Abstract
During vesicular trafficking and release of enveloped viruses, the budding and fission processes dynamically remodel the donor cell membrane in a protein- or a lipid-mediated manner. In all cases, in addition to the generation or relief of the curvature stress, the buds recruit specific lipids and proteins from the donor membrane through restricted diffusion for the development of a ring-type raft domain of closed topology. Here, by reconstituting the bud topography in a model membrane, we demonstrate the preferential localization of cholesterol- and sphingomyelin-enriched microdomains in the collar band of the bud-neck interfaced with the donor membrane. The geometrical approach to the recapitulation of the dynamic membrane reorganization, resulting from the local radii of curvatures from nanometre-to-micrometre scales, offers important clues for understanding the active roles of the bud topography in the sorting and migration machinery of key signalling proteins involved in membrane budding.
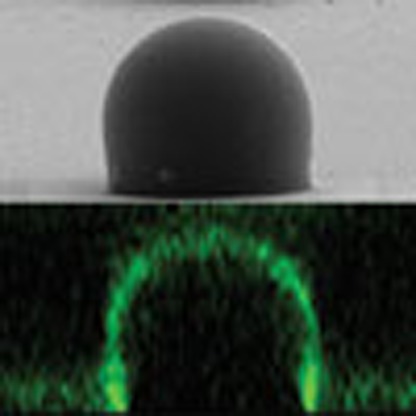 Budding processes on cell membranes involve distortions of surface geometry, driven by the sorting of membrane components. Here, the authors model a budding process and observe spontaneous reordering of materials in the areas of high curvature, especially around bud-necks, producing ring-raft type structures.
Budding processes on cell membranes involve distortions of surface geometry, driven by the sorting of membrane components. Here, the authors model a budding process and observe spontaneous reordering of materials in the areas of high curvature, especially around bud-necks, producing ring-raft type structures.
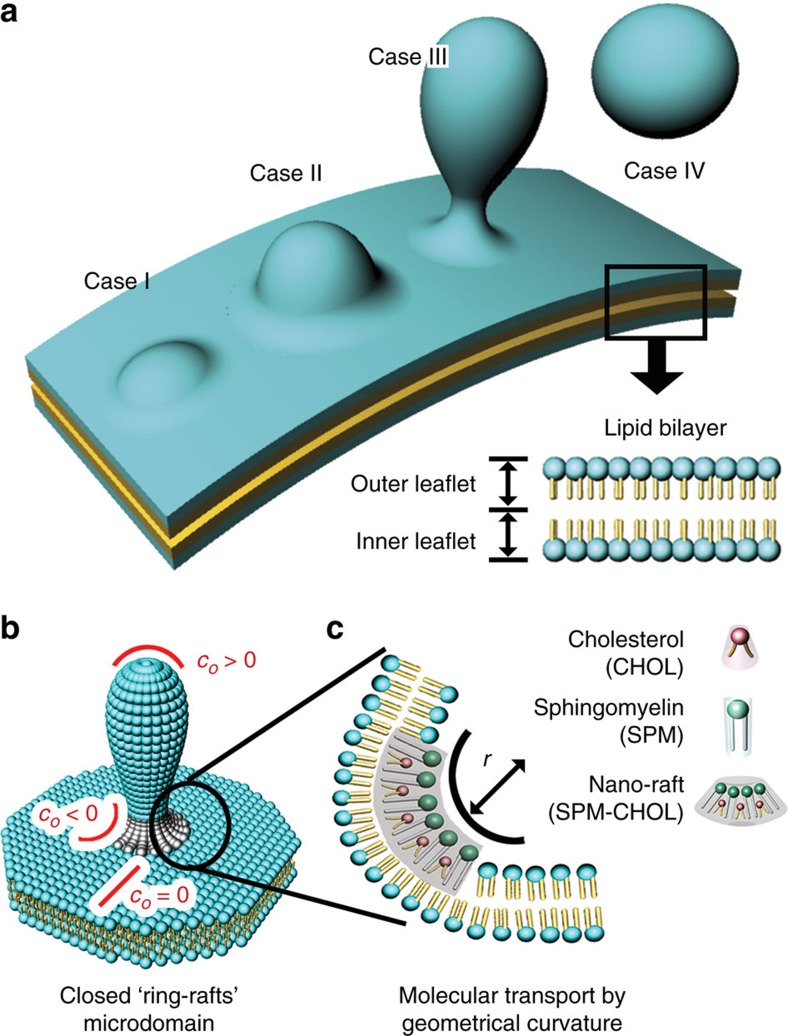
Membrane budding, a key step in a host of biological processes involving the vesicle transport1, the multivesicle body biogenesis and the release of enveloped viruses2,3, is associated with morphological deformations of an essentially planar patch of the donor membrane into a spherical bud (Fig. 1a, case I to case IV). The free energy cost of such structural transformation, in the absence of the intrinsic membrane curvature and before pinch-off, is approximately 8πκ where κ represents the mean bending rigidity4. For a typical lipid bilayer (10kBT<κ<20kBT where kB represents the Boltzmann constant and T the temperature)5, the energetic penalty for any size (about 250 to 500 kBT) is too large for the spontaneous bud formation and must be offset by active membrane processes. Indeed, many protein-mediated6,7 budding events involve the curvature-scaffolding process resulting from the membrane association of coat proteins (for example, BAR domain proteins, clathrin and COP II proteins), viral nucleocapsids or even cytoskeletal elements—all of which act to reduce the energy needed for the membrane bending7. Alternatively, the lateral or transverse inhomogeneity in the sorting dynamics and/or the equilibrium distribution of membrane constituents may contribute to the stabilization of the local membrane curvature by lowering the energy required for budding8.
Figure 1. Membrane fission and ring-raft formation in bud-mimicking architecture.
(a) Schematic representation of key budding steps (case I–case IV). (b) Closed ring-raft domain located around a bud-neck region with a negative curvature (c0<0; radius r=1/c0). (c) Formation of a nano-raft enriched with CHOL and SPM molecules in the outer leaflet by the bud-neck curvature.
In the process of membrane budding, both the size and the shape of the bud are highly variable even when produced through single mechanisms, either involving protein association or a compositional bias. Besides sorting specific proteins (for example, the endosomal sorting complexes required for transport (ESCRT) machinery during viral budding9 and the dynamin10 self-assembly in vesicle scission), membrane budding events have been speculated to sort specific lipid types (for example, cholesterol (CHOL) in synaptic vesicles and raft components in viral envelopes) owing to the curvature generation within the bud-neck region2,3,6,11,12,13, which is also thought to aid vesicle scission14. Such curvature-dependent lipid sorting is quite well-established in model membranes15 and has been observed in response to the membrane pore (100–200 nm diameter) formation in the single-celled organism Tetrahymena thermophila during mating behaviour16,17. Since the forces responsible for these events are oppositely directed (for example, ESCRT-mediated virus budding and dynamin driven vesicle invaginations), the sorting process of membrane lipids might be a consequence of the physical curvatures produced during budding. From a geometrical point of view, the curvature niche in the vicinity of a late-stage bud is quite complex8. The curvature pattern consists of (1) bidirectional positive principal curvatures in the bud-pit region, (2) a negative curvature at the neck and a positive curvature at the interface with the donor membrane and (3) a positive curvature and a zero curvature within the neck (in matured buds)—together generating a pattern of Gaussian curvatures18,19 in the lipid bilayer topology. Thus, it appears reasonable that this unique curvature landscape in the bud niche might play an essential role in dictating the local membrane composition20,21. It is noted, however, that the reduction in the curvature energy alone is generally insufficient to overcome the entropic penalty of sorting individual lipids22. This leads to the assertion that in addition to the lateral segregation of lipids into domains of differing spontaneous curvatures23 and/or bending rigidities24 due to the non-ideal mixing, any significant interleaflet compositional asymmetry can provide an effective driving force for sorting to occur. The compositional gradients creating the local molecular environment may facilitate the self-assembly of dynamins in a ring structure during the fission of membrane buds3. However, the spontaneous appearance of the compositional inhomogeneity due to the non-ideal mixing or the generation of the transverse compositional asymmetry of the lipids around the bud-neck region has not been experimentally confirmed so far.
Since the mechanochemical considerations invoked above are purely physical in nature, in vitro model membranes can be used for recapitulating them to clarify the fundamental mechanisms for the compositional inhomogeneity produced by the bud-neck topography. Here, using a lipid bilayer model supported on a complex surface topography that mimics the membrane curvature patterns generated during a budding event, we report on the sorting dynamics and the equilibrium distribution of membrane lipids, that is, higher-order spatial organizations of CHOL- and sphingomyelin (SPM)-enriched raft microdomains25,26 producing ring-rafts in the collar band of a bud.
Results
Architecture of bud-mimicking topography
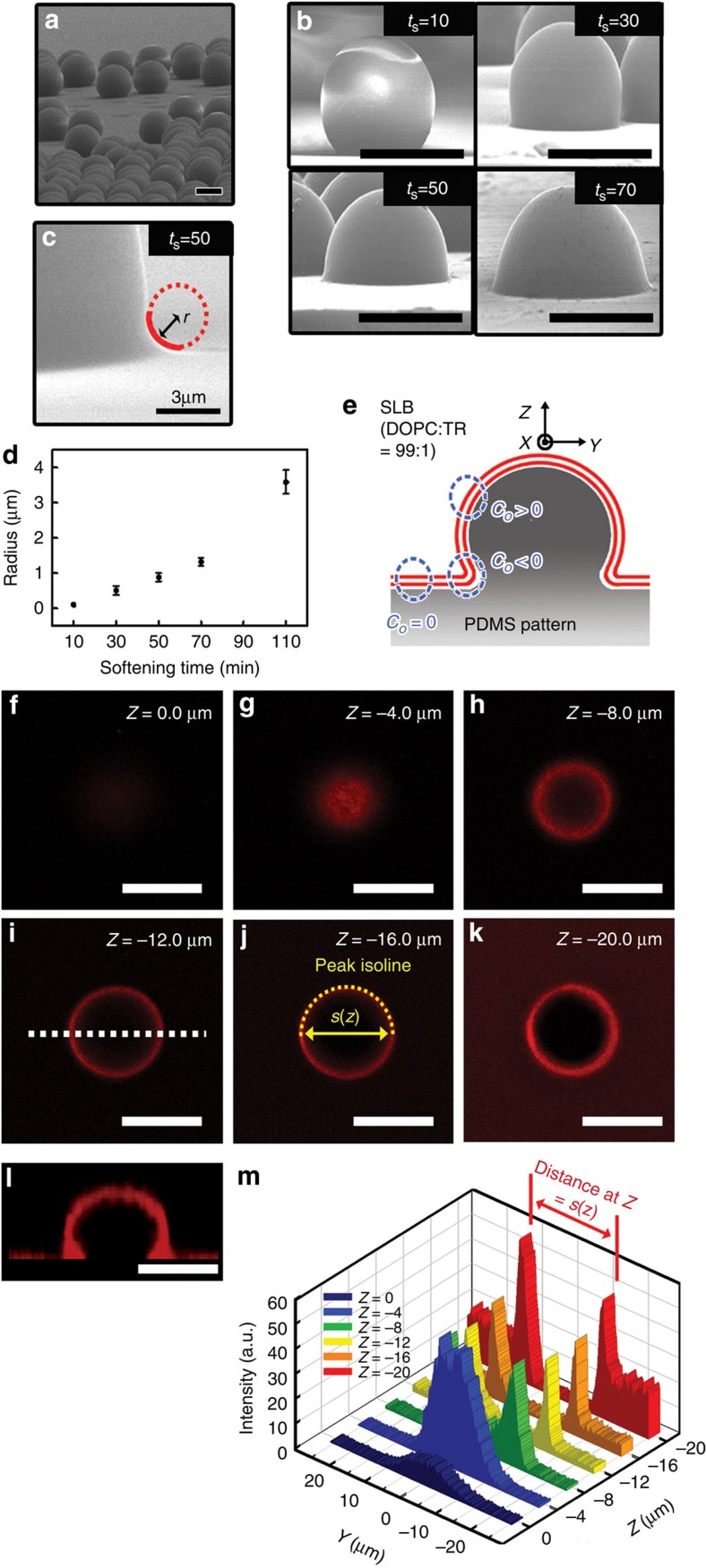
Let us begin with the design of a hydrophilic, polydimethylsiloxane (PDMS) elastomer with curvature patterns that represent a matured, late-stage bud (case III of Fig. 1a). The surface patterns (Fig. 1b) consist of a negative curvature (c0<0) representing the characteristic topography of the bud-neck region, a positive curvature (c0>0) of the bud-pit structure and an essentially zero curvature (c0=0) mimicking the donor membrane around the bud junction. The methodology of creating the local radii of curvatures on nanometre-to-micrometre scales was based on a combination of colloidal, soft and photolithographic tools in conjunction with a thermally triggered colloidal softening process (See Supplementary Fig. 1 for details). Briefly, polystyrene (PS) particles (25 μm in diameter) were assembled on a glass substrate through a convective self-assembly process27, transferred onto the PDMS plate by micro-contact printing and then treated thermally for softening and fixation of the PS particles (Supplementary Fig. 1a–d). Depending on the softening process, the curvature radius at the PS–PDMS interface was varied. Using such PDMS plate, a mould of a photo-curable polymer (NOA65), having negative replica patterns of the PS particles, was produced after the removal of the PS particles (Supplementary Fig. 1e,f). A negatively curved PDMS (nc-PDMS) substrate with the bud-neck mimicking topography was routinely fabricated with the help of the photo-curable polymer mould (Supplementary Fig. 1g, Fig. 2a). By simply adjusting the softening time (ts), a range of the curvature radius (r=1/c) at the bud-neck in the collar region, representing the degree of the bud maturity, was varied from about 100 nm to several micrometres (Fig. 2b,d). For instance, for ts=50 min, r≈900 nm (Fig. 2c). Before lipid deposition, the PDMS substrates were treated with standard plasma so as to render them hydrophilic. Since the plasma treatment removed backbone methyl (-CH3) groups from the exposed PDMS molecules by oxidation and newly formed polar silanol (Si-OH) groups were cross-linked in a depth-wise, self-limiting manner, a stiff, hydrophilic silica-like skin (about 5-nm thick) was readily produced on the top of the elastic PDMS foundation28. The hydrophilic nature of the nc-PDMS surface was confirmed from the fact that the water contact angle29 was varied from 120° to well below 20°. This is critical for self-assembling the lipid molecules into a uniform bilayer structure30.
Figure 2. Bud-mimicking topography with different curvatures and 3D reconstitution of the SLB.
(a) The array of the bud mimicking the negatively curved PDMS (nc-PDMS) patterns fabricated using the PS particles of 25 μm in diameter. (b) The SEM images of the nc-PDMS patterns varied with the softening time (ts) for the PS particles. (c) The magnified SEM image of a bud-neck region of the nc-PDMS pattern with the radius of curvature r≈900 nm at ts=50 min. (scale bar, 3 μm) (d) The mean value of the radius of curvature (r) as function of the softening time (ts; the error bar represents the s.d. for the mean value averaged over 10 different samples at each time scale). (e) Schematic diagram of the nc-PDMS pattern (r≈500 nm at ts=30 min) covered with the SLB of the B1 mixture. (f–k) The height-resolved confocal fluorescence profiles of the TR at the scanning depth z=0 μm (f), z=−4 μm (g), z=−8 μm (h), z=−12 μm (i), z=−16 μm (j) and z=−20 μm (k), respectively. (l) The side-view of confocal image of the PDMS pattern covered with the SLB. (m) The light intensity along the white dotted line (in i) in the y–z plane. The distance between two edges of the ‘peak isoline’ for given z was defined as the ring size, s(z). Unlabelled scale bars, 20 μm.
3D reconstitution of SLB
We first determined whether the three-dimensional (3D) closed topology representing the complex pattern of curvatures in the nc-PDMS substrate was transcribed with high fidelity into adhering lipid bilayers (Fig. 2e). For this purpose, we constructed a supported lipid bilayer (SLB) of a single prototypical component and characterized it using a height-resolved confocal fluorescence microscopy. The SLB was prepared by incubating an aqueous suspension of small unilamellar vesicles (SUVs; B1 membrane in Table 1) consisting of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) stained with fluorescent Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TR) on the nc-PDMS substrate. The radius of the curvature at the bud-neck was about 500 nm. A selected set of the height-resolved images obtained using a confocal fluorescence microscope, shown in Fig. 2f–l, revealed the 3D distribution of the TR molecules in the nc-PDMS topography. The lateral fluorescence profile seen in Fig. 2l (side-view), generated from confocal slices, was virtually indistinguishable from the physical shape of the topography observed in the scanning electron microscope (SEM) image (Fig. 2b; ts=30 min) of the nc-PDMS substrate. The height-resolved fluorescence profiles in Fig. 2f–k provided the spatial distribution of the TR molecules in 3D. Note that the fluorescence is barely detectable at the centre of the bud around the scanning depth z=0 μm (Fig. 2f), whereas the circular fluorescence emission profiles are clearly seen at arbitrary scanning depths (Fig. 2g–k), indicating that the SLB was uniformly covered on the nc-PDMS substrate in 3D. The uniform coverage of the SLB membrane over the entire nc-PDMS substrate was further confirmed by the 3D representation of the fluorescence profiles shown in Fig. 2m. Defining the ring size s(z) of the fluorescence as the diameter of the circular ‘peak isoline’ at z along the white line in Fig. 2i, the ring size s(−12)≈21.4 μm at z=−12 μm. This agrees well with the diameter of a single bud observed in the SEM image (Fig. 2b; ts=30 min). Similarly, the fact that s(−20)≈18.7 μm at z=−20 μm is also consistent with the SEM data (Supplementary Fig. 2). Within the confocal resolution in our experiments, however, establishing the exact quantitative correspondence between the physical topography of the bud and the geometry of the adhered SLB was difficult because of the reduction in resolution (~2 μm) near the bud-neck region. This results from the 3D curved topography of the substrate together with the lower axial or z-resolution in our confocal configuration.
Table 1. The compositions of different SLB mixtures studied.
| SLB types | Composition of SLB |
|---|---|
| B1 | 99% DOPC, and 1% TR |
| B2 | 66% DOPC, 33% CHOL and 1% TR |
| B3 | 66% DOPC, 33% SPM and 1% TR |
| B4 | 98% DOPC, 1% GM1 and 1% TR |
| B5 | 65% DOPC, 33% SPM, 1% GM1 and 1% TR |
| B6 | 65% DOPC, 33% CHOL, 1% GM1 and 1% TR |
| B7 | 33% DOPC, 33% SPM, 33% CHOL and 1% TR |
| B8 | 33% SPM, 32.5% CHOL, 32.5% DOPC, 1% GM1 and 1% TR |
CHOL, cholesterol; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; SPM, sphingomyelin; TR, Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine.
Next, we measured the lateral mobility of the B1 membrane (the simplest SLB) on a hydrophilic, nc-PDMS substrate with the 3D topography. The fluorescence recovery after photobleaching (FRAP) measurements31 were carried out to determine the diffusion coefficients of the lipids on the flat region and the bud-pit region with r≈100 nm. A representative set of the time-lapse fluorescence images taken from both the flat region (Supplementary Fig. 3a–c) and the bud-pit region (Supplementary Fig. 3d–f) indeed confirmed the lateral fluidity and the structural contiguity of the B1 membrane over the 3D topography. Note that the FRAP tests were performed 3 days after the preparation of the SLBs to check the temporal stability of the uniform coverage. The mean value of the diffusion coefficients was 1.4±0.1 μm2 s−1 (the error represents the s.d. for the mean value averaged over five different measurements) and nearly identical in the flat and bud-pit regions. This value is comparable to those reported previously for the lipid bilayers supported on glass31,32 and oxidized PDMS substrates33. The formation of the uniform and laterally fluidic SLB over the 3D complex topography can be readily understood from a mechanical point of view: the favourable adhesion energy34 between the SLB and the hydrophilic silica-like surface enables the SLB to overcome the bending energy penalty associated with the elastic distortions of the SLB in mimicking the topography of a late-stage bud in the neck and the surrounding collar region.
Composition dependence of the spatial inhomogeneity of 3D SLB
In establishing a generic principle of the spatial distribution of the SLB domains in the bud-neck of closed topology in a composition-dependent manner, we prepared a series of mixtures with different compositions of the membrane components spanning from two to five (Table 1). The primary lipids we tested were combinatorial mixtures of DOPC, CHOL and SPM. Depending on the composition and temperature35,36, microscopic phase separation characterized by two co-existing membrane phases is known to occur in several of these mixtures: a liquid-ordered (Lo) or ‘raft’ phase representing a dense phase enriched with SPM and CHOL and a liquid-disordered (Ld) phase consisting primarily of unsaturated DOPC. During the vesicle fusion process, the SLB samples were maintained at room temperature for more than 36 h to ensure the long-term stability of the phase-separated domains before characterization. For one of the mixtures (the B7 membrane, exhibiting the raft phase), diffusion coefficients at different elapsed times (0 h, 18 h and 36 h) after vesicle fusion were determined from the intensity profiles of the FRAP (Supplementary Fig. 4). Similar values (the mean value of about 0.9 μm2 s−1) for the three cases suggest that the lateral fluidity and the structural contiguity of the SLB were well maintained over 36 h.
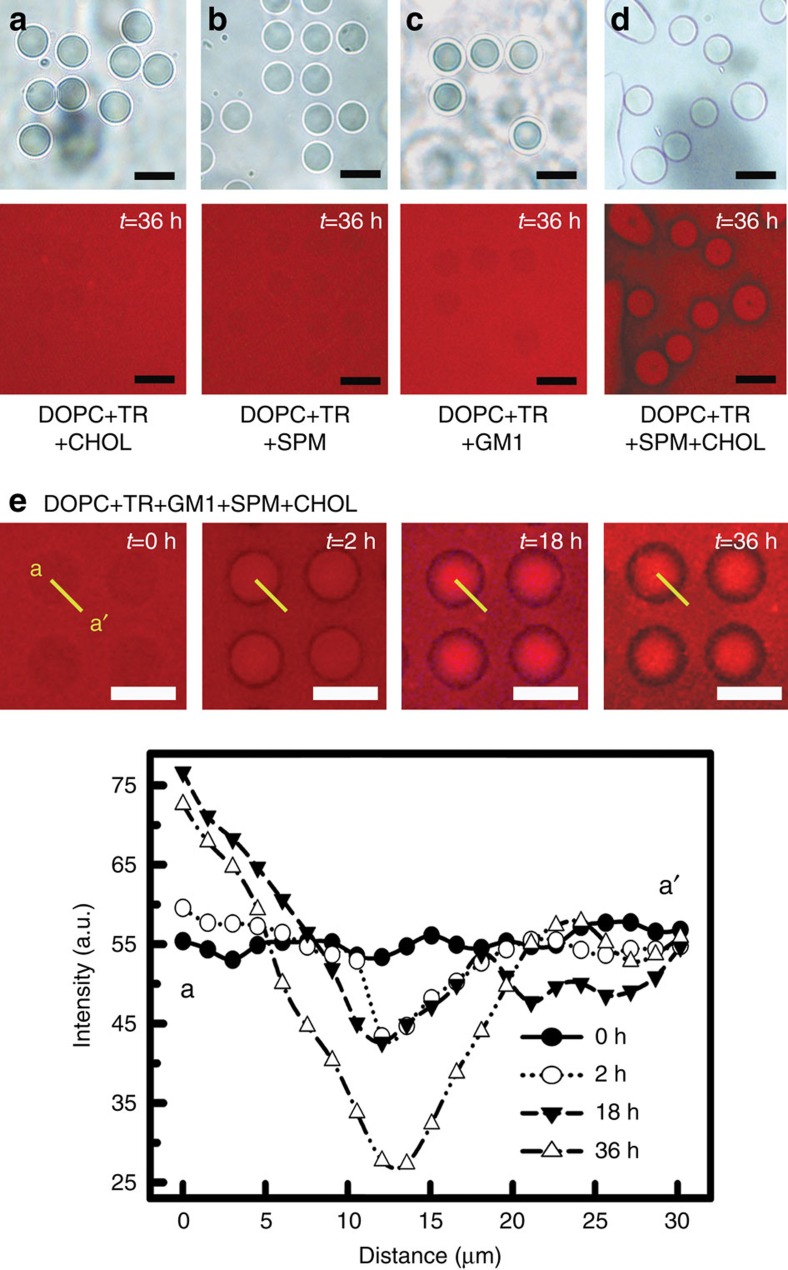
Let us start with three ternary systems (B2, B3 and B4) having one of the three components (CHOL, SPM, or GM1) mixed individually in the background of DOPC doped with a phase-sensitive fluorescent probe (TR). The wide-field fluorescence data in Fig. 3 were obtained using epifluorescence microscopy for the nc-PDMS substrate having the curvature of 900 nm−1 at the bud-neck (the substrate obtained for ts=50 min). As clearly seen from the corresponding fluorescence images (t=36 h) in Fig. 3a–c, no phase separation was optically detectable. This is not surprising because only one among the three essential components was present in these mixtures25,37. Among the three quaternary systems of SPM-GM1 (B5), CHOL-GM1 (B6) and SPM-CHOL (B7), only the B7 membrane exhibited the phase separation around the bud-necks and produced a raft domain of closed topology in 3D as shown in Fig. 3d. Note that a quinary system (B8) containing all the three components (SPM, CHOL, and GM1) showed the development of such raft domain in any case.
Figure 3. Time-lapse spatial inhomogeneity of the SLBs in bud-mimicking 3D topography.
(a–d) A ternary system consisting of CHOL (a), SPM (b), GM1 (c) and a quaternary system composed of CHOL and SPM (d) in the background of DOPC and TR at the time of 36 h after vesicle fusion. The corresponding optical microscopic textures (top) and the fluorescence images (bottom) taken using an epifluorescence microscopy were shown. (e) The time-lapse fluorescence micrographs and the fluorescence intensity profiles along the line a–a' across the topographic pattern. Scale bar, 50 μm.
The time-lapse fluorescence images shown in Fig. 3e reveal the dynamics of the ring-raft formation. Over the course of 36 h, the initial homogeneous distribution of the TRs was gradually transformed into a ring-type distribution. Two distinct features were evidently observed: (1) a dark ring appeared in the bud-neck region due to the depletion of the TRs and (2) the bright fluorescence was seen in the central bud-pit region. From the fluorescence profiles measured along the yellow lines (a-a') in Fig. 3e, the dark ring at t=36 h became dimmer and wider as the Lo phase became larger. This is due to the tendency for the exclusion of the TRs from the Lo phase3,38,39. The precise width of the ring-raft structure, however, cannot be ascertained from these measurements alone because of the lower lateral resolution in our experimental geometry and the 3D topography of the bud feature. Note that although the raft-partitioning membrane components accumulate around the bud-neck, not all of them can be localized in the ring-raft domain because the excess molecules cannot be accommodated within the area of the ring-raft. From all the results for the wide range of the combinatorial lipid mixtures (Fig. 3a–e) above, the two raft-partitioning components, CHOL and SPM, must be contained for the formation of a ring-raft of closed topology around the bud-neck. This shares the common features with the cases for planar SLBs and giant unilamellar vesicles where both CHOL and SPM are required to produce Lo phase domains40,41. Another point is that the ring-raft domains were substantially disrupted at elevated temperatures above the effective phase transition of the membrane composition in reconstructed Lo domains in the prescribed regions with local curvatures (See Supplementary Fig. 5).
Reconstitution of ring-rafts and ligand-receptor binding
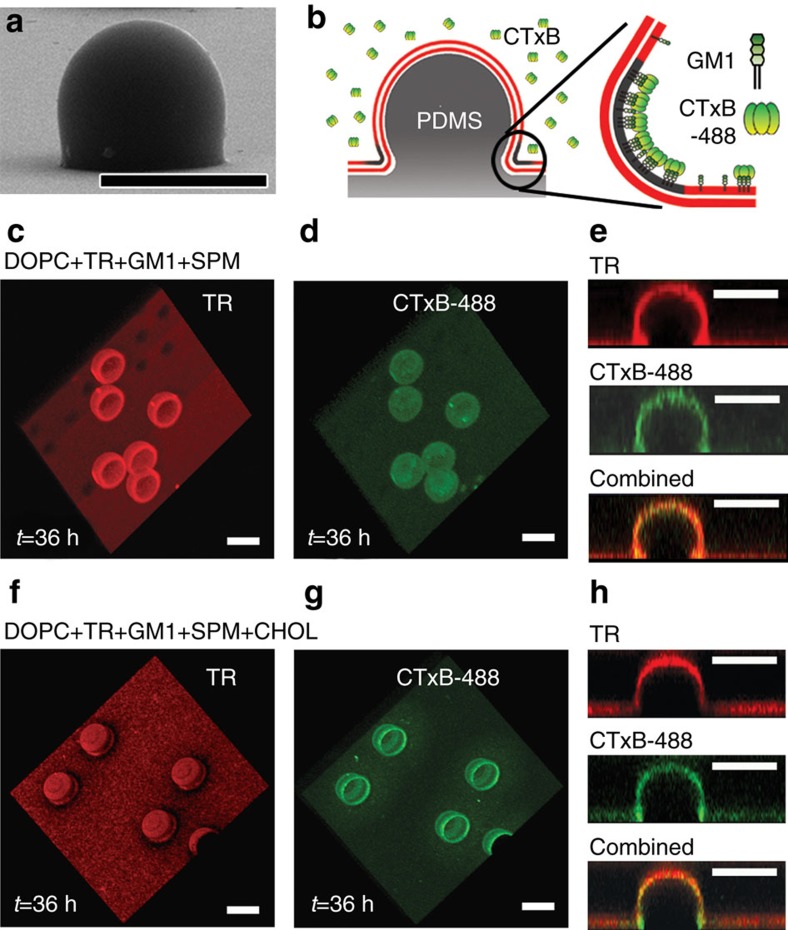
In exploring the essential role of one of the raft-partitioning components, CHOL, on the compositional inhomogeneity around the bud-neck, the ligand-receptor (cholera toxin–GM1) binding events were monitored in two model systems: one exhibiting a ring-raft structure (CHOL-doped) and the other showing no raft (devoid of CHOL). The cholera toxin, a member of the AB5 family of bacterial toxins, is a well-known marker to identify a lipid raft because it binds bio-specifically to raft-partitioning GM1 with high affinity38,42. For the B5 mixture where no CHOL is present, no raft domain was observed and the TRs were uniformly distributed across the entire nc-PDMS substrate as shown in Fig. 4c. Similarly, no preferential distribution of GM1 bound with the Alexa Fluor 488-labelled cholera toxin B subunit (CTxB-488) was detected during 36 h as shown in Fig. 4d (Supplementary Movie 1). Figure 4e shows the cross-sectional micrographs for the 3D distributions of target lipids (TR; top), proteins (CTxB-488; middle), and the two components (TR+CTxB-488; bottom). In contrast, for the B8 lipid mixture, a ring-raft was clearly seen around the bud-necks (Fig. 4f). On toxin binding, the fluorescence emission from CTxB-488 was observed as shown in Fig. 4g (Supplementary Movie 2). The cross-sectional micrographs showing the localization of protein binding were presented in Fig. 4h. The correspondence between the TR-devoid rings and the patterns obtained on CTxB binding43 suggests that the binding event is irrelevant to the ring-raft formation in our case.
Figure 4. Reconstitution of closed ring-rafts and protein binding.
(a) The SEM image of a single nc-PDMS pattern (r≈500 nm). (b) Schematic illustration of the localization of CTxB protein binding to glycolipid receptor GM1 with highly affinity in the model membrane. (c–e) The confocal micrographs of the target lipid (TR) only, protein (CTxB-488) only, and the mixture (TR+CTxB-488) for the B5 membrane (CHOL-free). (f–h) The confocal micrographs of the target lipid (TR), protein (CTxB-488), and the mixture (TR+CTxB-488) for the quinary B8 membrane. Scale bar, 25 μm.
Membrane compartmentalization by ring raft
Membrane compartmentalization by ring raft has been speculated that the raft domains at the bud boundaries might serve to spatially segregate the membrane components (for example, proteins) in achieving the compositional enrichment of selected lipids14 such as found in viral envelopes and synaptic vesicles6. Of particular interest is to assess whether a topologically closed ring-raft we observed indeed behaves as a barrier for the free, thermally excited lipid exchange between the bud membrane and the donor membrane across the bud-neck. On the basis of the electric field-directed diffusion test39, the diffusion of the lipids in the bud membrane across the ring-raft was found to be essentially prohibited (Supplementary Fig. 6), thus suggesting that ring-raft serves as an effective barrier for the compartmentalization of membrane components within the bud-pit.
Discussion
Two major mechanochemical issues must be invoked to appreciate the formation of ring-raft domains at the bud-neck-collar interface. The first issue is that the Lo phase is thicker than the Ld phase since the extended chains of the saturated lipids become more ordered and denser due to the tight packing with the CHOLs. Such height mismatch at the phase boundary is energetically unfavourable. Thus, the line tension is produced between the two phases and the phase-separated Lo domain tends to adopt a circular geometry. Beyond a critical size, however, the line tension energy of a single Lo domain exceeds the curvature-dependent Helfrich-type energy4 and consequently, the membrane is deformed out of plane and is in favour of budding to reduce the line tension energy. If this is the case, the raft-partitioning components would be concentrated in both the bud-pit region and the surrounding collar. However, this is not observed. The second issue is that the line tension effect is coupled with the membrane curvature as well as the bending rigidity difference between the two phases in developing the ring-raft morphologies. A preliminary analysis44 based on the Helfrich-free energy description for the membranes45 suggests that the large negative spontaneous curvature of the Lo domain in relative to the Ld domain (cLo=−1/68 Å−1 and cLd=−1/160 Å−1) (ref. 44) together with the difference in the bending stiffness (kLo/kLd=1.25) can account for the accumulation of the unit rafts in the collar band around the bud-neck (Supplementary Fig. 7). This requires the preferential localization of the raft domains in the outer leaflet of the SLB in the curvature niche imposed by the nc-PDMS substrate. However, a complete picture of the budding scenario remains to be acquired from more systematic experiments.
The generic principle of reconstituting the ring-rafts reported here can be further extended toward understanding the correlations between the physicochemical variables, such as the curvature6,46,47 and the lipid compaction48, and the macroscopic phenomena including consequential protein migration and clustering43,49,50. The geometrical deformations of the SLB were found to allow for the collective sorting of specific types of the membrane molecules (that is, raft-partitioning CHOL, SPM and GM1) into an unusual ring shape, which is in contrast to the morphology of a stripe or a bubble developed commonly in a phase-separating system with competing interactions44,46,51,52. Finally, our reconstitution of the ring-raft of 3D closed topology in vitro might not only serve as the machinery for manipulating and realizing the curvature-associated raft activities, for example, the endocytic vesicle scission14,53 but also provide an insight into the peculiar nature of the self-assembly of the ubiquitous membrane division protein such as dynamin10,54 whose migration into the bud-neck regions remains poorly understood.
Methods
Fabrication of bud-mimicking PDMS patterns with 3D curvature
Commercially available glass substrates were first sonicated in acetone for more than 30 min and then in deionized water for 10 min. The cleaned substrates were dried under a gentle stream of nitrogen and subsequently coated with a standard negative photoresist material (SU-8 2015, MICROCHEM) according to the vendor’s instructions (url: http://microchem.com). The thickness of the SU-8 photoresist layer was about 15 μm. Using a mask aligner (MA-6, EVG), the glass substrates coated with the SU-8 were exposed to ultraviolet (UV) light through a predefined photomask with an array of grid patterns of 50 μm in each side, and developed to leave out only desired SU-8 patterns. The SU-8 patterns produced on the glass substrates were capable of periodically assembling monodisperse PS particles. Typically, the PS particles of 25 μm in diameter were used in our experiments.
After the colloidal PS particles in water were prepared on the glass substrate with the SU-8 patterns by spin-coating, a clean, flat glass slide was used as a top cover for the PS particles. The separation between the bottom glass substrate and the top cover slide was uniformly maintained using 150-μm-thick plastic spacers. The slow evaporation of water at room temperature for about 1 day allowed to assemble the PS particles into the SU-8 grid patterns through a well-established convective assembly process27 (Supplementary Fig. 1a,b). The transfer of the PS particles onto a flat PDMS plate by micro-contact printing and the thermal treatment for softening and fixation of the PS particles were then successively carried out (Supplementary Fig. 1c,d). The PDMS plate with the PS particles was placed on a hotplate for the thermal treatment at 200 °C for 5–25 min, depending on the desired radius of curvature, to produce the nc-topography at the interface between the PS particle and the PDMS plate (Supplementary Fig. 1d). The radius of curvature was systematically varied as r=100, 500 and 900 nm for the softening time t=10, 30 and 50 min, respectively. A photo-curable (under the UV light) polymer (NOA65, Norland Products, Inc.) was coated on the PDMS plate with the thermally treated PS particles, followed by UV exposure for 2 min (Supplementary Fig. 1e). After the photo-cured NOA65 layer together with the PS particles was peeled off, it was immersed in toluene for 1 min to remove the PS particles from the NOA65 layer (Supplementary Fig. 1f). The Sylgard 184 silicone elastomer (Dow Corning Corp.), mixed with the curing agent in a 10:1 ratio, was then poured into the PS vacancies and thermally cured for 3 h at 80 °C (Supplementary Fig. 1f). The NOA65 layer was then removed in a mixed solution (dichloromethane: methanol=100:15) for about 1 day and the PDMS substrate having bud-mimicking patterns (the nc-PDMS substrate; Supplementary Fig. 1g) was finally produced. As the last step, the nc-PDMS substrate was annealed on a hotplate at 200 °C for more than 1 h to remove any residual solution adsorbed or entrapped within the PDMS.
The SLB formation
DOPC was used as a main lipid component for all bilayer mixtures. For imaging by fluorescence, a small concentration (~1 mol%) of a negatively charged lipid-probe, TR (Molecular Probes, Eugene, Oregon), was mixed with DOPC. Other lipids, including SPM (brain, porcine), CHOL, ganglioside GM1 (brain, ovine-ammonium salt) were purchased from Avanti Polar Lipids (Birmingham, Alabama). The Alexa Fluor 488-labelled cholera toxin B subunit (CTxB-488, Molecular Probe, Eugene, Oregon) was used as a raft maker at a concentration of 10 μg ml−1 in Tris buffer (100 mM NaCl and 10 mM Tris at pH.=8.0). All lipids were dissolved in chloroform. The rapid solvent exchange method55 was employed to evaporate chloroform and to hydrate with Tris buffer (100 mM NaCl and 10 mM Tris at pH. 8.0) simultaneously. SUVs were produced by extruding 20 times through a 50 nm-pore filter. The residual SUVs in the Tris buffer solution were removed with deionized water after the SLB was formed through the exposure of the substrate to a solution of the SUVs for 5 min. Unlike giant vesicles and floating bilayers29, the SLB experienced relatively strong adhesion on the nc-PDMS surface, and the Lo and Ld phases were fully phase separated in the SLB in the course of about 36 h as reported previously56.
Fluorescence microscopy measurements
Fluorescence microscopy measurements were carried out in both epi- (Eclipse E600-POL, Nikon) and confocal (Carl Zeiss LSM710, Carl Zeiss) geometries. Texas Red and Alexa-Fluor 488 probes were fluorescently excited at appropriate wavelengths of He-Ne lasers and collected with appropriate excitation/emission filters (Ex: 590 nm, Em: 610 nm for Texas Red and Ex: 500 nm, Em: 526 nm for Alexa Fluor 488).
FRAP measurements and data analysis
The diffusion coefficients of the probe lipids in the SLB samples were determined from the intensity profiles obtained by the microscopy-based FRAP measurements (Supplementary Fig. 4). For the FRAP measurements, a circular region (30–50 μm in diameter) in the fluorescent sample was illuminated continuously at the excitation wavelength for the fluorophore through a × 60 (NA, 0.70) objective lens for about 3 min. After photobleaching, the fluorescent recovery was monitored using a probe beam along the illumination path through a × 20 objective lens. The time-lapse sequences of the wide-field fluorescence micrographs of the bleached sample were recorded in time (typically, every 5 s). The FRAP data were analysed using numerical solutions to the two-dimensional diffusion equation. Data analysis was performed with the help of an image analysis software available in publicly (ImageJ, NIH, USA, url: http://rsb.info.nih.gov/ij/).
Preparation of substrates for EM measurements and data analysis
The bud-mimicking PDMS template was coated with a 3-nm-thick layer of Pt using a sputter coater (SCD 500, Bal-Tec) before imaging with a scanning electron microscope (S-4800, Hitachi). The accelerating voltage was 15 kV and the current was 10 μA. The radii of curvatures (r’s) at each softening time (ts) were measured using Image J software in the magnified bud-neck region. A typical example of determining the radius of curvature was shown in Fig. 2c.
Author contributions
Y.-S.R. and S.-D.L. initiated and designed the experiments, analysed the data and wrote the initial draft of the manuscript. Y.-S.R. and J.-H.S. performed vesicular experiments. I.-H.L. and S.C.P. performed colloidal particle experiments. S.O., B.L., L.R.J., N.J.W., S.-H.O. and N.L.J. were involved in confocal microscopy measurements and contributed to data analysis. S.-D.L. and A.N.P. provided scientific and technical guidance, interpreted the data and finalized the manuscript. All authors agreed on its final content.
Additional information
How to cite this article: Ryu, Y.-S. et al. Reconstituting ring-rafts in bud-mimicking topography of model membranes. Nat. Commun. 5:4507 doi: 10.1038/ncomms5507 (2014).
Supplementary Material
Supplementary Figures 1-7 and Supplementary References
The 3-D distribution of fluorescence emission from CTxB-488 in the SLB (DOPC:TR:GM1:SPM=65:1:1:33) draped the bud patterns.
The 3-D distribution of fluorescence emission from CTxB-488 in the SLB (DOPC:TR:GM1:SPM:CHOL=33:1:1:32.5:32.5) draped the bud patterns.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the government of Korea (MSIP; No. 2007-0054847). Contribution by A.N.P. was supported by a grant from the U.S. Department of Energy under the award number DE-FG02-04ER46173. Contribution by L.R.J., N.J.W., S.-H.O. was supported by grants from the U.S. National Institutes of Health (NIH R01 GM092993) and the National Science Foundation (CAREER, DBI 1054191).
References
- Rothman J. E. & Orci L. Budding vesicles in living cells. Sci. Am. 274, 70–75 (1996). [DOI] [PubMed] [Google Scholar]
- Chazal N. & Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67, 226–237 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votteler J. & Sundquist W. I. Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich W. Elastic properties of lipid bilayers - theory and possible experiments. Z Naturforsch. C 28, 693–703 (1973). [DOI] [PubMed] [Google Scholar]
- Marsh D. Elastic curvature constants of lipid monolayers and bilayers. Chem. Phys. Lipids 144, 146–159 (2006). [DOI] [PubMed] [Google Scholar]
- Huttner W. B. & Zimmerberg J. Implications of lipid microdomains for membrane curvature, budding and fission - Commentary. Curr. Opin. Cell Biol. 13, 478–484 (2001). [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Boura E., Carlson L. A. & Rozycki B. Membrane budding. Cell 143, 875–887 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H. T. & Gallop J. L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596 (2005). [DOI] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J. & Hurley J. H. Membrane scission by the ESCRT-III complex. Nature 458, 172–177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J. E. & Schmid S. L. Dynamin self-assembles into rings suggesing a mechanism for coated vesicle budding. Nature 374, 190–192 (1995). [DOI] [PubMed] [Google Scholar]
- Chernomordik L. V. & Kozlov M. M. Protein-lipid interplay in fusion and fission of biological membranes. Ann. Rev. Biochem. 72, 175–207 (2003). [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Soe T. T. & Maxfield F. R. Endocytic sorting of lipid analogues differing solely in the chemistry of their hydrophobic tails. J. Cell Biol. 144, 1271–1284 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L. & Mayor S. Induced domain formation in endocytic invagination, lipid sorting, and scission. Cell 142, 507–510 (2010). [DOI] [PubMed] [Google Scholar]
- Liu J., Sun Y., Drubin D. G. & Oster G. F. The mechanochemistry of endocytosis. PLoS Biol. 7, 1–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurczy M. E., Mellander L. J., Najafinobar N. & Cans A.-S. Composition based strategies for controlling radii in lipid nanotubes. PLoS ONE 9, e81293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurczy M. E. et al. Mass spectrometry imaging of mating Tetrahymena show that changes in cell morphology regulate lipid domain formation. Proc. Natl Acad. Sci. USA 107, 2751–2756 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski S. G., Van Bell C. T., Winograd N. & Ewing A. G. Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science 305, 71–73 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A. R. et al. α-Synuclein induces both positive mean curvature and negative Gaussian curvature in membranes. J. Am. Chem. Soc. 134, 2613–2620 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt N. W. et al. Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J. Am. Chem. Soc. 133, 6720–6727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart T., Hess S. T. & Webb W. W. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature 425, 821–824 (2003). [DOI] [PubMed] [Google Scholar]
- Kumar P. B. S., Gompper G. & Lipowsky R. Budding dynamics of multicomponent membranes. Phys. Rev. Lett. 86, 3911–3914 (2001). [DOI] [PubMed] [Google Scholar]
- Tian A. & Baumgart T. Sorting of lipids and proteins in membrane curvature gradients. Biophys. J. 96, 2676–2688 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert U. Curvature-induced lateral phase segregation in two-component vesicles. Phys. Rev. Lett. 70, 1335–1338 (1993). [DOI] [PubMed] [Google Scholar]
- Baumgart T., Das S., Webb W. & Jenkins J. Membrane elasticity in giant vesicles with fluid phase coexistence. Biophys. J. 89, 1067–1080 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K. & Ikonen E. Functional rafts in cell membranes. Nature 387, 569–572 (1997). [DOI] [PubMed] [Google Scholar]
- Lingwood D. & Simons K. Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010). [DOI] [PubMed] [Google Scholar]
- Dimitrov A. S. & Nagayama K. Continuous convective assembling of fine particles into two-dimensional arrays on solid surfaces. Langmuir 12, 1303–1311 (1996). [Google Scholar]
- Efimenko K. et al. Nested self-similar wrinkling patterns in skins. Nat. Mater. 4, 293–297 (2005). [DOI] [PubMed] [Google Scholar]
- Ginn B. T. & Steinbock O. Polymer surface modification using microwave-oven-generated plasma. Langmuir 19, 8117–8118 (2003). [Google Scholar]
- Cremer P. S. & Boxer S. G. Formation and spreading of lipid bilayers on planar glass supports. J. Phys. Chem. B 103, 2554–2559 (1999). [Google Scholar]
- Yee C. K., Amweg M. L. & Parikh A. N. Membrane photolithography: Direct micropatterning and manipulation of fluid phospholipid membranes in the aqueous phase using deep-UV light. Adv. Mater. 16, 1184–1189 (2004). [Google Scholar]
- Kaizuka Y. & Groves J. T. Bending-mediated superstructural organizations in phase-separated lipid membranes. New J. Phys. 12, 11 (2010). [Google Scholar]
- Sanii B., Smith A. M., Butti R., Brozell A. M. & Parikh A. N. Bending membranes on demand: fluid phospholipid bilayers on topographically deformable substrates. Nano Lett. 8, 866–871 (2008). [DOI] [PubMed] [Google Scholar]
- Chen I. J. & Lindner E. The stability of radio-frequency plasma-treated polydimethylsiloxane surfaces. Langmuir 23, 3118–3122 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R., Yu C.-h. & Groves J. T. Curvature-modulated phase separation in lipid bilayer membranes. Langmuir 22, 5095–5099 (2006). [DOI] [PubMed] [Google Scholar]
- Veatch S. L. & Keller S. L. Miscibility phase diagrams of giant vesicles containing sphingomyelin. Phys. Rev. Lett. 94, 148101 (2005). [DOI] [PubMed] [Google Scholar]
- Korade Z. & Kenworthy A. K. Lipid rafts, cholesterol, and the brain. Neuropharmacology 55, 1265–1273 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy A. K., Petranova N. & Edidin M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol. Biol. Cell 11, 1645–1655 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oudenaarden A. & Boxer S. G. Brownian ratchets: molecular separations in lipid bilayers supported on patterned arrays. Science 285, 1046–1048 (1999). [DOI] [PubMed] [Google Scholar]
- de Almeida R. F. M., Fedorov A. & Prieto M. Sphingomyelin/phosphatidylcholine/cholesterol phase diagram: boundaries and composition of lipid rafts. Biophys. J. 85, 2406–2416 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida R. F., Loura L., Fedorov A. & Prieto M. Lipid rafts have different sizes depending on membrane composition: a time-resolved fluorescence resonance energy transfer study. J. Mol. Biol. 346, 1109–1120 (2005). [DOI] [PubMed] [Google Scholar]
- Merritt E. A. et al. Crystal-structure of cholera-toxin B-pentamer bound to receptor G(M1) pentasaccharide. Protein Sci. 3, 166–175 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A. et al. Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc. Natl Acad. Sci. USA 102, 6320–6325 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C., Lee S.-W., Yoon T.-Y. & Lee S.-D. Water meniscus-directed organization of liquid-ordered domains in lipid monolayer. J. Nanosci. Nanotechnol. 11, 4527–4531 (2011). [DOI] [PubMed] [Google Scholar]
- Safran S. A. Statistical Thermodynamics of Surfaces, Interfaces, and Membranes Addison-Wesley Publishing Company (1994). [Google Scholar]
- García-Sáez A. J., Chiantia S. & Schwille P. Effect of line tension on the lateral organization of lipid membranes. J. Biol. Chem. 282, 33537–33544 (2007). [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Taniguchi T. & Imai M. Pore formation in a binary giant vesicle induced by cone-shaped lipids. Biophys. J. 99, 472–479 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun Ü. & Simons K. Membrane rafting: from apical sorting to phase segregation. FEBS Lett. 584, 1685–1693 (2010). [DOI] [PubMed] [Google Scholar]
- Ewers H. et al. GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 12, 11–18 (2010). [DOI] [PubMed] [Google Scholar]
- Windschiegl B. et al. Lipid reorganization induced by Shiga toxin clustering on planar membranes. PLoS ONE 4, e6238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seul M. & Andelman D. Domain shapes and patterns-The phenomenology of modulated phases. Science 267, 476–483 (1995). [DOI] [PubMed] [Google Scholar]
- Henderson R. M., Edwardson J. M., Geisse N. A. & Saslowsky D. E. Lipid rafts: feeling is believing. News Physiol. Sci. 19, 39–43 (2004). [DOI] [PubMed] [Google Scholar]
- Liu J., Kaksonen M., Drubin D. G. & Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proc. Natl Acad. Sci. USA 103, 10277–10282 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh P., Horner T., Witkiewicz H. & Schnitzer J. E. Endothelin induces rapid, dynamin-mediated budding of endothelial caveolae rich in ET-B. J. Biol. Chem. 287, 17353–17362 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buboltz J. T. & Feigenson G. W. A novel strategy for the preparation of liposomes: rapid solvent exchange. Biochim. Biophys. Acta 1417, 232–245 (1999). [DOI] [PubMed] [Google Scholar]
- Yoon T.-Y. et al. Topographic control of lipid-raft reconstitution in model membranes. Nat. Mater. 5, 281–285 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-7 and Supplementary References
The 3-D distribution of fluorescence emission from CTxB-488 in the SLB (DOPC:TR:GM1:SPM=65:1:1:33) draped the bud patterns.
The 3-D distribution of fluorescence emission from CTxB-488 in the SLB (DOPC:TR:GM1:SPM:CHOL=33:1:1:32.5:32.5) draped the bud patterns.