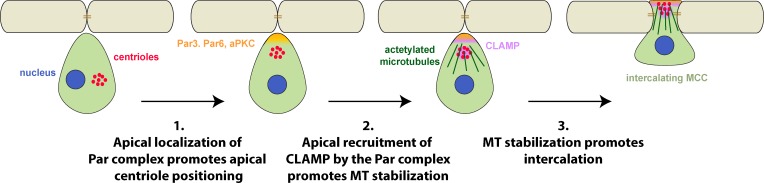
During radial intercalation of epithelial cells, Par3 and aPKC promote the apical positioning of centrioles, whereas CLAMP stabilizes microtubules along the axis of migration.
Abstract
The directed movement of cells is critical for numerous developmental and disease processes. A developmentally reiterated form of migration is radial intercalation; the process by which cells move in a direction orthogonal to the plane of the tissue from an inner layer to an outer layer. We use the radial intercalation of cells into the skin of Xenopus laevis embryos as a model to study directed cell migration within an epithelial tissue. We identify a novel function for both the microtubule-binding protein CLAMP and members of the microtubule-regulating Par complex during intercalation. Specifically, we show that Par3 and aPKC promote the apical positioning of centrioles, whereas CLAMP stabilizes microtubules along the axis of migration. We propose a model in which the Par complex defines the orientation of apical migration during intercalation and in which subcellular localization of CLAMP promotes the establishment of an axis of microtubule stability required for the active migration of cells into the outer epithelium.
Introduction
The directed movement of cells is a fundamental aspect of tissue morphogenesis. Cells migrating in vivo often must invade tissue barriers and ultimately form new cellular contacts in their target tissues. Furthermore, in many epithelial tissues proliferation occurs via a population of basal stem cells that divide below the apical surface and move apically to join the epithelium (Rock and Hogan, 2011). This epithelialization requires that cells move in a specific direction and that they incorporate into the epithelium without disrupting its integrity.
The skin of Xenopus laevis embryos represents an excellent model to address the invasive behavior of migrating cells (Fig. 1 A). Multiciliated cells (MCCs) and ionocytes (ICs) are specialized cell types that differentiate in a subapical layer of the epidermis (Drysdale and Elinson, 1992; Deblandre et al., 1999; Dubaissi and Papalopulu, 2011; Quigley et al., 2011; Kim et al., 2012). These cells then move in a directed manner toward the outer epithelial cells (OCs; Fig. 1 A). As they meet the apical junctions of the OCs, intercalating cells selectively localize to vertices consisting of three or more cells (Stubbs et al., 2006). Next, these cells undergo radial intercalation where they push through the vertices and form new apical junctions with the OCs, forming a complete intact epithelium (Fig. 1 A).
Figure 1.
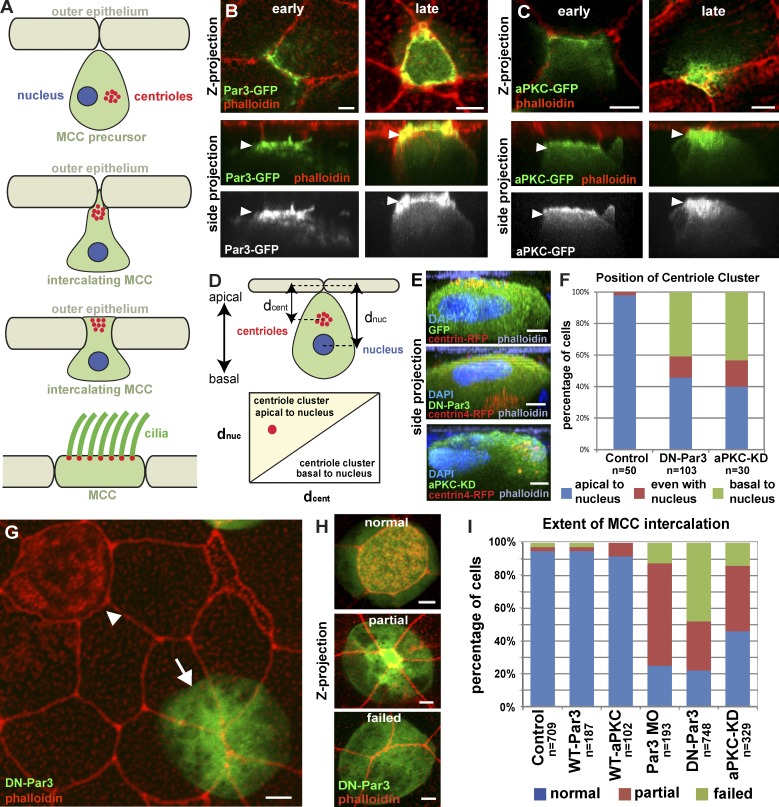
Apical positioning of centrioles and intercalation requires the Par complex. (A) Schematic representation of MCC intercalation. (B and C) Apical localization (arrowheads) of Par3-GFP (B) and aPKC-GFP (C) at different stages of intercalation (early and late). (D) Schematic of the quantification of centriole positioning in MCCs. (E and F) Representative z stack cross-section images (E) and quantification (F) of centriole position in control, DN-Par3, and aPKC-KD MCCs. (G) Mosaic image showing wild-type MCC that has intercalated (arrowhead) and a DN-PAR3-GFP (arrow) MCC that failed to intercalate. (H) Images illustrating the three different phenotypes used for scoring MCC intercalation. (I) Quantification of intercalation in MCCs expressing GFP, Par3-GFP, aPKC-GFP, Par-3 MO, DN-Par3-GFP, or aPKC-KD-GFP. For all experiments, cells from a total of at least five embryos from at least two independent experiments were quantified unless specified otherwise. Quantification in F is based on at least 10 cells each from a total of at least five embryos from at least two independent experiments. Quantifications of DN-Par3 and aPKC-KD phenotypes in F and I are statically significantly different from controls (P < 0.0001, see Table S1). Side projection refers to side views of projection along the x axis in all figures. See Fig. S1. Bars, 5 µm.
The Par complex is involved in a wide variety of cellular processes including the establishment of apical-basal polarity, the formation of adherens junctions, and directed cell migration (Chen and Macara, 2005; Chihara and Nance, 2012; Tepass, 2012). To accomplish these functions, the core proteins Par3, Par6, and the atypical PKC (aPKC) interact with an assortment of effector proteins (Goldstein and Macara, 2007). Of particular relevance is the role of the Par complex in establishing force-generating interactions between centrioles, microtubules (MTs), and the cell cortex (Labbé et al., 2003; Munro, 2006; Feldman and Priess, 2012). In numerous cell types, the centrosome is positioned between the leading edge and the nucleus, creating an enrichment of MTs along the axis of migration (Gomes et al., 2005; Luxton and Gundersen, 2011). This is mediated by Par3’s interaction with the dynein light intermediate chain 2 (LIC2), which can modulate MT dynamics at the cell cortex (Schmoranzer et al., 2009). Additionally, in neurons Par3 has been reported to directly bind to and stabilize MTs, which suggests that the Par complex can regulate MT stability in a variety of ways (Chen et al., 2013).
MTs are essential for a wide range of biological functions, and numerous MT-associated proteins (MAPs) have context-dependent roles in regulating their stability and dynamics. The poorly characterized clamp/Spef1 gene encodes a MAP and is expressed in several tissues and cell types. Most notably, it is highly up-regulated in ciliated epithelia (Chan et al., 2005; Dougherty et al., 2005; Stubbs et al., 2008). EM studies in mice have revealed CLAMP localization to sperm flagellum, and immunofluorescence studies in mammalian cells have shown that CLAMP localizes to the MT lattice (Chan et al., 2005; Dougherty et al., 2005). Additionally, CLAMP overexpression protects MTs from depolymerization during cold shock treatment, which suggests a role for CLAMP in stabilizing MTs (Dougherty et al., 2005). The localization and overexpression analysis suggests a role in regulating MT dynamics; however, that role is unclear, as no loss-of-function analysis has been reported. Here we report an essential role for members of the Par complex and CLAMP during radial intercalation.
Results and discussion
The Par complex is required for apical positioning of centrioles and intercalation of MCCs
As MCCs undergo centriole amplification, they generate a cluster of centrioles at the level of the nucleus (Klos Dehring et al., 2013). These centrioles move apically before apical cell migration and intercalation, and will ultimately form the basal bodies of the motile cilia in mature MCCs (Fig. 1 A). The Par complex has been implicated in both the generation of apical-basal cell polarity and centriole positioning in migrating cells (Goldstein and Macara, 2007; Luxton and Gundersen, 2011). We observe that GFP fusions of Par3, aPKC, and Par6 are all positioned at the leading edge of MCCs as they initiate their apical migration (Fig. 1, B and C; and Fig. S1, early), and associate with the apical surface as the cells begin to intercalate into the outer epithelium (Fig. 1, B and C; and Fig. S1 A, late).
A truncated version of Par3 (DN-Par3) containing only the CR1 domain acts as a dominant negative when overexpressed, compromising both Par3 localization and function (Mizuno et al., 2003). Widespread developmental defects result from a tissue-level loss of Par function. We therefore drove expression of DN-Par3 specifically in postmitotic MCCs using the α-tubulin promoter (αtub), allowing us to address the cell-autonomous role of Par3 during intercalation (Stubbs et al., 2006). Expression of DN-Par3-GFP resulted in mispositioning of centrioles relative to the apical-basal axis (Fig. 1 E). We quantified this by measuring the distance of the centriole cluster and nucleus to the apical surface and showed that in control cells, but not DN-Par3 cells, the centriole cluster is always positioned closer to the apical surface (Fig. 1, D and F; and Fig. S1, C and D).
We next tested whether Par3 regulated apical migration during intercalation by comparing MCCs expressing DN-Par3-GFP, full-length Par3-GFP, or GFP alone (e.g., Fig. 1 H, normal, partial, or failed). In control cells, failure to intercalate is a rare event. However, we observed that ∼80% of DN-Par3-GFP–positive cells exhibit intercalation defects (Fig. 1, G and I). To further quantify the extent of the intercalation defect, we measured the size of the apical surface and found an 80% decrease in DN-Par3-GFP cells relative to controls (Fig. S1 B, P < 0.0001). We observed similar defects in centriole position and intercalation when we overexpressed a kinase-dead version of aPKC (aPKC-KD), which has also been found to disrupt Par complex function (Fig. 1, E, F, and I; and Fig. S1, B and E; 55% intercalation defect, 61% reduction in apical surface, P < 0.0001; Sotillos et al., 2004). Finally, the intercalation defect can be replicated using a Par3-specific morpholino (Figs. 1 I and S1 B; Moore et al., 2013). These results suggest that the Par complex is required for the apical positioning of centrioles and the directed movement of MCCs into the outer epithelium.
MTs promote intercalation downstream of the Par complex
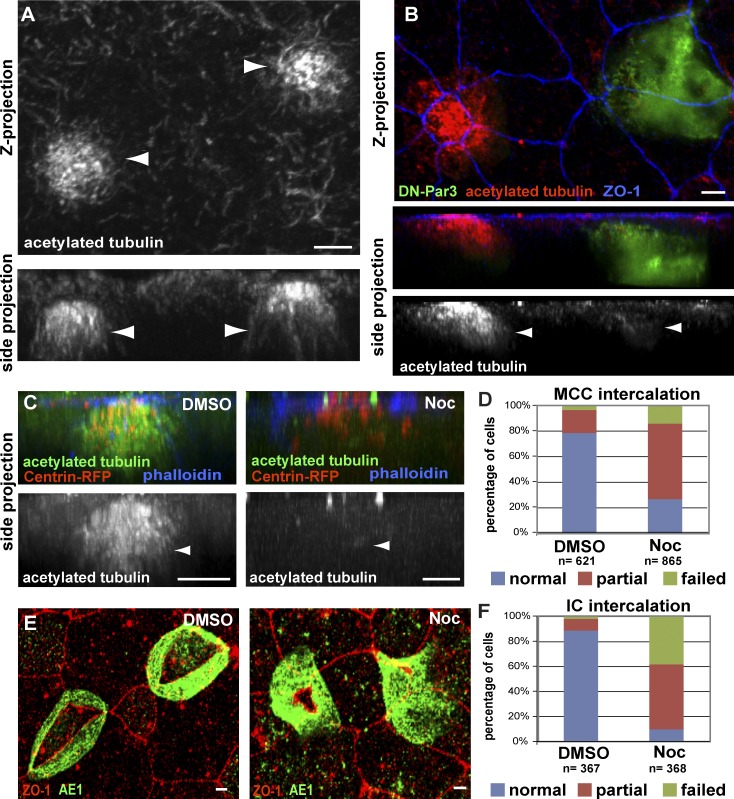
In cultured cells, the position of centrioles between the nucleus and the leading edge promotes the reorganization of the MT network (Luxton and Gundersen, 2011). In intercalating MCCs, we observed an apical enrichment of stable MTs using an antibody against posttranslationally modified acetylated α-tubulin (Fig. 2 A; Chu and Klymkowsky, 1989). Importantly, mosaic embryos, generated by injecting only two cells at the four-cell stage, exhibit a substantial loss of stable acetylated MTs in Par-defective cells when compared with neighboring wild-type cells (Fig. 2 B). To determine whether stabilized MTs are required for intercalation, we treated embryos with 1 µM of the MT depolymerizing drug Nocodazole during the window of intercalation (stage 13–24; Fig. 2 C). In cells with no detectable apical MTs, we observed severe intercalation defects and a 62% reduction in apical surface area (Fig. 2 D, P < 0.0001). We also quantified a similar defect in ICs using an anti-AE1 antibody that recognizes the IC protein, anion exchange factor 1 (Fig. 2, E and F; Quigley et al., 2011). Collectively, these data show that MTs accumulate at the apical side of intercalating cells in a Par-dependent manner and that MT stability is critical during intercalation.
Figure 2.
Stabilized apical MTs are required for radial intercalation of MCCs and ICs. (A and B) Anti-acetylated tubulin antibody staining of wild-type MCCs (A, arrowheads) and of a mosaic tissue showing a control and a DN-Par3-GFP–expressing MCC (B, arrowheads) during intercalation. Maximum-intensity projection (top) and corresponding cross section (bottom) are shown. (C and D) Images (C) of MCCs stained with an antibody marking acetylated α-tubulin in DMSO (left, arrowhead), which is lost following treatment with 1 μM Nocodazole (right, arrowhead), along with quantification of intercalation (D). (E and F) Images of IC marker AE1 in DMSO (left)- and Nocodazole (right)-treated ICs (E), along with quantification of intercalation (F). Quantifications of the Nocodazole phenotypes in D and F are statistically significantly different from controls (P < 0.0001, see Table S1). Bars, 5 µm.
CLAMP is a MT regulator that interacts with aPKC
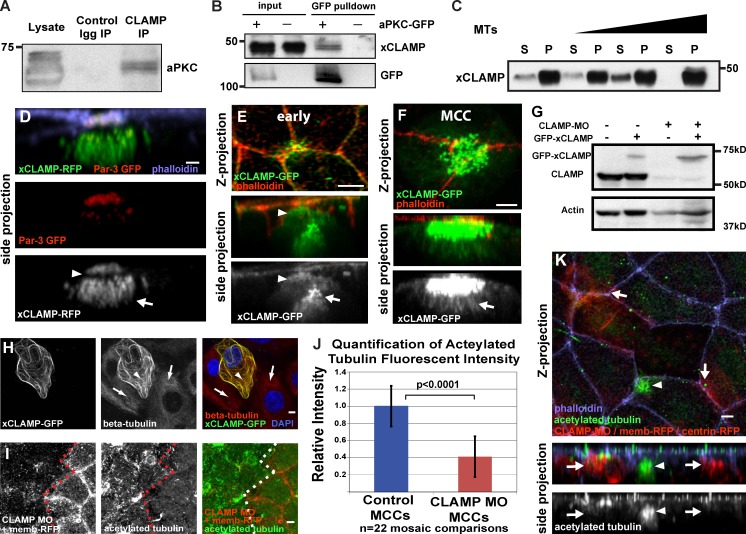
The Par complex’s wide variety of cellular functions suggests an equally wide assortment of interacting proteins that mediate these downstream functions. The common theme of Par-mediated changes in MT dynamics suggests that MAPs could be downstream effectors of Par function (Schmoranzer et al., 2009). We identified CLAMP as a protein that co-immunoprecipitates with endogenous aPKC in embryo extracts (Fig. 3 A). Furthermore, we can use GFP-aPKC expressed explicitly in intercalating MCCs to pull down endogenous CLAMP, which suggests that aPKC and CLAMP interact during intercalation (Fig. 3 B). Using a GFP-tagged version of Xenopus CLAMP (xCLAMP) and a monoclonal antibody (CLAMP mouse monoclonal antibody [mAb]), we observed CLAMP at the leading edge of intercalating cells, partially colocalizing with components of the Par complex (Fig. 3, D and E; and Fig. S2, A and C). Additionally, we see localization of CLAMP to the striated rootlet, to the centrioles and associated MTs, and at apical cell contacts (Fig. 3, D–F; and Fig. S2, A, B, H, and I; Park et al., 2008; Mitchell et al., 2009; Brooks and Wallingford, 2012). Similar to Homo sapiens CLAMP (hCLAMP), we observed GFP-xCLAMP localizing to cytoplasmic MTs in human RPE-1 cells (Fig. S2, D and E).
Figure 3.
CLAMP interacts with aPKC and stabilizes MTs. (A) Western blot of a coimmunoprecipitation using anti-CLAMP mAb probed with anti-aPKC antibody. (B) Western blot probed with anti-CLAMP mAb of a GFP pull-down using NHS-GFP beads with wild-type lysates (−) or lysates from embryos expressing aPKC-GFP (+). (C) Western blot using anti-CLAMP mAbs of an MT pelleting assay with recombinant GST-tagged CLAMP. S, supernatant fraction; P, pellet. MT concentrations are 0, 0.3, 1.2, and 2.4 µM. (D and E) Localization of CLAMP to the leading edge (arrowheads) colocalized with Par3-GFP (D) and to the centriole cluster (arrows). (F) Image highlighting CLAMP localization to the apical surface and to the associated MTs (arrow). (G) Western blot using anti-CLAMP mAb on lysates from control embryos (lane 1) and embryos injected with GFP CLAMP (lanes 2 and 4) and CLAMP MO (lanes 3 and 4). (H) Staining with anti–β-tubulin and DAPI of RPE-1 cells expressing xCLAMP-GFP (arrowheads) compared with neighboring non-CLAMP-expressing cells (arrows) after cold-induced MT depolymerization. (I) Acetylated MT staining in mosaic embryos with CLAMP morphant OCs marked with membrane-RFP. (J and K) Quantification (J) and representative image (K) of acetylated MT staining during intercalation in mosaic embryo CLAMP morphant MCCs (arrows), and wild-type MCCs (arrowheads). Membrane-RFP and centrin-RFP were injected to identify CLAMP MO–containing MCCs. The distinct staining pattern of acetylated tubulin in MCCs allows for the differentiation control MCCs from other intercalating cell types. Quantification in J is statistically significant (P < 0.0001, t test). Error bars represent standard deviations. See Fig. S2. Bars, 5 µm.
CLAMP dynamically interacts with and stabilizes MTs
Because CLAMP can localize to MTs in vivo, we asked whether bacterially expressed CLAMP can bind directly to MTs in vitro. We found that xCLAMP co-pellets with increasing concentrations of purified MTs, which indicates a direct interaction (Fig. 3 C). We further determined that this interaction was dynamic. The GFP-hCLAMP signal at MTs quickly recovers after photobleaching (Fig. S2 F) by freely exchanging from the cytoplasmic pool similar to other MAPs (Breuzard et al., 2013; Video 1). This is consistent with photoactivatable CLAMP-GFP (PA-GFP-hCLAMP), which quickly spreads throughout the cytoplasm and labels MTs in the entire cell (Fig. S2 G and Video 2).
Similar to what has been reported with hCLAMP, the overexpression of xCLAMP results in protection against cold-induced MT depolymerization (Fig. 3 H; Dougherty et al., 2005). To address the role of CLAMP-mediated MT stabilization in vivo, we generated a CLAMP-specific morpholino oligo (MO). Using the CLAMP mAb, we confirmed that xCLAMP is successfully depleted in CLAMP morphants (Fig. 3 G). We next tested the effect of CLAMP depletion on MTs in mosaic embryos in which CLAMP MO cells have been coinjected with a membrane-RFP tracer. We observed a clear decrease in overall acetylated MTs in OCs that contain the CLAMP MO (Fig. 3 I). Importantly, this loss of acetylated MTs is even more profound in intercalating MCCs, where we observed a 60% decrease in acetylated MT staining when directly compared with neighboring wild-type MCCs (Fig. 3, J and K), similar to what we observed in MCCs with compromised Par function (Fig. 2 B). These data provide evidence that CLAMP can stabilize MTs by directly and dynamically associating with the MT lattice.
Knockdown of CLAMP inhibits intercalation of both MCCs and ICs in a cell-autonomous manner
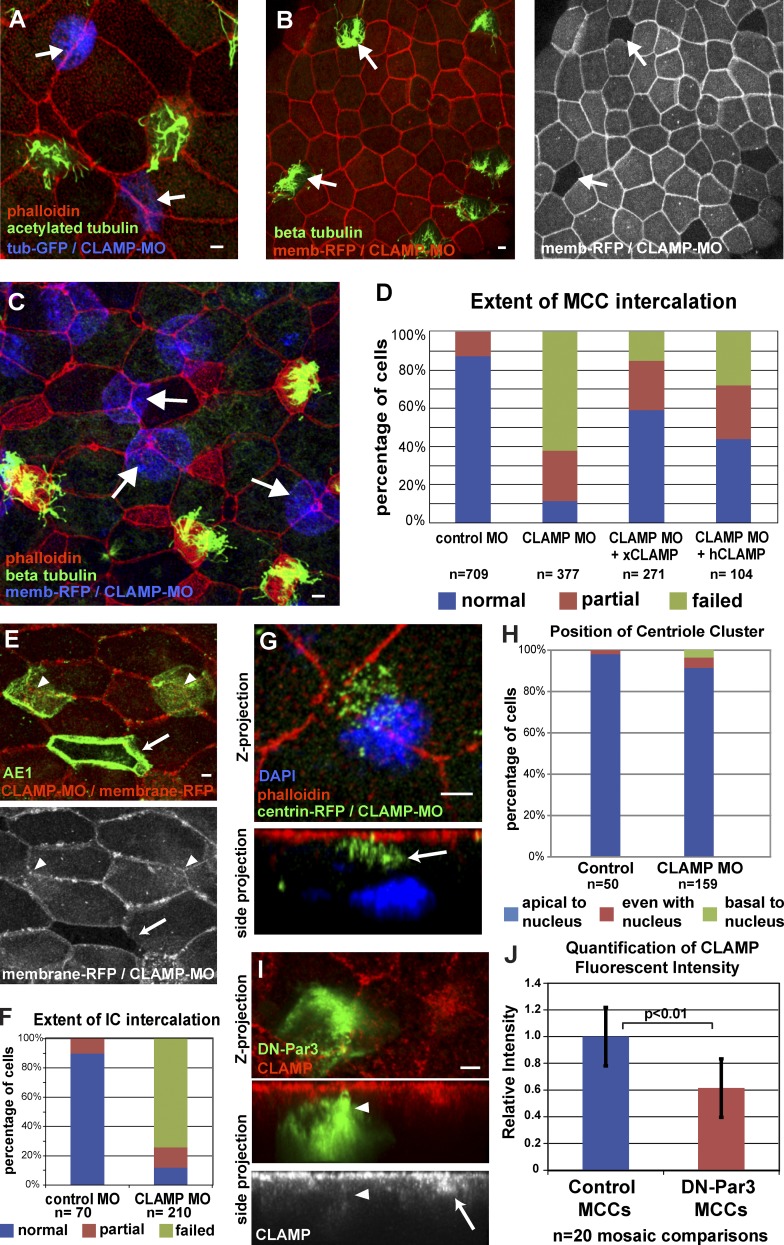
We next determined the functional significance of the loss of CLAMP-mediated acetylated MTs. Loss of xCLAMP significantly reduces the fluid flow on the surface of embryos due to a reduction in the number of MCCs (Fig. S3 A). The postdifferentiation expression of the αtub-GFP and the presence of numerous centrioles indicates that MCC specification is not compromised in CLAMP morphants (Fig. 4, A and G). MCCs are present but remain below the apical surface, which is consistent with a defect in intercalation (Fig. 4, A, C, and G). Similar to Par-defective cells, close to 90% of CLAMP morphant MCCs exhibit intercalation defects, with an 83% reduction in apical surface area (Figs. 4 D and S3 D, P < 0.0001). Importantly, this defect can be partially rescued by injecting mRNA of GFP-xCLAMP or hCLAMP, confirming the specificity of the MO (Figs. 4 D and S3, C and D).
Figure 4.
CLAMP is required cell autonomously for intercalation. (A) Mosaic embryos coinjected with CLAMP-MO and αtub-GFP (arrows) stained with anti-acetylated tubulin and phalloidin. (B) Mosaic embryos coinjected with CLAMP-MO and membrane-RFP stained with anti–β-tubulin and phalloidin (wild-type MCCs still intercalate, arrows). (C) Mosaic embryo coinjected with CLAMP-MO and membrane-RFP (blue), showing MCC intercalation defects (arrows). (D) Quantification of intercalation in MCCs injected with control MO, CLAMP MO, or CLAMP MO rescued with xCLAMP or hCLAMP. (E and F) Representative image (E) and quantification (F) of intercalation defect in ICs (green, AE1 staining) containing CLAMP MO (arrowheads) versus wild-type (arrow, lack of red). (G and H) Image (G) and quantification (H) of centriole position in CLAMP morphant MCCs (the arrow highlights the centriole cluster). (I and J) Representative image (I) and quantification (J) of mosaic embryos showing the loss of CLAMP staining in DN-Par3–expressing MCCs (arrowheads) compared with wild-type (arrow). Quantification in H is based on at least 10 cells, each from a total of at least five embryos from at least two independent experiments. Quantifications of intercalation defects in CLAMP morphants in D and F are significantly different from controls, as are morphant embryos from rescue (P < 0.0001, see Table S1). A χ2 test shows no statistical significant difference in H (P = 0.236, see Table S1). Quantification of reduction in apical CLAMP levels in DN-Par–expressing cells is significantly different from controls (P < 0.01, see Table S1). See Fig. S3. Error bars represent standard deviations. Bars, 5 µm.
To determine if CLAMP is required cell autonomously during intercalation or whether it is required in OCs to accommodate intercalation, we again created mosaic embryos. We observed that morphant MCCs fail to intercalate into wild-type OCs (Fig. 4, A and C), whereas wild-type MCCs intercalate normally into morphant OCs (Fig. 4 B). This indicates that CLAMP promotes radial intercalation cell autonomously. Additionally, we tested whether the role of CLAMP was restricted to MCCs or was more generally involved in intercalation. Indeed, we observed that, similar to MCCs, the ability of CLAMP-depleted ICs to intercalate was dramatically compromised (Fig. 4, E and F).
The Par complex promotes CLAMP localization
The similarity of intercalation defects observed in DN-Par3, aPKC-KD, and CLAMP morphant cells, together with CLAMP’s ability to stabilize MTs and the physical interaction between CLAMP and aPKC, led us to test whether CLAMP was functioning downstream of the Par complex during intercalation. To determine whether the loss of acetylated MTs in CLAMP morphants was caused by a role for CLAMP in apical positioning of the centriole cluster, we coinjected embryos with CLAMP MO and centrin4-RFP. Unlike Par-defective cells, we observed that centrioles are still localized on the apical side of the cell relative to the nucleus (Fig. 4, G and H; and Fig. S3 B). This suggests that the Par-mediated apical positioning of the centriole cluster, while required, is not sufficient to generate the enriched axis of stable MTs. During intercalation, CLAMP colocalizes at the leading edge with components of the Par complex (Figs. 3 D and S2 C). Strikingly, MCCs expressing DN-Par3 or aPKC-KD have a substantial loss of CLAMP localization (Fig. 4, I and J; and Fig. S3, E and F). Additionally, overexpression of CLAMP-RFP in MCCs fails to rescue the intercalation defect caused by DN-Par3-GFP expression (Fig. S3 G). These results suggest that in addition to positioning the centriole cluster, the Par complex is required to position CLAMP apically, which in turn leads to an accumulation of stable acetylated MTs at the leading edge that is required for intercalation.
In this study, we have shown that Par complex components localize at the prospective apical surface and are essential to the directed movement of MCCs into the outer epithelium because they promote the apical positioning of centrioles and CLAMP, which together create an enrichment of stable MTs. Asymmetric protein localization coupled with changes in cytoskeletal dynamics is thought to underlie most forms of cell polarization, including directed migration. Here we provide evidence that the Par complex mediates the subcellular positioning of the MT-stabilizing protein CLAMP during the directed migration of cells undergoing intercalation in the Xenopus skin. This positioning of CLAMP leads to a dramatic increase in stable acetylated MTs along that axis of migration, which is essential for intercalation (Fig. 5). These results suggest that CLAMP could be an important mediator of both cell polarity and directed cell migration and furthermore that the subcellular localization of MAPs may be a common mechanism to achieve cellular asymmetry.
Figure 5.
Model for the steps involved in regulating MT stability during radial intercalation. We propose that the Par complex mediates both apical positioning of centrioles (1) and asymmetric accumulation of CLAMP (2). CLAMP asymmetry leads to asymmetric stabilization of MTs (2) along the axis of migration that promotes intercalation (3).
Materials and methods
Plasmids and mRNA
Genes used were Par3 (Xl.16888), Par6 (Xl.626), aPKC (Xl.967), xCLAMP (Xl.26316), hCLAMP (Hs.72620), and centrin4 (Xl.50437). We used pCS2 plasmids containing membrane-RFP as a tracer for injections, and the pCS2 vector containing the α-tubulin promoter (TUBA1A-B on Scaffold 127187, pCS2tub) to restrict expression to MCCs has been described previously (Stubbs et al., 2006). hCLAMP (Spef1 Gene ID 25876) was cloned into pCS2+ and fused to GFP at the N or C terminus or to photoactivatable GFP at the N terminus. Photoactivatable GFP was cloned into PCS2+ using BglII and BamH1. To make full-length and dominant-negative Par3, the entire coding sequence (cDNA clone 5084932; Thermo Fisher Scientific) or the CR1 domain of ParD3 (xl.16888, amino acids 1–178), respectively, were cloned into pCS2tub fused to GFP at the N terminus. Full-length aPKC-Iota (xl.967) was fused to GFP at the N terminus and cloned into pCS2tub. aPKC-KD (K to E, aa 276) was generated using the QuikChange Mutagenesis kit (Agilent Technologies) of the kinase domain. For bacterial expression of recombinant GST-CLAMP, CLAMP was cloned in pGEX4T. pCS2 plasmids containing centrin4-RFP and GFP-xCLAMP are the same as described previously (Werner et al., 2011). In vitro transcription was described previously, and mRNA was purified using an RNA isolation kit (QIAGEN; Sive et al., 2007).
Embryo injections and drug treatments
All Xenopus experiments were performed using previously described techniques (Werner and Mitchell, 2013). In brief, Xenopus embryos were obtained by in vitro fertilization using standard protocols (Sive et al., 2007) approved by the Northwestern University Institutional Animal Care and Use Committee. Embryos were injected at the two- or four-cell stage with 40–250 pg mRNA or 10–20 pg of plasmid DNA into all four blastomeres. To create mosaic embryos, only two blastomeres were injected at the four cell stage. We designed a start site MO (Gene Tools, LLC) for CLAMP with the sequence 5′-TCTCCTCATCAAACTCCACCGCCAT-3′. A previously published Par3 MO with the sequence 5′-TCCCAAAGCTCACCGTCACCTTCAT-3′ was used (Moore et al., 2013). 10–20 ng per cell of MO was injected into two blastomeres at the four cell stage together with membrane-RFP RNA as a tracer. Standard scrambled morpholinos were used as controls in all experiments (Gene Tools, LLC). Nocodazole treatments were performed as described between stages 13 and 24 (Werner et al., 2011). In brief, embryos were peeled and incubated in the presence of 1 µM Nocodazole or DMSO starting at stage 13 until control embryos reached stage 24. Embryos were fixed immediately thereafter.
Cell culture, plasmid transfection, and cold shock treatment
Human telomerase immortalized retinal pigment epithelial (RPE-1) cells were cultured in DMEM supplemented with 10% FBS 1% penicillin-streptomycin, and 2 mM l-Glutamine. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were imaged 24–48 h after transfection. For live imaging, cells were grown in 35-mm microwell glass-bottom dishes (MatTek Corporation). For cold-shock treatment, 24 h after transfection cells were incubated at 4°C for 60 min and fixed immediately with 100% ice-cold methanol.
Immunofluorescence
For most stainings, embryos were fixed with 3% PFA in 80 mM K+ Pipes, pH 6.8, containing 2 mM MgCl2 and 5 mM EDTA for 2 h (Werner and Mitchell, 2013). For AE1, acetylated tubulin, γ-tubulin, and CLAMP mAb staining, embryos were fixed in 100% ice-cold methanol followed by rehydration in EtOH. RPE cells were fixed with 3% PFA in 80 mM K+ Pipes, pH 6.8, containing 2 mM MgCl2 and 5 mM EDTA for 10 min or 100% ice-cold methanol for 20 min. RPE cells and embryos were blocked with 5% normal donkey serum in PBS with 0.1% Triton X-100 for 1 h. The following primary antibodies were used according to the manufacturer’s recommended dilutions: mouse anti–β-tubulin (7–10) and mouse anti-AE1 (IVF12; both from the Developmental Studies Hybridoma Bank), mouse anti-acetylated α-tubulin (T7451; Sigma-Aldrich), mouse anti–γ-tubulin (GTU-88; Sigma-Aldrich), rabbit anti–ZO-1 (61-7300; Invitrogen), and Cy-2–, Cy-3–, or Cy-5–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.). Phalloidin 568 or 647 (Invitrogen) were used to visualize actin. Embryos were mounted between two coverslips using Fluoro-Gel (Electron Microscopy Sciences; Werner and Mitchell, 2013).
Microscopy
All microscopy was performed on a laser-scanning confocal microscope (A1R; Nikon) using a 60× oil Plan-Apochromat objective lens with a 1.4 NA. Embryos were imaged at room temperature. RPE cells were imaged at 37°C. Nikon Elements Software was used for all acquisition and image processing. For all images, multiple z planes were visualized every 0.3–0.5 µm. Images are maximum-intensity projections of z stacks. For FRAP experiments, RPE cells exhibiting similar levels of GFP-hCLAMP expression were selected and a small region of the cell was bleached with a 405-nM laser for 5 s. FRAP was visualized by time-lapse acquisition of GFP fluorescence every 2 s. For photoactivation experiments, 24 h after transfection a small area of a cell was illuminated with a 405-nM laser for 1 s followed by time-lapse acquisition every 2 s. Fluid flow was recorded with a microscope (M165FC; Leica) using fluorescent microbeads (FluoSpheres, polystyrene microspheres 10 µm; Invitrogen) as described previously (Werner et al., 2011; Werner and Mitchell, 2013). In brief, stage 29 embryos are immobilized using vacuum grease in 0.1× MMR, and diluted FluoSpheres were added drop-wise over the embryonic surface. Movement of spheres was imaged every 100 ms and the speed of bead movement was quantified using Elements Software (Nikon).
Image processing and quantification
To determine the position of the nucleus and basal body cluster, the relative distance of the center of the nucleus and the center of the basal body cluster relative to the apical surface was measured, and data were plotted in a scatter plot. Alternatively, cells were categorized into three categories: centriole cluster closer to the apical surface (apical to nucleus), centriole cluster and nucleus equidistant to the apical surface (even with nucleus), or nucleus closer to the apical distance (basal to nucleus) before plotting the distribution. At least 10 cells, each from a total of at least five embryos from at least two independent experiments, were analyzed. For control embryos, centriole positioning was determined in stage 20 embryos. For morphant embryos or embryos expressing DN-Par3, basal body position was determined at stage 28 when all MCCs in wild-type embryos have completed intercalation. MCCs were identified by GFP expression under the control of the MCC-specific α-tubulin promoter, whereas ICs were identified by AE1 staining. To quantify intercalation defects, apical cell area was measured using Elements software (Nikon) and results were averaged and normalized to the mean cell area of GFP control or control MO expressing stage 28 MCCs. To illustrate the distribution of individual intercalation defects, individual cells were assigned to three distinct categories depicted in Fig. 1 H. For all experiments, cells from a total of least five embryos from at least two independent experiments were quantified.
To quantify anti-acetylated tubulin staining and anti-CLAMP mAb staining levels in CLAMP MO and DN-Par3–expressing cells, respectively, total fluorescence intensity was measured in a 10 × 1 × 1 µm area at the apical surface of MCCs at stage 20 using Elements software (Nikon). Fluorescent intensities were compared between manipulated MCCs and neighboring wild-type MCCs using the same settings and the same plane of view. FRAP was quantified by measuring the amount of total fluorescence within the photobleached area over time and normalizing the fluorescence intensity against total fluorescence in an equal size area in the same cell that has not been photobleached to account for changes in fluorescence due to photobleaching during time-lapse acquisition. Represented are averages of three independent FRAP experiments, with error bars indicating standard deviations. To determine statistical significance, t tests or χ2 tests were performed.
Bacterially expressed recombinant protein
GST-tagged xCLAMP was expressed in BL21 bacteria. Transfected BL21 cells were incubated for 3 h after induction before being harvested. Cells were lysed using a combination of lysozyme treatment and sonication in 20 mM Hepes, pH 7.7, 50 mM NaCl, 0.1% Triton X-100, and bacterial proteinase inhibitors. After a 30-min spin at 13,000 rpm, lysates were incubated in the presence of Glutathione Sepharose (BioWorld) for 4 h. GST-xCLAMP was eluted with 5 mM Glutathione in 100 mM Tris, pH 8.0, followed by dialysis. Purified MTs were a gift from S. Rice (Feinberg School of Medicine, Northwestern University, Chicago, IL). For the MT pelleting assay, MTs were polymerized in BRB80 buffer in the presence of 10% DMSO, 10 mM GTP, and 2 µM Taxol (Seeger and Rice, 2010). 0.1 µM of purified GST-xCLAMP was incubated with 0, 0.3, 1.2, and 2.4 µM of polymerized tubulin in BRB80 in the presence of 50 mM NaCl and 2 µM Taxol for 10 min. The binding reaction was loaded on a sucrose cushion and spun at 100,000 rpm for 20 min at room temperature. After acetone precipitation, supernatant and pellet fraction were loaded on a polyacrylamide gel. Because GST-xCLAMP and tubulin monomers exhibit similar molecular weights, GST-CLAMP in the supernatant and pellet fraction were detected on Western blots using monoclonal anti-CLAMP antibodies.
Generation of monoclonal antibody
Bacterially expressed GST-xCLAMP was used to inoculate mice for the productions of monoclonal antibodies. Monoclonal clones were preselected by ELISA, and positive candidates were screened by immunofluorescence and Western blotting for clone recapitulating GFP localization in immunofluorescence as well as recognizing both endogenous xCLAMP and GFP-tagged exogenous xCLAMP on Western blots from embryo extracts.
Embryo extracts and pull-down experiments
To generate embryonic cell extracts, animal caps were removed at stage 10 and incubated at 16°C for 24 h until control embryos exhibit the desired stage. Caps were lysed in 50 mM Tris, pH 8.0, 150 mM NaCl, 0.8% NP-40, and 1× NEM, followed by centrifugation at 3,000 rpm for 3 min. For pull-down experiments, bacterially expressed GFP-binding protein (GFP-BP) was covalently linked to NHS-activated Sepharose fast flow (GE Healthcare). Lysates from animal caps expressing GFP-tagged proteins were incubated in the presence of GFP-BP–bound NHS Sepharose for 4 h. For coimmunoprecipitation experiments, anti-CLAMP mAb was preincubated with protein–G Sepharose (GE Healthcare) for 1 h before incubation in the presence of lysate for 4 h. After five washes, beads were resuspended in Laemmli buffer and processed for Western blot analysis. Antibodies used for Western blot analysis were mouse monoclonal anti-CLAMP antibody (1:300, see “Generation of monoclonal antibody”), chicken anti-GFP (1:1,000; Aves Laboratories), rabbit anti-PKCζ (C-20; sc-216; 1:1,000; Santa Cruz Biotechnology, Inc.), and mouse anti-actin antibody (JLA20; 1:300; Developmental Studies Hybridoma Bank). Secondary antibodies used were peroxidase-conjugated AffinityPure goat anti–mouse or anti–chicken (Jackson ImmunoResearch Laboratories, Inc.). Western blots were developed using Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific), and chemiluminescence was visualized using a LAS4000 Mini imager (FujiFilm Life Science).
Online supplemental materials
Fig. S1 shows Par6-GFP localization during intercalation as well as cell surface area measurement corresponding to Fig. 1 I, and scatter plot data showing the relative basal body nucleus position for individual cells for data shown in Fig. 1 F. Fig. S2 shows endogenous CLAMP localization in intercalating and mature MCCs as well as in ICs. Additionally, Fig. S2 shows colocalization of CLAMP with aPKC in intercalating MCCs as well as colocalization of CLAMP with MTs in intercalating ICs and in RPE cells. Finally, Fig. S2 shows the dynamics of CLAMP interaction with the MT lattice based on FRAP (see Video 1) and photoactivation (see Video 2) experiments. Fig. S3 shows quantification of the loss of fluid flow in CLAMP morphant embryos as well as scatter plot data showing centriole position for individual cells for data shown in Fig. 4 H. Furthermore, Fig. S3 shows a representative image and quantification of apical surface of rescue of the intercalation defect in CLAMP morphants by xCLAMP-GFP expression (see Fig. 4 D). Finally, Fig. S3 shows the loss of apical CLAMP localization in MCCs expressing aPKC-KD-GFP. Video 1 shows a representative FRAP experiment of hCLAMP-GFP in RPE-1 cells. Video 2 shows a representative photoactivation experiment in PA-GFP-hCLAMP–expressing RPE-1 cells. Table S1 shows p-values based on either χ2 or a t tests for all figures. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201312045/DC1.
Supplementary Material
Acknowledgments
We would like to thank Chris Kintner for discussions and Chad Pearson for critically reading an early draft of the manuscript.
M.E. Werner was supported by a fellowship from the American Heart Association. This work was support by a pilot grant from the Northwestern University Skin Disease Research Center (5P30AR057216-04) and from National Institutes of Health National Institute of General Medical Sciences to B.J. Mitchell (1R01GM089970-01).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- aPKC
- atypical PKC
- aPKC-KD
- kinase-dead aPKC construct
- hCLAMP
- Homo sapiens CLAMP
- IC
- ionocyte
- mAb
- mouse monoclonal antibody
- MAP
- MT-associated protein
- MCC
- multiciliated cell
- MO
- morpholino oligo
- MT
- microtubule
- OC
- outer epithelial cell
- xCLAMP
- Xenopus laevis CLAMP
References
- Breuzard, G., Hubert P., Nouar R., De Bessa T., Devred F., Barbier P., Sturgis J.N., and Peyrot V.. 2013. Molecular mechanisms of Tau binding to microtubules and its role in microtubule dynamics in live cells. J. Cell Sci. 126:2810–2819 10.1242/jcs.120832 [DOI] [PubMed] [Google Scholar]
- Brooks, E.R., and Wallingford J.B.. 2012. Control of vertebrate intraflagellar transport by the planar cell polarity effector Fuz. J. Cell Biol. 198:37–45 10.1083/jcb.201204072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W., Fowler K.J., Choo K.H., and Kalitsis P.. 2005. Spef1, a conserved novel testis protein found in mouse sperm flagella. Gene. 353:189–199 10.1016/j.gene.2005.04.025 [DOI] [PubMed] [Google Scholar]
- Chen, X., and Macara I.G.. 2005. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat. Cell Biol. 7:262–269 10.1038/ncb1226 [DOI] [PubMed] [Google Scholar]
- Chen, S., Chen J., Shi H., Wei M., Castaneda-Castellanos D.R., Bultje R.S., Pei X., Kriegstein A.R., Zhang M., and Shi S.H.. 2013. Regulation of microtubule stability and organization by mammalian Par3 in specifying neuronal polarity. Dev. Cell. 24:26–40 10.1016/j.devcel.2012.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara, D., and Nance J.. 2012. An E-cadherin-mediated hitchhiking mechanism for C. elegans germ cell internalization during gastrulation. Development. 139:2547–2556 10.1242/dev.079863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, D.T., and Klymkowsky M.W.. 1989. The appearance of acetylated α-tubulin during early development and cellular differentiation in Xenopus. Dev. Biol. 136:104–117 10.1016/0012-1606(89)90134-6 [DOI] [PubMed] [Google Scholar]
- Deblandre, G.A., Wettstein D.A., Koyano-Nakagawa N., and Kintner C.. 1999. A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development. 126:4715–4728 [DOI] [PubMed] [Google Scholar]
- Dougherty, G.W., Adler H.J., Rzadzinska A., Gimona M., Tomita Y., Lattig M.C., Merritt R.C. Jr, and Kachar B.. 2005. CLAMP, a novel microtubule-associated protein with EB-type calponin homology. Cell Motil. Cytoskeleton. 62:141–156 10.1002/cm.20093 [DOI] [PubMed] [Google Scholar]
- Drysdale, A.J., and Elinson R.P.. 1992. Cell Migration and Induction in the Development of the Surface Ectodermal Pattern of the Xenopus laevis Tadpole. Dev. Growth Differ. 34:51–59 10.1111/j.1440-169X.1992.00051.x [DOI] [PubMed] [Google Scholar]
- Dubaissi, E., and Papalopulu N.. 2011. Embryonic frog epidermis: a model for the study of cell-cell interactions in the development of mucociliary disease. Dis. Model. Mech. 4:179–192 10.1242/dmm.006494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, J.L., and Priess J.R.. 2012. A role for the centrosome and PAR-3 in the hand-off of MTOC function during epithelial polarization. Curr. Biol. 22:575–582 10.1016/j.cub.2012.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, B., and Macara I.G.. 2007. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 13:609–622 10.1016/j.devcel.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, E.R., Jani S., and Gundersen G.G.. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Kim, K., Lake B.B., Haremaki T., Weinstein D.C., and Sokol S.Y.. 2012. Rab11 regulates planar polarity and migratory behavior of multiciliated cells in Xenopus embryonic epidermis. Dev. Dyn. 241:1385–1395 10.1002/dvdy.23826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos Dehring, D.A., Vladar E.K., Werner M.E., Mitchell J.W., Hwang P., and Mitchell B.J.. 2013. Deuterosome-mediated centriole biogenesis. Dev. Cell. 27:103–112 10.1016/j.devcel.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé, J.C., Maddox P.S., Salmon E.D., and Goldstein B.. 2003. PAR proteins regulate microtubule dynamics at the cell cortex in C. elegans. Curr. Biol. 13:707–714 10.1016/S0960-9822(03)00251-3 [DOI] [PubMed] [Google Scholar]
- Luxton, G.W., and Gundersen G.G.. 2011. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr. Opin. Cell Biol. 23:579–588 10.1016/j.ceb.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, B., Stubbs J.L., Huisman F., Taborek P., Yu C., and Kintner C.. 2009. The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr. Biol. 19:924–929 10.1016/j.cub.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, K., Suzuki A., Hirose T., Kitamura K., Kutsuzawa K., Futaki M., Amano Y., and Ohno S.. 2003. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J. Biol. Chem. 278:31240–31250 10.1074/jbc.M303593200 [DOI] [PubMed] [Google Scholar]
- Moore, R., Theveneau E., Pozzi S., Alexandre P., Richardson J., Merks A., Parsons M., Kashef J., Linker C., and Mayor R.. 2013. Par3 controls neural crest migration by promoting microtubule catastrophe during contact inhibition of locomotion. Development. 140:4763–4775 10.1242/dev.098509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, E.M.2006. PAR proteins and the cytoskeleton: a marriage of equals. Curr. Opin. Cell Biol. 18:86–94 10.1016/j.ceb.2005.12.007 [DOI] [PubMed] [Google Scholar]
- Park, T.J., Mitchell B.J., Abitua P.B., Kintner C., and Wallingford J.B.. 2008. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat. Genet. 40:871–879 10.1038/ng.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley, I.K., Stubbs J.L., and Kintner C.. 2011. Specification of ion transport cells in the Xenopus larval skin. Development. 138:705–714 10.1242/dev.055699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, J.R., and Hogan B.L.. 2011. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 27:493–512 10.1146/annurev-cellbio-100109-104040 [DOI] [PubMed] [Google Scholar]
- Schmoranzer, J., Fawcett J.P., Segura M., Tan S., Vallee R.B., Pawson T., and Gundersen G.G.. 2009. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr. Biol. 19:1065–1074 10.1016/j.cub.2009.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger, M.A., and Rice S.E.. 2010. Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J. Biol. Chem. 285:8155–8162 10.1074/jbc.M109.068247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive, H.L., Grainger R.M., and Harland R.M.. 2007. Xenopus laevis In Vitro Fertilization and Natural Mating Methods. CSH Protoc. 2007:t4737. [DOI] [PubMed] [Google Scholar]
- Sotillos, S., Díaz-Meco M.T., Caminero E., Moscat J., and Campuzano S.. 2004. DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. J. Cell Biol. 166:549–557 10.1083/jcb.200311031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs, J.L., Davidson L., Keller R., and Kintner C.. 2006. Radial intercalation of ciliated cells during Xenopus skin development. Development. 133:2507–2515 10.1242/dev.02417 [DOI] [PubMed] [Google Scholar]
- Stubbs, J.L., Oishi I., Izpisúa Belmonte J.C., and Kintner C.. 2008. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat. Genet. 40:1454–1460 10.1038/ng.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass, U.2012. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28:655–685 10.1146/annurev-cellbio-092910-154033 [DOI] [PubMed] [Google Scholar]
- Werner, M.E., and Mitchell B.J.. 2013. Using Xenopus skin to study cilia development and function. Methods Enzymol. 525:191–217 10.1016/B978-0-12-397944-5.00010-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, M.E., Hwang P., Huisman F., Taborek P., Yu C.C., and Mitchell B.J.. 2011. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J. Cell Biol. 195:19–26 10.1083/jcb.201106110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.