During stationary phase growth in yeast, a feed-forward loop exists in which lipophagy stimulates vacuolar microdomain formation, which, in turn, further promotes lipophagy.
Abstract
Stationary phase (stat-phase) is a poorly understood physiological state under which cells arrest proliferation and acquire resistance to multiple stresses. Lipid droplets (LDs), organelles specialized for cellular lipid homeostasis, increase in size and number at the onset of stat-phase. However, little is known about the dynamics of LDs under this condition. In this paper, we reveal the passage of LDs from perinuclear endoplasmic reticulum association to entry into vacuoles during the transition to stat-phase. We show that the process requires the core autophagy machinery and a subset of autophagy-related (Atg) proteins involved in selective autophagy. Notably, the process that we term stat-phase lipophagy is mediated through a sterol-enriched vacuolar microdomain whose formation and integrity directly affect LD translocation. Intriguingly, cells defective in stat-phase lipophagy showed disrupted vacuolar microdomains, implying that LD contents, likely sterol esters, contribute to the maintenance of vacuolar microdomains. Together, we propose a feed-forward loop in which lipophagy stimulates vacuolar microdomain formation, which in turn promotes lipophagy during stat-phase.
Introduction
Lipid droplets (LDs) are ubiquitous organelles that store triacylglycerol and sterol esters for use by cells to produce energy and membranes (Martin and Parton, 2006; Walther and Farese, 2012). LDs emerge from the ER (Kassan et al., 2013; Pol et al., 2014), and their number, size, and distribution vary under different growth conditions (Yang et al., 2012). LDs are capable of interacting with many organelles (Liu et al., 2008; Pu et al., 2011), which is thought to facilitate lipid transfer. However, the exact mechanisms through which LDs form, grow, and mobilize their contents remain largely unknown.
The breakdown of storage lipids within LDs is attributed to various lipid hydrolase activities (Ducharme and Bickel, 2008). Recent evidence suggests that autophagy provides an alternative route for LD breakdown in the hepatocyte (Singh et al., 2009; Singh and Cuervo, 2012). This lipophagy process uses macroautophagy machinery that is mediated by core autophagy-related (Atg) proteins (Xie and Klionsky, 2007; Mizushima et al., 2011) to sequester LDs into forming autophagosomes that subsequently fuse with lysosomes. Lipophagy was also found in yeast. When triacylglycerol is overloaded into LDs by oleate induction, autophagy machinery targets LDs to vacuoles for lipid mobilization (van Zutphen et al., 2014). The yeast lipophagy morphologically resembles microautophagy, which involves a direct modification on the vacuolar membrane that engulfs LDs. Although it seems likely that lipophagy is a selective process, the exact LD targets for lipophagy remain elusive.
Most cells, including yeast, spend most of life dealing with many faces of stresses and nutrient deficiency in quiescence. Mimicking the conditions by culturing cells in nutrient-rich medium for extended periods has been a valuable model to study cell physiology during stationary phase (stat-phase; Werner-Washburne et al., 1993). Herein, we analyze LD distribution and find that LDs enter the vacuole lumen along with a unique, hydrolase-resistant, membrane during stat-phase. The membrane is the liquid-ordered (Lo) vacuolar microdomain that forms only when cells enter stat-phase (Toulmay and Prinz, 2013). Our data reveal that LD entry into the vacuole requires vacuolar microdomain formation and that the pathway contributes to the vacuolar microdomain maintenance during stat-phase.
Results and discussion
LD distribution during the transition from log to stat-phases
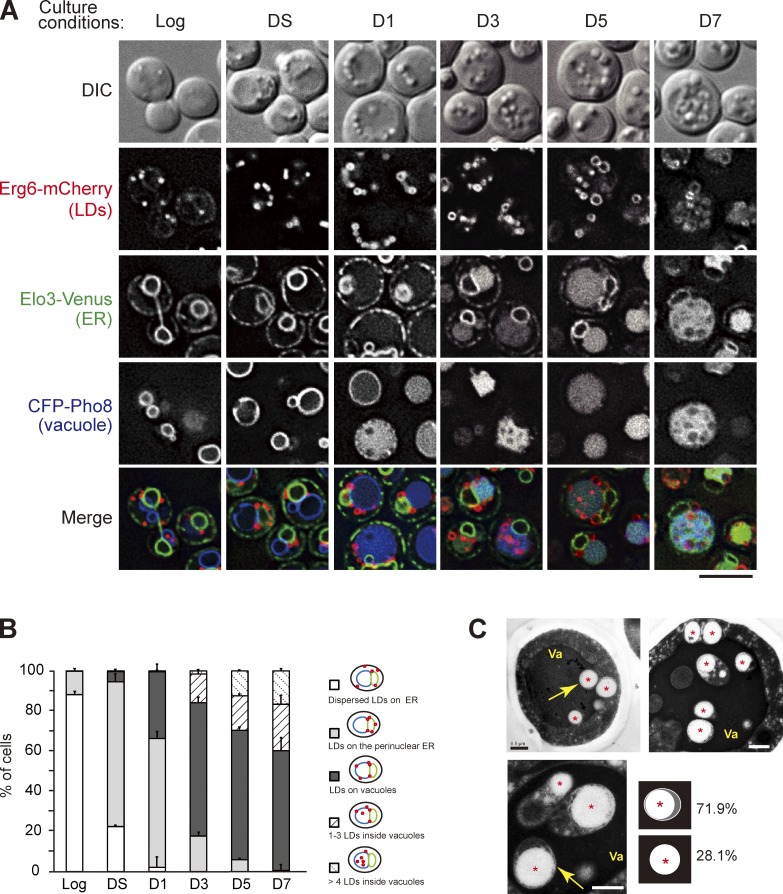
To study the dynamics of LDs, we followed mCherry-tagged Erg6, a sterol biosynthetic enzyme on LDs, in cells containing ER marker Elo3-Venus and vacuole marker CFP-Pho8. Direct imaging of cells in various growth conditions shows that LDs spread throughout the cortical and perinuclear ER in the log phase (Fig. 1, A and B). When cells entered diauxic shift (DS), characterized by growth rate decline, LDs enlarged and concentrated on perinuclear ER and the junctions between perinuclear ER and vacuoles. After day 1 (D1), when vacuoles enlarge and CFP-Pho8 signals dispersed in the lumen rather than vacuolar membranes, LDs gradually moved from the ER to vacuoles and appeared to sink into the vacuole membranes, indicative of a close contact. Starting from D3, cumulative LDs were found inside the vacuole lumen and moved in random patterns. EM results further indicated that the majority of LDs in the vacuole lumen were enclosed by membranes (Fig. 1 C), explaining why the internal Erg6-mCherry signals were not readily accessible to vacuole hydrolases. The nature of the membrane is unknown, except for the fact that it seems to be more resistant to vacuolar hydrolases. Curiously, the internal vesicles containing LDs in the vacuole lumen resemble the autophagic bodies seen when vacuolar hydrolase activities are compromised (Takeshige et al., 1992).
Figure 1.
The dynamics of LDs during transition to stat-phase. (A) Cells expressing the indicated proteins grown in SC medium to log phase, diauxic shift (DS), or the indicated days after log phase (D1–D7) were subjected to fluorescence microscopy. DIC, differential interference contrast. Bar, 5 µm. (B) Quantification of data in A for the indicated localization patterns from three independent experiments was plotted as mean ± SD. (C) The thin-sectioned EM pictures of wild-type cells grown in SC medium to D4. Va, vacuole. Asterisks show LDs. Bars, 0.5 µm. Yellow arrows indicate the LDs in the vacuole lumen. The LDs (n = 96) inside the vacuole lumen with or without outer membranes as depicted were quantified, and the percentage is shown.
The LD entry into vacuoles is mediated through microautophagy
To monitor autophagy, we examined LD distribution by staining vacuoles with the lipophilic dye FM4-64 (Vida and Emr, 1995) at DS and followed LDs with BODIPY staining (Fig. S1 A and Video 1), in comparison with cells expressing the bulk autophagy marker GFP-Pho8Δ60 (Klionsky, 2007), a cytosolic version of truncated Pho8, and the peroxisome marker GFP-SKL (Fig. S1, B and C; Monosov et al., 1996). We found that free GFP that is resistant to vacuolar proteases accumulated when cells were grown into D1 and reached the plateau around D5 (Fig. S1 C). Similarly, free GFP accumulation was also seen when the ER-phagy marker GFP-Elo3 (Fig. S1 D), the mitophagy marker Om45-GFP (Fig. S1 E), and the phagophore marker GFP-Atg8 (Fig. S1 D) were monitored, confirming that multiple forms of autophagy were activated during stat-phase.
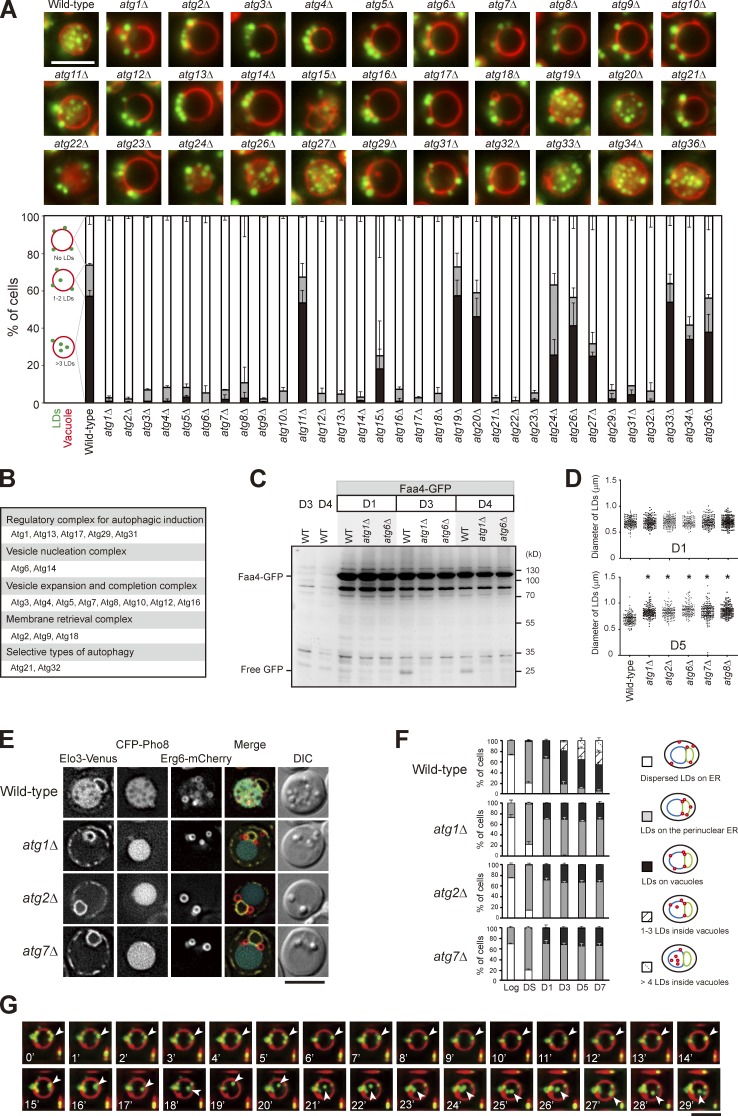
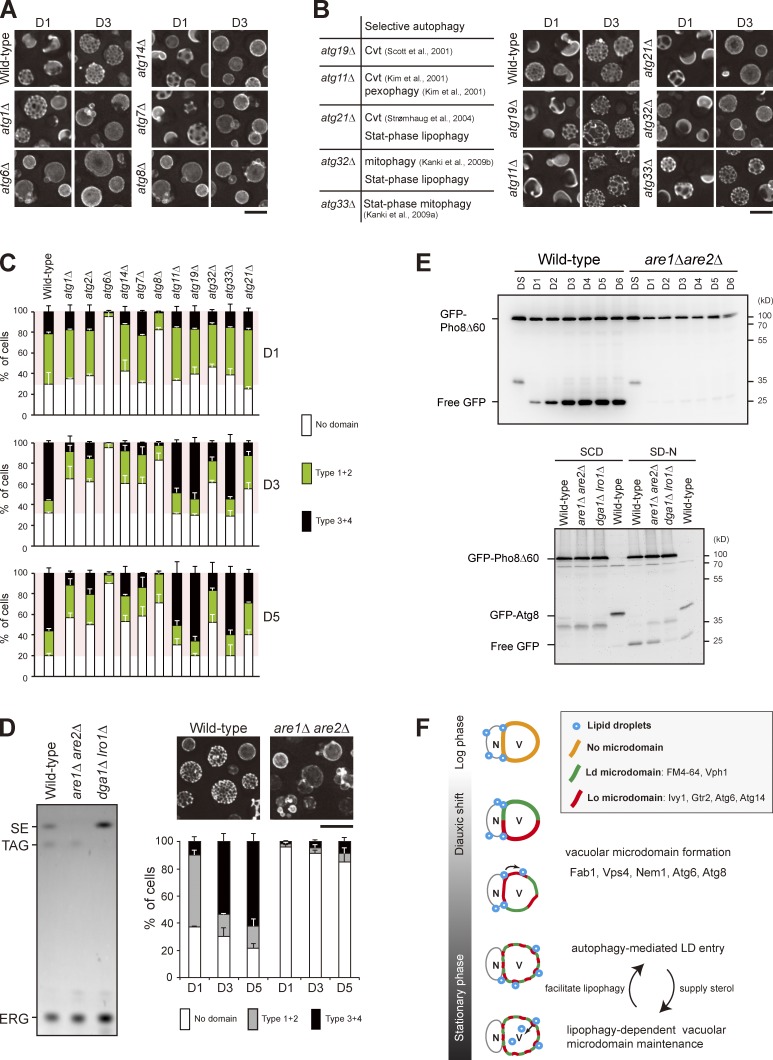
We next analyzed the entire set of atg mutants and found that the LD entry into vacuoles requires all of the core ATG genes (Fig. 2, A and B; Xie and Klionsky, 2007; Mizushima et al., 2011). In addition, wild type, but not atg1Δ and atg6Δ, expressing the LD marker Faa4-GFP accumulated free GFP after D3 (Fig. 2 C), supporting an autophagy-dependent LD breakdown, termed “stat-phase lipophagy.” Interestingly, LDs in the atg mutants appeared larger (Fig. 2 D), indicative of aberrant neutral lipid mobilization. Besides, cells lacking Atg21 and Atg32, but not Atg11 (Kim et al., 2001), Atg19 (Scott et al., 2001), Atg20 (Nice et al., 2002), and Atg33 (Kanki et al., 2009a), for selective autophagy blocked stat-phase lipophagy (Fig. 2, A and B). Although how Atg21, a protein specific for cytoplasm to vacuole–targeting pathway (Strømhaug et al., 2004), and Atg32, a mitochondria protein that initiates mitophagy (Kanki et al., 2009b), act during stat-phase lipophagy remains unclear, it seems likely that stat-phase lipophagy is a selective form of autophagy.
Figure 2.
Translocation of LDs into the vacuole lumen is mediated by a microautophagy mechanism. (A) Representative images of LDs and vacuoles in the indicated strains grown to D5. Quantification of localization based on the indicated patterns. Three independent data were plotted as mean ± SD. (B) Summary of A based on their roles in autophagy. (C) Wild type (WT) and various strains as indicated expressing Faa4-GFP were grown in SC medium to the indicated growth conditions. Cells were lysed, and the lysates were analyzed by immunoblotting with the anti-GFP antibody. (D) The comparison of LD diameter in the indicated strains grown to D1 and D5. The data shown are from one experiment (n = 100 for each cell type). *, P < 0.01. (E) Cells expressing the indicated proteins were grown in SC medium to D5 and imaged by fluorescence microscopy. DIC, differential interference contrast. (F) Quantification of data in E for the indicated localization patterns from three independent experiments, which were plotted as mean ± SD. (G) Wild-type cells stained with FM4-64 (vacuole) at DS and BODIPY (LDs) on D3 were subjected to time-lapse fluorescence microscopy. Images were taken every 1 min. Arrowheads denote an LD during its translocation into the vacuole lumen. Bars, 5 µm.
By following Erg6-mCherry, we found that LDs in core atg mutants tend to stay with perinuclear ER, and many localized to the border between perinuclear ER and vacuoles (Fig. 2, E and F), suggesting a defect in lateral movement. We suspect that stat-phase lipophagy may use a microautophagy mechanism, similar to the nitrogen starvation-induced lipophagy described recently in yeast (van Zutphen et al., 2014). By time-lapse microscopy, we found that LDs entered vacuole lumen through the vacuole surface in stat-phase, and the internalization process often completed within a few minutes (Fig. 2 G). However, unlike the previous study, FM4-64–containing lipids in our analyses did not seem to move along with LDs during translocation into the vacuole lumen (Fig. 2 G), and the membrane-enclosed LDs (Fig. 1 C) are distinct findings. Thus, it seems likely that yeast has more than one type of microlipophagy that are operated by distinct mechanisms.
Stat-phase lipophagy involves the vacuolar microdomains
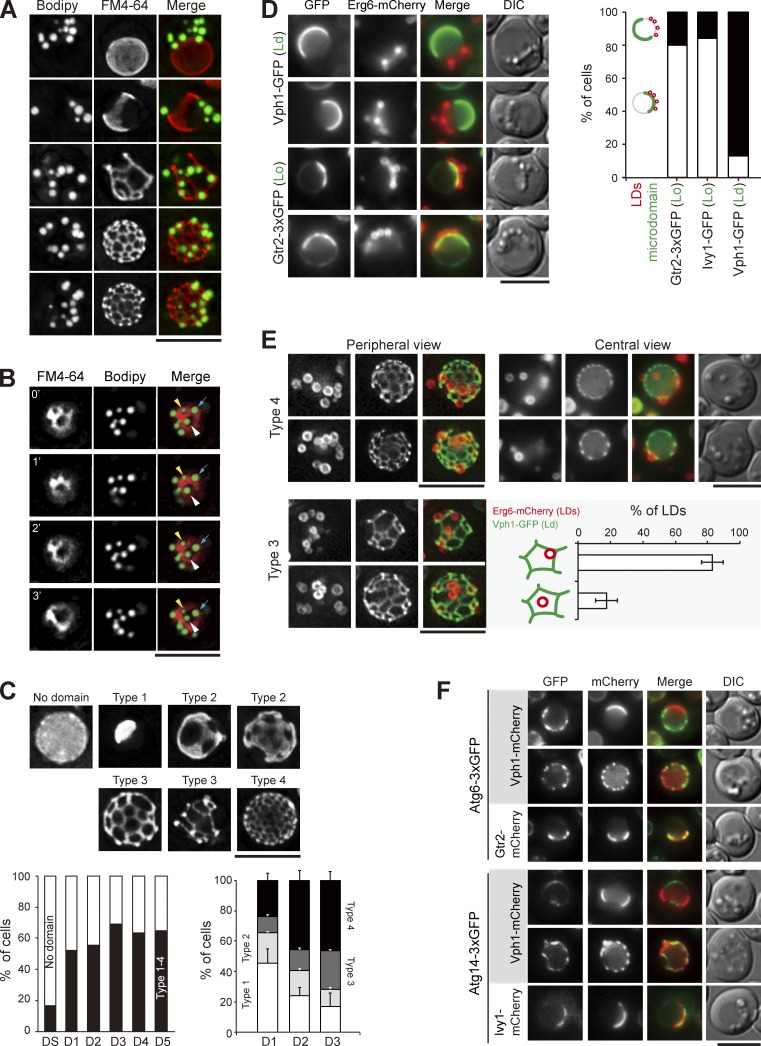
We suspect that a portion of the vacuolar membrane is internalized together with LDs into vacuoles during stat-phase lipophagy. To decipher the relationship between the FM4-64–containing lipids and LDs, we imaged the periphery of vacuoles in wild-type cells during stat-phase. Strikingly, LDs only associated with the regions devoid of FM4-64 signals (Fig. 3 A). Recent evidence suggests that stable vacuolar microdomains form when yeast cells reach stat-phase and that the FM4-64 staining displays a pattern of a liquid-disordered (Ld) microdomain (Toulmay and Prinz, 2013). Consistent with the idea that the microdomains are stable structures, vacuole-associated LDs were not mobile in stat-phase. In some cases, when the FM4-64 network evolved, LDs moved but remained in areas lacking the FM4-64 signals (Fig. 3 B).
Figure 3.
LDs associate with the sterol-enriched vacuolar microdomain Lo. (A) Wild-type cells stained with FM4-64 (vacuole) and BODIPY (LDs) were imaged by fluorescence microscopy. Images were processed by deconvolution followed by maximal projection. (B) Same as A, except that the images were acquired by a time interval of every 1 min. The yellow and white arrowheads and blue arrows define three independent LDs. (C) Cells expressing Vph1-mCherry were imaged and processed as in A. Representative images of various Vph1-mCherry patterns are shown. The plots show quantification of these patterns under various growth conditions. (left) The data shown are from a single representative experiment (n > 200 for each indicated condition) out of three repeats. (right) The data shown are from three independent experiments and plotted as mean ± SD. (D) Cells expressing Erg6-mCherry (LDs) and microdomain (Ld or Lo) markers as indicated were imaged by fluorescence microscopy on D1. LDs and microdomain patterns as indicated were quantified and plotted. The data shown are from a single representative experiment (n > 60 for each cell type) out of three repeats. (E) Cells expressing Erg6-mCherry and Vph1-GFP were imaged for periphery and center on D3. The localization of types 3 and 4 of Vph1-GFP patterns were compared. The LDs in type 3 images were further quantified based on the two indicated patterns from three independent experiments. Error bars show mean ± SEM. (F) Cells expressing proteins as indicated were imaged by fluorescence microscopy. DIC, differential interference contrast. Bars, 5 µm.
To understand how LDs undergo lateral movement from the ER to vacuoles with respect to vacuolar microdomain formation, we imaged LDs and proteins previously known to partition into distinct vacuolar microdomains. Quantification results indicated that the vacuolar microdomains formed after DS and reached the plateau on D1 (Fig. 3 C), in agreement with the previous findings (Toulmay and Prinz, 2013). Furthermore, the Ld microdomain marker Vph1-mCherry appeared mostly as sheets (type 1 and 2) on D1 and gradually evolved into reticular network (type 3 and 4) on D3 (Fig. 3 C). Interestingly, LDs on D1 often associated with the hemisphere labeled with the Lo microdomain markers Ivy1-GFP and Gtr2-GFP but not with Vph1-GFP (Ld; Fig. 3 D). As D1 is the time when LDs start to travel from ER to vacuoles, LDs may interact with the forming Lo microdomain during the passage via the ER/vacuole junctions. By imaging Erg6-mCherry and Vph1-GFP at the vacuolar periphery and center on D3 when stat-phase lipophagy occurs, we found that most LDs on vacuoles were bound by the Vph1-GFP reticular network (Fig. 3 E). When the type 3 microdomain with larger mesh size was used to quantify the distribution of LDs unambiguously, we found that LDs preferentially associated with Lo next to the three-way junctions of the Ld reticular network (Fig. 3 E). Thus, the forming Ld microdomain may create a boundary to restrict LDs, thereby preventing LD retrieval to the ER.
The phosphatidylinositol (PtdIns)-3 kinase complex I consisting of Atg6, Atg14, Vps15, and Vps34 nucleates the engulfment structures during autophagy (Kihara et al., 2001; Cao and Klionsky, 2007). Interestingly, Atg6 and Atg14 enriched at the preautophagosomal structure in log phase (Fogel et al., 2013) appeared as highly dynamic structures localized within the Lo (Gtr2 and Ivy1), but not Ld (Vph1), vacuolar microdomain during stat-phase (Fig. 3 F). As LDs associate with Lo microdomain, the PtdIns-3 kinase complex I may nucleate phagophore formation during stat-phase lipophagy by modifying the Lo microdomains. The majority of Atg proteins did not show stable association with, although they were occasionally found around, LDs (Fig. S2). Detailed mechanisms for stat-phase lipophagy await further analyses to determine.
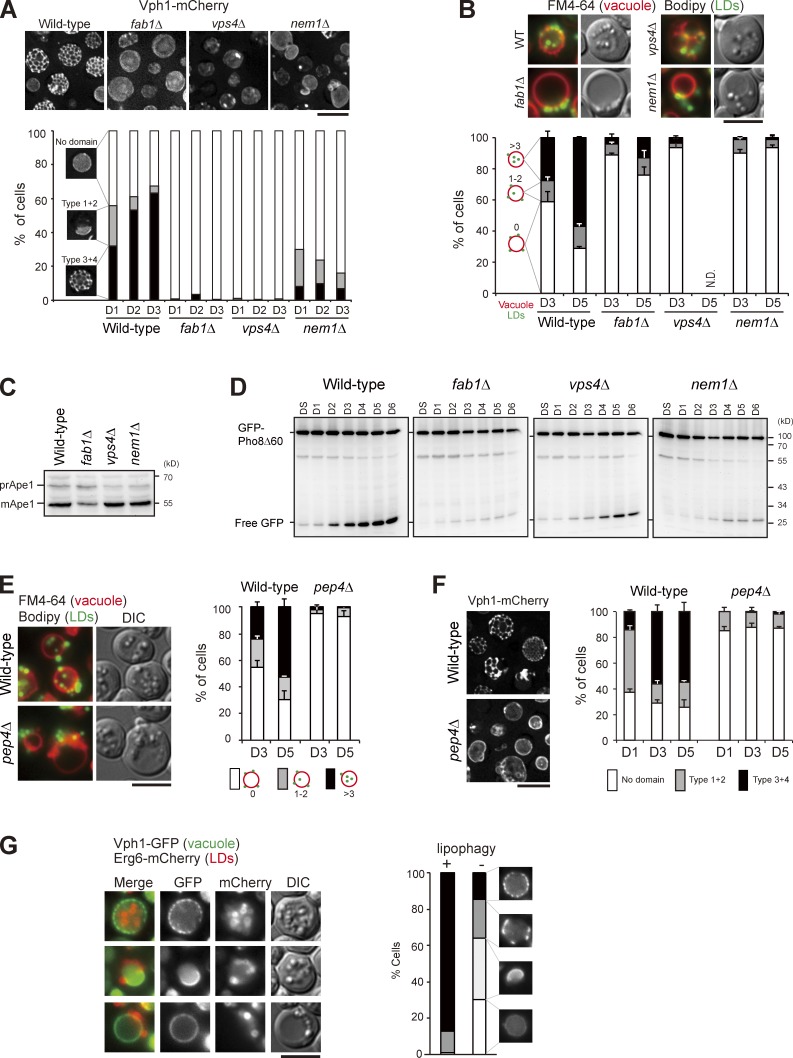
We next examined the vacuolar microdomain-deficient mutants fab1Δ, vps4Δ, and nem1Δ (Fig. 4 A; Toulmay and Prinz, 2013) and found that stat-phase lipophagy was blocked in all of them (Fig. 4 B). The vacuolar aminopeptidase Ape1 accumulated mostly as the mature form during nitrogen starvation (Fig. 4 C), indicative of functional autophagy in the mutants. In addition, free GFP accumulated in the domain-deficient mutants expressing GFP-Pho8Δ60 during stat-phase (Fig. 4 D), albeit at a reduced level in fab1Δ, ruling out an autophagy defect. Cells lacking the vacuolar protease Pep4 accumulated autophagic bodies inside the vacuole lumen (Takeshige et al., 1992). However, the pep4Δ cells blocked stat-phase lipophagy at the internalization step, rather than accumulated LD-containing autophagic bodies inside the vacuole lumen (Fig. 4 E). Interestingly, pep4Δ cells did not form vacuolar microdomains during stat-phase (Fig. 4 F), consistent with the notion that vacuolar microdomains are needed for stat-phase lipophagy. It would be interesting to examine whether other types of microautophagy, such as micropexophagy in Pichia pastoris (Farré and Subramani, 2004) that requires Pep4, also involve vacuolar microdomain formation.
Figure 4.
Vacuolar microdomain organization is crucial for stat-phase lipophagy. (A) Cells as indicated expressing Vph1-mCherry were imaged by fluorescence microscopy, and data were quantified based on the three indicated patterns. The data shown are from a single representative experiment (n > 200 for each condition) out of three repeats. (B) Cells as indicated stained with FM4-64 (vacuole) at DS and BODIPY (LDs) on D3 were imaged by fluorescence microscopy. Data were quantified based on the indicated patterns and plotted as mean ± SD from three independent experiments. WT, wild type. (C) Cells as indicated grown in SC medium were shifted to SD-N for 3 h. Cells were lysed, and the lysates were analyzed by immunoblotting with the anti-Ape1 antibody. pr, precursor; m, mature. (D) Cells expressing GFP-Pho8Δ60 under growth conditions as indicated were lysed, and the lysates were analyzed by immunoblotting with the anti-GFP antibody. (E) Same as B, except that wild type and pep4Δ were compared. (F) Same as A, except that wild type and pep4Δ were compared. Error bars show mean ± SEM from three experiments. (G) Cells expressing Vph1-GFP and Erg6-mCherry were imaged by fluorescence microscopy on D4. The indicated patterns of Vph1-GFP were quantified in cells with (+) and without (−) Erg6-mCherry inside vacuole lumen (lipophagy). The data shown are from a single representative experiment (n > 80 for each condition) out of three repeats. DIC, differential interference contrast. Bars, 5 µm.
The pattern of vacuolar Ld microdomains evolved from sheets to tubules during stat-phase. We next asked whether the cells undergoing extensive stat-phase lipophagy might be different from those containing no LDs inside the vacuole lumen. Interestingly, stat-phase lipophagy was often seen in the population of cells showing the reticular Vph1-GFP pattern (Fig. 4 G). In contrast, the population of cells lacking LDs in vacuoles showed either no microdomains or sheet-like Vph1-GFP patterns (Fig. 4 G). Thus, the reticular microdomain correlates well with stat-phase lipophagy. Although ∼25% of cells on D1 already exhibited the reticular microdomains, LDs were not engulfed by vacuoles until D3, suggesting that microdomains are necessary but probably not sufficient for stat-phase lipophagy. In addition, autophagy activation by rapamycin was not sufficient to induce microdomain formation and lipophagy (Fig. S3 A). Cells treated with oleate, a condition known to activate triacylglycerol but inhibit sterol ester synthesis (Connerth et al., 2010), accumulated LDs without inducing vacuolar domain formation and lipophagy (Fig. S3 B). Intriguingly, higher temperature induced the reticular microdomain formation but not LD translocation (Fig. S3 C). Thus, neither vacuolar microdomains nor autophagy is sufficient to activate LD translocation. Some unidentified signals are likely also required to activate stat-phase lipophagy.
Proper organization of vacuolar microdomains requires stat-phase lipophagy
The neutral lipids stored within LDs might be mobilized to support viability and/or to maintain membrane integrity during stat-phase. A previous study shows that autophagy is not needed for the establishment of vacuolar microdomains (Toulmay and Prinz, 2013). Indeed, the majority of core atg mutants, including atg1Δ, atg2Δ, atg7Δ, and atg14Δ, exhibited normal vacuolar microdomain patterns on D1 (Fig. 5, A and C). Strikingly, these mutants gradually lost their microdomains and showed very little if any microdomains after D3, indicating that autophagy is needed for vacuolar microdomain maintenance during stat-phase. Intriguingly, atg6Δ and atg8Δ mutants did not show microdomains even on D1 (Fig. 5, A and C). The defect in atg6Δ is likely caused by the compromised Atg6-containing PtdIns-3 kinase complex II activity for vacuolar protein sorting (Kihara et al., 2001). The defect in atg8Δ is probably unrelated to Atg8 lipidation because atg7Δ that blocked Atg8 lipidation (Ichimura et al., 2000) did not display the same phenotype.
Figure 5.
Stat-phase lipophagy is needed for vacuolar microdomain maintenance. (A) Cells as indicated expressing Vph1-mCherry were imaged by fluorescence microscopy on D1 and D3. (B, left) A table lists selective autophagy defects for the indicated strains. Cvt, cytoplasm-to-vacuole targeting. (right) Same as A. (C) Cells as indicated were quantified based on the Vph1-mCherry patterns and culture conditions as indicated. Data from three independent experiments were plotted as mean ± SD. (D, left) Lipids in strains as indicated were analyzed by TLC. SE, sterol ester; TAG, triacylglycerol; ERG, ergosterol. (right) Wild-type and are1Δ are2Δ cells expressing Vph1-mCherry were imaged by fluorescence microscopy, and three independent data were quantified. Error bars show mean ± SEM from three experiments. (E, top) Cells as indicated expressing GFP-Pho8Δ60 were grown in SC medium to various growth conditions. Cells were lysed, and the lysates were analyzed by immunoblotting with the anti-GFP antibody. (bottom) Same as top, except that cells expressing either GFP-Pho8Δ60 or GFP-Atg8 were grown in SC medium to log phase (SCD) or shifted to SD-N for 3 h from log phase. (F) Working model for stat-phase lipophagy. N, nucleus; V, vacuole. During DS, vacuolar proteins start to sort into either one of the vacuolar microdomains, Ld or Lo. Several proteins, Fab1, Vps4, Nem1, Atg6, and Atg8, are needed for microdomain formation at the stage. The pattern of quasisymmetrical microdomains appears later in the stationary phase. During this stage, LDs are engulfed by the Lo microdomain and feed vacuoles with sterols to maintain lipid phase partitioning, which in turn stimulates stat-phase lipophagy. Bars, 5 µm.
Among the selective autophagy mutants, only atg21Δ and atg32Δ for stat-phase lipophagy displayed aberrant vacuolar microdomains on D3 and D5 (Fig. 5, B and C). Given that the Lo microdomain is enriched in sterol, we asked whether are1Δ are2Δ, which fails to synthesize sterol esters (Zweytick et al., 2000), affects vacuolar microdomain formation. Unlike dga1Δ lro1Δ deficient in triacylglycerol synthesis, which died quickly in transition to stat-phase, are1Δ are2Δ grew normally but did not exhibit vacuolar microdomains (Fig. 5 D). Interestingly, the processing of GFP-Pho8Δ60 into free GFP in are1Δ are2Δ cells was defected during stat-phase but not during nitrogen starvation (Fig. 5 E). Thus, these results imply that storage sterol is crucial for vacuolar microdomain formation/maintenance and for autophagy during stat-phase.
In this study, we find that LDs require vacuolar microdomains for stable docking and for subsequent engulfment by vacuoles (Fig. 3 and Fig. 4). We propose that LDs on the perinuclear ER interact with the forming sterol-enriched Lo microdomains for lateral movement toward vacuoles, whereas the Ld microdomain constrains LDs onto vacuoles. (Fig. 5 F). In the absence of intact vacuolar microdomains, LDs fail to associate with vacuoles stably and therefore stay with the ER. However, we cannot exclude the possibility that retention of LDs on the ER is related to altered membrane lipid metabolism, as mutants, such as nem1Δ, defective in phosphatidic acid homeostasis displayed aberrant ER morphology (Siniossoglou et al., 1998). It is conceivable that lipid phase partitioning during vacuolar microdomain formation may change composition or distribution of protein complexes that attribute to the establishment of contact sites. LDs enlarge during the transition to stat-phase, and their contents are important for survival and for membrane maintenance during stat-phase. One such physiological relevance for stat-phase lipophagy is to maintain vacuolar microdomain integrity, probably via sterol catabolism, which in turn stimulates lipophagy (Fig. 5 F). As the vacuolar microdomains are likely modified by autophagy machinery to confer stat-phase lipophagy, the integrity of vacuolar domains probably works to monitor cellular conditions, perhaps helping cells to efficiently recycle cellular contents, including LDs, to survive in chronological aging or to extend life span.
Materials and methods
Strains, growth conditions, and reagents
Strains and plasmids used in this study are described in Table S1 and Table S2. Cells were grown at 30°C in synthetic complete (SC) medium (0.67% yeast nitrogen base, amino acids, and 2% glucose). The log phase culture is when cells were grown to OD = 0.6 starting from OD = 0.15. The DS stage is when the same cultures of log phase were grown for another 3 h to OD = ∼1.7. The D1, 2, 3, 4, 5, 6, and 7 are defined as when the same cultures of log phase were grown at 30°C for additional 24, 48, 72, 96, 120, 144, and 168 h, respectively. To monitor autophagy, cells grown in SC medium at 30°C were shifted to SD-N (0.17% yeast nitrogen base and 2% glucose) for 3 h. BODIPY 493/503 and FM4-64 were purchased from Invitrogen, and all other reagents were obtained from Sigma-Aldrich. The anti-Ape1 (Saccharomyces cerevisiae) antibody was obtained from D. Klionsky (University of Michigan, Ann Arbor, MI). The homemade anti-GFP (Aequorea victoria) antibody was a gift from Y.-C. Chang (National Taiwan University, Taipei, Taiwan) followed by affinity purification in our laboratory. We also used an anti-GFP antibody (ab290) purchased from Abcam.
Fluorescence microscopy
Cells for microscopy were either prepared in SC medium or in 50 mM Tris-HCl, pH 7.5. The images were acquired at 30oC by an imaging system (DeltaVision; Applied Precision) with a Plan Apochromat 100× objective lens, NA 1.4, and a charge-coupled device camera (CoolSNAP HQ2; Photometrics) controlled by softWoRx suite (Applied Precision). The filters used with the DeltaVision system were GFP (525/50 nm), mCherry (632/60 nm) YFP (559/38 nm), and CFP (470/24 nm). To image the microdomains of vacuoles, cells were imaged from the periphery by taking nine optical section images spaced at 0.3 µm. Images were further processed by deconvolution and maximal projection by softWoRx suite. To image LDs entering vacuoles by 4D (3D time lapse), we used the DeltaVision system equipped with a 512 × 512–pixel electron-multiplying charge-coupled device camera (Evolve 512; Photometrics) or CoolSNAP HQ2 charge-coupled device camera controlled by softWoRx suite.
EM
Yeast cells grown in SC medium for 4 d were harvested and subjected to EM as described previously (Wang and Lee, 2012). In brief, cells were fixed by the high pressure freezing system (EM HPM100; Leica) followed by freeze substitution by EM AFS2 (Leica). Cells were then embedded within LR gold resin. Ultrathin sections of 70–90 nm were stained with 5% uranyl acetate and 0.5% lead citrate. EM was performed by a microscope (Tecnai G2 Spirit TWIN; FEI Company) equipped with a charge-coupled device camera (Orius; Gatan), and images were processed by Photoshop (Adobe).
Screening of atg mutants for LD entry
3 ml culture grown in SC medium to DS was centrifuged, and the resulting cell pellet was resuspended in 120 µl of used SC medium. 3 µl of 16-mM FM4-64 was then added to stain vacuoles in the dark for 20 min at 30°C. Cells were washed twice with sterile water, and the rest of the used SC medium was added back to resuspend the cells. When cells were grown to D5, 300 µl culture was added with 3 µl BODIPY 494/503 (0.5 µg/ml) to stain LDs in the dark for 10 min, washed twice with 50 mM Tris-HCl, pH 7.5, and immediately subjected to fluorescence microscopy.
Immunoblotting
At the indicated time points, 1 ml culture was treated with a final concentration of 10% TCA on ice for 30 min, washed with cold acetone, and resuspended in 100 µl MURB (50 mM sodium phosphate, 25 mM MES, pH 7.0, 1% SDS, 3 M urea, and 5% β-mercaptoethanol). Glass beads were then added and vortexed to break the cells. Samples were heated to 55°C for 10 min before subjecting to SDS-PAGE followed by immunoblot analyses. The signal acquisition, processing, and quantification were performed by use of the imager (ChemiDoc-It; UVP) and ChemiDoc-It software.
Lipid analysis
Lipids were prepared as described previously (Folch et al., 1957). In brief, cells were resuspended in chloroform/methanol (2:1) and lysed with glass beads. A salt solution was added to separate lipids into organic phase. Lipids were then dried by a centrifuge (SpeedVac; Thermo Fisher Scientific) and dissolved in chloroform/methanol (2:1). For TLC analysis of neutral lipids, lipids were applied to a silica gel 60 plate as previously described (Leber et al., 1994) using the solvent systems of light petroleum/diethyl ether/acetic acid (25:25:1) followed by light petroleum/diethyl ether (49:1). Lipids were stained by 0.5% MnCl2, 3.2% H2SO4, and 48% methanol and heated at 170°C for ∼4 min. TLC plates were scanned by the ChemiDoc-It imager and software.
Online supplemental material
Fig. S1 shows that various forms of autophagy were activated during stat-phase. Fig. S2 shows the localization of various fluorescence-tagged Atg proteins in log and stat-phase. Fig. S3 shows that rapamycin, oleate, and higher temperature treatments did not stimulate LD entry into vacuoles. Table S1 lists strains used in this study. Table S2 lists plasmids used in this study. Video 1 provides time-lapse and z-section videos for LD dynamics of wild-type yeast cells in various growth conditions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201404115/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Rey-Huei Chen (Institute of Microbial Biology [IMB], Academia Sinica) for sharing laboratory resources and reading our manuscript, Miss Su-Ping Lee and Su-Ping Tsai (IMB, Academia Sinica) for transmission EM, and Miss Mei-Jane Fang (Institute of Plant and Microbial Biology, Academia Sinica) for advice on live imaging.
This study was supported by an intramural fund from Academia Sinica and a grant (NSC 101-2311-B-001-028-MY3) from the Ministry of Science and Technology, Taiwan.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Atg
- autophagy related
- DS
- diauxic shift
- LD
- lipid droplet
- Ld
- liquid disordered
- Lo
- liquid ordered
- PtdIns
- phosphatidylinositol
- SC
- synthetic complete
References
- Cao, Y., and Klionsky D.J.. 2007. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 17:839–849 10.1038/cr.2007.78 [DOI] [PubMed] [Google Scholar]
- Connerth, M., Czabany T., Wagner A., Zellnig G., Leitner E., Steyrer E., and Daum G.. 2010. Oleate inhibits steryl ester synthesis and causes liposensitivity in yeast. J. Biol. Chem. 285:26832–26841 10.1074/jbc.M110.122085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducharme, N.A., and Bickel P.E.. 2008. Lipid droplets in lipogenesis and lipolysis. Endocrinology. 149:942–949 10.1210/en.2007-1713 [DOI] [PubMed] [Google Scholar]
- Farré, J.C., and Subramani S.. 2004. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol. 14:515–523 10.1016/j.tcb.2004.07.014 [DOI] [PubMed] [Google Scholar]
- Fogel, A.I., Dlouhy B.J., Wang C., Ryu S.W., Neutzner A., Hasson S.A., Sideris D.P., Abeliovich H., and Youle R.J.. 2013. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol. Cell. Biol. 33:3675–3688 10.1128/MCB.00079-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch, J., Lees M., and Sloane Stanley G.H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497–509 [PubMed] [Google Scholar]
- Ichimura, Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., et al. . 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408:488–492 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Kanki, T., Wang K., Baba M., Bartholomew C.R., Lynch-Day M.A., Du Z., Geng J., Mao K., Yang Z., Yen W.L., and Klionsky D.J.. 2009a. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell. 20:4730–4738 10.1091/mbc.E09-03-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki, T., Wang K., Cao Y., Baba M., and Klionsky D.J.. 2009b. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 17:98–109 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassan, A., Herms A., Fernández-Vidal A., Bosch M., Schieber N.L., Reddy B.J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. . 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203:985–1001 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, A., Noda T., Ishihara N., and Ohsumi Y.. 2001. Two distinct Vps34 phosphatidylinositol 3–kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152:519–530 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kamada Y., Stromhaug P.E., Guan J., Hefner-Gravink A., Baba M., Scott S.V., Ohsumi Y., Dunn W.A. Jr, and Klionsky D.J.. 2001. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153:381–396 10.1083/jcb.153.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J.2007. Monitoring autophagy in yeast: the Pho8Delta60 assay. Methods Mol. Biol. 390:363–371 10.1007/978-1-59745-466-7_24 [DOI] [PubMed] [Google Scholar]
- Leber, R., Zinser E., Zellnig G., Paltauf F., and Daum G.. 1994. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast. 10:1421–1428 10.1002/yea.320101105 [DOI] [PubMed] [Google Scholar]
- Liu, P., Bartz R., Zehmer J.K., Ying Y., and Anderson R.G.. 2008. Rab-regulated membrane traffic between adiposomes and multiple endomembrane systems. Methods Enzymol. 439:327–337 10.1016/S0076-6879(07)00424-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S., and Parton R.G.. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378 10.1038/nrm1912 [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Yoshimori T., and Ohsumi Y.. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Monosov, E.Z., Wenzel T.J., Lüers G.H., Heyman J.A., and Subramani S.. 1996. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J. Histochem. Cytochem. 44:581–589 10.1177/44.6.8666743 [DOI] [PubMed] [Google Scholar]
- Nice, D.C., Sato T.K., Stromhaug P.E., Emr S.D., and Klionsky D.J.. 2002. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277:30198–30207 10.1074/jbc.M204736200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, A., Gross S.P., and Parton R.G.. 2014. Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 204:635–646 10.1083/jcb.201311051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu, J., Ha C.W., Zhang S., Jung J.P., Huh W.K., and Liu P.. 2011. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell. 2:487–496 10.1007/s13238-011-1061-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S.V., Guan J., Hutchins M.U., Kim J., and Klionsky D.J.. 2001. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 7:1131–1141 10.1016/S1097-2765(01)00263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., and Cuervo A.M.. 2012. Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012:1–12 10.1155/2012/282041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., and Czaja M.J.. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., Santos-Rosa H., Rappsilber J., Mann M., and Hurt E.. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17:6449–6464 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strømhaug, P.E., Reggiori F., Guan J., Wang C.W., and Klionsky D.J.. 2004. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 15:3553–3566 10.1091/mbc.E04-02-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige, K., Baba M., Tsuboi S., Noda T., and Ohsumi Y.. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301–311 10.1083/jcb.119.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay, A., and Prinz W.A.. 2013. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 202:35–44 10.1083/jcb.201301039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutphen, T., Todde V., de Boer R., Kreim M., Hofbauer H.F., Wolinski H., Veenhuis M., van der Klei I.J., and Kohlwein S.D.. 2014. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 25:290–301 10.1091/mbc.E13-08-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida, T.A., and Emr S.D.. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., and Farese R.V. Jr. 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81:687–714 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.W., and Lee S.C.. 2012. The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J. Cell Sci. 125:2930–2939 10.1242/jcs.100230 [DOI] [PubMed] [Google Scholar]
- Werner-Washburne, M., Braun E., Johnston G.C., and Singer R.A.. 1993. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57:383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., and Klionsky D.J.. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Yang, H., Galea A., Sytnyk V., and Crossley M.. 2012. Controlling the size of lipid droplets: lipid and protein factors. Curr. Opin. Cell Biol. 24:509–516 10.1016/j.ceb.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Zweytick, D., Leitner E., Kohlwein S.D., Yu C., Rothblatt J., and Daum G.. 2000. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 267:1075–1082 10.1046/j.1432-1327.2000.01103.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.