Abstract
Neutralizing antibodies (inhibitors) to replacement Factor-VIII impair the effective management of hemophilia-A1. Individuals with hemophilia-A due to major F8 gene disruptions lack antigenically cross-reactive material in their plasma (CRM-negative) and prevalence of inhibitors is >60%. Conversely, subjects with missense mutations are CRM-positive and the prevalence of inhibitors is <10%2. Individuals with the intron-22-inversion (~50% of individuals with severe hemophilia-A) should be in the former group based on the genetic defect. Although these individuals are CRM-negative, only 20% of them develop inhibitors3. Here we demonstrate the presence of comparable levels of F8 mRNA and intracellular Factor-VIII protein in B-lymphoblastoid cells and liver biopsies from healthy controls and subjects with the intron-22-inversion. These results support the hypothesis that most individuals with the intron-22-inversion are tolerized to Factor-VIII and thus do not develop inhibitors. Furthermore we developed a pharmacogenetic algorithm that permits the stratification of inhibitor risk for sub-populations by predicting immunogenicity using, as input, the number of putative T-cell epitopes in the infused FVIII and the competence of MHC-Class-II molecules to present such epitopes. The algorithm exhibited significant accuracy in predicting inhibitors in 25 unrelated individuals with the intron-22-inversion (AUC = 0.890; P = 0.001).
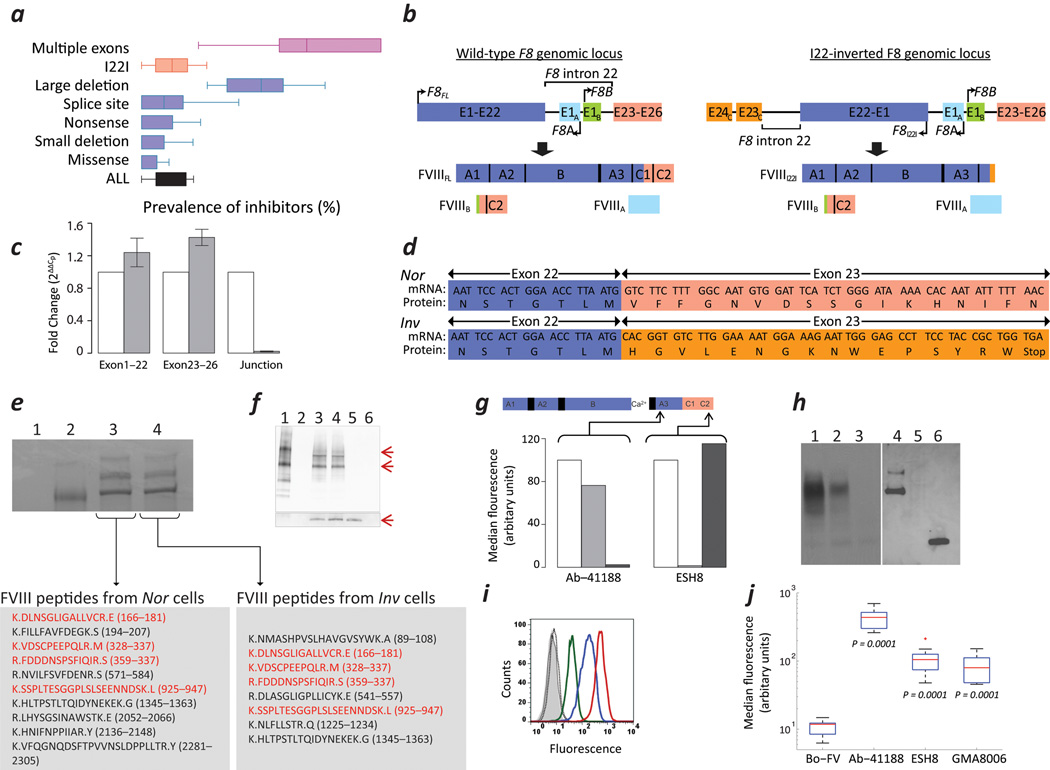
With improvements in technology and the increased use of recombinant Factor VIII (FVIII), product related risk-factors for immunogenicity have been minimized. Clinical studies have provided evidence that genetic variables, particularly the HA-causing F8-mutation-type, are important determinants of individual responses vis-à-vis immunogenicity3 (Fig. 1a). Synthesis of the FVIII polypeptide chain is necessary for inducing central tolerance; for example individuals with hemophilia-A (HA) who have a missense mutations in the F8 gene have a <10% life-time prevalence of inhibitors whereas prevalence of inhibitors in individuals with large gene deletions can be as high as 88%2. Interestingly, individuals with the I22I-mutation have a much lower than expected prevalence of inhibitors based on the type of genetic mutation and the clinical observation that these individuals exhibit CRM-negative plasma. Thus, a recent systematic review and meta-analysis of data from 5,385 subjects with severe HA showed that the individuals with large F8 deletions involving more than one exon developed inhibitors far more often than individuals with the I22I (pooled odds ratio: 3.6; 95% confidence interval: 2.3–5.7)3.
Figure 1.
Expression of FVIII in cells derived from subjects with HA. (a) Prevalence of inhibitors in individuals with HA due to various types of F8 gene defects3. (b) Structure of the wild-type (left) and I22-inverted F8 (right) and the predicted protein products. (c) mRNA expression of exons 1–22, exons 23–26, and the junction between exons 22 and 23 in the Nor (empty bars) and Inv (filled bars) cells (mean ± SD; n=3). (d) Sequence of cDNA obtained from Nor and Inv cells at the junction of exons 22 and 23. (e) Commassie Blue stained SDS-PAGE gel following immuno-precipitation of the following: (1) Negative control, immuno-precipitation in the absence of rFVIII or cell lysates. (2) ~100 ng purified FVIII. (3) lysate of > 50 × 106 Nor cells. (4) lysate of > 50 × 106 Inv cells. Human-FVIII specific peptides identified by mass spectrometric analysis of bands co-migrating with the purified rFVIII are shown in the lower panels. Peptides in red are those identified in both Nor and Inv cells. (f) Immunoblot following immuno-precipitation using a cocktail of mAbs to FVIII followed by immuno-blotting using a sheep FVIII-specific pAb: (1) ~100 ng purified FVIIIFL. (2) Blank lane. (3) Lysate of > 50 × 10 6Nor cells. (4) Lysate of >50 × 106 Inv cells. (5) ~100 ng of the purified C2 domain of FVIII. (6) Negative control, immuno-precipitation in the absence of rFVIII or cell lysates. The lower panel depicts the lower molecular weight bands overexposed using a more sensitive chemiluminescent substrate. (g) The MFI of NIH-3T3 cells transfected with F8FL (unfilled), F8I22I (light grey) and F8B (dark grey) stained with either Ab-41188 or ESH8. Arrows indicate site specificity of mAbs to FVIII molecule. (h) Immunoblots of cell lysates of HuH-7 cells transiently transfected with F8FL (Lanes 1 & 4), F8I22I (Lanes 2 & 5) and F8B (Lanes 3 & 6); domain-specific mAbs to FVIII, Ab-41188 (Lanes 1–3) and GMA8006 (Lanes 4–6) were used to probe a Western Blot of cell lysates. (i) Intracellular expression of FVIII in permeabilized Inv cells detected by flow cytometry using isotype controls IgG1 and IgG2a (grey filled and grey solid line respectively), Bo-FV (black, dotted line) and the mAbs to FVIII; Ab-41188 (red, solid), ESH8 (blue, solid) or GMA8006 (green, solid). (j) Bar and whisker plots showing the MFI of PBMCs obtained from individuals with the HA due to the I22I and labelled with the indicated Abs.
Based on the structure of the I22-inverted F8, we hypothesize that individuals with I22I translate two F8-exon-containing mRNAs (here referred to as F8I22I & F8B), which together include all 26 F8 exons of the full-length mRNA (F8FL). Furthermore, F8I22I and F8B together express the entire primary amino acid sequence of FVIII as two non-secreted polypeptide chains, FVIIII22I and FVIIIB (Figs. 1b & Supplementary Fig. 1).
To explore this possibility, we used a quantitative RT-PCR-based assay to detect and estimate the levels of F8-exon-containing mRNAs in EBV-immortalized B-lymphoblastoid cells obtained from a normal man (Nor) and from a subject with HA with the I22I (Inv). We used three sets of primers (Supplementary Table 1) to amplify exonic regions upstream and downstream of intron-22 (exons 1–22 and 23–26) and across the junction between exons 22 and 23. The primer set designed to generate cDNA spanning this junction detected F8FL transcripts, which encode the wild-type full-length FVIII protein (FVIIIFL), in Nor cells but not in Inv cells (Fig. 1c) while the primer sets designed to generate cDNAs spanning exons 1–22 and exons 23–26 showed comparable levels of F8FL, F8I22I and F8B mRNAs in Nor (F8FL and F8B) and Inv (F8I22I and F8B) cells. The intra-chromosomal homologous recombination that gives rise to the I22I-mutation can involve one of two ~10 kb segments of Xq28 chromosomal DNA, of these recombination at the distal of the two segments is more common4, and was detected in the subject that donated the Inv cells (Supplementary Fig. 2).
We predicted that the mRNA sequence of F8I22I obtained from Inv cells would yield a translated polypeptide containing 2,159 amino acid residues, with the N-terminal 2,143 residues being identical to those of the wild-type FVIII protein (Fig. 1d). The 16 additional non-FVIII amino acids at the C-terminal end of FVIIII22I, are encoded by exon-23C. Similarly, we bi-directionally sequenced a full-length cDNA of the F8B mRNA and performed an amino acid sequence alignment of the wild-type full-length FVIII protein (FVIIIFL) with the FVIIII22I and FVIIIB polypeptides, which are predicted to be encoded by the F8I22I and F8B mRNA sequences from Inv cells (Supplementary Fig. 3). Analogous to the 16 non-FVIII amino acid residues at the C-terminus of FVIIII22I, FVIIIB has eight additional residues at its N-terminal end that are not found in the FVIIIFL. Nonetheless, the polypeptides FVIIII22I and FVIIIB together comprise the entire primary sequence of F8FL (Supplementary Fig. 3).
To demonstrate that the F8inv gene does indeed synthesize the protein product, we used a human-FVIII-specific pAb to immuno-precipitate the FVIII protein. To confirm the identity of the bands that aligned with purified FVIII in a SDS-PAGE gel and subjected them to mass spectrometric analysis (Fig. 1e). These data demonstrate that both Nor and Inv cells synthesize human-FVIII polypeptides. The FVIIII22I and FVIIIB polypeptides were also identified in Inv cells by immunoprecipitation followed by an immunoblot (Fig. 1f).
We used HEK-293 cells that transiently express FVIIIFL, FVIIII22I or FVIIIB to demonstrate that the monoclonal antibodies (mAbs) Ab-41188 and ESH8 can discriminate between FVIIII22I and FVIIIB in a flow cytometry assay. The mAb Ab-41188 (A3-domain), detects FVIIIFL and FVIIII22I, but not FVIIIB while mAb ESH85–7 (C2-domain), detects FVIIIFL and FVIIIB, but not FVIIII22I (Fig. 1g). Furthermore, in all cell-lines investigated, both mAbs show a significant increase in fluorescence intensity in cells transfected with F8FL compared to the non-transfected negative control cells (Supplementary Figs. 4a,b). Moreover, the detection of FVIII polypeptides is dose dependent vis-à-vis the concentration of the F8-cDNA containing expression vector used during transfection (Supplementary Fig. 4c). Using the FVIII-specific mAbs Ab-41188 and GMA8006 (specific to the C2-domian) we also characterized the protein products of the F8FL, F8I22I and F8B genes transfected in the well differentiated human liver cell line HuH-7. In an immunoblot, as expected, both mAbs identify FVIIIFL while, Ab-41188 (A3-domain) identifies FVIIII22I but not FVIIIB and GMA8006 (C2-domain) identifies FVIIIB but not FVIIII22I (Fig. 1h).
We used the flow cytometry assay and mAbs validated above to detect FVIII polypeptides in permeabilized and non-permeabilized Nor or Inv cells (immortalized from an individual with the I22I, see above). Only a minimal amount of FVIII reactivity was observed in non-permeabilized Nor or Inv cells using A3 domain (Ab41188) or C2 domain (ESH8) specific Abs. However, when Nor and Inv cells were permeabilized, both mAbs detected their targets (Fig. 1i and Supplementary Fig. 5a), i.e. the fluorescent intensity associated with the FVIII-specific mAb was 5–10 fold higher compared to the isotype control. Moreover, a statistical analysis of four independent experiments shows that the median fluorescence intensity (MFI) of cells labeled with the antibody to Bovine Factor V (Bo-FV) used as a negative control was comparable to that of cells labeled with the isotype control. However, cells labeled with the FVIII-specific Abs showed a significantly greater fluorescence intensity compared to the Bo-FV (one sided unpaired t-test P = 0.0003 to < 0.0001) (Supplementary Fig. 5b). Similarly, FVIII could be detected in PBMCs obtained from individuals with the I22I F8, using either Ab-41188, GMA8006 or ESH8 (Fig. 1j). The MFI of cells labeled with an Bo-FV Ab was not significantly greater than that of cells labeled with the isotype control Ab (one sided unpaired t-test P = 0.1489). On the other hand the MFI values of cells labeled with mAbs Ab-41188, GMA8006 or ESH8 were significantly greater than that of cells labeled with the Bo-FV Ab (one sided unpaired t-test P < 0.0001).
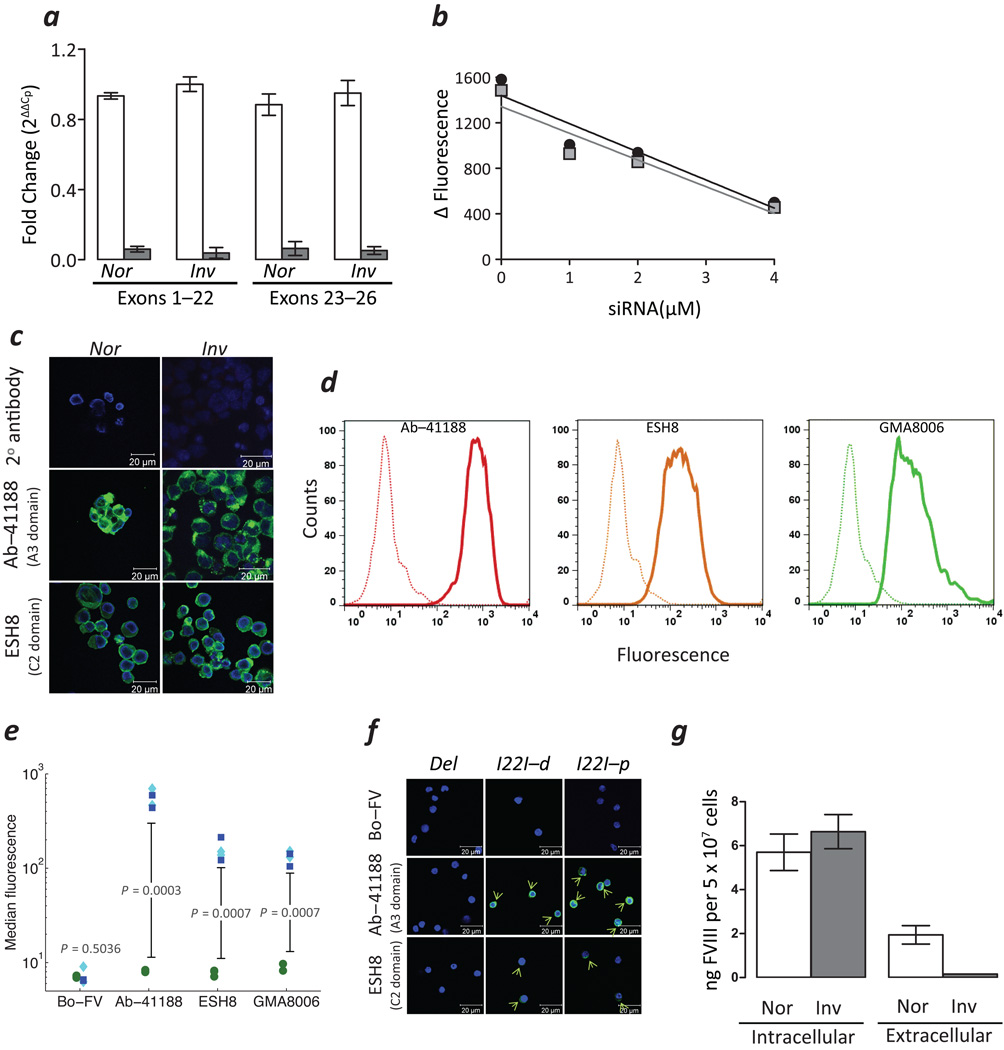
Although we have demonstrated the site-specificity of the mAbs used in transient expression systems (Figs. 1g,h and Supplementary Fig. 4), we provide additional evidence for the specificity of these mAbs in detecting FVIII-polypeptides in Nor and Inv cells by using Smart Pool FVIII targeted siRNA in a knockdown experiment. We demonstrate almost complete disappearance of the F8FL, F8I22I and F8B mRNAs (Fig. 2a) as well as a substantial decrease in the levels of FVIIIFL, FVIIII22I and FVIIIB (Fig. 2b) in cells transfected with FVIII specific (F8) siRNA but not in cells transfected with the scrambled (NT) siRNA. Moreover, there is a dose-dependent decrease in the FVIII signal as a function of siRNA concentration (Fig. 2b) and the highest concentration of siRNA reduced the FVIII levels by approximately 70%.
Figure 2.
FVIII expression in subjects with HA. (a) Expression of indicated mRNAs (mean ± SD; n = 3) in Nor and Inv cells treated with Smart Pool siRNA specific to FVIII (F8-siRNA, unfilled bars) or scrambled non-specific siRNA (NT-siRNA, filled gray bars). (b) Relative expression of FVIII estimated in Inv cells treated with indicated concentrations of F8-siRNA by flow cytometry using mAbs, Ab-41188 (grey squares) and ESH8 (black circles). (c) Confocal images of permeabilized Nor (left panels) and Inv cells (right panels) labelled with the indicated mAbs. Areas stained by the Abs to FVIII are depicted in green and the DAPI stained nuclei in blue. (d) Detection of intracellular FVIII in a flow cytometry assay using the indicated mAbs in PBMCs obtained from individuals with HA due to either a deletion in the F8 gene (dotted lines) or the I22I (solid lines). (e) The flow cytometry assay depicted in Fig. 2d was carried out on samples from multiple subjects. The MFI of cells obtained from subjects with the I22I (blue squares, proximal; cyan diamonds, distal) were compared with those obtained from subjects with the large deletion (green circles) when detected with the indicated mAbs. (f) Confocal images of permeabilized PBMCs from subjects with a large deletion and I22I (both distal (I22I-d) and proximal (I22I-p)) labelled with the indicated mAbs. Areas stained by the Abs to FVIII are depicted in green and the DAPI stained nuclei in blue. Green arrows show cells stained by FVIII-specific mAbs. (g) FVIII levels in the conditioned medium (extracellular) and lysates (intracellular) of Nor and Inv cells estimated by ELISA (mean ± SD; n = 3).
Confocal microscopic imaging of permeabilized Nor and Inv cells labelled with Ab-41188 or ESH8 also show the presence of FVIII polypeptides in intracellular compartments (Fig. 2c). In addition to using isotype-control Abs and a Bo-FV Ab as negative controls we also used PBMCs from two subjects with a large deletion (exons 23–26) in whom no FVIII-activity or protein has been detected. Permeabilized PBMCs from subjects with the large deletion or I22I were labelled with the Ab-41188, ESH8 and GMA8006 monoclonal antibodies (mAb) (Fig. 2d). All three mAbs show a significantly higher fluorescent signal in cells obtained from individuals with the I22I. Multiple flow cytometry assays on PBMCs obtained from 4 individuals with the I22I (two proximal and two distal) and two individuals with large multi-exon deletions are summarized in Fig. 2e. The MFI is significantly higher when PBMCs from subjects with the I22I are labelled with the mAbs Ab-41188, ESH8 and GMA8006 (one sided unpaired t-test P = 0.0003, 0.0007 and 0.0007 respectively). There was no statistically significant difference in the fluorescence when the cells were labeled with the Bo-FV Ab (one sided unpaired t-test P = 0.5036). Similarly, by using confocal microscopy we could detect FVIII protein in PBMCs from a subject with the I22I but not in cells from the subject with the large deletion (Fig. 2f). Finally, to test our hypothesis that individuals with the I22I synthesize but do not secrete the FVIIII22I or the FVIIIB polypeptides, we evaluated the intracellular and extracellular levels of FVIII in the two cell lines. Whereas FVIIIFL could be detected in both the cell lysate and the extracellular medium of Nor cells, FVIIII22I and FVIIIB could only be detected in the cell lysate of Inv cells (Fig. 2g).
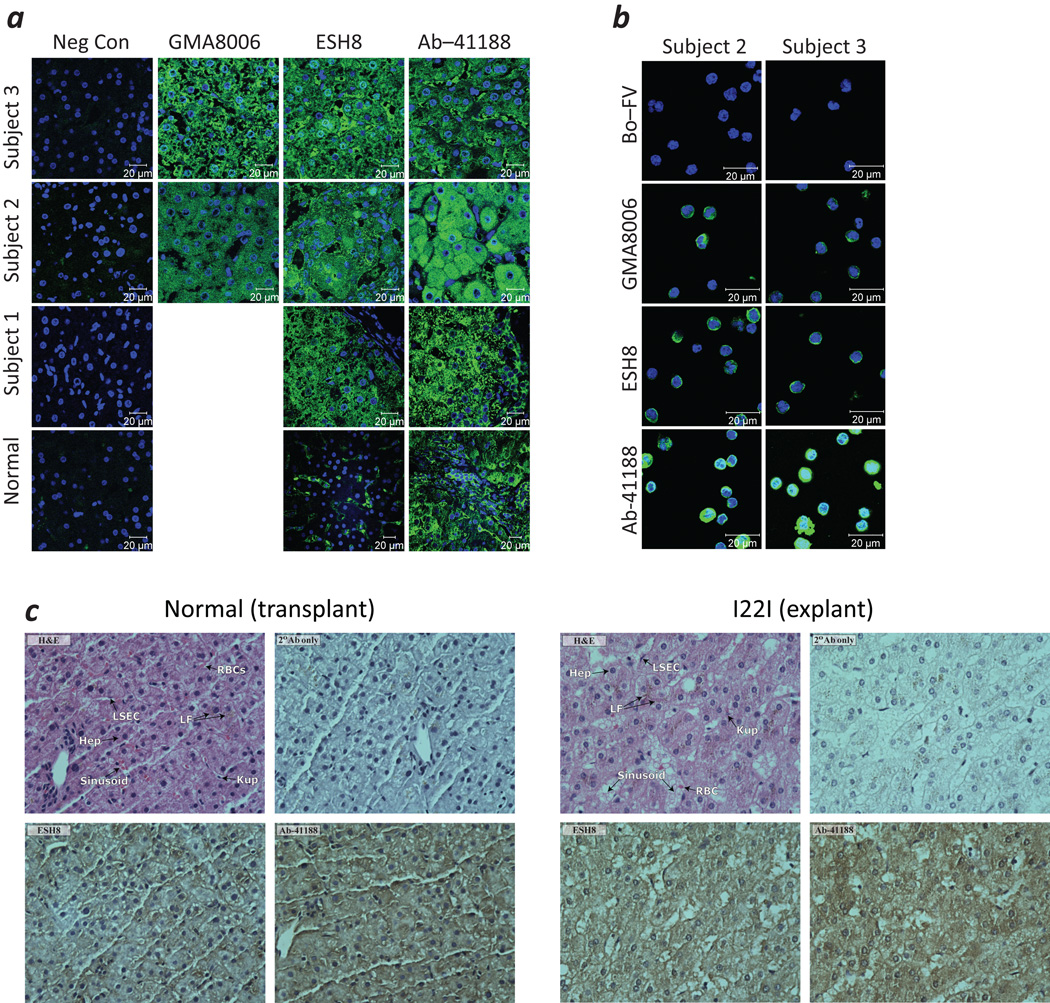
Although F8 has long been known to be transcribed by lymphoid cells, as observed in past analyses of lymph node and splenic tissues8, 9, most studies have determined that the primary site of FVIII expression is the liver9, 10, and liver transplant is curative11, 12. An immunofluorescence-based histological evaluation of tissue sections from the cirrhotic liver explanted from the subject with the I22I who was used to generate the Inv cell line as well as for 2 additional subjects revealed diffuse cytoplasmic staining of hepatocytes and liver-sinusoidal endothelia cells whether using mAbs GMA8006, ESH8 or Ab-41188, which is consistent with the expression of both FVIIII22I and FVIIIB (Fig. 3a). PBMCs from subjects 2 and 3 were also frozen at the time of liver transplant and these cells also show intracellular expression of the FVIII transcripts (Fig. 3b). Similar results were obtained from a histological examination of serial sections of the explanted liver using immunohistochemistry. ESH8-stained sections of the transplanted (normal) liver demonstrated the presence of FVIIIFL ‘and’ or ‘or’ FVIIIB within the cytoplasm of both hepatocytes and liver sinusoidal endothelial cells (LSECs) (Fig. 3c, left panel). A comparable staining pattern was seen in the explanted liver from the subject with the I22I, presumably due to the presence of FVIIIB alone (Fig. 3c, right panel). When sections of the transplanted (normal) liver were stained with Ab-41188, whose epitope resides within the A3-domain, the FVIIIFL protein was observed to be expressed by both hepatocytes and LSECs. In the explanted liver (from the subject with the I22I), a similar pattern with more pronounced staining was seen, presumably due to enhanced expression of FVIIII22I alone.
Figure 3.
FVIII expression in liver tissue obtained from individuals with HA (a) Confocal images of histologic sections obtained from the explanted liver of the individual with the I22I whose cells were used to generate the Inv cells (Sample 1), biopsies of his transplanted liver (Normal), and explanted livers from two additional subjects with the I22I (Subjects 2 & 3) immuostained with the indicated mAbs. The areas stained by the Abs specific to FVIII are depicted in green and the DAPI stained nuclei in blue. (b) Confocal images of PBMCs which were frozen at the time of transplant for Subjects 2 and 3 (Fig. 3a) stained with the indicated mAbs. Areas stained by the Abs specific to FVIII are depicted in green and the DAPI stained nuclei in blue. (c) Immunohistochemistry stained sections from a biopsy of the normal liver after its transplantation into the individual with the I22I (left) and the explanted liver from the same individual (right). Clockwise from upper left: (i) H&E stained sections; (ii) negative control sections stained with only the HRP-conjugated goat anti-mouse-IgG-Fc-secondary antibody; (iii) positive Ab-41188 stained sections and (iv) positive ESH8 stained sections.
The data presented above demonstrate that F8I22I and F8B direct the synthesis of the entire FVIII primary structure within two polypeptide chains, FVIIII22I and FVIIIB. Because the results from this comprehensive characterization of FVIII expression by the I22-inverted F8 were derived from experiments performed on samples from a single subject, with a distal inversion, we evaluated the expression of the two pertinent I22-inverted F8 loci in additional samples. We detected the F8I22I and F8B transcripts in all total RNA isolates of peripheral-blood sampled from a collection of 25 unrelated subjects with the I22I, 4 with proximal inversions and 21 with distal. For reasons described below, this cohort was comprised of subjects who self-identified as African American. Moreover, following RT-PCR and cDNA sequencing, we demonstrated that the F8I22I and F8B transcripts in these subjects exhibited sequences at their 5’- and 3’-ends, as well as throughout, which were identical to those observed in the Inv cells.
Re-sequencing all 26 exonic regions of the I22-inverted-F8 in any given subject can identify sets of foreign peptides (“putative T-cell epitopes”) contained in a candidate FVIII-therapeutic. In all subjects with the I22I, a set of 14 overlapping 15-mer peptides (derived from the FVIII-therapeutic) that span the residues at M2124 and V2125, which are encoded by the last codon of exon-22 and the first codon of exon-23, respectively would be foreign (Supplementary Fig. 6). An individual with the I22I would be exposed to additional sets of putative T-cell epitopes if the FVIII replacement product were derived from a haplotype that did not match the subject’s endogenous F8, or was a B-domain-deleted (BDD)-rFVIII product.
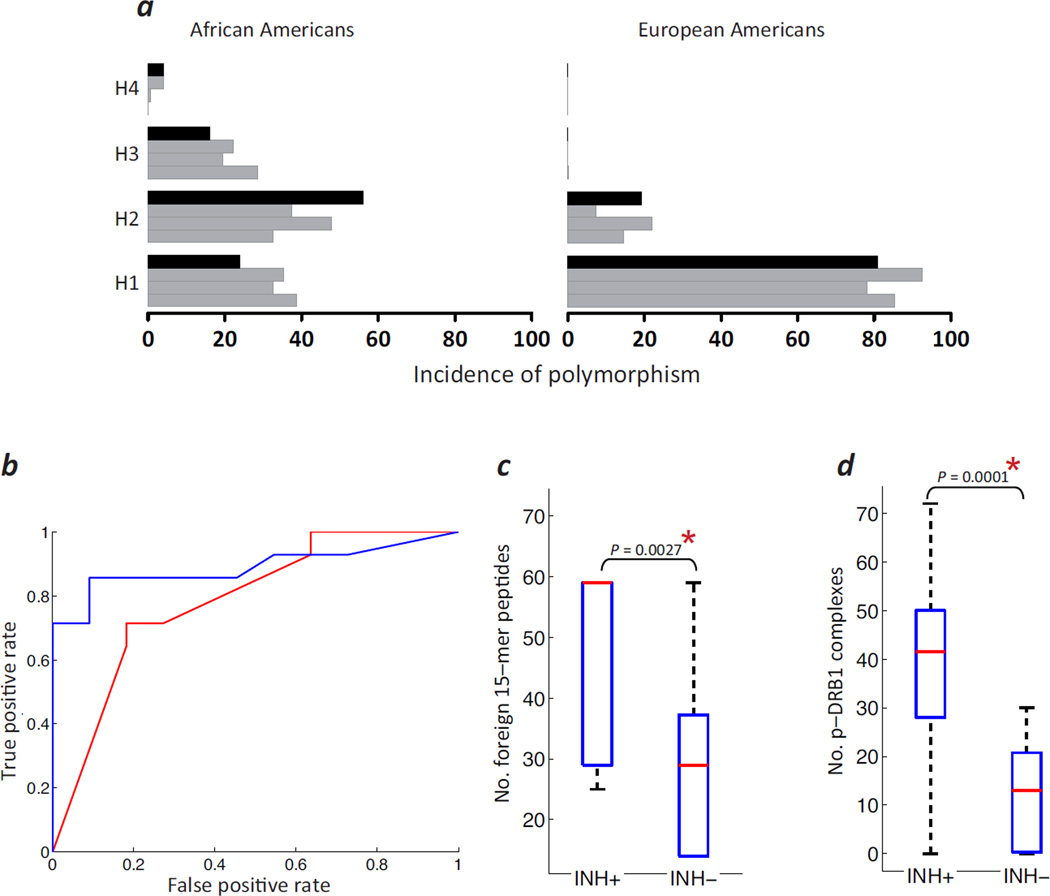
In 25 African American individuals with HA who had the I22I in F8, we obtained a history of their past FVIII-infusions and life-time inhibitor status, and determined their F8-nonsynonymous polymorphisms and DRB1* high-resolution MHC-II genotypes. This group of subjects has a distribution of polymorphisms comparable to that found in the three previous reports13–15 (Fig. 4a, left panel, grey bars). The right panel in Figure 4a makes it clear that with typically sized HA studies the European American population cannot provide a sufficient number of individuals with the different F8-nonsynonymous polymorphisms to allow an adequately powered statistical analysis and thus, selection of African American subjects with the I22I is a non-biased way of studying individuals with a larger distribution of such polymorphisms.
Figure 4.
A pharmacogenomic algorithm for personalized assessment of inhibitor risk in subjects with HA. (a) Incidence of four common FVIII variants (H1, wild-type; H2, D1241E; H3, D1241E & M2238V; H4, R484H & D1241E) in African American (left panel) and European American (right panel) individuals with HA who were evaluated in three previous studies13–15 (grey bars) and this study (black bars). (b) ROC curves comparing the number of foreign peptides (red line) and number of binders, i.e. peptide-MHC-II affinity ≤500 nM (blue line), as predictors of the development of inhibitors in individual subjects with HA. (c) The number of foreign 15-mer peptides each HA subject was exposed to in the inhibitor positive (INH+) and inhibitor negative (INH−) groups are depicted as box plots where the central red mark is the median and the edges represent the 25th and 75th percentiles. (d) Box plots (see c) depicting the number of peptide-DRB1 (p-DRB1) complexes in the INH+ and INH− groups.
We have hypothesized previously that a mismatch between the endogenous FVIII synthesized by an individual with HA and the infused protein drug can be a risk-factor for immunogenicity14, 16,17. A work-flow for analyzing personalized responses to FVIII infusions in this sub-population of subjects with the I22I-mutation and a diverse set of nonsynonymous polymorphisms is illustrated in Supplementary Fig. 6. We first used FVIII sequence information to determine all regions of potential mismatch between the endogenous and infused FVIII proteins. Sources of mismatch are: (i) The region that spans the junction between the exons 22 and 23 of all infused rFVIII proteins in subjects with the I22I; (ii) Nonsynonymous polymorphic positions where the drug’s sequence differs from the subject’s (in the case of plasma derived [pd-] FVIII products we have assumed that all four common haplotypes, H1, H2, H3 and H4, might be infused in the individual with HA); (iii) When the subject was treated with a BBD-rFVIII product, the non-naturally occurring sequence of the linker. We then identified, for each subject, all overlapping 15-mer peptides that span these mismatched positions and determined the total number of such foreign peptides. Again, note that as all individuals in this sub-set carry the I22I-mutation, no differences between individuals due to the HA-causing mutation per se are expected. We evaluated the association between the numbers of foreign peptides to which an individual is potentially exposed and whether or not that subject developed inhibitors. A Receiver Operator Characteristic (ROC)-curve analysis (Fig. 4b, red line) shows that this measure is a reasonably good predictor of immunogenicity (AUC = 0.727, P = 0.012). Similarly, we show that the average number of foreign peptides is significantly greater in individuals who develop inhibitors (one sided unpaired t-test P = 0.0027) (Fig. 4c).
Foreign peptides are potential T-cell epitopes; however, to elicit an immune response these peptides must be presented by the individual’s MHC-II repertoire16, 18. We therefore performed high-resolution genotyping of the structural locus encoding the HLA-class-II (HLA-II) protein DRB1 (DRB1) in all subjects and computed the affinity of all potential foreign peptides generated in each individual to that individual’s own HLA-DRB1* alleles. For each subject we estimated the number of stable peptide-DRB1 complexes (i.e., with affinity ≤ 500 nM, see19) which are likely to advance the immune response. The ROC-curve shows that the predictive value of this number (for developing inhibitors) is better than the number based solely on total number of foreign peptides (Fig. 4b, blue line, AUC = 0.890, P = 0.001 [compare to AUC = 0.727, P = 0.012; see above]). Similarly, the average number of peptide-HLA-II complexes show a more significant (one sided unpaired t-test P = 0.0001) difference between subjects with and without inhibitors (Fig. 4d; [compare to P = 0.0027; see above]).
Our results based on a sub-set of HA subjects with the I22I suggest that, consistent with a previous report14, polymorphisms in the F8 gene of African American subjects with HA could be a risk factor for immunogenicity. Larger data sets of geno- and HLA-typed patients are likely to emerge in the next few years (http://bit.ly/12jqnEz) and will be useful in validating these findings. Similarly, the identification of peptides presented by MHC-II molecules derived from individuals with HA that have developed inhibitors will provide direct evidence for the hypotheses. At this time additional support for our hypothesis also comes from a larger group (n = 313) of subjects with HA for who only race, the lifetime prevalence of inhibitors and status vis-à-vis the I22I are known. Broadly, our findings are comparable to previous reports suggesting a higher incidence of inhibitors among African American individuals with HA; 34% (46/135) of self-identified black individuals (either African Americans or black South Africans) developed inhibitors compared to 23% (41/178) of European Americans. However, we also separately analyzed subjects with the I22I and subjects with all other mutations. Individuals with the I22I showed a statistically significant increase in the prevalence of both high- and low-titer inhibitors in black HA subjects compared to European American (P = 0.052 & 0.017 for high and high+low titer inhibitors respectively) (see Supplementary Fig. 7 and Methods). However no statistically significant difference between the black and European American populations could be discerned in individuals with HA who had mutations other than the I22I (Supplementary Fig. 7). These results further support the hypothesis that the greater underlying sequence diversity of black populations may be the driver for higher prevalence of inhibitors. Sequence diversity, in terms of single nucleotide polymorphisms are increasingly being shown to be consistent with the so-called serial founder effect with a single origin in sub-Saharan Africa. Among modern populations, the sub-Saharan African populations are located at the root of the tree with sequential branches of North African, Middle Eastern, Europe, South or Central Asia, Oceania, America and East Asia20,21. The two populations we discuss in our study are almost at extreme ends of this tree and thus show sharp differences in sequence diversity which in turn has clinical consequences. This suggests that the demographic and biological factors that have shaped human diversity could have important consequences in the practice of medicine and the development of new drugs.
ONLINE METHODS
Human subjects and tissue preparation
Two white American men, one with severe HA caused by the intron-22 (I22)-inversion (I22I) mutation and the other a healthy control subject without HA (designated “Nor”), were voluntarily consented under an approved study based at the Georgia Health Sciences University (IRB# 04 01 206), Augusta, Georgia, USA. Lymphoblastoid cells were generated from both subjects at the Texas Biomedical Research Institute, San Antonio, Texas, USA. The same subject with HA consented to the release from the University of California at San Francisco (San Francisco, California, USA) of paraffin embedded tissue blocks containing representative sections of his explanted cirrhotic liver, in which all cells are hemizygous for the I22I, and needle biopsies of his transplanted liver, in which all cells carry a wild-type non-inverted F8 locus as the donor was a healthy male without HA. Paraffin-embedded tissue blocks of the explanted livers of two additional individuals with HA with the I22I as well as frozen PBMCs isolated from these subjects at the time of transplant were also obtained from the same source.
The experimental and clinical data used to establish and validate the novel prototype pharmacogenomic algorithm for personalized inhibitor risk prediction described here was derived from an additional 25 unrelated individuals who had the I22I, presented with severe HA who identified themselves as black. These subjects were voluntarily consented at one of 14 U.S. hemophilia treatment centres (HTCs) participating in the on-going PATH (Personalized Alternative Therapies for Hemophilia) study, an international multicentre observational human subjects investigation based at and approved by the institutional review boards (IRBs) of the Veterans Affairs Greater Los Angeles Healthcare System (IRB approval #: 2009-091280; project #: 0001) and the Puget Sound Blood Center (Seattle Children’s Hospital IRB approval #: 13018). Each subject was originally recruited and consented to participate in the PATH study during the course of a regularly scheduled annual visit to their HTC; prior to initiating subject enrolment, each HTC obtained approval for this study from their local IRB. Nucleic acid sequence data from the F8FL, F8I22I, F8B and MHC-II loci as well as clinical data pertinent to the risk of development of FVIII inhibitors was obtained from each subject.
Cells
Human lymphoblastoid cells were cultured in RPMI containing 10% heat inactivated fetal bovine serum (Invitrogen; CA, USA), 1% penicillin streptomycin (Invitrogen) and 1% glutamine (Invitrogen) at 37°C with a humidified 5% CO2 atmosphere.
Quantification of F8FL, F8I22I & F8B mRNAs by RT PCR
Total RNA from Nor and Inv cells was isolated using RNeasy Mini Kit (QIAGEN; CA, USA). Reverse transcription was performed on 0.5 µg of total RNA using a First Strand cDNA Synthesis Kit (Roche Diagnostics; IN, USA). The resulting cDNA fragments were PCR amplified using Syber green Inv master mix (Roche Diagnostics) in the Light Cycler 480 (Roche Diagnostics). Primer sets for specific F8 regions are given in Supplementary Table 1.
Eukaryotic vectors for the expression of F8FL, F8I22I & F8B in mammalian cell-lines
The three recombinant F8-minigene-based expression vectors employed in the transfection-based assay to validate the mouse FVIII-specific mAbs used in this study for their site-specificity and lack of cross-reactivity were developed by genOway (Lyon, France) and were purified using CsCl density gradient centrifugation (Loftstrand Laboratories; MD, USA). The backbone pUC plasmid contained ampicillin and neomycin resistance cassettes. The F8 mini-genes were cloned as cDNAs under the transcriptional control of a CMV promoter; a heterologous human growth hormone-polyadenylation cassette (hGH-polyA) was used to direct efficient and accurate mRNA 3’-end formation. The F8FL cDNA contained in the expression vector designated HOW1 FVIII WT has the H3 background haplotype of the wild-type human F8 locus and as such encodes the fully functional H3 version of the full-length FVIII protein (FVIIIFL). The F8I22I cDNA contained in the expression vector designated HOW2 FVIII-I22I has the H3 background haplotype of the I22-inverted human F8 locus and as such is predicted to encode the H3 version of a non-functional, slightly truncated FVIII protein (FVIIII22I), which has never been identified. The translated FVIIII22I polypeptide is predicted to contain 2,159 total amino acid residues that would include the same 19 residue leader-peptide found in the wild-type FVIII protein followed by 2,124 additional FVIII residues, which represent the N-terminal most portion of the mature circulating form of the wild-type FVIII protein, and then a unique C-terminal stretch of 16 non-FVIII residues. Consequently, during its synthesis, the nascent FVIIII22I polypeptide chain would be expected to undergo co-translational insertion into the rough-endoplasmic reticulum followed by removal of the 19 residue leader-peptide. The F8B cDNA contained in the expression vector designated HOW3 FVIII-B has the H3 background haplotype of the “wild-type” human F8B locus and as such is predicted to encode the H3 version of FVIIIB, an “orphan” protein with no known function. FVIIIB is predicted to contain 216 total amino acid residues that would include an N-terminal stretch of 8 non-FVIII residues, which are unique (i.e., not found in other known proteins) and do not resemble known leader-peptides, and a major C-terminal stretch of 208 residues, which are the same C-terminal 208 residues in the wild-type FVIII protein (FVIIIFL).
Mammalian cell-lines, tissue culture and transfection
Several well-characterized mammalian cell-lines, viz. the NIH 3T3 murine fibroblast cell-line, the Chinese hamster ovary (CHO) cell-line, the HEK-293 human embryonic kidney cell-line and the HuH-7 a well differentiated hepatocyte derived cellular carcinoma cell line were cultured in DMEM supplemented with 10% fetal bovine serum (Invitrogen) at 37°C in 5% CO2. Sixteen hours following GenJet transfection (Sigmagen Lab; USA) in a DNA GeneJet (3:1) complex ratio, cells were harvested to perform flow cytometry or lysed and the cell lysates subjected to immunoprecipitation, gel electrophoresis and immunoblotting.
Flow cytometry
Cells were grown overnight, harvested, fixed and permeabilized with IntraPrepTM permeabilization regent (Beckman Coulter; Marseille, France). Non-permeabilized cells were used as controls. Anti human-FVIII mAbs specific to different domains of the human FVIII protein (see Figure Legends and Results sections for details) were used for labeling. Anti mouse-IgG1 and -IgG2a or non-transfected cells served as negative controls. The primary Abs were detected using an Alexa-Fluor-488 labeled goat anti mouse-IgG secondary antibody (Invitrogen). Cells were then analyzed using Becton Dickinson FACS caliber and a median value of fluorescence intensity (MFI) was determined using the Cell Quest software.
Immunoblotting
Lymphoblastoid cells or cell-lines in which F8 constructs were transiently transfected were lysed in RIPA cell-lysis buffer (Pierce; IL, USA) with 1mM phenylmethylsulfonyl fluoride and 1% aprotinin (Sigma; MO, USA) and cell-lysates were collected as the supernatant following centrifugation at 10,000 g for 10 minutes at 4°C. Cell-lysates and purified full length FVIII (Advate®, Baxter; IL, USA) as well as the purified C2-domain (kind gift of Drs. Michael J. Lenardo and Kimberly Shafer-Weaver (National Institute of Allergy and Infectious Diseases) were subjected to immunoprecipitatation. Exosome Immunoprecipitation Protein A coated Dynabeads (Invitrogen) were coated with a cocktail of five previously characterized FVIII-specific mouse mAbs from Green Mountain Antibodies. Four of these mAbs (GMA8003, GMA8006, GMA8008, and GMA8014) are specific to the C2 domain and one (GMA8015) is specific to the A2 domain of FVIII. Following immunoprecipitation, the protein was eluted in 2X SDS-PAGE loading dye, heated at 100°C for 10 min, diluted 1X with H2O followed by electrophoreses and Western transfer to nitrocellulose. The blots were probed with an affinity purified sheep FVIII-specific pAb. The signal was developed and imaged by consecutively using the chemiluminescent substrates of increasing sensitivity (SuperSignal West Pico and SuperSignal West Dura, respectively).
Identification of proteins in Commassie Blue stained bands following SDS-PAGE
The protein bands were subjected to in-gel trypsin digestion. Briefly, the cysteines were reduced with DTT and alkylated with iodoacetamide. The protein was then digested with 13 ng µl−1 trypin (Sigma) in 10 mM ammonium bicarbonate for 16 h at 30 °C. The peptides were then extracted and dried by vacuum-evaporation. The dried peptides were resuspended in water containing 2% acetonitrile, 0.5% acetic acid. They were then injected onto a 0.2 × 50 mm Magic C18AQ reverse phase column (Michrom Bioresources, Inc.) using the Paradigm MS4 HPLC (Michrom Bioresources, Inc.). Peptides were separated at a flow rate of 2 µL min−1 followed by online analysis by tandem mass spectrometry using an LTQ ion trap mass spectrometer (Thermo Scientific) equipped with an ADVANCE CaptiveSpray ion source (Michrom Bioresources, Inc.). Peptides were eluted into the mass spectrometer using a linear gradient from 95% mobile phase A (2% acetonitrile, 0.5% acetic acid, 97.5% water) to 65% mobile phase B (10% water, 0.5% formic acid, 89.5% acetonitrile) over 20 minutes followed by 95% mobile phase B over 5 minutes. Peptides were detected in positive ion mode using a data-dependent method where ions on mass list derived from tryptic analysis of purified protein detected in an initial survey scan were selected for MS/MS analysis. If the ions on the parent mass list were not detected in the survey screen, the most abundant ions were instead selected for MS/MS analysis. The MS/MS spectra were searched against the human database using TurboSEQUEST in BioWorks v. 3.3.1 SP1 (ThermoElectron, San Jose, CA).
Southern blot analysis for I22I
Southern blot analysis was performed on genomic DNA samples that were extracted from the (i) Nor and Inv cells or (ii) EDTA-anticoagulated blood specimens from the 25 black American I22I subjects with HA (collected at the time of their enrollment into the PATH study) using the QIAamp-DNA-Mini Kit or -Blood Maxi Kit (QIAGEN; MD, USA), respectively, according to the manufacturer’s instructions.
Confocal microscopy
Cells were grown overnight, harvested and washed 3× with PBS + 0.2% BSA. Fixation was performed in 4% paraformaldehyde (20 minutes at room temperature) followed by permeabilization with 0.2% triton X100 (5 minutes at room temperature). FVIII or FVIII domain-containing proteins were labeled using mAbs specific to different FVIII domains (see Fig. 3c for details) and detected using an anti mouse-IgG secondary antibody conjugated with Alex-Fluor-488®. Nuclear counter-staining was performed with Vectashield mounting medium with DAPI (Vector Laboratories; CA, USA). Confocal images were acquired with Zeiss AM software on a Zeiss LSM 510 confocal microscope system (Carl Zeiss, Inc; NY, USA) with a Zeiss axiovert 100M inverted microscope.
Knock-down of FVIII protein with Si-RNA
FVIII expression in lymphoblastoid cells (1 × 106 mL−1) was knocked-down using Smart Pool FVIII targeted Si-RNA (Dharmacon; IL, USA) at a concentration of 1 4 µM in Accel-medium using the Accel-delivery system (Dharmacon). The F8 mRNA and FVIII protein levels were estimated using quantitative (q)-RT PCR and flow cytometry as described above.
Modified ELISA to detect FVIII proteins in cell-lysates and media
An anti human FVIII mAb (Ab 41188, Abcam; MA, USA) was cross linked to magnetic beads conjugated with protein A (Invitrogen). Conjugated beads were incubated overnight at 4°C with either the concentrated cell supernatants or with cell-lysates prepared in low salt lysis buffer and then washed (3×) with 0.05% Tween 20 in PBS. Bound FVIII was detected using HRP conjugated sheep anti human FVIII antibody (Affinity Biologicals; Hamilton, Canada). The bead protein complexes were incubated with premade TMB substrate (Affinity Biologicals) for 5 minutes and the reaction was stopped using 0.2 M H2SO4. The absorbance was read at 450 nm.
Immunohistochemical staining of liver sections
Immunohistochemistry was performed on 6 8 µm serial sections from biopsies of the normal transplanted liver and the liver explanted from the subject with HA. Sections were deparaffinized, rehydrated and heated in antigen unmasking solution (H 3300, Vector Laboratories) to enhance antigen retrieval. The sections were then treated with 2% H2O2 to inactivate endogenous peroxidase activity. After treating with blocking buffer, the sections were incubated overnight at 4°C with mouse monoclonal primary Abs (diluted 1:50). Subsequently, sections were incubated for 1 hour at room temperature with HRP conjugated goat anti mouse IgG. Binding of primary Abs was assessed with the ImmPACT DAB peroxidase substrate (Vector Laboratories) leading to a brown reaction product, counterstained with hematoxylin (Vector Laboratories) and mounted with VectaMount mounting medium (Vector Laboratories). A contiguous section was stained with hematoxylin and eosin (H&E).
Pharmacogenomic algorithm for personalized immunogenicity risk assessment in individuals with HA
An example of the workflow for the analysis in an individual subject is depicted in Figure S-5. We first determined the collection of foreign peptides each individual with HA had been exposed to based on their pertinent medical history data and the sequence of their endogenously synthesized FVIIII22I and FVIIIB polypeptide chains (Step 1, Fig. S-5). To that end, we extracted all (overlapping) 15 amino acid long peptides (each off-set by 1 residue) within their FVIIII22I and FVIIIB proteins and, similarly, within their administered (“exogenous”) FVIII protein-drug(s) and identified 15-mers that appeared solely in the latter set as foreign (Step 2, Fig. S-5). Plasma derived (pd-) FVIII products were considered to be a mixture of the four common polymorphic forms that occur in the U.S. population, H1 H4. Subjects under the age of 20 for whom the medical history was not known (N = 2), were assumed to be treated with the H1 rFVIII protein, Kogenate®; for older subjects (N = 2), pd-FVIII was assumed to be administered.
Thus, for each subject with HA, the set of infused-FVIII-derived foreign peptides included: (a) peptides that span the junction between exons 22 and 23 encoded between the M2124 and V2125 residues; (b) peptides that span nonsynonymous polymorphic positions where the drug’s peptide sequence is mismatched with the corresponding internal peptide sequence in the subject’s FVIIII22I or FVIIIB polypeptides; and (c) when the subject was treated with a BDD rFVIII product, peptides spanning the non-naturally occurring linker sequence [1]. Next, we used NetMHCIIpan (version 2.0) to predict the binding affinity of each foreign 15 mer peptide to the subject’s DRB1s protein(s) (Step 3, Fig. S-5), and considered peptides with affinity ≤ 500 nM as binders (Red text Step 3, Fig. S-5). Most individuals carry two distinct alleles of the DRB1 locus [DRB1] four of the 25 African American subjects with the I22I evaluated were DRB1 homozygous. We then compared the number of foreign 15 mer peptides and the number of predicted foreign-peptide DRB1 complexes across the entire cohort using ROC analysis.
Statistical analysis of clinical data
To test if subjects with HA who carried the I22I showed statistically significant increase in the prevalence of inhibition, we first estimated the probability of inhibition under the assumption there was no bias in the ratio of inhibitors in each group to all subjects in the group (e.g. for high+low titer inhibitors of African American subjects p = 46/135 = 0.34). In all estimations of subjects with high titer inhibitors only, we removed all low titer subjects from the analysis. Then we used a Binomial test to calculate the probability of observing at least a given number of inhibitors in the I22I group given the size of the I22I group. For the African American population and high+low titer inhibitors the probability of observing 22 cases or more in 50 samples (assuming the probability of a random observation to be 0.34) is 0.039. For the African American population high titer the probability of observing 11 cases or more in samples is 0.053. The same analysis for the European American population gives probability 0.092 and 0.132 for high+low and high titer respectively. Thus the increased prevalence of inhibition in the I22I African American is statistically significant while we could not reject the null hypothesis of no difference for the I22I European American population. To directly test whether the difference of the prevalence of the inhibition in I22I subjects in African American versus European American populations was statistically significant we used Fisher exact test obtaining respectively P 0.017 and 0.052 for subjects with high+low and high titers.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank P. Matzinger, B. Golding, J. Lusher, J. Lozier, D. Levin and V. La Terza for helpful comments and J. Sauna for the graphics. We are grateful to S.V. Ambudkar (US National Cancer Institute, Bethesda MD) for the use of facilities, to M.J. Lenardo and K. Shafer-Weaver (US National Institute of Allergy and Infectious Diseases) for the gift of the purified C2 domain of FVIII and to P. Stock (University of California, San Francisco) for liver tissue samples. These samples were obtained under the University of California University-Wide AIDS Research Program (TP-99-SF-001), CHR: H8024-23454 and the Solid Organ Transplantation in HIV: Multi-Site Study (AI052748) funded by the US National Institute of Allergy and infectious Diseases, CHR: 10-01178. Research conducted in the laboratory of ZES is funded by the modernization of science program of the Center for Biologics Evaluation and Research, Food and Drug Administration. Research conducted in the laboratory of TEH is funded by grants from the National Heart, Lung and Blood Institute, NIH (1RC2-HL101851, HL-71130 and HL-72533) as well as from the: Bayer Healthcare Corporation, Bayer Hemophilia Awards Program, Baxter Healthcare Corporation, and the Clinical Translational Science Institute at the University of Southern California. The work of TMP is fully supported by the Intramural Program of the US National Library of Medicine, NIH.
Footnotes
PATH study investigators:
Jeanne Lusher14, Meera Chitlur14, Afshin Ameri15, Kavita Natarajan15, Rathi V. Iyer16, Alexis A. Thompson17, Raymond G. Watts18, Christine L. Kempton19, Craig Kessler20, John C. Barrett21, Erica J. Martin21, Nigel Key22, Rebecca Kruse-Jarres23, Dr. Cindy Lessinger23, Kathleen P. Pratt24, Neil Josephson24, Kevin McRedmond25, Janice Withycombe25, Christopher Walsh26, Dana Matthews27, Johnny Mahlangu28, Amanda Krause28, Rosemary Schwyzer28, Rajendra Thejpal29, Nadine Rapiti29, Yasmin Goga29, Marius Coetzee30, David Stones30, Kenneth Mann31, Saulius Butenas31, Laura Almasy32, John Blangero32, Mel Carless32, Rajalingam Raja33, Elaine Reed33
14Wayne State University, Detroit, MI, USA.
15Georgia Health Sciences University, Augusta, GA, USA.
16University of Mississippi Medical Center, Jackson, MS, USA.
17Northwestern University, Chicago, IL, USA.
18University of Alabama, Birmingham, AL, USA.
19Emory University, Atlanta, GA, USA.
20Georgetown University, Washington, DC, USA.
21Virginia Commonwealth University, Richmond, VA, USA.
22University of North Carolina, Chapel Hill, NC, USA.
23Tulane University, New Orleans, LA, USA.
24Puget Sound Blood Center, Seattle, WA, USA.
25Palmetto Health, Columbia, SC, USA.
26Mount Sinai University, New York, NY, USA.
27Seattle Children’s Hospital, Seattle, WA, USA.
28University of Witwatersrand, Johannesburg, Gauteng, South Africa.
29King Edward VIII Hospital, Durban, KwaZulu Natal, South Africa.
30University of the Free State, Bloemfontein, Free State, South Africa.
31Department of Biochemistry, University of Vermont, Burlington, VT, USA.
32Department of Genetics, Texas Biomedical Research Institute, San Antonio, TX, USA.
33Immunogenetics Center, University of California, Los Angeles, CA, USA.
DISCLAIMER
The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy.
AUTHOR CONTIBUTIONS
G.S.P. executed experiments, analyzed results and generated the first draft of the manuscript. C.Y. performed computational assessments of peptide-MHC-II binding, analyzed the result and helped edit the manuscript. L.M. M-J performed the mass spectrometric analysis. S.G. provided the confocal microscopy images and advice on sample preparation. S.A.C, J.E.C. and E.K.M immortalized cells from the male donor with HA and sequenced the F8 gene in cell lines and in cells from subjects with HA. N.R and D.L. performed the intron-22-inversion assays on patient samples and reported the results. V.S. and C.K-S performed the experiments related to the transient expression of F8 constructs in different cell lines. K.V. helped with statistical analysis of clinical data. T.M.P. analyzed clinical data, provided advice on statistical analysis and helped with editing early drafts of the manuscript. G.F.P. provided the liver samples, and was involved with study design from the earliest stages and provided advice on clinical aspects of this study. T.E.H. conceptualized the hypothesis, provided all sequencing data and helped with the study design. Z.E.S was responsible for the study design, co-ordinated the different aspects of the study, analyzed results and wrote the final draft of the paper. PATH investigators obtained IRB approvals and provided all the human samples used in the study and analysis.
REFERENCES
- 1.Lillicrap D. Improvements in factor concentrates. Curr. Opin. Hematol. 2010;17:393–397. doi: 10.1097/MOH.0b013e32833c06c6. [DOI] [PubMed] [Google Scholar]
- 2.Wight J, Paisley S. The epidemiology of inhibitors in haemophilia A: a systematic review. Haemophilia. 2003;9:418–435. doi: 10.1046/j.1365-2516.2003.00780.x. [DOI] [PubMed] [Google Scholar]
- 3.Gouw SC, et al. F8 gene mutation type and inhibitor development in patients with severe hemophilia A: systematic review and meta-analysis. Blood. 2012;119:2922–2934. doi: 10.1182/blood-2011-09-379453. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins PV, et al. Analysis of intron 22 inversions of the factor VIII gene in severe hemophilia A: implications for genetic counseling. Blood. 1994;84:2197–2201. [PubMed] [Google Scholar]
- 5.Egler C, et al. Kinetic parameters of monoclonal antibodies ESH2, ESH4, ESH5, and ESH8 on coagulation factor VIII and their influence on factor VIII activity. J. Mol. Recognit. 2009;22:301–306. doi: 10.1002/jmr.947. [DOI] [PubMed] [Google Scholar]
- 6.Griffin BD, Micklem LR, McCann MC, James K, Pepper DS. The production and characterisation of a panel of ten murine monoclonal antibodies to human procoagulant factor VIII. Thromb. Haemost. 1986;55:40–46. [PubMed] [Google Scholar]
- 7.Scandella D, et al. Some factor VIII inhibitor antibodies recognize a common epitope corresponding to C2 domain amino acids 2248 through 2312, which overlap a phospholipid-binding site. Blood. 1995;86:1811–1819. [PubMed] [Google Scholar]
- 8.Do H, Healey JF, Waller EK, Lollar P. Expression of factor VIII by murine liver sinusoidal endothelial cells. J. Biol. Chem. 1999;274:19587–19592. doi: 10.1074/jbc.274.28.19587. [DOI] [PubMed] [Google Scholar]
- 9.Lenting PJ, van Mourik JA, Mertens K. The life cycle of coagulation factor VIII in view of its structure and function. Blood. 1998;92:3983–3996. [PubMed] [Google Scholar]
- 10.Wion KL, Kelly D, Summerfield JA, Tuddenham EG, Lawn RM. Distribution of factor VIII mRNA and antigen in human liver and other tissues. Nature. 1985;317:726–729. doi: 10.1038/317726a0. [DOI] [PubMed] [Google Scholar]
- 11.Lewis JH, Bontempo FA, Spero JA, Ragni MV, Starzl TE. Liver transplantation in a hemophiliac. N. Engl. J. Med. 1985;312:1189–1190. doi: 10.1056/NEJM198505023121812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stel HV, van der Kwast TH, Veerman EC. Detection of factor VIII/coagulant antigen in human liver tissue. Nature. 1983;303:530–532. doi: 10.1038/303530a0. [DOI] [PubMed] [Google Scholar]
- 13.Miller CH, et al. F8 and F9 mutations in US haemophilia patients: correlation with history of inhibitor and race/ethnicity. Haemophilia. 2012;18:375–382. doi: 10.1111/j.1365-2516.2011.02700.x. [DOI] [PubMed] [Google Scholar]
- 14.Viel KR, et al. Inhibitors of Factor VIII in Black Patients with Hemophilia. N. Engl. J. Med. 2009;360:1618–1627. doi: 10.1056/NEJMoa075760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwarz J, et al. F8 haplotype and inhibitor risk: results from the Hemophilia Inhibitor Genetics Study (HIGS) Combined Cohort. Haemophilia. 2012;[Epub ahead of print] doi: 10.1111/hae.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanover C, Jain N, Pierce G, Howard TE, Sauna ZE. Pharmacogenetics and the immunogenicity of protein therapeutics. Nat. Biotechnol. 2011;29:870–873. doi: 10.1038/nbt.2002. [DOI] [PubMed] [Google Scholar]
- 17.Pandey GS, Yanover C, Howard TE, Sauna ZE. Polymorphisms in the F8 Gene and MHC-II Variants as Risk Factors for the Development of Inhibitory Anti-Factor VIII Antibodies during the Treatment of Hemophilia A: A Computational Assessment. PLoS Comput. Biol. 2013;9:e1003066. doi: 10.1371/journal.pcbi.1003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarski CA, et al. The kinetic stability of MHC class II: Peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen M, et al. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput. Biol. 2008;4:e1000107. doi: 10.1371/journal.pcbi.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 21.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.