Abstract
The p53 transcription factor is a major tumor suppressor, whose diverse activities serve to ensure genome stability and inhibit neoplastic processes. In recent years, it is becoming increasingly clear that p53 also plays a broader role in maintaining cellular homeostasis, as well as contributing to tissue homeostasis in a non-cell-autonomous fashion. Chronic inflammation is a potential cancer-promoting condition, and as such is also within the radar of p53, which mounts a multifaceted attempt to prevent the escalation of chronic tissue imbalance into neoplasia. Recent understanding of the p53 pathway and other family members reveals a broad interaction with inflammatory elements such as reactive oxygen and nitrogen species, cytokines, infectious agents and major immune-regulatory pathways like nuclear factor-kappaB. This complex cross talk is highly dependent on p53 status, as different p53 isoforms and p53 mutants can mediate different responses and even promote chronic inflammation and associated cancer, acting in the tumor cells as well as in the stromal and immune compartments.
The evolution of p53 knowledge: from oncogene to stress-responsive transcription factor
Since the discovery of p53 more than 30 years ago, the field of p53 research has undergone several major conceptual changes. At first, the fact that p53 is overexpressed in the majority of tumors led to the conjecture that p53 is an oncogene, a notion that seemed to be supported by the demonstration that cancer-derived p53 DNA clones exhibited a variety of transforming activities in vitro and in vivo (reviewed in refs 1,2). However, within a few years, it became evident that although mutated forms of p53 (which accumulate in cancer cells) can confer transformed features when overexpressed, the wild-type (WT) p53 actually acts as a tumor suppressor (3, reviewed in refs 4,5).
Further research revealed that p53 operates primarily as a transcriptional regulator, whose cellular activity is finely tuned by a plethora of diverse stress signals (4–7). Already very early on, it was demonstrated that cellular p53 levels are induced by DNA damage and that this induction serves to arrest the cell cycle, presumably enabling the cells to attempt a successful resolution of the damage (8,9). Subsequently, it was shown that p53 can actually induce a much broader spectrum of cell fate changes, and in cases of severe damage, it would often favor the apoptotic execution of the damaged cell or push it into replicative senescence (10–13). The common denominator of practically all documented activities of p53 is that they prevent the emergence and persistence of cells with damaged genomes, rightfully earning p53 the title of ‘guardian of the genome’ (14).
Being such a cardinal hub for a large multitude of signals, it is not surprising that p53 is surrounded by a complex molecular weave composed of multilayered regulators and effectors (15,16). The most prominent negative regulator of p53 is Mdm2. Mdm2 is an E3 ubiquitin ligase that restricts p53 levels and functionality in the absence of stress stimuli but is incapacitated under stress conditions in order to enable the swift nuclear accumulation and activation of p53 (17). This interplay is governed by ubiquitylation and involves also the Mdm2-related Mdmx/Mdm4 protein (17). In reality, however, the picture is not that simple, since a number of additional E3 ligases have also been shown to target p53 and modulate its stability and activity (18). Furthermore, ubiquitylation is only one of many post-translational modifications affecting the outcome of p53 induction; others include phosphorylation, acetylation, lysine methylation, arginine methylation, sumoylation, neddylation and more, all of which affect gene expression as part of the cellular machinery sensing a stress signal and dispatching a specific message to the p53 protein (19). Excitingly, subsequent years of p53 research have seen the rise of a richer palette of cellular processes governed by this transcription factor (13,20). What began as a straightforward sensing of DNA damage and alarming the cell to respond appropriately eventually became a central hub converting a comprehensive range of signals into a context-adjusted transcriptional profile. In recent years, the multifaceted involvement of p53 in the regulation of cellular metabolism has been receiving growing attention (21–24).
For instance, WT p53 can restrict glucose uptake, inhibit glycolysis and favor mitochondrial oxidative phosphorylation, all of which counteract the tumor-associated ‘Warburg effect’ (22). p53 can also promote cell death in response to severe or prolonged endoplasmic reticulum stress that involves excessive accumulation of misfolded proteins (25,26). Furthermore, the involvement of p53 in autophagy is unquestionable, even though its exact impact on that process is still controversial. Thus, p53 has been reported to either induce autophagy or inhibit it, probably depending on its subcellular location (27,28). Most probably, p53 can regulate autophagy in many context-dependent ways, the common denominator being that it enables more effective and more dynamic adaptation to changing environmental conditions (29). Another example of the expanding biological portfolio of p53 is its role in embryonic development and differentiation control, where p53 was shown to maintain the differentiated state and restrain dedifferentiation (30,31) since lower p53 protein levels gave rise to higher reprogramming efficiency of somatic cells into induced pluripotent stem cells (32).
Together, all the long known as well as recently discovered functions of p53 have a central common denominator: p53 exploits its entire biochemical repertoire to preserve cellular homeostasis, preventing the escalation of chronic imbalance into neoplastic transformation of the cell. Transcriptional activation and transcriptional repression, as well as direct protein–protein interactions, are all p53 modes of action striving to protect the cell from being channeled toward a course that endangers the organism’s well-being (33).
One of the most profound conditions that have been increasingly linked to stress-promoting carcinogenesis is the chronic inflammatory response (34,35). In essence, the attempt of the host tissue to respond to harmful stimuli such as irritation and pathogens might be swayed into a chronic state leading to constant stress and through it to derangement of normal tissue homeostasis and to cancer. This review will discuss the roles that p53 plays in addressing such stress. We will particularly focus on the interactions of the WTp53 tumor suppressor, as well as of cancer-derived mutant p53 isoforms and p53 family members, with different elements of the inflammatory ensemble.
Cancer-associated inflammation
Inflammation is highly conserved throughout evolution (36) and serves as a first line of defense of the host against injury and infection. When managed properly, the inflammatory arm has the power to execute an acute outburst in response to toxins, irritants, pathogens and chemicals, orchestrating an arsenal of effectors fighting to return the tissue to its original state. Yet, it is the chronic exposure of the tissue to inflammation that interrupts this delicate balance, resulting in greater damage to the host.
The acute inflammatory phase is generally initiated by the activation of tissue-resident sentinel cells, such as macrophages and dendritic cells that, once stimulated, rapidly secrete a set of cytokines and chemokines as well as additional soluble effectors such as vasoactive peptides and specific lipids (37–39). As a result, vascular permeability is increased and additional innate immune cells like neutrophils are recruited to the site, where they assist in the phagocytosis of bacteria and dead host cells while secreting bactericidal agents. The transition from such acute to chronic phase involves the adaptive immune system as antigen presenting cells traffic to lymphoid tissues. The subsequent wave of antigen-specific effector T cells infiltrating the site of inflammation produces cytokines such as interferon (IFN)-γ and interleukin (IL)-17, which enhance the activation of phagocytic cells along with bactericidal effects that prolong their inflammatory functions. Once this process is compromised, it might lead to an imbalance and result in an overly inflamed tissue. Of note, a chronic response might arise also in the absence of an acute trigger and has the potential to persist for years, slowly accumulating and gradually smoldering the tissue.
The contribution of inflammation to tumorigenesis might be grossly divided into two main categories, where the first is associated with the initiation of the cancerous process, whereas the second impacts cancer progression. Conditions of chronic inflammation leading to cancer could be attributed to infectious pathogens (Helicobacter pylori in colorectal cancer, hepatitis B or C viruses in liver cancer), imbalanced immune regulation (inflammatory bowel disease in colorectal cancer), chemical irritants (tobacco smoke or asbestos in lung cancer) and even dietary habits (obesity in liver cancer) (40–43). A single epithelial cell sustaining several mutational hits may acquire selective advantages, allowing it to prosper due to hyperproliferative capabilities or evasion of cell death. In the inflammatory microenvironment, the propensity of such mutations to accumulate in the genome of transformed cells increases due to their exposure to a constant flux of reactive oxygen and nitrogen species (ROS and RNS), produced by activated immune cells (44,45). ROS and RNS are thought to induce genomic instability mainly by modifying the C8 position of guanine to form the highly mutagenic 7,8-dihydro-8-oxoguanine and also by contributing to single- and double-strand breaks (46,47). These mechanisms are added to the solid evidence for the link between preceding inflammation and subsequent carcinogenesis. Such correlation is provided by the numerous pathologies associating chronic inflammation with an increased risk for cancer development.
In addition to its role in tumor initiation, chronic inflammation may be exploited by the cancer cells to remodel their immediate microenvironment, favoring the tumor’s ability to prosper. Cancer cells express and secrete an array of tumor-promoting cytokines and chemokines that summon specific leukocytes; the latter cells, in turn, secrete additional effectors. Thus, the tumor as a ‘foreign’ tissue takes advantage of wound-healing features exerted by specific subtypes of immune cells, nourishing and contributing to tumor maintenance. For example, after an initial growth stage, many solid tumors suffer from lack of blood supply and are in need of angiogenic support. The newly recruited inflammatory cells will not only provide a consistent supply of growth factors but will also secrete neoangiogenic agents, allowing the vascular network to catch up with the growth of the malignant tissue (48–50).
Oncogenic mechanisms in chronic inflammation
For a cancer cell to become an invading and metastasizing tumor, it has to rely on a sequence of genetic and epigenetic alterations that will promote its ability to escape inhibition by tumor suppressors like p53, evade surveillance by the immune system and overcome physical barriers. Putting powerful oncogenes in the driver’s seat is, therefore, crucial for malignant progression. Some of those oncogenic alterations were reported to become hyperactive upon prolonged exposure to an inflammatory microenvironment. For instance, patients with chronic pancreatitis have an over 10-fold increased risk of developing pancreatic cancer, a cancer characterized by a very high rate of mutations in the K-Ras oncogene. It was shown that although pancreatic acinar cells are intrinsically highly refractory to K-Ras-induced transformation, they will readily yield pancreatic intraepithelial neoplasias and ductal adenocarcinomas when exposed to limited periods of non-acute pancreatitis (51). Interestingly, these inflammatory conditions were found to promote neoplastic transformation by suppressing K-Ras-induced senescence (51). Such interactions may also be governed by a feedback loop involving nuclear factor-kappaB (NF-κB), which, in turn, increases K-Ras to pathological levels (52). Another wide-scale mechanism affected by chronic exposure to inflammation involves the mismatch repair machinery. Pelvic inflammatory disease is associated with an increased risk of epithelial ovarian cancer by inducing mismatch repair abnormalities, manifested by decreased expression of human mutL homolog 1 and human mutS homolog 2 proteins (53). This mechanism is extremely prominent in colorectal cancer, where microsatellite instability resulting from defects in the mismatch repair machinery is abundant and considered to be a tumorigenic driving force exacerbated when associated with inflammatory bowel disease (54,55). An additional oncogenic feature mediated by the chronically inflamed microenvironment is DNA methylation alterations. DNA methyltransferases such as DNMT1 can be regulated by viral oncoproteins (56) and are overexpressed in premalignant tissues overloaded with inflammatory effectors, as reported for peripheral pancreatic ductal epithelia (57) and lung tissues of non-small cell lung cancer in smoking patients (58). Furthermore, proinflammatory cytokines like IL-15 may induce DNA hypermethylation, which precedes large granular lymphocytic leukemia, by activating an NF-κB-Myc axis (59).
Epithelial to mesenchymal transition is yet another manner by which oncogenic mechanisms are manifested under inflammatory conditions (60). Without a permissive environment, epithelial to mesenchymal transition is unlikely to occur even if cancer cells present aberrant stem cell signaling pathways like Wnt or Notch. Importantly, transforming growth factor (TGF)-β1 contributes to such epithelial to mesenchymal transition-permissive tumor microenvironment by serving as a chemotactic factor for neutrophils and macrophages. Moreover, TGF-β1 can also affect the protumoral polarization of such recruited cells once they reside within the tumor, resulting in induction of tumor-favoring M2-like macrophages, N2-like neutrophils and a Th2 cell phenotype (61,62).
This tight cross talk between oncogenesis and inflammation is actually bidirectional. Thus, many oncogenes possess the ability, when activated, to initiate a signaling cascade resulting in an inflammatory response in the immediate surroundings of the cells that harbor those oncogenes. This is exploited by the cancer cells to their selective advantage. One good example is that of the major proinflammatory cytokine IL-8, which can be activated by Ras in vitro and in vivo and can contribute to hyperactive inflammation, neovascularization and accelerated tumor growth (48). Such tumorigenesis-supporting milieu can also be provided by other cytokines such as IL-6 and the mouse IL-8 orthologs KC and MIP-2, all featuring in a Ras-induced secretory phenotype (63,64). Additional oncogenes, including Myc, BRAF and RET, have also similarly been implicated in induction of proinflammatory cytokine and chemokine secretion.
Molecular traits of inflammation-driven cancer
The specific molecular signatures of cancer-promoting inflammation vary between different body sites and different biological contexts, depending on the exact nature of the immune cell content and the secreted effectors. Nevertheless, some common features are often encountered. Typically, the response will involve activated general pathways such as NF-κB, activator protein 1 and signal transducer and activator of transcription (STAT), taking place in the transformed epithelial cells, immune cells and other stromal components. Together, these form a tumor-promoting microenvironment that is largely dependent on the secreted cytokine and chemokine profile building up in the tissue. The shift from tumoricidal mediators like IL-12 and IFN-γ to cytokines like IL-8, IL-17, IL-23 and IL-13 allows the cancer cells to escape immune surveillance and utilize the system for their own good (65). These imbalanced inflamed conditions are governed by a variety of molecules that normally play significant roles in cell signaling but become aberrantly induced in the chronic inflammatory setting. This is exemplified by the ROS family of molecules: low ROS levels regulate kinase-driven pathways in response to cellular stimuli such as nutrient deprivation and hypoxia (66), whereas high acute ROS/RNS outbursts provide destructive power required for microbial elimination. However, abnormal persistence of high ROS levels results in oxidation of nuclear and mitochondrial DNA, proteins and lipids, impairing their structure and function and driving the aberrant expression and deregulation of oncogenic pathways (67).
One protein that plays a pivotal role in orchestrating alterations in cellular oxidative and nitrative state is inducible nitric oxide synthase 2 (NOS2). Unlike the two constitutively expressed NOS enzymes (eNOS and nNOS), which are not thought to be associated with immune regulation, the inducible isoform produces NO for prolonged periods and at varying levels. Such variable release dictates NO-modulated microenvironments within a tissue, with many different outcomes (68). Overproduction of NO can lead to the formation of 8-nitroguanine, an indicator of nitrative DNA damage. Both 8-nitroguanine and 8-oxoguanine are formed in various inflammation-related cancers and precancerous lesions in an NOS2-dependent manner. Such nitrative and oxidative DNA damage can induce various types of genome instability as well as epigenetic changes, underpinned by tumor necrosis factor alpha (TNF-α) and IL-6 activities associated with NF-κB and STAT3 regulation (69).
The deregulation of cyclooxygenase-2 (COX-2; also known as prostaglandin synthase 2 or PTGS2), leading to increased abundance of its principal metabolic product, prostaglandin E2, is an additional characteristic of many inflammation-related malignancies. Similar to NOS2, COX-2 is inducible, becoming detectable in many tissues only after exposure to stimuli such as proinflammatory conditions. Prostaglandin E2 promotes cell survival and proliferation and was also linked to invasion and metastasis (70–72) in a COX-2-dependent manner. Imbalanced COX-2 expression is mechanistically associated with aberrations in the Wnt, NF-κB and TGF-β pathways, primarily in monocytes but also in epithelial cells.
Further to the above signaling molecules that play a prominent microenvironmental role in promoting carcinogenesis, secreted cytokines and chemokines are of paramount importance in that process. The impact of those ILs and chemotactic molecules is greatly affected by their concentration gradient within a defined microenvironment, the identity of the secreting cells, the responding cells and the tissue type. The classic, but rather crude, dichotomous classification of this diverse group into pro- and anti-inflammatory cytokines essentially recapitulates their respective association with cancer. Among the proinflammatory cytokines, TNF-α, IL-6 and IL-8 have been abundantly described in animal models and human tissues as mediators of increased tumor growth, DNA damage, angiogenesis and metastatic load (73). Conversely, anti-inflammatory effectors like IL-10, IL-4 and TGF-β are often considered as antitumorigenic and even tumor suppressors. However, in reality, the picture is much more complicated. Thus, relatively low levels of TNF-α or IL-6 may lead to immunoprotective and antipathogenic activities, whereas chronic secretion of anti-inflammatory cytokines like IL-4 and IL-13 might give rise to tumor-associated macrophages that can promote tumor growth. In that regard, TGF-β is particularly notorious for its dual role in carcinogenesis, inhibiting tumor growth at initial stages but acting as a driver of malignancy in advanced and metastatic stages (74). The growing field of non-coding RNA molecules, presently represented primarily by microRNAs (miRs), adds another tier to the molecular interactions underpinning the inflammatory response. The expression of these non-coding RNA molecules is often induced or repressed in response to different stimuli. miRs typically reduce the translation, and often also the steady state levels, of the messenger RNA species targeted by them, in both cases leading to reduced amounts of the corresponding proteins. Numerous miRs have been reported to participate in innate as well as adaptive immune responses, coordinating and fine-tuning the expression of many cellular components of both arms (75,76). Specifically, miRs such as miR-155, miR-196, miR-210, miR-21 and miR-125b were shown to mediate inflammation-related carcinogenesis by multiple mechanisms. Essentially all major signaling pathways involved in the chronic inflammatory response, including NF-κB, activator protein 1 and STAT3, are associated with subsets of regulatory miRs. Notably, many of those miRs were implicated in a causative role in infectious agent-induced cancer, involving bacteria and viruses. miR-155, for instance, is a central regulator of the immune system and is highly pertinent to many of the lymphoid tissues. Deregulation of miR-155 results in malignant development; indeed, miR-155 is overexpressed in many cancers, including those of the hematopoietic lineages (77). miR-21, another prominent highly expressed in a broad set of tumor types, was implicated in an epigenetic switch connecting NF-κB, miR-181b-1 and STAT3 to promote cell transformation (78) as well as in the context of oncogene addiction (79). Furthermore, miR-21 was also suggested to be responsible for the aberrant TGF-β response that contributes to the induction of cancer-associated fibroblasts (80).
WT p53 protects against inflammatory stress
Inside this intricate molecular network, p53 is caught in the cross fire, responding to the oxidative and nitrosylative stress (81). The cross talk between p53 and cellular ROS is characterized by a remarkable dualism. On the one hand, under conditions of severe stress, p53 employs high levels of ROS as a downstream tool to execute its apoptotic agenda (82). However, in non-stressed cells or upon mild stress, p53 monitors ROS levels and maintains them in tight control, thereby preventing excessive ROS accumulation that might endanger genome integrity. In this capacity of guardian of redox homeostasis, p53 acts as an antioxidant transcription factor elevating the expression of genes like ALDH4, PIG12, sestrin 1 and 2 and GPX1, all of which restrict ROS levels and prevent endogenous oxidative stress (82–85). In that context, NO produced by NOS2 in the colon tissues of ulcerative colitis patients have been shown to be responsible for p53 activation (86). Such an accumulation of p53 can actually lead to a negative feedback loop in which p53 protects from NO-induced DNA damage (86). Notably, nitrosylation and oxidation of tyrosine and cysteine residues are detectable in cells subjected to inflammation-induced stress (87,88). The interaction between p53 and COX-2 probably also contributes to prevention of tumorigenesis. Even though Cox-2 is induced by p53-mediated activation, its induction leads to antiapoptotic mechanisms that abate cellular stresses associated with p53 induction (89). COX-2 may benefit from p53 downregulation and mediate cell proliferation as well as differentiation of stem cells (90). Moreover, in many malignant tissues, COX-2 overexpression is enabled in the wake of p53 mutations, implying that such mutations provide a possible escape mechanism of the COX-2 pathway from its normal physiological regulation; furthermore, there appears to be an interplay between this pathway and specific mutant forms of p53 (91).
The best-documented connection of p53 to the miR world is through the miR-34 family, extensively shown to be directly transcriptionally upregulated by p53 and to contribute to p53-mediated biological effects such as apoptosis, G1 arrest and senescence (92). Interestingly, miR-34 levels were found to decrease in lungs of mice expressing a combination of mutant K-Ras and mutant p53, in association with a severe inflammatory response subsequent to tumor development; in parallel, oncogenic miRs such as miR-155 and miR-21 became upregulated (93). Furthermore, miR-34 was able to prevent both tumor development and the infiltration of immune cells when animals were infected with a lentivirus driving miR-34 expression (93).
Hints of a potential link of p53 with inflammation were already provided very early on, when the tight relationship between several viruses and p53 was uncovered. Of note, viruses are very common instigators of inflammation. Infection with a variety of viruses can elicit a protective cellular response, wherein the cell is driven to exit the cell cycle or undergo apoptosis as a preemptive measure aimed at preventing the production and release of mature infectious viral particles. p53 is a key player in this protective response, and its loss can often facilitate effective viral replication. As a countermeasure, several virus families have evolved molecular mechanisms that incapacitate p53 and get it out of their way, often through the action of viral-encoded proteins that bind and inactivate p53. In fact, the discovery of p53 was due to its direct targeting by such viral protein, the SV40 large T antigen (1). In addition, various viral proteins can achieve a similar outcome by inactivating downstream targets of p53. Although such mechanisms do not necessarily imply a direct p53-inflammation association, more recent work demonstrated that p53 is an active participant in the type I IFN antiviral defense mechanism (94). IFN-inducible genes such as IRF9, IRF5, ISG15 and TLR3 were identified as direct transcriptional targets of p53, indicating its key role in fighting viral infection. An even broader link between p53 and the innate arm of the immune system is manifested by its ability to regulate many members of the Toll-like receptor (TLR) family (95,96). Thus, in both T-lymphocytes and alveolar macrophages, a set of TLR genes is stimulated and activated in a p53-dependent manner, in a process that appears to be unique for primates. This might underpin a positive feedback loop, wherein p53 is stabilized due to various types of stress; as a consequence, TLR levels will also increase, resulting in their ligands generating an inflammatory response that may push p53 levels even higher (96). This and additional findings (97) reveal that, in the context of antipathogen defense, p53 can modulate inflammatory processes to the benefit of the host.
Evidence for an inflammation-antagonistic effect of p53 could be deduced from the analysis of p53 knockout mice, which displayed severe outbursts of autoimmune pathologies like arthritis and diabetes (98–100). Strikingly, a considerable fraction of these animals die before any evidence of cancer is detected; this is mostly attributable to severe inflammatory manifestations such as abscesses, gastroenteritis and myocarditis (101), underscoring the crucial impact of p53 on the proper regulation of innate immunity.
The complex interplay between cellular senescence and cancer and the pivotal role of p53 in cellular senescence have been extensively studied in recent years (reviewed in refs 102–104). A recent study (105) tries to settle the apparent quandary created by the fact that senescence, an anticancerous process in its essence, coexists typically with chronic inflammatory signaling that might promote cancer. The authors describe a novel low-grade response dubbed ‘parainflammation’, which is triggered by senescence in the colonic epithelium and does not rely on the recruitment of typical inflammatory cells. In the presence of functional p53 and its target, p21, this type of inflammation does not elicit neoplastic consequences; however, p53 loss converts the parainflammatory response into a tumor-promoting mechanism, resulting in accelerated tumor growth and invasion. This further highlights the importance of p53 in ensuring that inflammatory responses are properly contained.
p53 and NF-κB: a context-dependent interaction
At large, both p53 and NF-κB are transcription factors that date back to early invertebrates. Both serve as major cellular hubs collecting a variety of signals and translating those into a transcriptional outcome. Although p53 is supervising intrinsic stimuli such as DNA damage and hypoxia that threaten the fidelity of proper cell division, NF-κB is mostly entrusted with responding to extrinsic stimuli frequently bound to defend against pathogenic insults, where its activation is tightly involved with immune regulation.
In the context of cancer, the rather different modes of action of p53 and NF-kB position them on two opposite sides of the barricade. The fact that deregulated cell clones produced as a consequence of stress are eliminated through induction of cell cycle arrest, apoptosis and senescence, underlies a significant part of p53’s tumor suppressive capabilities, although p53’s contribution to metabolic homeostasis also appears to be crucial in that regard (106). On the other hand, part of the basic agenda of NF-κB upon identification of an infectious attack is geared toward accelerated division of immune cells, in order to augment clonal selection and thereby enhance the specificity and overall extent of the protective response. It is not surprising then that when this response gets out of balance, it might acquire cancer-promoting features (107).
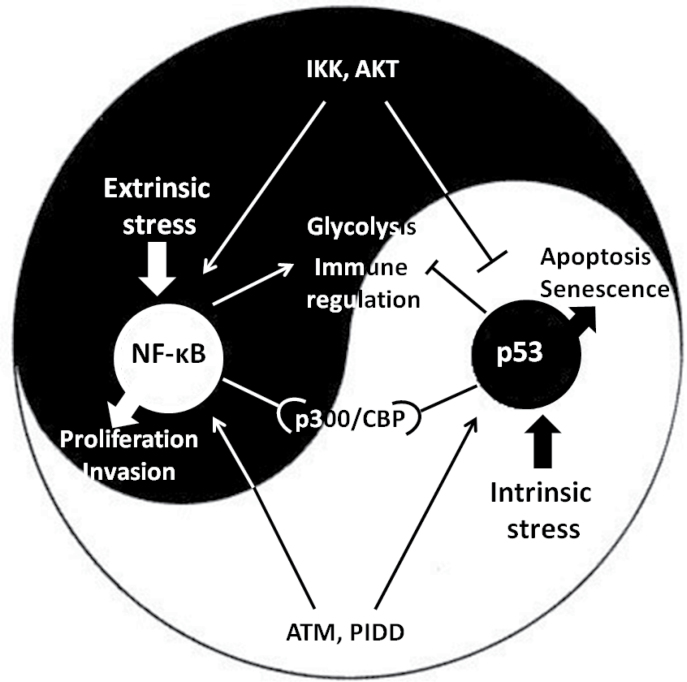
The cross talk between NF-κB and WT p53 is typically described as a rivalry between two forces trying to navigate the affected cell toward opposite fates. One of the central battlefields is cell metabolism. For example, NF-κB transcriptional activity will favor increased glucose uptake as a fundamental prerequisite for rapid cell proliferation, whereas p53 will act to slow down glucose uptake (108). Such mutually exclusive roles have probably favored an evolutionary process leading to a counter-dominant negative relationship between NF-κB and p53, enabling each of them to repress the other in particular biological contexts. As an undesirable byproduct, this might cause the cell to become more vulnerable to carcinogenic processes when p53 is inhibited by excessive NF-κB activity, whereas excessive NF-κB repression by p53 might potentially spell immune deficiencies. Several molecular mechanisms can control and tilt the p53/NF-κB balance, depending on the nature of the transduced signal. For example, the IκB kinase (IKK) complex activates NF-κB by phosphorylating its inhibitor IκB, which otherwise retains NF-κB mostly in the cytoplasm and restricts the transcriptional activity of those NF-kB molecules that do manage to get into the nucleus. However, IKK can also phosphorylate p53 on specific residues, making it susceptible to ubiquitylation by β-TrCp and subsequent degradation by the proteasome (109,110). In an additional mechanism, both transcription factors compete for common cofactors (111). Thus, the ability to induce rapid transcriptional responses is highly dependent on inducible changes in the chromatin; this renders histone modifiers such as the acetyltransferases p300 and CBP crucial components in the efficient initiation of signal-induced transcription. The availability of the p300 protein is limited, providing an opportunity for a competition between p53 and NF-κB whose outcome is dictated by the immediate transcriptional needs of the cell (109). Overall, such competitive interactions have contributed to the common perception of the p53-NF-κB cross talk as a mutually exclusive antagonistic relationship.
However, the picture is far from being as simplistic as that. In fact, a growing body of evidence supports the existence of mutual interactions where p53 and NF-κB actually join forces rather than antagonize each other. This is perhaps not all that surprising: although NF-κB is commonly portrayed as a potent inhibitor of cell death, it can actually promote cell death under specific conditions (112–114). Conversely, although p53 is well known for its proapoptotic features, it is also able to promote cell survival in a variety of settings (115–117). Hence, the early finding that NF-κB can sometimes be required for p53-mediated apoptosis (117), although at first glance counterintuitive, is perhaps not as unexpected as may have seemed initially.
The DNA damage response is a good example of how the cellular perturbations induced by genotoxic stress might recruit NF-κB to serve as a downstream effector of the Ataxia telangiectasia mutated complex, thereby bringing it to play in the ‘homecourt’ of p53 (118,119). Thus, in response to double-strand DNA breaks, p53 and NF-kB are coordinately activated and contribute together to the resultant changes in gene expression (120). It is still unclear whether p53 deflects NF-κB away from its proinflammatory course toward activities required for dealing with the genotoxic insult. p53 and NF-kB also cooperate in additional settings, such as in the induction of oncogene-induced senescence, where NF-κB actually displays features of a non-cell-autonomous tumor suppressor and promotes the clearance of oncogene-expressing cells by the immune system (121).
A possible positive link between p53 and NF-κB may occur through the p53-induced death domain protein complex, which is considered a downstream effector with a pivotal role in determining the cellular response to genotoxic stress. p53-induced death domain protein expression is upregulated by p53 and is positively correlated with p53-dependent cell death (118). At the same time, it forms an axis together with receptor interacting protein 1 and NF-kappa-B essential modulator upstream of NF-κB signaling as part of the NF-κB response to DNA damage; in several p53-induced death domain protein-deficient cellular systems, the NF-κB response to genotoxic stress was shown to be defective (122). The cross talk between p53 and NF-κB may also be modulated by a common polymorphism in the human TP53 gene, resulting in either proline or arginine (31,123) at position 72 of the p53 protein. Transgenic mice knocked in with the proisoform displayed increased NF-κB-dependent inflammatory gene expression, with increased chromatin association of the p65 subunit of NF-κB and enhanced response to an lipopolysaccharide challenge (123). Figure 1 depicts the ambivalent relationship between p53 and NF-κB. It is likely that these two ancient complementary cellular mechanisms have been trained and adjusted throughout evolution to respond to the complex ensemble of signals in a manner that depends on their nature, extent and origin of those signals. Both p53 and NF-κB follow rigorous lines of regulation, which, when compromised, might result in carcinogenic effects; chronic inflammatory conditions are just one facet of such compromised regulation.
Fig. 1.
The complex cross talk between p53 and NF-κB. Both transcription factors respond to a set of activating signals that accumulate and determine the interaction that varies from profound corepression to mutual activation, highly dependent on the cellular context.
The extended p53 family: other members, isoforms and mutants and their inflammatory links
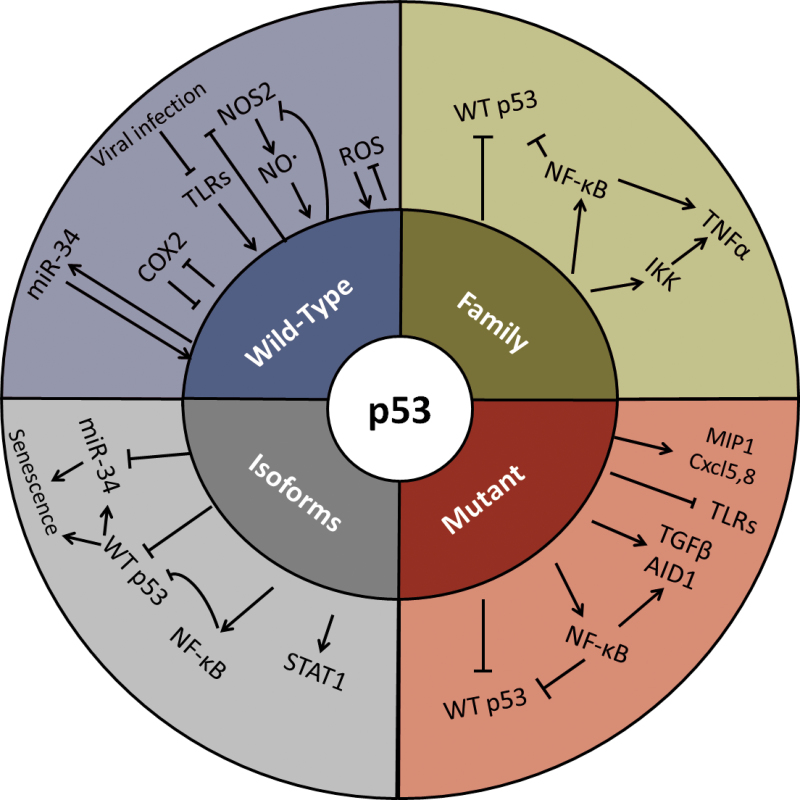
An increasing number of studies have clearly established that besides the full-length ‘classical’ WTp53, other p53 isoforms as well as p53-related proteins are fundamental and important components of this pathway (124,125). Both animal and cellular models suggest roles for these proteins in all biological activities regulated by WTp53, as well as in many additional activities (126). The TP53 gene, which consists of 11 exons, can give rise to a multitude of isoforms owing to alternative promoters, alternative splicing and multiple translational initiation sites (reviewed in ref. 127). In partial analogy with what has previously been found for the p53-related proteins p63 and p73 (see below), the p53 isoforms can be divided into subcategories, depending on whether they retain sequence-specific DNA binding and whether they retain at least part of the transactivation domain (TAD). Several of the p53 isoforms can exert dominant negative effects over coexpressed WTp53, thereby promoting oncogenic activity. It is thus reasonable to expect that at least some of them might also be capable of driving increased inflammation. Evidence in support of such conjecture was provided by the analysis of mice expressing Δ122p53; this N-terminally truncated mouse p53 is analogous to Δ133p53, one of the most common human p53 isoforms that is overexpressed in a variety of human tumors. The Δ122p53 mice display a wide range of inflammatory manifestations, including lymphocyte aggregates in different organs and numerous inflammatory pathologies such as hepatitis, vasculitis and augmented Peyer’s patches in the bowel (128). This p53 isoform was further shown to play a significant role in autoimmune conditions, where it triggers increased proinflammatory reaction owing to STAT1 activation (129). Interestingly, the inhibitory effects of different isoforms toward WTp53 were proposed to be modulated by external factors such as bacteria: interaction of H. pylori with human gastric epithelial cells induced the Δ133p53, Δ153p53 and Δ1160p53 isoforms, which were found to inhibit p53 while inducing NF-κB and increasing survival of infected cells (130).
In addition to the numerous p53 isoforms, the extended p53 family includes also two p53 paralogues, p63 and p73 (131). These proteins, which were reported to precede p53 in the evolutionary track going back to invertebrate species (132), share many structural and biochemical features with p53 and are capable of binding to p53 responsive elements via their DNA-binding domains, whose amino acid sequences are remarkably similar to the p53 DNA-binding domain. Besides sharing overlapping functions with p53, reinforcing its tumor suppressor agenda, p63 and p73 each also possess unique biochemical activities and biological functions, including prominent roles in differentiation and development. This repertoire is further enlarged through the production of a diversity of alternatively initiated and alternatively spliced isoforms of each protein, which are often more prominent that the p53 isoforms and therefore have been discovered early on. p73-deficient mice tend to develop tumors spontaneously, but on top of that they also display deregulated inflammatory responses, manifested by augmented neutrophil production and infiltration (133). The full-length, TAD-containing p73 also activates IL-4R alpha and regulates cytokine signalling in a p53-independent manner (134). TAD-deficient ΔNp73 is overexpressed in precancerous lesions in the esophagus associated with gastric reflux and, importantly, is induced by the proinflammatory cytokines IL-1β and TNF-α in an IKK-dependent manner (135). Like the p73 gene, the p63 gene also contains a separate promoter giving rise to TAD-deficient ΔNp63, which can act as a dominant negative inhibitor. Interestingly, IKKβ—the major NF-κB-activating kinase—interacts with ΔNp63 and induces it degradation and, by doing so, promotes stress-dependent cell death (136). On the other hand, ΔNp63α was shown to form a complex with the c-Rel subunit of NF-κB, which binds and represses the promoter of the p53 hallmark gene p21 in both human squamous cell carcinoma cells and normal keratinocytes; the resultant reduction in p21 expression promotes uncontrolled proliferation (137). The ΔNp63α:c-Rel complex was later reported to be induced by proinflammatory TNF-α, also inactivating TAD-containing p73 functions and enhancing cell survival (138). The cross talk of WTp53 and other isoforms and family members with inflammation is depicted in Figure 2.
Fig. 2.
p53 in inflammation. Significant reported molecular interactions of p53 and its derivatives affecting the inflammatory response.
TP53 gene mutations occur very frequently in human tumors (TP53 IARC database; http://p53.iarc.fr/). Initially, this was believed to contribute to cancer merely by abrogating the tumor suppressive functions of WTp53. However, it was then found that the majority of cancer-associated TP53 mutations actually result in production of full-length mutant proteins. These mutant p53 proteins, typically harboring only a single amino acid substitution, accumulate in the tumor cells and often reach highly elevated levels. This gave rise to the conjecture that such mutant p53 proteins may gain new oncogenic functions, as indeed proven by subsequent studies. To date, it is well accepted that mutant p53 (mutp53) gain of function is tightly involved in a wide range of cancer development aspects including inhibition of cell death, promotion of genomic instability, cell invasion and abnormal metabolism (139,140). Therefore, it is not surprising that mutp53 gain of function was also suggested as a possible mechanism driving chronic inflammation toward cancer.
Increased sensitivity to cigarette smoke, mediated by upregulation of immune response signature genes like MIP-1α/β, was observed in mice harboring a temperature-sensitive p53 mutation, which endows the mutant p53 with gain of function activities at body temperature (141). An NF-κB:mutp53 connection was observed both in cultured cells and in mouse models, suggesting that specific mutations in p53 not only abolish the anti-inflammatory properties of WT p53 but also actively intensify and prolong the inflammatory response. For example, elevated expression of CXCL5, CXCL8 and CXCL12 was found in cells expressing several p53 mutants, in conjunction with cell migration that was dependent on the p52/NF-κB2 subunit (142). Notably, expression of NF-κB2 can be induced by various p53 mutants (143). Additionally, NF-κB transcriptional activity was augmented by the presence of mutp53 in cancer cells stimulated with TNF-α, whereas depletion of endogenous mutp53 shifted the TNF-α response toward a proapoptotic phenotype (144). Mechanistically, mutp53 seems to join forces with p65 at consensus κB sites within the chromatin, presumably facilitating transcriptional activation by contributing its TAD (145,146).
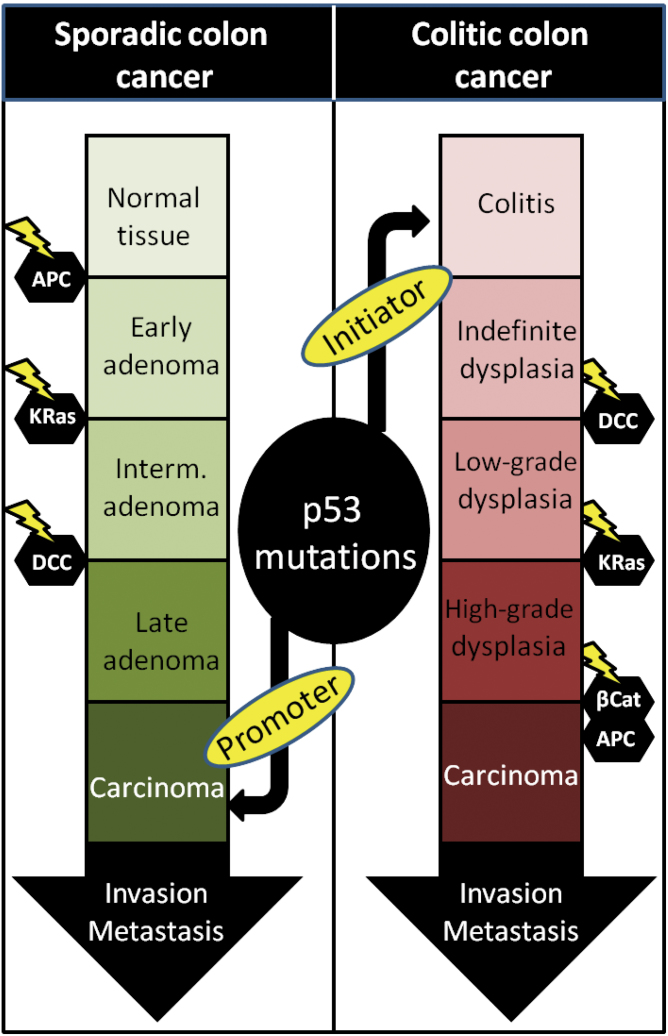
In that context, the role of p53 mutations may be significantly different when inflammation is a constant feature of the microenvironment. Importantly, TP53 mutations are a highly prevalent part of the molecular sequence of carcinogenic events in the colon epithelium. Strikingly though, in sporadic colorectal cancer, they are considered the ‘last nail in the coffin’, associated with the transition of the tumor toward an invasive, malignant state, whereas in colitis-associated colorectal cancer, p53 mutations are often observed before any other genetic alteration can be detected, suggesting a tumor-initiating role in colitis-associated colorectal cancer versus a tumor-promoting role in colorectal cancer (147,148). Notably, mutp53 was shown to augment and prolong NF-κB activity and intensify chronic colitis in mice, exposing them to a high risk of developing invasive colon carcinoma (146). The fact that early mutations in p53 will occur readily on the colitic background brings out the importance of this molecular event in the context of inflammation (Figure 3). Another mechanism linking gastric and hepatic inflammation to p53 mutations and NF-κB is the upregulation of activation-induced cytidine deaminase, which is induced either by TGF-β or by other proinflammatory cytokines in an NF-κB-dependent manner (149). Activation-induced cytidine deaminase, known to induce genomic instability and to increase mutation probability during error-prone joining of double-stranded DNA breaks, was shown to be aberrantly expressed in gastric epithelial cells infected with H.pylori, resulting in increased rates of p53 mutations. Finally, several different p53 mutants can dramatically affect the expression of the TLR family of genes, abolishing their transcriptional response to stress and adding up yet another means by which mutp53 enables the cancer cell to evade immune surveillance (150).
Fig. 3.
The shift of p53 mutations in colitis-associated colorectal cancer. Being a late mutational event in sporadic and familial colon cancer, the p53 mutation appearance is changed when chronic inflammation is a preceding condition in the tissue, becoming an early event. Such mutations switch their tumor-promoting properties to tumor-initiating properties, leading to colon tumorigenesis through a different path.
p53 in non-epithelial cells
The field of p53 research has evolved throughout more than three decades, focusing almost exclusively on its cell-autonomous properties as ‘guardian of the genome’ (14). Nevertheless, the finding that p53 possesses a broader array of functions and is also deeply involved in microenvironmental and inflammatory processes suggests possible paracrine effects where p53 expression in a stromal or immune cell may dictate epithelial cell fate and vice versa. Within cancer stromal cells, p53 was suggested to exert paracrine activities that inhibit tumor growth and delay malignant progression (151–153). It was found that p21 is upregulated in the tumor-neighboring fibroblasts in various malignant tissues and antiangiogenic p53 targets are also overexpressed in such stromal components (154,155). Conversely, the cancer cells that thrive under these potentially inhibitory conditions may acquire abilities to abolish stromal p53 activity, either directly or through pressure for selection of stromal cells with compromised p53 function (156,157). Interestingly, and consistent with p53’s antioxidant properties, p53-deficient fibroblasts display accumulation of oxidative products, alterations in glutathione synthesis enzymes and upregulation of redox-regulated cytokines mediated via eNOS (158). Such findings highlight the microenvironmental protective influence conveyed by p53 in the bystander cells. A fascinating example for such possible link in immune cells was demonstrated when lungs of p53− /− mice were found to display a genome-wide transcriptional induction of NF-κB response element-enriched proinflammatory genes (97). It was noted that in the p53− /− lungs, microbicidal activities and bacterial clearance were enhanced specifically in p53-deficient neutrophils and alveolar macrophages. Despite these protective elements, p53-deficient mice remained more prone to lung injury and tumorigenesis, implying that the absence of p53 in the epithelia overrides its absence in the immune compartment.
The relative contribution of p53 within different tissues and organs can be studied through the use of somatic bone marrow chimeras, where lethally irradiated mice of either p53− /− or WTp53 background are reconstituted with bone marrow of the inverse genotype. Such chimeric model was used to study the recovery of kidneys from ischemic injury (159). Mice lacking p53 only in their leukocytes sustained severe injury similar to p53-null mice, whereas comparably severe injuries were not observed in WTp53 mice. Lymphocyte p53 was suggested to promote an anti-inflammatory M2-like macrophage polarization phenotype, associated with cytokine production. Such cell type-specific linkage was also detected in the liver, another tissue where chronic injury results in inflammation and fibrosis that is limited by a p53-mediated senescence program (160). Cell-specific p53 activities in hepatic stellate cells were shown not only to restrain fibrosis but also to decrease epithelial tumorigenesis. Of note, the p53 status of the hepatic stellate cells affected the macrophage polarization in the tissue, tilting it toward the M1 proinflammatory program in the presence of WTp53 while favoring the M2 tumor-promoting phenotype in the absence of p53 (161). This study highlighted a non-cell-autonomous cross talk between p53 and NF-κB, extending this ever-growing complex dual even more. A non-cell-autonomous interplay between p53 and NF-κB may also occur in the context of the mutp53-NF-κB axis, as chimeric mice harboring mutp53 solely in the bone marrow-derived compartment were more susceptible to DSS-induced colitis than WT animals (146). Taken together, these studies reveal that in addition to the well-documented cell-autonomous functions of p53, p53-based intercellular communication can also contribute to tumor suppression, partly through modulation of inflammatory responses.
Concluding remarks
Over the last decade, ample evidence for a solid link between p53 and the inflammatory response has been accumulated. Chronic inflammation has been acknowledged as a worldwide health problem; its association with cancer is just part of a plethora of pathological consequences, positioning anti-inflammatory treatments high in the priority list of modern medicine. Keeping in mind the manifold recently discovered physiological activities of p53, its involvement in the regulation of inflammation is not surprising.
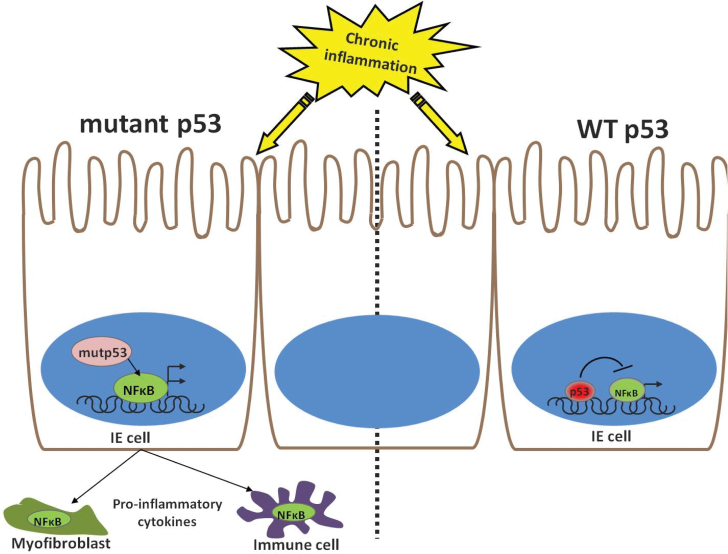
The general term ‘inflammation’ seems, however, too broad in this context. Categorization into subtypes of inflammatory processes is inevitable when one aims to understand the exact nature of the cross talk with the p53 pathway. To illustrate this need better, it was described herein that mutations in p53 may boost NF-κB activity to prolong and intensify chronic inflammation and promote tumorigenesis (Figure 4). Such interactions also take place in other overly inflamed conditions, such as rheumatoid arthritis (162), where high rates of p53 mutations are found in the synovial tissue. Unexpectedly though, the increased chronic inflammation combined with the oncogenic driving force of mutp53 do not suffice to push that tissue into cancer. This simple example recapitulates the complexity of the relationship between the extended p53 family and the multifaceted inflammatory arm. In order to reap therapeutic benefits from this cross talk, a deeper and more precise discrimination of inflammatory subtypes and their specific interplay with different forms of p53 and p53 family members remains to be applied.
Fig. 4.
A model for the mutp53:NF-κB interplay in the inflamed colon. The response of intestinal epithelial cells to stress in the form of chronic inflammation is dictated by the p53 status. In WTp53 cells, both p53 and NF-κB will be activated but will operate opposing transcriptional profile. However, when p53 is mutated, not only that NF-κB agenda is not interrupted, but it is tailwinded by the mutp53, presumably yielding a paracrine effect on neighboring stromal and immune cells.
Funding
Intramural Research Program of the National Institutes of Health, National Cancer Institute ; Dr. Miriam and Sheldon G. Adelson Medical Research Foundation; a Center of Excellence grant (1779/11) from the Israel Science Foundation; a Center of Excellence grant from the Flight Attendant Medical Research Institute (FAMRI); National Cancer Institute (R37 CA40099); Robert Bosch Foundation.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- COX-2
cyclooxygenase-2
- IFN
interferon
- IKK
IκB kinase
- IL
interleukin
- miR
microRNA
- mutp53
mutant p53
- NF-κB
nuclear factor-kappaB
- NOS
nitric oxide synthase
- ROS
reactive oxygen species
- STAT
signal transducer and activator of transcription
- TAD
transactivation domain
- TGF
transforming growth factor
- TLR
Toll-like receptor
- TNF-α
tumor necrosis factor alpha
- WT
wild-type.
References
- 1. Levine A.J., et al. (2009). The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer, 9, 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lane D., et al. (2010). p53 Research: the past thirty years and the next thirty years. Cold Spring Harb. Perspect. Biol., 2, a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nigro J.M., et al. (1989). Mutations in the p53 gene occur in diverse human tumour types. Nature, 342, 705–708 [DOI] [PubMed] [Google Scholar]
- 4. Dittmer D., et al. (1993). Gain of function mutations in p53. Nat. Genet., 4, 42–46 [DOI] [PubMed] [Google Scholar]
- 5. Freed-Pastor W.A., et al. (2012). Mutant p53: one name, many proteins. Genes Dev., 26, 1268–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bieging K.T., et al. (2012). Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol., 22, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sullivan K.D., et al. (2012). The p53 circuit board. Biochim. Biophys. Acta, 1825, 229–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maltzman W., et al. (1984). UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol. Cell. Biol., 4, 1689–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kastan M.B., et al. (1992). A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell, 71, 587–597 [DOI] [PubMed] [Google Scholar]
- 10. Yonish-Rouach E., et al. (1991). Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature, 352, 345–347 [DOI] [PubMed] [Google Scholar]
- 11. Serrano M., et al. (1997). Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell, 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 12. Wang Y., et al. (1998). Induced p53 expression in lung cancer cell line promotes cell senescence and differentially modifies the cytotoxicity of anti-cancer drugs. Oncogene, 17, 1923–1930 [DOI] [PubMed] [Google Scholar]
- 13. Carvajal L.A., et al. (2013). Another fork in the road—life or death decisions by the tumour suppressor p53. EMBO Rep., 14, 414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lane D.P. (1992). Cancer. p53, guardian of the genome. Nature, 358, 15–16 [DOI] [PubMed] [Google Scholar]
- 15. Vogelstein B., et al. (2000). Surfing the p53 network. Nature, 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 16. Reinhardt H.C., et al. (2012). The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet., 28, 128–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wade M., et al. (2013). MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer, 13, 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Love I.M., et al. (2012). It takes 15 to tango: making sense of the many ubiquitin ligases of p53. Genes Cancer, 3, 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taira N., et al. (2012). Post-translational modifications of p53 tumor suppressor: determinants of its functional targets. Histol. Histopathol., 27, 437–443 [DOI] [PubMed] [Google Scholar]
- 20. Vousden K.H., et al. (2009). Blinded by the light: the growing complexity of p53. Cell, 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 21. Goldstein I., et al. (2012). Regulation of lipid metabolism by p53—fighting two villains with one sword. Trends Endocrinol. Metab., 23, 567–575 [DOI] [PubMed] [Google Scholar]
- 22. Johnson R.F., et al. (2012). Nuclear factor-κB, p53, and mitochondria: regulation of cellular metabolism and the Warburg effect. Trends Biochem. Sci., 37, 317–324 [DOI] [PubMed] [Google Scholar]
- 23. Shen L., et al. (2012). The fundamental role of the p53 pathway in tumor metabolism and its implication in tumor therapy. Clin. Cancer Res., 18, 1561–1567 [DOI] [PubMed] [Google Scholar]
- 24. Liu J., et al. (2013). Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. pii: S0304-3835(13)00863-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dioufa N., et al. (2012). p53 antagonizes the unfolded protein response and inhibits ground glass hepatocyte development during endoplasmic reticulum stress. Exp. Biol. Med. (Maywood), 237, 1173–1180 [DOI] [PubMed] [Google Scholar]
- 26. Thomas S.E., et al. (2013). p53 and translation attenuation regulate distinct cell cycle checkpoints during endoplasmic reticulum (ER) stress. J. Biol. Chem., 288, 7606–7617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maiuri M.C., et al. (2010). Autophagy regulation by p53. Curr. Opin. Cell Biol., 22, 181–185 [DOI] [PubMed] [Google Scholar]
- 28. Lorin S., et al. (2013). Autophagy regulation and its role in cancer. Semin. Cancer Biol., 23, 361–379 [DOI] [PubMed] [Google Scholar]
- 29. Scherz-Shouval R., et al. (2010). p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proc. Natl Acad. Sci. USA, 107, 18511–18516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Molchadsky A., et al. (2010). p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis, 31, 1501–1508 [DOI] [PubMed] [Google Scholar]
- 31. Paskulin D., et al. (2012). The TP53 fertility network. Genet. Mol. Biol., 35 (4 suppl.), 939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kawamura T., et al. (2009). Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature, 460, 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sengupta S., et al. (2005). p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol., 6, 44–55 [DOI] [PubMed] [Google Scholar]
- 34. Grivennikov S.I., et al. (2010). Immunity, inflammation, and cancer. Cell, 140, 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ben-Neriah Y., et al. (2011). Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat. Immunol., 12, 715–723 [DOI] [PubMed] [Google Scholar]
- 36. Kimbrell D.A., et al. (2001). The evolution and genetics of innate immunity. Nat. Rev. Genet., 2, 256–267 [DOI] [PubMed] [Google Scholar]
- 37. Ingersoll M.A., et al. (2011). Monocyte trafficking in acute and chronic inflammation. Trends Immunol., 32, 470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alessandri A.L., et al. (2013). Resolution of inflammation: mechanisms and opportunity for drug development. Pharmacol. Ther., 139, 189–212 [DOI] [PubMed] [Google Scholar]
- 39. Fox L., et al. (2010). Natural killer T cells: innate lymphocytes positioned as a bridge between acute and chronic inflammation? Microbes Infect., 12, 1125–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boffetta P., et al. (2003). Contribution of environmental factors to cancer risk. Br. Med. Bull., 68, 71–94 [DOI] [PubMed] [Google Scholar]
- 41. Azer S.A. (2013). Overview of molecular pathways in inflammatory bowel disease associated with colorectal cancer development. Eur. J. Gastroenterol. Hepatol., 25, 271–281 [DOI] [PubMed] [Google Scholar]
- 42. Sonnenberg A., et al. (2013). Helicobacter pylori is a risk factor for colonic neoplasms. Am. J. Gastroenterol., 108, 208–215 [DOI] [PubMed] [Google Scholar]
- 43. Sun B., et al. (2013). Inflammation and liver tumorigenesis. Front. Med., 7, 242–254 [DOI] [PubMed] [Google Scholar]
- 44. Hofseth L.J., et al. (2003). Nitric oxide in cancer and chemoprevention. Free Radic. Biol. Med., 34, 955–968 [DOI] [PubMed] [Google Scholar]
- 45. Vendramini-Costa D.B., et al. (2012). Molecular link mechanisms between inflammation and cancer. Curr. Pharm. Des., 18, 3831–3852 [DOI] [PubMed] [Google Scholar]
- 46. Shibutani S., et al. (1991). Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434 [DOI] [PubMed] [Google Scholar]
- 47. Bindra R.S., et al. (2005). Genetic instability and the tumor microenvironment: towards the concept of microenvironment-induced mutagenesis. Mutat. Res., 569, 75–85 [DOI] [PubMed] [Google Scholar]
- 48. Sparmann A., et al. (2004). Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell, 6, 447–458 [DOI] [PubMed] [Google Scholar]
- 49. Soucek L., et al. (2007). Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat. Med., 13, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 50. Cardona S.M., et al. (2013). The fine balance of chemokines during disease: trafficking, inflammation, and homeostasis. Methods Mol. Biol., 1013, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guerra C., et al. (2011). Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell, 19, 728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Daniluk J., et al. (2012). An NF-κB pathway-mediated positive feedback loop amplifies Ras activity to pathological levels in mice. J. Clin. Invest., 122, 1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fuseya C., et al. (2012). Involvement of pelvic inflammation-related mismatch repair abnormalities and microsatellite instability in the malignant transformation of ovarian endometriosis. Hum. Pathol., 43, 1964–1972 [DOI] [PubMed] [Google Scholar]
- 54. Fleisher A.S., et al. (2000). Microsatellite instability in inflammatory bowel disease-associated neoplastic lesions is associated with hypermethylation and diminished expression of the DNA mismatch repair gene, hMLH1. Cancer Res., 60, 4864–4868 [PubMed] [Google Scholar]
- 55. Schulmann K., et al. (2005). Molecular phenotype of inflammatory bowel disease-associated neoplasms with microsatellite instability. Gastroenterology, 129, 74–85 [DOI] [PubMed] [Google Scholar]
- 56. Burgers W.A., et al. (2007). Viral oncoproteins target the DNA methyltransferases. Oncogene, 26, 1650–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peng D.F., et al. (2005). Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci., 96, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eguchi K., et al. (1997). DNA hypermethylation at the D17S5 locus in non-small cell lung cancers: its association with smoking history. Cancer Res., 57, 4913–4915 [PubMed] [Google Scholar]
- 59. Mishra A., et al. (2012). Aberrant overexpression of IL-15 initiates large granular lymphocyte leukemia through chromosomal instability and DNA hypermethylation. Cancer Cell, 22, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. De Craene B., et al. (2013). Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer, 13, 97–110 [DOI] [PubMed] [Google Scholar]
- 61. Maeda H., et al. (1996). TGF-beta contributes to the shift toward Th2-type responses through direct and IL-10-mediated pathways in tumor-bearing mice. J. Immunol., 156, 73–78 [PubMed] [Google Scholar]
- 62. Torroella-Kouri M., et al. (2009). Identification of a subpopulation of macrophages in mammary tumor-bearing mice that are neither M1 nor M2 and are less differentiated. Cancer Res., 69, 4800–4809 [DOI] [PubMed] [Google Scholar]
- 63. Wislez M., et al. (2006). High expression of ligands for chemokine receptor CXCR2 in alveolar epithelial neoplasia induced by oncogenic kras. Cancer Res., 66, 4198–4207 [DOI] [PubMed] [Google Scholar]
- 64. Ancrile B., et al. (2007). Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes Dev., 21, 1714–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Candido J., et al. (2013). Cancer-related inflammation. J. Clin. Immunol., 33 (suppl. 1), S79–S84 [DOI] [PubMed] [Google Scholar]
- 66. Meng T.C., et al. (2002). Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo . Mol. Cell, 9, 387–399 [DOI] [PubMed] [Google Scholar]
- 67. Nowsheen S., et al. (2009). Accumulation of oxidatively induced clustered DNA lesions in human tumor tissues. Mutat. Res., 674, 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kawanishi S., et al. (2006). Oxidative and nitrative DNA damage in animals and patients with inflammatory diseases in relation to inflammation-related carcinogenesis. Biol. Chem., 387, 365–372 [DOI] [PubMed] [Google Scholar]
- 69. Hiraku Y., et al. (2010). The role of iNOS-mediated DNA damage in infection- and asbestos-induced carcinogenesis. Ann. NY Acad. Sci., 1203, 15–22 [DOI] [PubMed] [Google Scholar]
- 70. Williams C.S., et al. (2000). Host cyclooxygenase-2 modulates carcinoma growth. J. Clin. Invest., 105, 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gupta G.P., et al. (2007). Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature, 446, 765–770 [DOI] [PubMed] [Google Scholar]
- 72. Qualtrough D., et al. (2007). Prostaglandin F(2alpha) stimulates motility and invasion in colorectal tumor cells. Int. J. Cancer, 121, 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lippitz B.E. (2013). Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol., 14, e218–e228 [DOI] [PubMed] [Google Scholar]
- 74. Meulmeester E., et al. (2011). The dynamic roles of TGF-beta in cancer. J. Pathol., 223, 205–218 [DOI] [PubMed] [Google Scholar]
- 75. Schetter A.J., et al. (2010). Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis, 31, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chen C.Z., et al. (2013). Regulation of immune responses and tolerance: the microRNA perspective. Immunol. Rev., 253, 112–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen Z., et al. (2013). The pivotal role of microRNA-155 in the control of cancer. J. Cell. Physiol., 229, 545–550 [DOI] [PubMed] [Google Scholar]
- 78. Iliopoulos D., et al. (2010). STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell, 39, 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Medina P.P., et al. (2010). OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature, 467, 86–90 [DOI] [PubMed] [Google Scholar]
- 80. Li Q., et al. (2013). MiR-21/Smad 7 signaling determines TGF-beta1-induced CAF formation. Sci. Rep., 3, 2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Staib F., et al. (2005). The p53 tumor suppressor network is a key responder to microenvironmental components of chronic inflammatory stress. Cancer Res., 65, 10255–10264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Polyak K., et al. (1997). A model for p53-induced apoptosis. Nature, 389, 300–305 [DOI] [PubMed] [Google Scholar]
- 83. Tan M., et al. (1999). Transcriptional activation of the human glutathione peroxidase promoter by p53. J. Biol. Chem., 274, 12061–12066 [DOI] [PubMed] [Google Scholar]
- 84. Budanov A.V., et al. (2004). Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science, 304, 596–600 [DOI] [PubMed] [Google Scholar]
- 85. Yoon K.A., et al. (2004). Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J. Hum. Genet., 49, 134–140 [DOI] [PubMed] [Google Scholar]
- 86. Hofseth L.J., et al. (2003). Nitric oxide-induced cellular stress and p53 activation in chronic inflammation. Proc. Natl Acad. Sci. USA, 100, 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Forrester K., et al. (1996). Nitric oxide-induced p53 accumulation and regulation of inducible nitric oxide synthase expression by wild-type p53. Proc. Natl Acad. Sci. USA, 93, 2442–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chazotte-Aubert L., et al. (2000). Nitric oxide nitrates tyrosine residues of tumor-suppressor p53 protein in MCF-7 cells. Biochem. Biophys. Res. Commun., 267, 609–613 [DOI] [PubMed] [Google Scholar]
- 89. Han J.A., et al. (2002). P53-mediated induction of Cox-2 counteracts p53- or genotoxic stress-induced apoptosis. EMBO J., 21, 5635–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Khajeniazi S., et al. (2013). Changes in COX-2 and oxidative damage factors during differentiation of human mesenchymal stem cells to hepatocyte-like cells is associated with downregulation of p53 gene. Biol. Chem., 394, 1213–1222 [DOI] [PubMed] [Google Scholar]
- 91. Niki T., et al. (2002). Frequent co-localization of Cox-2 and laminin-5 gamma2 chain at the invasive front of early-stage lung adenocarcinomas. Am. J. Pathol., 160, 1129–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hünten S., et al. (2013). The p53/microRNA network in cancer: experimental and bioinformatics approaches. Adv. Exp. Med. Biol., 774, 77–101 [DOI] [PubMed] [Google Scholar]
- 93. Kasinski A.L., et al. (2012). miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res., 72, 5576–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rivas C., et al. (2010). Dual role of p53 in innate antiviral immunity. Viruses, 2, 298–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tomso D.J., et al. (2005). Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proc. Natl Acad. Sci. USA, 102, 6431–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Menendez D., et al. (2011). The Toll-like receptor gene family is integrated into human DNA damage and p53 networks. PLoS Genet., 7, e1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Madenspacher J.H., et al. (2013). p53 integrates host defense and cell fate during bacterial pneumonia. J. Exp. Med., 210, 891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yamanishi Y., et al. (2002). Regulation of joint destruction and inflammation by p53 in collagen-induced arthritis. Am. J. Pathol., 160, 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zheng S.J., et al. (2005). Tumor suppressor p53 inhibits autoimmune inflammation and macrophage function. Diabetes, 54, 1423–1428 [DOI] [PubMed] [Google Scholar]
- 100. Park J.S., et al. (2013). p53 controls autoimmune arthritis via STAT-mediated regulation of the Th17 cell/Treg cell balance in mice. Arthritis Rheum., 65, 949–959 [DOI] [PubMed] [Google Scholar]
- 101. Gudkov A.V., et al. (2011). Inflammation and p53: a tale of two stresses. Genes Cancer, 2, 503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Campisi J. (2013). Aging, cellular senescence, and cancer. Annu. Rev. Physiol., 75, 685–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Menendez D., et al. (2013). Interactions between the tumor suppressor p53 and immune responses. Curr. Opin. Oncol., 25, 85–92 [DOI] [PubMed] [Google Scholar]
- 104. Rufini A., et al. (2013). Senescence and aging: the critical roles of p53. Oncogene, 32, 5129–5143 [DOI] [PubMed] [Google Scholar]
- 105. Pribluda A., et al. (2013). A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell, 24, 242–256 [DOI] [PubMed] [Google Scholar]
- 106. Li T., et al. (2012). Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell, 149, 1269–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ak P., et al. (2010). p53 and NF-kappaB: different strategies for responding to stress lead to a functional antagonism. FASEB J., 24, 3643–3652 [DOI] [PubMed] [Google Scholar]
- 108. Kawauchi K., et al. (2008). p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol., 10, 611–618 [DOI] [PubMed] [Google Scholar]
- 109. Ikeda A., et al. (2000). p300/CBP-dependent and -independent transcriptional interference between NF-kappaB RelA and p53. Biochem. Biophys. Res. Commun., 272, 375–379 [DOI] [PubMed] [Google Scholar]
- 110. Xia Y., et al. (2009). Phosphorylation of p53 by IkappaB kinase 2 promotes its degradation by beta-TrCP. Proc. Natl Acad. Sci. USA, 106, 2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wadgaonkar R., et al. (1999). CREB-binding protein is a nuclear integrator of nuclear factor-kappaB and p53 signaling. J. Biol. Chem., 274, 1879–1882 [DOI] [PubMed] [Google Scholar]
- 112. Campbell K.J., et al. (2004). Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol. Cell, 13, 853–865 [DOI] [PubMed] [Google Scholar]
- 113. Janssens S., et al. (2006). Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ., 13, 773–784 [DOI] [PubMed] [Google Scholar]
- 114. Strozyk E., et al. (2006). Differential effects of NF-kappaB on apoptosis induced by DNA-damaging agents: the type of DNA damage determines the final outcome. Oncogene, 25, 6239–6251 [DOI] [PubMed] [Google Scholar]
- 115. Lassus P., et al. (1996). Anti-apoptotic activity of low levels of wild-type p53. EMBO J., 15, 4566–4573 [PMC free article] [PubMed] [Google Scholar]
- 116. Jänicke R.U., et al. (2008). The dark side of a tumor suppressor: anti-apoptotic p53. Cell Death Differ., 15, 959–976 [DOI] [PubMed] [Google Scholar]
- 117. Ryan K.M., et al. (2000). Role of NF-kappaB in p53-mediated programmed cell death. Nature, 404, 892–897 [DOI] [PubMed] [Google Scholar]
- 118. Lin Y., et al. (2000). Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat. Genet., 26, 122–127 [DOI] [PubMed] [Google Scholar]
- 119. Perkins N.D. (2007). Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol., 8, 49–62 [DOI] [PubMed] [Google Scholar]
- 120. Elkon R., et al. (2005). Dissection of a DNA-damage-induced transcriptional network using a combination of microarrays, RNA interference and computational promoter analysis. Genome Biol., 6, R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Chien Y., et al. (2011). Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes Dev., 25, 2125–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Janssens S., et al. (2005). PIDD mediates NF-kappaB activation in response to DNA damage. Cell, 123, 1079–1092 [DOI] [PubMed] [Google Scholar]
- 123. Frank A.K., et al. (2011). The codon 72 polymorphism of p53 regulates interaction with NF-{kappa}B and transactivation of genes involved in immunity and inflammation. Mol. Cell. Biol., 31, 1201–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Bourdon J.C., et al. (2005). p53 isoforms can regulate p53 transcriptional activity. Genes Dev., 19, 2122–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Surget S., et al. (2013). Uncovering the role of p53 splice variants in human malignancy: a clinical perspective. Onco Targets Ther., 7, 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Marcel V., et al. (2011). Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ., 18, 1815–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Surget S., et al. (2013). Uncovering the role of p53 splice variants in human malignancy: a clinical perspective. Onco Targets Ther., 7, 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Slatter T., et al. (2011). Hyperproliferation, cancer, and inflammation in mice expressing a Delta133p53-like isoform. Blood, 117, 5166–5177 [DOI] [PubMed] [Google Scholar]
- 129. Campbell H.G., et al. (2012). Does Delta133p53 isoform trigger inflammation and autoimmunity? Cell Cycle, 11, 446–450 [DOI] [PubMed] [Google Scholar]
- 130. Wei J., et al. (2012). Pathogenic bacterium Helicobacter pylori alters the expression profile of p53 protein isoforms and p53 response to cellular stresses. Proc. Natl Acad. Sci. USA, 109, E2543–E2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Allocati N., et al. (2012). p63/p73 in the control of cell cycle and cell death. Exp. Cell Res., 318, 1285–1290 [DOI] [PubMed] [Google Scholar]
- 132. Dötsch V., et al. (2010). p63 and p73, the ancestors of p53. Cold Spring Harb. Perspect. Biol., 2, a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Yang A., et al. (2000). p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature, 404, 99–103 [DOI] [PubMed] [Google Scholar]
- 134. Sasaki Y., et al. (2003). Identification of the interleukin 4 receptor alpha gene as a direct target for p73. Cancer Res., 63, 8145–8152 [PubMed] [Google Scholar]
- 135. Zaika E., et al. (2013). Proinflammatory cytokines and bile acids upregulate ΔNp73 protein, an inhibitor of p53 and p73 tumor suppressors. PLoS One, 8, e64306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chatterjee A., et al. (2010). Regulation of p53 family member isoform DeltaNp63alpha by the nuclear factor-kappaB targeting kinase IkappaB kinase beta. Cancer Res., 70, 1419–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. King K.E., et al. (2008). The p53 homologue DeltaNp63alpha interacts with the nuclear factor-kappaB pathway to modulate epithelial cell growth. Cancer Res., 68, 5122–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Lu H., et al. (2011). TNF-alpha promotes c-REL/DeltaNp63alpha interaction and TAp73 dissociation from key genes that mediate growth arrest and apoptosis in head and neck cancer. Cancer Res., 71, 6867–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Oren M., et al. (2010). Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol., 2, a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Muller P.A., et al. (2013). p53 mutations in cancer. Nat. Cell Biol., 15, 2–8 [DOI] [PubMed] [Google Scholar]
- 141. Izzotti A., et al. (2004). Gene expression in the lung of p53 mutant mice exposed to cigarette smoke. Cancer Res., 64, 8566–8572 [DOI] [PubMed] [Google Scholar]
- 142. Yeudall W.A., et al. (2012). Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis, 33, 442–451 [DOI] [PubMed] [Google Scholar]
- 143. Scian M.J., et al. (2005). Tumor-derived p53 mutants induce NF-kappaB2 gene expression. Mol. Cell. Biol., 25, 10097–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Weisz L., et al. (2007). Mutant p53 enhances nuclear factor kappaB activation by tumor necrosis factor alpha in cancer cells. Cancer Res., 67, 2396–2401 [DOI] [PubMed] [Google Scholar]
- 145. Schneider G., et al. (2010). Cross talk between stimulated NF-kappaB and the tumor suppressor p53. Oncogene, 29, 2795–2806 [DOI] [PubMed] [Google Scholar]
- 146. Cooks T., et al. (2013). Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell, 23, 634–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Hussain S.P., et al. (2000). Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: a cancer-prone chronic inflammatory disease. Cancer Res., 60, 3333–3337 [PubMed] [Google Scholar]
- 148. Ullman T.A., et al. (2011). Intestinal inflammation and cancer. Gastroenterology, 140, 1807–1816 [DOI] [PubMed] [Google Scholar]
- 149. Matsumoto Y., et al. (2007). Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med., 13, 470–476 [DOI] [PubMed] [Google Scholar]
- 150. Shatz M., et al. (2012). The human TLR innate immune gene family is differentially influenced by DNA stress and p53 status in cancer cells. Cancer Res., 72, 3948–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Moskovits N., et al. (2006). p53 attenuates cancer cell migration and invasion through repression of SDF-1/CXCL12 expression in stromal fibroblasts. Cancer Res., 66, 10671–10676 [DOI] [PubMed] [Google Scholar]
- 152. Kiaris H., et al. (2005). Evidence for nonautonomous effect of p53 tumor suppressor in carcinogenesis. Cancer Res., 65, 1627–1630 [DOI] [PubMed] [Google Scholar]
- 153. Addadi Y., et al. (2010). p53 status in stromal fibroblasts modulates tumor growth in an SDF1-dependent manner. Cancer Res., 70, 9650–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Nishizaki M., et al. (1999). Recombinant adenovirus expressing wild-type p53 is antiangiogenic: a proposed mechanism for bystander effect. Clin. Cancer Res., 5, 1015–1023 [PubMed] [Google Scholar]
- 155. Trimis G., et al. (2008). Expression of p21waf1/Cip1 in stromal fibroblasts of primary breast tumors. Hum. Mol. Genet., 17, 3596–3600 [DOI] [PubMed] [Google Scholar]
- 156. Hill R., et al. (2005). Selective evolution of stromal mesenchyme with p53 loss in response to epithelial tumorigenesis. Cell, 123, 1001–1011 [DOI] [PubMed] [Google Scholar]
- 157. Bar J., et al. (2009). Cancer cells suppress p53 in adjacent fibroblasts. Oncogene, 28, 933–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Trachootham D., et al. (2013). Loss of p53 in stromal fibroblasts promotes epithelial cell invasion through redox-mediated ICAM1 signal. Free Radic. Biol. Med., 58, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Sutton T.A., et al. (2013). p53 is renoprotective after ischemic kidney injury by reducing inflammation. J. Am. Soc. Nephrol., 24, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Krizhanovsky V., et al. (2008). Senescence of activated stellate cells limits liver fibrosis. Cell, 134, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Lujambio A., et al. (2013). Non-cell-autonomous tumor suppression by p53. Cell, 153, 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Yamanishi Y., et al. (2002). Regional analysis of p53 mutations in rheumatoid arthritis synovium. Proc. Natl Acad. Sci. USA, 99, 10025–10030 [DOI] [PMC free article] [PubMed] [Google Scholar]