Abstract
The Mediator, a conserved multisubunit protein complex in eukaryotic organisms, regulates gene expression by bridging sequence-specific DNA-binding transcription factors to the general RNA polymerase II machinery. In yeast, Mediator complex is organized in three core modules (head, middle and tail) and a separable ‘CDK8 submodule’ consisting of four subunits including Cyclin-dependent kinase CDK8 (CDK8), Cyclin C (CycC), MED12, and MED13. The 3-D structure of human CDK8-CycC complex has been recently experimentally determined. To take advantage of this structure and the improved theoretical calculation methods, we have performed molecular dynamic simulations to study dynamics of CDK8 and two CDK8 point mutations (D173A and D189N), which have been identified in human cancers, with and without full length of the A-loop as well as the binding between CDK8 and CycC. We found that CDK8 structure gradually loses two helical structures during the 50-ns molecular dynamic simulation, likely due to the presence of the full-length A-loop. In addition, our studies showed the hydrogen bond occupation of the CDK8 A-loop increases during the first 20-ns MD simulation and stays stable during the later 30-ns MD simulation. Four residues in the A-loop of CDK8 have high hydrogen bond occupation, while the rest residues have low or no hydrogen bond occupation. The hydrogen bond dynamic study of the A-loop residues exhibits three types of changes: increasing, decreasing, and stable. Furthermore, the 3-D structures of CDK8 point mutations D173A, D189N, T196A and T196D have been built by molecular modeling and further investigated by 50-ns molecular dynamic simulations. D173A has the highest average potential energy, while T196D has the lowest average potential energy, indicating that T196D is the most stable structure. Finally, we calculated theoretical binding energy of CDK8 and CycC by MM/PBSA and MM/GBSA methods, and the negative values obtained from both methods demonstrate stability of CDK8-CycC complex. Taken together, these analyses will improve our understanding of the exact functions of CDK8 and the interaction with its partner CycC.
Introduction
Cyclin-dependent kinases (CDKs) are critical regulators of cell-cycle progression and gene transcription, whose activities require association with their specific binding partners called cyclins. Because of the fundamental roles of CDKs in controlling cell proliferation, differentiation and apoptosis and dysregulation of these processes in human cancers, CDKs have been considered as attractive targets for development of novel anticancer drugs [1, 2]. Based on the sequence similarity, the human genome contains 21 genes encoding CDKs and five additional genes encoding a more distant group of proteins known as CDK-like (CDKL) kinases [3]. Instead of directly regulating cell-cycle progression, CDK7, CDK8, and CDK9 are thought to play more important roles in regulating gene transcription compared to other CDKs [4].
CDK8 is particularly interesting because both CDK8 and its regulatory partner Cyclin C (CycC) are found to be either mutated or amplified in a variety of human cancers [5]. CDK8 and CycC are two subunits of the Mediator complex [6], and together with MED12 and MED13, these four subunits form the “CDK8 submodule” [7]. Biochemical analyses have identified two distinct Mediator complexes: a “small” complex of over 20 polypeptides that activates transcription of polymerase II-dependent genes, and a “large” complex forms that is generally considered to represses transcription after the small complex is associated with the “CDK8 submodule” [8–12]. The CDK8 submodule may suppress transcription either directly blocking the interaction between the small Mediator complex and the RNA Pol II, or indirectly phosphorylating certain transcription factors or components of the general transcription machinery such as RNA polymerase II in yeast and human cyclin H (CycH) [5, 13, 14]. The CDK8 module was postulated to regulate initiation and re-initiation of transcription [15]. Interestingly, the CDK8 module can also positively regulate transcription [16]. For example, CDK8 has been reported as a positive regulator of p53-activated gene transcription [17], as well as genes involved in the serum response network [18] and cellular response to hypoxia [19]. CDK8 was identified as a potent oncogene in colon cancerogenesis, since CDK8 positively regulates β-catenin-dependent gene expression [20–22]. In addition, CDK8 was shown to promote the proliferation of melanoma cancer cells and the expression of CDK8 gene is up-regulated because of mH2A deletion in melanoma cancer cells [23]. Depletion of CDK8 in CDK8-amplified colorectal cancer cells or CDK8-overexpressed melanoma cells potently blocked their proliferation, suggesting that gain of CDK8 drives tumorigenesis in these two types of cancers [17, 20, 24].
To understand the function of the CDK8-CycC complex, a 2.2-Å crystal structure of the human CDK8-CycC pair in complex with the inhibitor sorafenib has been recently determined by X-ray crystallography [25]. By comparing to other CDK-cyclin family members, including human CDK4-cyclin D (CycD), CDK9-cyclin T (CycT), CDK5-p25, CDK6-V-Cyclin (Herpesvirus saimiri Cyclin), CDK6/INK4s, CDK7, cyclin K (CycK), cyclin T (CycT)/TAT/TAR, and CycH, as well as the yeast CycC homologue SRB11 (for a detailed overview, see [26]), the CDK8-CycC complex displays additional potential recognition surfaces for interactions, possibly for the recognition of MED12, MED13, or the substrates of CDK8 [25].
When examining the CDK8-CycC crystal structure, we noticed that the part of the structure containing 16 amino acids within the activation loop (or A-loop) of CDK8 was not experimentally determined. For full activation, most protein kinases require the phosphorylation of Thr, Ser, or Tyr residues within the A-loop, also known as the regulatory T-loop that contains the predicted residues for both ATP and cyclin binding. It is thought that the A-loop is flexible: without cyclin, the A-loop blocks the ATP binding site because the positions of several key amino acid residues are not optimal for ATP binding; in the presence of cyclin, however, two α helices change their positions to permit ATP binding [27]. Interestingly, two Asp residues (D173 and D189) are located within the A-loop of CDK8. This is important because D173A mutation causes loss of CDK8 kinase activity (kinase dead CDK8), and D189N has been reported in several types of human cancers [14, 20, 28]. However, how these point mutants affect CDK8 activity or its binding with CycC remains unknown.
With the continuously improved computation powers coupled with increased number of protein structures being solved experimentally [29], computational methods, particularly simulation research, present an attractive and complementary approach to understand the molecular function and regulation of many proteins. Specifically, the new 2.2-Å crystal structure of the human CDK8-CycC complex has provided an opportunity for computational methods to better understand the function and dynamics of CDK8, CDK8 point mutations and the CDK8-CycC interactions. Proteins are highly dynamic, and are constantly switching between different conformations. Molecular dynamics (MD) simulations can provide atomically detailed views of protein motions in vacuum or in solutions, sampling multiple timescales ranging from nanoseconds to milliseconds using supercomputing resources [29].
Therefore, to understand the A-loop and effect of this loop on overall structure of CDK8, we have employed molecular modeling approach to build the structure of human CDK8 with a complete A-loop, and used MD simulations to optimize and study dynamic changes of human CDK8 in solution. Additionally, we have generated theoretical structures of human CDK8 with the point mutations of either D173A or D189N to study the effects of the mutations on the A-loop and overall structures of CDK8. Furthermore, we have investigated physical interactions between CDK8 and CycC by yeast two-hybrid approach, and calculated binding energy between CDK8 and CycC by Molecular Mechanics/Poisson Boltzmann Surface Area (MM/PBSA) [30] and Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) [31] methods to understand the dynamics of CDK8-CycC complex.
Methods
Molecular modeling of human CDK8
The molecular modeling procedure was based on the method described by Liu et al [32]. The sequences were aligned by using the ClustalW pairwise alignment algorithm of the Vector NTI software [33]. The alignment data were imported into MODELLER version 9.9 [34] to generate the structures of human CDK8 with or without full length of the A-loop or with the point mutations using the experimentally solved structures as the templates by searching Protein Data Bank (PDB).
Molecular dynamics simulations of human CDK8
Molecular dynamics simulations were based on the procedure described by Simmerling et al [35]. All computations for human CDK8, including the initial energy minimization, heating, and MD simulations, were performed under fully unrestrained conditions using the supercomputers at Louisiana Optical Network Initiative (LONI). All calculations used AMBER’s all-atom force field (ff99SB) as implemented in Amber 11 software [36]. The SANDER and PTRAJ modules of AMBER, respectively, were used for computation and analysis. A total of 1,000 steps of initial energy minimization, including 500 steepest descent steps (ncyc=500) followed by 500 conjugate gradient steps (maxcyc-ncyc) and using a large cutoff (cut=999 angstroms) and non-periodic simulation (ntb=0), were performed to adjust the human CDK8 structures created by MODELLER 9v9. To avoid kinetic entrapment in local minima, the minimized system was slowly heated from 0 to 325 Kelvin (K) in seven increments of 50 K over 50 pico second (ps) (5 ps for the first six steps and 20 ps for the seventh step). The production MD simulation was conducted for 50 nano second (ns) at a constant 325 K in ten 5-ns stages. The generalized Born solvent model was used in the heating and equilibration phases. Structure images were prepared by using the Visual Molecular Dynamics (VMD) package [37].
CDK8-CycC binding energy calculations by molecular dynamics simulations
We have followed the procedure in the Amber advanced tutorial 3 provided by Ross Walker to calculate CDK8-CycC binding energy. The first step was to split the CDK8-CycC pdb file (PDB code: 3RGF) into the separate structure files: cdk8-cycc.pdb, cdk8.pdb and cycc.pdb. These three structures were then used to create three gas phase prmtop and inpcrd file pairs for the MM-PBSA and MM-GBSA calculations as well as one for the solvated complex, which were used to run the MD simulation. Next, the solvated complex was equilibrated by carrying out a short minimization, 50 ps of heating and 50 ps of density equilibration with weak restraints on the complex followed by 500 ps of constant pressure equilibration at 300 K. All simulations were run with shake on hydrogen atoms, a 2 femto (fs) time step and langevin dynamics for temperature control. Finally, a total of 2 ns production simulation was run with recording the production coordinates every 10 ps, and the binding free energy was calculated following the instructions of the Perl script MMPBSA.pl.
Study the interactions between CDK8 and CycC by the yeast two-hybrid approach
We have cloned Drosophila CDK8 (dCDK8) and CycC (dCycC) into pGBKT7 and pGADT7, yeast two-hybrid vectors (Clontech, Mountain View, CA, USA), to investigate physically interactions between pGBKT7 and pGADT7 in yeast cells. The sense and anti-sense primers of Drosophila CycC are: 5’-CAGAATTCATGGCGGGCAATTTTTGGCAGAGTTC-3’ (dCycC sense primer) and 5’-CAGGATCCCTAACGCTGAGGCGGTGGTTTC-3’ (dCycC anti-sense primer). An EcoR I site and a BamH I site were introduced into the 5’ end of Drosophila CycC sense and anti-sense primers to subclone dCycC into pGBKT7 and pGADT7. The sense and anti-sense primers of Drosophila CDK8 are: 5’-CACCCGGGAATGGACTACGATTTCAAGATGAAAACGC-3’ (dCDK8 sense primer) and 5’-CACTCGAGTCAGTTGAAGCGCTGGAAGTTCTG-3’ (dCDK8 anti-sense primer). A Sma I site and a Xho I site were added at the 5’ end of Drosophila CDK8 sense and anti-sense primers respectively to subclone dCDK8 into pGADT7 vector. Similarly, the sense and anti-sense primers of Drosophila CDK8 are: 5’-CACCATGGATGGACTACGATTTCAAGATGAAAACGC-3’ (dCDK8 sense primer) and 5’-CAGTCGACGTTGAAGCGCTGGAAGTTCTGC-3’ (dCDK8 anti-sense primer). A Nco I site and a Sal I site were designed at the 5’ end of Drosophila CDK8 sense and anti-sense primers respectively to subclone dCDK8 into pGBKT7 vector. The full lengths of CDK8 and CycC were amplified by PCR, the CDK8 and CycC PCR products were first cloned into pGEM-T easy vector (Promega, Masison, WI, USA), and then subcloned into pGBKT7 and pGADT7. The correctness of all the constructs was confirmed by DNA sequencing. The two-hybrid assays were performed in yeast strain Mav203 (Genotype: MATα; leu2–3,112; trp1–901; his3Δ200; ade2–101; cyh2R; can1R; gal4Δ; gal80Δ; GAL1::lacZ; HIS3UASGAL1::HIS3@LYS2; SPAL10::URA3, Life Technologies, Grand Island, USA). The interaction of each pair of Drosophila CDK8 and CycC was tested by sequential transformation of the pGBKT7-dCDK8 (pGBKT7-dCycC) plasmids encoding the Gal4 DNA binding domain (DBD) fusions, and then with the pGADT7-dCycC (pGADT7-dCDK8) plasmids encoding the Gal4 transactivation domain (TAD) fusions. Transformants were selected on SC-Trp or SC-Leu plates and then on SC-Leu-Trp plates. β-galactosidase activity was revealed by quantitative liquid o-nitrophenyl-β-D-galactopyranoside (ONPG) assay performed according to the manufacturer’s instructions (Life Technologies).
Results
Overall structure of human CDK8 with the A-loop
The predicted A-loop of human CDK8 has 27 amino acids. The crystal structure of human CDK8 (PDB code: 3RGF) contains only seven amino acids at the N-terminal and four amino acids at the C-terminal end of the A-loop, thus leaving the 16 amino acids in the middle of the A-loop not determined by X-ray crystallography [25]. To understand the role of the A-loop and consequence of point mutations within the A-loop, we have employed molecular modeling approach to build a theoretical structure of human CDK8 with its full length of the A-loop using two experimentally solved structures as the templates. Specifically, human CDK8 crystal structure (PDB code: 3RGF) was to build CDK8 structure except 16 missing amino acids in the middle of the A-loop, and human CDK7 crystal structure (PDB code: 1UA2) was used to build the coordinates of the 12 missing amino acids in the A-loop. The structure of the rest missing four amino acids was modeled by MODELLER without a template (Figure 1).
Figure 1. The sequence alignment of human and drosophila CDK8 and human CDK7 (PDB code: IUA2, A chain).
Sixteen amino acids in the middle of the human CDK8 A-loop are missing in the crystal structure of human CDK8 (PDB code: 3RGF, A chain).
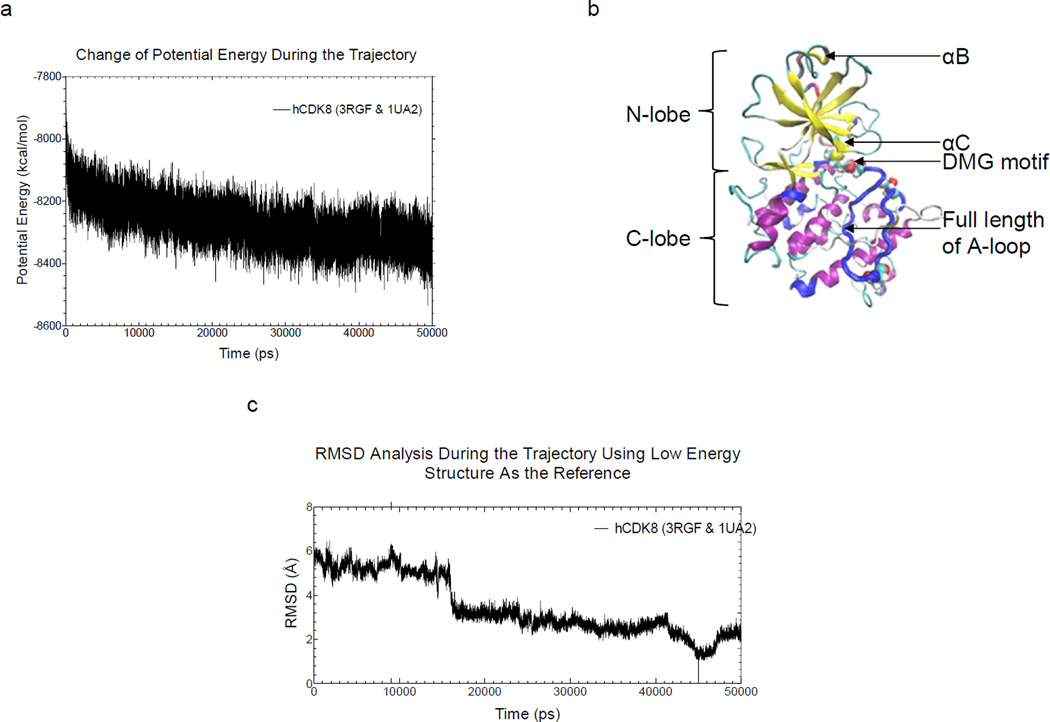
First, we sought to optimize and study dynamics of the theoretical structure of human CDK8 by using MD simulations. Molecular dynamic simulations revealed that the heating phase displayed a gradual increase in temperature with a few abrupt elevations (Supplementary Figure 1a); temperature at the equilibration phase was approximately 325 K, as expected (Supplementary Figure 1b). The energy profile (kinetic energy, potential energy, and total energy) is shown in Supplementary Figure 1c. The kinetic energy was stable, as predicted by the stability of the temperature plot, which is directly proportional to the kinetic energy. The total and potential energy plots showed a gradual decrease, implying that the system had achieved an energy state that is more stable than the starting structure (Supplementary Figure 1c and Figure 2a). The potential energy plot (Figure 2a), which showed the trend of potential energy change of the structures, and from which, the structure with the lowest potential energy, the most stable conformation, was extracted (Figure 2c) and its coordinates are provided in the supplemental file. The backbone root-mean-square deviation (RMSD) plot, generated by using the lowest-energy structure as the reference, showed a decrease in RMSD during the simulation trajectory (Figure 2b).
Figure 2. The theoretical structure of human CDK8 based on molecular modeling and 50-ns molecular dynamics simulation.
a, Change of potential energy as a function of time during the MD simulation; b, The theoretical structure with the lowest potential energy of human CDK8, which occurred at 49,408 ps during the MD simulation. The N-lobe, C-lobe, aB, aC, DMG motif, T196, A-loop and D189 are labeled; c, The backbone RMSD during the MD simulation is compared to the lowest potential energy conformation.
Serine/threonine-specific protein kinases (such as CDKs) and tyrosine-specific protein kinases share a catalytic domain of −290 residues. The active site of the catalytic domain is sandwiched between an N-terminal lobe composed of a β-sheet and a single α-helix (the “C helix”) and a larger C-terminal lobe, connected by a linker. The C-terminal lobe is predominantly α-helical and includes the activation loop, a region of 20–35 residues located between a conserved DFG (using the single letter amino acid codes) motif and a less conserved APE (using the single letter amino acid codes) motif [38, 39]. In the active conformation, the C helix (αC) packs against the N-terminal lobe, and the aspartate of the DFG chelates an Mg2+ ion to orientate the ATP substrate. In the inactive conformation, however, this latter interaction is often disrupted, and the phenylalanine of the DFG motif is turned in toward the ATP site [40].
CDKs were considered difficult to target with small molecules because structures of CDKs in their active and inactive states showed that the activation loop adopts an exclusively DFG-in conformation [41]. The published structure of the ternary complex of CDK8-CycC with CDK8 inhibitor sorafenib was the first to show a DFG-out conformation (DMG-out in CDK8) for a CDK in complex with a small-molecule inhibitor [25, 42]. The recent kinetic study indicated that the structure of CDK8-CycC implicates specificity in the CDK-cyclin family and reveals interaction with a deep pocket binder. This encouraging result shows that kinases for which Type II inhibition was previously believed to be unattainable can undergo DFG motif rearrangements, thus opening the door to the design of more potent and selective CDK8 kinase inhibitors [43].
The structure of human CDK8 built by MODELLER 9v9 is without its partner CycC and the sorafenib. Sorafenib (co-developed and co-marketed by Bayer and Onyx Pharmaceuticals as Nexavar) is a drug approved for the treatment of primary kidney cancer (advanced renal cell carcinoma) and advanced primary liver cancer (hepatocellular carcinoma) [44]. To understand dynamics of CDK8 in solution and the effects of CycC and sorafenib on overall CDK8 structure, we have analyzed our CDK8 structures generated from the MD simulations. The CDK8 structure with the lowest potential energy during 50-ns MD simulations, designated as the “CDK8 structure No. 5” (Figure 2b), shows the N-terminal lobe predominately composed of a β-sheet and a larger C-terminal lobe containing α helices without a single β-sheet. We also did not observe any α helices in the N-terminal lobe.
When comparing our CDK8 structure No. 5 with the crystal CDK8 structure in the CDK8-CycC complex with sorafenib, we found that the major differences are located within the N-terminal lobe. The CDK8 crystal structure has two α helices (αB and αC) in the N-terminal lobe, while there is no obvious α helix in the N-terminal lobe of CDK8 structure No. 5. The N-terminal helix, αB, which is unique to CDK8 and explains discrimination against CDK3 and even other transcriptional CDK-cyclin complexes, such as CDK7-CycH and CDK9-CycT, makes extensive additional interactions with CycC besides the common binding surface [25]. αC of CDK8 becomes an active conformation to relieve the steric block to the catalytic cleft when CDK8 is associated with CycC [25], which is in accordance with the common CDK activation mechanism. The αC helix is expected to stabilize the conformations of a certain CDKs once they are activated by cyclins [45, 46].
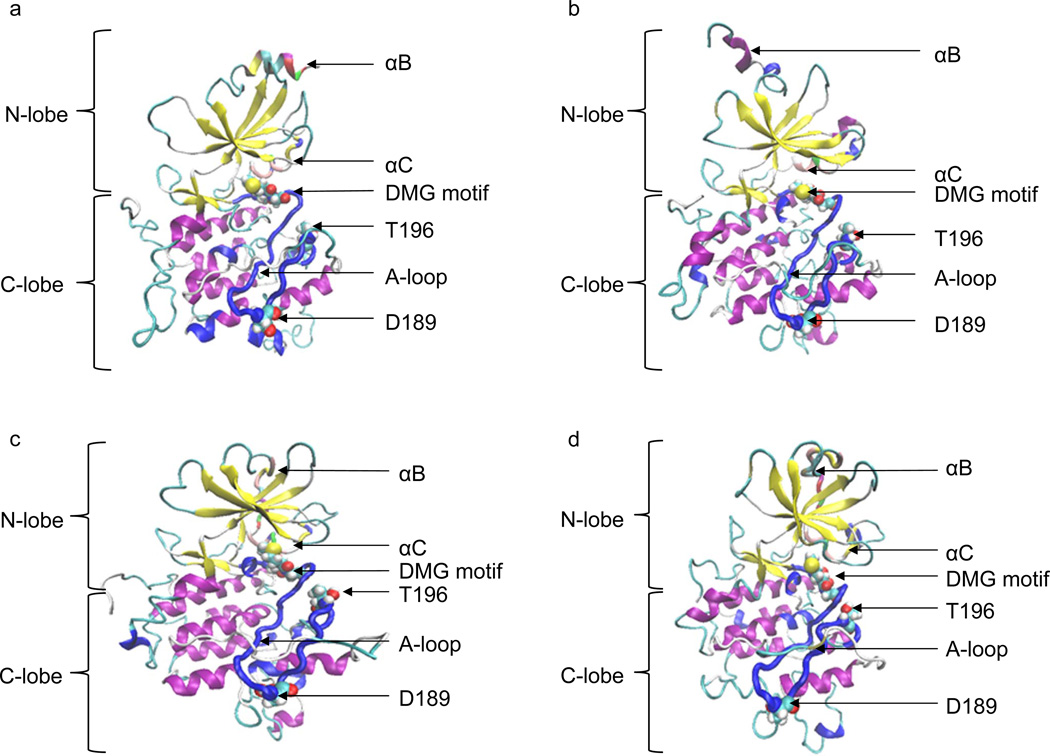
In most of the other CDKs, but probably not in CDK8, a second step of kinase activation requires phosphorylation of the A-loop [47, 48]. To further understand dynamics of two α helices: αB and αC, during our simulations, we have obtained the CDK8 structures with the lowest potential energies in the first 10-ns, 20-ns, 30-ns and 40-ns MD simulations, which were designated as CDK8 structure No. 1, No. 2, No. 3 and No. 4, respectively. Our MD simulations of CDK8 structure demonstrated that αB and αC gradually lost their helical structural conformation (Figure 3a, 3b, 3c and 3d), suggesting their helical structures requires presence of its partner CycC or full length of the A-loop can affect their helical structures.
Figure 3. The molecular dynamic study of human CDK8.
a, The CDK8 structure with the lowest potential energy during the 10-ns MD simulation; b, The CDK8 structure with the lowest potential energy during the 20-ns MD simulation; c, The CDK8 structure with the lowest potential energy during the 30-ns MD simulation; d, The CDK8 structure with the lowest potential energy during the 40-ns MD simulation.
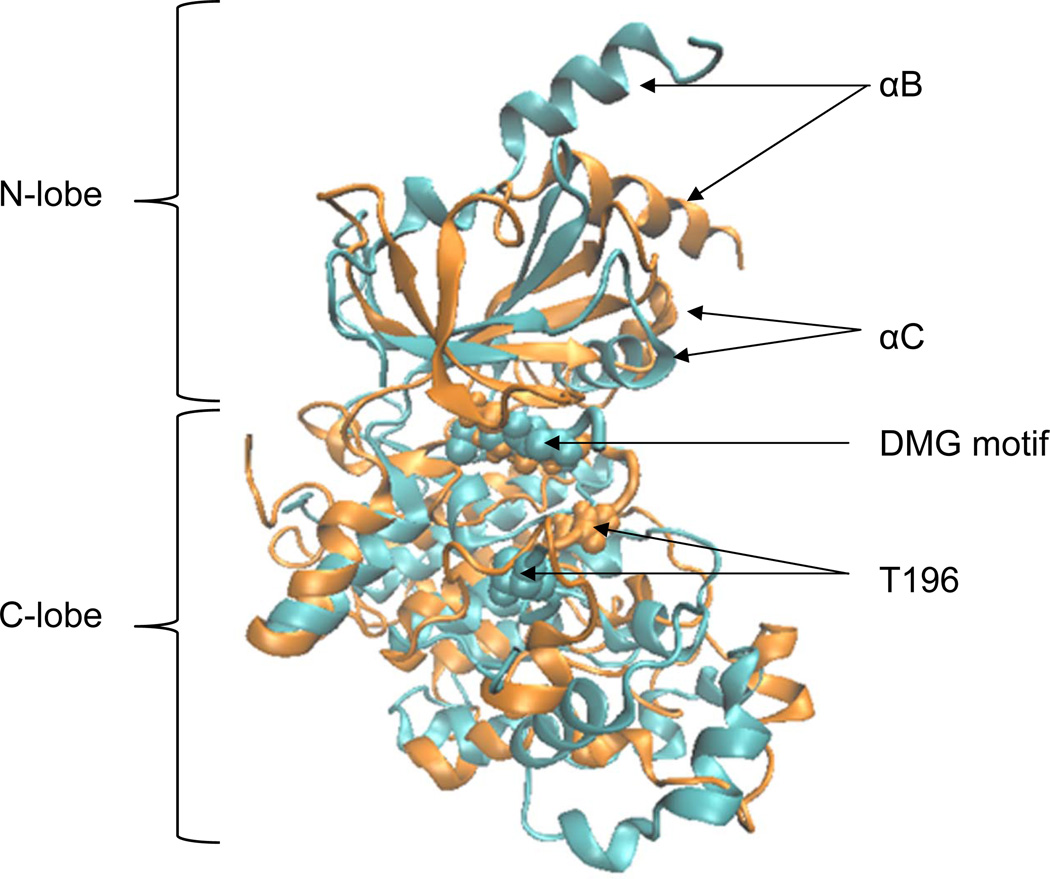
To further investigate the cause of loss of two helical structures, we have performed an independent MD simulation of the CDK8 crystal structure (PDB code: 3RGF A chain) using the identical simulation procedure. The structural differences between CDK8 crystal structure and the CDK8 structure with the lowest potential energy during the 50-ns MD simulation, designated as the “CDK8 MD structure” hereafter, are shown in Figure 4. Two αB and αC of the CDK8 MD structure still maintain their helical structures during the 50-ns simulations, although their relative local positions have been shifted. Since the CDK8 MD structure has the A-loop with the deletion of 16 amino acids, it is reasonable to conclude that presence of CDK8 partner CycC is not necessary to maintain their helical structures and full length of the A-loop may affect the helical structures of αB and αC.
Figure 4. Comparison of the CDK8 crystal and MD Structures.
Cyan: crystal CDK8 structure; Orange: CDK8 MD structure
The structures and dynamics of human CDK8 A-loop
The A-loop plays a critical role in controlling the activation state of the kinases. The A-loop is often partially disordered in inactive kinases. In many kinases, adoption of the catalytically competent conformation to form the peptide-binding platform is triggered by phosphorylation. The side chain of the aspartate in the DFG motif (DMG motif in CDK8) in the N-terminal region of the A-loop points towards the phosphate groups of ATP and plays a critical role in coordinating a magnesium ion, which is required for ATP binding, while the C-terminal region of the A-loop adopts an open conformation, serving as a platform for docking the substrate peptides.
Conformational change of DFG motif in the A-loop is reported to be important for kinase catalysis. Long-time scale MD simulations of the Abl kinase have yielded insights into the mechanism of the DFG conformational changes [49]. If one considers just the peptide segment spanning the DFG motif, the preferred conformation corresponds to the DFG-out conformation, with the aspartate out of the active site. This is because the backbone φ and ψ values in the DFG-in conformation are in an entropically disfavored region of the Ramachandran diagram, whereas the DFG-out conformation is in a more favorable region. The DFG-in conformation resembles a coiled spring that is ready to flip to a more favorable DFG-out conformation. The nucleotide-free DFG-out conformation, which is more flexible than the DFG-in conformation, might facilitate nucleotide release and the rebinding of ATP [49]. The DFG flip results in the polar DFG-aspartate entering the hydrophobic environment previously occupied by the DFG-phenylalanine, which carries a high energetic penalty. Protonation of the DFG-aspartate, driven by the increase of its pKa upon ATP hydrolysis and the release of ADP and a magnesium ion from the active site, might decrease the associated cost in free energy [49].
Due to the conservation of the DFG motif across the kinase family tree, this protonation-dependent switch might represent a general mechanism that facilitates the release of ADP from the active site [50]. The MD simulations of the Abl kinase also point to a potential role of the CDK/Src-like inactive conformation as an intermediate in DFG flipping that would facilitate the DFG-flip by enabling the bulky hydrophobic DFG-phenylalanine to move towards the position previously occupied by the DFG-aspartate [49, 51]. One interesting aspect of this analysis is that intermediate structures in the computational trajectories correlate with the crystal structures of different kinases. That is, intermediate steps in the DFG flip can be reconstructed by linking the experimentally determined structures of different kinases [50].
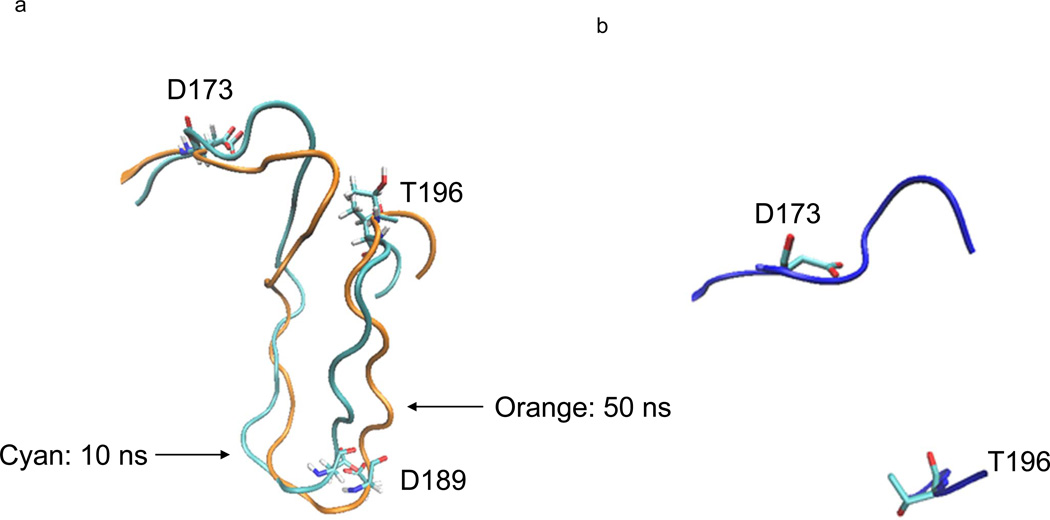
The A-loop in the CDK8 MD structures are different from the A-loop of crystal CDK8 structure (Figure 5a and 5b), although this loop appears not undergo dramatic changes over the entire 50-ns simulation time scale by comparing the 10-ns structure with the one of 50-ns (Figure 5a). However, the DMG stays at outuring the 50-ns simulation. Thus to understand the dynamic changes, we analyzed hydrogen bond formation and breakage of A-loop during the simulation.
Figure 5. Dynamics of human CDK8 A-loop during the 50-ns molecular dynamic simulation.
a, The two conformations of human CDK8 A-loop during the 50-ns MD simulation. The conformation with cyan color was from the structure with the lowest potential energy during the first 10-ns simulation, and the conformation with orange color was from the structure with the lowest potential energy during the 50-ns simulation; b, The conformation of the A-loop of human CDK8 crystal structure (PDB code: 3RGF). The 16 amino acids in the middle of the A-loop are not solved in the X-ray crystallography.
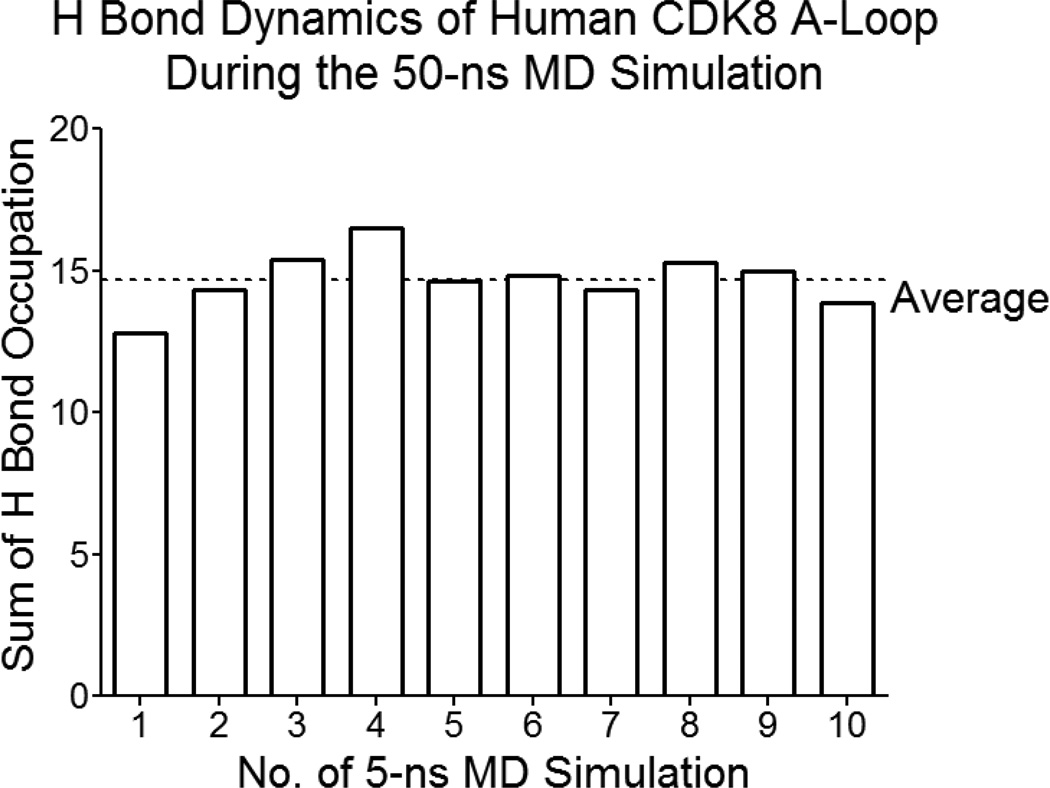
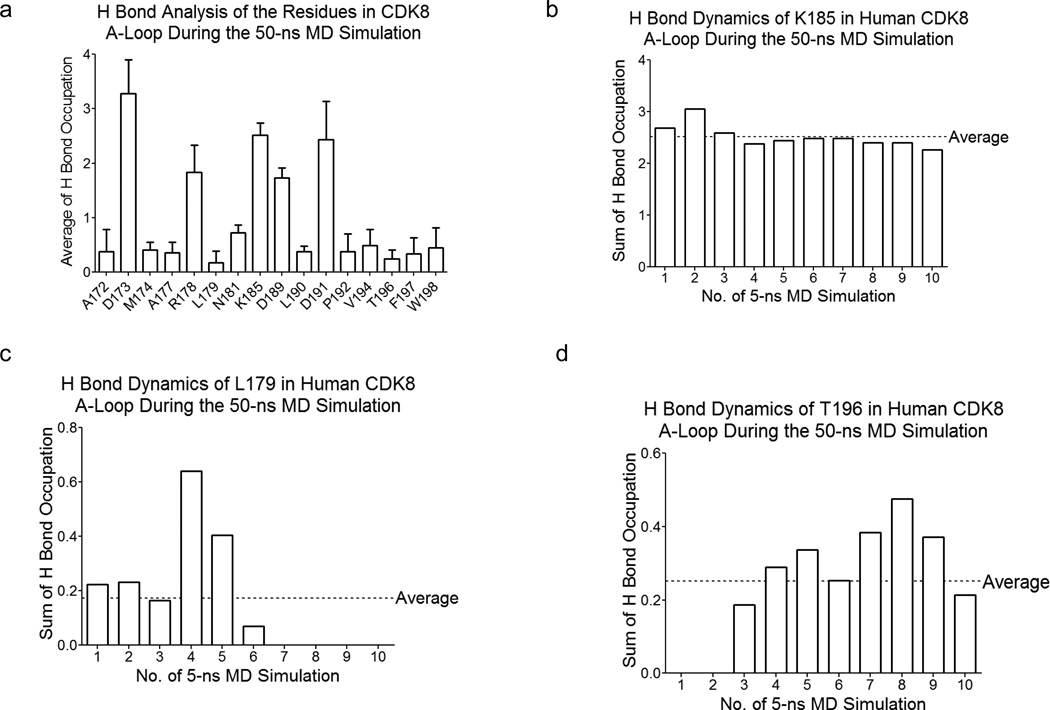
The hydrogen bond occupation gradually increases in the first 20-ns simulation, and stays relatively stable in the later 30-ns simulation (Figure 6). Next, we analyzed hydrogen bond dynamics of each residue in the A-loop. Our analysis shows three categories of hydrogen bond interactions: (i) high hydrogen bond interactions including D173, R178, K185, D189 and D191; (ii) low hydrogen bond interactions: A172, M174, A177, L179, N181, L190, P192, V194, T196, F197 and W198 (Figure 7a); (iii) no or nearly no hydrogen bond interactions: G175, F176, F180, S182, P183, L184, P186, L187, A188, V193 and V195. In addition, we also found three types of dynamic processes of hydrogen bond formation and breakage: steady, decrease, increase. For example, K185 stays steady during the simulation (Figure 7b); while there is hydrogen occupation for the first 30-ns simulation for L179 and then hydrogen bond interactions disappeared in the later 20-ns simulation (Figure 7c). In contrast, T196 has no hydrogen bond interaction in the beginning of the simulation (the first 10-ns), and then hydrogen bonding stays at relative low level in the later 40-ns simulation (Figure 7d).
Figure 6. Hydrogen bond dynamics of human CDK8 A-loop during the 50-ns MD simulation.
Hydrogen bond occupation of human CDK8 A-loop gradually increases in the first 20-ns MD simulation and stays relative stable during later 30-ns simulation.
Figure 7. Hydrogen bond analysis of the residues in human CDK8 A-loop during the 50-ns MD simulation.
a, Average hydrogen bond distribution per residue (the residues were selected with >10% average hydrogen bond occupation) in human CDK8 A-loop; b, Hydrogen bond dynamics of K185 in human CDK8; c, Hydrogen bond dynamics of Leu179 in human CDK8 A-loop; d, Hydrogen bond dynamics of T196 in human CDK8 A-loop.
The effect of point mutations of D173A and D189N in the A-loop on the overall structures
Asp in the DMG motif of CDK8 is highly conserved, and it is predicted to bind phosphate group of ATP. It has been experimentally shown that the D173A point mutation of CDK8 completely inactivates its kinase activity [14, 20], but this D173A mutation does not affect the transcription repression function of CDK8 for certain genes [15]. Our sequence alignment study showed that Asp173 is highly conserved from yeast to man [5]. A CDK8 point mutation (D189N) was found in diverse tumor samples [28], but the functional consequence of this point mutation remains unknown. The alignment studies of CDK8 sequences identified a conserved Thr196 in the A-loop from yeast to human [52] and also across all CDKs [5]. The experimental data demonstrated that Thr172 of CDK4, Thr170 of CDK7, and Thr186 of CDK9, corresponding to Thr196 of CDK8, are phosphorylated [27, 53, 54]. The phosphorylated Thr170 (pT170) in CDK7 makes one hydrogen bond through its phosphate group to the main chain nitrogen of Gln22 from the glycine loop between β1/β2 and approaches the γ phosphate of ATP [27], suggesting the importance of Thr phosphorylation in ATP binding, or its kinase activity, or both. Phosphorylated Thr172 (pT172) must be accessible to γ phosphate in the pT172CDK4-CycD3 complex present in this sample, further supporting that unphosphorylated CDK4-CycD3, unlike unphosphorylated CDK2-CycA [55], lacks any detectable activity [53]. Phosphorylation of Thr186 (pT186) in the activation segment is important for CDK9 kinase activity [54]. Unlike other CDKs, Thr186 is not phosphorylated by the CDK-activating kinase CAK (CDK7/CycH/Mat1) [56], instead it has been suggested that it is an autophosphorylation site [54].
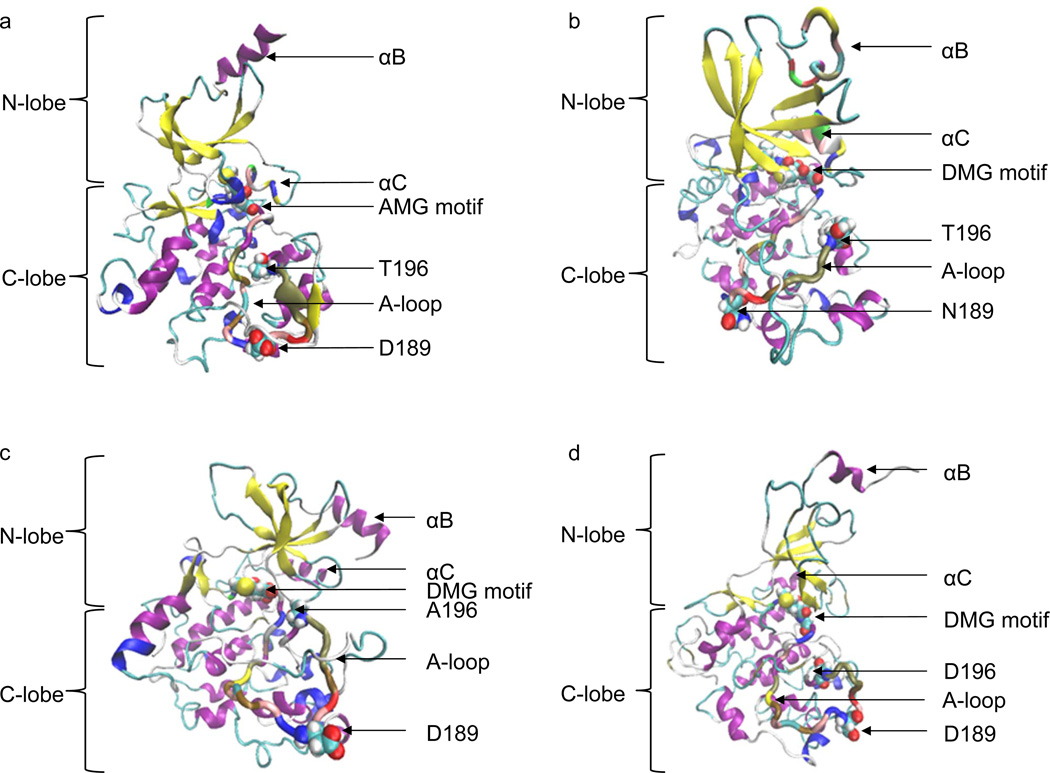
To understand effects of the point mutations on CDK8 overall structure, we have generated the structures of CDK8 D173A, D189N, T196A and T196D. Phosphorylated Thr196 parameter file has not been developed, so we generated T196D to use Asp for mimicking phosphorylation of Thr196 and T196A as the phosphorylation-site mutant. We found that the N-lobe and the C-lobe are not separated well in CDK8 D173A, T196A and T196D mutations, while CDK8 D189N mutation did not influence overall structure of N-lobe and C-lobe (Figure 8a, 8b, 8c and 8d). Interestingly, CDK8 D173A and T196A did not significantly affect the helical structures of αB and αC. In contrast, CDK8 D189N and T196D affect their helical structures.
Figure 8. The molecular dynamics of CDK8 D173A, D189N, T196A and T196D structures.
a, The CDK8 D173A structure with the lowest potential energy during the 50-ns MD simulation; b, The CDK8 D189N structure with the lowest potential energy during the 50-ns MD simulation; c, The CDK8 D196A structure with the lowest potential energy during the 50-ns MD simulation; d, The CDK8 T196D structure with the lowest potential energy during the 50-ns MD simulation.
Next, we evaluated potential energy changes. The potential energy change of CDK8 decreases gradually and smoothly during the 50-ns MD simulations. In contrast, none of the point mutations has a gradual and smooth trend of decreasing (Supplementary Figure 2a, 2b, 2c and 2d). CDK8 D173A (−8192.73 kcal/mol) has higher average potential energy than those of CDK8 (−8268.68 kcal/mol), CDK8 D189N (−8296.61 kcal/mol) and CDK8 T196A (−8236.07 kcal/mol), which are slightly higher than that of CDK8 T196D (−8326.55 kcal/mol) over the 50-ns MD simulations. By averaging the entire 50-ns simulations, CDK8 T196D has the lowest energy potential as well. The effect of the point mutations on the lowest potential energy follows the similar trend as the average potential energy: CDK8 D173A (−8399.94 kcal/mol at 28,603 ps) > CDK8 D189N (−8480.03 kcal/mol 23,717 ps), CDK8 T196A (−8461.10 kcal/mol at 48,044 ps) > CDK8 (−8536.06 kcal/mol at 49,408 ps) and CDK8 T196D (−8532.34 kcal/mol at 41,687 ps).
To further validate the effect of the point mutation on potential energy, we independently generated CDK8 D173A, T196A and T196D solely using CDK8 crystal structure as the template by molecular modeling, and then performed independent 50-ns MD simulations for each mutation along with the crystal structure as the control. We note that those point mutations have the A-loops with the 16-amino acid deletion and D189 is located within the deleted region, thus preventing us from studying effect of D189N mutation using this approach. The simulation results confirmed that the D173A mutation has higher potential energy (the lowest potential energy, −7808.91 kcal/mol at 47,003 ps) and T196D has lower potential energy (the lowest potential energy, −7901.92 kcal/mol at 10,168 ps), suggesting T196D is thermodynamically more stable and D173A is less stable.
Molecular dynamic simulation of CDK8-CycC complex
Computational methods that combine molecular mechanics energy and implicit solvation models, such as Molecular Mechanics/Poisson Boltzmann Surface Area (MM/PBSA) [30] and Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) [31], have been widely exploited in free energy calculations. The crystal structure of human CDK8-CycC complex has revealed the residues involved in the physical binding between these two proteins [25]. The residues of CDK8 interacting with CycC are localized only in the N-terminal region (~150 amino acids) (Supplementary Figure 3a), while the residues of CycC participating the interactions are from the N-terminal, middle and C-terminal regions (Supplementary Figure 3b). The major contacts between CDK8 and CycC include electrostatic, hydrogen bond and van der Waals interactions (Supplementary Figure 3a and 3b).
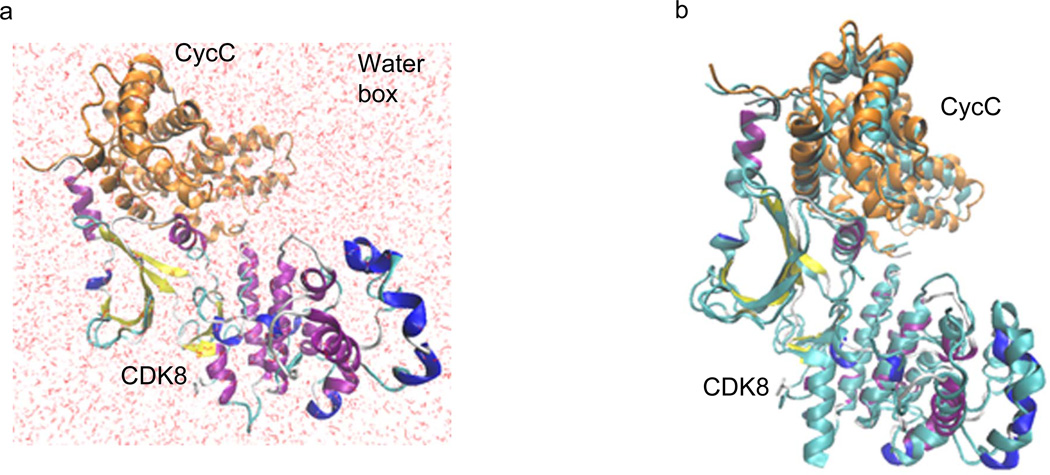
To further evaluate dynamics and binding energy of the CDK8-CycC complex in solution, MM-PBSA and MM/GBSA methods as implemented in AMBER 11 were used to calculate the binding free energy for the association of CDK8 and CycC. As summarized in Table 1, the binding free energies of CDK8-CycC complex are −105.20 kcal/mol by MM/GBSA method and −103.28 kcal/mol by MM/PBSA method, suggesting binding of CDK8 and CycC is thermodynamically favored. The binding energies between CDK8 and CycC calculated by either MM/GBSA method or MM/PBSA method are comparable. In addition, the major favorable contributors to the binding of CDK8 and CycC of both methods are van der Waals energies (VDWAALS) and electro-static energies (EEL). The structure of CDK8-CycC complex with the lowest potential energy in solution during the MD simulation was extracted (Figure 9A). By comparing this structure with the crystal structure, minor structural changes were observed (Figure 9B), demonstrating a relative stable CDK8-CycC complex in solution.
Table 1.
The Binding Energy Calculation of Human CDK8-CycC Complex by MM/PBSA and MM/GBSA Methods.
| Methods | Energy Component |
CDK8-CycC Complex (kcal/mol) |
CDK8 (kcal/mol) |
CycC (kcal/mol) |
Difference (Complex – CDK8 – CycC) (kcal/mol) |
|---|---|---|---|---|---|
| GENERALIZED BORN |
VDWAALS | −4989.5033 ± 29.1219 | −2548.0729 ± 22.9616 | −2259.4853 ± 19.3454 | −181.9451 ± 6.9641 |
| EEL | −42816.1127 ± 138.5376 | −23548.6335 ± 106.0965 | −18986.0963 ± 74.4193 | −281.3829 ± 35.8417 | |
| EGB | −7494.7055 ± 106.5943 | −4487.4092 ± 87.8479 | −3391.7071 ± 55.1357 | 384.4108 ± 35.2996 | |
| ESURF | 216.1872 ± 2.1345 | 138.0664 ± 1.8321 | 104.4015 ± 1.1693 | −26.2807 ± 0.5023 | |
| G gas | −47805.6160 ± 141.5653 | −26096.7064 ± 108.5528 | −21245.5816 ± 76.8926 | −463.3280 ± 36.5120 | |
| G solv | −7278.5183 ± 106.6157 | −4349.3428 ± 87.8670 | −3287.3056 ± 55.1481 | 358.1301 ± 35.3032 | |
| TOTAL | −55084.1343 ± 78.4694 | −30446.0492 ± 52.0191 | −24532.8872 ± 50.4504 | −105.1979 ± 7.7940 | |
| POISSON BOLTZMANN |
VDWAALS | −4989.5033 ± 29.1219 | −2548.0729 ± 22.9616 | −2259.4853 ± 19.3454 | −181.9451 ± 6.9641 |
| EEL | −42816.1127 ± 138.5376 | −23548.6335 ± 106.0965 | −18986.0963 ± 74.4193 | −281.3829 ± 35.8417 | |
| EPB | −7358.6092 ± 107.4503 | −4397.9726 ± 86.7258 | −3339.6900 ± 54.8069 | 379.0534 ± 37.7017 | |
| ECAVITY | 160.2009 ± 1.6427 | 102.2377 ± 1.4619 | 76.9701 ± 0.9047 | −19.0068 ± 0.3807 | |
| G gas | −47805.6160 ± 141.5653 | −26096.7064 ± 108.5528 | −21245.5816 ± 76.8926 | −463.3280 ± 36.5120 | |
| G solv | −7198.4083 ± 107.4629 | −4295.7350 ± 86.7381 | −3262.7199 ± 54.8144 | 360.0466 ± 37.7036 | |
| TOTAL | −55004.0243 ± 81.6833 | −30392.4414 ± 54.5076 | −24508.3015 ± 50.1600 | −103.2814 ± 11.0789 |
Figure 9. The theoretical structure of human CDK8-CycC complex with the lowest potential energy in solution during the molecular dynamic simulation.
a, The theoretical structure of human CDK8-CycC complex with the lowest potential energy in the water box; b, Overlap of MD simulated CDK8-CycC complex with the crystal CDK8-CycC complex. CDK8: cyan represents crystal structure and structure presentation represents MD structure. CycC: cyan represents crystal structure and orange represents MD structure.
Physical interactions between Drosophila CDK8 and CycC by using yeast two-hybrid assay
To examine the direct interactions between the fly CDK8 and CycC, CDK8 or CycC was fused to the yeast Gal4 DNA-binding or transactivation domain, and then their physical interaction was tested in the yeast two-hybrid system. As shown in Supplementary Figure 4, there is no detectable interaction between dCDK8 and dCycC when dCDK8 was fused to Gal4 BDB and dCycC was fused to Gal4 TAD. In contrast, we observed a weak interaction between dCDK8 and dCycC when dCDK8 was fused to Gal4 TAD and dCycC was fused to Gal4 BDB. These observations are consistent to the previous report that used the LexA yeast two-hybrid assay [57]. In addition, we found that dCycC-BDB was self-active, suggesting dCycC is able to physically interact with yeast proteins to initiate transcription in yeast cells.
Discussion and Conclusion
We have employed molecular modeling and simulations to investigate, with atomistic details, CDK8 and CDK8-CycC interactions in the rigor state. Although our 50-ns MD simulations on CDK8 and CDK8 mutations are more extensive than previous simulations, they remain relatively short compared with the functionally relevant time scales for CDK8 folding in solution. Additionally, we have carried out 2-ns MD simulation for CDK8-CycC complex that is short time scale for CDK8-CycC binding. Longer simulations will be needed to assess the convergence of our results, which will be pursued in the future.
The phosphate group of phosphorylated kinase usually promotes closure of the two lobes of the domain and the correct conformation of the activation segment for substrate binding. For example, the phosphothreonine 197 of protein kinase A (PKA) acts as an organizing center and hydrogen bonds to the side chains of H87 from the C helix, R165 located immediately at the N-terminal to the catalytic aspartate, and K189 from the activation segment [58]. In the CDK9-CycT complex, the phosphoresidue was predicted to serve as anchor to adjust the orientation of three important arginines, thereby inducing an open conformation of the activation segment involving two of the three proposed arginines [59]. Specifically, Arg65CDK9 (αC helix) and Arg148CDK9 (A-loop), interact with pThr186CDK9, while Arg172CDK9 (C-lobe) is just too distant [59]. In other kinases [e.g., phosphorylase kinase (PhK), PDB code: 1PHK; epidermal growth factor receptor (EGFR), PDB code: 2GS2; CDK5, PDB code: 1H4L], the activation segment does not require phosphorylation for activity and is able to adopt the correct conformation through other interactions [40].
In human CDK8, two activation segment distant phosphorylation sites (pThr410CDK8 and pSer413CDK8) have been identified in its C-terminus [60], but no functional role has been described for these yet. Thr196 in the A-loop is conserved from yeast to human [52] and also across all CDKs [5]. However, there is no experimental data to date demonstrating that CDK8 Thr196 is phosphorylated. The CDK8 T196D mutation has the lowest average potential energy in our 50-ns MD simulation, which prompted us to investigate the hydrogen bonds of CDK8 T196 and D196. In the CDK8 crystal structure, T196 forms a hydrogen bond with a water molecule (Water ID: 486) (Supplementary Figure 5a), and T196 forms a hydrogen bond with R140 in the structure with the lowest potential energy during 50-ns MD simulation (Supplementary Figure 5b). With full-length of the A-loop, T196 has a hydrogen bond with R200 (Supplementary Figure 5c) and D196 has a hydrogen bond with Q251 and is close to R237 (Supplementary Figure 5d). It thus appears that full length of the A-loop has an impact on T196 hydrogen bonding, and the T196D mutation completely changes the hydrogen bond interactions (between T196 and R140/R200).
The interaction between Drosophila CDK8 and CycC has been tested previously using pAS2 and pACT2 vectors (LexA yeast two-hybrid system) in yeast strain Y190 by yeast two-hybrid approach [57]. It appears that dCycC alone is not active in their assay. However, an interaction between dCDK8 and dCycC is only observed in the systems when dCycC is fused with DBD and dCDK8 is fused with TAD, not in the systems when dCDK8 is fused with DBD and dCycC is fused with TAD, which is consistent in both the LexA [57] and Gal4 yeast two-hybrid assay systems (Supplementary Figure 4). The results from both of the assay systems demonstrate the interaction between dCDK8 and dCycC is direct, but relatively weak.
In summary, our molecular dynamic simulations of the 3-D structures of human CDK8 and CDK8 point mutations (D173A and D189N) with full-length of the A-loop in solution and associated binding mode shed light on CDK8 structure dynamics and interaction between CDK8 and CycC. Our results provide a means to evaluate the function of CDK8, its A-loop and its interaction with CycC. These results will help to improve our understanding of the exact biochemical and physiological functions and regulation of CDK8 and CycC.
Supplementary Material
Highlights.
-
➢
CDK8 structure gradually loses two helical structures during the MD simulations.
-
➢
Hydrogen bond occupation of the CDK8 A-loop increases and then stays stable.
-
➢
The A-loop residues exhibit three changes: increasing, decreasing, and stable.
-
➢
CDK8 D173A has the highest average potential energy, while T196D has the lowest.
-
➢
Binding energy of CDK8 and CycC was calculated by MM/PBSA and MM/GBSA methods.
Acknowledgements
This work was supported by the grants of Enhancement Program of Louisiana Board of Reagents (LEQSF(2012-13)-ENH-TR-32) (awarded to WX) and the sub-award from NIH LEQSF(2010-15)-INBRE program (awarded to WX). XJX and JYJ are supported by a grant from NIH (NIDDK 1R01DK095013) (awarded to JYJ). The MD simulation and visualization of human CDK8 and CycC were performed at the Louisiana Optical Network Initiative (LONI) and Louisiana Immersive Technologies Enterprise (LITE). The authors thank the LONI support team, especially to Alexander Pacheco.
Glossary
Abbreviations footnote
- CDK8
Cyclin-dependent kinase 8
- CycC
Cyclin C
- MD
molecular dynamics
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24(11):1770–1783. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz GK, Shah MA. Targeting the cell cycle: a new approach to cancer therapy. J Clin Oncol. 2005;23(36):9408–9421. doi: 10.1200/JCO.2005.01.5594. [DOI] [PubMed] [Google Scholar]
- 3.Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, Morgan DO, Tsai LH, Wolgemuth DJ. Cyclin-dependent kinases: a family portrait. Nat Cell Biol. 2009;11(11):1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher RP. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J Cell Sci. 2005;118(Pt 22):5171–5180. doi: 10.1242/jcs.02718. [DOI] [PubMed] [Google Scholar]
- 5.Xu W, Ji JY. Dysregulation of CDK8 and Cyclin C in tumorigenesis. J Genet Genomics. 2011;38(10):439–452. doi: 10.1016/j.jgg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013;48(6):575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casamassimi A, Napoli C. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie. 2007;89(12):1439–1446. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 9.Mittler G, Kremmer E, Timmers HT, Meisterernst M. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2001;2(9):808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 11.Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M, et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell. 2004;14(5):553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30(5):256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2(1):43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 14.Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407(6800):102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 15.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23(4):439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galbraith MD, Donner AJ, Espinosa JM. CDK8: a positive regulator of transcription. Transcription. 2010;1(1):4–12. doi: 10.4161/trns.1.1.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27(1):121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17(2):194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153(6):1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455(7212):547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455(7212):552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firestein R, Hahn WC. Revving the Throttle on an oncogene: CDK8 takes the driver seat. Cancer Res. 2009;69(20):7899–7901. doi: 10.1158/0008-5472.CAN-09-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, Leroy G, Vidal CI, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468(7327):1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiano C, Casamassimi A, Rienzo M, de Nigris F, Sommese L. Napoli C: Involvement of Mediator complex in malignancy. Biochim Biophys Acta. 2013;1845(1):66–83. doi: 10.1016/j.bbcan.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Schneider EV, Bottcher J, Blaesse M, Neumann L, Huber R, Maskos K. The Structure of CDK8/CycC Implicates Specificity in the CDK/Cyclin Family and Reveals Interaction with a Deep Pocket Binder. J Mol Biol. 2011;412(2):251–266. doi: 10.1016/j.jmb.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 26.Lolli G. Structural dissection of cyclin dependent kinases regulation and protein recognition properties. Cell Cycle. 2010;9(8):1551–1561. doi: 10.4161/cc.9.8.11195. [DOI] [PubMed] [Google Scholar]
- 27.Lolli G, Lowe ED, Brown NR, Johnson LN. The crystal structure of human CDK7 and its protein recognition properties. Structure. 2004;12(11):2067–2079. doi: 10.1016/j.str.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 28.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwier MC, Chong LT. Reaching biological timescales with all-atom molecular dynamics simulations. Curr Opin Pharmacol. 2010;10(6):745–752. doi: 10.1016/j.coph.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J Am Chem Soc. 2001;123(22):5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 31.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Gallo AA, Bajpai RK, Chistoserdov A, Nelson AT, Segura LN, Xu W. The diversity and molecular modelling analysis of BSUB alignright12-dependent and BSUB alignright12-independent glycerol dehydratases. International Journal of Bioinformatics Research and Applications. 2010;6:484–507. doi: 10.1504/IJBRA.2010.037988. [DOI] [PubMed] [Google Scholar]
- 33.Lu G, Moriyama EN. Vector NTI, a balanced all-in-one sequence analysis suite. Briefings in Bioinformatics. 2004;5(4):378–388. doi: 10.1093/bib/5.4.378. [DOI] [PubMed] [Google Scholar]
- 34.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 35.Simmerling C, Strockbine B, Roitberg AE. All-atom structure prediction and folding simulations of a stable protein. J Am Chem Soc. 2002;124(38):11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 36.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr., Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. Journal of computational chemistry. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 38.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109(3):275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 39.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15(5):661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 40.Endicott JA, Noble ME, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81:587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- 41.Betzi S, Alam R, Martin M, Lubbers DJ, Han H, Jakkaraj SR, Georg GI, Schonbrunn E. Discovery of a potential allosteric ligand binding site in CDK2. ACS Chem Biol. 2011;6(5):492–501. doi: 10.1021/cb100410m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider EV, Bottcher J, Huber R, Maskos K, Neumann L. Structure-kinetic relationship study of CDK8/CycC specific compounds. Proc Natl Acad Sci U S A. 2013;110(20):8081–8086. doi: 10.1073/pnas.1305378110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norman RA, Toader D, Ferguson AD. Structural approaches to obtain kinase selectivity. Trends Pharmacol Sci. 2012;33(5):273–278. doi: 10.1016/j.tips.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 44.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356(2):125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 45.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376(6538):313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 46.Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol. 1999;287(5):821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- 47.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A. 2006;103(47):17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noble M, Barrett P, Endicott J, Johnson L, McDonnell J, Robertson G, Zawaira A. Exploiting structural principles to design cyclin-dependent kinase inhibitors. Biochim Biophys Acta. 2005;1754(1–2):58–64. doi: 10.1016/j.bbapap.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 49.Shan Y, Seeliger MA, Eastwood MP, Frank F, Xu H, Jensen MO, Dror RO, Kuriyan J, Shaw DE. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci U S A. 2009;106(1):139–144. doi: 10.1073/pnas.0811223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42(1):9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levinson NM, Kuchment O, Shen K, Young MA, Koldobskiy M, Karplus M, Cole PA, Kuriyan J. A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 2006;4(5):e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36(12):3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takaki T, Echalier A, Brown NR, Hunt T, Endicott JA, Noble ME. The structure of CDK4/cyclin D3 has implications for models of CDK activation. Proc Natl Acad Sci U S A. 2009;106(11):4171–4176. doi: 10.1073/pnas.0809674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive PTEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280(31):28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 55.Hagopian JC, Kirtley MP, Stevenson LM, Gergis RM, Russo AA, Pavletich NP, Parsons SM, Lew J. Kinetic basis for activation of CDK2/cyclin A by phosphorylation. J Biol Chem. 2001;276(1):275–280. doi: 10.1074/jbc.M007337200. [DOI] [PubMed] [Google Scholar]
- 56.Kim JB, Sharp PA. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J Biol Chem. 2001;276(15):12317–12323. doi: 10.1074/jbc.M010908200. [DOI] [PubMed] [Google Scholar]
- 57.Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, Bourbon HM. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J. 2007;26(4):1045–1054. doi: 10.1038/sj.emboj.7601566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng J, Trafny EA, Knighton DR, Xuong NH, Taylor SS, Ten Eyck LF, Sowadski JM. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr D Biol Crystallogr. 1993;49(Pt 3):362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 59.Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27(13):1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8(7):1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.