Abstract
Bacteriophages have an essential gene kit that enables their invasion, replication, and production. In addition to this “core” genome, they can carry “accessory” genes that dramatically impact bacterial biology, and presumably boost their own success. The content of phage genomes continue to surprise us by revealing new ways that viruses impact bacterial biology. The genome of a Clostridium difficile myovirus, phiCDHM1, contains homologs of three bacterial accessory gene regulator (agr) genes. The agr system is a type of quorum sensing (QS), via which the phage may modify C. difficile interactions with its environment. Although their mechanism of action is unknown, mutants in bacterial versions of these genes impact sporulation and virulence. To explore how phage QS genes may influence C. difficile biology, we examine the main categories of bacterial behavior that phages have been shown to influence and discuss how interactions via QS could influence behavior at a wider level.
Keywords: bacteriophage, bacterial physiology, sporulation, Clostridium difficile, accessory gene regulator, microbial ecology, microbial interactions, quorum sensing, autoinducing peptide, accessory gene
Introduction
Bacterial phenotypic conversion resulting from phage infection has been the subject of much scientific research, encompassing diverse bacterial phyla and microbial systems. This conversion can occur either via generalized transduction or by the introduction of phage-encoded proteins, whose expression results in changes to their host’s phenotype or activity.1 Phages have acquired these genes from their bacterial host in partial transduction events and have continued to evolve within the phage genome, e.g.2 These “accessory” genes can govern the biology of their bacterial hosts and fine tune the way in which bacteria interact with their environments. Such observations have been made apparent due to our ability to sequence phage genomes, and the information serves as a starting point for further study to determine how phage infection can contribute to their bacterial host’s physiology, endurance, and evolution.
In an effort to determine how phages contribute to the evolution and virulence of the human pathogen Clostridium difficile, we sequenced a temperate phage (induced from a natural lysogen) which has lytic capacity on another C. difficile strain. Its genome architecture was similar to previously sequenced C. difficile phages, but within the predicted lysogenic conversion module we identified three putative CDSs in a cassette that are homologous to bacterial agr genes. These genes encode proteins that are involved in the accessory gene regulator (agr) quorum sensing (QS) system. The agr system is a peptide based signaling pathway in which a pre-peptide, AgrD, is processed by the enzyme AgrB to produce the autoinducing peptide (AIP) that is released from the bacterial cell.3 The extracellular AIP is detected by a histidine kinase, AgrC, which phosphorylates the response regulator, AgrA. The phage phiCDHM1 has homologs of agrC, agrD, and agrB, but not agrA. Previously in C. difficile, two types of agr loci were identified that contain different agr genes.4 Our phylogenetic and bioinformatic analysis determined that this phage cassette forms a third type of agr locus, agr3, which is also present in some C. difficile strains.
While lysogenization by phiCDHM1 may aid in the dissemination of these genes throughout a susceptible population of C. difficile, they also are likely to have a functional role during infection. Work to determine this is ongoing in our laboratory. The amino acid sequences of the phage genes share ~60–70% similarity to their closest bacterial homologs in C. difficile strains NAP07 and NAP08. Following phylogenetic analyses of host and phage genes we have hypothesized that they have evolved separately within the phage genome after their acquisition. Without a phage-encoded equivalent of the response regulator (AgrA), we hypothesize that the signal is transferred on to evoke a host mediated response. In C. difficile, the agr2 system has been experimentally shown to effect toxin regulation and sporulation.5 Whether the different C. difficile agr loci are also functionally distinct remains to be determined, however we think it is likely that they all will influence bacterial behavior.
Phage-mediated changes in bacterial phenotype may result from the products of known “accessory” genes such as the examples given in this review, but there are many cases where the physiological changes are attributed to other factors including phage “core” genes, non-CDS elements, and where the genes responsible or the exact mechanisms have not been determined.
In this addendum to our cited paper, we aim to put our observation of agr genes in a phage genome into the context of known examples of phages modulating bacterial behavior. We have summarized the types of ways that phages have been shown to impact bacterial biology, and subsequent animal or plant interactions, in Figure 1 and in Table 1, and the modes of impact of host QS genes in Figure 2. There is a clear overlap between the known impacts of bacterial QS processes, and of known phage influences on bacteria. Therefore by reviewing our findings in the context of known phage influences on bacterial biology, we hope to provide a useful contextual framework as to what types of bacterial processes phage QS genes are likely to impact. Phage QS genes could provide an additional way for phage to influence one, or multiple categories of host behavior, and their sphere of influence is likely to be wider than at the individual cell level as they have the potential to elicit these changes at the community level.
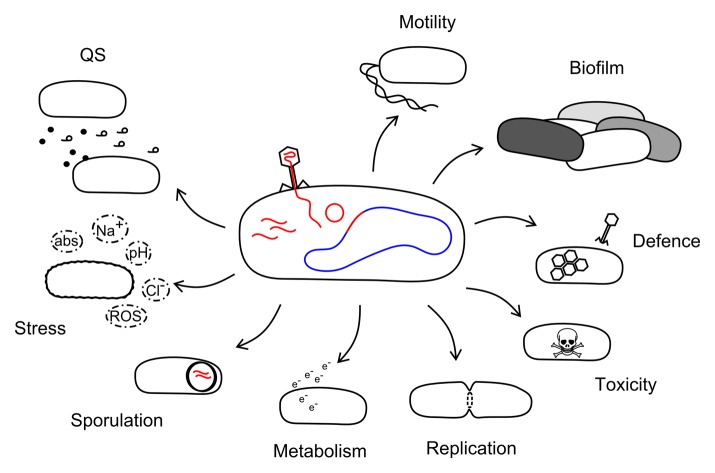
Figure 1. Phage influences on bacterial behaviors. Diagram illustrating the various behaviors phage infection has been found to modulate and include motility, biofilm formation, defense, toxicity, replication, metabolism, sporulation, stress response, and quorum sensing. Abbreviations: ROS = reactive oxygen species, abs = antibiotics, QS = quorum sensing.
Table 1. The types of ways that phages have been shown to impact bacterial biology, and subsequent animal or plant interactions.
| Behavior | Species | Phages | Mechanism | Predicted outcome |
|---|---|---|---|---|
| Toxicity | V. cholerae | CTXφ | ctxAB operon | Production of CT toxin |
| H. defensa | ASPE phages | cdtAB | Production of CDT toxin | |
| C. difficile | φCD38–2 | Unknown | Increased production of TcdB and TcdA | |
| C. toxicus | NCPPB 3778 | Unknown | Associated with corynetoxin production in ARGT | |
| E. coli | STX-2 | Stx genes | Production of Shiga toxin | |
| Phage defense | E. coli | Lambda | CI repressor | Inhibit secondary phage infection |
| C. difficile | φC2 | AbiF-like gene | Unknown mechanism | |
| C. difficile | CD630 prophages | CRISPR arrays | cRNA production | |
| Replication | C. crescentus | φCbK-like | GcrA | Upregulation of DNA replication machinery |
| B. cenocepacia; Mycobacterium; M. xanthus; Synechococcus | KL1 and AH2; phage Mx8; phage L5; S-PM2 | MazG | Cell death regulation and ppGpp regulation | |
| Changing environments and colonization | B. anthracis | Wip1, Wip2, Wip4, Wip5, Frp1, Frp2, Htp1, Slp1, Wβ, Φ1615, Φ1047, and Bcp1 | Sigma factors and bacterial host factor | Production of exopolysaccharides - biofilm, soil survival, growth rate |
| E. faeclis; S. mitis | V583 pp1, pp4 and pp6; φSM1 | PblA and PblB | Platelet binding proteins | |
| E. coli | STX-2 | Unknown | Increase motility and acid stress | |
| E. coli | Nine K-12 BW2511 prophages | Unknown | Biofilm formation | |
| Cell metabolism and energy production | Synechococcus | S-PM2 | psbA and psbD; cpeT | Photosynthesis apparatus |
| Noxious environments | E. coli | K-12 BW2511 prophages; various depending on ab | Include dicB and kilR | Quinolone and β-lactam resistance |
| Nine K-12 BW2511 prophages | Unknown | Osmotic stress | ||
| K-12 BW2511 prophage CPS-53 | yfdK, yfdO, and yfdS | Oxidative stress | ||
| B. anthracis | Phage γ | gp41 | Fosfomycin resistance | |
| Sporulation | Bacillus subtilis | PMB12 and SP10 | Unknown | Enhance or induce sporulation |
| B. anthracis | Wip4, Wip5, Frp1, Htp1, and Bcp1 | Sigma factors | Block sporulation | |
| B. anthracis | Wip1, Wip2, Wβ, Frp2, and Slp1 | Sigma factors | Promote sporulation | |
| Communication | C. difficile | phiCDHM1 | agrD, agrB, and agrC | |
| Clostridium tyrobutyricum | φCTP1 | LuxR homolog | ||
| Pseudomonas | LytTR homologs | |||
| Iodobacter | ɸPLPE | Acylhydrolase |
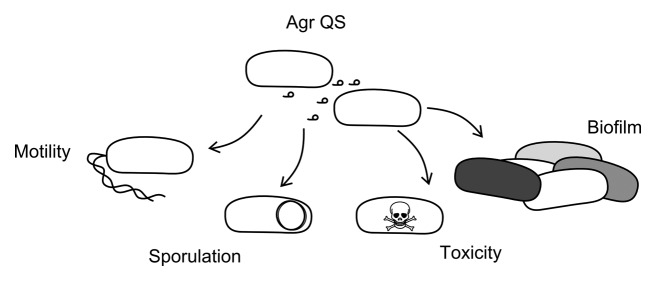
Figure 2. Quorum sensing and bacterial behavior. Diagram illustrating the impact the agr QS system has been found to influence and include biofilm formation, sporulation, toxicity, and motility. Abbreviations; QS = Quorum sensing and Agr = accessory gene regulatory system.
Host Communication
By accessing quorum sensing pathways in bacteria, phages may be able to manipulate bacteria by effecting one or multiple behaviors, as several bacterial processes have been shown to be induced or modified by QS (Fig. 2). There are two main types of QS systems in bacteria; the Lux-type which utilizes chemical signaling molecules, N-Acyl homoserine lactones, and a type that utilizes a short peptide as the signal.6 Both forms of bacterial communication are based on the detection of the secreted signal which reflects the state of the surrounding bacterial population and elicits a phenotypic response in the detecting cell.
There are known links between phages and QS: for example, the induction and release of temperate phages has been observed following exposure to the N-Acyl homoserine lactone of the LuxS system.7 Phage genomes have also been shown to contain homologs of response regulators, e.g., that of Clostridium tyrobutyricum phage φCTP1, and so may respond to signaling via these gene products.8 Likewise, three phages encoding response regulators associated with the agr system have been described which infect Pseudomonas (see ref. 9). In contrast, the Iodobacteria phage ɸPLPE encodes a predicted acylhydrolase which could in effect block the Lux signal by degrading the signaling molecule.10
The phage phiCDHM1 encodes both the signal and detection units of the agr system, and we predict the message is transferred to a host-encoded response regulator, but the response remains to be established. We outline other ways by which phage alter their host’s physiology in the remainder of the article, and it may be that, via the QS pathway, this phage could influence a diverse set of bacterial behaviors.
Host Defense Against Predation
The relationship between phage and their bacterial hosts has been the subject of many excellent reviews, for example see Brüssow.11 One area where phages appear to have a clear impact on bacterial biology is in preventing the destruction of their host bacterial cell, thus promoting the long-term success of the bacteria and therefore the phage. One complex but well characterized example of this is the interplay between phage, bacteria, and multicellular hosts in the symbiotic relationship between the aphid Acyrthosiphon pisum and resident bacterium Hamiltonella defensa.12 The H. defensa phage ASPE-2 encodes a toxin which protects the aphid from parasitism by the larvae of the Aphidius ervi wasp which promotes the bacterial survival and, consequently, that of the phage.
Although not as well studied, another example of downstream protection from predation, and a fascinating example of how phages effect in whole ecosystems, can be seen in a “consortium” where a temperate phage NCPPB 3778 in Clavibacter toxicus has been suggested to modify bacterial toxin production levels.13 The bacterium infects a nematode that is not susceptible to the toxin and which in turn infects the seeds of the ryegrass Lolium rigidum. Consumption of the infected rye grass containing the toxin can poison grazing livestock, and we see how phage infection can indirectly have significant ecological ramifications.
Host Defense Against Phage Infection
Clearly phage infection is not always beneficial to bacteria, and they have evolved a multitude of mechanisms to resist phages. These include altering receptor molecules and producing proteins that lead to abortive infection.14 Many prokaryotes also have a form of adaptive immunity, the CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR) system to protect them against sources of foreign DNA, including phages.15 Phages have, in turn, evolved counter strategies to overcome bacterial resistance efforts; for example, there is a subset of V. cholerae phages which has acquired CRISPR/Cas machinery from their hosts that targets a phage inducible chromosomal island (PICI)-like element (PLE) in the chromosome of V. cholerae. Successful lytic infection requires the action of this phage-encoded CRISPR/Cas system with spacers that match to the PLE.16
In addition to hijacking bacterial defense machinery to permit their own replication, phages can add to host defenses by inhibiting secondary infection. Prophages in the bacterial cell prevent infection with related phages by conferring superinfection immunity. This is best characterized in the model phage lambda, where the resident prophage’s CI repressor blocks the replication of invading phages. Two further examples where phage may prevent secondary phage infection, but by using host derived mechanisms can be seen in C. difficile. C. difficile prophages harbor CRISPR arrays containing spacers17 which have been shown to be expressed and processed to mature CRISPR RNA (cRNA).18 These spacer sequences share identity with the genomes of other C. difficile phages19 and prophage content could alter phage immunity across this species. In the second example, the C. difficile phage phiC2 encodes a homolog of AbiF.20 This is a bacterial abortive infection protein which is usually carried on a plasmid and inhibits phage infection.21 Neither example has been experimentally verified, but each highlights the different ways phages can potentially use host mechanisms to wage inter-phage wars.
Host Virulence, Colonization, and Transmission
The converse of stopping predation is to increase virulence, colonization or transmission, and several species of bacteria can be converted from non-pathogenic to pathogenic strains by prophages.22 A classic example of this is in V. cholerae, where the gene operon encoding the CT endotoxin is present on the phage CTXφ 1. Production of this toxin results in diarrhea, and contributes to the transmission success of V. cholerae.23 Even if phages do not encode toxins directly, they can influence toxin production: for example, C. difficile phages can modulate toxin production of several C. difficile strains.24-26 One of these phages, φCD38-2, can upregulate the production of bacterial host toxins TcdA and TcdB.26 These toxins contribute to the onset of diarrhea in C. difficile infection and their increased production could worsen the severity of disease and, again, promote bacterial transmission.
In addition to modulating the bacterial toxicity, phages can influence colonization by altering host motility and providing resistance to environmental stress, as reported for the E. coli STX-2 phages27 and E. coli cryptic prophages.28 While the mechanisms of how these responses are mediated have not been fully determined, both effects have implications for the infection of mammalian hosts.
Lastly, phages are also thought to add to the plasticity of the bacterial phenotype during changing environmental conditions.29 An example are prophages of Enterococcus faecalis and Streptococcus mitis which carry genes encoding platelet-binding-like proteins, PblA and PblB.30,31 These have been shown to be expressed on the cell surface of S. mitis during lysogenic infection and it has been suggested that in the case of S. mitis these proteins contribute resulting endocarditis.31 Another complex scenario is presented by the changing life stages of Bacillus anthracis, which can live as a soil bacterium as well as colonize the guts of earthworms. Phage-encoded sigma factors alter gene expression and thus the ability of bacteria to survive different conditions while in these environments.32
Host Replication
An obvious place for phages to redirect host resources away from their host’s replication and toward phage propagation. Several DNA metabolism and replication machinery genes have been observed in phage genomes, and the ways in which phages manipulate host replication can vary extensively. A striking example of phages manipulating host replication can be seen in the dimorphic bacterium Caulobacter crescentus. This has stalked (sessile) and free swimming or “swarmer” cells which results in a complex replication cycle. The stalked cell divides to produce a swarmer cell, with the bacterial cell cycle regulator protein GcrA only present in the stalked cells. Several φCbK-like phages that infect this organism have been found to encode homologs of GcrA.33 The phages preferentially infect the better disseminated swarmer cells, and the phage-encoded GcrA is thought to upregulate DNA replication in these cells, and so enhance their own replication.
Another example of the ability of phages to influence their host’s replicative process, is the phage-encoded homologs of MazG. This is a regulator of cell death in E. coli and its expression influences bacterial replication in nutrient limited environments.34 Homologs of MazG have been found in phages infecting several diverse bacterial species including several cyanophages, Burkholderia cenocepacia phages KL1 and AH2, Myxococcus xanthus phage Mx8, and Mycobacterium phage L5.35 Their bacterial hosts are present in contrasting environments; nutrient “poor” oceans (the cyanobacteria) and nutrient “rich” sewage and soil (B. cenocapacia, M. xanthus). Although there has been no experimental work on phage-encoded MazG, its appearance in phage genomes has led to suggestions that it may function by re-directing metabolism in nutrient deprived host bacteria and thus enhances phage propagation.
Host Metabolism, Energy, and Nutrient Acquisition
In many ways cyanophages can be viewed as champions in “host” gene acquisition, and their interactions with their bacterial hosts can point at ways that other bacterial groups may also be manipulated. The best characterized cyanophages are the T4-like myoviruses which encode a range of genes with known, predicted, and novel functions.36 The most studied “host acquired” genes encode homologs of bacterial photosynthetic apparatus, psbA and psbD. These are widespread in cyanophages and have been shown to maintain cyanobacterial photosynthesis and energy acquisition during infection by expressing the phage genes and downregulating the host versions.37,38
Cyanophages are the subject of much research due to their abundance, ecological significance and the interconnectedness of their genomes with their bacterial hosts.2,39 Several other genes involved with photosynthesis have also been identified, including those predicted to manipulate and modify cyanobacterial pigments, carbon, nitrogen and phosphate metabolism, and utilization. Genes are also carried that assist the cyanobacteria during light or nutrient stress and they likely maintain the host cyanobacteria to boost phage production. Recent work has added to some of these hypotheses, for example, cpeT elongates the cyanobacterial antennae during infection40 and that CP12 appears to redirect energy resources from the Calvin cycle to increase dNTP synthesis.41 Metagenomic approaches have confirmed that phage-carried versions of bacterial genes constitute a vast proportion of the total viral genes observed; suggesting their presence is significant in natural settings.42,43
Host Survival in Noxious Environments
Phages can also play a role in allowing bacteria to survive exposure to noxious agents such as antibiotics (during a treated infection) or oxidative stress (mediated by an immune response in human infections, etc.). As mentioned previously, cryptic prophages of E. coli have been shown to have roles in resistance against antibiotic, oxidative, osmotic, and acid stresses.28 The deletion of all nine prophages in strain K-12 BW2511 decreased its resistance to oxidative stress 245-fold. In vitro growth assays using the deletion mutants for each, and all, nine prophages demonstrated that while none of the prophage genes were essential, the KO mutants did exhibit decreased growth rates.
There is clear clinical relevance to these findings as the prophages also conferred increased resistance to six first and second generation quinolones and to 11 β-lactams. However, antibiotics are often associated with natural bacterial populations, especially in the soils due to the production of many by the Streptomycetes, so these phage relationships are likely to predate more recent exposure to the overuse and misuse of antibiotics. An example of this is the Bacillus cereus phage γ that carries a gene, gp41, which confers resistance against fosfomycin, a product of some Streptomycetes. Therefore phage infection may promote B. cereus survival in the soil environment.44 From these examples, we see that infection by specific phages can support bacterial growth in adverse conditions, and may then confer evolutionary fitness in microbial competition within different environments.
Additionally, several bacterial genera can form endospores under conditions that are unfavorable to vegetative cell growth. These specialist structures are highly resistant to heat and chemical degradation and permit the bacteria to persist until conditions are favorable once more. Phages infecting the endospore forming Bacillus can modulate their own replication relative to the sporulation state of the cell; for example ɸ29 can suppress its lytic cycle depending on the host cell cycle.45 However, other phages can enhance or induce sporulation, e.g., B. subtilis phages PMB12 and SP10.46 Packaging of phage DNA into a spore may offer an evolutionary advantage as when the spore outgrows, phage replication can resume. Phages that can enhance sporulation therefore increase their protection in adverse conditions. Interestingly although several phages could increase sporulation rates, other phages were found to block sporulation, illustrating the complex interactions that occur in phage ecology.32
Summary
Clearly, phages direct several aspects of bacterial behavior and, considering the diversity and prevalence of the means described to date, we predict that the discovery of novel ways in which phages shape biological interaction networks will continue. As phages have been found that can impact carbon acquisition and metabolism, we also suggest that phages most likely have roles in other key metabolic bacterial processes, such as nitrogen fixing. Lastly, the ability of phages to alter bacterial survival in adverse conditions leads us to surmise that they may have roles in other specific behaviors such as chemotaxis and other adaptations in challenging environments, e.g., to heavy metal poisoning and exposure to irradiation.
As we have discussed, phages can modify bacterial behaviors either by directly introducing additional functions or by indirectly by regulating bacterial genes, e.g., by phage-encoded sigma factors. We present several examples of how phages could increase cells defense, virulence, or survival in particular environments. The agr gene cassette of phiCDHM1 provides an additional mechanism of how a phage may alter bacterial behavior in a non-localized manner, and evoke a response in neighboring cells. The agr QS system is known to promote many of the behaviors and phenotypes discussed in this article, including toxin production, biofilm formation, motility, and sporulation. Alternatively, the phage QS genes may be a form of informing an expanded population of the same lysogen, (and therefore the phage), of its density within the environment with implications for induction and phage dynamics.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was funded by an MRC Centenary Fellowship (University of Leicester) awarded to KRH, A-base funding from the Laboratory for Foodborne Zoonoses to AMK and an MRC New Investigator Award (G0700855), and a Wellcome Trust Institutional Strategic Support Fund grant, awarded to MRJC.
References
- 1.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–4. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 2.Clokie MRJ, Millard AD, Mann NH. T4 genes in the marine ecosystem: studies of the T4-like cyanophages and their role in marine ecology. Virol J. 2010;7:291. doi: 10.1186/1743-422X-7-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–64. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 4.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin MJ, Clare S, Goulding D, Faulds-Pain A, Barquist L, Browne HP, Pettit L, Dougan G, Lawley TD, Wren BW. The agr locus regulates virulence and colonization genes in Clostridium difficile 027. J Bacteriol. 2013;195:3672–81. doi: 10.1128/JB.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh D, Roy K, Williamson KE, Srinivasiah S, Wommack KE, Radosevich M. Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl Environ Microbiol. 2009;75:7142–52. doi: 10.1128/AEM.00950-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer MJ, Payne J, Gasson MJ, Narbad A. Genomic sequence and characterization of the virulent bacteriophage phiCTP1 from Clostridium tyrobutyricum and heterologous expression of its endolysin. Appl Environ Microbiol. 2010;76:5415–22. doi: 10.1128/AEM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hargreaves KR, Kropinski AM, Clokie MRJ. What does the talking?: quorum sensing signalling genes discovered in a bacteriophage genome. PLoS One. 2014;9:e85131. doi: 10.1371/journal.pone.0085131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leblanc C, Caumont-Sarcos A, Comeau AM, Krisch HM. Isolation and genomic characterization of the first phage infecting Iodobacteria: ϕPLPE, a myovirus having a novel set of features. Environ Microbiol Rep. 2009;1:499–509. doi: 10.1111/j.1758-2229.2009.00055.x. [DOI] [PubMed] [Google Scholar]
- 11.Brüssow H. The not so universal tree of life or the place of viruses in the living world. Philos Trans R Soc Lond B Biol Sci. 2009;364:2263–74. doi: 10.1098/rstb.2009.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–4. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ophel KM, Bird AF, Kerr A. Association of bacteriophage particles with toxin production by Clavibacter toxicus, the causal agent of annual ryegrass toxicity. Phytopathology. 1993;83:676–81. doi: 10.1094/Phyto-83-676. [DOI] [Google Scholar]
- 14.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 15.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 16.Seed KD, Lazinski DW, Calderwood SB, Camilli A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–91. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–86. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 18.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppée JY, et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013;9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hargreaves KR, Flores CO, Lawley TD, Clokie MRJ. Abundant and diverse CRISPR spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. MBio. 2014 doi: 10.1128/mBio.01045-13. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goh S, Ong PF, Song KP, Riley TV, Chang BJ. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology. 2007;153:676–85. doi: 10.1099/mic.0.2006/002436-0. [DOI] [PubMed] [Google Scholar]
- 21.Garvey P, Fitzgerald GF, Hill C. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl Environ Microbiol. 1995;61:4321–8. doi: 10.1128/aem.61.12.4321-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abedon ST, Lejeune JT. Why bacteriophage encode exotoxins and other virulence factors. Evol Bioinform Online. 2005;1:97–110. [PMC free article] [PubMed] [Google Scholar]
- 23.Organisation WHO Fact Sheet N°107 - Cholera. 2014
- 24.Goh S, Chang BJ, Riley TV. Effect of phage infection on toxin production by Clostridium difficile. J Med Microbiol. 2005;54:129–35. doi: 10.1099/jmm.0.45821-0. [DOI] [PubMed] [Google Scholar]
- 25.Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J Virol. 2009;83:12037–45. doi: 10.1128/JVI.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekulovic O, Meessen-Pinard M, Fortier LC. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J Bacteriol. 2011;193:2726–34. doi: 10.1128/JB.00787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su LK, Lu CP, Wang Y, Cao DM, Sun JH, Yan YX. [Lysogenic infection of a Shiga toxin 2-converting bacteriophage changes host gene expression, enhances host acid resistance and motility] Mol Biol (Mosk) 2010;44:60–73. [PubMed] [Google Scholar]
- 28.Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK. Cryptic prophages help bacteria cope with adverse environments. Nat Commun. 2010;1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–8. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matos RC, Lapaque N, Rigottier-Gois L, Debarbieux L, Meylheuc T, Gonzalez-Zorn B, Repoila F, Lopes MdeF, Serror P. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 2013;9:e1003539. doi: 10.1371/journal.pgen.1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell J, Siboo IR, Takamatsu D, Chambers HF, Sullam PM. Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol Microbiol. 2007;64:844–57. doi: 10.1111/j.1365-2958.2007.05703.x. [DOI] [PubMed] [Google Scholar]
- 32.Schuch R, Fischetti VA. The secret life of the anthrax agent Bacillus anthracis: bacteriophage-mediated ecological adaptations. PLoS One. 2009;4:e6532. doi: 10.1371/journal.pone.0006532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gill JJ, Berry JD, Russell WK, Lessor L, Escobar-Garcia DA, Hernandez D, Kane A, Keene J, Maddox M, Martin R, et al. The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genomics. 2012;13:542. doi: 10.1186/1471-2164-13-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross M, Marianovsky I, Glaser G. MazG -- a regulator of programmed cell death in Escherichia coli. Mol Microbiol. 2006;59:590–601. doi: 10.1111/j.1365-2958.2005.04956.x. [DOI] [PubMed] [Google Scholar]
- 35.Lynch KH, Stothard P, Dennis JJ. Comparative analysis of two phenotypically-similar but genomically-distinct Burkholderia cenocepacia-specific bacteriophages. BMC Genomics. 2012;13:223. doi: 10.1186/1471-2164-13-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mann NH, Cook A, Millard A, Bailey S, Clokie M. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature. 2003;424:741. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- 38.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–9. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan MB, Huang KH, Ignacio-Espinoza JC, Berlin AM, Kelly L, Weigele PR, DeFrancesco AS, Kern SE, Thompson LR, Young S, et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol. 2010;12:3035–56. doi: 10.1111/j.1462-2920.2010.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan J, Jia Y, Clokie MRJ, Mann NH. Infection by the ‘photosynthetic’ phage S-PM2 induces increased synthesis of phycoerythrin in Synechococcus sp. WH7803. FEMS Microbiol Lett. 2008;283:154–61. doi: 10.1111/j.1574-6968.2008.01148.x. [DOI] [PubMed] [Google Scholar]
- 41.Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, Chisholm SW. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci U S A. 2011;108:E757–64. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuch R, Fischetti VA. Detailed genomic analysis of the Wbeta and gamma phages infecting Bacillus anthracis: implications for evolution of environmental fitness and antibiotic resistance. J Bacteriol. 2006;188:3037–51. doi: 10.1128/JB.188.8.3037-3051.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonenshein AL. Bacteriophages: how bacterial spores capture and protect phage DNA. Curr Biol. 2006;16:R14–6. doi: 10.1016/j.cub.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 46.Silver-Mysliwiec TH, Bramucci MG. Bacteriophage-enhanced sporulation: comparison of spore-converting bacteriophages PMB12 and SP10. J Bacteriol. 1990;172:1948–53. doi: 10.1128/jb.172.4.1948-1953.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]