Key Points
Methylation analysis at ZAP-70 CpG+223 in CLL provides superior prognostic information vs IGHV status or CD38 or ZAP-70 expression.
A pyrosequencing method for the feasible assessment of CpG+223 methylation in CLL samples is provided.
Abstract
ZAP-70 methylation 223 nucleotides downstream of transcription start (CpG+223) predicts outcome in chronic lymphocytic leukemia (CLL), but its impact relative to CD38 and ZAP-70 expression or immunoglobulin heavy chain variable region (IGHV) status is uncertain. Additionally, standardizing ZAP-70 expression analysis has been unsuccessful. CpG+223 methylation was quantitatively determined in 295 untreated CLL cases using MassARRAY. Impact on clinical outcome vs CD38 and ZAP-70 expression and IGHV status was evaluated. Cases with low methylation (<20%) had significantly shortened time to first treatment (TT) and overall survival (OS) (P < .0001). For TT, low methylation defined a large subset of ZAP-70 protein-negative cases with significantly shortened TT (median, 8.0 vs 3.9 years for high vs low methylation; hazard ratio [HR] = 0.43; 95% confidence interval [CI], 0.25-0.74). Conversely, 16 ZAP-70 protein-positive cases with high methylation had poor outcome (median, 1.1 vs 2.3 years for high vs low methylation; HR = 1.62; 95% CI, 0.87-3.03). For OS, ZAP-70 methylation was the strongest risk factor; CD38 and ZAP-70 expression or IGHV status did not significantly improve OS prediction. A pyrosequencing assay was established that reproduced the MassARRAY data (κ coefficient > 0.90). Thus, ZAP-70 CpG+223 methylation represents a superior biomarker for TT and OS that can be feasibly measured, supporting its use in risk-stratifying CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is the most prevalent adult leukemia, and despite the introduction of new therapies, remains a serious clinical challenge. Based upon the lack of survival advantage observed to date with early treatment,1 therapy for CLL is not initiated until symptoms or clinical signs develop.2 The increasing availability and diversity of novel agents makes it even more important to identify CLL patients who are more likely to require aggressive intervention or who are more likely to respond to a given therapy. The ability to predict time to first treatment (TT) has improved with the identification of cytogenetic abnormalities [del(17p13.1), del(11q22.3), or complex karyotype], immunoglobulin heavy chain variable region (IGHV) gene mutation status, or expression of 70-kDa ζ-chain associated protein (ZAP-70) or CD38.3 One of the earliest biomarkers identified in CLL was ZAP-70 expression, which is associated with unmutated IGHV4 and shorter TT and overall survival (OS).5-7 The abnormal presence of ZAP-70 protein in CLL cells impacts their biology via enhancement of B-cell receptor (BCR) signaling, proliferation, and migration toward the tumor microenvironment.8-10 The success to date of the BCR pathway-targeting agents idelalisib11 and ibrutinib12 further underscores the importance of ZAP-70 in CLL.
Despite the strong interest in measuring ZAP-70 expression to predict TT, reliable clinical assays for this are problematic, making their use inapplicable to routine CLL patient care. Previously, we identified differential methylation in CLL tumor cells of a single CpG dinucleotide 223 bp downstream of the transcriptional start site (CpG+223) in exon 1 of ZAP-70. Methylation of this site was significantly associated with a lack of ZAP-70 protein expression as well as increased TT, progression-free survival (PFS), and OS.13 However, only a subset of patients in that study had ZAP-70 expression data available, preventing firm conclusions regarding the relative prognostic value of the 2 different ZAP-70 measures.13 Additionally, the previous application of the MassARRAY technique for DNA methylation assessment, although powerful and highly quantifiable,14 is not readily translatable to a clinical laboratory. Addressing these issues is necessary to bring ZAP-70 methylation analysis to practical use in CLL management. Herein, we not only confirm our initial findings with CpG+223 methylation but substantially extend them to demonstrate the relative superiority of this parameter for prognostication over ZAP-70 protein expression, IGHV mutational status, and CD38 expression. Finally, we describe a simplified pyrosequencing assay that can be applied for routine clinical use.
Methods
Patients and cells
Peripheral blood was obtained from asymptomatic, untreated CLL patients. All patients were enrolled on a prospective natural history study of the CLL Research Consortium (CRC). All patients provided written, informed consent for this study under institutional review board–approved protocols according to the Declaration of Helsinki. Samples used were derived from 295 patients reported previously for prognostic significance of ZAP-70 protein expression.15,16 Samples were considered to be ZAP-70 and CD38 positive if the percentage of ZAP-70– or CD38-expressing CLL cells exceeded 20%. Here we used a 20% cut point for CD38, as this is a standard cut point with most flow cytometric antigens assessed and has been previously published.17 However, we also report results of sensitivity analyses using the previously reported cut points of 7%18 and 30%.19 IGHV mutational analysis was performed as described.15 Cases with sequence homology of <98% vs germline sequence were considered mutated. Patient demographics are summarized in Table 1.
Table 1.
Demographic and molecular features for all patients and by ZAP-70 CpG+223 methylation
| CLL marker data | Overall, N = 295 | Methylation <20%, n = 197 | Methylation ≥20%, n = 98 | P |
|---|---|---|---|---|
| CpG+223 methylation, % | NA | |||
| Median | 9 | 6 | 49.5 | |
| Range | 2-86 | 2-19 | 24-86 | |
| Age, y | .57 | |||
| Median | 55 | 55 | 56 | |
| Range | 26-82 | 26-79 | 33-82 | |
| Sex, N (%) | .50 | |||
| Male | 204 (69) | 139 (71) | 65 (66) | |
| Female | 91 (31) | 58 (29) | 33 (34) | |
| IGHV, N (%) | < .0001 | |||
| Mutated | 91 (31) | 21 (11) | 70 (71) | |
| Unmutated (≥98%) | 204 (69) | 176 (89) | 28 (29) | |
| CD38, N (%) | < .0001 | |||
| Negative | 139 (47) | 74 (38) | 65 (66) | |
| Positive (≥20%) | 156 (53) | 123 (62) | 33 (34) | |
| ZAP-70, N (%) | < .0001 | |||
| Negative | 136 (46) | 57 (29) | 79 (81) | |
| Positive (≥20%) | 159 (54) | 140 (71) | 19 (19) |
NA, not applicable.
Quantitative high-resolution DNA methylation analysis by MassARRAY
DNA methylation was assessed at single CpG units (consisting of 1 or more CpG dinucleotides) using the MassCleave assay (Sequenom) as described.13
Quantitative DNA methylation analysis by pyrosequencing
Genomic DNA (500 ng) was bisulfite-treated using the EZ DNA Methylation-Gold kit (Zymo). Bisulfite-treated DNA (50 ng) was amplified in a polymerase chain reaction (PCR) with primers: ZAP70_FW: 5′-TGGGAGATTTGGTAGAGGATGAA-3′; ZAP70_RV: 5′-GTGCCAGGCTCAGGCCCTCCTAACTCCCAATTAATATTCTATCTT-3′; Universal: 5′-Biotin-ATCTGTGCCAGGCTCAGGC-3′.
Thermal cycler conditions were: 95°C, 15 minutes; then 49 cycles of 95°C, 30 seconds; 57.5°C, 30 seconds; 72°C, 30 seconds; then 72° for 5 minutes using HotStarTaq (Qiagen). Pyrosequencing was performed on a Pyromark Q96 MD instrument (Qiagen) with setting CDT0003 using 8 µL of PCR product and sequencing primer ZAP70_SEQ 5′-ATGAGTGAGAAATTTTGG-3′ (0.3 µM) following the standard Qiagen protocol with Qiagen Pyromark reagents, GE Streptavidin Sepharose beads (GE Healthcare), and the PyroMark Q96 Vacuum Workstation. Nucleotides were diluted 1:2 in water. This same protocol was used in an independent laboratory to confirm transferability of this method.
Clinical laboratory improvement amendment validated assay development
Pyrosequencing is becoming more commonly applied in the clinical setting for detection of disease-related gene mutations or methylation (eg, MGMT, EGFR).20,21 For this validated assay in a third, clinical laboratory, the instrument used was a Pyromark ID (setting ID/MA0008) and 37 µL of PCR was sequenced. Also, nucleotides in the pyrosequencing run for the ID instrument were used undiluted, sepharose beads were increased from 2 µL to 3 µL, and sequencing primer was increased from 0.3 µM to 0.4 µM. All other conditions remained the same (detailed protocol in supplemental Materials, available on the Blood Web site).
Statistical analysis
Associations between demographic and molecular features with ZAP-70 CpG+223 methylation levels (<20% vs ≥20%) were tested using the Wilcoxon rank sum and Fisher exact tests for continuous and categoric variables, respectively. TT was measured from the date of diagnosis until the date of first treatment. Patients who were confirmed to have remained untreated were censored at the date last known to be treatment-free. OS was measured from the date of diagnosis until the date of death, censoring patients alive at last follow-up. Estimates of treatment-free proportions and OS were obtained by the Kaplan-Meier method, and the log-rank test compared differences between survival curves. Proportional hazards models were used to analyze the association of CpG+223 methylation with TT and OS when adjusting for other molecular markers (IGHV status, expression of CD38 and ZAP-70). All tests were 2-sided and statistical significance was set at α = 0.05. Concordance in methylation values measured by pyrosequencing vs MassARRAY is shown graphically using Bland-Altman plots and quantified using Lin’s concordance correlation coefficient.22,23 The degree of agreement in classification of low and high methylation levels between the 2 methods was evaluated using the Cohen κ coefficient.24
Results
ZAP-70 CpG+223 methylation correlates with extended TT and OS
Among the 295 patients included in this study, 263 were followed for TT and included in the analysis. With a median follow-up of 3.9 years in 50 patients who had not yet started treatment, the estimated median TT was 3.5 years (95% confidence interval [CI], 2.9-4.0). All patients were followed for OS. With a median follow-up of 5.6 years among the 196 patients still alive, the estimated median OS was 11.0 years (95% CI, 9.1-12.5). We confirmed that methylation of CpG+223 as a continuous variable was significantly associated with both TT and OS (P < .0001 for both end points), as we reported previously.13
Using the data-driven cut point of 15% previously identified,13 we formed 2 prognostic groups based upon methylation of CpG+223: <15% and ≥15%. As seen in supplemental Figure 1A, methylation of CpG+223 significantly discriminated TT (P < .0001), where the estimated median TT for those with low and high methylation, respectively, was 2.9 years (95% CI, 2.0-3.3) vs 6.1 years (95% CI, 4.0-7.6). Likewise, the estimated median OS was 9.0 years (95% CI, 7.9-10.1) in those with low methylation (supplemental Figure 1B), significantly shorter than the 16.7 years (95% CI, 11.0-22.7) estimated for those with high methylation. Collectively, we confirmed our previous findings that methylation of CpG+223 was highly predictive of TT and OS for early-stage untreated CLL.
Cut point for CpG+223 methylation
In this data set, the median CpG+223 methylation value by MassARRAY was 9% and levels of methylation ranged from 2% to 86%. When examining the distribution of CpG+223 methylation levels for the entire cohort of patients in this study and in the previously published studies13 (supplemental Figure 2A-B), a gap in the methylation values occurred between 20% and 25%, with sparseness in values also between 55% and 65%. As CLL cell genomes are generally diploid,25 we hypothesized that this distinct trimodal distribution of ZAP-70 methylation values reflects the stable allelic methylation state in the entire CLL cell population at this particular locus, as has been shown to occur in the CLL genome at large.26 Thus, the first distinct break in the methylation values implies biological significance and may separate cases with both alleles robustly unmethylated in almost all CLL cells from those with monoallelic or biallelic methylation. Given that a data-driven, optimal cut point is subject to change depending on the data set, a cut point with potential biological significance and robustness is appealing. Using a 20% cut point based on the allelic methylation state of CpG+223, the 2 larger previously published data sets still showed a strong significant association between methylation and TT, PFS, and OS (P < .0001; supplemental Figure 3A-D). Therefore, this cut point was used in subsequent analyses presented herein.
Patient demographics are shown in Table 1. Neither age nor sex was associated with methylation status. IGHV status and CD38 and ZAP-70 protein expression were measured on all patients and all were associated with ZAP-70 CpG+223 methylation levels. IGHV unmutated disease, CD38 expression, and ZAP-70 protein expression were more commonly and significantly observed in the ZAP-70 unmethylated group (P < .0001 for all comparisons).
Prognostic value of CpG+223 methylation relative to IGHV status and CD38 and ZAP-70 protein expression with respect to TT
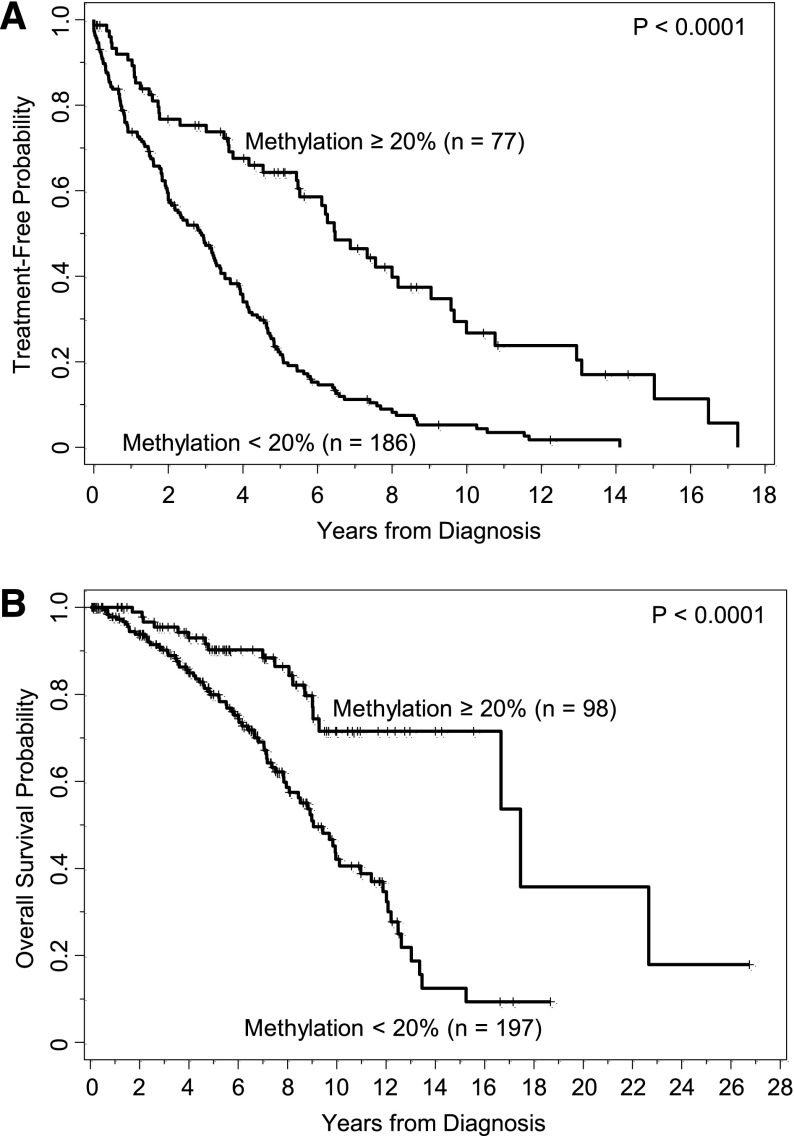
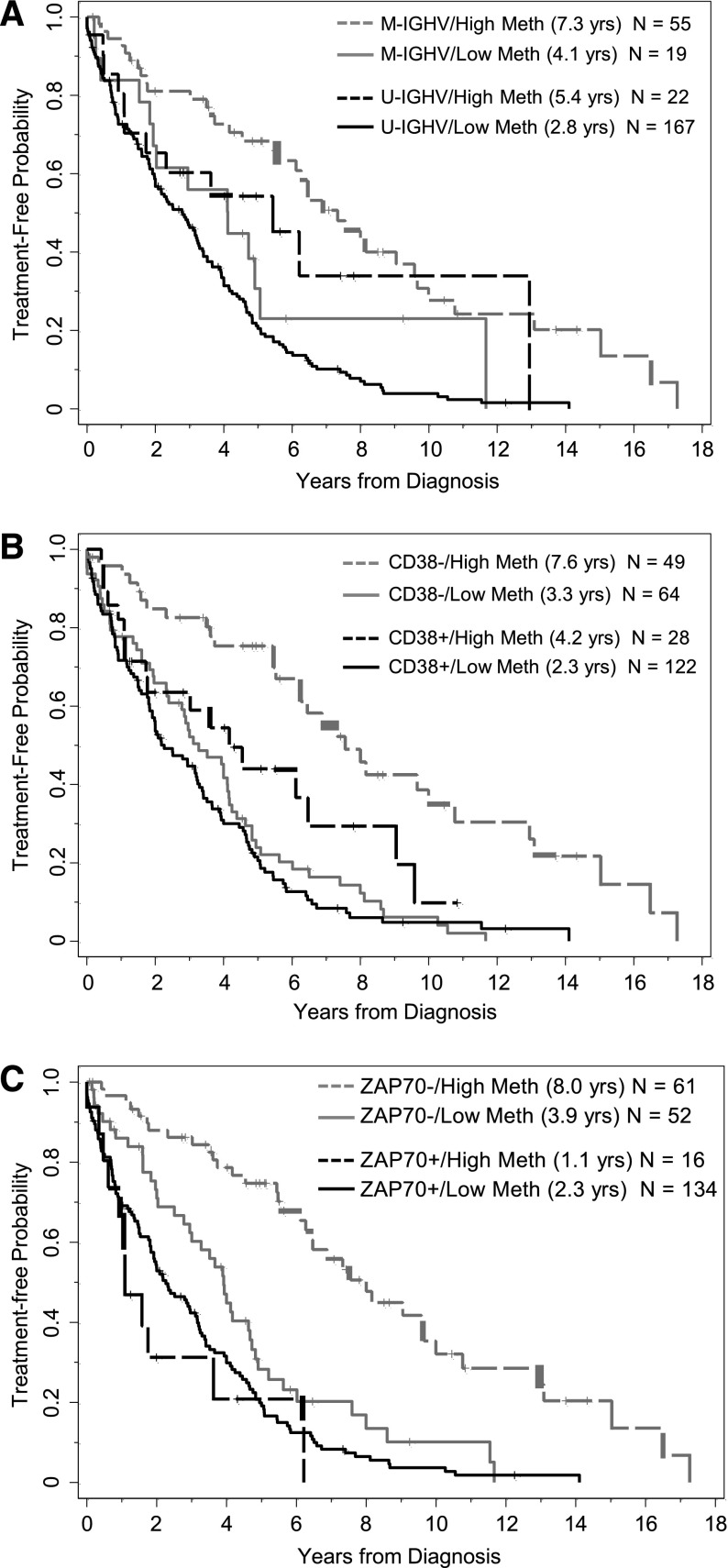
When applying the ZAP-70 methylation cutoff of 20% to form 2 prognostic groups, TT was significantly longer for those with higher vs lower methylation levels (P < .0001, Table 2; Figure 1A), where the median TT was 6.5 (95% CI, 5.4-9.0) and 2.9 (95% CI, 2.0-3.3) years, respectively. Similarly, patients with IGHV mutated disease had longer TT compared with those with IGHV unmutated disease (P < .0001), patients who were negative for CD38 expression had longer TT than those positive for CD38 (P < .0001), and patients who were negative for ZAP-70 expression had longer TT compared with those who were positive (P < .0001). Because all 4 of these biomarkers were strongly and significantly associated with TT, we next evaluated the prognostic impact that CpG+223 methylation had on TT in the context of the other 3 established biomarkers in the subgroups segregated by IGHV status and CD38 and ZAP-70 protein expression. Among each of the IGHV and CD38 subgroups (mutated/unmutated; positive/negative), those cases with higher CpG+223 methylation values had significantly extended TT compared with those with lower methylation values (Figure 2A-B). In each setting, knowledge of CpG+223 methylation status revealed a subgroup of patients who are typically considered lower risk (IGHV mutated or CD38 negative), but who actually fared poorly with low methylation. Conversely, knowledge of CpG+223 methylation status revealed a subgroup of patients who are typically considered high risk (IGHV unmutated or CD38 positive), but who actually had an intermediate prognostic profile with high methylation. A different pattern was observed when TT according to methylation status was examined by ZAP-70 protein expression. In cases lacking ZAP-70 expression, high methylation was again strongly protective (P < .0001, Figure 2C) with a median TT of 8.0 years (95% CI, 6.3-10.0) compared with only 3.9 years (95% CI, 2.8-4.7) in those with low methylation. However, methylation had no significant effect in those with ZAP-70 protein expression (P = .27, Figure 2C), where TT was short regardless of methylation levels. A multivariable model for TT was fit including IGHV status, CD38 positivity and the interaction between CpG+223 methylation and ZAP-70 positivity. With all variables in the model, only the interaction effect between CpG+223 methylation and ZAP-70 protein status was statistically significant (P = .0007), whereas IGHV status and CD38 expression did not provide additional prognostic information (2 degrees of freedom test, P = .27) (supplemental Table 1). When alternative cut points (7% and 30%) for CD38 expression were investigated in sensitivity analyses, the importance of the CpG+223 methylation and ZAP-70 positivity interaction effect did not change (P < .001); however, CD38 provided additional prognostic information independent of the interaction effect when using these cut points (P = .046 and P = .043 for 7% and 30%, respectively).
Table 2.
Kaplan-Meier estimates for TT and OS by risk factor
| Biomarker | TT | OS | ||
|---|---|---|---|---|
| Median, y | 95% CI | Median, y | 95% CI | |
| Methylation, <20% | 2.9 | 2.0-3.3 | 9.1 | 8.0-11.1 |
| Methylation, ≥20% | 6.5 | 5.4-9.0 | 17.5 | 16.7-NR |
| M-IGHV | 6.3 | 4.5-8.0 | 16.7 | 12.1-22.7 |
| U-IGHV | 2.9 | 2.1-3.4 | 9.1 | 8.0-11.0 |
| CD38− | 4.4 | 3.7-5.6 | 12.0 | 9.1-16.7 |
| CD38+ | 2.8 | 1.9-3.4 | 9.7 | 8.4-12.2 |
| ZAP-70− | 5.5 | 4.2-6.9 | 16.7 | 12.1-22.7 |
| ZAP-70+ | 2.1 | 1.7-2.9 | 8.8 | 7.8-9.9 |
M-IGHV, IGHV mutated; NR, not reached; U-IGHV, IGHV unmutated.
Figure 1.
DNA methylation at CpG+223 segregates patients with different clinical outcome using a potentially biologically meaningful cutoff of 20%. (A) Kaplan-Meier curves illustrate the time from diagnosis to first treatment of patients who have CLL cells with lower levels of methylation (<20%) compared with those patients who have CLL cells with higher levels of methylation (≥20%). (B) Kaplan-Meier curves illustrate the overall survival duration of patients who have CLL cells with lower vs higher levels of methylation.
Figure 2.
Effect of methylation (<20% vs ≥20%) on time from diagnosis to first treatment in subgroups of patients segregated by IGHV mutational status, CD38 expression, and ZAP-70 protein expression. (A) Methylation levels segregate patients who are M-IGHV (P = .02) or U-IGHV (P = .02). (B) Methylation levels segregate patients who are CD38 negative (P < .0001) or positive (P = .03). (C) Methylation levels segregate patients who are ZAP-70 negative (P < .0001) but not patients who are ZAP-70 positive (P = .27). M-IGHV, IGHV mutated; U-IGHV, IGHV unmutated.
Prognostic value of CpG+223 methylation for OS relative to IGHV status and CD38 and ZAP-70 protein expression
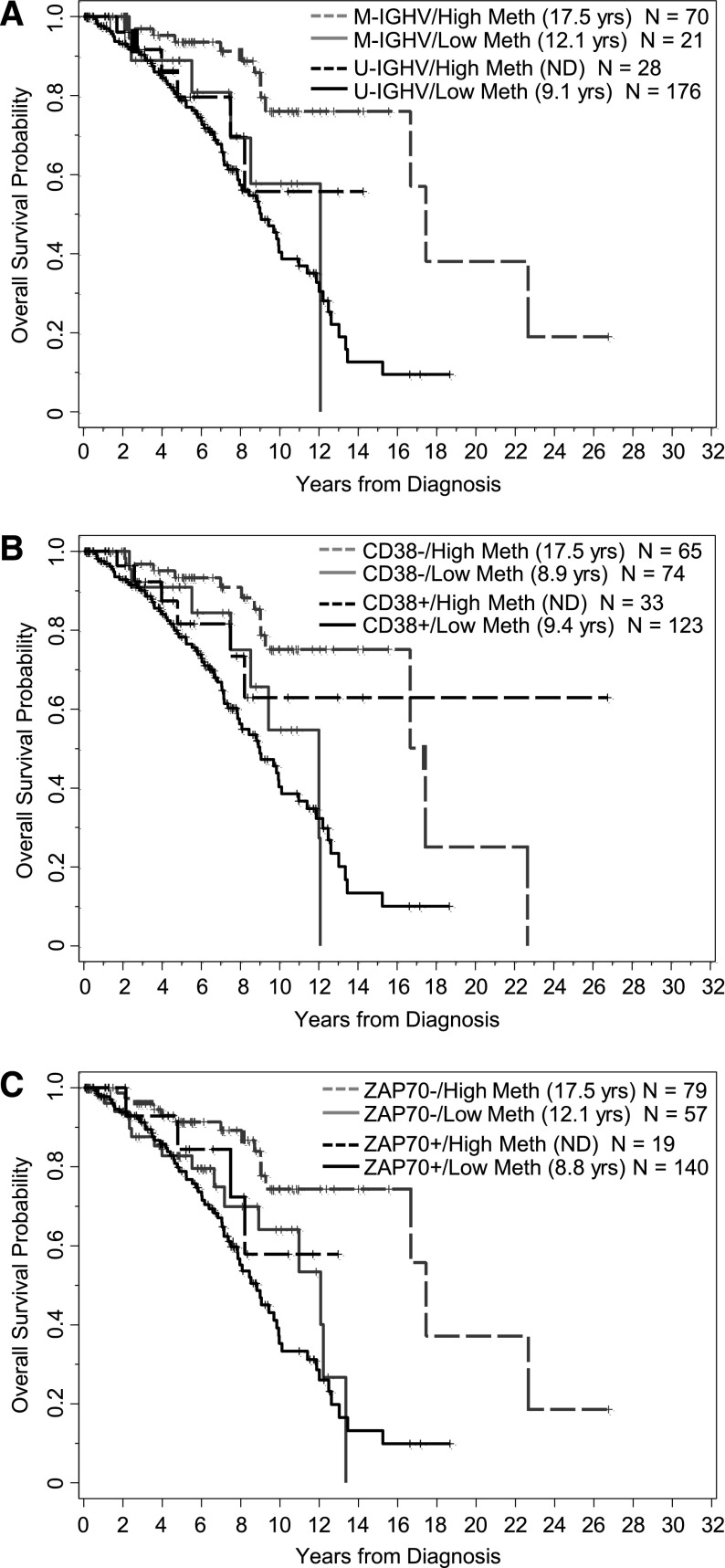
The analysis conducted for TT was repeated for OS. As shown in Table 2, OS from diagnosis was significantly longer with higher methylation (P < .0001), where the median OS was 17.5 (95% CI, 16.7–not reached) and 9.1 (95% CI, 8.0-11.1) years for those with higher and lower methylation levels, respectively (Figure 1B). Likewise, OS was longer for IGHV mutated patients (P < .0001), those lacking CD38 expression (P = .13), and those lacking ZAP-70 protein expression (P < .0001). The impact of methylation levels for each subgroup of patients segregated by IGHV status, CD38 expression, and ZAP-70 positivity are shown in Figure 3A. In the IGHV and CD38 subgroups, similar patterns seen for TT were observed for OS, where higher methylation levels tended to extend OS compared with lower levels. In contrast to what was observed with TT, the effect of methylation on OS was not significantly different according to ZAP-70 protein positivity (P = .57). Rather, methylation of CpG+223 was beneficial in both ZAP-70–negative and –positive patients, albeit more so in the negative subgroup (Figure 3C).
Figure 3.
Effect of methylation (<20% vs ≥20%) on OS from time of diagnosis in subgroups of patients segregated by IGHV mutational status, CD38 expression, and ZAP-70 protein expression. (A) Methylation levels tend to segregate patients who are M-IGHV (P = .03) or U-IGHV (P = .21). (B) Methylation levels tend to segregate patients who are CD38 negative (P < .0001) or CD38 positive (P = .11). (C) Methylation levels tend to segregate patients who are ZAP-70 negative (P = .006) or ZAP-70 positive (P = .19).
In a multivariable model for OS, the 4 biomarkers jointly explained a significant amount of variability in survival (overall test of significance, P < .0001); however, the prognostic information is shared among the related biomarkers, leading to nonsignificant signals for each marker in the model individually (supplemental Table 2). Given that CpG+223 methylation as a sole prognostic marker for OS was highly statistically significant and is also the strongest of the 4 biomarkers associated with OS in the multivariable model (hazard ratio [HR] = 0.52; 95% CI, 0.26-1.07; P = .07), it could be considered the dominant biomarker of the 4 in predicting OS. Furthermore, with CpG+223 methylation in the model, IGHV status, CD38 positivity, and ZAP-70 protein expression did not provide additional significant prognostic information (3 degrees of freedom test, P = .31). Similar results were obtained regardless of the cut point used for CD38 in sensitivity analyses.
Development of a feasible, quantifiable assay to measure CpG+223 methylation in clinical samples
We next sought to convert the quantitative ZAP-70 methylation detection assay from MassArray to pyrosequencing. We designed and validated anappropriate pyrosequencing protocol and compared pyrosequencing data obtained in 3 different laboratories (one clinical laboratory improvement amendment [CLIA] approved) to each other and to data obtained previously by MassARRAY analysis. Direct comparisons are shown in supplemental Figure 4A-C, whereas differences in the 2 measures can be seen more clearly in Bland-Altman plots, shown in supplemental Figure 4D-F. In the CLIA-accredited and College of American Pathologists-certified clinical molecular diagnostic laboratory, 5 replicates at 15 dilutions were analyzed. Methylation values by pyrosequencing tended to be overestimated at low dilutions and underestimated at higher dilutions, but the average difference was near 0 (0.69), with a standard deviation of 5%. In general, the concordance was substantial (concordance correlation coefficient, ρc = 0.98). Importantly, at a dilution level of 20%, the pyrosequencing values did not differ by >3%, with the lowest value at 17% and the highest at 23%.
Concordance with pyrosequencing data for the patient samples was only moderate in laboratories 2 and 3 (ρc = 0.92 and ρc = 0.90, respectively) due to a small number of samples that had very different methylation values by the 2 methods (supplemental Figure 4E-F). When categorized as having high and low methylation levels based on the 20% cut point, both laboratories had high measures of agreement between the 2 methods (second laboratory, κ coefficient = 0.95; third laboratory, κ coefficient = 0.91). In the second laboratory, 98% of the samples were in agreement, with only 6 samples examined by pyrosequencing miscalled as being more (n = 2) or less (n = 4) than 20% as initially identified on MassARRAY. In the third laboratory, 96% of the samples were in agreement, with only 12 samples examined by pyrosequencing miscalled as being more (n = 4) or less (n = 8) than 20% on MassARRAY (these 12 included the 6 miscalled in the first laboratory). In summary, despite minor limitations in the quantification of methylation as a continuous measure by pyrosequencing, the vast majority of patient samples were correctly categorized as having high (≥20%) or low (<20%) levels of methylation in 2 independent laboratories. Further studies are needed to assess whether this high level of correct classification is maintained across a broader range of laboratories.
Discussion
Here, we investigated a large CLL patient cohort not only to validate initial findings that ZAP-70 CpG+223 methylation shows strong prognostic relevance with respect to TT and OS,13 but also, importantly, to extend these studies to demonstrate the prognostic value of ZAP-70 methylation relative to established CLL prognostic factors. The data set used here includes samples derived from one of the reference studies examining protein expression of ZAP-70, along with the surrogate markers CD38 expression and IGHV mutation status, in conjunction with TT and OS.15,16 In this larger sample set, we identified a break in the methylation levels at 20% that provided similar and consistent results with our previous work.13 CLL cell populations generally exhibit a remarkably low level of overall intratumor heterogeneity of DNA methylation patterns, which is associated with the presence of a high degree (up to 10% of total CpGs) of allele-specific methylation occurring genome-wide.26 Thus, the observed distinct, trimodal distribution is very likely a consequence of an allelic pattern of DNA methylation at this particular locus, which is stably maintained in the entire CLL cell population, and the break at 20% separates CLL patients lacking CpG+223 methylation from those who have monoallelic or biallelic methylation. Indeed, CLL methylation patterns are highly stable over time, even following treatment.26,27 The presence of such a stable DNA methylation pattern at the ZAP-70 promoter locus suggests a robust clonal event occurring early during leukemogenesis. In practical terms, this permits reliable analysis of archival samples, or those with suboptimal cell viability as frequently occurs in CLL sample procurement.
Investigation of this large, characterized sample set allowed us to demonstrate that ZAP-70 CpG+223 methylation discriminates clinical outcome as measured by TT and OS. Furthermore, this new study also identified a large subset of ZAP-70 expression-negative cases (ie, expected to have a better prognosis) that had low CpG+223 methylation levels and shortened TT and OS comparable to ZAP-70 expression-positive cases. Thus, assessment of CpG+223 methylation improves risk stratification within what would be considered a favorable risk group. A small group (6%) of patients with higher levels of methylation continued to express ZAP-70 protein and had particularly short TT. For OS, the significant modification of ZAP-70 protein expression on the impact of CpG+223 methylation was not observed. Here, ZAP-70 methylation was the factor most strongly predictive of OS in a multivariable model that included IGHV status and expression of CD38 and ZAP-70.
Although ZAP-70 methylation results typically coincide with ZAP-70 expression data, discordant cases do occur as noted in “Results.” We observed a large proportion of cases with low ZAP-70 protein expression that had low methylation levels and reduced TT; conversely, a small group of cases with highly methylated CpG+223 continued to express ZAP-70 and had short TT. Reasons for discordant ZAP-70 protein expression and methylation are uncertain, but could include alternative promoter regulation, divergent CLL clones, hydroxymethylation of this site, or histone modifications,28 and are currently under investigation. Regardless, in practice, these data support first assessing methylation status, and then considering protein analysis (if possible) for those cases exhibiting high levels of methylation; however, the difficulties of assessing ZAP-70 protein expression levels in clinical samples unfortunately remain.
Corcoran and colleagues were the first to identify the relevance of ZAP-70 methylation.29 Methylation of the specific site in the Corcoran study was assessed using combined bisulfite restriction analysis, which is dependent upon restriction enzyme activity at a single site, and accurate quantification is difficult. Additionally, Chantepie et al performed pyrosequencing analysis of ZAP-70 methylation in CLL investigating several alternative sites in intron 1,30 reporting that methylation correlates with CD38 expression, IGHV mutation status, TT, and OS. Our findings significantly extend these observations and refine the prognostic significance, focusing on a distinct CpG dinucleotide. To increase the applicability of ZAP-70 methylation analysis, we also developed a new pyrosequencing method for ZAP-70 CpG+223, as pyrosequencing is becoming increasingly available in clinical molecular laboratories. Transferability of this pyrosequencing assay was verified, strong agreement in methylation classification into low and high groups was confirmed, and standards for a CLIA-approved diagnostic test were developed that can be used clinically for risk stratification.
Importantly, our prognostic abilities as well as our understanding of CLL biology will continue to evolve as newly reported markers such as CD49d expression31 are further validated and functionally characterized. Additionally, the dramatic increase in discovery of novel mutations relevant to CLL disease pathology is allowing improved disease classification and prognostication via the integration of different genetic parameters, as proposed by Rossi et al.32 These ongoing developments will require continued re-evaluation of the prognostic value of ZAP-70 methylation in the context of other abnormalities, ideally to develop tailored treatment strategies for each patient. As novel therapies targeting BCR signaling (eg, ibrutinib, idelalisib) become more widely available, it will also be important to reinvestigate the prognostic significance of ZAP-70 methylation compared with established prognostic markers, such as genomic abnormalities detected by fluorescence in situ hybridization, in this context. Although it will likely be many years before survival data are available with these targeted agents, we envision that ZAP-70 assessment will become even more relevant in the era of BCR-directed therapies, as ZAP-70 expression is likely to be tightly connected to BCR signaling. Similarly, it will be valuable to investigate the molecular mechanism by which CpG+223 methylation impacts ZAP-70 expression, and whether such a mechanism may constitute a therapeutic target of its own. ZAP-70 methylation assessment has already been included as part of a phase 1b/2 trial with ibrutinib,33 and is now being integrated into the risk-classification process for an ongoing phase 3 trial involving ibrutinib (NCT0188687234) to allow its formal evaluation relative to response to this highly effective agent.
In conclusion, we have validated and extended our previous study of the prognostic value of ZAP-70 methylation at CpG+223, demonstrated its overall superiority over ZAP-70 protein expression assessment, and adapted an assay that is transportable to clinical application. These results strongly support assessment of ZAP-70 CpG+223 methylation as part of prospective analyses and risk stratification of CLL.
Acknowledgments
The authors are grateful to all of the patients who contributed samples for this analysis.
R.C. receives a fellowship from the German Research Society (DFG) and is supported by the German Cancer Aid (Max Eder stipend, DKH 110461). C.P. is supported by the US National Institutes of Health, National Cancer Institute grant PO1 CA101956, the German Cancer Research Center, and the German Cancer Consortium. This work was supported by the US National Institutes of Health, National Cancer Institute (grants P50 CA140158, PO1 CA95426, PO1 CA81534, 1K12 CA133250, and RO1 CA95241), as well as The Leukemia & Lymphoma Society, The Harry Mangurian Foundation, and The D. Warren Brown Foundation.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.C. and D.M.L. designed and performed or directed the research and wrote the manuscript; A.S.R. performed statistical analyses and wrote the manuscript; K.E.W. and K.P. performed the research; D.W. and P.S. provided critical guidance on conducting and interpreting the research; M.Z. and S.G. provided important input on the statistical analysis and interpretation; C.C.O. provided data interpretation and wrote the manuscript; A.W.G. and L.Z.R. collected, processed, analyzed, and organized patient samples and data; K.E.W. and K.P. optimized and validated the pyrosequencing assay under the guidance of W.Z.; W.G.W., J.R.B., J.G.G., J. C. Barrientos, K.R.R., N.E.K., T.J.K., M.R.G., and J. C. Byrd contributed essential samples and patient data; J.G.G. and N.E.K. provided important input on the research analysis and interpretation; C.P. and J. C. Byrd co-directed the project, interpreted the data, and wrote the manuscript; and all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John C. Byrd, 410 West 12th Ave, Room 455, Columbus, OH 43210; e-mail: john.byrd@osumc.edu.
References
- 1.Dighiero G, Maloum K, Desablens B, et al. French Cooperative Group on Chronic Lymphocytic Leukemia. Chlorambucil in indolent chronic lymphocytic leukemia. N Engl J Med. 1998;338(21):1506–1514. doi: 10.1056/NEJM199805213382104. [DOI] [PubMed] [Google Scholar]
- 2.Hallek M, Cheson BD, Catovsky D, et al. International Workshop on Chronic Lymphocytic Leukemia. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cramer P, Hallek M. Prognostic factors in chronic lymphocytic leukemia-what do we need to know? Nat Rev Clin Oncol. 2011;8(1):38–47. doi: 10.1038/nrclinonc.2010.167. [DOI] [PubMed] [Google Scholar]
- 4.Wiestner A, Rosenwald A, Barry TS, et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003;101(12):4944–4951. doi: 10.1182/blood-2002-10-3306. [DOI] [PubMed] [Google Scholar]
- 5.Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348(18):1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 6.Del Poeta G, Del Principe MI, Consalvo MA, et al. The addition of rituximab to fludarabine improves clinical outcome in untreated patients with ZAP-70-negative chronic lymphocytic leukemia. Cancer. 2005;104(12):2743–2752. doi: 10.1002/cncr.21535. [DOI] [PubMed] [Google Scholar]
- 7.Dürig J, Nückel H, Cremer M, et al. ZAP-70 expression is a prognostic factor in chronic lymphocytic leukemia. Leukemia. 2003;17(12):2426–2434. doi: 10.1038/sj.leu.2403147. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Apgar J, Huynh L, et al. ZAP-70 directly enhances IgM signaling in chronic lymphocytic leukemia. Blood. 2005;105(5):2036–2041. doi: 10.1182/blood-2004-05-1715. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Widhopf G, Huynh L, et al. Expression of ZAP-70 is associated with increased B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2002;100(13):4609–4614. doi: 10.1182/blood-2002-06-1683. [DOI] [PubMed] [Google Scholar]
- 10.Richardson SJ, Matthews C, Catherwood MA, et al. ZAP-70 expression is associated with enhanced ability to respond to migratory and survival signals in B-cell chronic lymphocytic leukemia (B-CLL). Blood. 2006;107(9):3584–3592. doi: 10.1182/blood-2005-04-1718. [DOI] [PubMed] [Google Scholar]
- 11.Coutre SE, Leonard JP, Barrientos JC, et al. Clinical activity of idelalisib (GS-1101), a selective inhibitor of PI3Kδ, in phase 1 and 2 trials in chronic lymphocytic leukemia (CLL): effect of Del(17p)/TP53 mutation, Del(11q), IGHV mutation, and NOTCH1 mutation [abstract]. Blood. 2013;122(21) Abstract 1632. [Google Scholar]
- 12.Byrd JC, Furman RR, Coutre SE, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claus R, Lucas DM, Stilgenbauer S, et al. Quantitative DNA methylation analysis identifies a single CpG dinucleotide important for ZAP-70 expression and predictive of prognosis in chronic lymphocytic leukemia. J Clin Oncol. 2012;30(20):2483–2491. doi: 10.1200/JCO.2011.39.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claus R, Wilop S, Hielscher T, et al. A systematic comparison of quantitative high-resolution DNA methylation analysis and methylation-specific PCR. Epigenetics. 2012;7(7):772–780. doi: 10.4161/epi.20299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351(9):893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 16.Rassenti LZ, Jain S, Keating MJ, et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112(5):1923–1930. doi: 10.1182/blood-2007-05-092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dürig J, Naschar M, Schmücker U, et al. CD38 expression is an important prognostic marker in chronic lymphocytic leukaemia. Leukemia. 2002;16(1):30–35. doi: 10.1038/sj.leu.2402339. [DOI] [PubMed] [Google Scholar]
- 18.Kröber A, Seiler T, Benner A, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100(4):1410–1416. [PubMed] [Google Scholar]
- 19.Rassenti LZ, Kipps TJ. Clinical utility of assessing ZAP-70 and CD38 in chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2006;70(4):209–213. doi: 10.1002/cyto.b.20129. [DOI] [PubMed] [Google Scholar]
- 20.Preusser M, Berghoff AS, Manzl C, et al. Clinical Neuropathology practice news 1-2014: pyrosequencing meets clinical and analytical performance criteria for routine testing of MGMT promoter methylation status in glioblastoma. Clin Neuropathol. 2014;33(1):6–14. doi: 10.5414/NP300730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahnane N, Gueli R, Tibiletti MG, et al. Pyrosequencing for EGFR mutation detection: diagnostic accuracy and clinical implications. Diagn Mol Pathol. 2013;22(4):196–203. doi: 10.1097/PDM.0b013e3182893f55. [DOI] [PubMed] [Google Scholar]
- 22.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. [PubMed] [Google Scholar]
- 23.McBride GB. A proposal for strength-of-agreement criteria for Lin’s concordance correlation coefficient. NIWA Client Report: HAM2005-062. May 2005. MedCalc Web site. http://www.medcalc.org/download/pdf/McBride2005.pdf. Accessed March 2013.
- 24.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20(1):37–46. [Google Scholar]
- 25.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oakes CC, Claus R, Gu L, et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 2014;4(3):348–361. doi: 10.1158/2159-8290.CD-13-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cahill N, Bergh AC, Kanduri M, et al. 450K-array analysis of chronic lymphocytic leukemia cells reveals global DNA methylation to be relatively stable over time and similar in resting and proliferative compartments. Leukemia. 2013;27(1):150–158. doi: 10.1038/leu.2012.245. [DOI] [PubMed] [Google Scholar]
- 28.Amin S, Walsh M, Wilson C, et al. Cross-talk between DNA methylation and active histone modifications regulates aberrant expression of ZAP70 in CLL. J Cell Mol Med. 2012;16(9):2074–2084. doi: 10.1111/j.1582-4934.2011.01503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corcoran M, Parker A, Orchard J, et al. ZAP-70 methylation status is associated with ZAP-70 expression status in chronic lymphocytic leukemia. Haematologica. 2005;90(8):1078–1088. [PubMed] [Google Scholar]
- 30.Chantepie SP, Vaur D, Grunau C, et al. ZAP-70 intron1 DNA methylation status: determination by pyrosequencing in B chronic lymphocytic leukemia. Leuk Res. 2010;34(6):800–808. doi: 10.1016/j.leukres.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 31.Bulian P, Shanafelt TD, Fegan C, et al. CD49d is the strongest flow cytometry-based predictor of overall survival in chronic lymphocytic leukemia. J Clin Oncol. 2014;32(9):897–904. doi: 10.1200/JCO.2013.50.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi D, Rasi S, Spina V, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121(8):1403–1412. doi: 10.1182/blood-2012-09-458265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Brien S, Furman RR, Coutre SE, et al. Ibrutinib as initial therapy for elderly patients with chronic lymphocytic leukaemia or small lymphocytic lymphoma: an open-label, multicentre, phase 1b/2 trial. Lancet Oncol. 2014;15(1):48–58. doi: 10.1016/S1470-2045(13)70513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rituximab and bendamustine hydrochloride, rituximab and ibrutinib, or ibrutinib alone in treating older patients with previously untreated chronic lymphocytic leukemia. www.clinicaltrials.gov. Accessed March 28, 2014.