent protein45 (Table 1). pDsRed.122TS in addition to the elements of pDsRed plasmid also contains 4 miR-122 target sites located in the 3′UTR. pLacZ is a β-galactosidase expressing plasmid kindly provided by Pontus Blomberg (Vecura, Karolinska University Hospital Huddinge, Sweden). The plasmid peGFPLuc (Clontech, BD Bioscience, San Jose, CA, USA) expresses the enhanced green fluorescent protein in fusion with firefly Luciferase and modified as described previously.25 This vector contains the human cytomegalovirus (CMV) immediate-early enhancer and promoter; it lacks modifications in the 3′UTR and was used as an internal control by cotransfection into Huh7 cells. In the pUbC.Luc, the CMV promoter and enhancer were replaced with the ubiquitin C (UbC) promoter and enhancer.24 pUbC.Luc.142TS is the same plasmid with the addition of four target sites of miR-142-3p in the 3′UTR, while pUbC.Luc.122TS.142TS contains an additional four target sites for miR-122 in the 3′-UTR. pLive.Luc has the same eGFPLuc-fusion as pUbC.Luc, cloned into the multiple cloning site of the pLive vector (Mirus Bio LLC), and pLive.Luc.122TS contains four target sites of miR-122 in the 3′UTR. The antagomir directed against miR-122 (AmiR122) is a 15mer DNA-LNA mixmer complementary to the endogenous miR-122.21 As a control oligonucleotide, we used a 15mer DNA-LNA mixmer complementary to the endogenous miR-142-3p (AmiR142) (Eurogentec S.A.).
Cell lines and transfections. The Huh7, human hepatoma cell line expressing high levels of miR-122 and the human leukemic monocyte lymphoma cell line, U937, expressing high levels of miR-142-3p were used for in vitro assays. The 293T cell line does not express miR-122 and was used as a control. Transfections of AmiR122 and plasmids in Huh7 and 293T cell lines were done using Lipofectamine 2000, (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. The U937 cells were nucleotransfected using Amaxa's protocol (Lonza, Lonza Cologene AG, Germany). Gene expression was assayed after 48 hours.
Real-time PCR quantification of different mRNAs. cDNA was synthesized using 0.2 µg of Trizol-extracted liver total RNA in 20 µl reaction mixture, containing oligo(dT)12–18 primers and Superscript II reverse transcriptase (GIBCO Invitrogen), according to the manufacturer's instructions. Real-time quantitative RT-PCR Taqman assays were performed using the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). All reactions were done in quadruplicates with the use of a predeveloped gene expression assay mix (Applied Biosystems) containing primers and a probe for the mRNA of interest. Additional reactions for each experiment included predeveloped gene expression assay mix for HPRT1 for normalizing the RNA input. All data were analyzed with 7300 System SDS Software v 1.4.
Animal experiments. Hydrodynamic tail vein injections of plasmids in physiological 0.9% NaCl solution were carried out as previously described.26,27 Briefly, 8% v/w was introduced by tail vein injection over a period of 5 seconds to inbred NMRI or Balb/c adult mice. Mice were anesthetized with 4% isofluran and injected intraperitoneally with 150 mg/kg D-luciferin in 500 µl sterile PBS. Anesthetized animals were imaged for 1 second up to 5 minutes, depending on the intensity of the luminescence signal, using an intensified CCD camera (IVIS Imaging System, Xenogen, Alameda, CA). During imaging, the mice were kept under anesthesia using 2.3% isofluran. Images are comprised of pseudocolored images representing intensity of emitted light (red most intense and blue least intense) superimposed on grayscale reference images for orientation. Data analysis was performed using a Living Image 3.2. software (Caliper LifeSciences, Xenogen). All animal experiments were approved by the local ethical committee in Stockholm, Sweden.
Luciferase-based assays. For most animal experiments, we used an in vivo luminescence-based assay (IVIS 100, Xenogen). For titration of the AmiR122 dose, liver extracts were measured for level of luciferase expression (Luciferase assay Kit A, Biothema AB) and related to the expression of β-galactosidase to compensate for different transfection efficiency. The β-galactosidase was measured using Luminescent β-Gal kit (Clontech) following the manufacturer's protocol. These samples where analyzed using the GloMax instrument (Promega, Sweden).
Statistical analysis. Pair-wise comparisons among treatments were made using a Student's t-test. Comparisons among multiple treatments were made using one-way analysis of variance, followed by the SNK test. A P value of <0.05 was considered as significant difference.
SUPPLEMENTARY MATERIAL Figure S1. Effects of miR-122 and miR-142-3p on plasmid expression in transfected cells. Figure S2. Effect of AmiR122 administration on expression from plasmids containing miR-122 target sites. Figure S3. In vivo luciferase expression profiles from pUbC.Luc plasmids containing, or lacking, target sites for miR-142-3p. Figure S4. Toxicity assay for mice treated with either AmiR142 or AmiR122.
Acknowledgments
This work was supported by grants from the Swedish Research Council, the Swedish Cancer Society, The Swedish Childhood Cancer Foundation, Crown Princess Margareta's Foundation for the Visually Impaired, King Gustaf V Jubilee Foundation, Stockholm Cancer Society, the Swedish County Council (ALFproject), Aroseniusfonden and Karolinska Institutet. The authors have declared that no conflict of interest exists.
Supplementary Material
Effects of miR-122 and miR-142-3p on plasmid expression in transfected cells.
Effect of AmiR122 administration on expression from plasmids containing miR-122 target sites.
In vivo luciferase expression profiles from pUbC.Luc plasmids containing, or lacking, target sites for miR-142-3p.
Toxicity assay for mice treated with either AmiR142 or AmiR122.
References
- Gossen M, Bonin AL, Freundlieb S, Bujard H. Inducible gene expression systems for higher eukaryotic cells. Curr Opin Biotechnol. 1994;5:516–520. doi: 10.1016/0958-1669(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, O'Malley BW, Jr, Tsai SY, O'Malley BW. A regulatory system for use in gene transfer. Proc Natl Acad Sci USA. 1994;91:8180–8184. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola JR, El-Andaloussi S, Oprea II, Smith CI. Non-viral nanovectors for gene delivery: factors that govern successful therapeutics. Expert Opin Drug Deliv. 2010;7:721–735. doi: 10.1517/17425241003716810. [DOI] [PubMed] [Google Scholar]
- Weber W, Fussenegger M. Inducible product gene expression technology tailored to bioprocess engineering. Curr Opin Biotechnol. 2007;18:399–410. doi: 10.1016/j.copbio.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- Grigg P, Titong A, Jones LA, Yilma TD, Verardi PH. Safety mechanism assisted by the repressor of tetracycline (SMART) vaccinia virus vectors for vaccines and therapeutics. Proc Natl Acad Sci USA. 2013;110:15407–15412. doi: 10.1073/pnas.1314483110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrell L, Di Scala M, Blanco L, Otano I, Gil-Farina I, Baldim V, et al. Development of a liver-specific Tet-on inducible system for AAV vectors and its application in the treatment of liver cancer. Mol Ther. 2011;19:1245–1253. doi: 10.1038/mt.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth PA, St Onge L, Böger H, Gruss P, Gossen M, Kistner A, et al. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc Natl Acad Sci USA. 1994;91:9302–9306. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- Brown BD, Cantore A, Annoni A, Sergi LS, Lombardo A, Della Valle P, et al. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. 2007;110:4144–4152. doi: 10.1182/blood-2007-03-078493. [DOI] [PubMed] [Google Scholar]
- Annoni A, Brown BD, Cantore A, Sergi LS, Naldini L, Roncarolo MG. In vivo delivery of a microRNA-regulated transgene induces antigen-specific regulatory T cells and promotes immunologic tolerance. Blood. 2009;114:5152–5161. doi: 10.1182/blood-2009-04-214569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorpp M, Jäger R, Schellander K, Schenkel J, Wagner EF, Weiher H, et al. The human ubiquitin C promoter directs high ubiquitous expression of transgenes in mice. Nucleic Acids Res. 1996;24:1787–1788. doi: 10.1093/nar/24.9.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin KE, Ge R, Svahn MG, Törnquist E, Leijon M, Brandén LJ, et al. Cooperative strand invasion of supercoiled plasmid DNA by mixed linear PNA and PNA-peptide chimeras. Biomol Eng. 2004;21:51–59. doi: 10.1016/j.bioeng.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- Nishikawa M, Nakayama A, Takahashi Y, Fukuhara Y, Takakura Y. Reactivation of silenced transgene expression in mouse liver by rapid, large-volume injection of isotonic solution. Hum Gene Ther. 2008;19:1009–1020. doi: 10.1089/hum.2008.020. [DOI] [PubMed] [Google Scholar]
- Magami Y, Azuma T, Inokuchi H, Kokuno S, Moriyasu F, Kawai K, et al. Cell proliferation and renewal of normal hepatocytes and bile duct cells in adult mouse liver. Liver. 2002;22:419–425. doi: 10.1034/j.1600-0676.2002.01702.x. [DOI] [PubMed] [Google Scholar]
- Wooddell CI, Reppen T, Wolff JA, Herweijer H. Sustained liver-specific transgene expression from the albumin promoter in mice following hydrodynamic plasmid DNA delivery. J Gene Med. 2008;10:551–563. doi: 10.1002/jgm.1179. [DOI] [PubMed] [Google Scholar]
- Wolff LJ, Wolff JA, Sebestyén MG. Effect of tissue-specific promoters and microRNA recognition elements on stability of transgene expression after hydrodynamic naked plasmid DNA delivery. Hum Gene Ther. 2009;20:374–388. doi: 10.1089/hum.2008.088. [DOI] [PubMed] [Google Scholar]
- No D, Yao TP, Evans RM. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Sellmyer MA, Contag CH, Wandless TJ, Thorne SH. Chemical control of protein stability and function in living mice. Nat Med. 2008;14:1123–1127. doi: 10.1038/nm.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GJ, Mostoslavsky G, Kotton DN, Mulligan RC. Exogenous control of mammalian gene expression via modulation of translational termination. Nat Med. 2006;12:1093–1099. doi: 10.1038/nm1376. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15:31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- Kojima K, Takata A, Vadnais C, Otsuka M, Yoshikawa T, Akanuma M, et al. MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat Commun. 2011;2:338. doi: 10.1038/ncomms1345. [DOI] [PubMed] [Google Scholar]
- Li A, Qian J, He J, Zhang Q, Zhai A, Song W, et al. Modulation of miR122 expression affects the interferon response in human hepatoma cells. Mol Med Rep. 2013;7:585–590. doi: 10.3892/mmr.2012.1233. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Takata A, Otsuka M, Kishikawa T, Kojima K, Yoshida H, et al. Silencing of microRNA-122 enhances interferon-α signaling in the liver through regulating SOCS3 promoter methylation. Sci Rep. 2012;2:637. doi: 10.1038/srep00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenler S, Wiklander OP, Badal-Tejedor M, Turunen J, Nordin JZ, Hallengärd D, et al. Micro-minicircle Gene Therapy: Implications of Size on Fermentation, Complexation, Shearing Resistance, and Expression. Mol Ther Nucleic Acids. 2014;2:e140. doi: 10.1038/mtna.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh JS, Lee JY, Choi YS, Chung CP, Chong PC, Park YJ. Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomaterials. 2013;34:4347–4359. doi: 10.1016/j.biomaterials.2013.02.039. [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, Murata M, Sato Y, Takahashi M, Minakawa N, Matsuda A, et al. The systemic administration of an anti-miRNA oligonucleotide encapsulated pH-sensitive liposome results in reduced level of hepatic microRNA-122 in mice. J Control Release. 2014;173:43–50. [PubMed] [Google Scholar]
- Schievenbusch S, Strack I, Scheffler M, Nischt R, Coutelle O, Hösel M, et al. Combined paracrine and endocrine AAV9 mediated expression of hepatocyte growth factor for the treatment of renal fibrosis. Mol Ther. 2010;18:1302–1309. doi: 10.1038/mt.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Carrasco N, Chandler RJ, Chandrasekaran S, Venditti CP. Liver-directed recombinant adeno-associated viral gene delivery rescues a lethal mouse model of methylmalonic acidemia and provides long-term phenotypic correction. Hum Gene Ther. 2010;21:1147–1154. doi: 10.1089/hum.2010.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen JE, Mohamed AJ, Nore BF, Smith CI. Inducible H1 promoter-driven lentiviral siRNA expression by Stuffer reporter deletion. Oligonucleotides. 2005;15:139–144. doi: 10.1089/oli.2005.15.139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of miR-122 and miR-142-3p on plasmid expression in transfected cells.
Effect of AmiR122 administration on expression from plasmids containing miR-122 target sites.
In vivo luciferase expression profiles from pUbC.Luc plasmids containing, or lacking, target sites for miR-142-3p.
Toxicity assay for mice treated with either AmiR142 or AmiR122.