Abstract
Burkholderia pseudomallei is the causative agent of melioidosis, an often fatal infection of humans and animals. The virulence of this pathogen is thought to depend on a number of secreted proteins, including the MprA metalloprotease. We observed that MprA is produced upon entry into the stationary phase, when the cell density is high, and this prompted us to study cell density-dependent regulation in B. pseudomallei. A search of the B. pseudomallei genome led to identification of a quorum-sensing system involving the LuxI-LuxR homologs PmlI-PmlR. PmlI directed the synthesis of an N-acylhomoserine lactone identified as N-decanoylhomoserine lactone. A B. pseudomallei pmlI mutant was significantly less virulent than the parental strain in a murine model of infection by the intraperitoneal, subcutaneous, and intranasal routes. Inactivation of pmlI resulted in overproduction of MprA at the onset of the stationary phase. A wild-type phenotype was restored following complementation with pmlI or addition of cell-free culture supernatant. In contrast, there was no significant difference between the virulence of a B. pseudomallei mprA mutant and the virulence of the wild-type strain. These results suggest that the PmlI-PmlR quorum-sensing system of B. pseudomallei is essential for full virulence in a mouse model and downregulates the production of MprA at a high cell density.
Burkholderia pseudomallei, a gram-negative bacterium, is the causative agent of melioidosis in humans and animals (9, 13, 45). Melioidosis is endemic in tropical areas of Southeast Asia and northern Australia, but it is also sporadically found in many other countries. This highly pathogenic microorganism is deemed a potential agent of bioterrorism (listed as category B by the Centers for Disease Control and Prevention). The clinical manifestations of melioidosis vary greatly, ranging from an asymptomatic state to acute septicemia, pulmonary forms, and chronic granulomatous lesions (34). The latency period of the disease is between 2 days and 26 years (21, 34). Successful treatment is difficult because B. pseudomallei is inherently resistant to a wide range of antibiotics and relapse is common (43). Although melioidosis was first described in 1912 (44), the virulence determinants of B. pseudomallei have not been well characterized. Studies of B. pseudomallei pathogenicity have mostly concentrated on exoproducts, which include a protease, a lipase, a phospholipase C, and a hemolysin (3, 34, 45). The regulatory circuits governing the production of exoproducts in B. pseudomallei remain unknown. However, many gram-negative pathogens regulate the production of extracellular virulence factors by quorum sensing, a cell-density-dependent mechanism. Quorum sensing is the process of producing and responding to high intracellular concentrations of N-acylhomoserine lactone (AHL) autoinducers and relies on two proteins: (i) an AHL synthase belonging to the LuxI family, which directs the synthesis of AHL; and (ii) a transcriptional regulator belonging to the LuxR family, which, after binding of AHL, is thought to activate or repress transcription of targeted genes (11, 22, 29). In Burkholderia cepacia, which is phylogenetically related to B. pseudomallei (13), the CepI-CepR quorum-sensing system positively regulates protease production and represses synthesis of the siderophore ornibactin (19).
There has been much interest in the contribution of the B. pseudomallei protease to virulence. This enzyme was purified from culture supernatants as a 36-kDa metalloenzyme and appeared to be necessary for full virulence in a rat model of lung infection (32). In contrast, we found no correlation between virulence and the level of protease activity after intraperitoneal infection of mice with B. pseudomallei (12). These results suggested that the role played by the protease in pathogenesis could be dependent on the route of infection. Recently, the mprA gene of B. pseudomallei was cloned and was found to encode a 50-kDa serine metalloprotease, the only protease produced by B. pseudomallei (17). However, the discrepancy with the size determined by Sexton et al. (32) has not been explained yet; perhaps the difference is due to proteolysis or the use of different techniques (17). Our previous investigations on the B. pseudomallei protease (12, 25) led us to examine whether enzyme production is regulated by quorum sensing.
The role of quorum-sensing systems in the pathogenicity of different gram-negative bacteria has been demonstrated previously (28, 35). The existence of such a system in B. pseudomallei could provide new data on the virulence determinants of this organism.
Here we describe identification of the PmlI-PmlR quorum-sensing system of B. pseudomallei. This system is crucial for full virulence of B. pseudomallei in a murine model of infection and modulates the production of the MprA protease.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains used in this study were obtained from the collection of the Centre de Recherches du Service de Santé des Armées Emile Pardé (La Tronche, France). The prototype strain B. pseudomallei 008 was isolated in 1993 from a patient at the Grenoble-La Tronche Hospital in La Tronche, France. All experiments involving live bacteria were conducted in a biosafety level 3 facility. Escherichia coli S17-1 (pro thi recA hsdR chromosomal RP4-2; Tn1::ISR1 Tc::Mu Km::Tn7) (33) and E. coli Sm10 (thi thr leu chromosomal RP4-2; Tc::Mu) (33) carried the transfer genes of plasmid RP4 integrated into the chromosome and allowed mobilization of cloning vectors in which the Mob (oriT) region of pRP4 was cloned. Strains were routinely grown at 37°C in tryptic soy broth (TSB) or on tryptic soy agar. Cell density was monitored at 600 nm with a colorimeter (model 6061; Jenway, Dunmow, Essex, United Kingdom) and was expressed as optical density at 600 nm (OD600). When necessary, antibiotics were added at the following concentrations for E. coli and B. pseudomallei: tetracycline, 20 μg ml−1; trimethoprim, 100 μg ml−1; and carbenicillin, 100 μg ml−1. Kanamycin was added at a concentration of 50 μg ml−1 to E. coli cultures and at a concentration of 200 μg ml−1 to B. pseudomallei cultures.
The mobilizable plasmid pSUP401 (33) was used as a cloning vector in E. coli and as a suicide vector in B. pseudomallei. The mobilizable plasmid pUCP28T (31) was used as a cloning vector in E. coli and B. pseudomallei. The tetracycline cassette, used for gene disruption, was PCR amplified from pACYC184 (6) with primers TC-3 (AATTTATCTCTTCAAATGTAGCAGCTGAAGTCAGCCCC) and TC-4 (ATGCGCCGCGTGCGGCAGCTGGAGATGGCGGAC) and was used to construct B. pseudomallei 008 (mprA::Tc) and B. pseudomallei 008 (pmlI::Tc) mutants (see below).
DNA manipulations.
The methods employed for making constructs and manipulating recombinant DNA were essentially the methods described by Sambrook et al. (30). Plasmid DNA was prepared as described by Birnboim and Doly (4). Transformation was performed in E. coli S17-1 and in E. coli Sm10 by the CaCl2 method (30). When required, homologous recombination between the disrupted gene in pSUP401 derivatives and the corresponding wild-type gene on the B. pseudomallei 008 chromosome was performed by allelic exchange as described previously (40). Double recombination was further confirmed by PCR analysis by using the appropriate primers.
PCR was performed with a PTC 200 thermocycler (MJ Research, Waltham, Mass.) by using Ready To Go PCR beads (Amersham Pharmacia Biotech Inc, Piscataway, N.J.) according to the manufacturer's instructions.
Construction of a B. pseudomallei mprA mutant.
A fragment containing the mprA gene and promoter (17) was amplified from B. pseudomallei 008 DNA by using primers MprA-1 (ACGGAAGACGAATTCTCCGGCTCGCGCAGCCGG) and MprA-2 (GCAACGCCCGAATTCGCTCACTGCGCGGCGGCG), both of which were designed to contain an EcoRI restriction site at the 5′ end. The 1.8-kb amplified fragment was cloned into pSUP401, a suicide vector in B. pseudomallei. To insertionally inactivate the mprA gene in pSUP401, a tetracycline cassette was inserted into the internal EcoRV site of the gene. This construct was used to transform E. coli S17-1, and the resulting strain was mated with B. pseudomallei 008. Transconjugants that had undergone homologous recombination between the inactivated gene on pSUP401 and the corresponding wild-type gene on the B. pseudomallei chromosome were first selected on plates containing carbenicillin and tetracycline. The clones were then tested for kanamycin sensitivity and loss of the vector. As expected, most clones were kanamycin sensitive. The double recombination event was confirmed by PCR with primers MprA-1 and MprA-2. One of the mutants, designated B. pseudomallei 008 (mprA::Tc), was selected for further study. For complementation analysis, the amplified mprA gene was cloned into pUCP28T, yielding pMprA, and this recombinant plasmid was transferred by conjugation from E. coli Sm10 to B. pseudomallei 008 (mprA::Tc).
Construction of a B. pseudomallei pmlI mutant.
The pmlI gene was amplified by PCR with primers PmlI-1 (GGCGCTAAATTGAAGCTTGGCGTCTTGCCAGCG) and PmlI-8 (ATCGAACGTAGGAAGCTTCGCGCGAAATACCG), cloned into pSUP401, and then interrupted by the tetracycline cassette inserted into the NruI site of the pmlI open reading frame (ORF). This recombinant plasmid was transferred by conjugation from E. coli S17-1 into B. pseudomallei 008. The double recombination event in carbenicillin- and tetracycline-resistant but kanamycin-sensitive transformants was confirmed by PCR analysis by using primers PmlI-1 and PmlI-8. One of the transformants, B. pseudomallei 008 (pmlI::Tc), was selected for further analysis. In parallel, the amplified pmlI gene was cloned into pUCP28T, yielding pPmlI, which was transferred into B. pseudomallei 008 (pmlI::Tc) for complementation analysis.
Measurement of protease activity.
Strains were grown at 37°C in 100 ml of TSB shaken at 180 rpm in 250-ml glass culture flasks. Media were inoculated with cell pellets from 5-ml cultures shaken overnight in TSB at 37°C. At selected times, 2 ml of culture was removed, and the bacteria were harvested by centrifugation (10,000 × g for 10 min). Culture supernatants were carefully collected, filtered through 0.2-μm-pore-size filters, and supplemented with 10% (final concentration) sodium dodecyl sulfate (SDS). The bacterial pellets were washed twice in TSB and resuspended in 2 ml of TSB. The cells were lysed with 10% SDS as described previously (25) and filtered through 0.2-μm-pore-size filters, which yielded cell lysates. Total extracts were obtained by adding 10% (final concentration) SDS to 2 ml of total culture. Protease activity was assessed by using an EnzChek protease assay kit (Interchim, Rockford, Ill.) as recommended by the manufacturer. This activity was determined at different stages of the growth curve. Briefly, samples were diluted 50-fold in 1× digestion buffer. Then 100-μl portions of a BODIPY FL casein solution were added to 100-μl portions of diluted samples in microplates with black walls. After the microplates were incubated in the dark at 37°C for 18 h, fluorescence was measured with a microplate reader (FL800; Bio-Tek Instruments, Winooski, Vt.). One protease unit was defined as the activity that increased the fluorescence by 1 relative fluorescence unit per OD600 unit. Mean values were calculated by using the results of at least three independent assays, and the standard deviations were less than 10% of the reported values.
Preparation and use of cell-free conditioned supernatants.
Cell-free supernatants were prepared as described previously (7). Briefly, B. pseudomallei 008 (mprA::Tc) and E. coli TG1 (30) were shaken in TSB for 18 h at 37°C. Cell-free supernatants were prepared by centrifugation, sterilized with 0.2-μm-pore-size filters, and stored at 4°C. To support growth, 10% (vol/vol) fresh TSB was added to the sterile conditioned supernatant. B. pseudomallei 008 or B. pseudomallei 008 (pmlI::Tc) was first grown overnight in TSB, and then 1 ml of the culture was removed and the bacteria were harvested by centrifugation. The pellet was used to inoculate 100 ml of conditioned medium.
Identification of the B. pseudomallei AHL.
Spent supernatants (500 ml) from stationary-phase cultures of B. pseudomallei grown in minimal medium M9 were passed through a 0.2-μm-pore-size filter and extracted with 200 ml of dichloromethane. The extract was concentrated by rotary evaporation at room temperature. The residue was reconstituted in 1.5 ml of acetonitrile and applied to a C8 reverse-phase analytical high-performance liquid chromatography (HPLC) column (Spheri 5 RP8; 220 by 4.6 mm; Perkin-Elmer, Norwalk, Conn.). It was then eluted with a 5 to 70% acetonitrile gradient in water at a flow rate of 1.5 ml per min. The autoinducer-containing fractions, as determined by the E. coli VJS533(pHV200I−), E. coli MG4(pVKDT17) (24), and Ralstonia solanacearum(p395B) bioassays (10), were pooled and subjected to chromatography again by using a 20 to 100% acetonitrile gradient in water. Synthetic AHLs (C8-AHL, C10-AHL, and C12-AHL) (Fluka, Saint Quentin Fallavier, France), used as standards, were subjected to chromatography on the same column as described above.
Mice and experimental infection.
Female Swiss mice were purchased from the Centre d'Elevage R. Janvier (Le Genest St. Isle, France). Animals were given sterilized U.A.R. chow (U.A.R., Villemoisson sur Orge, France) and sterile water ad libitum. For all experiments, animals were used when they were 8 weeks old. The Centre de Recherches du Service de Santé des Armées Emile Pardé Animal Care Committee approved all in vivo studies described below (file numbers 34-2001 and 26-2002). Bacteria were streaked on tryptic soy agar plates and incubated overnight at 37°C. TSB was inoculated with single colonies and shaken for 18 h at 37°C. Mice were inoculated with 10-fold dilutions of each culture in sterile saline. The numbers of viable bacteria in the dilutions used were determined by plate counting. To establish the 50% lethal dose (LD50), groups of six mice were inoculated with 200-μl aliquots of the various dilutions via the intraperitoneal or subcutaneous route or with 50-μl aliquots via the intranasal route. Death was recorded over the following 6 weeks. The LD50 was calculated as described by Reed and Muench (27). In another experiment, we assessed the survival rates of mice infected by the intraperitoneal, subcutaneous, and intranasal routes. To do this, groups of 20 mice were inoculated with ca. 1 LD50 of B. pseudomallei 008, as determined for each challenge route. Death was recorded daily for 6 weeks. Kaplan-Meier survival curves were constructed, and the significance of differences between groups was assessed by the log-rank test (26).
RESULTS
Activity of MprA in relation to the growth phase.
We first measured the activity of the MprA protease in supernatants of B. pseudomallei 008 grown in TSB. The MprA activity was low during the exponential phase (7,429 U after 4 h) and then drastically increased upon entry into the stationary phase (27,936 U after 8 h) (Fig. 1). To examine whether MprA was produced but not secreted during the exponential phase, we measured the MprA activity in cell lysates and total extracts during growth (Fig. 1). Like the protease activities in culture supernatants, the protease activities in cell lysates and total extracts were low during the exponential phase. Thus, the MrpA protease appeared to be poorly produced during the exponential growth phase. The protease activity in total extracts increased at the beginning of the stationary phase, whereas it remained nearly constant in cell lysates. After 24 h, 85% of the MprA activity was present in the culture supernatant and the remaining 15% was cell associated (Fig. 1). Thus, production and secretion of MprA are growth phase dependent, increasing markedly upon entry into the stationary phase.
FIG. 1.
Growth phase modulation of MprA protease activity. B. pseudomallei 008 was grown at 37°C in TSB. The OD600 (dashed line) and the MprA activity in culture supernatants (open bars), cell lysates (striped bars), and total extracts (solid bars) were measured every 4 h.
Characterization of a B. pseudomallei mprA mutant.
MprA activity was determined in total extracts during growth of the B. pseudomallei 008 (mprA::Tc) mutant and of its derivative harboring pMprA. Total extracts of the mprA::Tc mutant contained very low levels of protease activity throughout growth (Fig. 2). Complementation of B. pseudomallei 008 (mprA::Tc) with pMprA restored production of the protease, and moreover, this production was growth phase dependent (Fig. 2), as observed for the parental strain (Fig. 1). These results demonstrated that the protease activity detected in culture supernatants of B. pseudomallei 008 requires an intact mprA gene and that the MprA protein is probably the only protease produced by B. pseudomallei 008 in our experimental conditions.
FIG. 2.
Effect of mprA mutation on protease production in B. pseudomallei. Bacterial strains were grown at 37°C in TSB. The OD600 (dashed lines) and the MprA activity (bars) in total extracts were measured every 4 h. ▴ and solid bars, B. pseudomallei 008 (mprA::Tc) mutant; ▪ and striped bars, B. pseudomallei 008 (mprA::Tc) mutant harboring pMprA.
Identification of the PmlI-PmlR quorum-sensing system in B. pseudomallei.
To identify a putative quorum-sensing system in B. pseudomallei, we carried out a BLAST search of the B. pseudomallei genome using the assembly contigs generated by the B. pseudomallei genome sequencing project (www.sanger.ac.uk/Projects/B_pseudomallei). This search allowed us to identify two ORFs, which we designated pmlI and pmlR and which exhibited high levels of sequence identity to B. cepacia cepI and cepR, respectively. The pmlI and pmlR sequences were analyzed further with the programs available at the www.ncbi.nlm.nih.gov/BLAST/ website. The pmlI ORF is predicted to encode a 203-amino-acid protein with a calculated molecular mass of 22,156 Da. The PmlI protein exhibited 98% sequence identity to the BpsI protein described by Lumjiaktase et al. (accession no. AF501236), 97% sequence identity to the AHL synthase of Burkholderia mallei, and 78% sequence identity to the B. cepacia CepI protein. The characteristic domain of the LuxI family members (accession no. pfam00765) is highly conserved in PmlI. The pmlR ORF is predicted to encode a 239-amino-acid protein with a calculated molecular mass of 26,668 Da. The PmlR protein exhibited 98% sequence identity to the AHL-binding regulator of B. mallei and 79% sequence identity to the CepR protein of B. cepacia. Both the AHL-binding domain and the helix-turn-helix motif of the LuxR family members (accession no. pfam03472) are present in PmlR. pmlR is separated from pmlI by 741 bp and is divergently transcribed. This genetic organization is similar to that found in B. cepacia (19) and in Burkholderia vietnamiensis (8). Together, all these data strongly suggest that pmlI and pmlR constitute a quorum-sensing system in B. pseudomallei.
Construction and characterization of a B. pseudomallei pmlI mutant.
We constructed a pmlI mutant of B. pseudomallei 008 as described in Materials and Methods. We then measured the MprA activities in culture supernatants and total extracts of the parental strain, the pmlI::Tc mutant, and the pmlI::Tc mutant complemented with pPmlI throughout growth. Inactivation of the pmlI gene had no obvious effect on protease activity during the exponential phase (Fig. 3). In contrast, the protease activity in the culture supernatant was 1.5- to 2-fold higher upon entry into the stationary phase for the pmlI::Tc mutant than for the parental strain (Fig. 3). Consistent with the results obtained with the wild-type strain, approximately 90% of the total activity was present in the culture supernatant of the pmlI::Tc mutant (data not shown), indicating that MprA was efficiently secreted by this mutant. When the mutant strain was complemented with pPmlI, the MprA protease activity of this strain was restored to a level similar to that of the parental strain (Fig. 3). These results strongly suggested that B. pseudomallei MprA production is limited by the PmlI-PmlR quorum-sensing system at a high cell density.
FIG. 3.
Modulation of MprA protease activity by the PmlI-PmlR quorum-sensing system. Bacterial strains were grown at 37°C in TSB. The OD600 (dashed lines) and MprA activity (bars) in culture supernatants were determined every 4 h. ▴ and solid bars, parental strain B. pseudomallei 008; ▪ and stippled bars, B. pseudomallei 008 (pmlI::Tc) mutant; ♦ and striped bars, B. pseudomallei 008 (pmlI::Tc) mutant harboring pPmlI.
Characterization of the B. pseudomallei AHL.
Assuming that PmlI is an AHL synthase, we looked for autoinducer activity in culture supernatants of B. pseudomallei. Cell-free supernatant of E. coli TG1 or B. pseudomallei 008 (mprA::Tc), which contained very low levels of protease activity (Fig. 2), was added prior to inoculation of B. pseudomallei 008 and B. pseudomallei 008 (pmlI::Tc), the latter of which overproduced the MprA protease (Fig. 3). As a control, the E. coli TG1 cell-free supernatant did not affect bacterial growth or the protease activity of total extracts from B. pseudomallei 008 (pmlI::Tc) (data not shown). Additionally, growth of B. pseudomallei 008 (pmlI::Tc) was not modified by B. pseudomallei 008 (mprA::Tc) supernatant (Fig. 4). Therefore, the presence of B. pseudomallei 008 (mprA::Tc) culture supernatant in the growth medium did not affect the protease activity in total extracts of B. pseudomallei 008 (Fig. 1 and 4). In contrast, addition of B. pseudomallei 008 (mprA::Tc) cell-free supernatant greatly reduced the MprA activity in total extracts of B. pseudomallei 008 (pmlI::Tc) and restored the parental phenotype to the pmlI::Tc mutant (Fig. 4). Thus, soluble signaling molecules absent from the E. coli TG1 supernatant are involved in negative control of MprA protease production by the PmlI-PmlR quorum-sensing system in B. pseudomallei.
FIG. 4.
Effects of cell-free culture supernatants on MprA protease activity. Bacterial strains were grown at 37°C in normal or conditioned TSB. OD600 (dashed lines) and MprA activity (bars) of total extracts were measured every 4 h. ▴ and solid bars, parental strain B. pseudomallei 008 grown in conditioned TSB; ▪ and spotted bars, B. pseudomallei 008 (pmlI::Tc) mutant grown in normal TSB; ♦ and striped bars, B. pseudomallei 008 (pmlI::Tc) mutant grown in conditioned TSB.
To identify these signaling molecules, an acetonitrile extract of B. pseudomallei 008 culture supernatant was fractionated by C8 reverse-phase HPLC. Only one peak of AHL activity was found when each fraction was assayed by using three AHL bioassays. The E. coli VJS533(pHV200I−) bioassay, which can detect AHL molecules with acyl groups containing five to eight carbons (24), responded very weakly to the compound produced by B. pseudomallei 008. Both the E. coli MG4(pVKDT17) and R. solanacearum(p395B) bioassays, which can detect AHL with acyl groups containing eight or more carbons (10, 24), gave positive responses (data not shown). This suggested that B. pseudomallei 008 produces an AHL with an acyl group that is longer than eight carbon atoms. As expected, the acetonitrile extract of the B. pseudomallei 008 (pmlI::Tc) mutant did not contain detectable levels of autoinducer activity, as shown by the E. coli MG4(pVKDT17) and R. solanacearum(p395B) bioassays. Fractions containing the active compound were pooled and further separated by HPLC as described in Materials and Methods. B. pseudomallei autoinducer was eluted at a position very similar to that of synthetic N-decanoylhomoserine lactone (data not shown). These results were consistent with the hypothesis that pmlI encodes an autoinducer synthase that is required for the production of homoserine lactone in B. pseudomallei.
Virulence of B. pseudomallei pmlI and mprA mutants in mice.
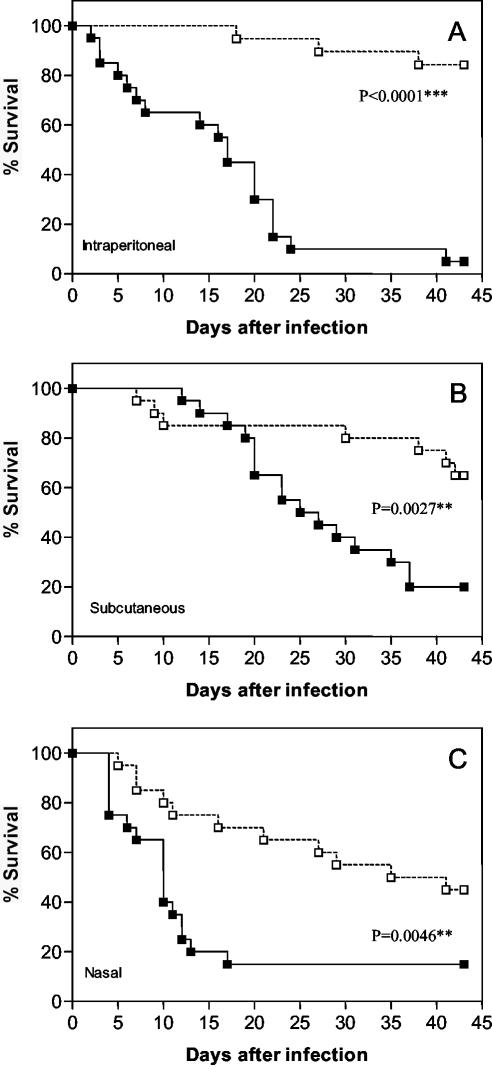
As quorum sensing regulates the production of virulence factors in several gram-negative species (11, 22, 29), we investigated the role of the PmlI-PmlR system in B. pseudomallei virulence using a murine model of infection. To do this, Swiss mice were infected with the wild-type B. pseudomallei 008 strain and its isogenic pmlI::Tc mutant, and the LD50s were determined. The LD50s were log10 5.6, 6.8, and 2.2 for B. pseudomallei 008 and 6.3, 7.9, and 3.6 for B. pseudomallei 008 (pmlI::Tc) after intraperitoneal, subcutaneous, and nasal infection, respectively. Thus, the LD50 of the pmlI mutant was about 5- to 25-fold higher than that of the parental strain. In agreement, there was a significant difference (P < 0.05, as determined by a log rank test) between the percentage of survival for the group of mice infected with B. pseudomallei 008 and the percentage of survival for the group of mice infected with B. pseudomallei 008 (pmlI::Tc) (Fig. 5). Only one, four, and three of the mice inoculated with B. pseudomallei 008 by the intraperitoneal, subcutaneous, and intranasal routes, respectively, survived the challenge. In contrast, 17, 13, and 10 mice survived after inoculation with B. pseudomallei 008 (pmlI::Tc). Thus, the PmlI-PmlR quorum-sensing system is essential for full virulence of B. pseudomallei in this mouse model.
FIG. 5.
Percentages of survival of Swiss mice after intraperitoneal (A), subcutaneous (B), and intranasal (C) inoculation with the parental strain B. pseudomallei 008 (▪) and the isogenic pmlI::Tc mutant (□). Twenty mice in each group were infected with ca. 1 LD50 of B. pseudomallei 008; the number of bacteria was dependent on the infection route. The P value is significant if it is <0.05.
To examine the contribution of MprA to virulence, we studied the survival of mice infected with parental strain B. pseudomallei 008 and its isogenic mprA::Tc mutant via the intraperitoneal, subcutaneous, and intranasal routes. Neither the percentage of survival nor the median survival time of mice infected by either of the strains was significantly affected by the infection route (data not shown). Thus, MprA is probably not a virulence determinant in this infection model.
DISCUSSION
Many gram-negative bacteria, including pathogens, sense population density and control the expression of many phenotypes by using members of the LuxI-LuxR family of quorum-sensing components (11, 22, 29). To date, two quorum-sensing systems have been characterized in the genus Burkholderia: the CepI-CepR system in B. cepacia (19) and the BviI-BviR system in B. vietnamiensis (8). Here, we identified a third LuxI-LuxR homolog, the PmlI-PmlR quorum-sensing system in B. pseudomallei. The pmlI gene appears to encode an autoinducer synthase as the PmlI protein is a member of the LuxI protein family and as a pmlI mutant did not produce detectable levels of AHL. HPLC analysis strongly suggested that the PmlI synthase directs the synthesis of N-decanoylhomoserine lactone, an AHL also produced by B. vietnamiensis (8). On the basis of a sequence comparison, we speculated that pmlR encodes a transcriptional regulatory protein belonging to the LuxR family, but further studies are required to confirm this hypothesis. The large region between pmlI and pmlR (741 bp) does not contain any putative ORF with significant similarity to any known gene. A similar observation was reported for the cepI-cepR intergenic region (727 bp) in B. cepacia (19).
The PmlI-PmlR system appears to negatively regulate MprA production in B. pseudomallei during the stationary phase. In the pmlI mutant, which did not produce detectable amounts of AHL signal molecules, MprA was overproduced upon entry into the stationary phase. Indeed, addition of cell-free culture supernatant of B. pseudomallei, which produced the autoinducer molecule, restored a wild-type phenotype to the pmlI mutant. Assuming that PmlR is a transcriptional regulator, it is tempting to speculate that PmlR represses mprA expression at high autoinducer concentrations.
Other quorum-sensing systems also repress the expression of their target genes. The CepI-CepR system negatively controls the production of ornibactin, a siderophore of B. cepacia, when the cell density is high (19). EsaR from Pantoea stewartii represses exopolysaccharide production at a low cell density, and derepression requires micromolar amounts of AHL produced by the EsaI synthase (41, 42).
Quorum sensing regulates not only exoproducts but also other types of virulence factors in pathogenic bacteria (11, 22, 29). For example, recent work has demonstrated that LuxO, a quorum-sensing regulator in Vibrio cholerae, is pivotal for biofilm formation, cholera toxin production, and protease secretion (39).
In the human opportunistic pathogen Pseudomonas aeruginosa, the secreted virulence factors are under control of two quorum-sensing systems, LasI-LasR and RhlI-RhlR (16, 23). The fact that a lasR mutant exhibits significantly reduced virulence in a mouse model of pneumonia demonstrates the importance of quorum sensing in the pathogenesis of P. aeruginosa infection (18, 38). Consistent with this, our results indicated that the PmlI-PmlR quorum-sensing system is essential for the pathogenesis of melioidosis, as a pmlI mutant of B. pseudomallei is less virulent than the wild-type strain in a murine model of infection when either the intraperitoneal, subcutaneous, or intranasal route is used. This work clearly demonstrated that the production of MprA is regulated by the PmlI-PmlR system but that this protein plays a minor role in B. pseudomallei virulence, unlike the role observed in respiratory infection by Burkholderia cenocepacia (36). Further studies are thus specifically needed to identify essential virulence determinants controlled by the PmlI-PmlR quorum-sensing system in B. pseudomallei (2). In P. aeruginosa, it has been demonstrated that the autoinducer has a direct effect on the host immune response and contributes to the pathogenicity in a pulmonary challenge (28, 35). Thus, the role of the PmlI-PmlR quorum-sensing system in B. pseudomallei could be explained by regulated expression of virulence factors and induced immunomodulation in the host. However, Cabrol et al. pointed out that the quorum-sensing gene lasR played a minor role in the pathogenesis of 50% of the P. aeruginosa strains that they studied (5). Similarly, it will be of interest to determine the role of other regulatory systems, such as alternative sigma factors (1) or posttranscriptional regulators (15), in the pathogenicity of B. pseudomallei.
In most of the previous studies quorum sensing of risk group 2 pathogens was examined. Jones and Blaser (14) and Taminiau et al. (37) described in vitro AHL production in the risk group 3 bacteria Bacillus anthracis and Brucella melitentis, respectively. To our knowledge, the present study is the first in vivo demonstration that a quorum-sensing system is indispensable for the full virulence of a risk group 3 pathogen.
As the treatment of melioidosis is long and difficult due to resistance to multiple antibiotics and the lack of an efficacious vaccine, the results described here indicate that inactivation of AHL cell-cell signaling might represent a novel strategy for therapy (20).
Acknowledgments
This work was supported by grant 99CO036 from Délégation Générale pour l'Armement (DSP/STTC).
We thank J. Croize for providing B. pseudomallei strain 008, J. P. Pearson and E. P. Greenberg for the generous gift of the strains used for the bioassays, D. E. Woods for providing pUCP28T, and F. Desor, I. Perrichon, D. Riou, and D. Cariou for helpful technical assistance.
REFERENCES
- 1.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (RpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, C., A. Friscina, G. Devescovi, M. Kojic, and V. Venturi. 2003. Identification of quorum-sensing-regulated genes of Burkholderia cepacia. J. Bacteriol. 185:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashdown, L. R., and J. M. Koehler. 1990. Production of hemolysin and other extracellular enzymes by clinical isolates of Pseudomonas pseudomallei. J. Clin. Microbiol. 28:2331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabrol, S., A. Olliver, G. B. Pier, A. Andremont, and R. Ruimy. 2003. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:7222-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from p15A cryptic miniplasmid. J. Bacteriol. 134:1141-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapon-Hervé, V., M. Akrim, A. Latifi, P. Williams, A. Lazdunski, and M. Bally. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169-1178. [DOI] [PubMed] [Google Scholar]
- 8.Conway, B. A., and E. P. Greenberg. 2002. Quorum-sensing signals and quorum-sensing genes in Burkholderia vietnamiensis. J. Bacteriol. 184:1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dance, D. 1991. Melioidosis: the tip of the iceberg? Clin. Microbiol. Rev. 4:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flavier, A. B., L. M. Ganova-Raeva, M. A. Schell, and T. P. Denny. 1997. Hierarchical autoinduction in Ralstonia solanacearum: control of N-acyl-homoserine lactone production by a novel autoregulatory system responsive to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:7089-7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 12.Gauthier, Y. P., F. M. Thibault, J. C. Paucod, and D. R. Vidal. 2000. Protease production by Burkholderia pseudomallei and virulence in mice. Acta Trop. 74:215-220. [DOI] [PubMed] [Google Scholar]
- 13.Howe, C., A. Sampath, and M. Spotnitz. 1971. The pseudomallei group: a review. J. Infect. Dis. 124:598-606. [DOI] [PubMed] [Google Scholar]
- 14.Jones, M. B., and M. J. Blaser. 2003. Detection of a luxS-signaling molecule in Bacillus anthracis. Infect. Immun. 71:3914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jude, F., T. Kohler, P. Branny, K. Perron, M. P. Mayer, R. Comte, and C. van Delden. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J. Bacteriol. 185:3558-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latifi, A., K. M. Winson, M. Foglino, B. W. Bycroft, G. S. A. B. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333-344. [DOI] [PubMed] [Google Scholar]
- 17.Lee, M. A., and Y. C. Liu. 2000. Sequencing and characterization of a novel serine metalloprotease from Burkholderia pseudomallei. FEMS Microbiol. Lett. 192:67-72. [DOI] [PubMed] [Google Scholar]
- 18.Lesprit, P., F. Faurisson, O. Join-Lambert, F. Roudot-Thoraval, M. Foglino, C. Vissuzaine, and C. Carbon. 2003. Role of the quorum-sensing system in experimental pneumonia due to Pseudomonas aeruginosa in rats. Am. J. Respir. Crit. Care Med. 167:1478-1482. [DOI] [PubMed] [Google Scholar]
- 19.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyon, G. J., and T. W. Muir. 2003. Chemical signaling among bacteria and its inhibition. Chem. Biol. 10:1007-1021. [DOI] [PubMed] [Google Scholar]
- 21.Mays, E., and E. Ricketts. 1975. Melioidosis: recrudescence associated with bronchogenic carcinoma twenty-six years following initial geographic exposure. Chest Aug. 68:261-263. [DOI] [PubMed] [Google Scholar]
- 22.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 23.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 24.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Percheron, G., F. Thibault, J. C. Paucod, and D. Vidal. 1995. Burkholderia pseudomallei requires Zn2+ for optimal exoprotease production in chemically defined media. Appl. Environ. Microbiol. 61:3151-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peto, R., M. C. Pike, P. Armitage, N. E. Breslow, D. R. Cox, S. V. Howard, N. Mantel, K. McPherson, J. Peto, and P. G. Smith. 1977. Design and analysis of randomized clinical trials requiring prolonged observation of each patient. II. Analysis and examples Br. J. Cancer 35:1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 28.Ritchie, A. J., A. O. M. Yam, K. M. Tanabe, S. A. Rice, and M. A. Cooley. 2003. Modification of in vivo and in vitro T- and B-cell-mediated immune responses by the Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxodecanoyl)-l-homoserine lactone. Infect. Immun. 71:4421-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salmond, G. P. C., B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1995. The bacterial ′enigma': cracking the code of cell-cell communication. Mol. Microbiol. 16:615-624. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Schweizer, H. P., T. Klassen, and T. Hoang. 1996. Improved methods for gene analysis and expression in Pseudomonas spp., p. 229-237. In T. Nakazawa, K. Furukawa, D. Haas, and S. Silver (ed.), Molecular biology of pseudomonas. American Society for Microbiology, Washington, D.C.
- 32.Sexton, M., A. Jones, W. Chaowagul, and D. Woods. 1994. Purification and characterization of a protease from Pseudomonas pseudomallei. Can. J. Microbiol. 40:903-910. [DOI] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 34.Smith, C. J., J. C. Allen, M. Noor Embi, O. Othman, N. Razak, and G. Ismail. 1987. Human melioidosis: an emerging medical problem. MIRCEN J. 3:343-366. [Google Scholar]
- 35.Smith, R. S., S. G. Harris, R. Phipps, and B. Iglewski. 2002. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 184:1132-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149: 3649-3658. [DOI] [PubMed] [Google Scholar]
- 37.Taminiau, B., M. Daykin, S. Swift, M. L. Boschiroli, A. Tibor, P. Lestrate, X. DeBolle, D. OCallaghan, P. Williams, and J. J. Letesson. 2002. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 70:3004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, H. B., E. Dimango, R. Bryan, M. J. Gambello, B. H. Iglewski, J. B. Goldberg, and A. Prince. 1996. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect. Immun. 64:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vance, R. E., J. Zhu, and J. J. Mekalanos. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virlogeux, I., H. Waxin, C. Ecobichon, and M. Y. Popoff. 1995. Role of the viaB locus in synthesis, transport and expression of Salmonella typhi Vi antigen. Microbiology 141:3039-3047. [DOI] [PubMed] [Google Scholar]
- 41.von Bodman, S. B., D. R. Majerczak, and D. L. Coplin. 1998. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl. Acad. Sci. 95:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Bodman, S. B., J. K. Ball, M. A. Faini, C. M. Herrera, T. D. Minogue, M. L. Urbanowski, and A. M. Stevens. 2003. The quorum sensing negative regulators EsaR and ExpREcc, homologues within the LuxR family, retain the ability to function as activators of transcription. J. Bacteriol. 185:7001-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]
- 44.Whitmore, A., and C. S. Krishnasawami. 1912. An account of the discovery of a hitherto undescribed infective disease occurring among the population of Rangoon. India Med. Gaz. 47:262-267. [PMC free article] [PubMed] [Google Scholar]
- 45.Woods, D. E., D. DeShazer, R. A. Moore, P. J. Brett, M. N. Burtnick, S. L. Reckseidler, and M. D. Senkiw. 1999. Current studies on the pathogenesis of melioidosis. Microbes Infect. 1:157-162. [DOI] [PubMed] [Google Scholar]