Abstract
Disruptions in DNA repair pathways predispose cells to accumulating DNA damage. A growing body of evidence indicates that tumors accumulate progressively more mutations in DNA repair proteins as cancers progress. DNA repair mechanisms greatly affect the response to cytotoxic treatments, so understanding those mechanisms and finding ways to turn dysregulated repair processes against themselves to induce tumor death is the goal of all DNA repair inhibition efforts. Inhibition may be direct or indirect. This burgeoning field of research is replete with promise and challenge, as more intricacies of each repair pathway are discovered. In an era of increasing concern about healthcare costs, use of DNA repair inhibitors can prove to be highly effective stewardship of R&D resources and patient expenses.
Keywords: clinical trials, DNA repair inhibition, DNA repair pathways, DNA repair targets, small-molecule inhibitors
Capitalizing on differences between cancerous and noncancerous cells to find more effective therapeutic solutions is an area of ongoing, intense research. Defects in DNA and/or DNA repair can cause cancer as well as promote its growth. As cancers become increasingly mutagenic, genetic streamlining leads to deficiencies in one or more DNA repair pathways – accompanied by compensatory activities that increase the levels of certain repair proteins in the same pathway or a different one [1]. This contributes to intrinsic or acquired cellular resistance to DNA-damaging agents [2]. Interrupting DNA repair in such a way that shuts down a tumor's compensatory repair mechanisms and induces cell death is the goal of all research surrounding DNA repair inhibition.
DNA repair inhibitors, particularly small-molecule inhibitors, hold great promise for damaging tumor cells. Their specificity can be honed to target a single step or single protein of a DNA repair pathway. Achieving that goal moves us closer to truly personalized medicine. However, the development of such inhibitors is offset by several real-world challenges.
Research has amassed much data regarding DNA repair protein(s) that are under- or over-expressed in cancers – but which ones do the cancers themselves uniquely require? In a perfect world, one altered gene or gene product would create a unique footprint that corresponded to just one repair pathway or would drive a rate-limiting or saturable reaction. Unfortunately, finding a target that fulfils that wish list is the exception, rather than the rule. A mutagenic phenotype is rarely the result of one under- or over-expressed protein, and molecular pathogenesis is not linked to an isolated step in oncogenic progression. In addition, the multifunctionality of many DNA repair proteins can complicate inhibition efforts with unexpected results. Pathways are complex and crosstalk between pathways occurs (Table 1) [3–5]. As mutagenesis increases, greater tumor heterogeneity from crosstalk and compensation further complicates identification of viable targets for inhibition. Finally, the relative lack of biomarkers to help sort this out is not unlike driving in uncharted territory without a map. Research is in its infancy in corralling the collective contributions of multiple proteins that constitute a mutagenic phenotype.
Table 1.
Overlap and crosstalk among DNA repair pathways.
| Pathway | Backup | Type of problem/provision for repair |
|---|---|---|
| DR | MMR | If MGMT fails to remove O6-methylguanine, the MMR pathway can recognize and fix O6-methylguanine misspairs |
| MMR | MMR can repair postreplicative guanine/thymine mismatches Futile MMR cycles create DSBs, which either induces apoptosis or activate repair by HR or NHEJ |
|
| NER | NER can repair larger adducts at the O6 position of guaninethat MGMT cannot repair | |
| BER | BER can repair mismatch pairs and other alkylation adducts that DR does not repair | |
|
| ||
| BER | NER | NER can serve as a backup for repairing minor oxidative damage |
| HR | If BER does not repair ssDNA breaks, they may lead to DSBs, which HR can repair | |
| HR/NHEJ | If BER fails to repair SSBs, HRcan repair them during replication (S phase). If signaling arrests the cell cycle at G1, then NHEJ can repair the breaks | |
|
| ||
| NHEJ | HR | HR can repair DNA DSBs that the NHEJ pathway fails to process |
BER: Base excision repair; DR: Direct repair; DSB: Double-stranded break; HR: Honnologous reconnbination; MMR: Misnnatch repair; NER: Nucleotide excision repair; NHEJ: Nonhonnologous end joining.
Data taken from [6].
Inhibition & synthetic lethality
Because preserving the genome is paramount, DNA repair is replete with alternate plans. If one pathway fails to repair a problem, another pathway can step in (Table 1) [3–5]. While that elegant plan helps maintain genomic stability under normal circumstances, it contributes to chemoresistance when repair mechanisms go awry. However, if the alternative pathway contains a mutation that makes the pathway dysfunctional or nonfunctional, then impairing a step in the main pathway can force repairs into the backup mode where repair will fail, causing the cells to self-destruct. That is the principle of synthetic lethality, and PARP inhibition is the leader in that principle.
PARP's primary activity is in the base excision repair (BER) pathway, where it gauges the extent of damage and functions as a scaffold or stabilizer for other BER proteins. PARP inhibition abrogates BER functionality, causing accumulation of unresolved single-stranded breaks (SSBs) that convert to double-stranded breaks (DSBs) during S phase. Because BRCA-deficient cancer cells cannot repair DSBs via the homologous recombination (HR) repair pathway, they attempt to so do via the error-prone nonhomologous end joining (NHEJ) pathway. However, recombinogenic lesions and other errors cause the collapse of replication forks and cell death when NHEJ attempts the repairs [7,8].
Successes & bumps in the road
The concept of `treating a weakness' to create a synthetic lethality [9] was launched in 2005 when two seminal papers demonstrated that PARP inhibitors (PARPis) could be used as single agents to treat BRCA-deficient cell lines [8]. The first clinical study that demonstrated the benefit of the PARPi olaparib as monotherapy in BRCA−/− patients was presented in 2007; final results were published in 2009 [10]. PARP's stunning success against BRCA1 and BRCA2 breast cancers led to an explosion of research surrounding PARP inhibition, including a quest for its use in broader clinical applications (Table 2). Since then, a plethora of clinical trials have studied PARPis both as monotherapy and combination therapy. However, in early 2011, that research suffered two blows.
Table 2.
Clinical trials in progress for PARP inhibitors.
| Compound (alt. name) | Manufacturer | Clinical trials in progress | Targeted cancers | NCT identifiers |
|---|---|---|---|---|
| Iniparib (SAR240550, BSI-201) | Sanofi-Aventis | Phase I | Brain metastases, advanced solid tumors | NCT01551680, NCT01213381 |
| Phase II | Primary and metastatic breast, advanced or recurrent uterine, stage IV NSCLC | NCT01045304, NCT00687687, NCT01086254 | ||
| Phase III | Metastatic TNBC | NCT00938652 | ||
|
| ||||
| Olaparib (AZD2281, KU-0069436) | AstraZeneca | Phase I | Ovarian, fallopian tube, breast, HER and PR breast, metastatic TNBC, pancreatic, advanced and metastatic solid tumors, melanomas, Ewing's sarcoma | NCT00516373, NCT00707707, NCT00516724, NCT00515866, NCT00516802 |
| Phase II | Breast, HER and PR breast, ovarian, fallopian tube, prostate, gastric, colorectal, Ewing's sarcoma | NCT00494234, NCT01116648, NCT00679783, NCT01078662, NCT01583543 | ||
| Phase III | Platinum-sensitive, BRCA-mutated, relapsed ovarian | NCT01874353 | ||
|
| ||||
| Niraparib | Tesaro | Phase III | Platinum-sensitive ovarian, HER2, BRCA-mutated, breast | NCT01847274 |
|
| ||||
| Rucaparib (CO-338, AG-014699, PF-O1367338) | Pfizer | Phase I | Advanced solid tumors | NCT01009190 |
| Phase II | Primary and metastatic breast and ovarian, BRCA-mutated, fallopian tube, peritoneal | NCT00664781, NCT01074970, NCT01482715 | ||
| Phase III | Ovarian, fallopian tube, peritoneal | NCT01968213 | ||
|
| ||||
| Veliparib (ABT-888) | Abbott | Phase I | BRCA-mutated, numerous leukemias, lymphoma, medulloblastoma, pontine glioma, ependymoma, astrocytoma, advanced solid tumors, numerous stage II–IV gynecological and peritoneal cavity cancers, TNBC, gastric, locally advanced rectal, metastatic melanoma | NCT01853306, NCT00740805, NCT00946335, NCT01514201, NCT01123876, NCT01063816, NCT01434316, NCT01749397, NCT01145430, NCT01012817, NCT00989651, NCT01589419, NCT01366144 |
| Phase II | Breast, metastatic breast, TNBC, ovarian, fallopian tube, BRCA-mutated, colorectal, non-Hodgkin's lymphoma, metastatic pancreatic, recurrent NSCLC, primary peritoneal | NCT01149083, NCT01506609, NCT01009788, NCT01306032, NCT01540565, NCT01690598, NCT01051596, NCT01326702, NCT01489865, NCT01585805 | ||
|
| ||||
| AZD2461 | AstraZeneca | Phase I | Refractory solid tumors (activity against olaparib-resistant tumors) | NCT01247168 |
|
| ||||
| BMN673 | BioMarin Pharmaceutical | Phase I | AML, CLL, mantle cell lymphoma, myelodysplastic syndrome, advanced solid tumors, NSCLC, prostate, pancreatic | NCT01399840, NCT01989546, NCT01286987 |
| Phase III | Breast, BRCA-mutated | NCT01945775 | ||
|
| ||||
| CEP-9722 | Cephalon | Phase I | Mantle cell lymphoma, advanced solid tumors | NCT01345357 |
|
| ||||
| E7016 | Esai | Phase I | Advanced solid tumors | NCT01127178 |
| Phase II | Stage III and IV melanoma | NCT01605162 | ||
|
| ||||
| INO-1001 | Inotek Pharmaceuticals | Phase I | Melanoma | NCT00272415 |
|
| ||||
| MK-4827 | Merck | Phase I | Advanced solid tumors, CLL, T-cell-prolymphocytic leukemia | NCT0079502 |
|
| ||||
| Methoxyamine | Sigma Aldrich | Phase I | Numerous lymphomas | NCT01658319 |
PARP inhibitor clinical trials in progress as of Novennber 2013: 115 (status is unknown on some trials; those were not included in this total).
alt: Alternative; AML: Acute myelogenous leukemia; CLL: Chronic lymphocytic leukemia, NSCLC: Non-small-cell lung cancer; TNBC:Triple-negative breast cancer.
Data taken from [11].
First, a Phase III trial of iniparib (BSI-201) to treat metastatic, triple-negative breast cancer (TNBC) failed to prolong patient survival, despite promising Phase II trial results [12]. TNBC is clinically and pathologically similarity to BRCA1/2-mutated breast cancers in that both have very aggressive profiles, poor prognosis and limited treatment options [13]. Subsequently, Phase III development of olaparib (AZD-2281) to treat hereditary BRCA1- and BRCA 2-associated breast cancer was halted [7,14].
These seeming `failures' spawned greater scrutiny of both the products themselves as well as protocol criteria. Iniparib was deemed technically not to be a PARPi but rather a cysteine-binding poison (and is still being pursued as a chemotherapeutic). Heavily pretreated patient cohorts and the phenotypical heterogeneity of some cancers, particularly TNBC, were deemed to have contributed to differences observed in Phase II versus III testing, as well as variations in patient outcomes within the same trial. Ultimately, this led to a redoubled effort to learn more about PARP's precise mechanisms of action and a redirection of some clinical trials in what patient populations to target [13].
Lessons learned from PARPis
The ups and downs of intense PARP research are helping scientists identify and anticipate bumps in the road for developing targeted inhibitors in general. To summarize, science has learned that:
PARP proteins are not solely involved in DNA repair. They also play roles in transcription, telomere replication, cellular transport, NF-κB regulation and HSP90 expression [15,16]. Hints at those (and yet-to-be discovered) functions are seen outside PARP's catalytic region. Although the catalytic domain is conserved among all 18 members of the PARP family, dissimilarities in PARP's automodification domain and DNA binding domain distinguish each PARP from one another [10];
PARPis do more than bind the catalytic domains of PARP1 and PARP2. Because PARP1 has multiple domains that bind DNA damage, enzymatic activity can, in theory, be blocked without interfering with the catalytic site itself and without affecting essential functions of other PARP family members. This could increase specificity and decrease treatment side effects [13];
Not all PARPis are created equal. PARP must be inhibited by >90% to detectably inhibit DNA repair [17]. However, some PARPis work on tumors that are resistant to other kinds of PARPis. For example, AZD2461 (first Phase I trial recently completed) shows clinical activity on olaparib-resistant tumors [18]. This is due in part to, but cannot be fully explained by, structural differences. First- and second-generation PARPis are nicotinamide analogs, benzamides or substituted benzamines; their specificity and potency vary greatly. Many third-generation PARPis are derived from the 3-aminobenzamide structure; others are polycyclic lactams; most are competitive inhibitors [10,16]. Ongoing research into the structural and mechanistic aspects of PARP will hopefully clarify the reasons for these differences;
The `poisoning potency' of a PARPi depends on its strength in stabilizing PARP-DNA complexes – irrespective of the compound's catalytic inhibition. Thus, the extent to which PARP trapping occurs has a greater clinical effect on cell killing than enzymatic inhibition of PARP activity [19,20]. This is one plausible explanation for why various PARPis perform differently, even on the same cohort of patients [16];
The synthetic lethality that PARP inhibition confers (capitalizing on a weakness in the HR repair pathway) is not limited to BRCA1- and 2-deficient cancers;
Genetic deficiencies that confer high sensitivity to PARPis include deficiencies in XRCC2, XRCC3, RAD54 and H2AX [20]. Cancers containing PTEN1 and ATM deficiencies and microsatellite instabilities (as seen in colorectal cancers) also respond well to PARPis [16].
These discoveries not only fuel the fire for broader therapeutic applications [16]; they also provide clues regarding how to approach the development and use of other types of DNA repair inhibitors.
Double-edged sword of inhibiting multifunctional repair proteins
PARP proteins, like many other DNA repair proteins, are multifunctional. That characteristic cuts both ways. While inhibiting a multi-functional protein can affect multiple pathways and theoretically increase its tumor-killing ability, it may produce unanticipated results and/or increased toxicities.
Similar challenges and opportunities exist with checkpoint proteins, the sentries of DNA damage response. Due to their ubiquitous nature and multitasking abilities, inhibiting them could either cause great good or great harm – unless research can pinpoint how and when such inhibition would have the greatest therapeutic effect. For a full discussion of checkpoint inhibitors as monotherapy or combination therapy, see [21].
Overview of each pathway & inhibitors in development
Direct repair pathway
The direct repair (DR) pathway is unique in that only one protein is involving in performing a solitary, nonenzymatic process that repairs instead of replacing a damaged base. The sole protein involved, MGMT, removes one alkyl group from the O6 position of a damaged guanine base, such as generated by treatment with the clinical alkylating agent temozolomide (TMZ), and transfers it to an internal Cys residue in MGMT. The non-reversible reaction, which is greatest before late G1 [22], culminates with inactivation and degradation of the MGMT molecule. The stoichiometric reaction is driven by cells' ability to continually make more MGMT. To a lesser extent, MGMT performs the same function on the O4 position of thymine. Without MGMT repair, alkyl adducts would cause thymine mispairings during replication, leading to erroneous G:C-to-A:T transitions or strand breaks – necessitating the recruitment of other pathways to perform more complex repairs [3].
Overactivity of MGMT is responsible for chemoresistance; for example, >90% of recurrent gliomas show no response to a second cycle of chemotherapy. Conversely, inhibition of MGMT renders cancer cells sensitive to TMZ. Addtionally, MGMT promoter alkylation is a significant determinant in the sensitivity of drugs such as TMZ. There is abundant evidence linking the methylation of the MGMT promoter to loss of protein expression resulting in increased sensitivity to chemotherapeutic agents and to the prognostic outcome of patients treated. However, the role of MGMT promoter methylation in tumorigenesis and its utility as a prognostic bio-maker still needs further attention. Similarly, low MGMT expression appears to be a biomarker for slower tumor progression [22].
DR inhibitors in development & on the market
Many compounds initially thought to be MGMT inhibitors have proved to be checkpoint inhibitors instead. Only one true MGMT inhibitor, O6 benzylguanine (O6-BG), is currently on the market. In vitro studies of O6-(4 bromothenyl) guanine (PaTrin-2 or PAT), a pseudosubstrate inactivator of MGMT, show greater potency than O6-BG. However, it causes dose-limiting toxicities when administered with TMZ [23].
More novel approaches to MGMT inhibition are also being tried (Box 1) [22,24–27]. A Phase I trial of extended low-dose administration of TMZ to deplete MGMT prior to standard TMZ dosing is showing promise for patients previously resistant to TMZ. Oncolytic viruses (Adenovirus E1A protein, mutant HSV G207) show preclinical activity in inhibiting MGMT transcription [22]. For more details on MGMT inhibitors in development, see [22].
Box 1. MGMt inhibitors in development and clinical use.
Function(s)
Removes alkyl groups from the O6 position of guanine, and, to a lesser extent, the O4 position of thymine
Why it is a good target for inhibition
A discrete protein with a highly specialized function in one repair pathway
Methylating MGMT's gene promoter sensitizes cells to alkylating agents and IR
Related: MGMT methylation status appears to be a good prognostic biomarker
Challenges
MGMT inhibition affects both healthy cells and tumor cells
MGMT levels vary greatly among cancers and even within tumor cells
MGMT inhibition does not work if a deficiency exists in the MMR pathway
Other pathways can repair methylating damage in the absence of MGMT
Compounds being investigated
In clinical use: O6-BG; first drug developed as a chemosensitizer
In clinical trials: O6-BG + many other drug combinations; PaTrin-2
Under investigation: oncolytic viruses; extended low-dose TMZ (an alkylator) to deplete MGMT levels before administering standard-dose TMZ
Other notes: KU-60019 turned out to be a checkpoint (ATM/ATR) inhibitor
IR: Ionizing radiation; MMR: Mismatch repair; O6-BG: O6-benzylguanine; PaTrin-2: O6-(4 bromothenyl)guanine; TMZ: Temozolomide. Adapted with permission from [28].
Base excision repair
The BER pathway corrects single-base (nonhelix-distorting) damage caused by oxidation, alkylation, deamination and ionizing radiation (IR). If left unchecked, such damage would cause incorrect base pairings that would become mutagenic if transcribed [4]. BER consists of two subpathways; the activation of one or the other is predicated by first the cause and type of damage, second the type of abasic (apurinic, apyrimidinic) (AP) site generated in the first repair step [4] and third the cell cycle phase in progress when the damage occurs [29]. The short-patch pathway quickly repairs single-base damage during the G1 phase; the long-patch pathway handles lengthier repairs during S or G2, when resynthesis of two to eight nucleotides surrounding the AP site is required [29]. For details of BER pathway mechanisms, see [30].
Alkylating agents, platinating agents, cytotoxic antibiotics and taxanes create DNA lesions that the BER pathway normally repairs [31]. Thus, BER inhibition holds promise for potentiating the effects of those treatments. Inhibitors of four BER proteins that are either unique to this pathway, have very specific BER functions or are otherwise attractive candidates for inhibition are in development: APE1, Pol β, FEN1 and PARP (Table 3) [32–39].
Table 3.
Base excision repair inhibitors in development and clinical use.
| BER protein | Function(s) | Rationales for inhibiting | Challenges | Compounds being investigated |
|---|---|---|---|---|
| APE1 | Repair: prepares AP site to receive new nucleotide(s) Redox: maintains genes in their active, reduced state |
Overexpressed in many cancers Associated with resistance to alkylating and oxidizing agents No substitute exists for this protein |
Achieving specificity to solely inhibit the repair or redox function | Repair: |
| • Nonspecific: methoxyamine (binds to the aldehyde in the AP site of DNA); lucanthone | ||||
| • Specific: AR03; lnhibitor-1; ML-199 and analogs | ||||
| Redox: | ||||
| • E3330and analogs | ||||
|
| ||||
| Pol β | Resynthesizes nucleotides Removes certain blocking residues |
A rate-limiting step in BER Often overexpressed in tumor cells Associated with resistance to IR, bleomycin, some alkylating agents, cisplatin |
Difficult to develop an inhibitor specific to the polymerase domain that would not also inhibit polymerases involved in DNA replication Inhibiting its lyase function may be more lucrative |
>60 inhibitors identified; most lack specificity |
| Most promising so far (cell studies only): | ||||
| • Oleanolicacid | ||||
| • Edgeworin | ||||
| • Betulinic | ||||
| • Stigmasterol | ||||
| •NCS-666715 | ||||
| •NSC-124854 | ||||
|
| ||||
| FEN1 | Progressivity factor Helps polymerases synthesize nucleotides efficiently and accurately | Elevated in many cancers Inhibition creates unligatable DNAand makes cells hypersensitive to alkylating agents | Importance of protein in pathway could prove difficult in identifying inhibitors with lowtoxicity to normal cells | Afewhydroxyurea-based FEN1 inhibitors (cell studies only) |
|
| ||||
| PARP1 | Surveillance/damage sensor Assesses extent of damage; determines whether to signal apoptosis Helps decondense chromatin Recruits repair proteins to the damage site Facilitates repairs |
Uses NAD+to transfer ADP-ribose polymers onto specific acceptor proteins including itself; this modifies the protein's properties Although not essential to BER, PARPI's absence causes an accumulation of DNA damage that certain cancers cannot repair Inhibitors potentiate the effects of alkylating agents, platinating agents, topoisomerase 1 poisons, IR | Secondary mutations can correct for this repair deficiency, causing resistance to PARPis | Already in clinical use: |
| • Iniparib | ||||
| • Olaparib | ||||
| In clinical trials: | ||||
| • ABT-888 | ||||
| • AZD2461 | ||||
| •BMN673 | ||||
| • E7449 | ||||
| • INO-1001 | ||||
| • CEP-9722 | ||||
| • MK-4827 | ||||
| • Rucaparib | ||||
| • Velaparib | ||||
| • >115 clinical trials in progress for newer-generation PARPis and broader use of first-generation inhibitors | ||||
| Cell studies only: | ||||
| • PJ-34 | ||||
| •DPQ | ||||
AP: Abasic (apurinic, apyrimidinic); BER: Base excision repair; IR: Ionizing radiation; PARPi: PARP inhibitor.
APE1 is the only DNA repair protein that also regulates reduction-oxidation (redox) activities. Its redox functions affect DNA repair indirectly and influence many transcription factors involved in cancer promotion and progression [34]. In the repair process, APE1 activity creates special ter-mini to prepare the abasic site so that a polymer-ase can insert the correct resynthesized base [40]. APE1's redox functions help maintain transcription factors in a reduced, activated state so they can fold properly, bind to DNA and produce proteins the cells need. APE1 does this directly through a thiol/sulfite exchange and indirectly through a `redox chaperone' function that is still being characterized [31].
APE1 overexpression confers chemo- and radio-resistance [31] and is associated with shorter time to progression and poorer prognosis [34]. APE1 is dysregulated or upregulated in many solid cancers, including hepatocellular, prostate, pancreatic, ovarian, cervical, germ cell tumor, rhabdomyosarcoma and colon cancers [4,31,38]. Many other characteristics of APE1 make it a highly desirable target for inhibitor development. APE1's diverse activities offer multiple opportunities for inhibitor development to modulate multiple repair and signaling pathways that represent multiple cancer survival mechanisms [4,38,41–42].
In preclinical studies, blockade of APE1's repair functions potentiates the cell-killing abilities of many anticancer agents, including methyl methane sulfonate (MMS), H2O2, bleomycin, TMZ, melphalan, cisplatin, IR and gemcitabine [4,43–44]. Blockade of APE1's redox functions has numerous antiproliferative and antiangiogenic effects [41,43–45]. Redox inhibition also alters the tumor microenvironment, including downregulation of HIF1α, AP-1 and NF-κB [34]. Treatment with an APE1 redox inhibitor could prevent DNA binding of cytokine signaling. Very recent study results show that blockade of APE1's redox function blocks phosphorylation (and thus transcription) of STAT3 [46]. Thus, treatment with a STAT3 inhibitor (which directly blocks the DNA binding region of STAT3) plus an APE1 redox inhibitor has been demonstrated to cause synthetic lethality in human pancreatic and glioblastoma cell lines. A bonus is that APE1 redox blockade controls the signaling crosstalk that occurs between the tumor and the tumor micro-environment [34,46]. This could eventually provide a new treatment paradigm for hard-to-treat cancers.
BER inhibitors in development & on the market
Inhibitors of both APE1's repair and redox functions are in various stages of preclinical development. Some inhibitors originally touted to be specific for APE1 have turned out to be more properly `BER inhibitors' because they bind to the aldehyde of the AP site on DNA. An example is methoxyamine (MX, or TRC102). However, MX continues to be studied as a component of combination treatment for a variety of cancers. At the time of this writing, one Phase I trial had been completed and three more were in progress [47]. Other nonspecific APE1 inhibitors are actually topoisomerase poisons [48]. A number of investigators have identified several specific APE1 repair inhibitors; however, research has not yet progressed much past the cell culture stage [36–38,49–51].
E3330 and newer analogs show promise for specifically inhibiting APE1's redox functions. Initial indications are that these will be used in adults and children with acute lymphoblastic leukemia and other cancers [52].
The second BER protein that is a candidate for inhibition is Pol β. It is an attractive target for inhibition for three reasons: it performs both DNA resynthesis and removal of the blocking 5′-deoxyribose-5-phosphate (5′-dRP) residue in both short- and long-patch BER; its associated lyase activity is often rate-limiting in BER; and it is upregulated in many cancers, which contributes to resistance to IR, bleomycin, monofunctional alkylating agents and cisplatin [31].
Research into scores of potential inhibitors showed that early candidates lacked specificity. Interestingly, four naturally occurring compounds (oleanolic acid, edgeworin, betulinic acid and stigmasterol) appear to affect DNA repair only – not scheduled replicative activity [39]. However, none of those compounds are very potent.
Two new compounds, NCS-666715 and NSC-124854, show high potency at very low concentrations. Both are being evaluated in murine models as chemosensitizers for colorectal cancers. Notably, both can block both short- and long-patch BER without affecting APE1, FEN1 or DNA Ligase I activity, which theoretically would minimize collateral damage to healthy cells. Studies to date show that combining TMZ with either of these compounds blocks the growth of both mismatch repair (MMR)-proficient and MMR-deficient colon cancer cells in vitro and causes antitumor activity in vivo [33,35].
Although the NCS compounds are far from moving into clinical trials, they underscore the interactivity of multiple DNA repair pathways – and how the research of DNA repair inhibitors must adopt a broader `systems' approach because of that. Many colon tumors become resistant to alkylating agents, either due to MGMT overexpression, MMR deficiency or both. Both BER and MMR can repair mismatch pairs and other alkylation adducts that DR (MGMT) does not repair. However, if BER is inhibited and 8-oxoguanine (8-oxoG) adducts accumulate, the damage becomes lethal to cells deficient in the MMR proteins MLH1 or MSH2.
FEN1 is critical to DNA repair and replication. FEN1 is the major human endonuclease that recognizes and cleaves 5′ DNA flaps in long-patch BER; it also removes Okazaki primers in lagging strand DNA synthesis – approximately 50 million per cell cycle [53]. To perform this endonuclease function imprecisely or inefficiently results in DNA that is not ligatable, which delays cell replication and necessitates postreplicative repairs that endanger genomic stability [53].
FEN1 is elevated in many cancers, including gastric, lung, prostate, pancreatic, breast and brain cancers [53]. Cell studies demonstrate that lack of the FEN1 gene makes cells hypersensitive to alkylating agents [31]. All these reasons make FEN1 an attractive target for inhibition. Although its potential for broad therapeutic application has been likened to that of PARP [54], development of any FEN1 inhibitors is in only the very earliest stages, as finding specific compounds with inhibitory capacity at nanomolar concentrations has been elusive (Table 3) [33].
Finally, for BER, many PARPis are already in clinical use; trials are ongoing for second- and third-generation PARPis, as discussed earlier in this article [3,16].
Mismatch repair
The MMR pathway is the cell's main repair mechanisms for correcting base–base mismatches and repairing insertion and/or deletion loops formed during DNA replication [55]. Before the damage can become permanent or duplicated in future cell cycles, MMR's postreplicative damage control removes the DNA immediately surrounding the mismatch and replaces it with a newly synthesized segment copied from the daughter strand as a template. The MSH2–MSH6 complex attends to the repair of base substitutions and small mismatched loops, while the MSH2–MSH3 complex repairs both small loops and large-loop mismatches. Varying recognition complexes are formed based on the type of mismatch to be repaired. Notably, the repair is completed specifically on the new strand [55].
Deficiencies in MMR increase mutation rates in cells up to 1000-fold [55,56]. Mutations in four MMR genes (MSH2, MLH1, PMS2 and MSH6) predispose cells to a range of cancers, including hereditary nonpolyposis colon cancer [55]. However, up to 20% of sporadic cancers are due to MMR defects as well [57].
MMR inhibitors in development
Paradoxically, impaired MMR functionality fosters damage tolerance, which contributes to increased mutagenicity, tumor heterogeneity and chemoresistance [56]. One way to exploit the lack of one or more MMR genes is to create a synthetic lethality – to ensure that the damage is truly beyond repair. Studies showing that a high accumulation of oxidative stress induced in MMR-deficient cells can create such a synthetic lethality. A Phase II clinical trial is underway to test the efficacy of methotrexate on MSH2-deficient cells [55,58]. Cell studies show that a Pol β inhibitor can create a synthetic lethality in MSH2-deficient cells. Similarly, a Pol γ inhibitor can create a synthetic lethality in cells lacking MSH2 (Table 4) [55,59]. Both polymerase inhibitors create an abundance of 8-oxoG lesions [60]. The BER pathway would normally repair such oxidative lesions; but because those polymerase inhibitors would also affect BER, a synthetic lethality is created.
Table 4.
Mismatch repair inhibitors in development.
| MMR protein | Function(s) | Rationales for inhibiting | Challenges | Compounds being investigated |
|---|---|---|---|---|
| MLH1 | Scaffolding protein Damage sensor Helps determine the specific strand error | Can be hypomethylated to restore functionality Inhibition in deficient cells could theoretically cause a synthetic lethality |
MMR deficiencies cause or increase chemoresistance Intact MMR function is crucial for proper cell cycle checkpoint control | To restore functionality: |
| • FdCyd | ||||
| To create synthetic lethality in MLH1-deficient cells (cell studies): | ||||
| • Pol γ inhibitor | ||||
|
| ||||
| MSH2 | Damage sensor | Its damage-sensing ability can be bypassed by inducing a synthetic lethality | MMR deficiencies cause or increase chemoresistance Intact MMR function is crucial for proper cell cycle checkpoint control | In clinical trials: |
| • Methotrexate | ||||
| In MLH2-deficient cells (cell studies): | ||||
| • Pol β inhibitor | ||||
MMR: Mismatch repair.
MMR proficiency increases cells' sensitivity to alkylating agents, antimetabolites and fluoropyrimidines by 2- to 100-fold – enabling other cellular processes to arrest the cell cycle in G2 and later trigger cell death pathways [61]. Because hypermethylation of the hMLH1 gene promoter reduces promoter expression in many sporadic cancers, hypomethylation could restore MMR function, sensitizing cells to those classes of chemotherapeutics. Cell studies of fluoropyrimidine derivative, 5-fluoro-2-deoxycytidine (FdCyd), have demonstrated this potential utility [57].
Nucleotide excision repair
NER repairs bulky, helix-distorting lesions caused by UV irradiation and chemical mutagens that crosslink adjacent purine bases and form intrastrand adducts. Deficiencies in NER render cells sensitive to platinating agents, which attempt to arrest the cell cycle at G2 [62]. The success of this drug class is demonstrated most dramatically in the 95% cure rate of testicular cancer treated with cisplatin [63]. However, intact NER activity contributes to chemoresistance because it can repair damage inflicted by cisplatin, carboplatin and oxaliplatin [62].
More than 30 proteins compose the NER pathway, four of which appear to be unique to NER or drive rate-limiting reactions in NER, making those proteins key targets for inhibition [63]. The XPA protein of NER opens the DNA helix, forming a bubble of approximately 30 base pairs around the lesion. RPA and XPA stabilize the opened structure and recruit other proteins to the site, including two endonucleases (XPG and XPF-ERCC1) to perform the incision step [62–64]. RPA also activates ATR to halt cell cycle progression [62]. In short, without XPA, NER cannot occur [62–64]. Damaged-strand incision is the rate-limiting step for the pathway; thus, XPG, XPF, XPA, RPA and ERCC1 are the focus of virtually all inhibitor research that targets NER. The first three proteins appear to be unique to NER; ERCC1 has functions in other repair pathways, including HR [64]. For more details of the mechanisms involving NER activity, see Table 5 and [63].
Table 5.
Nucleotide excision repair inhibitors in development.
| NER protein | Function(s) | Rationales for inhibiting | Challenges | Compounds being Investigated |
|---|---|---|---|---|
| NER pathway† | UCN-0l1 and trabectedin affect multiple NER proteins Inhibition decreases the interaction of XPA and ERCC1, as well as other proteins with kinase activity F11782 isatopoisomerase inhibitor with yet-to-be-defined NER activity |
Complexity and importance of pathway in normal cells could complicate development and determination of therapeutic window | Phase I and II trials: | |
| • UCN-01 | ||||
| • Trabectedin | ||||
| Cell studies: | ||||
| •F11782 | ||||
|
| ||||
| XPA | Stabilizer Positions endonucleases for excision | Its only known role is in NER Not highly expressed; likely a rate-limiting repair factor |
Complexity of the network of protein interactions involved Chemical similarities in binding pockets of proteins complicates the development of protein-specific inhibitors |
Indirectly; see XPF/ERCC1 discussion |
|
| ||||
| RPA | Scaffolding protein Stabilizer | Is essential to NER RPA mutations are linked to development of cancer |
Complexity of the network of protein interactions involved Chemical similarities in binding pockets of proteins complicates the development of protein-specific inhibitors |
Cell studies: |
| • MCI13E and MCI13F | ||||
| • TRDL-505 | ||||
|
| ||||
| XPF/ERCC1 | Endonuclease | Its only known role is in NER Overexpressed in cisplatin-resistant cancers |
Complexity of the network of protein interactions involved Chemical similarities in binding pockets of proteins complicates the development of protein-specific inhibitors |
In silico: |
| • NERI01 | ||||
Noncatalytic aspects of NER (protein-protein and protein-DNA interactions, nnodulation of transcription regulation) nnay be nnore viable targets for inhibition, as seen in the effect ofachimeric IgGI nnonoclonal antibody on reducing XPF/ERCC1 expression.
MuItiple proteins within NER are inhibited; which ones are still being researched.
NER: Nucleotide excision repair.
Adapted with permission fronn [28].
Mutations at the XPA binding site within ERCC1 can prevent the interaction between the two proteins; even more notably, this also prevents recruitment of XPF, which normally binds almost all of its residues to ERCC1. Upstream blockage of ERCC1-XPF activity via XPA inhibition has been shown to sensitize cancer cells to IR [64].
NER inhibitors in development
As exciting as these findings are, research into NER inhibition is still in early stages – and most so-called NER inhibitors appear to be rather nonspecific (Table 5) [62,64–65]. One compound originally thought to be a NER inhibitor (7-hydroxystaurosporine [UCN-01]) is actually a checkpoint inhibitor [64]. When the current article was written, 19 clinical trials (seven Phase II; 18 Phase I) had been completed to test UCN-01 as a monotherapy or combination therapy for a variety of recurrent or relapsed blood-based and solid tumors [66]. Another compound, F11782, is a topoisomerase I and II inhibitor, but it also appears to have some capacity to inhibit NER's helicase or incision step [67].
Another compound in clinical trials is trabectedin (also known as Ecteinascidin 743 or ET-743), which shows activity against the transcription-coupled (TC) NER subpathway (active during transcription) and HR pathway deficiencies. Although trabectedin's mode of action is still being elucidated, this alklyloid's DNA binding in the minor groove bends DNA toward the major groove in a fashion that seems unique to this compound. The net result is interference with several transcription factors, DNA binding proteins and repair pathways. Whether direct inhibition of a specific DNA protein occurs is in question. If trabectedin is specific to TC-NER, then it must affect that subpathway's unique damage sensors (CSA, CSB). However, to date, that has not been confirmed. We do know that TC-deficient cells and XPG-deficient colorectal cancer cells show resistance to trabectedin [68]. At the time of this writing, 43 clinical trials, including many Phase II trials, were underway for the use of trabectedin in numerous cancers [69].
MCI13E, a novel isoborneol haloacetate, shows promise in cell studies as an RPA inhibitor and a sensitizer to platinating agents. Although it irreversibly binds to RPA, the half-life of this experimental compound is relatively short-lived compared with platinating agents, which take at least 48 h to produce effects. So, current studies are pursuing a model of sequential treatment, administering cisplatin 24 h before adding the inhibitor [62]. TDRL-505, a reversible inhibitor that binds to a different site on RPA, is also under study. Both compounds are still being characterized to determine their mechanisms of action [62].
ERCC1 plays a unique role in NER. It is recruited by XPA, but ERCC1's functionality rests in its heterodimer form with XPF [65]. The ERCC1–XPF heterodimer excises the damaged stretch of DNA [33]. Thus, separating the protein–protein interactions of ERCC1 with XPA and XPF and identifying the domains essential to dimerization are topics of intense research. Blocking ERCC1–XPF is tantalizing, but the crystal structure for this domain of human XPF is yet unknown, and a number of endonucleases with similar divalent cation-based cleavage mechanisms complicate the search for specific inhibitors [70].
In silico models have uncovered a compound, NERI01, which is more flexible and establishes more hydrogen bonds with ERCC1 than all other compounds that generated to date from high-throughput screening [65]. Despite the essentially blank slate for ERCC1 inhibitors, research has correlated ERCC1 overexpression with cisplatin resistance [71]; and ERCC expression is emerging as a predictive biomarker of patients who will respond to platinum-based chemotherapeutics [64].
Repair of DSBs
A single unrepaired DSB is highly toxic and can lead to aneuploidy, genetic aberrations or cell death [72]. Such damage can occur naturally when topoisomerases uncoil DNA or be induced by IR or chemotherapeutics. The most common occurrence is when replication forks stall and break at the site of unrepaired DNA lesions [28]. Two main pathways repair DSBs. NHEJ inhibition performs repairs in less than a half hour; in contrast, the time-intensive work of HR takes many hours [73]. Inhibition of either or both is an important lynchpin in treating cancers, as unrepaired base damage and SSBs have their last hope for repair via one of these pathways. However, despite the plethora of repair proteins employed in each pathway, therapeutic inhibition poses many challenges.
Homologous recombination
The biggest gun in the arsenal of repairing DSBs is HR, as it accurately corrects the most serious and complicated forms of DSB damage. HR operates predominantly during the S and G2 phases of the cell cycle so that it can find a large area of homology on a sister chromatid to use as a template for resynthesizing damaged or lost bases [74,75].
HR repair can proceed via one of several subpathways [76], but they all follow the same general steps: resect DNA ends to generate an overhanging area of ssDNA; form a filament on the end of the overhang to search for a large area of homology on the sister chromatid; `invade' the filament onto the homologous area; form a DNA heteroduplex (a D-loop), created from displacing the invading strand; slide or migrate the D-loop to read the area of homology; extend the overhang as new nucleotides are generated past the original break point; resolve the Holliday junction that forms as the D-loop pushes along the border between hetero- and homo-duplex during resynthesis; migrate the repaired ends toward each other; and restore the duplex strand [28]. For a review of HR and its subpathways, see [72].
HR activity is dysregulated in many cancers. A defect in one or more of its proteins may impair or upregulate it, or loss of an upstream regulatory protein in another pathway of DNA damage recognition and repair may cause compensatory HR upregulation. This article has already discussed how loss of both BRCA1 or 2 alleles impairs HR; to compensate, NHEJ is used to repair DSBs. However, inhibition of PARP, which is not directly associated with either DSB repair pathway, can create an accumulation of damage that NHEJ cannot repair – effectively killing BRCA-deficient cancer cells [76].
Two crucial players in HR are the RPA complex and Rad51. RPA enters early in the repair process; it stimulates the unwinding of damaged DNA [72,77], then protects and stabilizes the segment that is resected as a single strand. However, RPA must be replaced by Rad51 in order for repairs to proceed [72]. Thus, defects, mutations in (or inhibition of) RPA can stall HR repair [78]. Because RPA has a common cleft for binding many types of proteins, early efforts are underway to inhibit RPA's protein–protein interactions without perturbing the binding of RPA to ssDNA [79].
Rad51 initiates strand exchange by complexing with other proteins to form a filament, the structure that finds and reads an area of homology so the right nucleotides can be resynethsized to repair the damage [56]. Post-translational modification of mediator proteins involved in HR repair highly regulate the balance of RPA and Rad51 so that repair is not inhibited and disassociation does not occur before Rad51's work is complete [72]. Rad51 overexpression is associated with breast and pancreatic cancers, non-small-cell lung cancer and leukemias [80].
cAbl is a nonreceptor tyrosine kinase that is activated by ATM. As such, it is not truly HR-specific, but the combination of ATM and cAbl affect Rad51 induction [81].
HR inhibitors in development & on the market
Efforts to modulate dysfunctional HR in malignancies are in their infancy. Many compounds investigated lack HR specificity. For example, mirin, which inhibits the MRE11–RAD50–NBS1 (MRN) complex (HR's damage sensor), also inhibits ATM and downregulates NHEJ [82]. With the exception of RI-1 (which has been tested in cell studies only) [82], small molecules that directly inhibit specific HR proteins are not in development (Table 6) [80,82–84].
Table 6.
Homologous recombination small-molecule inhibitors in development.
| HR protein | Function(s) | Rationales for inhibiting | Challenges | Compounds being investigated |
|---|---|---|---|---|
| Rad51 | Catalyzes the homology search Initiates DNA strand invasion and DNA strand exchange | The heart of HR activity | Incomplete characterization of Rad51's structure Determining whether direct or indirect inhibition would be more effective Unknowns regarding its activity (e.g., what triggers its nuclear translocation) | Cell studies only: |
| • RI-1 | ||||
|
| ||||
| RPA | Damage sensor, stabilizer Recruitment site for proteins involved in DNA replication, repair, recombination, and checkpoint activation | Essential for HR to happen | Common cleft/binding site for many proteins; difficult to isolate what would inhibit just DNA repair | Purified protein studies only: |
| • Compound 4 | ||||
|
| ||||
| Proteins that inhibit HR activity indirectly † | ||||
|
| ||||
| cAbl | Appears to be a decision maker, determining if damage is too extensive to be repaired | Inhibits Rad51's DNA strand exchange activity, which stalls HR Is activated by IR and alkylating agents A deletion or translocation on the gene promotes tumorigenesis cAbl-deficient cells are resistant to IR and other DNA-damaging agents |
Participates in many cellular processes Inhibitors of cAbl also inhibit cKIT and possibly other tyrosine kinases; not specific to HR Also interacts with DNA-PK (in NHEJ pathway) | In clinical use: |
| • Imatinib (Gleevec); available to treat CML since 2001; in trials for treating other cancers | ||||
| • Dasatinib | ||||
| • Nilotinib (Tasigna) | ||||
| • Bosutinib | ||||
|
| ||||
| PARP1 | Surveillance/damage sensor Assesses extent of damage; determines whether to signal apoptosis Helps decondense chromatin Recruits repair proteins to the damage site Facilitates repairs |
Uses NAD+ to transfer ADP-ribose polymers onto specific acceptor proteins including itself; this modifies the protein's properties PARP1 inhibition causes accumulation of DNA damage that collapses replication forks; cancers deficient in HR cannot repair such damage Inhibitors potentiate the effects of aklylating agents, platinating agents, topoisomerase 1 poisons, IR |
Secondary mutations can correct for this repair deficiency, causing a resistance to PARPis | PARPis are available to treat familial breast cancers and other BRCA-like cancers >110 clinical trials in progress for second- and third-generation PARPis and broader use of first-generation inhibitors |
|
| ||||
| HSP90 | Facilitates the correct folding, maturing and stabilizing of many proteins into their active form | Protects cells under stress conditions; upregulated in cancers Inhibition triggers ubiquitination Inhibition disrupts multiple pathways: blocks all major hallmarks of cancer; triggers ubiquitination; decreases Rad51 levels, thwarting HR |
Difficult to produce Inhibitory activity still being characterized; may inhibit one or more checkpoint kinases |
In Phase I and II trials: |
| • 17-DMAG | ||||
| • Alvespimycin (KOS-1022) | ||||
| • AT13387 | ||||
| • AUY922 | ||||
| • CNF2024 (BIIB021) | ||||
| • Debio 0932 (CUDC-305) | ||||
| • DS-2248 | ||||
| • Ganetespib (STA-9090) | ||||
| • KW-2478 | ||||
| • MPC-3100 | ||||
| • PU-H71 | ||||
| • Retaspimycin (IPI-504) | ||||
| • SNX-5422 | ||||
| • XL-888 | ||||
| In Phase III trials: | ||||
| • Tanespimycin (KOS-953; 17-AAG) | ||||
| Other candidates are in preclinical studies | ||||
Data on all PARP and HSP90 inhibitors in clinical trials are from [11].
No direct inhibitors of HR proteins have been found/developed yet. The proteins in this section inhibit HR activity indirectly by modulating DNAdamage response mechanisms, protein-protein interactions or other mechanisms.
CML: Chronic myelogenous leukemia; HR: Homologous recombination; IR: Ionizing radiation; NHEJ: Nonhomologous end joining.
Adapted with permission from [28].
Rad51 inhibition can be achieved either by directly blocking its recombinase activity or by attempting to interfere with Rad51's interactions with other proteins that form the filament. RI-1 appears to do the former by inhibiting the polymerization of Rad51 onto ssDNA, which, in turn inhibits monofilament formation. Studies with cancer cell lines indicate RI-1 has a synergistic effect on mitomycin activity. Other potential Rad51 inhibitors appear to work indirectly: for example, by activating the proapoptopic JNK and p38-MAPK pathways or by inhibiting ATPase activity. Such compounds have not progressed beyond identifying them from high-throughput screening of chemical libraries [80].
Three HR-related proteins show promise in indirectly inhibiting HR for anticancer activity: PARP (already discussed), cAbl and HSP90. Inhibitors of cAbl and HSP90 can arrest the cell cycle to the point that unrepaired damage triggers apoptosis.
Three classes of cAbl inhibitors exist, their primary distinction being where they bind to the kinase. Nine kinase inhibitors are currently US FDA approved, but not all of them act on cAbl [84,85]. Mutations at or distal to the drug-binding site can either cause direct loss of interaction with the drug or conformational changes that obviate kinase autoinhibition, respectively. Second-generation inhibitors, including dasatinib and bosutinib, are overcoming these problems (Table 6). However, a few mutations still do not respond to those drugs, either; so hybrids of the two drugs are in early development, as are other compounds that target alternative binding pockets [84].
HSP90 is a molecular chaperone that helps more than 200 proteins fold to their correct conformation; 48 of those proteins are associated directly with oncogenesis [86]. Inhibition of HSP90 decreases Rad51 activity and prevents multiple checkpoint proteins from being activated, both of which can delay cell cycle progression to the point where cumulative damage triggers apoptosis [83,87]. HSP90 inhibition can simultaneously disrupt tumors' growth signals, resistance to apoptosis, unregulated replication, neoangiogenesis and tissue invasion/metastasis [88].
Four classes of HSP90 inhibitors are in development today. Newer analogs of geldamycin, the first HSP90 inhibitor studied [83], include 17-DMAG, restapimycin (IPI-504), and tanespimycin. Of the purine and purine-like analogues, PU-H71 is also being tested as a predictive indicator of tumor dose response [83]. The third class of HSP90 inhibitors is resorcinol derivatives, including ganetespib, NVP-AUY922, KW-2478 and AT13387. Finally, SNX-5422, a dihydroindazolone derivative, is in Phase I and II trials (Table 6) [83].
Nonhomologous end joining
NHEJ represents the simplest and fastest mechanism for repairing DSBs [76]. Active throughout the entire cell cycle but especially in G0/G1, NHEJ directly rejoins the two severed DNA ends with minimal end processing, regardless of sequence homology. Such activity may be preferred in G0/G1, as much of the genome is non-coding – and HR's involvement at those phases could cause deletions, duplications, misalignments and crossovers [89]. Nonetheless, NHEJ can lose one to 20 nucleotides from either side of the DSB junction, causing erroneous repair [76]. Interestingly, NHEJ repairs the majority of damage that IR causes, even though IR damage rarely produces blunt, ligatable DNA [90].
Investigators have much to learn about NHEJ and its subpathways; however, all forms of NHEJ follow five general steps: encircle the DNA/detect and tether DSB ends together; process and remove end groups that cannot be ligated; process ends to make them ligatable; resynthesize nucleotides; and seal the break [76,86]. For details of all the proteins involved in NHEJ, see [91]. Within the framework of NHEJ's general steps, proteins in three steps appear to be unique to that pathway, and, as such, are potential targets for inhibition.
NHEJ inhibitors in development
Like MMR, diminished or defective NHEJ functioning leads to increased risk of cancer, particularly lymphoid malignancies [92]. Dysfunctional NHEJ also results in damage tolerance and chemoresistance [76]. Thus, counterintuitively, intact NHEJ function is linked to better prognosis or treatment response [92]. However, inhibiting NHEJ and thus forcing cells to perform DSB repair via the more time-intensive HR pathway could induce a synthetic lethality in HR-deficient tumors. This and other technical issues are hurdles yet to cross in today's forays into NHEJ inhibition. Indirectly modulating NHEJ activity through other means may ultimately prove to be at least as effective as a direct approach [92].
Regarding direct inhibition of NHEJ proteins, the most promising candidate is the catalytic subunit of DNA-PKcs (Table 7) [73,93]. Cells contain an abundance of DNA-PKcs, a nuclear serine/threonine kinase that is a prerequisite for establishing a functional NHEJ complex. Through complex autophosphorylations on 20 known sites, DNA-PKcs determines how/when to engage and disengage from DNA; regulates end access; modulates how damage is constrained to NHEJ [73]; is involved in telomere protection [94] and interacts with other DNA repair proteins, including cAbl, HSP90, PARP1 and H2AX, to name a few [90]. Overexpression of DNA-PKcs correlates with radioresistance in oral squamous cell carcinoma, lung carcinoma and esophageal cancer [95]. Thus, chemical inhibition of DNA-PK can enhance HR.
Table 7.
Nonhomologous end joining inhibitors in development.
| NHEJ protein | Function(s) | Rationales for inhibiting | Challenges | Compounds being investigated |
|---|---|---|---|---|
| DNA-PK | Processes incompatible ends so they can be ligated | Appears to be unique to NHEJ Essential for NHEJ activity | Increases DNA damage tolerance Causes chemoresistance Predisposes cells to autosomal recessive disorders and malignancies, especially lymphoid cancers | Nonspecific (inhibit PI3K): |
| • Wortmannin LY294002 DNA-PK-specific (cell studies only): | ||||
| • NU7441 | ||||
| • NU7026 | ||||
| Phase I studies: | ||||
| • CC-115 | ||||
| • CC-122 | ||||
|
| ||||
| PNKP | Processes incompatible ends so nucleotidescan be replaced and the termini ligated | Appears to be unique to NHEJ | Increases DNA damage tolerance Causes chemoresistance Predisposes cells to autosomal recessive disorders and malignancies, especially lymphoid cancers | Cell studies only: |
| • A12B4C3 | ||||
|
| ||||
| Ligase IV | Seals nicks in final repair step | A rate-limiting step Cannot work in the absence of XRCC4 | Increases DNA damage tolerance Causes chemoresistancePredisposes cells to autosomal recessive disorders and malignancies, especially lymphoid cancers | Murine models: |
| • SCR7 | ||||
Indirect ways of modulating NHEJ functionality are being explored as more viable options than direct inhibition. Examples: overexpression of a truncated form of XRCC4 can inhibit NHEJ by interfering with DNA Ligase IV; DNA-PKcs can be inhibited indirectly by inhibiting EGFR or by inhibiting ATM (by using miRNA or small-molecule inhibitors); topoisomerase inhibitors prevent NHEJ repair proteins from gaining access to areas of damage; and epigenetic factors (methylation of gene promoters) are under investigation as well. It is yet unknown how or if these findings can translate into future clinical use.
NHEJ: Nonhomologous end joining.
Adapted with permission from [28].
The first foray into DNA-PK inhibition started with wortmannin, a noncompetitive inhibitor of the related family of PI3Ks. The first nonspecific competitive PI3K inhibitor identified was LY294002, a morpholine derivative of quercitin [90]. Although its relative instability and high toxicity did not lend itself to use in humans, NU7441, a more potent and DNA-PK-specific inhibitor, was developed from it [96]. NU7026 likewise shows similar specificity in cell studies [94]. However, the only two DNA-PK inhibitors in clinical trials are dual-action inhibitors that potentially represent two first-in-class drugs. CC-115 is a DNA-PK and TOR inhibitor; CC-122 is a pleiotropic pathway modulator [11]. Additionally, the oral pan-class I PI3K inhibitior NVP-BEZ235 is in Phase I/II clinical trials [97].
PNKP, a protein active in NHEJ, processes incompatible termini to prepare them for nucleotide resynthesis and strand ligation. Interestingly, PNKP can process blunt-ended and 3′-overhanging termini; APE1 is relatively ineffective on the former and cannot process the latter, so PNKP can handle termini processing that the BER pathway cannot. In addition, emerging evidence exists for PNKP participation in an APE1-independent form of BER repair [92]. The one PNKP inhibitor currently being investigated is a polysubstituted imidopiperidine compound (A12B4C3) that is a noncompetitive but specific binder of PNKP. Cell studies indicate that A12B4C3 is a chemosensitizer to topoisomerase inhibitors [98].
Patients with intrinsically low levels of Ligase IV are radiosensitive. The one inhibitor being tested preclinically against it is SCR7; it appears to selectively interfere with Ligase IV–DNA binding [95]. Such disruption in sealing DSBs leads to accumulation of unrepaired breaks, activation of ATM and, ultimately, an intrinsic pathway of apoptosis. Other potential Ligase IV inhibitors have been modeled in silico [99]. See Table 7 for more details of all these inhibitors in development.
DNA inhibition as treatment for chemotherapy-induced peripheral neuropathy
Up to 90% of all cancer patients experience persistent chemotherapy-induced peripheral neuropathy (CIPN) [100,101]. The severity of CIPN can result in treatment delays, dose modifications or discontinuation of antineoplastic drugs [100,102]. A `perfect storm' of factors makes sensory neurons especially susceptible to damage: they are nondividing cells with high metabolic activity; by residing outside of the blood-brain barrier, they are exposed to higher amounts of agents that cause oxidative stress or direct DNA damage; and gene transcription and translation are much higher in neurons than other cells, so damage to mitochondrial DNA is particularly harmful to neurons. Collectively, this makes neurons very susceptible to functional damage [100,103].
Many potential treatments for CIPN, including antiepileptics and antidepressants, have had little to no effect in alleviating CIPN's symptoms, let alone reversing neuronal damage. Inconclusive or limited evidence exists for the efficacy of topical anesthetic creams, antioxidants, nutraceuticals, certain ion channel modulators or modalities (acupuncture, magnetic stimulation and electrostimulation) [100–101,104–105].
However, molecular characterization of how DNA-damaging agents affect neurons can reveal how to treat CIPN effectively [100,106]. Interestingly, the primary repair pathways in peripheral nerves are NER and BER [107–109]. Recent evidence suggests that modifying DNA repair pathways in CIPN models has an effect on a variety of neuropathic markers. Downregulation and inhibition of DNA repair elements may have adverse effects on sensory neurons [110] – but selectively upregulating a DNA repair protein could possibly alleviate CIPN.
Evidence for this exists in multiple forms. For example, in mouse models, the significant increase in thermal and nociceptive responses from oxaliplatin can be prevented by administration of antioxidants (flavonoids) [111]. The severity of CIPN correlates with dosing of platinating agents when NER is dysfunctional: mice deficient in XPA and XPC accumulate more platinum adducts in sensory neurons than wild-type mice given the same cisplatin dose [112]. However, cisplatin-induced increases in cell death and decreases in capsaicin-evoked release of CGRP in sensory neuronal cultures can be attenuated by overexpression of repair-competent APE1, an important BER endonuclease [113]. Other studies indicate that selectively enhancing the repair function BER's endonuclease, APE1, can prevent or alleviate CIPN [106].
However, modulating DNA repair components is not a clear-cut issue of selective upregulation. Inhibition of certain DNA repair elements may actually have a positive effect on sensory neurons. In a mouse model, concurrent administration of cisplatin or oxaliplatin with an experimental PARPi (Compound 4a) attenuated allodynia and hyperalgesia [114]. However, PARP may interfere with the activity of APE1 when significant DNA damage is present [115]. Additionally, PARP expression can stimulate or inhibit many hallmarks of cancer besides DNA repair [15].
The ongoing challenge is to find a laser focus for ameliorating CIPN without increasing tumor cells' survival capabilities. Modulation of DNA repair elements to treat CIPN is an emerging field. Ongoing studies are investigating diagnostic markers, molecular mechanisms, drug comparisons and potential treatments for CIPN [116–118]. Development of an effective small-molecule DNA repair inhibitor would be a first-in-class drug for neuropathic pain, which could change both survival and quality-of-life outcomes for many cancer patients.
Changing face of R&D in DNA repair inhibition
The potential clinical utility of DNA repair inhibitors is attractive, so several companies solely focused on DNA repair inhibition have emerged in recent years. Here are the ones that have been in the news for their research.
Inotek Pharmaceuticals Corporation (MA, USA; and Israel) is working to produce a line of PARPis, which are in late preclinical stages of development [119]. Its pipeline also includes a reactive oxygen species inhibitor and other classes of investigative drugs.
ApeX Therapeutics (IN, USA) is developing inhibitors to the DNA repair protein Ref1/Ape1, to treat cancers as well as other diseases involving pathological neovascularization [52]. It has several inhibitors in late preclinical development.
Tracon Pharmaceuticals (CA, USA) has three Phase I trials in progress for its lead compound, TRC102, which is showing promise in reversing BER-generated resistance to alkylator and antimetabolite chemotherapy. In addition, Tracon is developing TRC105, an antiangiogenic monoclonal antibody being tested as both monotherapy and combination therapy for a variety of solid tumors as well as macular degeneration (Phase I and II clinical trials) [120].
Sentinel Oncology (Cambridge, UK) is developing highly selective CHK1 and PI3K-mTOR inhibitors, as well as a `targeted synergy' vehicle for delivering and specifically activating drugs in the hypoxic microenvironment uniquely found in solid tumors [121].
The DNA Repair Company (MA, USA) is profiling all DNA repair pathways in tumor samples from registries. By determining which pathways are `on' or `off', they are generating antibodies that can be used in the future to test patients to determine what drugs they will most likely respond to. The company's longer-range goal is to screen for small-molecule inhibitors of all DNA repair pathways [122].
Mission Therapeutics (Cambridge, UK) concentrates on the `backside' of DNA damage response and repair: ubiquitination and related processes. In certain cancers, mutations in genes or activation of oncogenes (e.g., c-myc) prevent cells from disposing of misfolded or damaged proteins; the concentration of proteins involved in the cell cycle is also deregulated [123]. Among the dozen potential targets of ubiquitin inhibition that Mission Therapeutics is investigating are E2 conjugating enzymes, which affect the cyclins involved in cell cycle checkpoints, p53 (a tumor suppressor protein) and NF-κB (the transcription factor involved in inflammation and immune responses) [124]. E2s are relatively nonspecific, have many yet-unstudied functions and are not classically druggable (therefore, difficult to inhibit). Despite those challenges, the reversible process of ubiquitination makes inhibition of deubiquitinating enzymes a tantalizing target [123].
It is interesting that this is relatively short list. However, research efforts with DNA repair inhibitors within large multidivision pharmaceutical companies remain publicly unknown.
Future perspective
Emerging methods of molecular analysis to better manage a patient's disease or predisposition to disease are moving us closer to the reality of truly personalized medicine. Concurrent development of DNA repair inhibitors and companion testing for their targets is growing. In just the past 2 years, the FDA approved four cancer drugs for use in patients whose tumors have specific genetic characteristics that are identified by a companion diagnostic test [125].
Finding specific biomarkers poses multiple challenges because variant alleles may create different effects, based on tumor types, patient populations, treatment regimens and different stages of illness. Testing altered gene expression plus compensatory mechanisms can more accurately convey DNA repair activity in a given tumor [126].
Sifting through such massive data to determine whether a particular gene, its upstream regulators or mediators, or SNPs are clinically relevant as prognostic or predictive biomarkers is a daunting task. Next-generation genomic sequencing and accelerated data analyses of cancer tissues are providing ways to identify the molecular drivers of individual tumors. In the USA, Foundation Medicine [127] is doing that to provide actionable genomic information for physicians in choosing optimal treatments for individual tumors. Building a repository of such information may uncover new biomarkers as well as identify off-label treatments that could work when others fail (or when conventional treatment options are limited, such as with rare cancers).
On the international front, efforts such as the Gene-PARE project are underway. By analyzing its huge biorepository, it can validate tests that can predict which patients with specific genetic variants would be most likely to develop adverse responses to radiation treatment [128].
To offer widespread benefit, biomarker tests need to be reliable, readily available, able to be run using existing clinical laboratory technology [126], and fast enough to provide a timely turnaround without delaying the start of treatment. Advanced screening procedures and more careful selection of patient cohorts for clinical trials will help ensure that the outcomes of those trials accurately reflect the selected intervention's anticancer capabilities.
In the near future, comprehensive molecular characterization of DNA repair proteins will provide greater understanding of the crosstalk between DNA repair functionality and genetic stability. The REPAIRtoire database seeks to facilitate this. It is the first database of its kind to categorize all DNA repair proteins and repair intermediates, all diseases associated with DNA defects, all known DNA lesions linked to environmental mutagenic and cytotoxic agents, and all transformations that occur during the DNA damage response and repair process [129].
Better characterization of DNA repair proteins will lead to discoveries of more effective repair inhibitors and more efficient cancer-killing, synthetically lethal therapeutic combinations. Highly localized radiotherapy will also play an increasing role in combination or sequential treatment. For example, sequential treatment of multiple myeloma with a thymidylate synthase inhibitor followed by nanoirradiation by an Auger electron-emitting thymidine analog shows promise in eradicating multiple myeloma stem cells [130].
Also, treatment regimens will be more finely tuned to the stage of cancer. For example, as cancer progresses, its microenvironment becomes hypoxic, causing replicative stress, disrupting DNA synthesis and downregulating DNA repair [76], which makes cancer cells increasingly prone to DNA mutations. Thus, in advanced cancers, checkpoint inhibitors may be more effective than DNA repair inhibitors. Interestingly, chronic hypoxia often makes tumors more sensitive to radiotherapy [131].
Pinpointing what treatments will work best and when presents both opportunity and challenge: researchers must demonstrate not only that inhibitors are safe as single agents, but also that they augment DNA damage when given with chemotherapy or radiotherapy. After the efficacy of combination therapy is established in preclinical studies, the proper dosing and regimen in clinical trials is crucial to determine. With the addition of DNA repair inhibitors, standard chemotherapy could become more effective but also more toxic. The ultimate goal would be to be able to reduce the dose of chemotherapy in combination with DNA repair inhibitor and achieve more profound effects on tumor regression. Finding ways to fast-track combination-treatment clinical trials will be imperative in bringing discoveries from bench to bedside [24]. From an economic standpoint, researchers will also be increasingly involved in showing how the development of DNA repair inhibitors is highly effective stewardship of R&D resources and patient expenses [132].
Cancer cells can overcome genotoxic effects in at least three ways: by compensatory DNA repair mechanisms, by developing damage tolerance; and/or by reversing a genetic or epi-genetic defect. Epigenetic research is just starting to scratch the surface of the potential for more options for DNA repair inhibition, such as hyper- or hypo-methylation of gene promoters [131]. In the next 5–10 years, many relevant discoveries regarding cancer's behavior, thus, treatment are expected to emerge from epigenetic studies. In the meantime, intense research efforts to develop DNA repair inhibitors continue (Figure 1).
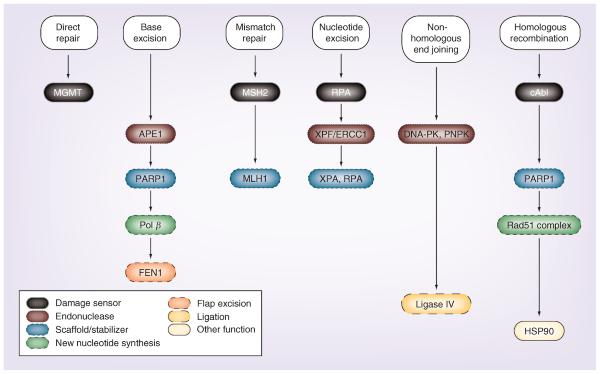
Figure 1.
current targets of nuclear DNA repair pathway proteins and their functions.
Conclusion & future perspective
Research and clinical trials have validated the therapeutic importance of using specific DNA inhibitors, primarily in combination with DNA-damaging drugs. Unanticipated issues can occur when trying to directly inhibit steps in some DNA repair pathways, but a greater understanding of pathway crosstalk is leading to `back-door' routes of inhibition. Globally, perhaps the biggest technical challenge in developing DNA repair inhibitors is target specificity, as a repair protein's active sites are often relatively small and are buried deep within the protein's tertiary structure [33]. In some cases, inhibitory binding to regions outside of the catalytic site can also modulate protein function, and some next-generation inhibitors (particularly kinase inhibitors) are seeking to exploit such divergent binding pockets. To provide the greatest therapeutic benefit from these therapies, the development of biomarkers is crucial in helping establish which patient populations are candidates for, and will respond best to, use of DNA repair inhibitors as part of their therapeutic regimen. The concurrent development of DNA repair inhibitors and biomarkers will enable earlier detection and treatment of cancers and will move us closer to delivering truly personalized medicine.
EXECUTIVE SUMMARY.
Inhibition & synthetic lethality
Development of DNA repair inhibitors is a burgeoning field of research.
Currently the most widely used DNA repair inhibitors block steps in the direct repair and base excision repair pathways.
Overview of each pathway & inhibitors in development
Multiple targets for DNA repair inhibition are discussed, as well as challenges and lessons learned in their development.
Pathway crosstalk, overlap and multifunctional proteins complicate the development of DNA repair inhibitors, but also provide opportunities for wider application, such as the tremendous success seen with PARP inhibitors.
Counterintuitively, the proficiency of some DNA repair pathways helps ensure sensitivity to chemo- and radio-therapeutics; for example, an intact mismatch repair pathway increases cells' sensitivity to many chemotherapeutics by 2–100-fold.
Although direct inhibition of a repair pathway is not always feasible, indirect inhibition and creating synthetic lethalities can be effective. Both approaches are discussed in this review.
Changing face of R&D in DNA repair inhibition
Concurrent development of biomarkers for cancers will aid in earlier detection and more effective treatment planning.
Acknowledgments
Financial & competing interests disclosure MR Kelley is the Chief Scientific Founder and consultant for ApeX Therapeutics, a company that has licensed intellectual property from his work. This work was supported by grants from the Riley Children's Foundation and NIH (grants NCI CA121168, CA167291) to MR Kelley and NCI grant CA167291 to ML Fishel. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Writing assistance was utilized in the production of this manuscript. The authors thank Lana Christian of CreateWrite Inc. for her expert editing assistance.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Kaelin WG., Jr Synthetic lethality: a framework for the development of wiser cancer therapeutics. Genome Med. 2009;1(10):99. doi: 10.1186/gm99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bapat A, Fishel ML, Kelley MR. Going ape as an approach to cancer therapeutics. Antioxid. Redox Signal. 2009;11(3):651–668. doi: 10.1089/ars.2008.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plummer R. Perspective on the pipeline of drugs being developed with modulation of DNA damage as a target. Clin. Cancer Res. 2010;16(18):4527–4531. doi: 10.1158/1078-0432.CCR-10-0984. [DOI] [PubMed] [Google Scholar]
- 4.Luo M, He H, Kelley MR, Georgiadis MM. Redox regulation of DNA repair: implications for human health and cancer therapeutic development. Antioxid. Redox Signal. 2010;12(11):1247–1269. doi: 10.1089/ars.2009.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst.) 2007;6(8):1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Kelley MR, Georgiadis MM, Fishel ML. DNA repair and redox signaling. In: Bagley R, editor. The Tumor Microenvironment. Springer; NY, USA: 2010. pp. 133–168. [Google Scholar]
- 7.De Lartigue J. New life for PARP inhibitors: emerging agents leave mark at ASCO. www.onclive.com/publications/oncology-live/2013/august-2013/new-life-for-parp-inhibitors-emerging-agents-leave-mark-at-asco/4.; • Documents the recent trials and tribulations in the use of PARP inhibitors with future directions.
- 8.Bryant HE, Schultz N, Thomas HD, et al. Specific tumor-killing of BRCA-deficient tumours with inhibitors of poly (ADP-ribose) polymerase. Nature. 2005;434(14):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 9.Underhill C, Toulmonde M, Bonnefoi H. A review of PARP inhibitors: from bench to bedside. Ann. Oncol. 2011;22(2):268–279. doi: 10.1093/annonc/mdq322. [DOI] [PubMed] [Google Scholar]
- 10.Leung M, Rosen D, Fields S, Cesano A, Budman DR. Poly(ADP-ribose) polymerase-1 inhibition: preclinical and clinical development of synthetic lethality. Mol. Med. 2011;17(7–8):854–862. doi: 10.2119/molmed.2010.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. ClinicalTrials.gov. ClinicalTrials.gov https://clinicaltrials.gov.
- 12.O'Shaughnessy J, Osborne C, Pippen JE, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N. Engl. J. Med. 2011;364(3):205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 13.De Lartigue J. The PARP inhibitors: down but not out. 2012 www.onclive.com/publications/oncology-live/2012/july-2012/the-parp-inhibitors-down-but-not-out/2.
- 14.Guha M. PARP inhibitors stumble in breast cancer. Nat. Biotechnol. 2011;29(5):373–374. doi: 10.1038/nbt0511-373. [DOI] [PubMed] [Google Scholar]
- 15.Weaver AN, Yang ES. Beyond DNA repair: additional functions of PARP-1 in cancer. Front. Oncol. 2013;3(290):1–11. doi: 10.3389/fonc.2013.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papao G, Casale E, Montagnoli A, Circla A. PARP inhibitors in cancer therapy: an update. Expert Opin. Ther. Pat. 2013;23(4):503–514. doi: 10.1517/13543776.2013.768615. [DOI] [PubMed] [Google Scholar]
- 17.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10(4):293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.AZD2461. 2013 www.selleckchem.com/products/azd2461.html.
- 19.Newswire G. BioMarin provides updated Phase 1/2 data on BMN 673 in breast cancer at the european cancer congress 2013. 2013;1:1. www.marketwatch.com/story/biomarin-provides-updated-phase-12-data-on-bmn-673-in-breast-cancer-at-the-european-cancer-congress-2013-2013-09-29. [Google Scholar]
- 20.Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNeely S, Beckmann R, Lin AB. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol. Ther. 2013;142(1):1–10. doi: 10.1016/j.pharmthera.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Fan CH, Liu WL, Cao H, Wen C, Chen L, Jiang G. O(6)-methylguanine DNA methyltransferase as a promising target for the treatment of temozolomide-resistant gliomas. Cell Death Dis. 2013;4:e876. doi: 10.1038/cddis.2013.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golding SE, Rosenberg E, Adams BR, et al. Dynamic inhibition of ATM kinase provides a strategy for glioblastoma multiforme radiosensitization and growth control. Cell Cycle. 2012;11(6):1167–1173. doi: 10.4161/cc.11.6.19576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxmen A. Beyond PARP inhibitors: agents in pipelines target DNA repair mechanisms. J. Natl Cancer Inst. 2010;102(15):1110–1111. doi: 10.1093/jnci/djq294. [DOI] [PubMed] [Google Scholar]
- 25.Golding SE, Rosenberg E, Valerie N, et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol. Cancer Ther. 2009;8(10):2894–2902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaina B, Muhlhausen U, Piee-Staffa A, et al. Inhibition of O6-methylguanine-DNA methyltransferase by glucose-conjugated inhibitors: comparison with nonconjugated inhibitors and effect on fotemustine and temozolomide-induced cell death. J. Pharmacol. Exp. Ther. 2004;311(2):585–593. doi: 10.1124/jpet.104.071316. [DOI] [PubMed] [Google Scholar]
- 27.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer. 2004;4(4):296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 28.Kelley MR, Fishel ML. DNA Repair and Cancer Therapeutics. In: Tan D, Lynch HT, editors. Principles of Molecular Diagnostics and Personalized Cancer Medicine. Lippincott Williams & Wilkins; PA, USA: 2013. pp. 566–579. [Google Scholar]
- 29.Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair. 2007;6(4):398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: base excision repair: the long and short of it. Cell. Mol. Life Sci. 2009;66(6):981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fishel ML, Vascotto C, Kelley MR. DNA base excision repair therapeutics: summary of targets with a focus on APE1. In: Madhusudan S, Wilson DM, editors. DNA Repair and Cancer: From Bench to Clinic. CRC Press; FL, USA: 2013. pp. 233–287. [Google Scholar]; •• Strong review of the DNA base excision repair (BER) pathway and implications for cancer therapies.
- 32.Scalia M, Satriano C, Greca R, Stella AM, Rizzarelli E, Spina-Purrello V. PARP-1 inhibitors DPQ and PJ-34 negatively modulate proinflammatory commitment of human glioblastoma cells. Neurochem. Res. 2013;38(1):50–58. doi: 10.1007/s11064-012-0887-x. [DOI] [PubMed] [Google Scholar]
- 33.Srinivasan A, Gold B. Small-molecule inhibitors of DNA damage-repair pathways: an approach to overcome tumor resistance to alkylating anticancer drugs. Future Med. Chem. 2012;4(9):1093–1111. doi: 10.4155/fmc.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley MR, Georgiadis MM, Fishel ML. APE1/Ref-1 role in redox signaling: translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Curr. Mol. Pharmacol. 2012;5(1):36–53. doi: 10.2174/1874467211205010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaiswal AS, Banerjee S, Aneja R, Sarkar FH, Ostrov DA, Narayan S. DNA polymerase beta as a novel target for chemotherapeutic intervention of colorectal cancer. PLoS ONE. 2011;6(2):e16691. doi: 10.1371/journal.pone.0016691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rai G, Vyjayanti VN, Dorjsuren D, et al. Small molecule inhibitors of the human apurinic/apyrimidinic endonuclease 1 (APE1) National Center for Biotechnology Information; MD, USA: 2010. p. 27. [PubMed] [Google Scholar]
- 37.Bapat A, Glass LS, Luo M, et al. Novel small-molecule inhibitor of apurinic/apyrimidinic endonuclease 1 blocks proliferation and reduces viability of glioblastoma cells. J. Pharmacol. Exp. Ther. 2010;334(3):988–998. doi: 10.1124/jpet.110.169128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abbotts R, Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat. Rev. 2010;36(5):425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Gao Z, Maloney DJ, Dedkova LM, Hecht SM. Inhibitors of DNA polymerase beta: activity and mechanism. Bioorg. Med. Chem. 2008;16(8):4331–4340. doi: 10.1016/j.bmc.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 40.Hegde ML, Hazra TK, Mitra S. Functions of disordered regions in mammalian early base excision repair proteins. Cell. Mol. Life Sci. 2010;67(21):3573–3587. doi: 10.1007/s00018-010-0485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou GM, Karikari C, Kabe Y, Handa H, Anders RA, Maitra A. The Ape-1/Ref-1 redox antagonist E3330 inhibits the growth of tumor endothelium and endothelial progenitor cells: therapeutic implications in tumor angiogenesis. J. Cell. Phys. 2009;219(1):209–218. doi: 10.1002/jcp.21666. [DOI] [PubMed] [Google Scholar]
- 42.Kelley MR, Fishel ML. DNA repair proteins as molecular targets for cancer therapeutics. Anticancer Agents Med. Chem. 2008;8(4):417–425. doi: 10.2174/187152008784220294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tell G, Wilson DM., 3rd Targeting DNA repair proteins for cancer treatment. Cell. Mol. Life Sci. 2010;67(21):3569–3572. doi: 10.1007/s00018-010-0484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid. Redox Signal. 2009;11(3):601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Strong review on the multifunctions of a critical DNA repair protein.
- 45.Fishel ML, Jiang Y, Rajeshkumar NV, et al. Impact of APE1/Ref-1 redox inhibition on pancreatic tumor growth. Mol. Cancer Ther. 2011;10(9):1698–1708. doi: 10.1158/1535-7163.MCT-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cardoso AA, Jiang Y, Luo M, et al. APE1/Ref-1 regulates STAT3 transcriptional activity and APE1/Ref-1-STAT3 dual-targeting effectively inhibits pancreatic cancer cell survival. PLoS ONE. 2012;7(10):e47462. doi: 10.1371/journal.pone.0047462. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Novel report of a critical transcription factor under redox control by a multifunctional DNA repair and redox signaling protein.
- 47.Methoxyamine clinical trials 2013 www.clinicaltrials.gov/ct2/results?term=methoxyamine+&Search=Search.
- 48.Reed AM, Fishel ML, Kelley MR. Small-molecule inhibitors of proteins involved in base excision repair potentiate the anti-tumorigenic effect of existing chemotherapeutics and irradiation. Future Oncol. 2009;5(5):713–726. doi: 10.2217/FON.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M, Wilson DM., 3rd Human apurinic/apyrimidinic endonuclease 1. Antioxid Redox Signal. 2013;20(4):678–707. doi: 10.1089/ars.2013.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sultana R, McNeill DR, Abbotts R, et al. Synthetic lethal targeting of DNA double-strand break repair deficient cells by human apurinic/apyrimidinic endonuclease inhibitors. Int. J. Cancer. 2012;131(10):2433–2444. doi: 10.1002/ijc.27512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson DM, 3rd, Simeonov A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell. Mol. Life Sci. 2010;67(21):3621–3631. doi: 10.1007/s00018-010-0488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.ApeX Therapeutics Research 2013 http://apextherapeutics.com/research.
- 53.Tsutakawa SE, Classen S, Chapados BR, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145(2):198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Pel DM, Barrett IJ, Shimizu Y, et al. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 2013;9(1):e1003254. doi: 10.1371/journal.pgen.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin SA, Lord CJ, Ashworth A. Therapeutic targeting of the DNA mismatch repair pathway. Clin. Cancer Res. 2010;16(21):5107–5113. doi: 10.1158/1078-0432.CCR-10-0821. [DOI] [PubMed] [Google Scholar]
- 56.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 57.Li YH, Wang X, Pan Y, Lee DH, Chowdhury D, Kimmelman AC. Inhibition of nonhomologous end joining repair impairs pancreatic cancer growth and enhances radiation response. PLoS ONE. 2012;7(6):e39588. doi: 10.1371/journal.pone.0039588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Methotrexate in Metastatic Colorectal Cancer With MSH2 Deficiency (MESH) http://clinicaltrials.gov/ct2/show/NCT00952016.
- 59.Martin SA, Hewish M, Sims D, Lord CJ, Ashworth A. Parallel high-throughput RNA interference screens identify PINK1 as a potential therapeutic target for the treatment of DNA mismatch repair-deficient cancers. Cancer Res. 2011;71(5):1836–1848. doi: 10.1158/0008-5472.CAN-10-2836. [DOI] [PubMed] [Google Scholar]; • Strong presentation of development of new therapeutic agents for a mismatch DNA repair target protein.
- 60.Dianov G. Repair pathways for processing of 8-oxoguanine in DNA by mammalian cell extracts. J. Biol. Chem. 1998;273(50):33811–33816. doi: 10.1074/jbc.273.50.33811. [DOI] [PubMed] [Google Scholar]
- 61.Kinsella TJ. Coordination of DNA mismatch repair and base excision repair processing of chemotherapy and radiation damage for targeting resistant cancers. Clin. Cancer Res. 2009;15(6):1853–1859. doi: 10.1158/1078-0432.CCR-08-1307. [DOI] [PubMed] [Google Scholar]
- 62.Neher TM, Bodenmiller D, Fitch RW, Jalal SI, Turchi JJ. Novel irreversible small molecule inhibitors of replication protein A display single-agent activity and synergize with cisplatin. Mol. Cancer Ther. 2011;10(10):1796–1806. doi: 10.1158/1535-7163.MCT-11-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair, from understanding mechanisms to influencing biology. Cell Res. 2008;18(1):64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barakat KH, Tuszynski JA. Nucleotide excision repair inhibitors: still a long way to go. In: Chen C, editor. New Research Directions in DNA Repair. InTech; AZ, USA: 2013. pp. 559–576. [Google Scholar]
- 65.Barakat KH, Jordheim LP, Perez-Pineiro R, Wishart D, Dumontet C, Tuszynski JA. Virtual screening and biological evaluation of inhibitors targeting the XPA-ERCC1 interaction. PLoS ONE. 2012;7(12):e51329. doi: 10.1371/journal.pone.0051329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.UCN-01 clinical trials. 2013 www.clinicaltrials.gov/ct2/results?term=UCN-01&pg=1.
- 67.Barret JM, Cadou M, Hill BT. Inhibition of nucleotide excision repair and sensitisation of cells to DNA cross-linking anticancer drugs by F 11782, a novel fluorinated epipodophylloid. Biochem. Pharmacol. 2002;63(2):251–258. doi: 10.1016/s0006-2952(01)00835-8. [DOI] [PubMed] [Google Scholar]
- 68.D'Incalci M, Galmarini CM. A review of trabectedin (ET-743): a unique mechanism of action. Mol. Cancer Ther. 2010;9(8):2157–2163. doi: 10.1158/1535-7163.MCT-10-0263. [DOI] [PubMed] [Google Scholar]
- 69.Trabectedin clinical trials. 2013 www.clinicaltrials.gov/ct2/results?term=trab ectedin+&Search=Search.
- 70.Mcneil EM, Melton DW. DNA repair endonuclease ERCC1-XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucleic Acids Res. 2012;40(20):9990–10004. doi: 10.1093/nar/gks818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Usanova S, Piee-Staffa A, Sied U, et al. Cisplatin sensitivity of testis tumour cells is due to deficiency in interstrand-crosslink repair and low ERCC1-XPF expression. Mol. Cancer. 2010;9:248. doi: 10.1186/1476-4598-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40(13):5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neal JA, Dang V, Douglas P, Wold MS, Lees-Miller SP, Meek K. Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol. Cell Biol. 2011;31(8):1719–1733. doi: 10.1128/MCB.01298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chernikova SB, Game JC, Brown JM. Inhibiting homologous recombination for cancer therapy. Cancer Biol. Ther. 2012;13(2):61–68. doi: 10.4161/cbt.13.2.18872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31(6):955–960. doi: 10.1093/carcin/bgq064. [DOI] [PubMed] [Google Scholar]
- 76.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8(3):193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 77.Yan H, Toczylowski T, McCane J, Chen C, Liao S. Replication protein A promotes 5′→ 3′ end processing during homology-dependent DNA double-strand break repair. J. Cell Biol. 2011;192(2):251–261. doi: 10.1083/jcb.201005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hass CS, Lam K, Wold MS. Repair-specific functions of replication protein A. J. Biol. Chem. 2012;287(6):3908–3918. doi: 10.1074/jbc.M111.287441. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Clearly written manuscript detailing a critical protein in nucleotide excision repair that could be a valuable target for cancer therapeutics.
- 79.Frank AO, Feldkamp MD, Kennedy JP, et al. Discovery of a potent inhibitor of replication protein a protein–protein interactions using a fragment-linking approach. J. Med. Chem. 2013;56(22):9242–9250. doi: 10.1021/jm401333u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Renodon-Cornire A, Weigel P, Le M, Fleury F. New potential therapeutic approaches by targeting Rad51-dependent homologous recombination. In: Chen C, editor. New Research Directions in DNA Repair. InTech; AZ, USA: 2013. pp. 467–488. [Google Scholar]; • Strong review of a target for homologous recombination (HR) and the potential for therapeutic indications.
- 81.Chen G, Yuan S-SF, Liu W, et al. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J. Biol. Chem. 1999;274(18):12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- 82.Budke B, Logan HL, Kalin JH, et al. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 2012;40(15):7347–7357. doi: 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jhaveri K, Taldone T, Modi S, Chiosis G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim. Bophys. Acta. 2012;1823(3):742–755. doi: 10.1016/j.bbamcr.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dinitto JP, Wu JC. Molecular mechanisms of drug resistance in tyrosine kinases cAbl and cKit. Crt. Rev. Biochem. Mol. Biol. 2011;46(4):295–309. doi: 10.3109/10409238.2011.578612. [DOI] [PubMed] [Google Scholar]
- 85.Dar AC, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Ann. Rev. Biochem. 2011;80:769–795. doi: 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- 86.Roberts RJ, Agius C, Saliba C, Bossier P, Sung YY. Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J. Fish Dis. 2010;33(10):789–801. doi: 10.1111/j.1365-2761.2010.01183.x. [DOI] [PubMed] [Google Scholar]
- 87.Mittelman D, Sykoudis K, Hersh M, Lin Y, Wilson JH. Hsp90 modulates CAG repeat instability in human cells. Cell Stress Chaperones. 2010;15(5):753–759. doi: 10.1007/s12192-010-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Messaoudi A, Peyrat JF, Brion JD, Alami M. Recent advances in Hsp90 inhibitors as antitumor agents. Anticancer Agents Med. Chem. 2008;8:761–782. doi: 10.2174/187152008785914824. [DOI] [PubMed] [Google Scholar]
- 89.Meek K, Dang V, Lees-Miller SP. Chapter 2 DNA-PK. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 90.Collis SJ, Deweese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24(6):949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 91.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18(1):114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 92.Weinfeld M, Lees-Miller SP. DNA double-strand break repair by nonhomologous end joining and its clinical relevance. In: Kelley MR, editor. DNA Repair in Cancer Therapy: Molecular Targets and Clinical Applications. Academic Press; MA, USA: 2012. pp. 161–181. [Google Scholar]
- 93.Davidson D, Amrein L, Panasci L, Aloyz R. Small molecules, inhibitors of DNA-PK, targeting DNA repair, and beyond. Front. Pharmacol. 2013;4:5. doi: 10.3389/fphar.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Outstanding review of aspects of DNA repair pathways and upstream signaling targets that could be strong targets for anti-DNA repair therapy.
- 94.Zhou X, Zhang X, Xie Y, Tanaka K, Wang B, Zhang H. DNA-PKcs inhibition sensitizes cancer cells to carbon-ion irradiation via telomere capping disruption. PLoS ONE. 2013;8(8):e72641. doi: 10.1371/journal.pone.0072641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Srivastava M, Nambiar M, Sharma S, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151(7):1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 96.Tavecchio M, Munck JM, Cano C, Newell DR, Curtin NJ. Further characterisation of the cellular activity of the DNA-PK inhibitor, NU7441, reveals potential cross-talk with homologous recombination. Cancer Chemother. Pharmacol. 2012;69(1):155–164. doi: 10.1007/s00280-011-1662-4. [DOI] [PubMed] [Google Scholar]
- 97.Mukherjee B, Tomimatsu N, Amancherla K, Camacho CV, Pichamoorthy N, Burma S. The dual PI3K/mTOR inhibitor NVPBEZ235 is a potent inhibitor of ATM- and DNA-PKCs-mediated DNA damage responses. Neoplasia. 2012;14(1):34–43. doi: 10.1593/neo.111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Freschauf GK, Karimi-Busheri F, Ulaczyk-Lesanko A, et al. Identification of a small molecule inhibitor of the human DNA repair enzyme polynucleotide kinase/phosphatase. Cancer Res. 2009;69(19):7739–7746. doi: 10.1158/0008-5472.CAN-09-1805. [DOI] [PubMed] [Google Scholar]
- 99.Chen X, Zhong S, Zhu X, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res. 2008;68(9):3169–3177. doi: 10.1158/0008-5472.CAN-07-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park SB, Goldstein D, Krishnan AV, et al. Chemotherapy-induced peripheral neurotoxicity: a critical analysis. CA Cancer J. Clin. 2013;63(6):419–437. doi: 10.3322/caac.21204. [DOI] [PubMed] [Google Scholar]
- 101.Fallon MT. Neuropathic pain in cancer. Br. J. Anaesth. 2013;111(1):105–111. doi: 10.1093/bja/aet208. [DOI] [PubMed] [Google Scholar]
- 102.Alberti P, Rossi E, Cornblath DR, et al. Physician-assessed and patient-reported outcome measures in chemotherapy-induced sensory peripheral neurotoxicity: two sides of the same coin. Ann. Oncol. 2013;25(1):257–264. doi: 10.1093/annonc/mdt409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duarte DB, Vasko MR. The role of DNA damage and repair in neurotoxicity caused by cancer therapies. In: Kelley MR, editor. DNA Repair in Cancer Therapy: Molecular Targets and Clinical Applications. Academic Press; MA, USA: 2012. pp. 283–299. [Google Scholar]
- 104.Schroder S, Beckmann K, Franconi G, et al. Can medical herbs stimulate regeneration or neuroprotection and treat neuropathic pain in chemotherapy-induced peripheral neuropathy? Evid. Based Complement. Alternat. Med. 2013;2013:423713. doi: 10.1155/2013/423713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schloss JM, Colosimo M, Airey C, Masci PP, Linnane AW, Vitetta L. Nutraceuticals and chemotherapy induced peripheral neuropathy (CIPN): a systematic review. Clin. Nutr. 2013;32(6):888–893. doi: 10.1016/j.clnu.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 106.Vasko MR, Guo C, Thompson EL, Kelley MR. The repair function of the multifunctional DNA repair/redox protein APE1 is neuroprotective after ionizing radiation. DNA Repair. 2011;10(9):942–952. doi: 10.1016/j.dnarep.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Englander EW. DNA damage response in peripheral nervous system: coping with cancer therapy-induced DNA lesions. DNA Repair. 2013;12(8):685–690. doi: 10.1016/j.dnarep.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fishel ML, Vasko MR, Kelley MR. DNA repair in neurons: so if they don't divide what's to repair? Mutat. Res. 2007;614(1–2):24–36. doi: 10.1016/j.mrfmmm.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 109.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol. Aspects Med. 2007;28(3–4):375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 110.Palanca A, Casafont I, Berciano MT, Lafarga M. Proteasome inhibition induces DNA damage and reorganizes nuclear architecture and protein synthesis machinery in sensory ganglion neurons. Cell. Mol. Life Sci. 2013;71:1961–1975. doi: 10.1007/s00018-013-1474-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Azevedo MI, Pereira AF, Nogueira RB, et al. The antioxidant effects of the flavonoids rutin and quercetin inhibit oxaliplatin-induced chronic painful peripheral neuropathy. Mol. Pain. 2013;9(1):53. doi: 10.1186/1744-8069-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dzagnidze A, Katsarava Z, Makhalova J, et al. Repair capacity for platinum-DNA adducts determines the severity of cisplatin-induced peripheral neuropathy. J. Neurosci. 2007;27(35):9451–9457. doi: 10.1523/JNEUROSCI.0523-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang Y, Guo C, Vasko MR, Kelley MR. Implications of apurinic/apyrimidinic endonuclease in reactive oxygen signaling response after cisplatin treatment of dorsal root ganglion neurons. Cancer Res. 2008;68(15):6425–6434. doi: 10.1158/0008-5472.CAN-08-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Another article in a string of publications implicating BER in chemotherapy-induced peripheral neuropathy and importance in understanding mechanisms of CIPN for future drug development.
- 114.Ta LE, Schmelzer JD, Bieber AJ, et al. A novel and selective poly (ADP-ribose) polymerase inhibitor ameliorates chemotherapy-induced painful neuropathy. PLoS ONE. 2013;8(1):e54161. doi: 10.1371/journal.pone.0054161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peddi SR, Chattopadhyay R, Naidu CV, Izumi T. The human apurinic/apyrimidinic endonuclease-1 suppresses activation of poly(adp-ribose) polymerase-1 induced by DNA single strand breaks. Toxicology. 2006;224(1–2):44–55. doi: 10.1016/j.tox.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 116.Kono T, Hata T, Morita S, et al. Goshajinkigan oxaliplatin neurotoxicity evaluation (GONE): a Phase 2, multicenter, randomized, double-blind, placebo-controlled trial of goshajinkigan to prevent oxaliplatin-induced neuropathy. Cancer Chemother. Pharmacol. 2013;72(6):1283–1290. doi: 10.1007/s00280-013-2306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gilchrist LS, Marais L, Tanner L. Comparison of two chemotherapy-induced peripheral neuropathy measurement approaches in children. Support. Care Cancer. 2013;22(2):359–366. doi: 10.1007/s00520-013-1981-6. [DOI] [PubMed] [Google Scholar]
- 118.Di Francia R, Siesto RS, Valente D, et al. Current strategies to minimize toxicity of oxaliplatin: selection of pharmacogenomic panel tests. Anticancer Drugs. 2013;24(10):1069–1078. doi: 10.1097/CAD.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 119.Genentech & Inotek Enter into Exclusive Global Strategic Alliance for the Discovery, Development and Commercialization of PARP Inhibitors for Cancer. 2006 www.gene.com/media/press-releases/9907/2006-07-25/genentech-inotek-enter-into-exclusive-gl.
- 120.Tracon Pharma Programs 2013 www.traconpharma.com/progrms.php.
- 121.Sentinel Oncology R&D Programmes 2013 www.sentineloncology.com/pipeline.
- 122.Allison M. Haseltine's DNA repair company steps out. 2007 www.xconomy.com/boston/2007/12/07/haseltines-dna-repair-company-steps-out/
- 123.Garber K. Mission therapeutics. Nat. Biotechnol. 2012;30(3):206. doi: 10.1038/nbt0312-206. [DOI] [PubMed] [Google Scholar]
- 124.Ubiquitination: labelling the proteins. 2008 www.sebiology.org/publications/Bulletin/March08/Ubiquitination.html.
- 125.US FDA . Paving the Way for Personalized Medicine. FDA's Role in a new Era of Medical Product Development. US Department of Health and Human Services; DC, USA: 2013. pp. 1–61. [Google Scholar]
- 126.Perry C, Sultana R, Madhusudan S. Personalized cancer medicine: DNA repair alterations are promising predictive biomarkers in cancer. In: Kelley MR, editor. DNA Repair in Cancer Therapy: Molecular Targets and Clinical Applications. Academic Press; MA, USA: 2012. pp. 257–275. [Google Scholar]
- 127.About Foundation Medicine 2013 www.foundationmedicine.com/about.php.
- 128.Ho AY, Atencio DP, Peters S, et al. Genetic predictors of adverse radiotherapy effects: the Gene-PARE project. Int. J. Radiat. Oncol. Biol. Phys. 2006;65(3):646–655. doi: 10.1016/j.ijrobp.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 129.Milanowska K, Krwawicz J, Papaj G, et al. REPAIRtoire – a database of DNA repair pathways. Nucleic Acids Res. 2011;39(Database issue):D788–D792. doi: 10.1093/nar/gkq1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Morgenroth A, Vogg AT, Zlatopolskiy BD, Siluschek M, Oedekoven C, Mottaghy FM. Breaking the invulnerability of cancer stem cells: two-step strategy to kill the stem-like cell subpopulation of multiple myeloma. Mol. Cancer Ther. 2013;13(1):144–153. doi: 10.1158/1535-7163.MCT-13-0240. [DOI] [PubMed] [Google Scholar]
- 131.Rowe BP, Glazer PM. Emergence of rationally designed therapeutic strategies for breast cancer targeting DNA repair mechanisms. Breast Cancer Res. 2010;12(203):1–11. doi: 10.1186/bcr2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Aziz K, Nowsheen S, Georgakilas AG. Nanotechnology in cancer therapy: targeting the inhibition of key DNA repair pathways. Curr. Mol. Med. 2010;10(7):626–639. doi: 10.2174/156652410792630599. [DOI] [PubMed] [Google Scholar]