Summary
Our GWAS of smoking and bladder cancer risk based on data from 5,424 cases and 10,162 controls suggest that exploring additive and multiplicative gene–environment interactions can identify novel susceptibility loci that are associated with risk for different subgroups.
Abstract
Bladder cancer is a complex disease with known environmental and genetic risk factors. We performed a genome-wide interaction study (GWAS) of smoking and bladder cancer risk based on primary scan data from 3002 cases and 4411 controls from the National Cancer Institute Bladder Cancer GWAS. Alternative methods were used to evaluate both additive and multiplicative interactions between individual single nucleotide polymorphisms (SNPs) and smoking exposure. SNPs with interaction P values < 5 × 10− 5 were evaluated further in an independent dataset of 2422 bladder cancer cases and 5751 controls. We identified 10 SNPs that showed association in a consistent manner with the initial dataset and in the combined dataset, providing evidence of interaction with tobacco use. Further, two of these novel SNPs showed strong evidence of association with bladder cancer in tobacco use subgroups that approached genome-wide significance. Specifically, rs1711973 (FOXF2) on 6p25.3 was a susceptibility SNP for never smokers [combined odds ratio (OR) = 1.34, 95% confidence interval (CI) = 1.20–1.50, P value = 5.18 × 10− 7]; and rs12216499 (RSPH3-TAGAP-EZR) on 6q25.3 was a susceptibility SNP for ever smokers (combined OR = 0.75, 95% CI = 0.67–0.84, P value = 6.35 × 10− 7). In our analysis of smoking and bladder cancer, the tests for multiplicative interaction seemed to more commonly identify susceptibility loci with associations in never smokers, whereas the additive interaction analysis identified more loci with associations among smokers—including the known smoking and NAT2 acetylation interaction. Our findings provide additional evidence of gene–environment interactions for tobacco and bladder cancer.
Introduction
Bladder cancer is a complex disease with established environmental and genetic risk factors (1). Bladder cancer provides an ideal setting in which to study the complex interplay of genes and the environment, because of the established causal roles of smoking and occupational exposures to aromatic amines, along with evidence of gene–environment interactions for these and other exposures (2–8). Although cigarette smoking is the most established risk factor for bladder cancer, the magnitude of association is not as strong as that for other cancers, such as lung cancer, suggesting that bladder cancer is a disease which may provide insights into the interaction between genes and smoking. From the numerous candidate gene studies focusing on enzymes that metabolize known carcinogens found in tobacco products, data have consistently shown a modest increased risk of bladder cancer associated with the NAT2 slow acetylation genotype among smokers (2,8,9), which is consistent with the role of NAT2 in aromatic amine metabolism.
The ‘agnostic’ approach, free of candidate hypotheses has been successfully applied in genome-wide association studies (GWAS) to identify new bladder cancer susceptibility alleles (6,10–18). The application of a comparable, agnostic approach to a genome-wide study of interaction of smoking and bladder cancer risk could identify new susceptibility loci, previously undetected by single-locus tests of GWAS, which modify the association between tobacco use and bladder cancer. We have recently shown that among identified bladder cancer susceptibility regions, NAT2 shows both multiplicative and additive interactions with tobacco use (19); individuals with NAT2 slow acetylation genotypes show higher relative risk (i.e. multiplicative interaction) and elevated absolute risk difference (i.e. additive interaction) from smoking, compared with those with rapid/intermediate acetylation genotypes. Other bladder cancer susceptibility loci, however, do not appear to modify the relative risk of smoking. Yet, 10 additional loci besides NAT2 show evidence of absolute risk difference with smoking (i.e. additive interaction), specifically, GSTM1 deletion, rs9642880, rs2294008 (PSCA), rs401681 (CLPTM1L- TERT), rs798766 (TMEM129- TACC3-FGFR3), rs1014871 (CBX6, APOBEC3A), rs8102137 (CCNE1), rs17863783 (UGT1A6), rs10775480/rs10853535 (SLC14A1) and rs10936599 (TERC-ACTRT3-MYNN-LRRC34) (18,19).
To further explore gene–environment interactions, we performed a genome-wide interaction study of smoking and bladder cancer risk based on data from 3002 cases and 4411 controls from the National Cancer Institute (NCI) Bladder Cancer GWAS (13) and evaluated promising single nucleotide polymorphisms (SNPs) in an independent dataset of 2422 bladder cancer cases and 5751 controls (18).
Materials and methods
Study populations and exposure assessment
We conducted our analyses using data from our previously reported primary scan of 591 637 SNPs in 3002 cases and 4411 controls from four studies that had enrolled both smokers and non-smokers, and which were included in the NCI Bladder Cancer GWAS, referred to here as NCI-GWAS1 (13), i.e. the Spanish Bladder Cancer Study (SBCS); New England Bladder Cancer Study, Maine and Vermont components (NEBCS-ME/VT); the American Cancer Society Cancer Prevention Study II Nutrition Cohort (CPS-II) and the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO) (Supplementary Table 1, available at Carcinogenesis Online).
NCI-GWAS2 genotype data has been recently described (18). In brief, the studies genotyped in this dataset consisted of cases and controls for the New Hampshire component of the New England Bladder Cancer Study (NEBCS-NH); cases and controls for four cohort studies, namely the European Prospective Investigation Into Cancer and Nutrition Study (EPIC), Women’s Health Initiative (WHI), Health Professionals Follow-up Study (HPFS) and Nurses’ Health Study I and II (NHS I and II) and cases for four case–control studies, namely the Los Angeles Bladder Cancer Study (LABCS), the French Center for Research on Prostate Diseases (CeRePP), the French Bladder Study (FBCS) and the Brescia Bladder Cancer Study (BBCS). For LABCS, CeRePP, FBCS and BBCS studies where we genotyped cases only, we created in silico study groups based on comparable geographic/demographic parameters, which resulted in three new ‘study groups’, specifically, Europe (which comprised data from EPIC, CeRePP and FBCS), MEC/LA (which comprised cases from LABCS and controls from the Multiethnic Cohort, MEC) and Italy (which comprised cases from BBCS and controls from the Environment And Genetics in Lung cancer Etiology Study, EAGLE). All subjects gave informed consent and each study was approved by the host institutions’ Institutional Review Boards.
After removal of SNPs genotyped with <90% completion and SNPs with minor allele frequencies <5%, 491 011 SNPs were available for analysis. Cases were defined as patients diagnosed with histologically confirmed primary carcinoma of the urinary bladder including carcinoma in situ (ICD-0–2 topography codes C67.0-C67.9 or ICD9 codes 188.0-188.9 and 2337). Cases and controls were of European background as determined by population substructure analyses with STRUCTURE 17 and principal component analysis as reported previously (13,18). Smoking histories were obtained by risk factor questionnaires administered at the time of enrollment into the studies. The primary interaction with smoking was modeled as a binary variable (smoker/never smoker). For case–control studies, data was collected at the time of diagnosis for cases and time of interview for controls (20,21). For prospective cohort studies, the time between questionnaire administered at enrollment and subsequent cancer diagnosis varied depending on how long after enrollment cases were diagnosed. In the SBCS, NEBCS, CPS-II, WHI, BBCS and LABCS, ‘never-smokers’ were defined as subjects who had smoked <100 cigarettes over their lifetime. In the PLCO, EPIC, CeRePP and FBCS studies, ‘never-smokers’ were defined as subjects who smoked <6 months in their lifetime.
Statistical analysis
We assessed gene–environment interactions on multiplicative and additive scales, which test two distinct hypotheses (22). Assessment of gene–environment interactions for both additive and multiplicative models tests whether the observed joint effects odds ratio (OR) for smoking and the genetic risk are significantly different than the expected joint effects OR. On a multiplicative scale, this evaluates whether the relative risk for smoking varies across levels of genetic risk, the expected joint effects are calculated as ORSNP × ORsmoking. On an additive scale, this evaluates whether the risk difference for smoking varies across levels of genetic risk, the expected joint effects are calculated as ORSNP + ORsmoking − 1. In particular, when the underlying risk factors have strong effects, which is the case for smoking and bladder cancer, the additive and the multiplicative model results can be quite distinct.
Multiplicative and additive interaction methods were tested using two alternative methods, with and without assuming independence between a gene and an environmental exposure in the underlying population. It is known that assuming gene–environment independence can increase power for detecting interactions (23–25), but it can lead to bias if the assumption is violated (26). For each of the four tests considered, we assessed whether we identified a significant excess number of interactions with P values < 5 × 10− 5 using a Chi2 test.
Multiplicative interactions
Tests for multiplicative interactions assessed whether relative measures of risk are modified by an exposure for a given genetic factor. The first method was based on a typical logistic regression analysis, which does not assume independence between a SNP and the exposure. A likelihood ratio test was performed by comparing two logistic regression models, one with and one without an interaction term for a SNP and smoking. The resulting likelihood ratio test has one degree of freedom as we assumed an additive genetic model for each SNP. The logistic regression models were adjusted for study, age (5-year categories) and gender and included an interaction term for smoking and an indicator variable for the PLCO study to account for stratified sampling of controls by smoking status.
The second method assumes independence between SNPs and smoking. Although case-only approaches have been proposed to test for multiplicative interaction under the independence constraint (23), these only allow inference on the interaction parameter of a logistic model. We used a more general approach that can exploit the assumption of gene–environment independence and yet use cases and controls, for efficient inference on all the parameters of a logistic regression model (25). A likelihood ratio test (1 df) was performed using an R package, CGEN, that implements the alternative approaches to analysis of case–control studies with or without the assumption of gene–environment independence (http://dceg.cancer.gov/bb/tools/genetanalcasecontdata).
Additive interactions
Tests for additive interaction assessed whether absolute measures of risk associated with an exposure are modified by a genetic factor. Our first method is based on standard prospective likelihood that does not impose any assumption of gene–environment independence. A likelihood ratio test was performed using logistic regression models comparing saturated and additive models (27); under the null hypothesis of the additive model, the OR for the combined effect of a given SNP and smoking status is constrained so that the risk difference associated with one exposure (e.g. smoking) is constant across levels of other exposure (e.g. SNP), or the reverse. All tests for additive interactions were performed using categorical variables (each SNP was coded as a dichotomous variable indicating the presence of any variant allele) to avoid complex numerical issues related to non-standard model fitting procedures when using continuous variables, such as log-additive effects of SNP alleles. The resulting likelihood ratio test has one degree of freedom and the logistic regression models were adjusted for the same set of covariates used for a multiplicative interaction test.
For testing additive interactions using gene–environment independence assumption, we used a method based on the retrospective likelihood as recently proposed by Han et al. (27), which is in the R package CGEN.
In silico replication of top hits from genome-wide interaction study
For the replication study, we selected those SNPs that showed interaction P values < 5 × 10− 5 based on multiplicative or additive models with or without assuming gene–environment independence in the original GWAS, referred to as NCI-GWAS1, and tested for the same interaction (i.e. multiplicative or additive interaction) using the same methods in a new set of 2422 bladder cancer cases and 5751 controls from six studies with GWAS data, referred to as NCI-GWAS2 (18).We performed pooled analysis from the NCI-GWAS1 and NCI-GWAS2 data to report overall evidence for significance for top SNPs.
Associations with bladder cancer risk stratified by smoking status
Logistic regression models were used to estimate ORs and 95% confidence intervals (CI) for SNPs identified as top hits from the genome-wide interaction scans and bladder cancer risk, stratified by smoking status (never, ever, former and current), cigarettes per day, duration of smoking in years and adjusted for study, age (5-year categories), gender and eigenvectors (EV 1, 2, 3, 4, 6 and 7) as described previously (18).
Results
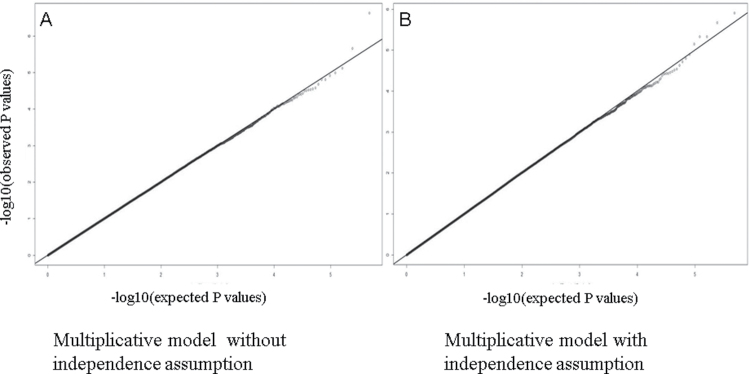
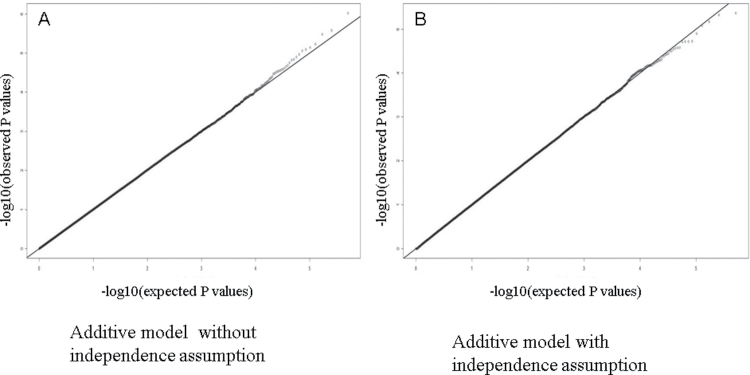
A total of 3002 cases and 4411 controls from NCI-GWAS1 were used for our genome-wide interaction study of smoking (Supplementary Table 1, available at Carcinogenesis Online). Inspection of the quantile–quantile plots for additive and multiplicative interaction tests for all genome-wide interaction scans of smoking suggests that there was no evidence for large-scale systematic bias or overdispersion (Figures 1 and 2).
Fig. 1.
Quantile–quantile plots for interaction P values from multiplicative models without and with independence assumption. Quantile–quantile plots for multiplicative interaction P values of smoking–SNP genome scan. P values were computed using two different methods to test for multiplicative interactions. The first method (A) used a likelihood ratio test performed by comparing two logistic regression models, one with and one without an interaction term for a SNP and smoking, did not assume independence between a SNP and smoking, and assumed an additive genetic model for each SNP. The logistic regression models were adjusted for study, age (5-year categories), gender and an interaction term for smoking and an indicator variable for the PLCO study to account for stratified sampling of controls by smoking status. The second method (B) assumed that SNP and smoking exposure are independent, using a retrospective likelihood, which exploits the gene–environment independence assumption in a general logistic regression framework.
Fig. 2.
Quantile–quantile plots for interaction P values from additive models without and with independence assumptions. Quantile–quantile plots for additive interaction P values of smoking–SNP genome scan. P values were computed using two different methods to test for additive interactions. The first method (A) does not assume gene–environment independence and was calculated using a likelihood ratio test using logistic regression models comparing saturated and additive models (27); under the null hypothesis of the additive model, the OR for the combined effect of a given SNP and smoking status is constrained so that the risk difference associated with one exposure (e.g. smoking) is constant across levels of other exposure (e.g. SNP), or the reverse, and models were adjusted for study, age (5-year categories), gender and an interaction term for smoking and an indicator variable for the PLCO study to account for stratified sampling of controls by smoking status. All tests for additive interactions were performed using categorical variables (each SNP was coded as a dichotomous variable indicating the presence of any risk allele) to avoid complex numerical issues related to non-standard model fitting procedures when using continuous variables, such as log-additive effect of SNP alleles. For testing additive interactions using a gene–environment independence assumption, we used a method proposed by Han et al. (27), which is based on the retrospective likelihood by Chatterjee et al. (25).
Genome-wide interaction study of smoking and bladder cancer risk in NCI-GWAS1-multiplicative interaction
In our analysis of multiplicative interaction, while 24 SNPs were expected by chance, we found 25 SNPs with P value < 5 × 10− 5 with the independence assumption (Chi P value = 0.86), and 22 without the independence assumption (Chi2 P value = 0.66). After removal of one of each pair of correlated SNPs with r 2 ≥ 0.20, we observed 32 independent SNP interactions based on either multiplicative test having a P value < 5 × 10− 5 (Supplementary Table 2, available at Carcinogenesis Online). In our multiplicative interaction genome scan, the ‘positive control’ SNP rs1495741, which tags NAT2 acetylation status and interacts with smoking (6), had P values of 0.009 (with independence assumption) and 0.006 (without independence assumption), ranked 4325 and 2666, based on P values of all tests conducted.
Genome-wide interaction study of smoking and bladder cancer risk in NCI-GWAS1-additive interaction
We used two tests for additive interaction with and without the SNP–smoking independence assumption in our genome-wide interaction study of smoking. Although 24 were expected by chance, we found 20 SNPs with P value < 5 × 10− 5 in additive models with the independence assumption (Chi2 P value = 0.40), and 28 without the independence assumption (Chi2 P value = 0.43). After removal of one of each pair of correlated SNPs with r 2 ≥ 0.20 we observed 29 independent SNPs with P values < 5 × 10− 5 (Supplementary Table 3, available at Carcinogenesis Online).
Validation of SNPs with evidence of multiplicative interaction in an independent dataset, NCI-GWAS2
Among the 32 SNPs identified above, we observed six with a multiplicative interaction P value < 0.10 (rs1711973, rs2969540, rs3752645, rs2411843, rs11692793 and rs11206140) in the NCI-GWAS2 dataset (Table I). However, two of these SNPs, rs11692793 and rs11206140 showed reverse patterns of interaction in the NCI-GWAS1 and NCI-GWAS2 datasets, and therefore, were not evaluated further. The rs1711973 (FOXF2) variant, was associated with bladder cancer risk limited to never smokers (combined OR = 1.34, 95% CI = 1.20–1.50, P value = 5.18 × 10− 7), with no association with bladder cancer risk among smokers (combined OR = 1.00, 95% CI = 0.93–1.07, P value = 0.90). The combined P interaction for rs1711973 (FOXF2) without independence assumption was 3.42 × 10− 6 and a P interaction with independence assumption was 7.18 × 10− 5. The rs2969540 (HTR5A-PAXIP1-INSIG1) variant was associated with bladder cancer risk among never smokers (combined OR = 1.44, 95% CI = 1.21–1.71, P value = 3.54 × 10− 5), and in contrast this variant showed a null association with bladder cancer risk among smokers (combined OR = 0.91, 95% CI = 0.81–1.01, P value = 0.08). The rs3752645 (PRKAR2B) variant was also associated with bladder cancer risk in never smokers (combined OR = 1.36, 95% CI = 1.16–1.60, P value = 1.80 × 10− 4), and in contrast showed a signficant inverse association with bladder cancer risk among smokers (combined OR = 0.87, 95% CI = 0.79–0.96, P value = 0.01). The rs3752645 (PRKAR2B) had a combined P interaction without independence assumption = 5.65 × 10− 6 and a P interaction with independence assumption = 1.22 × 10− 5. The last notable SNP with some suggestion of multiplicative interaction particularly without the independence assumption, was rs2411843 (HDAC4), and the variant was associated with bladder cancer risk in never smokers (combined OR = 1.17, 95% CI = 1.05–1.30, P value = 3.47 × 10− 3), with a null association among smokers (combined OR = 0.97, 95% CI = 0.91–1.03, P value = 0.36).
Table I.
NCI-GWAS1 and NCI-GWAS2 associations with bladder cancer risk stratified by smoking status (never, ever), for one known and ten new SNPs with evidence of interaction with smoking
| SNP | Cytoband | Gene region | MAF | Cases | Controls | Group | Smoke | OR | 95% CI | P value | P mult.pro | P mult.retro | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1711973 | 6p25.3 | FOXF2 | 0.26 | 550 | 1368 | NCI-GWAS1 | Never | 1.41 | 1.21 | 1.64 | 1.59E-05 | ||
| 484 | 2259 | NCI-GWAS2 | Never | 1.26 | 1.06 | 1.50 | 0.01 | ||||||
| 1034 | 3627 | Combined | Never | 1.34 | 1.20 | 1.50 | 5.18E-07 | ||||||
| 2418 | 3023 | NCI-GWAS1 | Ever | 0.94 | 0.86 | 1.03 | 0.16 | 6.38E-06 | 1.57E-04 | ||||
| 1815 | 3397 | NCI-GWAS2 | Ever | 1.10 | 0.98 | 1.23 | 0.10 | 0.08 | 0.08 | ||||
| 4233 | 6420 | Combined | Ever | 1.00 | 0.93 | 1.07 | 0.90 | 3.42E-06 | 7.18E-05 | ||||
| rs2969540 | 7q36.1 | HTR5A-PAXIP1- INSIG1 | 0.08 | 554 | 1368 | NCI-GWAS1 | Never | 1.50 | 1.19 | 1.89 | 5.16E-04 | ||
| 493 | 2260 | NCI-GWAS2 | Never | 1.38 | 1.06 | 1.81 | 0.02 | ||||||
| 1047 | 3628 | Combined | Never | 1.44 | 1.21 | 1.71 | 3.54E-05 | ||||||
| 2421 | 3019 | NCI-GWAS1 | Ever | 0.91 | 0.79 | 1.05 | 0.20 | 1.11E-03 | 1.73E-05 | ||||
| 1828 | 3396 | NCI-GWAS2 | Ever | 0.94 | 0.78 | 1.12 | 0.47 | 0.02 | 0.03 | ||||
| 4249 | 6415 | Combined | Ever | 0.91 | 0.81 | 1.01 | 0.08 | 1.38E-05 | 2.09E-06 | ||||
| rs3752645 | 7q22 | PRKAR2B | 0.09 | 553 | 1368 | NCI-GWAS1 | Never | 1.43 | 1.14 | 1.79 | 1.77E-03 | ||
| 492 | 2257 | NCI-GWAS2 | Never | 1.30 | 1.02 | 1.67 | 0.03 | ||||||
| 1045 | 3625 | Combined | Never | 1.36 | 1.16 | 1.60 | 1.80E-04 | ||||||
| 2425 | 3025 | NCI-GWAS1 | Ever | 0.83 | 0.74 | 0.94 | 3.83E-03 | 4.79E-05 | 5.56E-04 | ||||
| 1833 | 3399 | NCI-GWAS2 | Ever | 0.93 | 0.80 | 1.08 | 0.34 | 0.07 | 0.05 | ||||
| 4258 | 6424 | Combined | Ever | 0.87 | 0.79 | 0.96 | 0.01 | 5.65E-06 | 1.22E-05 | ||||
| rs2411843 | 2q37.3 | HDAC4 | 0.45 | 552 | 1369 | NCI-GWAS1 | Never | 1.18 | 1.02 | 1.36 | 0.03 | ||
| 493 | 2259 | NCI-GWAS2 | Never | 1.15 | 0.99 | 1.35 | 0.07 | ||||||
| 1045 | 3628 | Combined | Never | 1.17 | 1.05 | 1.30 | 3.47E-03 | ||||||
| 2423 | 3022 | NCI-GWAS1 | Ever | 0.97 | 0.90 | 1.05 | 0.49 | 0.03 | 4.98E-05 | ||||
| 1833 | 3397 | NCI-GWAS2 | Ever | 0.97 | 0.88 | 1.07 | 0.55 | 0.04 | 0.10 | ||||
| 4256 | 6419 | Combined | Ever | 0.97 | 0.91 | 1.03 | 0.36 | 2.43E-03 | 5.47E-05 | ||||
| P add.pro | P add.retro | ||||||||||||
| rs12216499 | 6q25.3 | RSPH3-TAGAP- EZR | 0.08 | 554 | 1369 | NCI-GWAS1 | Never | 1.26 | 0.96 | 1.64 | 0.09 | ||
| 492 | 2258 | NCI-GWAS2 | Never | 0.94 | 0.71 | 1.25 | 0.67 | ||||||
| 1046 | 3627 | Combined | Never | 1.10 | 0.91 | 1.33 | 0.33 | ||||||
| 2424 | 3023 | NCI-GWAS1 | Ever | 0.72 | 0.62 | 0.84 | 2.79E-05 | 1.47E-05 | 1.17E-03 | ||||
| 1828 | 3398 | NCI-GWAS2 | Ever | 0.80 | 0.67 | 0.96 | 0.02 | 0.04 | 0.03 | ||||
| 4252 | 6421 | Combined | Ever | 0.75 | 0.67 | 0.84 | 6.35E-07 | 1.41E-06 | 1.35E-04 | ||||
| rs1495741 | 8p22 | NAT2 | 0.22 | 552 | 1368 | NCI-GWAS1 | Never | 1.03 | 0.87 | 1.22 | 0.69 | ||
| 493 | 2260 | NCI-GWAS2 | Never | 0.90 | 0.74 | 1.10 | 0.31 | ||||||
| 1045 | 3628 | Combined | Never | 0.98 | 0.86 | 1.11 | 0.72 | ||||||
| 2423 | 3023 | NCI-GWAS1 | Ever | 0.81 | 0.74 | 0.89 | 4.69E-06 | 9.42E-06 | 5.16E-05 | ||||
| 1829 | 3398 | NCI-GWAS2 | Ever | 0.86 | 0.77 | 0.95 | 4.93E-03 | 0.20 | 3.44E-03 | ||||
| 4252 | 6421 | Combined | Ever | 0.83 | 0.77 | 0.89 | 8.55E-07 | 5.81E-04 | 1.36E-05 | ||||
| rs948798 | 18q22.3 | TIMM21- FBOX15-CYB5A | 0.09 | 554 | 1369 | NCI-GWAS1 | Never | 0.74 | 0.57 | 0.97 | 0.03 | ||
| 493 | 2259 | NCI-GWAS2 | Never | 1.13 | 0.86 | 1.48 | 0.38 | ||||||
| 1047 | 3628 | Combined | Never | 0.92 | 0.77 | 1.10 | 0.36 | ||||||
| 2427 | 3023 | NCI-GWAS1 | Ever | 1.28 | 1.13 | 1.45 | 9.78E-05 | 1.48E-05 | 1.33E-04 | ||||
| 1832 | 3399 | NCI-GWAS2 | Ever | 1.19 | 1.02 | 1.39 | 0.02 | 0.25 | 0.02 | ||||
| 4259 | 6422 | Combined | Ever | 1.22 | 1.10 | 1.35 | 1.87E-04 | 1.80E-04 | 5.71E-05 | ||||
| rs9502305 | 6q25.1 | FARS2 | 0.24 | 554 | 1369 | NCI-GWAS1 | Never | 0.97 | 0.82 | 1.14 | 0.69 | ||
| 493 | 2257 | NCI-GWAS2 | Never | 0.97 | 0.81 | 1.17 | 0.76 | ||||||
| 1047 | 3626 | Combined | Never | 0.97 | 0.86 | 1.10 | 0.66 | ||||||
| 2426 | 3024 | NCI-GWAS1 | Ever | 1.18 | 1.08 | 1.30 | 4.02E-04 | 1.40E-05 | 8.22E-05 | ||||
| 1834 | 3398 | NCI-GWAS2 | Ever | 1.10 | 0.98 | 1.23 | 0.12 | 0.07 | 0.03 | ||||
| 4260 | 6422 | Combined | Ever | 1.13 | 1.06 | 1.22 | 5.83E-04 | 4.12E-05 | 1.24E-05 | ||||
| rs846906 | 1q32-q41 | HSD11B1 | 0.15 | 554 | 1369 | NCI-GWAS1 | Never | 1.09 | 0.90 | 1.32 | 0.36 | ||
| 493 | 2259 | NCI-GWAS2 | Never | 0.90 | 0.72 | 1.13 | 0.37 | ||||||
| 1046 | 3609 | Combined | Never | 1.01 | 0.88 | 1.16 | 0.89 | ||||||
| 2421 | 3004 | NCI-GWAS1 | Ever | 0.84 | 0.76 | 0.93 | 9.23E-04 | 9.27E-04 | 1.60E-05 | ||||
| 1822 | 3374 | NCI-GWAS2 | Ever | 0.85 | 0.76 | 0.97 | 0.01 | 0.17 | 0.05 | ||||
| 4243 | 6378 | Combined | Ever | 0.87 | 0.80 | 0.95 | 1.38E-03 | 5.05E-03 | 1.41E-05 | ||||
| rs2380945 | 2q21.2 | LRP1B | 0.16 | 553 | 1357 | NCI-GWAS1 | Never | 1.19 | 0.98 | 1.44 | 0.08 | ||
| 492 | 2242 | NCI-GWAS2 | Never | 1.18 | 0.95 | 1.48 | 0.13 | ||||||
| 1045 | 3599 | Combined | Never | 1.19 | 1.03 | 1.37 | 0.01 | ||||||
| 2421 | 3011 | NCI-GWAS1 | Ever | 0.88 | 0.80 | 0.97 | 0.01 | 4.67E-05 | 3.18E-04 | ||||
| 1831 | 3382 | NCI-GWAS2 | Ever | 0.91 | 0.80 | 1.02 | 0.11 | 0.02 | 0.02 | ||||
| 4252 | 6393 | Combined | Ever | 0.90 | 0.82 | 0.97 | 0.01 | 5.49E-05 | 1.41E-05 | ||||
| rs1258767 | 15q13.3 | FMN1 | 0.25 | 551 | 1365 | NCI-GWAS1 | Never | 1.07 | 0.91 | 1.26 | 0.43 | ||
| 492 | 2257 | NCI-GWAS2 | Never | 1.11 | 0.92 | 1.35 | 0.26 | ||||||
| 1043 | 3622 | Combined | Never | 1.11 | 0.98 | 1.25 | 0.10 | ||||||
| 2421 | 3021 | NCI-GWAS1 | Ever | 0.90 | 0.83 | 0.98 | 0.01 | 1.05E-02 | 1.63E-05 | ||||
| 1832 | 3394 | NCI-GWAS2 | Ever | 0.92 | 0.84 | 1.02 | 0.13 | 0.04 | 0.04 | ||||
| 4253 | 6415 | Combined | Ever | 0.93 | 0.87 | 1.00 | 0.05 | 1.65E-04 | 6.70E-07 | ||||
Selected SNPs were those among 32 SNPs identified in multiplicative interaction scans or 28 SNPs identified in additive interaction scans with P values < 5.00 × 10− 5 (see Supplementary Tables 2 and 3, available at Carcinogenesis Online), which had an interaction P value < 0.10 in an independent dataset. We present per-allele OR and 95% CI and P values adjusted for age, gender, study and eigenvectors (EV 1, 2, 3, 4, 6 and 7, for combined data), the combined interaction P values for multiplicative (P mult.pro) or additive (P add.pro) interaction tests using a prospective likelihood without independence assumption, and for multiplicative (P mult.retro) or additive (P add.retro) interaction tests using a retrospective likelihood with independence assumption. The rs1495741 SNP tags NAT2 acetylation genotype, which has been shown previously to interact with smoking and bladder cancer risk (6,13), and the referent allele is slow acetylation status. MAF, minor allele frequency. Bold values indicate combined data for NCI-GWAS1 and NCI-GWAS2.
Further analysis by current/former smoking status showed the rs3752645 SNP to have a stronger inverse association with bladder cancer risk among current smokers (Supplementary Table 4, available at Carcinogenesis Online), whereas the other SNPs (rs1711973, rs2969540 and rs2411843) did not show any notable differences in association by current/former smoking status. Evaluation by cigarettes/day showed the rs1711973 variant to have a significant inverse association among subjects smoking 25+ cigarettes/day, and the rs3752645 variant a significant inverse association with risk in subjects that smoked 15–24 cigarettes/day (Supplementary Table 5, available at Carcinogenesis Online). Lastly, evaluation of years smoked showed the rs3752645 SNP to have a stronger inverse association with 40+ years of smoking, although a clear trend was not evident (Supplementary Table 6, available at Carcinogenesis Online).
Validation of SNPs with evidence of additive interaction in an independent dataset, NCI-GWAS2
Among the 28 SNPs identified above, we observed nine SNPs with additive interaction P values < 0.10 in NCI-GWAS2 (rs12216499, rs1495741, rs948798, rs9502305, rs846906, rs2380945, rs1258767, rs9927752 and rs1090292). However, two of these SNPs, rs9927752 and rs1090292 showed reverse patterns of interaction in the NCI-GWAS1 and NCI-GWAS2 datasets, and therefore, were not evaluated further (Table I). The rs12216499 (RSPH3-TAGAP-EZR) variant, was inversely associated with bladder cancer risk limited to ever smokers with a P value just approaching the genome-wide significance threshold of 5 × 10− 8 (never smokers, combined OR = 1.10, 95% CI = 0.91–1.33, P value = 0.33; ever smokers, combined OR = 0.75, 95% CI = 0.67–0.84, P value = 6.35 × 10− 7). The combined additive interaction P value for rs12216499 was 1.41 × 10− 6 without assuming independence and 1.35 × 10− 4 assuming independence (Table I). The rs1495741 SNP that tags NAT2 acetylation status showed the expected inverse association between the variant allele and bladder cancer risk limited to smokers (never smokers, combined OR = 0.98, 95% CI = 0.86–1.11, P value = 0.72; ever smokers, combined OR = 0.83, 95% CI = 0.77–0.89, P value = 8.55 × 10− 7), and the combined additive interaction P value was 5.81 × 10− 4 without assuming independence and 1.36 × 10− 5 assuming independence. Similar to the rs12216499 (RSPH3-TAGAP-EZR) and rs1495741 NAT2 tag SNP, the observed associations between the rs948798, rs9502305, rs846906, rs1258767 and SNPs and bladder cancer risk were limited to smokers and combined additive interaction P values were stronger when assuming independence (all P values < 5.00 × 10− 4, Table I), except for the rs12216499. Other notable interactions were rs2380945 and rs1258767.
Analysis by current and former smoking showed rs12216499, rs948798, rs9502305 and rs846906 SNPs to be stronger among former smokers, whereas the rs1495741 NAT2 tag SNP showed significant associations among current and former smokers (Supplementary Table 4, available at Carcinogenesis Online). Analysis by cigarettes/day and smoking duration showed rs12216499, rs1495741 and rs1258767 to have associations strongest for subjects who reported smoking 25+ cigarettes/day (Supplementary Table 5, available at Carcinogenesis Online). Analysis by smoking duration showed rs948798 as having the strongest association with bladder cancer risk in subjects who smoked 40+ years (Supplementary Table 6, available at Carcinogenesis Online).
Discussion
We conducted a a genome-wide interaction study of smoking and bladder cancer risk and found evidence of either additive or multiplicative interactions between tobacco use and 10 previously unindentified SNPs. Further, we found that two of these SNPs showed association that approached genome-wide significance among specific smoking subgroups. Specifically, rs1711973 (FOXF2) on 6p25.3 was a susceptibility SNP for never smokers, which was identified through our multiplicative interaction analysis, and rs12216499 (RSPH3-TAGAP-EZR) on 6q25.3 was a susceptibility SNP for ever smokers, which was identified through our additive interaction analysis. These findings also support evaluation of interaction on both the additive and multiplicative scales as a tool for identifying new susceptibility loci for important subgroups.
Our analysis of multiplicative interaction identified three promising SNPs (rs1711973, rs2969540, rs3752645). The two strongest findings, rs1711973 (FOXF2) and rs2969540 (HTR5A-PAXIP1-INSIG1), were associated with risk of bladder cancer only among never smokers. Using ENCODE resources (28), including HaploReg (29) and RegulomeDB (30), both of these SNPs (rs1711973, rs2969540) are in linkage disequilibrium with SNPs that may cause changes in transcription-binding regions. Interestingly, the rs1711973 SNP lies 3′ of the FOXF2 gene—a transcription factor and tumor suppressor gene (31). The rs3752645 SNP lies within the PRKAR2B gene, which encodes for a regulatory subunit for cyclic adenosine 3′,5′-monophosphate kinase and knockout studies in mice suggest that this subunit may play an important role in regulating energy balance and adiposity (32). Obesity and diabetes have each been associated with bladder cancer risk, particularly, among never smokers, suggesting a plausible mechanism by which this region may have some relevance to bladder cancer etiology and the potential opposite associations seen for never and ever smokers (33–36).
Through our additive interaction analysis we identified four new regions that were risk/protective factors for bladder cancer, specifically among ever smokers (rs12216499, rs948798, rs9502305, rs846906). It is noteworthy that the rs12216499 (RSPH3-TAGAP-EZR) SNP showed the strongest associations with bladder cancer risk among subjects reported to have the highest levels of smoking intensity; and the rs948798 SNP showed the strongest association with bladder cancer risk for those subjects who smoked 40+ years, providing some evidence that these associations are biologically plausible. According to ENCODE resources, the rs12216499 SNP is in high linkage disequilibrium with SNPs that result in missense mutations in RSPH3, a protein kinase anchoring protein for the mitogen-activated protein kinase, ERK2 (37). The nearest neighboring genes to rs948798 are TIMM21-FBXO15-CYB5A. Interestingly, CYB5A is a metabolic enzyme that can detoxify known bladder carcinogens (2-naphthylamine and 4-aminobiphenyl) and variation in this gene has been associated with variability in hydroxylamine reduction (38,39). If the association is replicated in other datasets, the rs948798S SNP near CYB5A maybe a potentially good candidate for functional work, given previous toxicological studies.
Cumulatively, our findings indicate that gene–smoking interactions with strong effects (ORs around 1.5) are unlikely. Further, our analysis found fewer than expected SNPs with P value < 5 × 10− 5 for our tests with multiplicative interaction and the expected number for additive interaction, and neither approach shows an excess of false-positive findings as shown in the Q-Q plots. Our analysis found that two of the SNPs identified, namely rs1711973 (FOXF2) and rs12216499 (RSPH3-TAGAP-EZR), approached the statistical evidence for GWAS in subgroups of ever or never smokers; however, independent datasets will be needed to replicate these findings. Although genetic factors associated with bladder cancer risk that are specific for smokers have been identified (e.g. NAT2 acetylation), it is likely that there are also genetic risk factors for bladder cancer that are specific to never smokers as compared with smokers. For example, recent work has identified several specific genetic variants related to lung cancer among never smokers that are distinct from variants found in smoking-related lung cancer (40), and our results suggest the rs1711973 (FOXF2) SNP a potential candidate. Therefore, future work in bladder cancer GWAS could exploit gene–environment interactions, in order to identify susceptibility factors for important subgroups of individuals, such as never smokers, which otherwise may not be found in current GWAS. In particular, our analysis suggest that the analysis of interaction, could be used to identify promising candidates for replication in other datasets, and identifying SNPs associated at the level of genome-wide significance is possible for important subgroups such as smokers. It is estimated that for a 1:2 case to control ratio, over 15 000 cases would be needed to detect the established NAT2 and smoking interaction (6,13) at genome-wide significance level using multiplicative models of interaction with 80% power. Our analysis suggests that additive interaction might be used to identify promising candidates for replication in other datasets, and identifying SNPs associated at the level of genome-wide significance is possible for important subgroups such as smokers. Regardless, very large sample sets with excellent exposure data will be required.
Strengths of our study include the use of an array of powerful statistical methods to explore hypotheses regarding interactions, the use of an independent dataset to replicate findings. Our results suggest that genome-wide interaction studies on both the multiplicative and additive scale, could provide clues to new regions of susceptibility for bladder cancer that may have specific effects with regard to smoking status and that there is no one test/model that is best for exploring gene–environment interactions. In our analysis of smoking and bladder cancer, the tests for multiplicative interaction seemed to more commonly identify susceptibility loci with associations in never smokers that were not observed for smokers, whereas the additive interaction analysis more commonly identified susceptibility loci with associations among smokers—including the known smoking–NAT2 acetylation interaction with bladder cancer risk. Since it is not yet clear what the standard should be for genome-wide interaction studies, we believe the application of novel methods for interaction, as presented in our current manuscript, and validation of potential loci in an independent dataset, should serve as an important reference to investigators evaluating gene–environment interactions. Specifically, our study is one of the first to explore how various methods for detecting statistical interaction perform in the context of bladder cancer risk—one of the few known cancers with established gene–environment interactions. Furthermore, in our analysis we performed additive and multiplicative tests for interaction with and without the gene–environment independence assumption. In theory, methods that assume gene–environment independence can be more powerful, but also can lead to false positives when the gene–environment independence assumption is violated. In practice, we found no single method could identify all of the SNPs with suggested interaction with smoking, which we believe to be promising. Thus, our analysis, which applies an array of different methods for interaction analysis in a GWAS scale, not only identifies promising SNPs for bladder cancers, but also provides an empirical demonstration of the need for application of different types of methods for future GWAS analysis of gene–environment interactions. Future investigations should include larger sample sizes, as well as studies in different populations of individuals with more variation in exposure, which might provide additional opportunities to detect gene–environment interactions and new susceptibility loci.
Supplementary material
Supplementary Tables 1–6 can be found at http://carcin.oxfordjournals.org/
Funding
EPIC (P.V.) - ICL - Europe Against Cancer Program of the European Commission (SANCO) ; IARC - International Agency for Research on Cancer; France - Ligue contre le Cancer Societe 3M, Mutuelle Generale de l’Education Nationale; Institut National de la Santé et de la Recherche Médicale (INSERM); Italy - Italian Association for Research on Cancer National Research Council; Spain - Health Research Fund (FIS) of the Spanish Ministry of Health; the CIBER en Epidemiología y Salud Pública (CIBERESP), Spain; ISCIII RETIC (RD06/0020); Spanish Regional Governments of Andalusia, Asturias, Basque Country, Murcia (N 6236) and Navarra and the Catalan Institute of Oncology; UK - Cancer Research UK Medical Research Council with additional support from the Stroke Association, British Heart Foundation, Department of Health, Food Standards Agency, the Wellcome Trust; The Netherlands - Dutch Ministry of Public Health, Dutch Prevention Funds LK Research Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF); Greece - Hellenic Ministry of Health, the Stavros Niarchos Foundation and the Hellenic Health Foundation; Germany - German Cancer Aid, German Cancer Research Center Federal Ministry of Education and Research (Grant 01-EA-9401); Sweden - Swedish Cancer Society, Swedish Scientific Council, Regional Government of Skane, Sweden; Denmark - Danish Cancer Society. FBCS (S.B.) - Ligue Contre le Cancer du Val-de-Marne; Fondation de France; Groupement d’Entreprises Françaises dans la Lutte contre le Cancer; Association pour la Recherche sur le Cancer, France. LABCS (M.P.) – National Institutes of Health grants (R01CA65726, R01CA114665, R01CA114665, 1P01CA86871). NEBCS (D.T.S.) - Intramural Research Program of the National Institutes of Health; National Cancer Institute; Division of Cancer Epidemiology and Genetics and intramural contract number (NCI N02-CP-01037). NHS & HPFS (I.D.V.) - CA055075, P01 CA87969, R01 CA49449, UM1 CA176726, R01 CA67262, UM1 CA167552. PLCO (M.P.P.) - The NIH Genes, Environment and Health Initiative (GEI) partly funded DNA extraction and statistical analyses (HG-06-033-NCI-01, RO1HL091172-01), genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438, NIH HHSN268200782096C) and study coordination at the GENEVA (N.C.). Coordination Center (U01HG004446) for the genotyping of the lung studies with controls from EAGLE study and part of the PLCO. Genotyping for the remaining part of PLCO and all ATBC and CPS-II samples were supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics. The PLCO is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, National Institutes of Health. SBCS (D.T.S.) - Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics and intramural contract number (NCI N02-CP-11015). FIS/Spain 98/1274, FIS/Spain 00/0745, PI061614, G03/174; Fundació Marató TV3; Red Temática Investigación Cooperativa en Cáncer (RTICC); Consolíder ONCOBIO, EU-FP7-201663; RO1-CA089715 and CA34627. WHI (C.K.) - The WHI program is funded by the National Heart, Lung, and Blood Institute; National Institutes of Health; U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, HHSN271201100004C.
Supplementary Material
Acknowledgements
M.Audouin, C.Gaffory, V.Ondet; P.Porter-Gill (NCI); L.Carroll (Information Management Services, Silver Spring, MD, USA); G.Castaño-Vinyals (Institut Municipal d’Investigació Mèdica, Barcelona, Spain); F.Fernández (Institut Municipal d’Investigació Mèdica, Barcelona, Spain); A.McIntosh (Westat, Inc., Rockville, MD, USA); F.Real [Spanish National Cancer Research Centre (CNIO), Madrid, Spain]; R.Saal (Westat, Rockville, MD, USA); M.Sala (Institut Municipal d’Investigació Mèdica, Barcelona, Spain); K.Snyder (Information Management Services, Inc., Silver Spring, MD); A.Taylor (Information Management Services, Inc., Silver Spring, MD); M.Torà (Institut Municipal d’Investigació Mèdica, Barcelona, Spain); J.Wang (Information Management Services, Silver Spring, MD, USA). WHI Program Office: (National Heart, Lung, and Blood Institute, Bethesda, MD) J.Rossouw, S.Ludlam, D.Burwen, J.McGowan, L.Ford and N.Geller. Clinical Coordinating Center: (Fred Hutchinson Cancer Research Center, Seattle, WA) G.Anderson, R.Prentice, A.LaCroix and C.Kooperberg. Investigators and Academic Centers: (Brigham and Women’s Hospital, Harvard Medical School, Boston, MA) J.E.Manson; (MedStar Health Research Institute/Howard University, Washington, DC) B.V.Howard; (Stanford Prevention Research Center, Stanford, CA) M.L.Stefanick; (The Ohio State University, Columbus, OH) R.Jackson; (University of Arizona, Tucson/Phoenix, AZ) C.A.Thomson; (University at Buffalo, Buffalo, NY) J.Wactawski-Wende; (University of Florida, Gainesville/Jacksonville, FL) M.Limacher; (University of Iowa, Iowa City/Davenport, IA) R.Wallace; (University of Pittsburgh, Pittsburgh, PA) L.Kuller; (Wake Forest University School of Medicine, Winston-Salem, NC) S.Shumaker. Women’s Health Initiative Memory Study: (Wake Forest University School of Medicine, Winston-Salem, NC) S.Shumaker. For a list of all the investigators who have contributed to WHI science, please visit: https://cleo.whi.org/researchers/SitePages/Write%20a%20Paper.aspx. We would like to thank the participants and staff of the Nurses’ Health Studies and Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. In addition, this study was approved by the Connecticut Department of Public Health (DPH) Human Investigations Committee. Certain data used in this publication were obtained from the DPH.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- BBCS
Brescia Bladder Cancer Study
- CeRePP
French Center for Research on Prostate Diseases
- EPIC
European Prospective Investigation Into Cancer and Nutrition Study
- FBCS
French Bladder Study
- GWAS
genome-wide association study
- LABCS
Los Angeles Bladder Cancer Study
- NEBCS
New England Bladder Cancer Study
- NCI
National Cancer Institute
- OR
odds ratio
- PLCO
Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial
- SNP
single nucleotide polymorphism.
References:
- 1. Silverman D.T., et al. (2006). Bladder cancer. In Fraumeni J.F., Jr, Schottenfeld D. (eds) Cancer Epidemiology and Prevention Third Edition. Oxford University Press, New York, NY, pp. 1101–1127 [Google Scholar]
- 2. García-Closas M., et al. (2005). NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet, 366, 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu X., et al. (2006). Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am. J. Hum. Genet., 78, 464–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stern M.C., et al. ; International Consortium of Bladder Cancer. (2009). Polymorphisms in DNA repair genes, smoking, and bladder cancer risk: findings from the international consortium of bladder cancer. Cancer Res., 69, 6857–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cantor K.P., et al. (2010). Polymorphisms in GSTT1, GSTZ1, and CYP2E1, disinfection by-products, and risk of bladder cancer in Spain. Environ. Health Perspect., 118, 1545–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. García-Closas M., et al. (2011). A single nucleotide polymorphism tags variation in the arylamine N-acetyltransferase 2 phenotype in populations of European background. Pharmacogenet. Genomics, 21, 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koutros S., et al. (2011). Hair dye use and risk of bladder cancer in the New England bladder cancer study. Int. J. Cancer, 129, 2894–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moore L.E., et al. (2011). GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis, 32, 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rothman N., et al. (2007). Commentary: Reflections on G. M. Lower and colleagues’ 1979 study associating slow acetylator phenotype with urinary bladder cancer: meta-analysis, historical refinements of the hypothesis, and lessons learned. Int. J. Epidemiol., 36, 23–28 [DOI] [PubMed] [Google Scholar]
- 10. Kiemeney L.A., et al. (2008). Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet., 40, 1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rafnar T., et al. (2009). Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet., 41, 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu X., et al. (2009). Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet., 41, 991–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rothman N., et al. (2010). A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet., 42, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia-Closas M., et al. (2011). A genome-wide association study of bladder cancer identifies a new susceptibility locus within SLC14A1, a urea transporter gene on chromosome 18q12.3. Hum. Mol. Genet., 20, 4282–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rafnar T., et al. (2011). European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Hum. Mol. Genet., 20, 4268–4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu Y.P, et al. (2012). Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc. Natl Acad. Sci. USA, 109, 4974–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang W., et al. (2012). Mapping of the UGT1A locus identifies an uncommon coding variant that affects mRNA expression and protects from bladder cancer. Hum. Mol. Genet., 21, 1918–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Figueroa J.D., et al. (2014). Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum. Mol. Genet., 23, 1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garcia-Closas M., et al. (2013). Common genetic polymorphisms modify the effect of smoking on absolute risk of bladder cancer. Cancer Res., 73, 2211–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baris D., et al. (2009). A case-control study of smoking and bladder cancer risk: emergent patterns over time. J. Natl Cancer Inst., 101, 1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Samanic C., et al. (2006). Smoking and bladder cancer in Spain: effects of tobacco type, timing, environmental tobacco smoke, and gender. Cancer Epidemiol. Biomarkers Prev., 15, 1348–1354 [DOI] [PubMed] [Google Scholar]
- 22. Rothman K.J., et al. (2008). Modern Epidemiology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 23. Piegorsch W.W., et al. (1994). Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat. Med., 13, 153–162 [DOI] [PubMed] [Google Scholar]
- 24. Umbach D.M., et al. (1997). Designing and analysing case-control studies to exploit independence of genotype and exposure. Stat. Med., 16, 1731–1743 [DOI] [PubMed] [Google Scholar]
- 25. Chatterjee N., et al. (2005). Semiparametric maximum likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika, 92, 399–418 [Google Scholar]
- 26. Albert P.S., et al. (2001). Limitations of the case-only design for identifying gene-environment interactions. Am. J. Epidemiol., 154, 687–693 [DOI] [PubMed] [Google Scholar]
- 27. Han S.S., et al. (2012). Likelihood ratio test for detecting gene (G)-environment (E) interactions under an additive risk model exploiting G-E independence for case-control data. Am. J. Epidemiol., 176, 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerstein M.B., et al. (2012). Architecture of the human regulatory network derived from ENCODE data. Nature, 489, 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ward L.D., et al. (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40(Database issue), D930–D934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boyle A.P., et al. (2012). Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ormestad M., et al. (2006). Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development, 133, 833–843 [DOI] [PubMed] [Google Scholar]
- 32. Cummings D.E., et al. (1996). Genetically lean mice result from targeted disruption of the RII beta subunit of protein kinase A. Nature, 382, 622–626 [DOI] [PubMed] [Google Scholar]
- 33. Lai G.Y., et al. (2013). The association between self-reported diabetes and cancer incidence in the NIH-AARP Diet and Health Study. J. Clin. Endocrinol. Metab., 98, E497–E502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koebnick C., et al. (2008). Body mass index, physical activity, and bladder cancer in a large prospective study. Cancer Epidemiol. Biomarkers Prev., 17, 1214–1221 [DOI] [PubMed] [Google Scholar]
- 35. Batty G.D., et al. (2005). Obesity and overweight in relation to organ-specific cancer mortality in London (UK): findings from the original Whitehall study. Int. J. Obes. (Lond)., 29, 1267–1274 [DOI] [PubMed] [Google Scholar]
- 36. Wolk A., et al. (2001). A prospective study of obesity and cancer risk (Sweden). Cancer Causes Control, 12, 13–21 [DOI] [PubMed] [Google Scholar]
- 37. Jivan A., et al. (2009). Radial spoke protein 3 is a mammalian protein kinase A-anchoring protein that binds ERK1/2. J. Biol. Chem., 284, 29437–29445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sacco J.C., et al. (2010). Cytochrome b5 and NADH cytochrome b5 reductase: genotype-phenotype correlations for hydroxylamine reduction. Pharmacogenet. Genomics, 20, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kurian J.R., et al. (2006). Reductive detoxification of arylhydroxylamine carcinogens by human NADH cytochrome b5 reductase and cytochrome b5. Chem. Res. Toxicol., 19, 1366–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lan Q., et al. (2012). Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat. Genet., 44, 1330–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.